Open Access Article
Open Access ArticleUltrasensitive detection of aromatic water pollutants through protein immobilization driven organic electrochemical transistors†
Subhankar
Sahu‡
 a,
Lokesh
Kumar‡
a,
Lokesh
Kumar‡
 b,
Sumita
Das
a,
Dipti
Gupta
b,
Sumita
Das
a,
Dipti
Gupta
 *b and
Ruchi
Anand
*b and
Ruchi
Anand
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology Bombay, Powai, Mumbai 400076, India. E-mail: ruchi@chem.iitb.ac.in
bDepartment of Metallurgical Engineering and Materials Science, Indian Institute of Technology Bombay, Powai, Mumbai 400076, India. E-mail: diptig@iitb.ac.in
First published on 13th December 2023
Abstract
Xenobiotic aromatic water pollutants pose an extreme threat to environmental sustainability. Due to the lack of detectable functional groups in these compounds and scarcity of selective bio-recognition scaffolds, easy-to-use sensing strategies capable of on-site detection remain unavailable. Herein, to address this lacune, we entail a strategy that combines biosensor scaffolds with organic electronics to create a compact device for environmental aromatic pollution monitoring. As proof of principle, a sensor module capable of rapid, economic, reliable, and ultrasensitive detection of phenol down to 2 ppb (0.02 μM) was designed wherein biosensing protein MopR was coupled with an organic electrochemical transistor (OECT). For effective interfacing of the sensing scaffold MopR, graphene oxide (GO) nanosheets were optimized as a host immobilization matrix. The MopR–GO immobilized sensor module was subsequently substituted as the gate electrode with PEDOT:PSS serving as an organic semiconductor material. The resulting OECT sensor provided a favourable microenvironment for protein activity, maintaining high specificity. Exclusive phenol detection with minimal loss of sensitivity (<5% error) could be achieved in both complex pollutant mixtures and real environmental samples. This fabrication strategy that amalgamates biological biosensors with organic electronics harnesses the potential to achieve detection of a host of emerging pollutants.
Introduction
Access to clean water is indispensable for maintaining a healthy ecosystem as it is the key element for the survival and sustenance of life. According to an estimation by the World Health Organization (WHO) around 844 million people lack basic drinking water services and most importantly roughly around 2 billion people have used polluted drinking water since 2015 around the globe.1 Specifically, the pollution caused by organic compounds such as benzene, chlorobenzene, phenol, catechol, and toluene produced from anthropogenic activities including catalytic reforming and cracking, petrochemical processing, petroleum refining, etc. leads to carcinogenicity and embryotoxicity.2,3 Traditional detection strategies such as GC-MS, LC-MS, or HPLC are typically employed for monitoring such mono-aromatic xenobiotics. However, despite being sensitive, these techniques are costly to operate and demand complicated sample-handling procedures.4 Additionally, it is hard to selectively monitor these compounds as they are essentially hydrophobic in nature and lack functional groups that can be exploited for detection. Thus, fast, sensitive, direct, and economically beneficial sensing alternatives remain scarce for these compounds.5 One of the potential choices to tackle this issue is to employ biosensing as an alternative. This is because due to the accumulation of these pollutants, interestingly a sub-set of bacteria in the environment has evolved to sense these xenobiotics and degrade them. Hence, these organisms possess natural sensors that can be used as a starting point to develop effective biosensing strategies.6,7Bioengineered sensing units amalgamated with electrochemical technologies serve as a lucrative prospect to create methods that possess several advantages such as ultra-sensitivity, ease of operation, on-site detection, and cost-effectiveness.8–11 Against this background, in the last decade, organic electrochemical transistor (OECT) based platforms have become pivotal as next-generation devices due to their exemplary biocompatibility, ability to operate in an aqueous medium, high transconductance, cheap fabrication, and portable nature.12–14 In particular OECTs have become extensively popular for probing biological interaction/reactions through the modulation of drain current in aqueous environments making them an impressive platform for biosensing. In the past few years, OECTs have been employed for sensing a wide spectrum of analytes ranging from small molecules such as ascorbic acid15 and dopamine16,17 to large virus particles such as influenza18 and SARS-CoV-2.19 However, this sensing platform has remained less chartered and elusive in the fields of water pollutant detection and environmental monitoring. Several limiting factors including a lack of suitable selective and sensitive biosensing scaffolds, difficulty in protein immobilization, noise issues from real samples, etc., have eluded its development.
Since there are several bacterial species that possess specific mechanisms for xenobiotic sensing, in recent times, whole cell biosensors (WCBs) have become a popular method for selective detection of mono-aromatics reaching highly sensitive LOD such as 1 ppb (∼0.01 μM) for phenol detection.20 In several instances, using synthetic biology approaches genetically modified organisms have been generated. However, it poses limitations as it is a less practical approach and accidental release of modified organisms poses ethical issues.21,22 Alternatively colorimetric detection using in vitro protein-based solution has also been employed but it is marred by issues pertaining to a low limit of detection (LOD). In parallel, electrochemical biosensors for detection of compounds such as phenol have been constructed using generic enzymes such as tyrosinase23 and laccase.24 Unfortunately, these enzymes lack selectivity and respond to a multitude of compounds with phenolic hydroxyl embedded in them.11,25,26 Recently, for phenol sensing, MopR, which is a very specific and selective protein, has found success as an excellent biosensing candidate.6 The MopR sensor scaffold is malleable and it has been shown that the phenol binding pocket can be engineered to create a series of selective sensors for a host of mono-aromatic pollutants such as benzene, toluene, and xylenol classes of compounds.27
The gap in MopR mediated biosensing remains twofold; first, the colorimetric detection system for in vitro sensing had a low LOD, and second, the setup was not in a translational-friendly compact version. Hence, here we have been successful in creating a compact and sensitive aromatic electrochemical biosensing platform using an OECT. As proof of principle, we show that phenol can be detected as low as 2 ppb (0.02 μM) in water, which is in the drinking water regime, thereby, surpassing the sensitivity achieved from the colorimetric MopR sensor by 5-fold. To create this setup a crucial step was to immobilize the protein on a suitable nano-support compatible with the OECT device. Graphene oxide (GO) was optimized as a host nano-interface to tackle protein immobilization. Since GO and its forms possess large surface area, abundant functional groups (such as epoxide, hydroxyl, and carboxylic groups), high dispersion ability in water, low toxicity under physiological conditions, etc.,28–30 they served as an excellent support matrix. The device employed PEDOT:PSS as both the channel and gate electrode material wherein the MopR–GO nano–bio module was immobilized onto the PEDOT:PSS gate electrode. The resulting electrochemical biosensor was specific for phenol and showed negligible response towards isostructural compounds such as benzene, toluene, and 2,3-dimethylphenol (2,3-DMP). The setup efficiently detected the target analyte even in a mixture of contaminants as well as in real environmental samples. Since the MopR scaffold is malleable, this approach can be extended to engineer a series of sensors for the xenobiotic class of aromatics such as benzene, toluene, etc., and has the potential to be extended as a generic method for biosensing-based environmental pollution monitoring.
