Open Access Article
Open Access ArticleBacterial elimination via cell membrane penetration by violet phosphorene peripheral sub-nanoneedles combined with oxidative stress†
Qiudi
Shen
a,
Jing
Kang
*a,
Xuewen
Zhao
b,
Wanqing
Lou
c,
Zhihao
Li
d,
Lihui
Zhang
b,
Bo
Zhang
 b,
Jinying
Zhang
b,
Jinying
Zhang
 *b,
Bailiang
Wang
*c and
Alideertu
Dong
*b,
Bailiang
Wang
*c and
Alideertu
Dong
 *a
*a
aCollege of Chemistry and Chemical Engineering, Engineering Research Center of Dairy Quality and Safety Control Technology, Ministry of Education, Inner Mongolia University, 235 University West Street, Hohhot, 010021, China. E-mail: dongali@imu.edu.cn
bState Key Laboratory of Electrical Insulation and Power Equipment, Center of Nanomaterials for Renewable Energy (CNRE), School of Electrical Engineering, Xi'an Jiaotong University, Xi'an, 710049, China
cSchool of Ophthalmology & Optometry, Eye Hospital, Wenzhou Medical University, Wenzhou, 325027, P. R. China
dDepartment of Chemistry, College of Sciences, Northeastern University, Shenyang, 110819, China
First published on 28th February 2024
Abstract
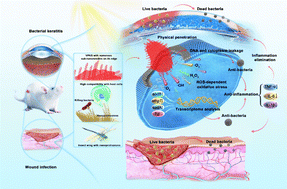
The effectiveness of an antibacterial agent is strongly influenced by its antibacterial mechanism, which, in turn, depends on the agent's topological structure. In the natural world, the nanoprotrusions on the surface of insect wings give them excellent antimicrobial properties through physical penetration while being compatible with host cells. Inspired by the novel nanostructure of insect wings, violet phosphorus (VP), a new member of the phosphorus family, has antibacterial potential due to the sub-nanoneedle on its edge. Here, we demonstrate that VP and its exfoliated product, violet phosphorene nanosheets (VPNSs), have superior antibacterial capability against pathogens via cell membrane penetration induced by peripheral sub-nanoneedles combined with oxidative stress. The results show that VPNSs can inactivate more than 99.9% of two common pathogens (Escherichia coli and Staphylococcus aureus) and more than 99.9% of two antibiotic-resistant bacteria (Escherichia coli pUC19 and methicillin-resistant Staphylococcus aureus), while showing almost no toxicity toward normal cells at a high concentration of 2.0 mg mL−1. Moreover, VPNSs can achieve effective treatment of induced skin wound infections and bacterial keratitis (BK) by Staphylococcus aureus and methicillin-resistant Staphylococcus aureus, respectively, showing promising potential for ocular and skin wound infection theragnostics.
Introduction
Nature is a versatile designer, sculpting various surprising micro-wonders such as insect wings whose surfaces have antibacterial and antibiofilm properties.1–3 These wings have dense surface protrusions at the nano- and micro-levels, which function as natural barriers to combat bacterial pathogens and prevent biofilm formation. The nanoprotrusions rupture microbial cells upon contact and lead to bacterial death, which is an effective physical mechanism for protecting insects from bacterial infection.4–7 Surprisingly, these nanoprotrusions have low toxicity and even promote eukaryotic cell proliferation.8,9 Several pieces of evidence suggested that the rigidity of the bacterial cell wall was significantly greater than that of the eukaryotic cell membrane.10–12 Eukaryotic cells could avoid membrane damage and cell death by stretching and twisting the shape to fit the nanostructure.13 Researchers inspired by the natural antibacterial structure have created new biomimetic surfaces with special nanostructured morphologies and self-cleaning antibacterial capacity.14–17 The nanotopographies and chemical configurations for fabricated biomimetic surfaces show a significant impact on their antibacterial functions. More efficient bactericidal and antibiofilm characteristics have been obtained using specially structured antibacterial surfaces that result in unique mechanisms.18,19Layered phosphorus and phosphorene have been demonstrated to be effective antibacterial agents, attracting growing interest in related fields due to their high bactericidal efficiency,20–24 biosafety,25–30 and on-demand degradability.31–34 Reactive oxygen species (ROS) are easily produced from phosphorus and phosphorene structures due to the lone pair electrons from phosphorus atoms.35–38 Layered phosphorus and phosphorene structures provide more active sites to produce ROS due to their high specific surface areas, which has been well demonstrated for black phosphorus (BP).39–42 The nanotopographies and chemical configurations of these layered structures may provide further ways to enhance their antibacterial activity in addition to ROS effects.43–46 The special structures of recently produced violet phosphorus (VP) and phosphorene nanosheets (VPNSs) feature numerous sub-nanoneedles on the flake periphery, whose high surface energy favors their interaction with the cell membrane of bacteria. The phenomenon is analogous to protrudes on the surface of an insect wing at the macro-scale, leading to bacterial death.47–49 Hence, both ROS-dependent oxidative stress and physical damage are the expected antibacterial mechanisms of VP and VPNSs.
Herein, we report on the antibacterial actions of VP and VPNSs and compare them with the antibacterial mechanism of BP to confirm the additional effects of physical penetration arising from the former structures. Meanwhile, Table S1† compares the antibacterial mechanism of VPNSs with some up-and-coming 2D nanomaterials. Higher antibacterial activity than BP is demonstrated by VP and VPNSs after a series of antibacterial tests against two common pathogens (Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus)) and two antibiotic-resistant bacteria (methicillin-resistant Staphylococcus aureus (MRSA) and Escherichia coli pUC19 (E. coli pUC19)). This is determined by the effect of the peripheral sub-nanoneedle induced VP and VPNSs penetrating into the cell membrane combined with oxidative stress to eliminate bacteria. Moreover, in vivo mouse experiments prove that VPNSs can achieve effective treatment of induced skin wound infections and bacterial keratitis (BK) by S. aureus and MRSA, respectively, showing promising potential for ocular and skin wound infection theragnostics (Scheme 1). This study offers a new 2D antibacterial agent to combat pathogens and proposes a new synergistic mechanism for eradicating pathogens.
