Open Access Article
Open Access ArticleLight and endogenous enzyme triggered plasmonic antennas for accurate subcellular molecular imaging with enhanced spatial resolution†
Shuwei
Chen
,
Yue
Yin
,
Xiaozhe
Pang
,
Congkai
Wang
,
Lei
Wang
,
Junqi
Wang
,
Jiangfei
Jia
,
Xinxue
Liu
,
Shenghao
Xu
 * and
Xiliang
Luo
* and
Xiliang
Luo
 *
*
Key Laboratory of Optic-Electric Sensing and Analytical Chemistry for Life Science, MOE, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, P. R. China. E-mail: xushenghao@qust.edu.cn; xiliangluo@qust.edu.cn
First published on 2nd December 2023
Abstract
Developing accurate tumor-specific molecular imaging approaches holds great potential for evaluating cancer progression. However, traditional molecular imaging approaches still suffer from restricted tumor specificity due to the “off-tumor” signal leakage. In this work, we proposed light and endogenous APE1-triggered plasmonic antennas for accurate tumor-specific subcellular molecular imaging with enhanced spatial resolution. Light activation ensures subcellular molecular imaging and endogenous enzyme activation ensures tumor-specific molecular imaging. In addition, combined with the introduction of plasmon enhanced fluorescence (PEF), off-tumor signal leakage at the subcellular level was effectively reduced, resulting in the significantly enhanced discrimination ratio of tumor/normal cells (∼11.57-fold) which is better than in previous reports, demonstrating great prospects of these plasmonic antennas triggered by light and endogenous enzymes for tumor-specific molecular imaging at the subcellular level.
Introduction
With the development of molecular biology and space biology technology, monitoring of physiological activities at the subcellular level has become a research hotspot.1,2In situ imaging of disease-related biomolecules at the subcellular level can provide more effective information such as positioning guidance for accurate and early clinical diagnosis.3–5 In view of this prominent prospect, accurate sensing and imaging of disease-related biomolecules including RNA,6 ions,7,8 small molecules9 and proteins,10 at the subcellular level has recently attracted considerable attention. Despite these significant advances achieved, accurate molecular imaging at the subcellular level still severely suffered from two non-negligible shortcomings: (1) most disease-related biomolecules are not only highly expressed in tumor cells, but also have non-negligible expression levels in normal cells,11,12 and the use of conventional molecular imaging strategies for biomolecule detection in tumor cells at the subcellular level still suffered from low signal-to-background ratios because of the signal responses in normal cells resulting from the “always-active” mode,13 resulting in “off-tumor” signal leakage and thus reducing the spatial resolution. (2) The insufficient sensitivity caused by photobleaching of fluorescent dyes will inevitably reduce subcellular imaging resolution. Therefore, engineering higher sensitive methods for accurate tumor-specific subcellular molecular imaging with spatial resolution still remains a formidable challenge.As for the problem of off-tumor signal leakage resulting from the conventional “always-active” sensing mode, endogenous activation strategies provide unprecedented opportunities for tumor-specific sensing to address this limitation.14 It is reported that endogenous apurinic/apyrimidinic endonuclease 1 (APE1) is highly expressed in the tumor cell cytoplasm but not in normal cells.13,15 Accordingly, endogenous activation strategies that are engineered based on the different expression levels of APE1 can reduce the leakage of tumor external signals.16–18 Despite these advances, the spatial resolution is still limited and subcellular imaging has not yet been achieved. Additionally, as for solving the problem of insufficient sensitivity at the subcellular level caused by photobleaching of fluorescent dyes, plasmon enhanced fluorescence (PEF) has been validated to be an efficient approach to solve this bottleneck.19–23 Therefore, combination of light and endogenous enzyme activation together with the PEF technique is highly desirable for accurate tumor-specific molecular imaging at the subcellular level with enhanced spatial resolution. Whereas, as far as we know, such promising attempts have yet to be explored.