Results and discussion
Graphene oxide as a biocompatible protein immobilization matrix for aromatic pollutant sensing
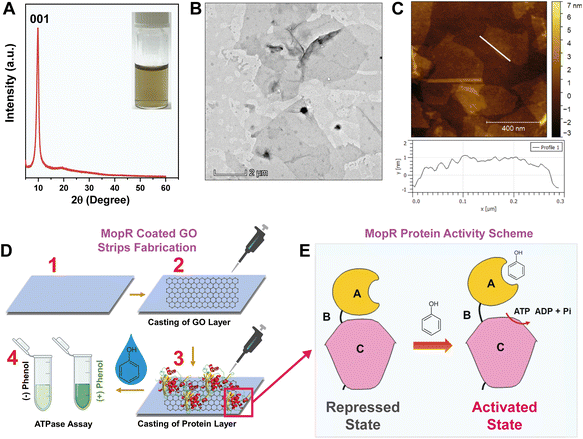
Biomolecule immobilization on nano-structures is one of the fundamental aspects of fabrication of ultrasensitive protein-based electrochemical sensors.31,32 As a first step, considering the views of rapid on-site detection process and potential for commercial translation, graphene oxide-based strips were designed for the effective immobilization of the protein (MopR) that is suitable for phenol detection without compromising protein activity. For this, GO was synthesized using modified Hummers' method33 as detailed in the Experimental section and a stable aqueous dispersion of graphene oxide flakes was obtained. The phase purity of the synthesized GO was first characterized by XRD analysis (Fig. 1A). The strong and sharp peak at 2θ = 9.8° corresponds to the characteristic 001 plane of GO suggesting the formation of a long-range ordered structure.34 The facile synthesis of GO yields a very thin and transparent sheet-like structure (Fig. 1B and S1A†) with lateral dimensions of several micrometres with an average height of around 1.4 nm (Fig. 1C) indicating the formation of mostly single-layer GO nanosheets. The functional groups identified for the synthesized GO nanosheets are O–H stretching (3412 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching (2135 cm−1), C
C stretching (2135 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(H2O) stretching (1650 cm−1), C–O–C stretching (1260 cm−1), and C–O stretching (1040 cm−1) vibrations as obtained from FT-IR study (Fig. S1B†).35 Furthermore, Raman analysis of the synthesized GO nanosheets also showed the characteristic D (1355 cm−1), G (1598 cm−1), 2D (2711 cm−1), and D + G (2916 cm−1) bands, respectively (Fig. S1C†). The D band originates from the structural defects present in the lattice whereas the G band corresponds to the sp2-bonded graphitic carbon network.36,37 The contact angle values of ∼12° (L) and ∼13° (R) confirm the hydrophilic nature of the synthesized GO (Fig. S1C-inset†). In addition, XPS survey analysis of GO nanosheets reveals the relative amounts of carbon and oxygen (Fig. S2A†). Subsequently, the collected core spectra of C 1s contain three distinct types of carbon peaks at around 284.8 eV (C–C/C
C(H2O) stretching (1650 cm−1), C–O–C stretching (1260 cm−1), and C–O stretching (1040 cm−1) vibrations as obtained from FT-IR study (Fig. S1B†).35 Furthermore, Raman analysis of the synthesized GO nanosheets also showed the characteristic D (1355 cm−1), G (1598 cm−1), 2D (2711 cm−1), and D + G (2916 cm−1) bands, respectively (Fig. S1C†). The D band originates from the structural defects present in the lattice whereas the G band corresponds to the sp2-bonded graphitic carbon network.36,37 The contact angle values of ∼12° (L) and ∼13° (R) confirm the hydrophilic nature of the synthesized GO (Fig. S1C-inset†). In addition, XPS survey analysis of GO nanosheets reveals the relative amounts of carbon and oxygen (Fig. S2A†). Subsequently, the collected core spectra of C 1s contain three distinct types of carbon peaks at around 284.8 eV (C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 287 eV (C–O–C), and 287.4 eV (C
C), 287 eV (C–O–C), and 287.4 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), respectively (Fig. S2B†)38 and Fig. S2C† represents the O 1s core spectra of GO sheets.
O), respectively (Fig. S2B†)38 and Fig. S2C† represents the O 1s core spectra of GO sheets.