Results and discussion
Synthesis and characterization of VPNSs and BPNSs
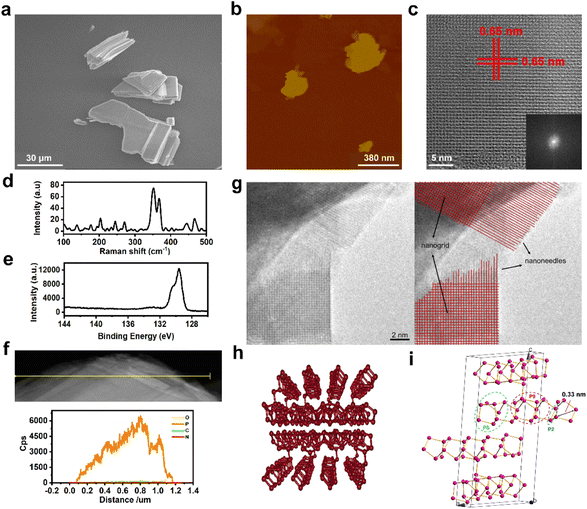
As illustrated in Fig. 1a and S1,† scanning electron microscopy (SEM) showed that VP has a multilayer structure. Then the VPNSs were obtained by a solvent exfoliation method from bulk VP under mild conditions. An atomic force microscope (AFM) and transmission electron microscope (TEM) showed that VPNSs showed a wide two-dimensional size distribution and their edges were irregular and rough after solution exfoliation (Fig. 1b, S2, and S3†). The structure of the as-obtained VPNSs was also characterized using high-resolution transmission electron microscopy (HRTEM) and X-ray diffraction (XRD). The HRTEM image showed that the lattice spacing was 0.65 nm and intersected at a 90° angle, corresponding to the (110) crystal planes of VPNSs (Fig. 1c). The selected area electron diffraction (SAED) pattern further revealed the interplanar spacing and angles of VPNSs (Fig. 1c inset). The XRD pattern was coherent with the reported phosphorus crystal structure (Fig. S4†), indicating that VPNSs were successfully synthesized via an exfoliation approach. The dispersivity and turbidity of VPNSs in water were also evaluated as a function of aging time. After standing for 90 min, the dispersed solution was still uniform and only a small part of it had settled, which was conducive to full contact between the bacteria and samples (Fig. S5†).Next, Raman spectra were used to explore the structural transformation that occurred during exfoliation (Fig. 1d and S6†). The Raman spectra of VPNSs shifted towards higher wavenumbers compared with VP, demonstrating that the thickness decreased after exfoliation.50 X-ray photoelectron spectra (XPS) were utilized to examine the degree of VPNS degradation during the exfoliation process. Three peaks located at 129.7, 130.4, and 133.7 eV, respectively, corresponded to 2P3/2, 2P1/2, and POx signals (Fig. 1e and S7†). A weak peak at 133.7 eV suggested that VP had no obvious oxidation or chemical degradation during the exfoliation process. Energy dispersive spectroscopy (EDS) and TEM mapping analyses revealed the existence and homogeneous distribution of elemental phosphorus across all samples, further indicating the high purity of the VPNSs (Fig. 1f and S8†). As the newest member in the layered phosphorus allotrope family, the structure of violet phosphorus has been confirmed by single-crystal XRD.51,52 The edges of the VPNSs are composed of serious sub-nanoneedles (Fig. 1g), and the sub-nanoneedles were tubular strands composed of –[P2][P8][P2][P9]– (Fig. 1h and i). The sub-nanoneedles were formed at the edges of VPNSs due to bond breakage during solvent exfoliation. The existence of sub-nanoneedles was also simulated for VPNSs (Fig. 1i and S9†). The length of the sub-nanoneedles varies from 2 nm to 8 nm, and each tubular sub-nanoneedle has a diameter of about 0.3–0.4 nm. The distance between each two sub-nanoneedles is about 0.6 nm. A series of sub-nanoneedle bundles help to penetrate the cell membrane. In comparison, each layer of BPNSs was made up of a zigzagging chain of phosphorus atoms, with the linked atoms forming a wrinkled sheet lacking the sub-nanoneedle structures in VPNSs (Fig. S10†). Meanwhile, we report for the first time that these sub-nanoneedle structures cannot be observed in most reported antibacterial two-dimensional materials.53–58
Antibacterial properties of VPNSs and BPNSs
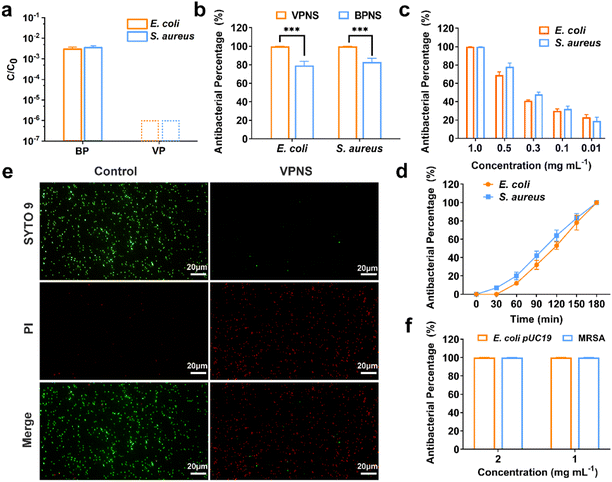
Knowing that the phosphorus allotrope BP and its phosphorene have proven successful at inactivating bacteria by photodynamic therapy (PDT) and photothermal therapy (PTT),59–62 we were inspired to examine the possibility of VP and VPNSs serving as antibacterial agents. We first evaluated the antibacterial action of VP against 107 colony-forming units per milliliter (CFU mL−1) of E. coli and S. aureus, with BP as a comparative using the colony-counting method. As shown in Fig. 2a, after exposure to 1.0 mg mL−1 of VP and BP for 3 hours under LED white light irradiation, the survival rate of E. coli and S. aureus in the presence of BP was significantly higher than in the presence of VP, and the corresponding digital photos further illustrated that VP had excellent antibacterial properties (Fig. S11†). Fig. S12† shows that there was no significant difference in the number of bacteria between the sample exposed to LED light for 180 min and that for 0 min, indicating no effect of long-term light irradiation on bacteria.We additionally explored the antibacterial properties of VPNSs and BPNSs under the same antibacterial conditions. Unexpectedly, a dramatic difference between the antibacterial effectiveness of VPNSs and BPNSs was observed. The antibacterial percentage of VPNSs reached >99.9% against E. coli and S. aureus, compared with 78% for E. coli and 85% for S. aureus in the presence of BPNSs, and this result illustrated that VPNSs had higher antibacterial activity than BPNSs (Fig. 2b). We also test the minimum inhibitory concentration (MIC) of VPNSs at 0.8 mg mL−1 against most of the bacteria and VPNSs can kill all the bacteria in this study efficiently with the MBC at 1.0 mg mL−1 (Fig. S13a and b†). As shown in Fig. 2c and d, dynamic testing also revealed that the antibacterial properties of VPNSs against E. coli and S. aureus were concentration- and time-dependent. Additionally, the photothermal efficacy of VPNSs was examined as a function of light time. The temperature of VPNSs was 26.2 °C even after irradiation for 180 min under LED white light, suggesting that photothermal action could not work on VPNSs (Fig. S14†).
To further assess the antibacterial action of VPNSs, E. coli and S. aureus in the absence and presence of VPNSs were assayed using a LIVE/DEAD Baclight Kit, and live bacteria were stained green with SYTO 9 and dead bacteria were stained red with PI. In Fig. 2e and S15,† intense green dots corresponded to the control group with live E. coli and S. aureus. After treatment with VPNS for 3 h under LED white light irradiation, intense red dots were detectable, revealing that E. coli and S. aureus had been killed. These data further verified VPNSs' high toxicity toward pathogens. We then investigated whether VPNSs worked on antibiotic-resistant bacteria using methicillin-resistant S. aureus (MRSA) and E. coli pUC19 as two models.63 As shown in Fig. 2f, VPNSs at concentrations of 1.0 and 2.0 mg mL−1 exhibited an antibacterial percentage of >99.9% for both models, indicating their effectiveness against drug-resistant bacteria. Additionally, the physical penetration effect of VPNSs on Gram-negative bacteria and Gram-positive bacteria was compared (Fig. S16†). A slightly lower antibacterial activity was observed for VPNSs against S. aureus after 3 h under dark conditions due to its thicker and more rigid cell wall compared to E. coli.