To explore this possibility, we report the design of light and endogenous APE1-triggered plasmonic antennas for accurate tumor-specific molecular imaging at the subcellular level with enhanced spatial resolution. Thanks to these endogenous activated plasmonic antennas, off-tumor signal leakage at the subcellular level was effectively reduced together with the significantly enhanced discrimination ratio of tumor/normal cells. As demonstrated in Scheme 1, a light-activated probe (LP) was prepared by modifying triphenylphosphine (TPP, mitochondrial targeting molecule) and the hybridization chains of L1 (consisting of the ATP aptamer sequence and activation sequence) and L2 (labeled with a PC-linker) onto the surface of upconversion nanoparticles (UCNPs). At this point, the sensing activity of the LP to the target (taking ATP as a model) was blocked. The enzyme-activated probe (EP) was prepared by modifying TPP and the hybridization chains of L3 (labeled with BHQ-3 and the AP-site) and L4 (labeled with Cy5.5) onto silica coated gold nanobipyramids (Au NBPs@SiO2), along with the quenched fluorescence owing to the fluorescence resonance energy transfer from Cy5.5 to BHQ-3. When the LP reaches the tumor cell mitochondria with the help of TPP, ultraviolet light (UV), converted from 808 nm near-infrared (NIR) light by UCNPs, cleaves the PC-linker, allowing the LP to bind to ATP and expose the activation sequence for subsequent reactions. The high expression of APE1 in the tumor cytoplasm will cleave the AP-site and hybridize the EP with the activation sequence to form plasmonic antennas, accompanied by the restored red fluorescence, and the best fluorescence enhancement can be acquired by managing the interval between Au NBPs and Cy5.5 by adjusting the thickness of the silica shell. It is noteworthy that the AP-site is specifically cleaved by APE1 in the tumor cytoplasm, allowing the signal responses to be confined within the tumor cell mitochondria region. Thus, through combining the NIR-light-mediated spatiotemporally controlled manner, enzyme-activated strategy and PEF technique, tumor-specific subcellular molecular imaging with significantly enhanced spatial resolution (discrimination ratio of tumor/normal cells) can be aquired.
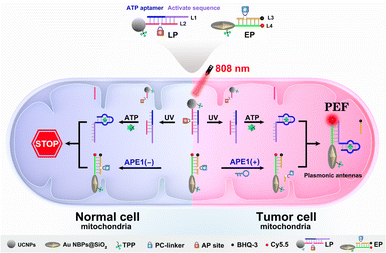 | ||
| Scheme 1 Schematic illustration of plasmonic antennas triggered by 808 nm NIR and endogenous APE1 for tumor-specific subcellular molecular imaging. | ||
Experimental
Preparation of the LP
TPP (20 μL, 10 mM), EDC (10 μL, 200 mM), LP (10 μL, 10 μM) and NHS (10 μL, 200 mM) were added to 50 μL of UCNPs (1 mg mL−1, detailed synthesis procedure of UCNPs is shown in the ESI†) and stirred at 37 °C overnight. Finally, the LP was acquired after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and re-dispersed in Tris–HCI (0.1 mL, 10 mM, pH = 7.4).
000 rpm and re-dispersed in Tris–HCI (0.1 mL, 10 mM, pH = 7.4).
Preparation of the EP
CTAB (10 μL, varying concentrations) was mixed with Au NBPs (1 mL, detailed synthesis procedure of Au NBPs is shown in the ESI†) and stirred for 1 day. Next, the pH was adjusted to alkaline and stirred for 30 min, followed by adding 3 μL of TEOS (VTEOS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vmethanol = 1
Vmethanol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) every 30 minutes with a total of 3 times. 1 day later, AuNBPs@SiO2 was acquired after centrifugation at 10
4) every 30 minutes with a total of 3 times. 1 day later, AuNBPs@SiO2 was acquired after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and re-dispersed in ethanol (1 mL). Afterwards, ammonium hydroxide (5 μL, 25%) was added and stirred for 30 min, followed by adding APTES (2.5 μL) and stirred at 35 °C for 180 min and then at 65 °C for 60 min. Then, Au NBP@SiO2–NH2 was acquired after centrifugation at 10
000 rpm and re-dispersed in ethanol (1 mL). Afterwards, ammonium hydroxide (5 μL, 25%) was added and stirred for 30 min, followed by adding APTES (2.5 μL) and stirred at 35 °C for 180 min and then at 65 °C for 60 min. Then, Au NBP@SiO2–NH2 was acquired after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and re-dispersed in Tris–HCI (900 μL, 10 mM, pH = 7.4), followed by the addition of TPP (40 μL, 10 mM), EDC (20 μL, 200 mM), LP (20 μL, 10 μM) and NHS (20 μL, 200 mM) and incubated for 6 h. Thus, the EP was acquired after centrifugation at 10
000 rpm and re-dispersed in Tris–HCI (900 μL, 10 mM, pH = 7.4), followed by the addition of TPP (40 μL, 10 mM), EDC (20 μL, 200 mM), LP (20 μL, 10 μM) and NHS (20 μL, 200 mM) and incubated for 6 h. Thus, the EP was acquired after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and re-dispersed in Tris–HCI (1.0 mL, 10 mM, pH = 7.4).