The design of MopR-coated GO strips and the steps adopted for the sensing strategy are entailed in Fig. 1D and S3.† Briefly, first, a uniform, air-dried, drop-casted layer of GO (Fig. S3A†) was created on a plastic surface. Subsequently, a pure MopR protein (Fig. S4A†) was immobilized on the GO nano-support by physical adsorption. Specifically, in our biosensor design, physical adsorption of the protein was preferred as MopR is reported to undergo dramatic reorganization and conformational changes during biosensing39 which might be impeded by alternative approaches such as covalent tethering. The strips were then further treated with synthetically prepared water samples spiked with phenol and a colorimetric malachite green dye-based ATPase assay40 was used to confirm the biosensing activity (Fig. S3B and S3C†). The amount of GO and the protein were both varied and an optimal concentration of both, where sensor performance was maximal, was finally chosen for further experimentation. By virtue of the biochemistry of the protein, phenol recognition by MopR is correlated with its ATP hydrolysis reaction. The MopR protein used here comprises two structural domains namely the A-phenol sensing domain and C-ATP hydrolysis domain connected by a B linker (Fig. 1E). The ATP hydrolysis reaction at the C domain is regulated and tightly controlled in a concentration dependent manner by phenol binding at the A domain.6 The hydrolysis of ATP results in production of ADP and inorganic phosphate. It is the phosphate generated by hydrolysis that complexes with malachite green producing the requisite signal (detailed in the Experimental section). It is important to note that the MopR biosensing scaffold employed for all phenol sensing assays is a truncated version of the full-length protein (which contains one additional DNA binding domain6 after the C domain), comprising only the A to C domain (first 500 amino acid residues). This engineered protein has enhanced thermostability and is hence an ideal candidate to pursue fabrication studies.41
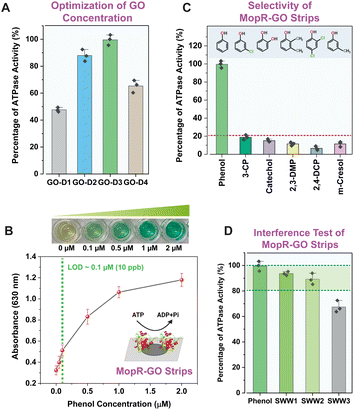
Fig. 2A clearly depicts that after sequential dilution of GO, GO-D3 strips with a GO concentration of around 0.2 mg mL−1 displayed significantly high sensor activity towards phenol detection compared to other dilutions of GO (GO-D1: 1.6 mg mL−1, GO-D2: 0.4 mg mL−1, and GO-D4: 0.1 mg mL−1). To enable robust phenol sensing under optimal assay conditions, the concentration of the functional nano-support is, therefore, critical. The circular dichroism (CD) spectra confirm that the protein is folded in the presence of GO (Fig. S4B†) inferring high biocompatibility of the synthesized GO nanosheets. Since the main biorecognition module is the MopR protein, the concentration of the active MopR species immobilized onto the GO surface is another crucial factor for phenol detection. Experiments show that a concentration of 2 μM protein exhibits the best phenol detection efficiency (Fig. S5†) and any protein concentration > 2 μM results in a poor dynamic range because of the high absorbance value beyond the linear range (higher than 2). The colorimetric limit of detection of the 2 μM MopR coated GO-D3 strips is 0.1 μM phenol (LOD ∼ 10 ppb) in water with an almost linear dynamic range of 0–1 μM (Fig. 2B). After optimization of the sensor design the specificity of the MopR–GO sensor strip was ascertained by testing a wide range of other isotypic phenolics such as 3-chlorophenol (3-CP), catechol, 2,3-di-methylphenol (2,3-DMP), 2,4-dichlorophenol (2,3-DCP) and m-cresol. The MopR–GO biosensing strip exhibited the highest activity response towards phenol in comparison to other phenolics (activity was less than 20%) (Fig. 2C). Thus, it can be concluded that the sensor possesses high sensitivity and selectivity in the developed strip format. The final optimized MopR-coated GO strips were similarly competent in detection of phenol (with 94% efficiency) when compared with the in vitro solution-based pure protein activity (Fig. S6 and S7†). Such an observation clearly emphasizes the success of GO as a host immobilization matrix for designing biosensing strips, as it can effectively retain the desired conformational plasticity of the protein. Furthermore, in order to screen the effect of surface functional groups on the sensor function, MopR immobilization on reduced graphene oxide (rGO) was also carried out. The results reveal that GO strips are a better nano-platform for sustaining the MopR protein activity (more details in the ESI†). Additionally, the MopR immobilized GO strip could also detect phenol with high confidence in simulated wastewater (SWW) samples containing other non-target pollutants (Fig. 2D). The response for SWW1 and SWW2 (comprising 1 μM and 5 μM of other aromatic pollutants respectively) was >80% thereby, showing negligible interference. SWW3 which contains a higher concentration, 10 μM, of other pollutants in the mixture also maintained a strong response of ∼70% demonstrating the robustness of the sensor design. Mop-GO strip parameters such as shelf-life, thermostability, and operating pH range (Fig. S8†) were also measured to demonstrate its environmental durability for field application.
Fabrication of OECTs for phenol monitoring
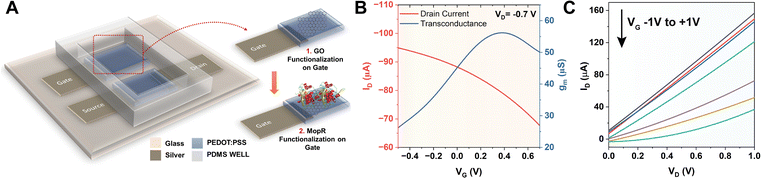
Biomolecule immobilization on different electrodes of OECTs is useful for sensing purposes as the device architecture is capable of monitoring biosensing reactions through current/voltage change as an output signal. The schematic of the designed OECT with the nano–bio module is presented in Fig. 3A, wherein the MopR-coated GO is immobilized on the gate electrode as a phenol sensing scaffold. The steps involved in device fabrication entail first the deposition process creating a screen printed PEDOT:PSS layer followed by optimization of nano–bio module immobilization on the gate electrode. PEDOT:PSS is used as a semiconducting channel between the source (S) and drain (D) and also as a gate (G) electrode. A similar kind of device architecture was exploited in the past by Gualandi et al. where the redox reaction at the PEDOT:PSS layer was monitored to detect ascorbic acid.42 However, here we aim to probe phenol via GO nanosheets coated with the protein. Briefly, the PEDOT:PSS gate electrode was primed for protein immobilization by first drop-casting and allowing physical adsorption of the GO layer onto the gate electrode before completing the functionalization. The integration of the nano–bio module on the gate electrode was next undertaken under well-controlled conditions of temperature and humidity to achieve the best quality of the electrode.After successful modulation of the gate electrode, the stability of the device was tested by immersing it in the buffer optimal for ATP hydrolysis of MopR. The device was then tested and it exhibited a drain current (ID) vs. gate voltage (VG) graph as typically observed for conventional OECT devices in depletion mode (Fig. 3B) at VD = −0.7 V. The maximum transconductance (gm) was achieved at VG = +0.4 V for the resulting OECT device (Fig. 3B). Increasing the VG influences the cations from the electrolyte to accumulate into the bulk of PEDOT:PSS and compensates for the negative sulfonate anions; this is equivalent to de-doping of the semiconducting material reducing the hole density and subsequently the ID. The output characteristics (IDvs. VD) of the designed OECT at varying VG of −1 V to +1 V are depicted in Fig. 3C. A typical decrease in the response of ID is observed for increasing values of VG which is a signature of depletion mode behaviour of PEDOT:PSS based OECTs.43 From the drain and transfer characteristics of the device, gate and channel bias were optimized and the ideal specifications for the operation of the OECT were set at VG +0.4 V and VD −0.7 V.