Antibacterial mechanism of VPNSs
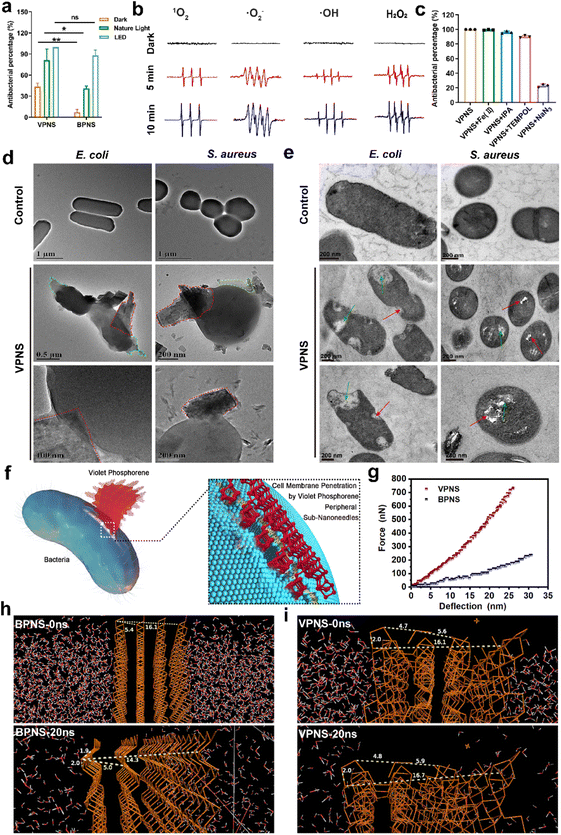
Given the notable discrepancy between the performance of VPNSs and BPNSs, we sought to explore the VPNS's antibacterial mechanism. First, we examined whether, like BPNSs, VPNSs's performance was affected by light irradiation. We conducted a contrast experiment by testing VPNSs's efficiency against E. coli in the dark, under natural light irradiation, and under LED white light irradiation. Fig. 3a showed that VPNS were more toxic under light (especially LED) than in the dark, indicating their light dependency. We therefore considered that VPNS attacked bacteria via ROS-dependent oxidative stress induced by PDT, a primary mechanism for BPNSs. Using electron spin resonance (ESR) spectra, we detected whether VPNSs generated ROS in the dark and under light irradiation. As presented in Fig. 3b, VPNSs produced four ROS signals under light irradiation: singlet oxygen (1O2), superoxide anion radical (˙O2−), hydrogen peroxide (H2O2), and hydroxyl radical (˙OH), respectively. On the contrary, no ROS were produced in the dark. Further, the four ROS signals strengthened as a function of light exposure time, suggesting that the generation of ROS depended on light irradiation time. In contrast, BPNSs can only generate 1O2, demonstrating that VPNSs produced more ROS species under the same testing conditions.64,65 Subsequently, a scavenging experiment was carried out to discover whether these four ROS species contributed to VPNSs' bactericidal activity under light irradiation.66–69 We used four trapping agents: 4-hydroxy-2,2,6,6 tetramethylpiperidinyloxy (TEMPOL) for ˙O2−, isopropyl alcohol (IPA) for ˙OH, ethylenediaminetetraacetic acid ferric sodium salt (Fe(II)) for H2O2, and sodium azide (NaN3) for 1O2. As illustrated in Fig. 3c, VPNSs exhibited almost the same antibacterial rates in the presence and absence of TEMPOL, IPA, and Fe(II), demonstrating that neither ˙O2−, ˙OH, nor H2O2 accounted for VPNSs' superior antibacterial performance. However, when NaN3 was used to scavenge 1O2, the antibacterial rate decreased to 21%, indicating that 1O2 played a key role in eliminating bacteria.In addition to light-dependent oxidative stress against bacteria, we also found that VPNSs also had a certain antibacterial ability (43%) in the dark and the activity of VPNSs was higher than that of BPNSs (7%) (Fig. 3a). Several other typical 2D nanomaterials can inflict physical damage on the cell membrane, including graphene oxide and graphdiyne.70–75 The interaction between VPNSs and bacteria is randomly orientated in solution. The VPNSs contacted with bacteria at all different orientations. The VPNSs with certain angles were able to penetrate the cell membrane of bacteria. This is an important reason to explain why the antimicrobial rate of VPNSs under dark conditions is only about 40%. The SEM and TEM images were taken to examine the morphology and membrane integrity of bacteria in the presence of VPNSs under LED irradiation conditions (Fig. 3d). The bacterial cells in control groups were observed to have smooth and intact cell membranes, without cytomembrane destruction or cytoplasmic leakage. However, significant wrinkles and depressions were observed to appear on the cytomembranes in the presence of VPNSs. Some VPNSs were also found to penetrate the cells (red dotted box in Fig. 3d), causing cytoplasm (green dotted box in Fig. 3d) and DNA (Fig. S17†) leakage and killing the bacteria. The damage of E. coli (Video 1†) and S. aureus (Video 2†) was also observed after VPNS treatment due to VPNS puncture combined with oxidative stress from TEM. The untreated bacteria were observed to have normal morphology and a dense intracellular matrix (Fig. 3e), while the VPNS-treated bacterial sections were observed to have bacterial rupture and disintegration (green arrow Fig. 3e) as well as cell wall and membrane damage caused by VPNS puncture (red arrow Fig. 3e). The morphology of bacteria treated with VPNSs has been demonstrated to show depressions, folds, and damage compared to the smooth and flat bacterial surface in the control group, indicating the death of bacteria of treated ones (SEM, Fig. S18†). Additionally, the morphology of bacteria treated with VPNSs under dark conditions was also determined to evaluate the physical destruction effect of VPNSs on bacteria. The VPNSs were observed to penetrate bacteria under the induction of sub-nanoneedles (Fig. S19†), resulting in bacterial death. The bacteria were observed to have many holes and damage on the cell wall and membrane after treatment of VPNSs (Fig. S20†). The VPNSs have been demonstrated to puncture bacteria and cause bacterial death under the induction of sub-nanoneedles. The length of sub-nanoneedles is usually shorter than the thickness of the phospholipid bilayer. The cell wall or membrane could not be pieced by the sub-nanoneedles from the edge of VPNSs alone. However, the sub-nanoneedles with sharp tips are favored to penetrate the bacteria like ‘gears’, driving entire VPNSs into the bacteria and eventually killing the bacteria (Fig. 3f). In addition, the dynamic light scattering (DLS) results demonstrated that the size of VPNSs was close to that of bacteria, which was conducive to sufficient contact between them for cell membrane penetration induced by the VPNS's peripheral sub-nanoneedles (Fig. S21†). The interaction between VPNSs and bacteria was further explored to understand the penetration of VPNSs to bacteria. A strong peak at 1032 cm−1 corresponding to the stretching vibration of P–OC and peaks at 2925 and 2857 cm−1 corresponding to the stretching vibrations of the CH2 and CH3 groups were detected from the Fourier transform infrared spectroscopy (FTIR) spectrum of VPNSs (Fig. S22†). The dangling phosphorus bonds of VPNSs have been demonstrated to be functionalized by the exfoliation solvent (anhydrous ethanol) to form stable –P–OCH2CH3 groups. The contact angle of VPNSs with water was also measured to be 92.8°, suggesting the hydrophobic feature of functionalized alkyl chains to VPNSs (Fig. S23†). Therefore, the hydrophobic chain of VPNSs has been demonstrated to further promote its insertion into bacteria through hydrophobic interaction with hydrophobic bacteria due to the presence of polysaccharides in the cell wall and phospholipids in the membrane.