000 rpm and re-dispersed in Tris–HCI (1.0 mL, 10 mM, pH = 7.4).
808 nm NIR light and APE1 synergistically activated detection of ATP in vitro
10 μL of LP was irradiated using an 808 nm laser (2 W cm−2) for 30 min, followed by the addition of APE1 (1.5 μL, 1.0 U mL−1), 150 μL of EP and ATP(40 μL, different concentrations) and incubated or 4 h. Lastly, fluorescence intensities were recorded, respectively.Confocal fluorescence imaging for discrimination of tumor/normal cells in mixed cell cultures
HeLa cells were seeded in a confocal dish and labeled through incubating with SiO2@Cy3 for 4 h, followed by washing three times using PBS, and then culture medium and L02 cells were added and further incubated overnight. Next, 20 μL LP and 20 μL EP were added and incubated for 4 h, followed by irradiation using an 808 nm laser for 0.5 h and further incubated for another 4 h. Lastly, confocal fluorescence microscopy images were obtained. It is worth noting that HeLa cells were located in the green fluorescence region, while L02 cells were located in the region without green fluorescence (Cy3 channel: laser Ar543nm, emission collection range: 500 to 650 nm. Cy5.5 channel: laser HeNe633nm, emission collection range: 650 to 750 nm).Results and discussion
Feasibility evaluation of the light and endogenous enzyme activated sensing design
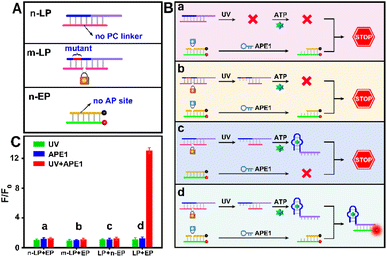
As indicated in Fig. 1A, due to the “always active” recognition manner of the traditional subcellular molecular imaging probe (TP), plasmonic antennas can be formed which are triggered equally by target ATP in both normal and tumor cell mitochondria. Therefore, “off-tumor” signal leakage together with the decreased spatial resolution will inevitably be generated. By contrast, plasmonic antennas can only be formed in tumor cell mitochondria, because the enzyme-activated subcellular molecular imaging probe (EP) can be only activated by the highly expressed cytoplasmic APE1 in tumor cells. Thus, “off-tumor” signal leakage can be efficiently shortened and tumor-specific molecular imaging at the subcellular level with significantly enhanced spatial resolution can be realized. Accordingly, the feasibility of the APE1-activated sensing design for the EP was evaluated. As shown in Fig. 1B and C, the TP exhibited obvious fluorescence recovery in the presence of target ATP, while the EP exhibited negligible fluorescence recovery, revealing that the sensing performance of the EP was inhibited. Moreover, only when UV and APE1 simultaneously coexisted can the EP produce obvious fluorescence recovery for ATP (Fig. S1†). Moreover, the result that target output can only be generated when UV and APE1 simultaneously coexisted was also proved by the native polyacrylamide gel electrophoresis (PAGE) results (Fig. 2D, lane 4 to lane 7). These results indicated that light activation can determine when to activate and endogenous enzyme activation can determine where to activate, such design of which can ensure the feasibility of tumor-specific molecular imaging at the subcellular level. In addition, the optimal APE1 concentration was optimized to be 1 U mL−1 (Fig. S2†).Furthermore, three control probes including n-LP (same sequences as LP but without a PC linker), m-LP (mutant sequence for the ATP aptamer that cannot recognize target ATP) and n-EP (same sequences as the EP but without the AP site) were designed to further validate the feasibility of the UV and endogenous APE1 activated sensing design (Fig. 2A). As shown in Fig. 2B and C, only the combination of LP and EP can produce significant fluorescence recovery of ATP when UV and endogenous APE1 simultaneously coexisted, further indicative of the feasibility of the UV and endogenous APE1 activated sensing design (Fig. 2B and C).