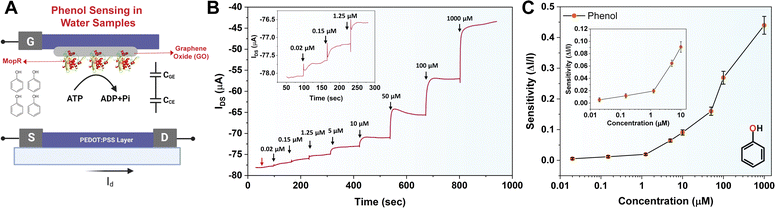
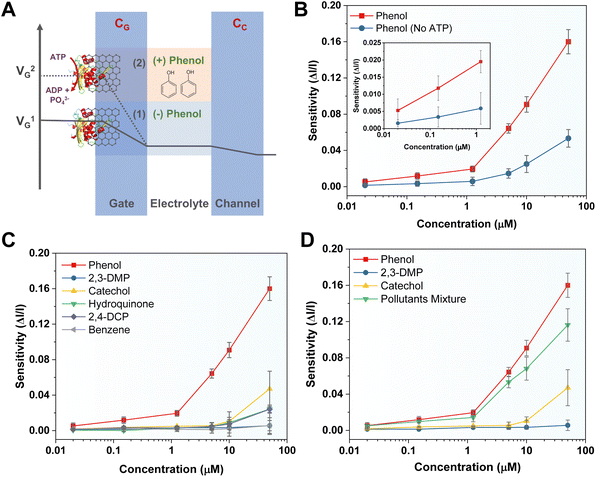
The working principle of the MopR-OECT biosensor is represented in Fig. 4A wherein the phenol concentration regulates the extent of ATP hydrolysis which in turn controls the negatively charged inorganic phosphate ions generated near the vicinity of the gate electrode (Fig. 4A). This influences the effective gate voltage (VeffG) and the proportional increase in the VG with the addition of phenol results in a reduction of ID. Immobilized MopR on the gate is inherently negatively charged in the electrolyte medium of pH ∼ 7.4 (MopR pI = 5.6). The negative charge arising from phosphate ions due to the ATPase activity alters the surface potential of the gate. In order to maintain a similar effective gate voltage, the VG increases resulting in a specific decrease in the ID (Fig. 5A). Moreover, the interplay of the capacitance at the gate/electrolyte (CGE) and channel/electrolyte (CCE) interface impacts the charge distribution at the two interfaces and thus influences the total current flow in the active channel.44 The amount of phenol in a particular sample is quantitated by employing this OECT device by serial addition of increasing concentrations of phenol and continuous monitoring of the ID. Real-time evolution of the ID as obtained showcases a staircase-like signal (Fig. 4B) which in general is perceived as a signature for the successful detection of analytes at the gate electrode.44 The decrease in the ID proportionally increases with increasing phenol concentration and the normalized current response is then converted to sensitivity. A titration of ATP concentrations yielded 5 mM as the optimal working concentration for the biosensor module (Fig. S9A†) and in the absence of ATP the device shows negligible sensitivity (Fig. 5B). Furthermore, it was established that no appreciable response was observed in the absence of the biosensing scaffold (Fig. S9B†). Moreover, Fig. 5B-inset, demonstrates that for 1 μM phenol the sensitivity of the device is around 4 times lower than that in the absence of ATP (sensitivity+ATP – 0.020 and sensitivity−ATP – 0.005).
Several other parameters such as device geometry and protein concentration were also tuned to optimize the biosensor device. Since PEDOT:PSS can take up the ions from the electrolyte into its bulk volume which results in volumetric doping, the optimization of the device geometry and thickness of the electrodes plays a crucial role in the sensitivity of the OECT geometry. Schematic of the optimized OECT geometry is shown in Fig. S10A† where the channel (LCH × WCH = 8 mm × 2 mm) and the gate (LG × WG = 8 mm × 2 mm) dimensions are the same and channel to gated lateral separation is kept at 2 mm. The surface topology of the screen printed PEDOT:PSS layer can be observed from a top view of FEG-SEM (Fig. S10B†) and the cross-section of the PEDOT:PSS layer indicated that it has an average thickness of ∼0.7 μm (Fig. S9C†). The GO layer with thickness ∼ 2.5 μm on PEDOT acts as an effective nano-platform for protein immobilization (Fig. S10C†). Furthermore, the EDS spectrum of the GO-coated gate indicates the desired functionalization of the PEDOT:PSS layer in combination with GO nanosheets (Fig. S10D†).
To achieve maximal sensing activity the protein concentration immobilized at the gate electrode was also optimized as clustering of the biomolecule at the nano-junction is a fundamental aspect of activity (schematic shown in Fig. S11A†). It was observed that a slightly higher protein concentration of 10 μM is optimum for achieving better sensitivity (Fig. S11B†). Here, it was noticed that for effective phenol probing through the OECT device, a higher protein concentration was required in comparison to the strip-based colorimetric biosensor. This could be because, in the case of the OECT, at least 10 μM of protein is required to produce sufficient phosphate anions to influence the VeffG. However, increasing the concentration further does not enhance device performance as the protein crowding in the interface45 might impact the conformational flexibility required by MopR affecting target molecule recognition and activity.