The cell membrane was easily penetrated induced by the peripheral sub-nanoneedles due to the extremely high 2D Young's modulus of VPNSs (Fig. 3g). The 2D Young's modulus of monolayer violet phosphorene (1512 ± 76 N m−1) has been deduced to be 4.4 times as high as that of graphene (340 N m−1) and much higher than that of BPNSs (214 N m−1).76 The mechanical properties were further confirmed using theoretical calculations, as shown in Fig. 3h, i, and S24.† Molecular dynamics (MD) simulation of the NPT (normal pressure and temperature) ensemble was employed to investigate the system changes under a pressure of 1.0 atm within 20 ns of simulation. The P–P bond lengths and layer distances in BPNSs and VPNSs were monitored during MD processing. At the starting point (0 ns), the distances between two layers and four layers of BPNSs were 5.4 angstroms and 16.1 angstroms, respectively, decreasing to 5.0 angstroms and 14.3 angstroms after 20 ns in the MD system. The conditions were different in the VPNS system. The distance from the top side to the bottom side and the distance between two layers changed slightly during 20 ns of MD processing, demonstrating that due to their 2D crisscross structure, VPNSs had a higher deformation resistance than BPNSs, making them more physically destructive toward bacteria. This effect of VPNSs on E. coli based on contact-triggered action was further confirmed by inhibition zone testing in the dark. As shown in Fig. S25,† unlike AgNO3, which exhibited an obvious inhibition ring due to the release of Ag+ ions, neither VPNSs nor BPNSs showed an aseptic area, confirming their antibacterial mechanism including physical penetration of the bacteria.
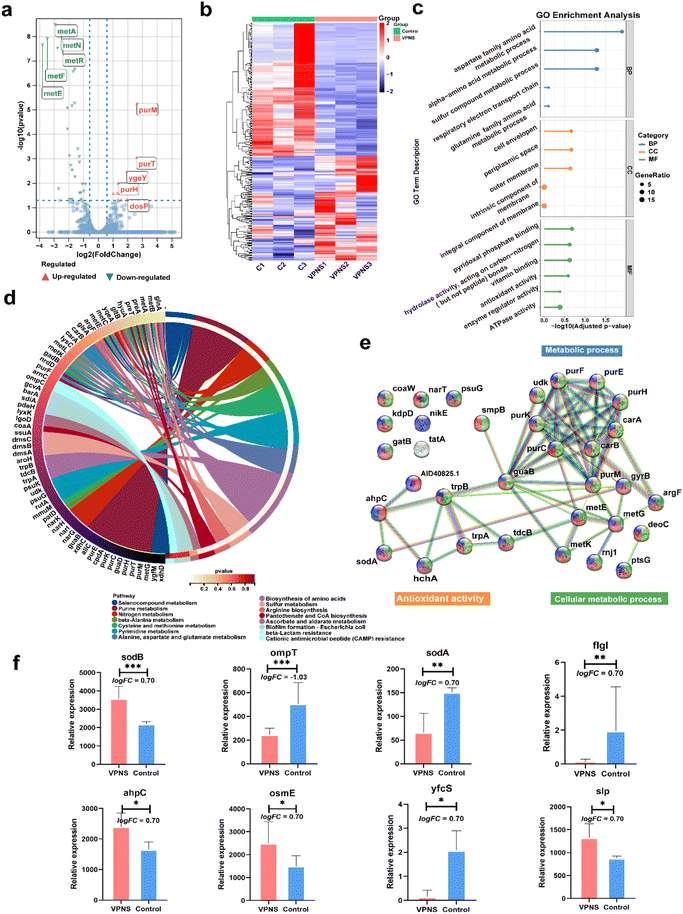
The above experiments demonstrated that VPNSs showed a significant inhibitory effect on bacterial growth. To further explore the molecular mechanism by which VPNSs inhibit bacterial growth, we attempted to assess the specific response at the mRNA level using RNA sequencing (RNA-Seq) (Fig. 4). The correlation of gene expression levels between samples, an important index to test the reliability of the experiment and the reasonableness of sample selection, was also conducted. The R2 values of both inter-group and intra-group samples were found to be greater than 0.8 (Fig. S26†), indicating a good biological repeatability. 4901 genes were identified, of which 374 were differentially expressed (|log2(fold change)| > 1 and P value < 0.05), including 210 upregulated genes and 164 downregulated genes. The top 5 most differentially upregulated and downregulated genes are displayed in the volcano plot (Fig. 4a and b). To further investigate the molecular mechanisms of differentially expressed genes, gene ontology (GO) enrichment analysis was performed. As shown in Fig. 4c, a total of 330 GO terms were enriched, including 183 biological process (BP), 21 cellular component (CC), and 126 molecular function (MF) terms. The following were noteworthy BP terms: aspartate family amino acid metabolic process (GO:0009066) and sulfur compound metabolic process (GO:000679). The periplasmic space (GO:0042597), cell envelope (GO:0030313), and outer membrane (GO:0019867) were the main distributed terms in CC ontology. With regard to the ontology of MF, the important categories were pyridoxal phosphate binding (GO:0030170), vitamin binding (GO:0019842), and hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds (GO:0016810).
Subsequently, the Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment analysis was employed to explore the possible molecular regulation pathways (Fig. 4d). In the ontology of KEGG, the main categories were gathered at terms: selenocompound metabolism, purine metabolism, nitrogen metabolism, beta-alanine metabolism, cysteine, methionine metabolism, pyrimidine metabolism, alanine, aspartate, and glutamate metabolism, biosynthesis of amino acids, sulfur metabolism, arginine biosynthesis, pantothenate, CoA biosynthesis etc. It suggested that VPNSs might play an important role in the antibacterial process by regulating those pathways. Additionally, to display the correlation of differential expression genes (DEGs) with each other, a protein–protein interaction (PPI) network was constructed (Fig. 4e). The majority of the identified DEGs were enriched by KEGG. The most interacted proteins were mainly concentrated in the following pathways: purine metabolism, biosynthesis of amino acids, and cellular anatomical entity. Based on the above result, the regulated genes associated with antimicrobial mechanisms are shown in Fig. 4f as bar graphs individually. All sodB, sodA, and ahpC were associated with oxidative stress and ompT, fIgI, osmE, yfcS, and slp associated with membrane destruction were up-regulated or down-regulated compared with the control group.