Construction and characterization of the LP and EP
Considering the inevitable shortcomings including phototoxicity and limited tissue penetration of UV, UCNPs that can convert NIR light with low phototoxicity and excellent tissue penetration to UV were chosen to construct the LP. As shown in Fig. 3A, the morphology of the UCNPs is hexagonal with an average size of ∼40 nm. Insets of the elemental mapping reveal that Tm, Nd, Gd and Yb distributed on UCNPs. Moreover, Nd is mainly distributed externally and Tm is mainly distributed internally (Fig. 3B). The upconversion fluorescence spectrum reveals that UCNPs exhibited upconverted emission peaks at 365 nm under 808 nm NIR light irradiation that can cleave the PC linker (Fig. 3C), allowing for remote activation of the LP at a specified time (such as when probes reach the subcellular organelle) by 808 nm NIR light. The photolysis efficiency caused by 808 nm NIR light was confirmed to be approximately 88% (Fig. S3†), which ensures the remote activation of the LP by 808 nm NIR light. As shown in Fig. S4†, almost no red fluorescence can be observed without 808 nm NIR irradiation, while obvious fluorescence can be observed with 808 nm NIR irradiation, revealing that remote activation of the LP at a specified time can be realized through 808 nm NIR light. The optimal time of light activation was optimized to be 30 min (Fig. S5†). Additionally, OA-free UCNPs were aquired by eliminating the OA ligand (validated using the contact angle shown in Fig. S6†) for subsequent modification and bioanalysis. Then, the successful preparation of the LP was confirmed by the decreased zeta potential (Fig. S7A†), increased hydrodynamic diameter (Fig. S7B†), the characteristic peak for TPP at 242 nm and DNA at 260 nm (Fig. S7C†) and the increased absorption peak intensity at 1650 cm−1 as well as the vibration absorption peak of the aromatic ring at 1450 cm−1 (Fig. S7D†).Of note, Au NBPs with the local electric field at sharper apexes (Fig. 3D) and a maximum plasmonic wavelength at ∼700 nm (Fig. S8A†) which matched well with the emission spectra of Cy5.5 (Fig. S8B†) were introduced to improve the detection sensitivity for tumor-specific molecular imaging at the subcellular level. In order to obtain the best fluorescence enhancement, the interval between Au NBPs and Cy5.5 is regulated by altering the thickness of the silica shells. It can be concluded that the best fluorescence enhancement (∼7.3-fold, Fig. 3F) was obtained when the maximum absorption wavelength of Au NBPs@SiO2 was 705 nm (Fig. S9†), and the corresponding TEM image is shown in Fig. 3E. The fluorescence enhancement factor was obtained by regulating the same concentration of Cy5.5 on SiO2 nanoparticles and Au NBPs@SiO2 nanoparticles, and then comparing the fluorescence intensity (detailed calculation process is provided in the ESI†). Additionally, successful preparation of the EP was also confirmed using zeta potentials (Fig. S10A†), FT-IR spectra (Fig. S10B†) and the absorption spectrum (Fig. S10C†), respectively.
Moreover, quantitative detection of ATP was performed based on the plasmonic antennas. As depicted in Fig. S11A,† elevated concentrations of ATP resulted in increased fluorescence intensities and the detection limit was calculated to be 0.34 μM (3σ/s, Fig. S11B and C†), which was comparable to, or even lower than the recent reports,24–26 and ensures highly sensitive in vitro detection of ATP. The linear range of the probe for ATP is from 0.001 mM to 4 mM, which basically covers the concentration of ATP in mitochondria (1–5 mM), and can realize semi-quantitative imaging of ATP in mitochondria. In addition, it can be seen that only ATP could induce fluorescence recovery, demonstrating the satisfactory specificity for ATP (Fig. S11D†).