The chronoamperometric response shows that the device can detect phenol as low as 0.02 μM (Fig. 4B, inset). The LOD of the device was confirmed to be 0.02 μM of phenol. A lower concentration of 0.008 μM (first arrow-red, Fig. 4B) doesn't manifest any significant ID response. The sensitivity (ΔI/I) response plot for different concentrations of the phenol (Fig. 4C) depicts that the biosensor exhibits a wide dynamic range. Previously Mayorga-Martinez et al.46 reported an electrochemical biosensor for phenol detection using tyrosinase enzyme immobilized on a pnictogen-based substrate and the device reports a LOD level of 0.5 μM (500 nM). A higher LOD in this case could be because despite the well-optimized design of the accompanying immobilization and sensing scaffold, the tyrosinase and laccase enzymes are less selective. In contrast, the MopR-based biosensing unit is highly selective for phenol. The MopR scaffold has additional advantages as the ligand binding pocket can be structurally engineered to bind a battery of compounds and resulting sensors exhibit selective sensing for an array of monoaromatic (both phenolic and non-phenolic) ligands.27,47 Thus not only can this sensor design be easily multiplexed in the future where each pollutant can be reported with high precision but also the optimized GO nano-platform achieves a LOD of 0.02 μM which is 25-fold better than that of the tyrosinase system under similar electrochemical-based readout setting making this a robust and versatile setup. The device stability is comparable to the stability exhibited by the MopR–GO strip-based colorimetric sensor (Fig. S12†).
Sensitivity and selectivity of the biosensor
The MopR–GO biosensing strips can detect phenol with high precision. This is apparent from its selectivity profile. A similar observation was also found in the case of the OECT device; isostructural compounds such as benzene, catechol, hydroquinone, 2,3-DMP, and 2,4-DCP show negligible sensitivity (Fig. 5C). Though at high concentrations (50 μM) of analytes such as catechol, hydroquinone, and 2,4-DMP, the device shows increased sensitivity ∼ 0.04, but it is significantly less than that for phenol (sensitivity ∼ 0.16 at 50 μM). The time-channel current response profile also validates these observations (Fig. S9C†). Furthermore, the OECT was tested for its ability to detect phenol in a simulated pollutant mixture. Samples comprising phenol, 2,3-DMP, and catechol (referred to as ‘Pollutant Mixture’) were tested and the MopR-based OECT device could detect phenol selectively without loss of sensitivity (less than 5% error) in a lower concentration range of 0.02 to 1.25 μM (Fig. 5D). At higher concentrations of pollutants (example 10 μM and 50 μM) the sensitivity is affected with respect to the water spiked with pure phenol samples with an error of 20%; however, the sensitivity of phenol remains substantially higher than that of the non-targeted pollutants. Next, in order to validate the operation of the electrochemical biosensor for environmental samples, a locally collected water sample (EVS) from Vihar Lake, Mumbai, was spiked with a similar concentration range of phenol and experimented on the device. The sensitivity of the OECT was very similar in comparison to distilled water spiked with lab-grade phenol (0.02–0.15 μM) with an error of less than 3% (Fig. S13†) inferring that the OECT device was responsive for real samples and didn't undergo any degradation. A comparison of the MopR-OECT prototype biosensor with other phenol-detecting biosensing platforms such as those constructed from tyrosinase and laccases (Table 1) clearly demonstrates the high sensitivity and specificity of the MopR-OECT-based detection module. As mentioned earlier, both laccase and tyrosinase display non-specific activity, often generating signals for various phenolic compounds such as catechol or chlorophenols,48 diminishing their selectivity. Conversely, the high selectivity for phenol, exceptional thermal stability, and biocompatibility of MopR make it conducive to integration within the transistor architecture. Thus, it positions the MopR–GO-OECT module as a more promising scaffold for probing mono-aromatics in water.| Biosensing protein | Details | Selectivity | LOD | Reference | |
|---|---|---|---|---|---|
| 1 | Tyrosinase | Electrochemical biosensor. A graphene-decorated gold nanoparticle/chitosan (Gr–Au–Chit/Tyr) nanocomposite-modified screen-printed carbon electrode (SPCE) is used for protein immobilization and detection of phenolic compounds | + | Phenol, 0.05 μM | 54 |
| 2 | Tyrosinase | Electrochemical biosensor. Screen-printed carbon electrodes were modified with electrodeposition of Au nanoparticles (AuNPs) and tyrosinase was covalently immobilized | ++ | Phenol, 0.5 μM | 55 |
| 3 | Tyrosinase | Electrochemical biosensor. A glassy carbon electrode (GCE) was modified with enzyme-immobilized carbon nanotubes (CNTs) | ++ | Phenol, 0.035 μM | 56 |
| 4 | Laccase & tyrosinase | Electrochemical biosensor. Tyrosinase and laccase on graphite screen printed electrodes modified with ferrocene | + | Phenol, 2 μM | 57 |
| 5 | Tyrosinase | Electrochemical biosensor. Two-dimensional pnictogen nano-sheets are used for protein immobilization on glassy carbon electrodes | +++ | Phenol, 0.5 μM | 58 |
| 6 | Tyrosinase | Electrochemical biosensor. The enzyme was immobilized using a glutaraldehyde linker on gold nanoparticle modified screen-printed electrodes (Tyr–AuNPS-SPCEs) | + | Phenol, 1.2 μM | 59 |
| 7 | Tyrosinase | Optical biosensor. Tyrosinase was immobilized on a chitosan film coated on microscopy glass slides | + | Phenol, 1 μM | 60 |
| 8 | Laccase | Optical biosensor. Auto-indicating optical properties of laccase including intrinsic molecular absorption and fluorescence are used for phenol sensing | ++ | Phenol, 9 μM | 61 |
| 9 | Tyrosinase | Optical biosensor. A long-period fiber grating coated with an enzyme-entrapped polyacrylamide gel is used | + | Phenol, 81 μM | 62 |
| 10 | MopR | Electrochemical biosensor. An organic electrochemical transistor (OECT) based device employing protein immobilized graphene oxide nano-sheets is used | +++ | Phenol, 0.02 μM | This work |
Conclusion