In addition to antibacterial activity, it is essential to evaluate the biocompatibility of VPNSs before the material can be applied in vivo. First, the cell viability and cytotoxicity of VPNSs were assessed using CCK-8 and MTT assays (Fig. S27 and S28†). The VPNSs showed no toxicity toward NIH 3T3 cells, Raw 264.7 cells, or L02 cells within a concentration range of 0.125–2.0 mg mL−1, suggesting that VPNSs have good biocompatibility. Hence, it was potential for use in clinical applications. In addition, Fig. S29 and S30† show that compared with PEI25K, L02 cells treated with different concentrations of VPNSs emitted strong green fluorescence and blue fluorescence, respectively, in live/dead cell staining and TUNEL cell staining assays, indicating that the majority of the cells remained viable. The results showed that no cytotoxicity or apoptosis was induced in the concentration range of 0.125–2.0 mg mL−1. The toxicity of VPNSs to human corneal epithelial cells (HCECs) associated with the eyeball has also been measured (Fig. S31†). Strong green fluorescence was detected in both live and dead HCECs after treating with VPNSs with different concentrations, indicating no toxicity of VPNSs in the concentration range of 0.125–2.0 mg mL−1 to HCECs.
In addition, we also performed hemolysis testing to further evaluate the erythrocyte compatibility of VPNSs, using Triton X-100 as the positive control. As shown in Fig. S32,† after incubation with the VPNSs, followed by centrifugation, the supernatants were transparent, indicating that no disruption of the erythrocytes had occurred and no hemoglobin was released. The hemolysis rate of the VPNSs was only 1.7% at 2.0 mg mL−1, much lower than the ASTM standard (ASTM F756-2008) for the hemolysis ratio of biomaterials (<5%). VPNSs showed high antibacterial activity and low cytotoxicity to normal cells may be due to the rigidity difference between bacterial and eukaryotic cell membranes. It was reported that Young's modulus of human mesenchymal stem cells was in the region of 0.09–49 kPa compared to 50–200 kPa for certain bacterial cell envelopes.77–79 Eukaryotic cells are able to avoid membrane damage and cell death by stretching and twisting to fit the sub-nanoneedle shape of VPNSs.80
In vivo wound infection treatment
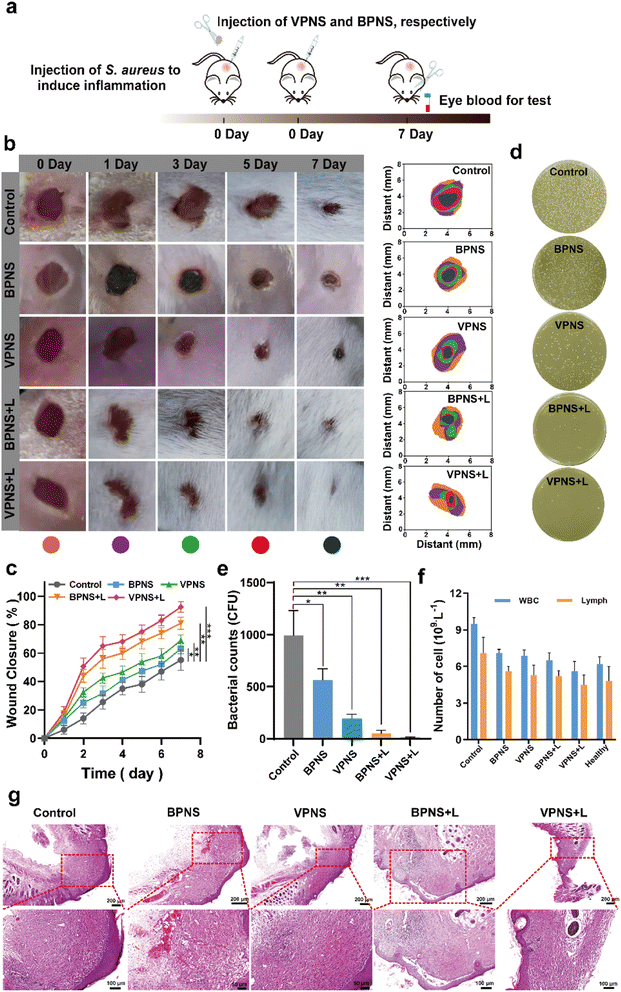
Encouraged by the excellent in vitro antibacterial properties of VPNSs, coupled with their biosafety, we then investigated in vivo antibacterial effectiveness of VPNSs in skin or soft tissue-related infections that induced severe inflammation. A wound-infection model in mice was established using S. aureus, and the efficiency of wound healing was observed before and after treatment with VPNSs and BPNSs (Fig. 5a). The infected mice were randomly divided into five groups and then treated with PBS (control), BPNSs, VPNSs, BPNSs with LED irradiation (BPNS + L), and VPNSs with LED irradiation (VPNS + L) within 7 days. As shown in Fig. 5b and c, the wounds treated with VPNSs and BPNSs exhibited significantly faster healing than those in the control group. Especially, those treated with VPNS + L healed the fastest and were almost completely gone within 7 days, whereas the wound area in the control group was still about 55% ± 7.3% of the original size. We also used the colony-counting method to evaluate bacterial survival at the infection site on day 7. Fig. 5d and e show that the bacterial survival numbers in wounds treated with VPNS + L and BPNS + L groups were obviously lower than for the other groups, with the VPNS + L group showing the lowest survival and thereby demonstrating that the material was active in vivo.Routine blood analysis was conducted on the treated mice, as shown in Fig. 5f. Compared with the control group, the experimental groups showed fewer white blood cells (WBCs) and lymphocytes (Lymph), demonstrating that VPNSs and BPNSs had anti-infection activity in vivo. Notably, the blood values of the VPNS + L-treated mice had almost returned to normal levels, showing that VPNSs were the most effective material. Finally, hematoxylin–eosin (H&E) staining of wound tissues revealed the wound-healing process in different treatment groups at the histological level. As shown in Fig. 5g, the control group presented obvious inflammatory cells, whereas none were found after treatment with BPNS, VPNS, VPNS + L, and BPNS + L groups and the wounds were filled with granulation tissue. Among the four sample types, the granulation tissue treated with VPNS + L was the most mature, indicating that VPNSs had the most positive impact on wound healing. Upon comparison, the VPNS + L group demonstrated greater healing efficiency than the other groups.