Light and endogenous enzyme activated accurate subcellular molecular imaging
To ensure subcellular molecular imaging, colocalization experiments were performed. As shown in Fig. 4 and S12,† after 4 h incubation, the Cy3 channel of the LP and the Cy5.5 channel of the EP anastomosed well with the green channel of MitoTracker Green, with a Pearson's correlation coefficient of ∼0.85 and ∼0.87, respectively, indicating the prominent localization of both the LP and EP in mitochondria. According to literature reports,27,28 TPP has good lipophilic and positive properties, which can not only help nanoparticles target mitochondria, but also help them escape from lysosomes. Accordingly, in order to verify lysosome escape, lysosome colocalization experiments were also performed. As shown in Fig. S13,† after 4 h incubation, the Cy3 channel of the LP and the Cy5.5 channel of the EP corresponded poorly with the green channel of LysoTracker, with a Pearson's correlation coefficient of ∼0.29 and ∼0.31, respectively, indicating that both the LP and EP escaped from lysosomes. In addition, cell viability assays verified that the LP and EP possessed low cytotoxicity and good biocompatibility (Fig. S14†), and the reaction time of 4 h was also optimized (Fig. S15†). To demonstrate the feasibility of the light and endogenous enzyme activated design for accurate subcellular molecular imaging, we investigated fluorescence responses of HeLa cells treated with four designed combination probes including the n-LP and EP, m-LP and EP, LP and n-EP, and LP and EP, respectively. As shown in Fig. 5, only LP and EP treated HeLa cells showed obvious red fluorescence, while feeble fluorescence can be seen for the other three combination probes (control groups), the results of which verified that the LP and EP allowed for subcellular molecular imaging based on the light and endogenous enzyme activated design. To further confirm the light and endogenous enzyme activated design, NCA (APE1 inhibitor) was introduced to down-regulate the expression level of APE1 to obtain different light and endogenous enzyme activated combinations. As indicated in Fig. S16,† only when both 808 nm NIR and APE1 (without NCA treated) simultaneously coexisted can the red fluorescence be observed, further confirming the light and endogenous enzyme activated design.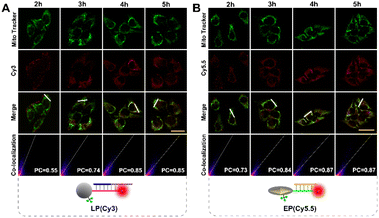 | ||
| Fig. 4 Confocal imaging of HeLa cells incubated with (A) LP (Cy3), Mito Tracker Green and (B) EP (Cy5.5), Mito Tracker Green signals at different times (scale bar: 50 μm), respectively. | ||
 | ||
| Fig. 5 Confocal images of HeLa cells incubated with (A) n-LP and EP, (B) m-LP and EP, (C) LP and n-EP, and (D) LP and EP, respectively (scale bar: 50 μm). | ||
Moreover, in order to achieve accurate molecular imaging with subcellular resolution, the ability of dynamic monitoring of ATP concentration in mitochondria was investigated. Here, the fluctuation of ATP expression levels is achieved by up-regulation with Ca2+ and down-regulation with oligomycin.25,29 As shown in Fig. 6, compared with the untreated control group, Ca2+ treatment significantly increased red fluorescence in response to mitochondria ATP, while oligomycin treatment significantly decreased red fluorescence. These results clearly indicated the ability of dynamic monitoring of ATP concentration in mitochondria, ensuring accurate molecular imaging with subcellular resolution.
Tumor-specific subcellular molecular imaging with enhanced spatial resolution
Owing to the non-negligible expression level of ATP in normal cell mitochondria, the TP with passive recognition mode will inevitably produce background signal interferences, compromising spatial specificity and resolution. By contrast, the sensing ability of the EP to ATP can be only activated by the highly expressed cytoplasmic APE1 in tumor cells, guaranteeing the tumor-specific subcellular molecular imaging (Fig. 7A). As displayed in Fig. S17,† HeLa cells incubated with the EP displayed evident red fluorescence but L02 cells incubated with the EP displayed negligible red fluorescence. However, both HeLa and L02 cells incubated with the TP exhibited obvious red fluorescence with poor cell-type selectivity. Since the cellular uptake of the EP by these two cells showed no obvious difference (Fig. S18†), the above different fluorescence results validated that the EP can realize tumor-specific subcellular molecular imaging.Since there is no obvious division between normal and tumor cells in tissues, such obtained results were still unconvincing and inadequate. Hence, whether the EP can realize tumor-specific subcellular molecular imaging was further explored in mixed cultivation cells. Of note, HeLa cells were first labeled with Cy3@SiO2 and then cross-mixed with L02 cells for co-culture. As shown in Fig. 7B and C, tumor cells are located in region 1 and normal cells are located in region 2, which is validated according to the labeled green fluorescence rather than cell morphology. Noticeably, TP treated HeLa and L02 cells displayed similarly red fluorescence (region 1). However, EP treated L02 cells displayed negligible red fluorescence compared to HeLa cells (region 2). Average fluorescence intensity based quantitative results indicated that the proposed EP/TP method displayed an ∼11.57-fold discrimination ratio (tumor/normal, Fig. 7C), much higher than the previous reported studies (∼2.6-fold and ∼3.44-fold, respectively.18,30) Therefore, these results further confirmed that “off-tumor” signal leakage can be efficiently shortened and tumor-specific molecular imaging at the subcellular level with significantly enhanced spatial resolution can be acquired. Moreover, other tumor cells including HepG2 and MCF-7 cells were also examined to confirm the universality of this strategy for tumor cell-specific imaging (Fig. S19†).
Conclusions
In summary, we propose light and endogenous APE1-triggered plasmonic antennas for accurate tumor-specific subcellular molecular imaging with enhanced spatial resolution. This system was constructed by the combination of the upconversion nanotechnology, enzyme-activated strategy, organelle localized strategy and PEF technique. Based on this design, off-tumor signal leakage at the subcellular level was effectively reduced together with the significantly enhanced discrimination ratio of tumor/normal cells (∼11.57-fold). As far as we know, this is the first example of using plasmonic antennas for accurate tumor-specific subcellular molecular imaging. In addition, by changing the aptamer sequence, other non-nucleic acid target analytes can be detected. Alternatively, changing the aptamer sequence to a complementary sequence of the target nucleic acid can also achieve the detection of nucleic acid targets (such as miRNA or mRNA). We expect that this work will afford available tools for designing activatable plasmonic antennas for precise subcellular molecular imaging.Author contributions
Shuwei Chen: Investigation, writing – original draft preparation, writing – reviewing and editing. Yue Yin, Xiaozhe Pang, Congkai Wang, Lei Wang: Data curation. Junqi Wang, Jiangfei Jia, Xinxue Liu: Methodology. Shenghao Xu, Xiliang Luo: Supervision.Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
The authors gratefully acknowledge the support from the National Natural Science Foundation of China (21505081 and 21974075), the Natural Science Foundation of Shandong Province of China (ZR2023MB110), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (2019KJC007) and the Taishan Scholar Program of Shandong Province, China (ts20110829).Notes and references
- G. Parlakgül, A. P. Arruda, S. Pang, E. Cagampan, N. Min, E. Güney, G. Y. Lee, K. Inouye, H. F. Hess, C. S. Xu and G. S. Hotamışlıgil, Regulation of liver subcellular architecture controls metabolic homeostasis, Nature, 2022, 603, 736–742 CrossRef PubMed
.
- H. Kobayashi, K. C. Cheveralls, M. D. Leonetti and L. A. Royer, Self-supervised deep learning encodes high-resolution features of protein subcellular localization, Nat. Methods, 2022, 19, 995–1003 CrossRef CAS
.
- C. I. Massengill, L. B. Edwards, C. C. Ceballos, E. R. Cebul, J. Cahill, A. Bharadwaj, E. Wilson, M. Z. Qin, M. R. Whorton, I. Baconguis, B. Ye, T. Y. Mao and H. N. Zhong, Sensitive genetically encoded sensors for population and subcellular imaging of cAMP in vivo, Nat. Methods, 2022, 19, 1461–1471 CrossRef CAS
.
- J. Y. Wei, C. L. Ji, Y. F. Wang, J. Tan, Q. Yuan and W. H. Tan, Nucleic Acid Probes for Single-Molecule Localization Imaging of Cellular Biomolecules, Chem. Biomed. Imaging, 2023, 1, 18–29 CrossRef CAS
.
- S. F. Yang, Y. Q. Wang, Q. Y. Wang, F. Y. Li and D. S. Ling, DNA-Driven Dynamic Assembly/Disassembly of Inorganic Nanocrystals for Biomedical Imaging, Chem. Biomed. Imaging, 2023, 1, 340–355 CrossRef CAS
.
- J. Zhao, Z. X. Li, Y. L. Shao, W. P. Hu and L. L. Li, Spatially Selective Imaging of Mitochondrial MicroRNAs via Optically Programmable Strand Displacement Reactions, Angew. Chem., Int. Ed., 2021, 60, 17937–17941 CrossRef CAS
.
- N. Narayanaswamy, K. Chakraborty, A. Saminathan, E. Zeichner, K. H. Leung, J. Devany and Y. Krishnan, A pH-correctable, DNA-based fluorescent reporter for organellar calcium, Nat. Methods, 2019, 16, 95–102 CrossRef CAS
.
- D. Y. Yi, H. Z. Zhao, J. Zhao and L. L. Li, Modular Engineering of DNAzyme-Based Sensors for Spatioselective Imaging of Metal Ions in Mitochondria, J. Am. Chem. Soc., 2023, 145, 1678–1685 CrossRef CAS
.
- S. Thekkan, M. S. Jani, C. Cui, K. Dan, G. L. Zhou, L. Becker and Y. Krishnan, A DNA-based fluorescent reporter maps HOCl production in the maturing phagosome, Nat. Chem. Biol., 2019, 15, 1165–1172 CrossRef CAS
.
- K. Dan, A. T. Veetil, K. Chakraborty and Y. Krishnan, DNA nanodevices map enzymatic activity in organelles, Nat. Nanotechnol., 2019, 14, 252–259 CrossRef CAS
.
- B. A. Badeau, M. P. Comerford, C. K. Arakawa, J. A. Shadish and C. A. DeForest, Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery, Nat. Chem., 2018, 10, 251–258 CrossRef CAS
.
- Y. D. Sun, L. Fang, Y. X. Han, A. B. Feng, S. J. Liu, K. Zhang and J. J. Xu, Reversible Ratiometric Electrochemiluminescence Biosensor Based on DNAzyme Regulated Resonance Energy Transfer for Myocardial miRNA Detection, Anal. Chem., 2022, 94, 7035–7040 CrossRef CAS
.
- Y. L. Shao, J. Zhao, J. Y. Yuan, Y. L. Zhao and L. L. Li, Organelle-Specific Photoactivation of DNA Nanosensors for Precise Profiling of Subcellular Enzymatic Activity, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS PubMed
.
- P. H. Zhang, D. Gao, K. An, Q. Shen, C. Wang, Y. C. Zhang, X. S. Pan, X. G. Chen, Y. F. Lyv, C. Cui, T. X. Z. Liang, X. M. Duan, J. Liu, T. L. Yang, X. X. Hu, J. J. Zhu, F. Xu and W. H. Tan, A programmable polymer library that enables the construction of stimuli-responsive nanocarriers containing logic gates, Nat. Chem., 2020, 12, 381–390 CrossRef CAS PubMed
.
- C. D. Mol, T. Izumi, S. Mitra and J. A. Tainer, DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination, Nature, 2000, 403, 451–456 CrossRef CAS
.
- Z. T. Fan, J. Zhao, X. Chai and L. L. Li, A Cooperatively Activatable, DNA-based Fluorescent Reporter for Imaging of Correlated Enzymatic Activities, Angew. Chem., Int. Ed., 2021, 60, 14887–14891 CrossRef CAS
.
- X. Huang, Y. F. Zhang, J. Chen, L. Zhang, Y. Z. Xu, W. Yin, Y. K. Shi, S. Y. Liu, X. Y. Zou and Z. Dai, Dual-Locked DNAzyme Platform for In Vitro and In Vivo Discrimination of Cancer Cells, Anal. Chem., 2022, 94, 12221–12230 CrossRef CAS
.
- J. H. Wang, Y. Y. Liu, T. T. Zhao, J. H. Shi, J. Chen, D. Li, Y. Y. Cui, S. H. Xu and X. L. Luo, Enzymatically Activated Autonomous-Motion DNAzyme Signal Amplification Strategy for Tumor Cell-Specific Molecular Imaging with Improved Spatial Specificity, Anal. Chem., 2023, 95, 9388–9395 CrossRef CAS
.
- T. Li, C. Wang, H. W. Zhao, F. W. Sun, H. Dong, K. Feng, P. Wang, G. R. He and G. T. Li, Rational Approach to Plasmonic Dimers with Controlled Gap Distance, Symmetry, and Capability of Precisely Hosting Guest Molecules in Hotspot Regions, J. Am. Chem. Soc., 2021, 143, 8631–8638 CrossRef
.
- T. T. Zhao, J. H. Shi, J. H. Wang, Y. Y. Cui, Y. F. Yang, S. H. Xu and X. L. Luo, Fluorescence-Enhanced Dual-Driven “OR-AND” DNA Logic Platform for Accurate Cell Subtype Identification, Anal. Chem., 2023, 95, 3525–3531 CrossRef CAS
.
- D. Semeniak, D. F. Cruz, C. Ashutosh and M. H. Mikkelsen, Plasmonic Fluorescence Enhancement in Diagnostics for Clinical Tests at Point-of-Care: A Review of Recent Technologies, Adv. Mater., 2023, 35, 210798 CrossRef PubMed
.
- Y. Y. Cui, C. Yuan, H. W. Tan, Z. B. Zhang, Y. J. Jia, N. Na and J. Ouyang, Plasmon-Enhanced Fluorescent Sensor based on Aggregation-Induced Emission for the Study of Protein Conformational Transformation, Adv. Funct. Mater., 2019, 29, 1807211 CrossRef
.
- J. H. Hwang, S. Park, J. Son, J. W. Park and J. M. Nam, DNA-Engineerable Ultraflat-Faceted Core–Shell Nanocuboids with Strong, Quantitative Plasmon-Enhanced Fluorescence Signals for Sensitive, Reliable MicroRNA Detection, Nano Lett., 2021, 21, 2132–2140 CrossRef CAS PubMed
.
- D. Samanta, S. B. Ebrahimi, N. Ramani and C. A. Mirkin, Enhancing CRISPR-Cas-Mediated Detection of Nucleic Acid and Non-nucleic Acid Targets Using Enzyme-Labeled Reporters, J. Am. Chem. Soc., 2022, 144, 16310–16315 CrossRef CAS PubMed
.
- S. N. Hong, X. T. Zhang, R. J. Lake, G. T. Pawel, Z. J. Guo, R. J. Pei and Y. Lu, A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells, Chem. Sci., 2020, 11, 713–720 RSC
.
- Y. Xiong, J. J. Zhang, Z. L. Yang, Q. B. Mou, Y. Ma, Y. H. Xiong and Y. Lu, Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets, J. Am. Chem. Soc., 2019, 142, 207–213 CrossRef PubMed
.
- X. J. Di, D. J. Wang, J. J. Zhou, L. Zhang, M. H. Stenzel, Q. P. Su and D. Y. Jin, Quantitatively monitoring in situ mitochondrial thermal dynamics by upconversion nanoparticles, Nano Lett., 2021, 21, 1651–1658 CrossRef CAS
.
- X. Zhao, N. Na and J. Ouyang, CRISPR/Cas9-based coronal nanostructures for targeted mitochondria single molecule imaging, Chem. Sci., 2022, 13, 11433–11441 RSC
.
- Z. Q. Zou, M. Pan, F. Y. Mo, Q. Y. Jiang, A. L. Feng, Y. Z. Zhou, F. A. Wang and X. Q. Liu, High-fidelity ATP imaging via an isothermal cascade catalytic amplifier, Chem. Sci., 2022, 13, 12198–12207 RSC
.
- Q. Liu, Y. Y. Huang, Z. P. Li, L. L. Li, Y. L. Zhao and M. Y. Li, An Enzymatically Gated Catalytic Hairpin Assembly Delivered by Lipid Nanoparticles for the Tumor-Specific Activation of Signal Amplification in miRNA Imaging, Angew. Chem., Int. Ed., 2022, 61, e202214230 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05728c |
| This journal is © The Royal Society of Chemistry 2024 |