In this work, an OECT-based device was fabricated for phenol monitoring by utilizing an MopR biosensing scaffold immobilized on the GO-modified gate electrode. Both the daunting aspects of protein immobilization and selective electrochemical sensing of hard-to-probe mono-aromatic xenobiotics were addressed. The resulting sensor yielded a LOD of less than 2 ppb towards phenol detection. In contrast to earlier reports, where enzymes such as tyrosinase and laccase were employed, MopR-based phenol biosensors were highly selective and could distinguish between isostructural ligands. The high thermal durability and biocompatibility allowed for integration with the OECT's modern architecture which is appealing due to the transistor-based amplification of the signal,49 making it more useful for probing biochemical interactions. In comparison to cyclic voltammetry and amperometry,50 OECTs are more suitable for bioelectronics and allow effective miniaturization of the sensing platform,51 which is useful for creation of low-cost and portable devices. Although the LOD of the MopR-OECT platform is comparable to that of synthetic biology inspired sensors,20 it circumvents most of the limitations associated with biocontamination, membrane transport and other biohazard risks, thereby, making the MopR-OECT design superior. The graphene oxide-based sensor strips developed here are also easy-to-fabricate, cheap, portable, and accurate for phenol detection in complex water samples without the need for any further modification to the sensor surface. Though the application of GO-based composite materials for bioremediation and sensing is widespread,52,53 the reported easy-to-fabricate MopR–GO design is versatile and is effective both in a colorimetric strip format as well as in an electrochemical sensing platform. Overall, the MopR-OECT platform established here holds the potential to be translated to other compounds and a panel of sensors that can individually sense a particular mono-aromatic can be designed. Thus, this design provides a future possibility of developing a water monitoring system that can accurately read multiple pollutants simultaneously. To conclude, here, a rational merging of MopR's biochemistry with an OECT device yielded a functional platform for ultrasensitive and selective detection of phenol. The work presented here opens doors for building systems that require amalgamation of bio-scaffolds with materials and device electronics. Our research sits at the crossroads of environmental sustainability and human health proving a promising approach for selective detection of aromatics.Experimental section
MopR–GO strip fabrication and colorimetric ATPase assay for phenol detection
The MopR protein was purified via a previously established protocol47 and detailed in the ESI.† Subsequently, different concentrations of the GO substrate were tested for MopR protein immobilization and activity testing. Plastic coverslips (Hampton Research, Cat. No. HR8-074) of 6 × 6 mm were used as a base and 10 μL of series of different concentrations of GO (GO-D1: 1.6 mg mL−1, GO-D2: 0.4 mg mL−1, GO-D3: 0.2 mg mL−1, and GO-D4: 0.1 mg mL−1) were drop-cast and dried for 30 min at 40 °C. The strips were then kept at room temperature for 5 min and 1 μL of 20 μM protein was drop-cast; the strips were incubated at RT for 20 min and finally, water samples containing the phenol samples were tested by colorimetric ATPase assay. After optimization of the GO concentration in the strip version, all further experiments were carried out using GO-D3 strips. Next, the MopR protein of different concentrations (1 μM, 2 μM, and 4 μM) was tested and the 2 μM protein was chosen due to its maximal performance. The ATPase assay employed herein is typically a malachite green dye based assay for the detection of phosphate in an in vitro solution.40 In an in vitro reaction, MopR protein, ATP, and water containing phenol are supplied for sensing (more details in the ESI†).Fabrication of OECTs
For making the OECT devices, the source, drain, and gate silver metal contacts (LADD, Cat. No. 60805) were screen printed on a cleaned glass slide. The substrate was printed with commercially available PEDOT:PSS (Sigma-Aldrich, Cat. No. 655201). Conductive screen printable ink was used as the active material (channel and gate) for the fabrication of the OECTs. The silver metal contact points are of dimensions 2 mm × 2 mm. The channel (LCH × WCH = 8 mm × 2 mm) and the gate (LG × WG = 8 mm × 2 mm) with a channel to gate lateral separation of 2 mm (D) were obtained by screen-printing PEDOT:PSS over a polyester film. For both steps, screen-printing was performed with a polyester 51T/130-71 PW mesh screen and 3M™ Screen Printing UV Ink for masking/patterning the mesh. Controlling the thickness of the PEDOT:PSS layer is crucial for the protein immobilization of the device. The average thickness of the PEDOT:PSS layer is optimized at ∼0.7 μM. For the thickness measurement, the PEDOT:PSS layer is coated on a silicon substrate (Sigma-Aldrich, Cat. No. 647675) and dried at 60 °C for 30 min. Then, at first the top image is captured showing the uniform formation of the PEDOT:PSS layer. Furthermore, the 0.2 mg per mL GO layer is coated on top of the PEDOT:PSS layer and the cross-section image is collected on a JEOL JSM-7600F FEG-SEM instrument. A Keithley 4200A parameter analyzer is used for measurement purposes. The sample testing and device operation are detailed in the ESI.†Further experimental details on protein purification, GO, rGO synthesis, and electrical characterization can be found in the ESI.†
Data availability
All the experimental data related to this study are available from the corresponding author upon reasonable request.Author contributions
Subhankar Sahu: conceptualization, data curation, formal analysis, methodology, investigation and writing – original draft. Lokesh Kumar: conceptualization, data curation, methodology, investigation and writing – original draft. Sumita Das: data curation, methodology, investigation and writing – original draft. Dipti Gupta: funding acquisition, supervision, resources and writing – review & editing. Ruchi Anand: project administration, funding acquisition, resources and writing – review & editing.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
R. A. acknowledges financial support from DST, Government of India [grant no. DST/TMD-EWO/WTI/2K19/EWFH/2019/48(G)] and DBT, Government of India [BT/PR18927/BCE/8/1376/2016]. D. G. acknowledges the funding support from the Indo-German Science and Technology Centre (IGSTC/WISER 2022/DG/46/2022-23/516) and SERB, Govt. of India (SPF/2021/000021). S. S. acknowledges funding agency Council of Scientific and Industrial Research (CSIR), Govt. of India for a PhD fellowship and IRCC, IIT Bombay for financial support. S. D. thanks IIT Bombay for a Post-Doctoral fellowship. L. K. acknowledges IIT Bombay for a PhD fellowship. The authors are also thankful to the Department of Chemistry, Metallurgical Engineering and Materials Science and Sophisticated Analytical Instrument Facility (SAIF), Indian Institute of Technology (IIT) Bombay, for the instrumental facility. The authors would also like to thank the IIT Bombay central facility at the Department of Physics for AFM and ESCA Lab for XPS characterization and TEM-300 kV MEMS facility of IIT Bombay for TEM characterization.References
- W. H. Organization and U. N. C. F. (UNICEF), Progress on drinking water, sanitation and hygiene: 2017 update and SDG baselines, World Health Organization, 2017 Search PubMed.