In vivo bacterial keratitis treatment
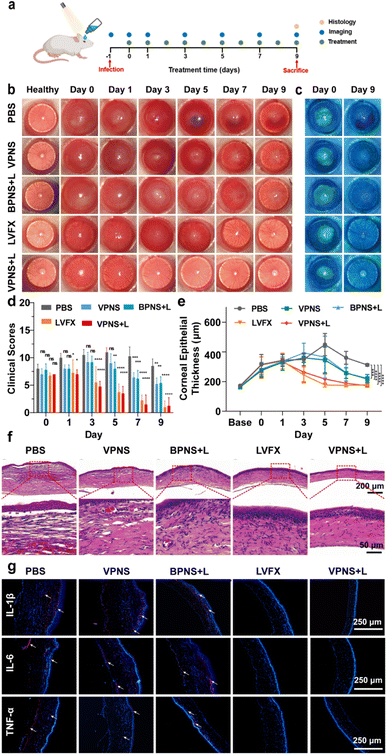
Based on the good therapeutic effect of VPNSs on skin wounds and their excellent antibacterial properties, a keratitis model of MRSA-infected SD rats was also established in this study to evaluate the therapeutic effect of VPNSs in vivo (Fig. 6a). The rats were randomly divided into five groups: (1) control (PBS), (2) VPNS, (3) BPNS with LED irradiation (BPNS + L), (4) VPNS with LED irradiation (VPNS + L), and (5) levofloxacin (LVFX), respectively. Then slit-lamp images were captured on the following 9 days to monitor the apparent morphology and inflammatory response characteristics of corneas (Fig. 6b and c). On day 0, similar corneal infiltrates and abscesses were observed in all groups, indicating the same infection level at the beginning of treatment. In the PBS-treated group, the eyes presented obvious corneal edema, inflammatory infiltration in the anterior chamber, and persistent infection. As a control, the development of corneal infections in VPNS and BPNS + L was well controlled, but the symptoms were not eliminated until day 9, leading to a longer recovery time. Surprisingly, the inflammation and infection were significantly reduced in the VPNS + L group on day 3, and the treatment efficiency of the VPNS + L group was even better than that of the levofloxacin group, which could be attributed to the rapid sterilization of VPNS-producing more ROS under LED irradiation. Correspondingly, the clinical scores of the eyes in PBS, VPNS, and BPNS + L groups showed no significant decrease over time. As a comparison, the eyes in the VPNS + L group showed the lowest clinical score on day 9 and the total clinical core was no higher than 1 (Fig. 6d). Furthermore, the increased corneal thickness revealed the lack of therapeutic effects of the VPNS group and BPNS + L group in treating refractory keratitis. It was also worth noting that the corneal thickness remained unchanged in the VPNS + L group compared with the healthy rats on day 9 (Fig. 6e).Furthermore, H&E staining and histological analysis were performed on the corneal tissues after 9 days of treatment. As shown in Fig. 6f, H&E staining images showed that the corneal structures of rats treated in PBS groups were destroyed with significant inflammatory cell infiltration and corneal edema. Compared with the PBS group, a decrease in corneal edema and infiltration of inflammatory cells was observed in the VPNS and BPNS + L groups. Only in VPNS + L and levofloxacin groups that showed uniform corneal layers without obvious inflammatory cell infiltration and corneal edema, the corneal epithelial cells regenerated well and were closely arranged. Next, immunofluorescence staining was also conducted to investigate the expression of proinflammatory factors 1L-1β, IL-6, and TNF-α associated with keratitis treatment. As seen in Fig. 6g, the lowest red fluorescence signal levels were observed in rats treated with the VPNS + L group, indicating much fewer inflammatory mediators expressed in this group. The above results fully demonstrated that VPNSs under LED irradiation had excellent antibacterial ability and good inflammatory mitigation ability, which provided new insight into healing refractory keratitis.
Conclusions
In summary, we reported the antibacterial activities of VP and VPNSs, which are able to inactivate more than 99.9% of common pathogens (E. coli and S. aureus) and antibiotic-resistant bacteria (E. coli pUC19 and MRSA). VPNSs were demonstrated to have higher antibacterial activity than BPNSs owing to the unique two-layer structure of crisscrossed rods that form a nanogrid with numerous sub-nanoneedles along the edge, which led to the extremely high deformation resistance of VPNSs and made the cell membrane easily penetrated. Given their excellent ability to inactivate bacteria via physical penetration of the cell membrane, combined with oxidative stress, VPNSs have also shown the potential to accelerate wound healing in mice and bacterial keratitis in rats. This study not only presents an emerging 2D antibacterial agent but also offers encouragement for future studies on different biomedical applications of VPNSs.Data availability
All the data supporting this article have been included in the main text and the ESI.†Author contributions
Q. S., J. K., and A. D. designed the research plan. Q. S., X. Z., W. L. and B. Z. performed all of the experiments and analyzed the data. Q. S., J. K., Z. L., and A. D. worked on the figures. Q. S. contributed to the writing of the manuscript. J. K., B. W., and A. D. refined the draft. J. Z. and L. Z. provided technical support and conceptual advice. All authors discussed the results and implications and edited the manuscript at all stages.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (22062017), the Natural Science Foundation of Inner Mongolia Autonomous Region (2023QN02011), the Ordos City Program for Key Science and Technology (2022YY003), the Program of Higher-Level Talents of Inner Mongolia University (10000-22311201/035), and the Research Program of science and technology at Universities of Inner Mongolia Autonomous Region (NJZZ23091). All cells were purchased from the National Collection of Authenticated Cell Cultures. Ethical permission for the cell experiment was obtained from Inner Mongolia University and Wenzhou Medical University.References
- J. Jenkins, J. Mantell, C. Neal, A. Gholinia, P. Verkade, A. H. Nobbs and B. Su, Nat. Commun., 2020, 11, 1626 CrossRef CAS PubMed
.
- E. P. Ivanova, J. Hasan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, V. A. Baulin, S. Pogodin, J. Y. Wang, M. J. Tobin, C. Lobbe and R. J. Crawford, Small, 2012, 8, 2489–2494 CrossRef CAS PubMed
.
- J. Hasan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, M. J. Tobin, G. Gervinskas, S. Juodkazis, J. Y. Wang, R. J. Crawford and E. P. Ivanova, Langmuir, 2012, 28, 17404–17409 CrossRef CAS PubMed
.
- T. Sun, G. Qing, B. Su and L. Jiang, Chem. Soc. Rev., 2011, 40, 2909–2921 RSC
.
- S. M. Kelleher, O. Habimana, J. Lawler, B. O. Reilly, S. Daniels, E. Casey and A. Cowley, ACS Appl. Mater. Interfaces, 2016, 8, 14966–14974 CrossRef CAS PubMed
.
- A. Tripathy, P. Sen, B. Su and W. H. Briscoe, Adv. Colloid Interface Sci., 2017, 248, 85–104 CrossRef CAS PubMed
.
- K. Modaresifar, S. Azizian, M. Ganjian, L. E. Fratila-Apachitei and A. A. Zadpoor, Acta Biomater., 2019, 83, 29–36 CrossRef CAS PubMed
.
- V. T. H. Pham, V. K. Truong, A. Orlowska, S. Ghanaati, M. Barbeck, P. Booms, A. J. Fulcher, C. M. Bhadra, R. Buividas, V. Baulin, C. J. Kirkpatrick, P. Doran, D. E. Mainwaring, S. Juodkazis, R. J. Crawford and E. P. Ivanova, ACS Appl. Mater. Interfaces, 2016, 8, 22025–22031 CrossRef CAS PubMed
.
- J. Butler, R. D. Handy, M. Upton and A. Besinis, ACS Nano, 2023, 17, 7064–7092 CrossRef CAS PubMed
.
- N. I. Nikolaev, T. Müller, D. J. Williams and Y. Liu, J. Biomech., 2014, 47, 625–630 CrossRef PubMed
.
- J. Butler, R. D. Handy, M. Upton and A. Besinis, ACS Nano, 2023, 17, 7064–7092 CrossRef CAS PubMed
.
- H. H. Tuson, G. K. Auer, L. D. Renner, M. Hasebe, C. Tropini, M. Salick, W. C. Crone, A. Gopinathan, K. C. Huang and D. B. Weibel, Mol. Microbiol., 2012, 84, 874–891 CrossRef CAS PubMed
.
- L. Hanson, Z. C. Lin, C. Xie, Y. Cui and B. Cui, Nano Lett., 2012, 12, 5815–5820 CrossRef CAS PubMed
.
- E. P. Ivanova, J. Hasan, H. K. Webb, G. Gervinskas, S. Juodkazis, V. K. Truong, A. H. Wu, R. N. Lamb, V. A. Baulin, G. S. Watson, J. A. Watson, D. E. Mainwaring and R. J. Crawford, Nat. Commun., 2013, 4, 2838 CrossRef PubMed
.
- D. L. Gonzalez Arellano, K. W. Kolewe, V. K. Champagne, I. S. Kurtz, E. K. Burnett, J. A. Zakashansky, F. D. Arisoy, A. L. Briseno and J. D. Schiffman, Sci. Rep., 2018, 8, 11618 CrossRef PubMed
.
- C. Wen, H. Guo, H. Bai, T. Xu, M. Liu, J. Yang, Y. Zhu, W. Zhao, J. Zhang, M. Cao and L. Zhang, ACS Appl. Mater. Interfaces, 2019, 11, 34330–34337 CrossRef CAS PubMed
.
- V. K. Truong, H. K. Webb, E. Fadeeva, B. N. Chichkov, A. H. Wu, R. Lamb, J. Y. Wang, R. J. Crawford and E. P. Ivanova, Biofouling, 2012, 28, 539–550 CrossRef CAS PubMed
.
- B. Ezraty, A. Gennaris, F. Barras and J. F. Collet, Nat. Rev. Microbiol., 2017, 15, 385–396 CrossRef CAS PubMed
.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed
.
- V. T. H. Pham, V. K. Truong, M. D. J. Quinn, S. M. Notley, Y. Guo, V. A. Baulin, M. A. Kobaisi, R. J. Crawford and E. P. Ivanova, ACS Nano, 2015, 9, 8458–8467 CrossRef CAS PubMed
.
- L. Tan, J. Li, X. Liu, Z. Cui, X. Yang, K. W. K. Yeung, H. Pan, Y. Zheng, X. Wang and S. Wu, Small, 2018, 14, 1703197 CrossRef PubMed
.
- Z. Xiong, X. Zhang, S. Zhang, L. Lei, W. Ma, D. Li, W. Wang, Q. Zhao and B. Xing, Ecotoxicol. Environ. Saf., 2018, 161, 507–514 CrossRef CAS PubMed
.
- W. Liu, Y. Zhang, Y. Zhang and A. Dong, Chem.–Eur. J., 2020, 26, 2478–2485 CrossRef CAS PubMed
.
- Z. Sun, Y. Zhang, H. Yu, C. Yan, Y. Liu, S. Hong, H. Tao, A. W. Robertson, Z. Wang and A. A. H. Padua, Nanoscale, 2018, 10, 12543–12553 RSC
.
- J. Ouyang, R. Y. Liu, W. Chen, Z. Liu, Q. Xu, K. Zeng, L. Deng, L. Shen and Y. N. Liu, J. Mater. Chem. B, 2018, 6, 6302–6310 RSC
.
- N. M. Latiff, W. Z. Teo, Z. Sofer, A. C. Fisher and M. Pumera, Chem.–Eur. J., 2015, 21, 13991–13995 CrossRef CAS PubMed
.
- G. Qu, T. Xia, W. Zhou, X. Zhang, H. Zhang, L. Hu, J. Shi, X. F. Yu and G. Jiang, Chem. Rev., 2020, 120, 2288–2346 CrossRef CAS PubMed
.
- X. Wang, J. Shao, M. Abd El Raouf, H. Xie, H. Huang, H. Wang, P. K. Chu, X. F. Yu, Y. Yang, A. M. AbdEl-Aal, N. H. M. Mekkawy, R. J. Miron and Y. Zhang, Biomaterials, 2018, 179, 164–174 CrossRef CAS PubMed
.
- J. Mo, Y. Xu, L. Zhu, W. Wei and J. Zhao, Angew. Chem., Int. Ed., 2021, 60, 12524–12531 CrossRef CAS PubMed
.
- W. Zhou, T. Pan, H. Cui, Z. Zhao, P. K. Chu and X.-F. Yu, Angew. Chem., Int. Ed., 2019, 58, 769–774 CrossRef CAS PubMed
.
- H. Wang and X. F. Yu, Small, 2018, 14, 1702830 CrossRef PubMed
.
- Q. Zhou, Q. Chen, Y. Tong and J. Wang, Angew. Chem., Int. Ed., 2016, 55, 11437–11441 CrossRef CAS PubMed
.
- J. Shao, H. Xie, H. Huang, Z. Li, Z. Sun, Y. Xu, Q. Xiao, X. F. Yu, Y. Zhao, H. Zhang, H. Wang and P. K. Chu, Nat. Commun., 2016, 7, 12967 CrossRef CAS PubMed
.
- L. Tong, Q. Liao, Y. Zhao, H. Huang, A. Gao, W. Zhang, X. Gao, W. Wei, M. Guan, P. K. Chu and H. Wang, Biomaterials, 2019, 193, 1–11 CrossRef CAS PubMed
.
- J. Plutnar, Z. Sofer and M. Pumera, ACS Nano, 2018, 12, 8390–8396 CrossRef CAS PubMed
.
- H. Wang, X. Yang, W. Shao, S. Chen, J. Xie, X. Zhang, J. Wang and Y. Xie, J. Am. Chem. Soc., 2015, 137, 11376–11382 CrossRef CAS PubMed
.
- H. Wang, S. Jiang, W. Shao, X. Zhang, S. Chen, X. Sun, Q. Zhang, Y. Luo and Y. Xie, J. Am. Chem. Soc., 2018, 140, 3474–3480 CrossRef CAS PubMed
.
- L. Wang, Y. Li, L. Zhao, Z. Qi, J. Gou, S. Zhang and J. Z. Zhang, Nanoscale, 2020, 12, 19516–19535 RSC
.
- D. Xia, Z. Shen, G. Huang, W. Wang, J. C. Yu and P. K. Wong, Environ. Sci. Technol., 2015, 49, 6264–6273 CrossRef CAS PubMed
.
- R. Lv, D. Yang, P. Yang, J. Xu, F. He, S. Gai, C. Li, Y. Dai, G. Yang and J. Lin, Chem. Mater., 2016, 28, 4724–4734 CrossRef CAS
.
- aW. Chen, J. Ouyang, H. Liu, M. Chen, K. Zeng, J. Sheng, Z. Liu, Y. Han, L. Wang, J. Li, L. Deng, Y. N. Liu and S. Guo, Adv. Mater., 2017, 29, 1603864 CrossRef PubMed
.
- T. Guo, Y. Wu, Y. Lin, X. Xu, H. Lian, G. Huang, J. Z. Liu, X. Wu and H. H. Yang, Small, 2018, 14, 1702815 CrossRef
.
- X. Zou, L. Zhang, Z. Wang and Y. Luo, J. Am. Chem. Soc., 2016, 138, 2064–2077 CrossRef CAS PubMed
.
- Q. Xin, H. Shah, A. Nawaz, W. Xie, M. Z. Akram, A. Batool, L. Tian, S. U. Jan, R. Boddula, B. Guo, Q. Liu and J. R. Gong, Adv. Mater., 2019, 31, e1804838 CrossRef PubMed
.
- Y. Chong, C. Ge, G. Fang, R. Wu, H. Zhang, Z. Chai, C. Chen and J. J. Yin, Environ. Sci. Technol., 2017, 51, 10154–10161 CrossRef CAS PubMed
.
- H. J. Jian, R. S. Wu, T. Y. Lin, Y. J. Li, H. J. Lin, S. G. Harroun, J. Y. Lai and C. C. Huang, ACS Nano, 2017, 11, 6703–6716 CrossRef CAS PubMed
.
- L. Zhang, H. Huang, B. Zhang, M. Gu, D. Zhao, X. Zhao, L. Li, J. Zhou, K. Wu, Y. Cheng and J. Zhang, Angew. Chem., Int. Ed., 2020, 59, 1074–1080 CrossRef CAS PubMed
.
- B. Zhang, Z. Wang, H. Huang, L. Zhang, M. Gu, Y. Cheng, K. Wu, J. Zhou and J. Zhang, J. Mater. Chem. A, 2020, 8, 8586–8592 RSC
.
- L. Zhang, M. Gu, L. Li, X. Zhao, C. Fu, T. Liu, X. Xu, Y. Cheng and J. Zhang, Chem. Mater., 2020, 32, 7363–7369 CrossRef CAS
.
- H. Wang, X. Yang, W. Shao, S. Chen, J. Xie, X. Zhang, J. Wang and Y. Xie, J. Am. Chem. Soc., 2015, 137, 11376–11382 CrossRef CAS PubMed
.
- B. Zhang, Z. Wang, H. Huang, L. Zhang, M. Gu, Y. Cheng, K. Wu, J. Zhou and J. Zhang, J. Mater. Chem. A, 2020, 8, 8586–8592 RSC
.
- L. Zhang, H. Huang, B. Zhang, M. Gu, D. Zhao, X. Zhao, L. Li, J. Zhou, K. Wu, Y. Cheng and J. Zhang, Angew. Chem., Int. Ed., 2020, 59, 1074–1080 CrossRef CAS PubMed
.
- H. Wang, S. Jiang, W. Shao, X. Zhang, S. Chen, X. Sun, Q. Zhang, Y. Luo and Y. Xie, J. Am. Chem. Soc., 2018, 140, 3474–3480 CrossRef CAS PubMed
.
- F. Perreault, A. F. d. Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226–7236 CrossRef CAS PubMed
.
- Y. Chen, Q. H. Ji, G. Zhang, H. J. Liu and J. H. Qu, Angew. Chem., Int. Ed., 2021, 60, 7744–7751 CrossRef CAS PubMed
.
- F. Q. Chen, Y. Luo, X. G. Liu, Y. F. Zheng, Y. Han, D. P. Yang and S. L. Wu, Adv. Healthcare Mater., 2022, 11, 2200360 CrossRef CAS PubMed
.
- L. Zhou, H. Zheng, Z. X. Liu, S. Q. Wang, Z. Liu, F. Chen, H. P. Zhang, J. Kong, F. T. Zhou and Q. Y. Zhang, ACS Nano, 2021, 15, 2468–2480 CrossRef CAS PubMed
.
- H. Q. Geng, X. M. Li, X. J. Gao, Y. Y. Cong, Q. Liu, J. J. Li, Y. Guan, L. M. Wang and W. W. He, Nano Today, 2023, 52, 101989 CrossRef CAS
.
- W. Chen, J. Ouyang, H. Liu, M. Chen, K. Zeng, J. Sheng, Z. Liu, Y. Han, L. Wang, J. Li, L. Deng, Y. Liu and S. Guo, Adv. Mater., 2017, 29, 1603864 CrossRef PubMed
.
- J. Zeng, C. Gu, X. Geng, K. Lin, Y. Xie and X. Chen, Biomaterials, 2023, 297, 122122 CrossRef CAS PubMed
.
- K. Hu, L. Xie, Y. Zhang, H. Masayuki, Z. Yang, K. Nagatsu, H. Suzuki, J. Ouyang, X. Ji, J. Wei, H. Xu, O. C. Farokhzad, S. H. Liang, L. Wang, W. Tao and M. Zhang, Nat. Commun., 2020, 11, 2778 CrossRef CAS PubMed
.
- F. Wang, Q. Wu, G. Jia, L. Kong, R. Zuo, K. Feng, M. Hou, Y. Chai, J. Xu, C. Zhang and Q. Kang, Adv. Sci., 2023, 2303911 CrossRef CAS PubMed
.
- M. Zhou, Y. Qian, J. Xie, W. Zhang, W. Jiang, X. Xiao, S. Chen, C. Dai, Z. Cong, Z. Ji, N. Shao, L. Liu, Y. Wu and R. Liu, Angew. Chem., Int. Ed., 2020, 59, 6412–6419 CrossRef CAS
.
- H. Wang, X. Yang, W. Shao, S. Chen, J. Xie, X. Zhang, J. Wang and Y. Xie, J. Am. Chem. Soc., 2015, 137, 11376–11382 CrossRef CAS PubMed
.
- W. Liu, Y. Zhang, Y. Zhang and A. Dong, Chem.–Eur. J., 2020, 26, 2478–2485 CrossRef CAS PubMed
.
- Y. Zhang, W. Liu, Y. Li, Y. W. Yang, A. Dong and Y. Li, iScience, 2019, 19, 662–675 CrossRef CAS PubMed
.
- Y. Chen, A. Lu, Y. Li, L. Zhang, H. Y. Yip, H. Zhao, T. An and P. K. Wong, Environ. Sci. Technol., 2011, 45, 5689–5695 CrossRef CAS PubMed
.
- K. Rasool, M. Helal, A. Ali, C. E. Ren, Y. Gogotsi and K. A. Mahmoud, ACS Nano, 2016, 10, 3674–3684 CrossRef CAS PubMed
.
- C. Li, Z. Sun, W. Zhang, C. Yu and S. Zheng, Appl. Catal., B, 2018, 220, 272–282 CrossRef CAS
.
- X. Lu, X. Feng, J. R. Werber, C. Chu, I. Zucker, J. H. Kim, C. O. Osuji and M. Elimelech, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E9793–E9801 CAS
.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS PubMed
.
- Y. Li, H. Yuan, A. von dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed
.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS PubMed
.
- T. Guo, S. Zhuang, H. Qiu, Y. Guo, L. Wang, G. Jin, W. Lin, G. Huang and H. Yang, Part. Part. Syst. Charact., 2020, 37, 2000169 CrossRef CAS
.
- W. Zhang, Y. Chen, T. Huynh, Y. Yang, X. Yang and R. Zhou, Nanoscale, 2020, 12, 2810–2819 RSC
.
- B. Zhang, L. Zhang, Z. Wang, Y. Li, Y. Cheng, L. Ma and J. Zhang, J. Mater. Chem. A, 2021, 9, 1385 Search PubMed
.
- N. I. Nikolaev, T. Müller, D. J. Williams and Y. Liu, J. Biomech., 2014, 47, 625–630 CrossRef PubMed
.
- J. Butler, R. D. Handy, M. Upton and A. Besinis, ACS Nano, 2023, 17, 7064–7092 CrossRef CAS PubMed
.
- H. H. Tuson, G. K. Auer, L. D. Renner, M. Hasebe, C. Tropini, M. Salick, W. C. Crone, A. Gopinathan, K. C. Huang and D. B. Weibel, Mol. Microbiol., 2012, 84, 874–891 CrossRef CAS PubMed
.
- L. Hanson, Z. C. Lin, C. Xie, Y. Cui and B. Cui, Nano Lett., 2012, 12, 5815–5820 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05517e |
| This journal is © The Royal Society of Chemistry 2024 |