- O. Al-Madanat, Y. Alsalka, W. Ramadan and D. W. Bahnemann, Catalysts, 2021, 11, 1–45 Search PubMed.
- L. G. C. Villegas, N. Mashhadi, M. Chen, D. Mukherjee, K. E. Taylor and N. Biswas, Curr. Pollut. Rep., 2016, 2, 157–167 CrossRef CAS.
- E. Matisová and S. Hrouzková, Int. J. Environ. Res. Public Health, 2012, 9(9), 3166–3196 CrossRef PubMed.
- S. Sahu and R. Anand, in The World Scientific Reference of Water Science: Volume 1 Molecular Engineering of Water Sensors, World Scientific, 2023, pp. 101–136 Search PubMed.
- S. Ray, M. J. Gunzburg, M. Wilce, S. Panjikar and R. Anand, ACS Chem. Biol., 2016, 11, 2357–2365 CrossRef CAS PubMed.
- R. Fernandez-López, R. Ruiz, F. de la Cruz and G. Moncalián, Front. Microbiol., 2015, 6, 1–21 Search PubMed.
- F. Arduini, S. Cinti, V. Scognamiglio, D. Moscone and G. Palleschi, Anal. Chim. Acta, 2017, 959, 15–42 CrossRef CAS PubMed.
- G. Hernandez-Vargas, J. Sosa-Hernández, S. Saldarriaga-Hernandez, A. Villalba-Rodríguez, R. Parra-Saldivar and H. Iqbal, Biosensors, 2018, 8, 29 CrossRef PubMed.
- G. K. Mishra, V. Sharma and R. K. Mishra, Biosensors, 2018, 8, 1–13 CrossRef PubMed.
- E. Moczko, G. Istamboulie, C. Calas-Blanchard, R. Rouillon and T. Noguer, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 2286–2292 CrossRef CAS.
- R. A. Picca, K. Manoli, E. Macchia, L. Sarcina, C. Di Franco, N. Cioffi, D. Blasi, R. Österbacka, F. Torricelli, G. Scamarcio and L. Torsi, Adv. Funct. Mater., 2020, 30, 1904513 CrossRef CAS.
- B. Burtscher, P. A. M. Urbina, C. Diacci, S. Borghi, M. Pinti, A. Cossarizza, C. Salvarani, M. Berggren, F. Biscarini, D. T. Simon and C. A. Bortolotti, Adv. Healthcare Mater., 2021, 10, 2100955 CrossRef CAS PubMed.
- J. Rivnay, S. Inal, A. Salleo, R. M. Owens, M. Berggren and G. G. Malliaras, Nat. Rev. Mater., 2018, 3, 17086 CrossRef CAS.
- I. Gualandi, M. Marzocchi, E. Scavetta, M. Calienni, A. Bonfiglio and B. Fraboni, J. Mater. Chem. B, 2015, 3, 6753–6762 RSC.
- I. Gualandi, D. Tonelli, F. Mariani, E. Scavetta, M. Marzocchi and B. Fraboni, Sci. Rep., 2016, 6, 1–10 CrossRef PubMed.
- C. Liao, M. Zhang, L. Niu, Z. Zheng and F. Yan, J. Mater. Chem. B, 2014, 2, 191–200 RSC.
- W. Hai, T. Goda, H. Takeuchi, S. Yamaoka, Y. Horiguchi, A. Matsumoto and Y. Miyahara, Sens. Actuators, B, 2018, 260, 635–641 CrossRef CAS.
- K. Guo, S. Wustoni, A. Koklu, E. Díaz-Galicia, M. Moser, A. Hama, A. A. Alqahtani, A. N. Ahmad, F. S. Alhamlan, M. Shuaib, A. Pain, I. McCulloch, S. T. Arold, R. Grünberg and S. Inal, Nat. Biomed. Eng., 2021, 5, 666–677 CrossRef CAS PubMed.
- R. Roy, S. Ray, A. Chowdhury and R. Anand, ACS Sens., 2021, 6, 1933–1939 CrossRef CAS PubMed.
- J. C. Gutiérrez, F. Amaro and A. MartÃ-n-González, Front. Microbiol., 2015, 6, 1–8 Search PubMed.
- S. Sahu, R. Roy and R. Anand, ACS Sens., 2022, 7, 704–715 CrossRef CAS PubMed.
- A. V. Bounegru and C. Apetrei, Nanomaterials, 2023, 13, 760 CrossRef CAS PubMed.
- M. C. Castrovilli, P. Bolognesi, J. Chiarinelli, L. Avaldi, P. Calandra, A. Antonacci and V. Scognamiglio, TrAC, Trends Anal. Chem., 2019, 119, 115615 CrossRef CAS.
- J. Adamski, P. Nowak and J. Kochana, Electrochim. Acta, 2010, 55, 2363–2367 CrossRef CAS.
- A. Tarasov, N. Stozhko, M. Bukharinova and E. Khamzina, Life, 2023, 13, 291 CrossRef CAS PubMed.
- S. Ray, S. Panjikar and R. Anand, ACS Sens., 2018, 3, 1632–1638 CrossRef CAS PubMed.
- K. Chaudhary, K. Kumar, P. Venkatesu and D. T. Masram, Adv. Colloid Interface Sci., 2021, 289, 102367 CrossRef CAS PubMed.
- G. Hernández-Cancel, D. Suazo-Dávila, A. J. Ojeda-Cruzado, D. García-Torres, C. R. Cabrera and K. Griebenow, J. Nanobiotechnol., 2015, 13, 70 CrossRef PubMed.
- M. Mathesh, B. Luan, T. O. Akanbi, J. K. Weber, J. Liu, C. J. Barrow, R. Zhou and W. Yang, ACS Catal., 2016, 6, 4760–4768 CrossRef CAS.
- V. Ganesh and A. Muthurasu, J. Phys.: Conf. Ser., 2012, 358, 012003 CrossRef.
- A. A. Homaei, R. Sariri, F. Vianello and R. Stevanato, J. Chem. Biol., 2013, 6, 185–205 CrossRef PubMed.
- K. Krishnamoorthy, M. Veerapandian, K. Yun and S. Kim, Carbon, 2012, 53, 38–49 CrossRef.
- D. C. Marcano, D. V. Kosynkin, J. M. Berlin, A. Sinitskii, Z. Sun, A. Slesarev, L. B. Alemany, W. Lu and J. M. Tour, ACS Nano, 2010, 4(8), 4806–4814 CrossRef CAS PubMed.
- H. Yuan, J. Ye, C. Ye, S. Yin, J. Li, K. Su, G. Fang, Y. Wu, Y. Zheng, M. Ge, R. Tang, G. Feng, Y. Qu and Y. Zhu, Chem. Mater., 2021, 33, 1731–1739 CrossRef CAS.
- Å. Björkman, Schweiz. Z. Hydrol., 1969, 31, 632–645 Search PubMed.
- M. A. Pimenta, G. Dresselhaus, M. S. Dresselhaus, L. G. Cançado, A. Jorio and R. Saito, Phys. Chem. Chem. Phys., 2007, 9, 1276–1290 RSC.
- W. Xu, Z. Jin, X. Pang, Y. Zeng, X. Jiang, Y. Lu and L. Shen, ACS Appl. Bio Mater., 2020, 3, 4263–4272 CrossRef CAS PubMed.
- J. Singh, R. Anand and A. Horovitz, J. Bacteriol., 2022, 204(8), e00179 CrossRef PubMed.
- A. A. Baykov, O. A. Evtushenko and S. M. Avaeva, Anal. Biochem., 1988, 171, 266–270 CrossRef CAS PubMed.
- S. Ray, T. Senapati, S. Sahu, R. Bandyopadhyaya and R. Anand, Anal. Chem., 2018, 90, 8960–8968 CrossRef CAS PubMed.
- I. Gualandi, M. Marzocchi, E. Scavetta, M. Calienni, A. Bonfiglio and B. Fraboni, J. Mater. Chem. B, 2015, 3, 6753–6762 RSC.
- N. Saraf, E. R. Woods, M. Peppler and S. Seal, Biosens. Bioelectron., 2018, 117, 40–46 CrossRef CAS PubMed.
- K. Tang, C. Turner, L. Case, A. Mehrehjedy, X. He, W. Miao and S. Guo, ACS Appl. Polym. Mater., 2022, 4, 2337–2345 CrossRef CAS.
- R. García-Álvarez and M. Vallet-Regí, Nanomaterials, 2021, 11, 888 CrossRef PubMed.
- C. C. Mayorga-Martinez, R. Gusmão, Z. Sofer and M. Pumera, Angew. Chem., Int. Ed., 2019, 58, 134–138 CrossRef CAS PubMed.
- S. Ray, S. Panjikar and R. Anand, ACS Sens., 2017, 2, 411–418 CrossRef CAS PubMed.
- P. A. Raymundo-Pereira, T. A. Silva, F. R. Caetano, L. Ribovski, E. Zapp, D. Brondani, M. F. Bergamini, L. H. Marcolino, C. E. Banks, O. N. Oliveira, B. C. Janegitz and O. Fatibello-Filho, Anal. Chim. Acta, 2020, 1139, 198–221 CrossRef CAS PubMed.
- D. Khodagholy, J. Rivnay, M. Sessolo, M. Gurfinkel, P. Leleux, L. H. Jimison, E. Stavrinidou, T. Herve, S. Sanaur, R. M. Owens and G. G. Malliaras, Nat. Commun., 2013, 4, 2133 CrossRef PubMed.
- G. Alarcon-Angeles, G. A. Álvarez-Romero and A. Merkoçi, in Encyclopedia of Interfacial Chemistry, Elsevier, 2018, pp. 140–155 Search PubMed.
- H. Liu, A. Yang, J. Song, N. Wang, P. Lam, Y. Li, H. K. Law and F. Yan, Sci. Adv., 2021, 7, 1–10 Search PubMed.
- D. Tyagi, H. Wang, W. Huang, L. Hu, Y. Tang, Z. Guo, Z. Ouyang and H. Zhang, Nanoscale, 2020, 12, 3535–3559 RSC.
- K. S. Obayomi, S. Y. Lau, M. Danquah, T. Chiong and M. Takeo, Environ. Nanotechnol., Monit. Manage., 2022, 17, 100647 CAS.
- F. Fartas, J. Abdullah, N. Yusof, Y. Sulaiman and M. Saiman, Sensors, 2017, 17, 1132 CrossRef PubMed.
- M. N. Karim and H. J. Lee, Talanta, 2013, 116, 991–996 CrossRef PubMed.
- Y. Wee, S. Park, Y. H. Kwon, Y. Ju, K.-M. Yeon and J. Kim, Biosens. Bioelectron., 2019, 132, 279–285 CrossRef CAS PubMed.
- M. R. Montereali, L. Della Seta, W. Vastarella and R. Pilloton, J. Mol. Catal. B: Enzym., 2010, 64, 189–194 CrossRef CAS.
- C. C. Mayorga-Martinez, R. Gusmão, Z. Sofer and M. Pumera, Angew. Chem., Int. Ed., 2019, 58, 134–138 CrossRef CAS PubMed.
- M. Cerrato-Alvarez, E. Bernalte, M. J. Bernalte-García and E. Pinilla-Gil, Talanta, 2019, 193, 93–99 CrossRef CAS PubMed.
- J. Abdullah, M. Ahmad, N. Karuppiah, L. Y. Heng and H. Sidek, Sens. Actuators, B, 2006, 114, 604–609 CrossRef CAS.
- J. Sanz, S. de Marcos and J. Galbán, Anal. Bioanal. Chem., 2012, 404, 351–359 CrossRef CAS PubMed.
- S. K. Mishra and K. S. Chiang, Opt. Laser Technol., 2020, 131, 106464 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional experimental details and supporting data. See DOI: https://doi.org/10.1039/d3sc03509c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |