Open Access Article
Open Access ArticleCognitive-enhancing effect of Cordia dichotoma fruit on scopolamine-induced cognitive impairment in rats: metabolite profiling, in vivo, and in silico investigations†
Hagar M. Hussein *a,
Mostafa A. Abdel Kawya,
Basma M. Eltananyb,
Laura Pontcd,
Fernando Benavente
*a,
Mostafa A. Abdel Kawya,
Basma M. Eltananyb,
Laura Pontcd,
Fernando Benavente c,
Ahmed M. Fayez
c,
Ahmed M. Fayez e,
Radwan Alnajjar
e,
Radwan Alnajjar fg,
Ahmed A. Al-Karmalawy
fg,
Ahmed A. Al-Karmalawy hi,
Azza R. Abdelmonema and
Engy Mohsena
hi,
Azza R. Abdelmonema and
Engy Mohsena
aDepartment of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt. E-mail: hagar.elhag@pharma.cu.edu.eg
bDepartment of Pharmaceutical Analytical Chemistry, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt
cDepartment of Chemical Engineering and Analytical Chemistry, Institute for Research on Nutrition and Food Safety (INSA-UB), University of Barcelona, Barcelona 08028, Spain
dSerra Húnter Program, Generalitat de Catalunya, Barcelona 08007, Spain
eSchool of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, New Administrative Capital, Cairo 11835, Egypt
fComputer-Aided Drug Design (CADD) Unit, Faculty of Pharmacy, Libyan International Medical University, Benghazi, Libya
gDepartment of Chemistry, Faculty of Science, University of Benghazi, Benghazi, Libya
hDepartment of Pharmaceutical Chemistry, College of Pharmacy, The University of Mashreq, Baghdad 10023, Iraq
iDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Horus University-Egypt, New Damietta 34518, Egypt
First published on 23rd December 2024
Abstract
Many plants are reported to enhance cognition in amnesic-animal models. The metabolite profile of Cordia dichotoma fruit methanolic extract (CDFME) was characterized by LC-QTOF-MS/MS, and its total phenolics content (TPC) and total flavonoids content (TFC) were determined. In parallel, its cognitive-enhancing effect on scopolamine (SCOP)-induced AD in rats was evaluated. The TPC and TFC were 44.75 ± 1.84 mg gallic acid equiv. g−1 sample and 5.66 ± 0.67 mg rutin equiv. g−1 sample, respectively. In total, 81 metabolites were identified, including phenolic acids, lignans, coumarins, amino acids, fatty acids, and their derivatives, fatty acid amides, polar lipids, terpenoids, and others. The most abundant metabolites identified were quinic acid, caffeoyl-4′-hydroxyphenyllactate, rosmarinic acid, and oleamide. CDFME (200 mg kg−1) was found to significantly enhance recognition memory in the novel object recognition test. Furthermore, it nearly corrected acetylcholinesterase (AChE), acetylcholine, noradrenaline, and dopamine hippocampal levels, which changed due to SCOP. Further in silico validation of the in vivo results was conducted, focusing on the most abundant metabolites. Molecular docking showed that rosmarinic acid, caffeoyl-4′-hydroxyphenyllactate, sebestenoid C, and sagerinic acid exhibited the greatest affinity for receptor binding against AChE. However, molecular dynamics and mechanics calculations clarified that the complex of caffeoyl-4′-hydroxyphenyllactate with AChE was the most stable one. This study represents the first comprehensive metabolite profiling of CDFME to assess its cognition-enhancing effect both in vivo and in silico. These results demonstrate that CDFME protects against SCOP-induced cognitive impairment. Thus, additional preclinical and clinical studies on CDFME may provide an attractive approach in pharmacotherapy and AD prophylaxis.
1. Introduction
Scopolamine (SCOP) is a non-selective post-synaptic muscarinic antagonist that disrupts cholinergic neurotransmission. Because it can penetrate the blood–brain barrier, SCOP is often used to cause cognitive deterioration in an experimental model to evaluate and discover anti-amnesic drugs. It impairs memory and learning in mice and humans, particularly the processes of learning acquisition and short-term memory. According to studies, Alzheimer's patients and SCOP-induced animals have similar memory problems.1,2There are about 50 million people who have dementia globally.3 Alzheimer's disease (AD) has become one of the century's biggest global health concerns, affecting millions of individuals worldwide. AD is currently considered the third leading cause of death in developed nations, behind cancer and cardiovascular disease. AD is a progressive and chronic neurodegenerative disorder that profoundly impacts cognitive functions such as memory, orientation, judgment, and reasoning.4 It is characterized by dementia resulting from various pathological features, including amyloid beta plaques, glutamate excitotoxicity, neurofibrillary tangles, and hyperphosphorylated tau protein.5 Further pathological characteristics include neuroinflammation, oxidative stress-induced cell damage, and cholinergic dysfunction. These conditions result in neurodegeneration and a loss of synapses and neurons, which results in the macroscopic atrophy of the brain areas related to memory, such as the hippocampus.4 Cholinergic neurons release acetylcholine (ACh), a neurotransmitter that has a role in signal transduction linked to memory and learning.6 Acetylcholinesterase (AChE) inhibitors raise synaptic ACh levels, allowing the signal to be potentiated, thus improving cognition and everyday functioning.4 So, methods that lower the high level of AChE are thought to be effective for treating memory problems.7
Additionally, noradrenaline (NA) and dopamine (DA) are crucial neuromodulators involved in cognitive functions and are related to the psychological and behavioral symptoms of dementia.8 Concerning AD treatments, current anti-AD drugs, such as donepezil (DON), rivastigmine, galantamine, and memantine, help alleviate clinical symptoms. However, these medications are associated with numerous side effects and are proven effective primarily in mild and moderate cases of AD.3 Moreover, the complexity of AD, characterized by multiple contributing factors, challenges their efficacy. As a result, research into developing novel, potent medications for AD has gained international attention. In this regard, several natural products have gained interest in the pharmaceutical industry and the scientific research community due to their neuroprotective effects, targeting different pathological pathways related to AD.4
Cordia dichotoma G. Forst. (Indian cherry), a flowering tree in the Boraginaceae family, presents various medicinal properties. C. dichotoma produces pulpy, edible fruits with single-seed kernels encased in a transparent, viscid, sweet pulp. The fruit pulp serves various culinary purposes, being consumed fresh or raw as a spice or condiment in a variety of foods.9 C. dichotoma is found in temperate regions worldwide, including South America, Central America, Mexico, West Africa, East Africa, South Asia, and Australia. C. dichotoma fruits are recognized for containing several bioactive phytochemicals such as pyrrolizidine alkaloids, coumarins, flavonoids, phenolic acids, carbohydrates, amino acids, tannins, saponins, terpenes, and sterols.10 Fruits have been used for their diverse medicinal properties, encompassing antioxidant, antiulcer, analgesic, anti-inflammatory, wound-healing, demulcent, astringent, diuretic, emollient, expectorant, hepatoprotective, analgesic, antidiabetic, antibacterial, anthelmintic, anti-fertility, and aphrodisiac effects. Mucilage from C. dichotoma fruits is traditionally used to treat cough, fever, chest disease, uterus, and urethra disorders, while fruit kernel is used to treat tinea.9
In addition, C. dichotoma has been employed to address behavioral and memory dysfunctions, as well as psychological symptoms of dementia.11 Cordia myxa leaves ethanolic extract significantly alleviated SCOP-induced cognitive impairment in mice, as measured by novel object recognition test (NORT) and passive avoidance test. Additionally, it decreases tau protein phosphorylation.12 Additionally, Cordia dichotoma and Cordia sebestena leaves methanolic extract are reported to have in vitro anticholinesterase effects.13 Cordia dichotoma leaves alleviate SCOP-induced AD type-dementia in mice by enhancing the cholinergic system, protecting against neuronal death in the brain, and anti-oxidant effect.14 However, no previous study reported the cognitive function-enhancing effect of Cordia dichotoma fruit extract, which emphasizes the novelty of our work.14 Given the multitude of interesting medicinal properties, there is a growing interest in employing up-to-date analytical tools to characterize such medicinal plants, aiming to understand their bioactivity and ensure their safety, quality, and efficacy. Presently, liquid chromatography-quadrupole time-of-flight tandem mass spectrometry (LC-QTOF-MS/MS) metabolomics stands out as one of the best alternatives for the untargeted metabolite profiling of plant extracts.15–17 The reason for this is the high separation efficiency of LC and the high scanning rate, mass accuracy, and resolution of QTOF mass spectrometers, enabling the acquisition of both MS and tandem MS (MS/MS) spectra with excellent sensitivity.18 Concerning current literature, no studies were found dealing with the elucidation and identification of the metabolite profile of CDFME and its anti-AD activity.
In this study, CDFME was profiled by untargeted LC-QTOF-MS-MS metabolomics in both negative and positive electrospray ionization (ESI) modes to characterize its bioactive compounds, and its total phenolics content (TPC) and total flavonoids content (TFC) were determined. In parallel, the cognitive-enhancing effect of CDFME on SCOP-induced cognitive impairment in rats was investigated. These alterations in rats were monitored through NORT and determinations of AChE and neurotransmitter levels in the hippocampus. To gain insights into the potential AChE inhibitory activity of the most abundant metabolites identified in CDFME, molecular docking studies were conducted, comparing their binding affinity for AChE with that of DON. Finally, the stability within the enzyme active site of the most potentially AChE inhibitory metabolites was evaluated using molecular dynamics and molecular mechanics methods.
2. Material and methods
2.1 Plant material and preparation of plant extracts
Fresh fruits (800 g) were collected from Mazhar Botanic Garden in Egypt. Fruits were authenticated by Prof. Dr Abd Haleem Abd El-Mogali, Head of the Department of Flora Researches and Plant Taxonomy at the Agriculture Research Centre, Giza, Egypt. A voucher specimen was deposited at the herbarium of the Faculty of Pharmacy, Cairo University, Cairo, Egypt (sample No. 7.9.2023). The fresh fruits were halved, and the seeds were removed. Subsequently, the pericarps were lyophilized using a Stellar laboratory freeze dryer (temp set at −40 °C, vacuum level of 100 mT) (Millrock Technology Inc., New York, NY, USA). The lyophilized dried pericarps (100 g) were extracted through the cold maceration method. The lyophilized dried pericarps were crushed with a blender (Toshiba, Tokyo, Japan) and then they were exhaustively extracted with 80% methanol (3 L) through repeated shaking and soaking (4 times) at room temperature over two weeks. After filtering the resultant extract, the solvent was evaporated using a rotary evaporator set to 40 °C and low pressure, yielding 6 g of brown semisolid residue of fruit extract. The dried fruit extract was kept in a freezer at −10 °C for further research.19 The percentage yield of fruit extract is presented in Table 1.| Plant | Solvent | Extraction method | % yield |
|---|---|---|---|
| CDFME | 80% methanol | Cold maceration | 6% |
2.2 Drugs and chemicals
Scopolamine hydrobromide was purchased from Merck (Darmstadt, Germany) and donepezil hydrochloride was obtained from Pfizer (New York, NY, USA). DON and SCOP were prepared in saline. Methanol (analytical grade) for extraction was provided by El-Gomhuria Company (Cairo, Egypt). Formic acid (≥95.0%), acetonitrile, water, and methanol (LC-MS grade) were provided by Merck.2.3 Total phenolics and flavonoids contents
The TPC was assessed by the Folin–Ciocalteu colorimetric method.20 Briefly, 10 μL of sample/standard was mixed with 100 μL of Folin–Ciocalteu reagent (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v) in a 96-well microplate. Following the addition of 80 μL of 1 M Na2CO3, the mixture was allowed to sit at room temperature (25 °C) in the dark for 20 minutes. The resulting blue color was measured at 630 nm using a spectrophotometric microplate reader (FLUOstar Omega, BMG LABTECH, Ortemberg, Germany). The TPC in samples was quantified as mg gallic acid equiv. g−1 extract, using an external gallic acid calibration curve (25, 50, 100, 200, 400, 600, 800, and 1000 μg mL−1).
10 v/v) in a 96-well microplate. Following the addition of 80 μL of 1 M Na2CO3, the mixture was allowed to sit at room temperature (25 °C) in the dark for 20 minutes. The resulting blue color was measured at 630 nm using a spectrophotometric microplate reader (FLUOstar Omega, BMG LABTECH, Ortemberg, Germany). The TPC in samples was quantified as mg gallic acid equiv. g−1 extract, using an external gallic acid calibration curve (25, 50, 100, 200, 400, 600, 800, and 1000 μg mL−1).
The TFC was determined according to the aluminum chloride method.21 In summary, 15 μL of the sample/standard was dispensed into a 96-well microplate, followed by the addition of 175 μL of methanol and 30 μL of 1.25% m/v AlCl3. After adding 30 μL of a 0.125 M C2H3NaO2 solution, the mixture was allowed to incubate for a duration of 5 minutes. The resulting yellow color was measured at 420 nm using the microplate reader. The TFC in samples was quantified as mg rutin equiv. g−1 extract using an external rutin calibration curve (1000, 500, 200, 125, 62.5, 31.4, 15.6, and 7.2 μg mL−1).
2.4 LC-QTOF-MS/MS analysis
For LC-QTOF-MS/MS analysis, CDFME sample was prepared by mixing one milliliter of methanol with 10 milligrams of the extract, after which the mixture was centrifuged for 10 minutes at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
×![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) g, and the result was filtered through a 0.22 μm nylon syringe filter. The LC-QTOF-MS/MS experiments were carried out using a 1260 Infinity liquid chromatograph connected to a 6546 LC/QTOF mass spectrometer with an orthogonal electrospray ionization (ESI) interface (Agilent Technologies, Waldbronn, Germany). CDFME samples were analyzed in both positive and negative ESI modes, using an Agilent Technologies Zorbax SB-C18 column with dimensions of 150 mm LT × 2.1 mm ID, 5 μm particle size, and 90 Å pore diameter, and an optimized acetonitrile: water gradient (both with 0.1% v/v of formic acid) that enabled the comprehensive profiling of CDFME compounds within 25 minutes. Detailed information about the separation conditions, MS, and MS/MS measurements, as well as data processing, was as described in a previous work.18,22
g, and the result was filtered through a 0.22 μm nylon syringe filter. The LC-QTOF-MS/MS experiments were carried out using a 1260 Infinity liquid chromatograph connected to a 6546 LC/QTOF mass spectrometer with an orthogonal electrospray ionization (ESI) interface (Agilent Technologies, Waldbronn, Germany). CDFME samples were analyzed in both positive and negative ESI modes, using an Agilent Technologies Zorbax SB-C18 column with dimensions of 150 mm LT × 2.1 mm ID, 5 μm particle size, and 90 Å pore diameter, and an optimized acetonitrile: water gradient (both with 0.1% v/v of formic acid) that enabled the comprehensive profiling of CDFME compounds within 25 minutes. Detailed information about the separation conditions, MS, and MS/MS measurements, as well as data processing, was as described in a previous work.18,22
Detected compounds were identified as metabolites based on their predicted molecular formula, accurate molecular mass, retention time (Rt), and MS/MS spectra, and by comparison with various databases (e.g., KEGG, https://www.genome.jp/kegg/compound/, the Phytochemical Dictionary of Natural Product Database, https://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml, Mass Bank of North America, https://mona.fiehnlab.ucdavis.edu/, and PubChem, https://pubchem.ncbi.nlm.nih.gov/), in addition to other literary sources. Therefore, the metabolites were identified at a high confidence level.
2.5 Animals
Forty adult male Wistar albino rats, weighing between 200 and 250 g (National Research Centre Giza, Egypt), were kept in cages made of polypropylene. The rats were retained together in a shared habitat with a moderate temperature of 25 ± 2 °C. Rats were given 7 days to acclimatize before starting the study. Throughout this period, regular rat pellet food and unlimited water were provided to the animals. The experiments were conducted in adherence with the “Guide for the Care and Use of Laboratory Animals” published by the US National Institutes of Health (NIH Publication No. 85-23, 2011). The research project has been approved by Cairo University's Faculty of Pharmacy Research Ethics Committee (approval number: MP (3257)).2.6 Acute toxicity study
The lethal dose (LD50) of CDFME in rats was determined adhering to the specified guidelines by the Organization for Economic Co-operation for Development (OECD, Test No. 420, 2002). Before the extract administration, rats underwent an overnight fast (12 hours) with unrestricted access to water. Just prior to the initial dosage, each rat's weight was noted. The rats were then divided into four groups of six. CDFME, suspended in saline, was orally administered to the rats at escalating doses (1000, 2000, and 4000 mg kg−1 body weight). The control group, kept under the same conditions, received a vehicle (saline). Over a 24 hours period, the animals were closely monitored for signs of toxicity, including draping, rising fur, excitability, tremors, salivation, twitching, and mortality. The mortality rates for each group were recorded.2.7 SCOP-induced cognitive impairment study
| DI = TN ÷ (TN + TF) × 100 |
2.8 Statistical analysis
GraphPad Prism software (version 8; GraphPad Software, Inc., San Diego, CA, USA) was used for graphical presentations and statistical analyses. The data are presented as mean ± standard deviation (SD). For data analysis, one-way ANOVA was applied, followed by Tukey's multiple comparison test. With the exception of how CDFME affects how long it takes to explore each object in the NORT, which was assessed using two-way ANOVA, followed by Sidak's multiple comparisons test. A probability level of <0.05 was deemed statistically significant for all tests conducted.2.9 Molecular docking
The 18 most abundant metabolites identified in CDFME were docked against the AChE target receptor using the Schrödinger 2021 suite.25 As reference standards, DON and the co-crystallized inhibitor were used. The chemical structures of these compounds were sketched with ChemDraw Professional 17.0 (Cambridge Soft Corporation., Cambridge, USA). Each structure was copied and pasted into the applied software working window, including correction of local partial charges and minimization to reach a possible local minimum energy state.26 All the prepared structures were assembled within the same database, prepared for the next docking step. On the other hand, the target AChE enzyme's X-ray structure was obtained from the Protein Data Bank (https://www.rcsb.org/structure/1OCE). The enzyme structure was subjected to correction, 3D hydrogenation, and energy minimization procedures.27A standardized docking procedure was conducted using the default program specifications.28 The ideal binding pose for each examined compound was selected based on criteria like root mean square deviation (RMSD), score, and binding mode. Additionally, a validation check for the applied force field was performed by redocking the co-crystallized ligand of AChE (DON) inside AChE binding site.29 The valid performance was verified by achieving a low RMSD value (<2 Å) and close similarities between the binding modes of both the native and redocked poses of the co-crystallized inhibitor of AChE. Finally, the binding scores for the identified metabolites were compared with those obtained for DON. Metabolites displaying the highest binding scores, indicative of their potential as AChE inhibitors, were further evaluated by molecular dynamics and molecular mechanics methods.
2.10 Molecular dynamics and molecular mechanics
Molecular dynamics simulations were conducted using the Desmond 2.2 package of the Schrödinger platform (Schrödinger, New York, NY, USA). The free energy of the bindings for the examined complexes was calculated with the same software, employing molecular mechanics with generalized Born and surface area solvation (MM-GBSA) methods. The complexes of rosmarinic acid, caffeoyl-4′-hydroxyphenyllactate, sebestenoid C, and sagerinic acid with the AChE receptor were subjected to a 200 ns simulation.30 As a reference, the co-crystallized inhibitor (DON) complex was used. The normal physiological conditions were kept to give more reproducible results. Further details about the aforementioned methods can be found in the ESI S1 and S2.†3. Results and discussion
3.1 Total phenolics and total flavonoids contents
Phenolic compounds are important plant components with significant redox properties. The hydroxyl groups in the structures of these compounds are essential in enabling the scavenging of free radicals, thereby contributing to their antioxidant activity.31 TPC and TFC values were determined to complement the metabolite profiling by LC-QTOF-MS/MS, as the phenolic contents of plant extracts have been linked to the enhancement of anti-AChE activity.9The TPC and TFC values in CDFME were 44.75 ± 1.84 mg gallic acid eq g−1 sample and 5.66 ± 0.67 mg rutin eq g−1 sample, respectively. These findings suggested that flavonoids constituted a minor proportion of the total phenolics in the fruit extract, suggesting that they would have a lesser impact on the bioactivity of CDFME. Graphs of gallic acid and rutin standard calibration curves were presented in Fig. S1.†
3.2 LC-MS/MS metabolite profiling
The CDFME chemical profile was analyzed using LC-QTOF-MS in both negative and positive ESI modes. The representative total ion chromatograms (TIC) of CDFME are shown in Fig. 2A and B, and the MS/MS spectra of some of the most abundant identified metabolites are illustrated in Supplementary Fig. S2–S20.† After data processing, 81 metabolites were tentatively identified depending on their Rt, predicted molecular formula, accurate molecular mass, MS/MS spectra, and cross-referencing with various databases and prior literature. Details of the annotated metabolites, including names, classes, molecular formulas, m/z of the detected molecular ions, and fragment ions are provided in Table 2.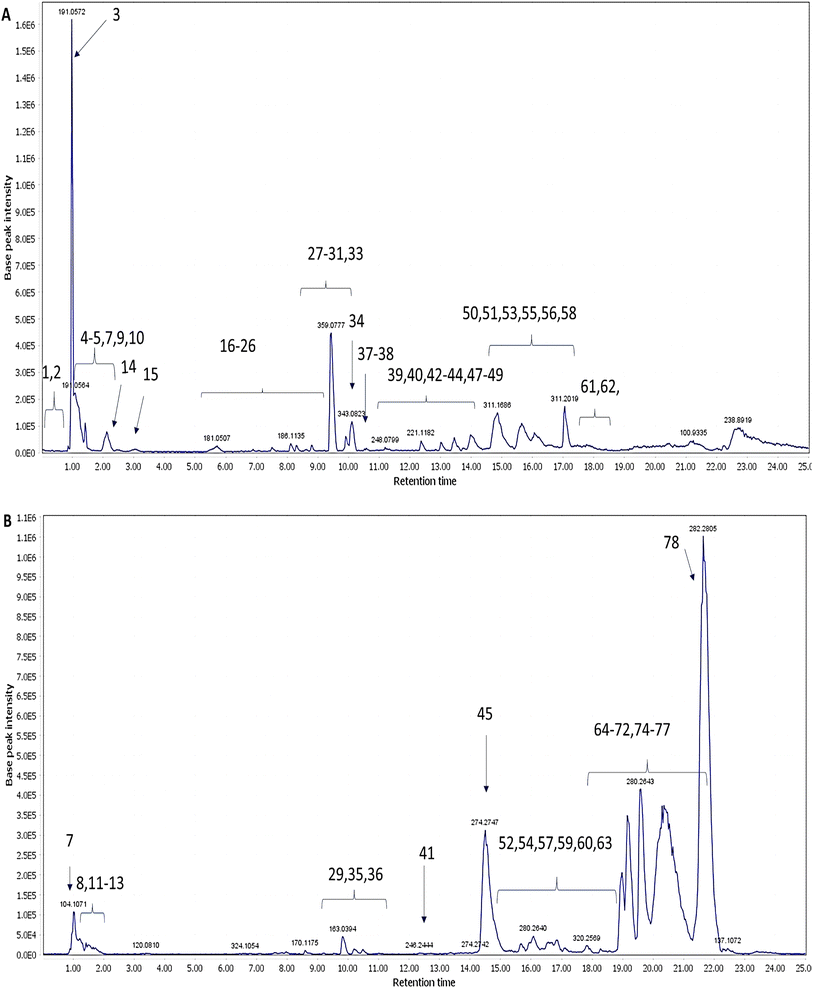 | ||
| Fig. 2 LC-QTOF-MS total ion chromatograms of CDFME in negative ESI mode (A) and positive ESI mode (B). The numbers of metabolites are indicated in Table 2. | ||
| No. | Rt (min) | [M + H]+ | [M–H]− | Error (ppm) | Formula | Proposed structure | MS/MS fragments |
|---|---|---|---|---|---|---|---|
| Hydroxybenzoic acid derivatives | |||||||
| 14 | 2.44 | — | 359.0982 | −0.4 | C15H20O10 | Glucosyringic acid | 197, 179, 135, 123, 72 |
| 15 | 3.01 | — | 197.0454 | −1.6 | C9H10O5 | Syringic acid | 179, 151, 135, 123, 117, 72 |
| 16 | 5.69 | — | 181.0506 | −1.1 | C9H10O4 | Syringaldehyde | 181, 163, 135, 134, 119, 107, 72 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Hydroxy cinnamic acid derivatives | |||||||
| 18 | 7.08 | — | 461.1654 | −2.2 | C20H30O12 | Verbasoside | 461, 161, 135, 113 |
| 19 | 7.50 | — | 179.0349 | −0.3 | C9H8O4 | Caffeic acid | 161, 135, 134, 133, 117, 107, 79 |
| 22 | 8.62 | — | 521.2023 | −1.2 | C24H26O13 | Rosmarinic acid hexoside | 359, 197, 179, 161, 135, 107 |
| 24 | 8.97 | — | 717.1449 | −1.6 | C36H30O16 | Salvianolic acid B | 339, 321, 295, 277, 197, 179, 109 |
| 25 | 9.42 | — | 197.0455 | −0.1 | C9H10O5 | Danshensu | 179, 151, 135, 134, 123, 109, 72 |
| 27 | 9.55 | — | 359.0774 | 0.5 | C18H16O8 | Rosmarinic acid | 197, 179, 161, 135, 133, 123 |
| 30 | 9.91 | — | 163.0401 | 0.3 | C9H8O3 | p-Coumaric acid | 119, 103, 93 |
| 31 | 9.92 | — | 337.0928 | −0.2 | C16H18O8 | p-Coumaroylquinic acid | 191, 163, 129, 119, 111, 101, 85 |
| 33 | 10.08 | — | 367.1033 | −0.4 | C17H20O9 | Feruloylquinic acid | 193, 191, 161, 134 |
| 34 | 10.10 | — | 343.0823 | 0 | C18H16O7 | Caffeoyl-4′-hydroxyphenyllactate | 181, 179, 163, 161, 135, 133, 119 |
| 37 | 10.52 | — | 685.1555 | −1.1 | C36H30O14 | Sebestenoid C | 339, 321, 295, 277, 185, 109 |
| 38 | 10.72 | — | 373.0932 | 0.9 | C19H18O8 | Methyl rosmarinate | 359, 197, 161, 150, 135, 123, 72 |
| 39 | 11.05 | — | 313.0719 | −0.1 | C17H14O6 | Nepetoidin B | 161, 151, 133, 123, 105 |
| 40 | 11.34 | — | 313.0717 | 0.5 | C17H14O6 | Nepetoidin B isomer | 161, 151, 133, 123, 105 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Organic acids | |||||||
| 3 | 0.97 | — | 191.0562 | −0.0001 | C7H12O6 | Quinic acid | 191, 173, 171, 127, 111, 109, 85 |
| 4 | 0.98 | — | 533.1722 | −0.2 | C19H34O17 | Quinic acid + hexose2 | 191, 171, 155, 127, 111, 85 |
| 5 | 1.00 | — | 383.1191 | −1.0 | C14H24O12 | Quinic acid derivative | 191, 173, 127, 85 |
| 7 | 1.03 | — | 133.0142 | −0.2 | C4H6O5 | Malic acid | 133, 115, 89, 71 |
| 9 | 1.41 | — | 191.0194 | −1.6 | C6H8O7 | Citric acid | 191, 173, 155, 127, 111, 87, 85 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Lignans | |||||||
| 20 | 8.29 | — | 521.2023 | −1.0 | C26H34O11 | Lariciresinol hexoside | 359, 329, 299, 205, 187, 175 |
| 21 | 8.32 | — | 567.2080 | −0.5 | C27H36O13 | Dehydrodehyrodiconiferyl glycoside derivative | 359, 341, 329, 299, 205, 187 |
| 26 | 9.42 | — | 719.1622 | −0.6 | C36H32O16 | Sagerinic acid | 359, 197, 179, 161, 135, 123, 72 |
| 28 | 9.72 | — | 717.1449 | −1.2 | C36H30O16 | Rabdosiin | 537, 339, 321, 295, 279, 249, 185 |
| 32 | 9.98 | — | 491.0976 | −1.5 | C26H20O10 | Globoidnan A | 491, 311, 267, 197, 135 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Coumarins | |||||||
| 29 | 9.82 | 163.0394 | 161.0242 | 2.5 | C9H6O3 | Hydroxycoumarin | 161, 133, 117, 115, 105, 77 |
| 163, 145, 135, 117, 107, 89, 77 | |||||||
| 35 | 10.28 | 147.0440 | — | 0.5 | C9H6O2 | Coumarin | 149, 119, 101, 91, 71, 65 |
| 36 | 10.46 | 177.0549 | — | 1.5 | C10H8O3 | Methoxy coumarin | 177, 162, 149, 145, 134, 135, 117, 89 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Amino acids | |||||||
| 8 | 1.40 | 280.1396 | — | 1.8 | C11H21NO7 | Hexosyl valine | 262, 244, 216, 198, 150, 118, 84, 72 |
| 10 | 1.41 | — | 290.0878 | −1.1 | C11H17NO8 | Hexosyl pyroglutamate | 200, 128, 101, 84 |
| 11 | 1.47 | 130.0502 | 128.0352 | 2.4 | C5H7NO3 | Pyroglutamic acid | 128, 102, 82, 79 |
| 130, 84, 71, 56 | |||||||
| 12 | 1.63 | 294.1553 | — | 1.9 | C12H23NO7 | N-Hexosyl isoleucine | 294, 230, 132, 212, 116, 112, 86, 69 |
| 13 | 1.71 | 132.1021 | — | 1.3 | C6H13NO2 | Isoleucine or leucine | 86, 69, 62, 56 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Fatty acids and their derivatives | |||||||
| 41 | 12.33 | 246.2432 | — | 0.3 | C14H31NO2 | N-Decyl diethanolamine | 246, 228, 106, 102, 88, 71, 70, 57 |
| 42 | 13.45 | — | 721.3646 | −0.8 | C34H58O16 | Palmitoleic linolenic glucoside | 397, 277, 255, 235 |
| 45 | 14.48 | 274.2748 | — | 2.6 | C16H35NO2 | N-Lauryldiethanolamine (N-Dodecyl diethanolamine) | 274, 256, 212, 106, 102, 88, 70, 57 |
| 46 | 14.62 | 230.2483 | — | 1.9 | C14H31NO | Lauramine oxide | 230, 212, 85, 71, 62, 58, 57 |
| 47 | 14.65 | — | 699.3782 | −3.8 | C32H60O16 | Palmitic oleic glucoside | 397, 255, 89, 71, 57 |
| 48 | 14.78 | — | 559.3121 | −1.7 | C28H48O11 | Dirhamosyl linolenic acid | 415, 277, 253, 101, 59 |
| 51 | 15.66 | — | 483.2724 | −5.6 | C29H40O6 | Palmitic acid derivative | 483, 255, 271, 227, 153 |
| 54 | 16.40 | 230.2485 | — | 1.0 | C14H31NO | Lauramine oxide isomer | 230, 212, 71, 62, 58, 57 |
| 57 | 16.81 | 296.2588 | — | 1.3 | C18H33NO2 | Linoleylhydroxamate | 251, 169, 109, 95, 93, 57, 55 |
| 59 | 17.13 | 296.2580 | — | 1.5 | C18H33NO2 | Linoleylhydroxamate isomer | 251, 169, 109, 95, 93, 67, 57, 55 |
| 62 | 18.38 | 279.2322 | 277.2169 | −1.4 | C18H30O2 | Linolenic acid | 277, 219, 188, 155, 127, 109, 83 |
| 279, 153, 149, 135, 123, 109, 83,69, 57 | |||||||
| 63 | 18.64 | 279.2321 | — | 0.8 | C18H30O2 | Linolenic acid isomer | 153, 135, 121, 109, 95, 67, 57 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Fatty acid amides | |||||||
| 52 | 16.08 | 280.2637 | — | 0.7 | C18H33NO | Linoleamide | 263, 245, 109, 95, 69, 55 |
| 60 | 18.29 | 278.2483 | — | 1.2 | C18H31NO | Linoleamide | 243, 149, 135, 109, 95, 81, 67, 55 |
| 64 | 18.95 | 228.2328 | — | 3.0 | C14H29NO | Myristamide (tetradecanamide) | 228, 172, 116, 102, 88, 74, 57 |
| 65 | 19.22 | 254.2484 | — | 2.1 | C16H31NO | Palmitoleamide | 237, 142, 149, 135, 128, 97, 83, 69, 55 |
| 66 | 19.58 | 280.2641 | — | 2.1 | C18H33NO | Linoleamide isomer | 280, 263, 245, 109, 95, 69, 55 |
| 67 | 19.84 | 242.2484 | — | 2.6 | C15H31NO | Pentadecanamide | 242, 200, 116, 102, 88, 57 |
| 68 | 20.00 | 284.2951 | — | 1.0 | C18H37NO | Octadecanamide | 284, 242, 228, 116, 102, 88, 71, 57 |
| 69 | 20.21 | 242.2832 | — | 1.8 | C15H31NO | Pentadecanamide isomer | 242, 200, 116, 102, 88, 57 |
| 71 | 20.28 | 268.2640 | — | 1.8 | C17H33NO | Heptadecenamide | 268, 233, 226, 149, 121, 97, 83, 69, 55 |
| 72 | 20.44 | 338.2679 | — | 3.1 | C22H43NO | Erucamide | 321, 149, 67, 57 |
| 75 | 21.11 | 310.3110 | — | 1.7 | C20H39NO | Eicosenamide | 310, 268, 149, 135, 121, 97, 83, 69, 57 |
| 76 | 21.32 | 256.2640 | — | 1.9 | C16H33NO | Palmitamide (hexadecanamide) | 256, 128, 116, 102, 88, 71, 57 |
| 77 | 21.45 | 284.2953 | — | 1.7 | C18H37NO | Octadecanamide isomer | 284, 242, 228, 116, 102, 88, 71, 57 |
| 78 | 21.89 | 282.2802 | — | 3.7 | C18H35NO | Octadecenamide (oleamide) | 265, 247, 149, 135, 121, 97, 83, 69, 57 |
| 79 | 21.81 | 537.5359 | — | 1.0 | C34H68N2O2 | Palmitamide derivative | 282, 256, 212, 135, 102, 97, 83, 69, 57 |
| 80 | 21.85 | 256.2642 | — | 2.7 | C16H33NO | Palmitamide (hexadecanamide) isomer | 256, 116, 102, 88, 71, 57 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Polar lipids | |||||||
| 43 | 13.49 | — | 555.2839 | −1.0 | C25H48O11S | SQMG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
555, 299, 255, 225, 207, 165, 81 |
| 53 | 16.32 | — | 815.4977 | −0.9 | C43H76O12S | SQDG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0; 18 0; 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
815, 559, 277, 255, 225, 207, 165, 81 |
| 56 | 16.80 | — | 833.5177 | −1.0 | C43H79O13P | Diacyl phosphatidyl-myoinositol (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
833, 279, 255, 241, 153, 81,79 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Terpenoids | |||||||
| 17 | 6.49 | — | 443.1921 | −0.3 | C21H32O10 | Penstemide | 443, 113, 101, 71, 59 |
| 55 | 16.64 | — | 347.1707 | −1.2 | C16H28O8 | Rhodiolosides A (monoterpene) | 301, 285, 217 |
| 73 | 20.44 | 621.3082 | — | 3.3 | C36H44O9 | Diterpene benzoate triacetate | 621, 561, 533, 515, 505, 461, 433, 193 |
| 81 | 21.89 | 621.3087 | — | 2.5 | C36H44O9 | Diterpene benzoate triacetate isomer | 621, 561, 533, 505, 461, 433, 193 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| Other compounds | |||||||
| 1 | 0.95 | — | 387.1139 | −1.3 | C13H24O13 | Disaccharide derivative (carbohydrate) | 191, 179, 119, 101, 89, 71, 59 |
| 2 | 0.96 | — | 341.1086 | −1.5 | C12H22O11 | Hexose-pentose | 127, 119, 101, 89, 71, 59 |
| 6 | 1.02 | 104.1071 | — | 0.8 | C5H13NO | Choline (nitrogenous compound) | 105, 104, 60, 59, 58 |
| 23 | 8.79 | — | 623.1613 | −0.7 | C28H32O16 | Isorhamnetin 3-O-rutinoside (narcissin) | 623, 315, 314, 299, 255, 151, 107 |
| 44 | 14.02 | — | 297.1526 | −1.2 | C16H26O3S | p-Decylbenzenesulfonic acid | 297, 225, 183, 182, 161, 119 |
| 49 | 14.82 | — | 311.1686 | 0 | C17H28O3S | Undecylbenzenesulfonic acid | 311, 225, 197, 183, 170, 119 |
| 50 | 15.64 | — | 325.1838 | −1.1 | C18H30O3S | Dodecylbenzenesulfonic acid | 325, 297, 183, 170, 119 |
| 58 | 17.05 | — | 311.2013 | −1.1 | C21H28O2 | Bisphenol G (hydrocarbons) | 183, 149, 134, 133 |
| 61 | 18.33 | 391.2849 | — | 1.5 | C24H38O4 | Dioctyl phthalate (hydrocarbons) | 291, 167, 149, 71 |
| 70 | 20.22 | 593.2769 | — | 0.7 | C35H36N4O5 | Pheophorbide A (chlorophyll derivatives) | 533, 505, 461, 447, 433 |
| 74 | 20.70 | 593.2759 | — | 2.2 | C35H36N4O5 | Pheophorbide A isomer (chlorophyll derivatives) | 593, 533, 505, 461, 447, 433 |
The annotated metabolites (Table 2) pertained to numerous classes encompassing: 17 phenolic acids, 5 organic acids, 5 lignans, 3 coumarins, 5 amino acids, 12 fatty acids and their derivatives, 16 fatty acid amides, 3 polar lipids, 4 terpenoids, and 11 other compounds.
3.2.1.1 Hydroxy benzoic acid derivatives. Syringic acid (15, Fig. S2†) and syringaldehyde (16, Fig. S3†) (m/z 197.0454 and 181.0506, respectively, [M–H]−) revealed an intense fragment ion at m/z 135, as a result of the loss of two methoxy groups [M–H–2*(31)]− and CO2 + H2O [M–H–28–18]−, respectively. Moreover, peak 14 showed a mass difference of 162 m/z with respect to peak 15, recommending a glucose moiety, hence was assigned as glucosyringic acid.35 Syringic acid and syringaldehyde were previously reported in C. dichotoma fruit,34 whereas glucosyringic acid was detected in the current investigation in CDFME for the first time.
3.2.1.2 Hydroxy cinnamic acid derivatives. A total of 14 hydroxycinnamic acid derivatives were eluted between 7 and 11 minutes (Fig. 2A and Table 2). The only non-conjugated phenylethanoid detected was annotated as verbasoside (18).36 Caffeic acid (19, Fig. S4†) (m/z 179.0349, [M–H]−) presented intense fragment ions at m/z 135, 117, and 107, resulting from the loss of CO2 [M–H–44]−, CO2 + H2O [M–H–44–18]−, and CO + CO2 [M–H–28–44]−, respectively. The mass spectra of peaks 34 (m/z 343.0810, [M–H]−) and 39 (m/z 313.0719, [M–H]−) presented an intense fragment ion at m/z 161 related to the dehydrated caffeic acid moiety, indicating a caffeic acid derivative. Peak 34 was annotated as caffeoyl-4′-hydroxyphenyllactate (Fig. S7†),37 while, peak 39 was annotated as nepetoidin B (Fig. S9†), which is a caffeic acid ester.38
Rosmarinic acid (27, Fig. S6†) (m/z 359.0774, [M–H]−) was readily characterized by its fragment ions at m/z 179 and 197, corresponding to caffeic acid and danshensu moieties, respectively. Peaks 22 and 38 presented a mass difference of 162 and 15 m/z in comparison to peak 27, indicating an extra hexose and methyl group moiety, respectively, hence they were annotated as rosmarinic acid hexoside and methyl rosmarinate, respectively.37
Regarding peak 24 (Fig. S5†) (m/z 717.1449, [M–H]−), it was identified as salvianolic acid B. Its mass spectrum revealed characteristic fragment ions at m/z 339 and 321 resulting from the loss of danshensu + caffeic acid [M–H–198–180]− and two danshensu moieties [M–H–2*198]−, respectively.37 Sebestenoid C (37, Fig. S8†) (m/z 685.1555, [M–H]−), presented fragment ions at m/z 321 [M–H–C18H18O7–H2O]− and 295 [M–H–C18H18O7-carboxy group]−.39 Finally, the rest of the phenolic acids were annotated as danshensu (25),37 p-coumaric (30) acid, p-coumaroylquinic acid (31), and feruloylquinic acid (33), respectively.40
It is noteworthy that rosmarinic acid and caffeoyl-4′-hydroxyphenyllactate were found to be the most abundant phenolic acids identified. Additionally, all of the previous phenolic acids, except caffeoyl-4′-hydroxyphenyllactate, salvianolic acid B, sebestenoid C, and nepetoidin B, had been previously reported in C. dichotoma fruit.34 Meanwhile, sebestenoid C and nepetoidin B were previously reported in Cordia sebestena fruit.39
Quinic acid (3, Fig. S10†), quinic acid derivatives (4 and 5),42 malic acid (7), and citric acid (9) were identified in CDFME (Fig. 2A and Table 2). Quinic acid was the most abundant organic acid identified. Their mass spectra revealed abundant fragment ions due to the loss of CH2, CO2, CO, and H2O groups (14, 44, 28, and 18 m/z, respectively).32,37
Five lignans (Fig. 2A and Table 2) (20, 21, 26, 28, and 38), not previously reported in C. dichotoma, were identified in negative ESI mode. Lariciresinol hexoside (20) (m/z 521.2023, [M–H]−) revealed fragment ions at m/z 359 and 329 after the loss of a hexose sugar [M–H–162]− and a hexose sugar plus two methyl groups [M–H–162–15–15]−, respectively.44 Neolignane dihydrodehyrodiconiferyl glycoside derivative (21) was also annotated.40 Neolignans had been previously isolated from Cordia americana.45 Sagerinic acid (26, Fig. S11†) (m/z 719.1622, [M–H]−), a possible dimer of rosmarinic acid, showed a fragment ion at m/z 359 after the loss of C18H16O8 [M–H–360]−, as reported in previous studies.46 Rabdosiin (28) (m/z 717.1449, [M–H]−), and globoidnan A (32, Fig. S12†) (m/z 491.0976, [M–H]−) with an arylnaphthalene-type lignan skeleton were also identified, which were previously reported in Cordia rufescens.47 They showed fragmentation patterns characterized by the neutral losses of caffeic acid (−180 m/z) or danshensu (−198 m/z) and carboxyl groups (−44 m/z), which are present in their structures.48
Sugar conjugates of amino acids, including hexosyl valine (8) (m/z 280.1396, [M + H]+), hexosyl pyroglutamate (10) (m/z 290.0878, [M–H]−), and hexosyl isoleucine (12) (m/z 294.1553, [M + H]+), exhibited similar fragmentation behavior, involving the loss of the attached sugar unit and formation of a base peak at m/z 118, 128, and 132, in metabolites 8, 10, and 12, respectively. Additionally, the typical amino acid fragments were observed, indicating the loss of NH3, H2O, CO, CO2, and HCOOH groups (17, 18, 28, 44, and 46 m/z units, respectively).32,54 These amino acid sugar conjugates are reported here for the first time in CDFME. Additionally, pyroglutamic acid (11, Fig. S15†) and isoleucine or leucine (13, Fig. S16†) were identified.32
Most fatty acid fragmentations revealed neutral water loss(es) followed by decarboxylation.32 Eight fatty acids and their derivatives were identified in positive and/or negative ESI mode (Fig. 2 and Table 2). They are namely palmitoleic linolenic glucoside (42), palmitic oleic glucoside (47),46 dirhamnosyl linolenic acid (48),55 palmitic acid derivative (51),46 linoleylhydroxamate (57), linoleylhydroxamate isomer (59),56 linolenic acid (62), and linolenic acid isomer (63).32
Unsaturated fatty acids, such as linolenic acid and its derivatives (42, 48, 57, 59, 62, and 63), are vital nutrients with a variety of physiological benefits, including neuroprotective, antioxidant, and anti-inflammatory actions.57 Additionally, four fatty acid nitrogenous derivatives were also identified, namely, N-decyl diethanolamine (41), N-decyl diethanolamine (45), lauramine oxide (46), and lauramine oxide isomer (54).54 Many studies have reported the presence of various fatty acids in C. dichotoma seeds, including linoleic acid, oleic acid, and palmitic acid.9 However, to the best of our knowledge, dirhamnosyl linolenic acid and linoleylhydroxamate, were detected here for the first time in CDFME.
Sixteen fatty acid amides were identified in positive ESI mode (Fig. 2B and Table 2). These compounds produced main fragment ions of [M + H–NH3]+ and [M + H–NH3–H2O]+, indicating ammonia and ammonia plus water losses at the carboxamide functional group. Additionally, all fatty acyl moieties displayed consecutive losses of 14 m/z units, indicative of an acyl chain.57 For instance, peak 65 (Fig. S17†) (m/z 254.2484, [M + H]+) revealed fragment ions at m/z 237 [M + H–NH3]+, 142 [M + H–C8H16]+, and 128 [M + H–C9H18]+ and was annotated as palmitoleamide. Similarly, the mass spectrum of octadecenamide (78, Fig. S18†) (m/z 282.2802, [M + H]+) showed fragment ions at m/z 265 [M + H–NH3]+ and 247 [M + H–NH3–H2O]+.
Finally, peaks 52, 60, 64, 66, 67, 68, 69, 71, 72, 75, 76, 77, 79, and 80 were annotated as linoleamide, linolenamide, myristamide, linoleamide isomer, pentadecanamide, octadecanamide, pentadecanamide isomer, heptadecenamide, erucamide, eicosenamide, palmitamide, octadecanamide isomer, palmitamide derivative, and palmitamide isomer, respectively.57,59 Fatty acid amides were found to be one of the most abundant class of metabolites detected in positive ESI mode and were identified here for the first time in CDFME.
Two sulfoglycolipids and one phospholipid were identified in the negative ESI mode (Fig. 2A and Table 2). Sulfolipids are a class of lipids in which a sulfated sugar (mostly quinovose or glucose) is bonded to acylglycerol. Peaks 43 and 53, with [M–H]− at m/z 555.2839 and 815.4977, were identified as sulfoquinovosyl-monoacylglycerol (SQMG) (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and sulfoquinovosyl-diacylglycerol (SQDG) (16
0) and sulfoquinovosyl-diacylglycerol (SQDG) (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0; 18
0; 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), respectively. Sulfolipids showed characteristic fragment ions at m/z 225 (sulfoquinovosyl-18) and 81 (sulfonate). SQMG (16
3), respectively. Sulfolipids showed characteristic fragment ions at m/z 225 (sulfoquinovosyl-18) and 81 (sulfonate). SQMG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) also featured a fragment ion at m/z 255 related to a palmitic acid moiety (16
0) also featured a fragment ion at m/z 255 related to a palmitic acid moiety (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0). Similarly, SQDG (16
0). Similarly, SQDG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0; 18
0; 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) displayed fragments at m/z 255 and 277 related to palmitic acid (16
3) displayed fragments at m/z 255 and 277 related to palmitic acid (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and linolenic acid moieties (18
0) and linolenic acid moieties (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), respectively.57 Regarding the identified phospholipid, peak 56 [m/z 833.5177, [M–H]−] was annotated as diacyl phosphatidyl-myoinositol. It revealed fragment ions at m/z 241 (phospho-myoinositol), 153 (phospho-glycerol), 81 (sulfonate), and 79 (phosphonate).18 To the best of our knowledge, polar lipids were detected here for the first time in CDFME.
3), respectively.57 Regarding the identified phospholipid, peak 56 [m/z 833.5177, [M–H]−] was annotated as diacyl phosphatidyl-myoinositol. It revealed fragment ions at m/z 241 (phospho-myoinositol), 153 (phospho-glycerol), 81 (sulfonate), and 79 (phosphonate).18 To the best of our knowledge, polar lipids were detected here for the first time in CDFME.
Four terpenoid compounds were identified in positive ESI (Fig. 2B and Table 2). Peaks 17 and 55 were annotated as penstemide (iridoid-type glucoside)56 and rhodioloside A,61 respectively. Peaks 73 and 81 (621.3082 and 621.3087, respectively, [M + H]+) were annotated as diterpene acetate ester isomers. Their fragmentation was characterized by the neutral loss of an acetic acid unit (−60 m/z), but their exact structure could not be conclusively determined.62
Several sulfonic acids (44, 49, and 50)35 and two chlorophyll derivatives [pheophorbide (70) and pheophorbide isomer (74)] were identified in our study.64 As far as we know, choline and these sulfonic acids were identified here for the first time in CDFME.
3.3 Acute toxicity study
The therapeutic application of plant medicines without scientific evidence about their toxicity profile may raise serious concerns. To ensure the safety of plant medicines for human use, toxicity testing is typically conducted on various animal models. No toxicity signs (such as hypoactivity, coma, convulsions, diarrhea, respiratory depression, perspiration, salivation, and alteration in the locomotor activity) or deaths were recorded following oral administration of CDFME at doses of 1000 and 2000 mg kg−1 compared to the control group. However, at a dose of 4000 mg kg−1, there was a 33.3% mortality rate. Overall, CDFME exhibited a low toxicity profile, and doses of 100 and 200 mg kg−1 were selected for further in vivo evaluation.3.4 Cognitive-enhancing effect of CDFME
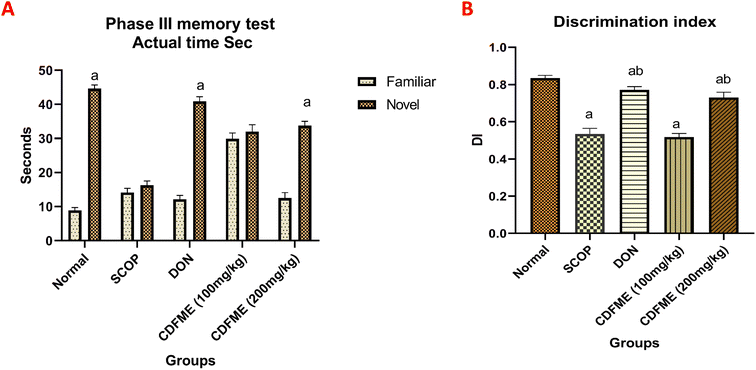
SCOP is an anticholinergic drug known to block muscarinic receptors and act as a muscarinic receptor antagonist. The antagonistic role of SCOP causes significant deficiencies in attention and memory. The SCOP-induced dementia model has been extensively used to assess possible treatment drugs for treating AD. AD is a brain disease that gradually decreases thinking and memory capacities, as well as the ability to do even the most fundamental tasks.2 To investigate the cognitive-enhancing effect of CDFME in rats, learning and memory were assessed using the NORT and various biochemical parameters related to memory function were measured. These included the measurement of the hippocampal levels of AChE enzyme and neurotransmitters (ACh, NA, and DA). Additionally, the results of these in vivo investigations were assessed in relation to the metabolite profile established by LC-QTOF-MS/MS. Table 3 shows the DI in the NORT, as well as the levels of AChE, ACh, NA, and DA in the different rat groups.| Group | DI | AChE (U mg−1) | ACh (nmol mg−1) | NA (pg mg−1) | DA (ng mg−1) |
|---|---|---|---|---|---|
| Normal | 0.83 ± 0.02 | 0.21 ± 0.02 | 5.100 ± 0.49 | 3.78 ± 0.74 | 16.49 ± 1.81 |
| SCOP (1.14 mg kg−1) | 0.53 ± 0.03 | 0.91 ± 0.01 | 1.200 ± 0.23 | 21.96 ± 1.05 | 45.06 ± 4.73 |
| DON (0.5 mg kg−1) + SCOP | 0.77 ± 0.17 | 0.17 ± 0.02 | 4.900 ± 0.20 | 6.53 ± 0.73 | 19.97 ± 1.51 |
| CDFME (100 mg kg−1) + SCOP | 0.52 ± 0.02 | 0.84 ± 0.02 | 1.200 ± 0.25 | 20.26 ± 0.98 | 41.58 ± 1.95 |
| CDFME (200 mg kg−1) + SCOP | 0.73 ± 0.03 | 0.40 ± 0.03 | 4.500 ± 0.76 | 10.20 ± 0.65 | 26.26 ± 2.34 |
Various phytoconstituents identified in CDFME may contribute to the observed cognitive enhancement effect. Specifically, phenolic acids, including salvianolic acid B, caffeic acid, rosmarinic acid, syringic acid, p-coumaric acid, and danshensu, have been reported to improve cognitive abilities in in vitro and in vivo animal models of AD.66,67 Essential amino acids have a potential role in enhancing human learning, memory, and neuro-cognitive performance.68 Additionally, administering oleamide to mice enhanced their learning and memory-related skills.69 Fatty acids like linolenic acid have also been reported to increase DI in rats in the NORT in in vivo animal models.70 Furthermore, phospholipids reversed SCOP-induced spatial memory deficits in mice.71
Various metabolites identified in CDFME have anti-AChE effect and restore ACh levels. Phenolic compounds, such as caffeic acid, syringic acid, lariciresinol,74 salvianolic acid B, and rosmarinic acid,67 were reported to have a potent anti-AChE effect. p-Coumaric acid mitigates LPS-induced brain damage and decreases AChE level in the brains of mice.66 Coumarins have the potential to treat AD and enhance cognitive performance by blocking AChE.75 Moreover, previous research showed that amino acids76 and organic acids such as quinic acid74 exhibited in vitro AChE inhibitory activity. Dietary polyunsaturated fatty acids77 and choline in the diet78 improved cholinergic transmission, increased ACh release in the aged brain, and inhibited AChE.
Several research works have emphasized the influence of classical neurotransmitter systems, like NA and DA, on cognitive processes such as concentration, learning, and memory.8 Neurochemical changes in aging and AD involve neurotransmitter systems like adrenergic and dopaminergic. In order to preserve functional stability, endogenous neurotransmitter secretion normally stays in a harmonious ratio and at a specified level. According to earlier research, an increase in NA and DA levels leads to AD symptoms.65 The association between AD progression and dysfunction in the noradrenergic system is not completely understood. An excess of NA may lead to production of toxic metabolites, contributing to disease progression.79 Drugs increasing NA levels in early AD stages may cause adverse effects and worsen cognitive symptoms due to altered adrenergic receptor function, making the decrease of NA level a potential therapeutic goal.79 Previous research suggested that SCOP increases NA and DA levels in both the cortex and hippocampus.65
As expected, in the SCOP control group, there was a significant increase in NA and DA levels by 481%, and 173%, respectively, compared to the normal control group (Table 3 and Fig. 4). DON decreased the elevated NA and DA levels by 70% and 55%, respectively, as compared with the SCOP control group. Likewise, as observed before for ACh, CDFME (200 mg kg−1) induced a similar neuroprotective effect to DON, resulting in significant reductions in NA and DA levels compared to the SCOP control group by 53% and 41%, respectively. These results suggested that CDFME may improve memory and learning capacities by modulating the metabolic pathway of monoamine neurotransmitters. To gain additional insights on the phytoconstituents responsible for the bioactivity of CDFME, molecular docking, molecular dynamics, and molecular mechanics studies were conducted.
The 2D and 3D interactions, along with the binding positioning for these four compounds, are depicted in Table 4. The co-crystallized inhibitor of AChE was stabilized by binding to Ser200, Ala201, Gly118, and Gly119 amino acids. In contrast, rosmarinic acid formed one H-bond with Glu199 and one pi–H bond with Gly118. Moreover, caffeoyl-4′-hydroxyphenyllactate bound Gly118 and Tyr334 with pi-H and pi–pi bonds, respectively. Sebestenoid C formed six H-bonds with Gly118, Glu199, Gln69, Tyr121, and Asp72 (2), along with one pi–pi bond with Trp279. Finally, sagerinic acid bound through three H-bonds with Tyr121, Asn85, and Gln74. It also formed two H2O-bridged H-bonds with Phe288 and Phe331 through the H2O892 molecule, and one pi–H bond with Gly118. Based on the above and considering that these four compounds occupied the same binding pocket of the co-crystallized inhibitor, a promising antagonistic activity could be inferred (Table 4).
The RMSD of the examined protein complexes as a function of the simulation time was compared to their initial values to examine their exact stability. Throughout the simulations, the RMSD values consistently remained below 2.5 Å, indicating highly stable behaviors. With the exception of the sebestenoid C-1OCE complex, all other complexes exhibited RMSD values around 2 Å, closely resembling those of the Co-1OCE reference (Fig. 5A).
Additionally, the ligands' RMSD analysis was conducted to clarify the degree of stability for rosmarinic acid, caffeoyl-4′-hydroxyphenyllactate, sebestenoid C, and sagerinic acid compared to that of the co-crystallized inhibitor (DON) (Fig. 5B). The RMSD values for the four compounds fluctuated along the simulation time up to 7.2, 4, 4.5, and 13.5 Å, respectively, compared to the co-crystallized DON (4.8 Å). As can be observed in Fig. 5B, rosmarinic acid showed moderate stability, where it fluctuated gradually from the start up to 7.2 Å. Meanwhile, sagerinic acid showed the least stable behavior due to its large fluctuations, especially from 0 to 100 ns, reaching an average of about 12 Å. Notably, both caffeoyl-4′-hydroxyphenyllactate and sebestenoid C described the most stable behaviors, which were very close to those of the co-crystallized ligand. Specifically, caffeoyl-4′-hydroxyphenyllactate achieved the highest stability, with fluctuations lower than 4 Å. This value was slightly lower than the value for the co-crystallized DON (4.8 Å). The subsequent discussion provides a detailed analysis of the caffeoyl-4′-hydroxyphenyllactate-1OCE complex in comparison to the Co-1OCE reference standard.
3.4.4.1 Protein-ligand interactions analysis (histogram and heat map). Both the histogram and heat map of the caffeoyl-4′-hydroxyphenyllactate-1OCE complex were compared to those of the Co-1OCE (Fig. 6A and 7A, respectively). The histograms and heat maps of rosmarinic acid, sebestenoid C, and sagerinic acid-1OCE complexes are provided as ESI (Fig. S22 and S23,† respectively).
 | ||
| Fig. 6 Histograms of (A) caffeoyl-4′-hydroxyphenyllactate and (B) DON (Co) ligands within the binding site of AChE(1OCE) during the simulation time of 200 ns. | ||
Regarding the histogram of the caffeoyl-4′-hydroxyphenyllactate-1OCE complex (Fig. 6A), it was evident that Tyr334 and Phe331 were the most contributing amino acids from the AChE receptor to the interaction with caffeoyl-4′-hydroxyphenyllactate. Tyr334 showed 140% total interactions, in the form of H-bonds (100%), hydrophobic interactions (10%), and H2O-bridges H-bonds (30%). Similarly, Phe331 described total interactions exceeding 130%, divided as H-bonds (90%), hydrophobic interactions (40%), and H2O-bridges H-bonds (∼5%). Additionally, the heat map of caffeoyl-4′-hydroxyphenyllactate-1OCE complex (Fig. 7A) showed that both Tyr334 and Phe331 amino acids of the AChE binding pocket involved in the interactions with the caffeoyl-4′-hydroxyphenyllactate ligand throughout the 200 ns of the simulation (indicated by the dark orange color).
 | ||
| Fig. 7 Heat maps of (A) caffeoyl-4′-hydroxyphenyllactate and (B) DON (Co) ligands within the binding site of AChE(1OCE) during the simulation time of 200 ns. | ||
In contrast to these results, the histogram of the Co-1OCE complex (Fig. 6B) represented that Ser81, Asn85, and Tyr70 amino acids were the most prominent residues involved in interactions with the co-crystallized inhibitor of AChE. Their total interactions fractions were >70%, 70%, and 60%, respectively. Ser81 interactions were in the form of H2O-bridges H-bonds (70%) and H-bonds (∼3%), while those of Asn85 were exclusively in the form of H2O-bridges H-bonds (70%). As for Tyr70, its interactions were classified as H2O-bridges H-bonds (58%) and hydrophobic bonds (∼2%). Additionally, the heat map of the Co-1OCE complex (Fig. 7B) highlighted that Ser81 interactions with the Co-1OCE throughout the 200 ns simulation, becoming more intense in the second half (>100 ns). On the other hand, Asn85 and Tyr70 showed more intense interactions from 30 to 200 ns of the simulation. Finally, Tyr70 interactions were clearer from 30 to about 185 ns of the simulation time.
| Complex | ΔG binding | Coulomb | Covalent | H-bond | Lipo | Bind packing | Solv_GB | VdW | SD |
|---|---|---|---|---|---|---|---|---|---|
| a Coulomb: coulomb energy; covalent: covalent binding energy; H-bond: hydrogen-bonding energy; Lipo: lipophilic energy; Solv_GB: generalized born electrostatic solvation energy; VdW: van der Waals energy; and SD: standard deviation. | |||||||||
| Rosmarinic acid-1OCE | −60.05 | 28.56 | 3.15 | −3.07 | −26.26 | −3.99 | −18.43 | −40.01 | 10.84 |
| Caffeoyl-4′-hydroxyphenyllactate-1OCE | −63.28 | 33.50 | 2.14 | −3.91 | −22.95 | −3.37 | −29.41 | −39.27 | 10.27 |
| Sebestenoid C-1OCE | −83.98 | −42.86 | 7.32 | −4.49 | −36.76 | −3.96 | 77.00 | −80.22 | 15.98 |
| Sagerinic acid-1OCE | −50.76 | 129.81 | 4.50 | −2.80 | −20.85 | −3.10 | −111.18 | −47.14 | 8.73 |
| Co-1OCE | −46.56 | −49.49 | 1.55 | −0.58 | −19.30 | 0 | 63.05 | −41.79 | 5.12 |
As can be observed in Table 5, all the examined complexes showed superior ΔG binding energies compared to that of the co-crystallized inhibitor complex (−46.56 kcal mol−1). Consistent with the findings from the molecular docking and molecular dynamics studies, both the caffeoyl-4′-hydroxyphenyllactate-1OCE and sebestenoid C-1OCE complexes presented the highest ΔG binding energies (−63.28 and −83.98 kcal mol−1, respectively). Additionally, the contributions from different components to the total energy in all cases were very close or superior to those of the Co-1OCE complex. This suggested a very promising perspective regarding the AChE inhibitory potential of the examined compounds from CDFME, particularly the phenolic acids; caffeoyl-4′-hydroxyphenyllactate and sebestenoid C.
4. Conclusion
In light of the aforementioned findings, CDFME demonstrated the presence of valuable phenolic compounds, coumarins, essential amino acids, and phospholipids, which were reported to have a neuroprotective effect. C. dichotoma fruit extract exhibited a cognition-enhancing and neuromodulatory effect by reversing SCOP-induced learning and memory deficits in rats. The protective effect was observed by an increase in visual recognition in the behavioral test. Also, the treatment improved cholinergic neuronal activity by decreasing AChE and increasing ACh levels in the rat hippocampus. Moreover, it showed a correction of neurotransmitter imbalances. Additionally, molecular docking revealed that among the most abundant metabolites, rosmarinic acid, caffeoyl-4′-hydroxyphenyllactate, sebestenoid C, and sagerinic acid were found to show the highest binding affinity towards AChE with binding scores of −7.57, −7.38, −8.47, and −7.78 kcal mol−1, respectively, compared to DON (−8.13 kcal mol−1) and the co-crystallized inhibitor (−7.88 kcal mol−1). Furthermore, molecular dynamics and molecular mechanics clarified that caffeoyl-4′-hydroxyphenyllactate-1OCE complex was the most stable one. Therefore, the study supports the use of CDFME in the food industries and pharmaceuticals for developing memory-enhancing nutraceuticals and functional foods. However, more detailed mechanistic studies are needed in the future to demonstrate cognitive function-enhancing effects of CDFME on other potential mechanisms involved in AD pathogenesis, such as inflammatory and glutamatergic pathways, and its antioxidant potential as well. Further research into bioassay-guided separation and quantification of potential active compounds in the fruit extract, as well as their therapeutic evaluation in AD, is advised. More detailed and conclusive in vivo and clinical studies are highly recommended to investigate the potential of. C. dichotoma fruit extract in treating patients with AD. As far as we are aware, our research is the first to characterize metabolic profile and investigate cognitive function-enhancing effects of C. dichotoma fruit extract.Abbreviations
| ACh | Acetylcholine |
| AChE | Acetylcholinesterase |
| AD | Alzheimer's disease |
| CDFME | C. dichotoma fruit methanolic extract |
| Co-1OCE | Co-crystallized ligand (inhibitor or DON) complex |
| DA | Dopamine |
| DI | Discrimination index |
| DON | Donepezil |
| ESI | Electrospray ionization |
| LC-QTOF-MS/MS | Liquid chromatography quadrupole time-of-flight mass spectrometry |
| MM-GBSA | Molecular mechanics with generalized Born and surface area solvation |
| NA | Noradrenaline |
| NORT | Novel object recognition test |
| RMSD | Root mean square deviation |
| SCOP | Scopolamine |
| TFC | Total flavonoids content |
| TPC | Total phenolics content |
Data availability
The authors confirm that the data supporting the finding of this study are available within the article and or its ESI.†Conflicts of interest
Authors declare no conflicts of interest.Acknowledgements
Basma M. Eltanany would like to thank the Egyptian Ministry of Higher Education for funding her postdoctoral research stay at the Bioanalysis group of the University of Barcelona.References
- V. Singh, A. Kahol, I. P. Singh, I. Saraf and R. Shri, J. Ethnopharmacol., 2016, 193, 490–499 CrossRef PubMed
.
- I. M. Ayoub, M. Y. George, E. T. Menze, M. Mahmoud, M. Botros, M. Essam, I. Ashmawy, P. Shendi, A. Hany, M. Galal, M. Ayman and R. M. Labib, Food Funct., 2022, 13, 2253–2268 RSC
.
- S. Bhatia, R. Rawal, P. Sharma, T. Singh, M. Singh and V. Singh, Curr. Neuropharmacol., 2021, 20, 675–692 CrossRef
.
- Y. Ma, M. W. Yang, X. W. Li, J. W. Yue, J. Z. Chen, M. W. Yang, X. Huang, L. L. Zhu, F. F. Hong and S. L. Yang, Front. Pharmacol., 2019, 10, 1355 CrossRef PubMed
.
- K. Randhawa, V. Singh, S. Kaur, R. Kaur, S. Kumar and R. Shri, Food Sci. Hum. Wellness, 2021, 10, 490–496 CrossRef
.
- S. Pruthi, K. Kaur, V. Singh and R. Shri, Metab. Brain Dis., 2021, 36, 901–910 CrossRef PubMed
.
- V. Singh, K. Kaur, S. Kaur, R. Shri, T. G. Singh and M. Singh, J. Ethnopharmacol., 2022, 295, 115438 CrossRef CAS PubMed
.
- X. Pan, A. C. Kaminga, P. Jia, S. W. Wen, K. Acheampong and A. Liu, Front. Aging Neurosci., 2020, 12, 1–12 CrossRef
.
- H. Hussein, E. Mohsen, A. Abdelmonem and M. Abdel Kawy, Egpt. J. Chem., 2023, 66, 437–459 Search PubMed
.
- D. Raghuvanshi, K. Sharma, R. Verma, D. Kumar, H. Kumar, A. Khan, M. Valko, S. Y. Alomar, S. H. Alwasel, E. Nepovimova and K. Kuca, Biomed. Pharmacother., 2022, 153, 113400 CrossRef CAS PubMed
.
- M. Yizibula, Z. Wusiman, N. Abudouhalike and B. Maimaitiming, Folia Neuropathol., 2021, 58, 365–376 CrossRef PubMed
.
- G. Kendir, H. J. Bae, J. Kim, Y. Jeong, H. J. Bae, K. Park, X. Yang, Y. jin Cho, J. Y. Kim, S. Y. Jung, A. Köroğlu, D. S. Jang and J. H. Ryu, BMC Complementary Med. Ther., 2022, 22, 215 CrossRef PubMed
.
- H. M. Hussein, E. Mohsen, A. R. Abdelmonem, M. A. A. Kawy, A. A. Al-Karmalawy and R. A. El-Shiekh, Egpt. J. Chem., 2024, 67, 225–237 Search PubMed
.
- R. Malik, S. Kalra, Pooja, G. Singh, Meenu, V. Gahlot, A. Kajal and Rimpy, Brain Res., 2024, 1822, 148616 CrossRef PubMed
.
- I. Y. Younis, M. S. Sedeek, A. F. Essa, A. M. Elgamal, B. M. Eltanany, Z. M. Goda, L. Pont, F. Benavente and E. Mohsen, Food Chem., 2024, 465, 141918 CrossRef PubMed
.
- A. M. Khalil, O. M. Sabry, H. El-Askary, S. M. El Zalabani, B. M. Eltanany, L. Pont, F. Benavente, A. F. Mohamed and N. M. Fayek, BMC Complementary Med.
Ther., 2024, 24, 379 CrossRef PubMed
.
- H. M. El-Sayed, D. M. Rasheed, E. A. Mahrous, B. M. Eltanany, Z. M. Goda, L. Pont, F. Benavente and E. Abdel-Sattar, J. Pharm. Biomed. Anal., 2025, 252, 116512 CrossRef PubMed
.
- A. M. Otify, S. A. ElBanna, B. M. Eltanany, L. Pont, F. Benavente and R. M. Ibrahim, Food Res. Int., 2023, 172, 113178 CrossRef PubMed
.
- N. M. Shofian, A. A. Hamid, A. Osman, N. Saari, F. Anwar, M. S. P. Dek and M. R. Hairuddin, Int. J. Mol. Sci., 2011, 12, 4678–4692 CrossRef PubMed
.
- E. Attard, Cent. Eur. J. Biol., 2013, 8, 48–53 Search PubMed
.
- M. Kiranmai, C. B. Mahendra Kumar and M. Ibrahim, Res. J. Pharm. Biol. Chem. Sci., 2011, 2, 254–261 Search PubMed
.
- R. M. Ibrahim, B. Eltanany, L. Pont, F. Benavente, S. AbdelSalam ElBanna and A. M. Otify, Food Res. Int., 2023, 168, 112742 CrossRef
.
- A. Ennaceur and J. Delacour, Behav. Brain Res., 1988, 31, 47–59 CrossRef PubMed
.
- N. A. Shaif, D. H. Chang, D. Cho, S. Kim, D. B. Seo and I. Shim, Biomedicines, 2018, 6, 108 CrossRef
.
- M. F. Elshal, N. M. Eid, I. El-Sayed, W. El-Sayed and A. A. Al-Karmalawy, Pharm. Sci., 2021, 28, 76–85 Search PubMed
.
- M. Khattab and A. A. Al-Karmalawy, Future Med. Chem., 2021, 13, 1623–1638 CrossRef PubMed
.
- A. A. Elmaaty, K. M. Darwish, A. Chrouda, A. A. Boseila, M. A. Tantawy, S. S. Elhady, A. B. Shaik, M. Mustafa and A. A. Al-Karmalawy, ACS Omega, 2021, 7, 875–899 CrossRef
.
- M. A. Ragab, W. M. Eldehna, A. Nocentini, A. Bonardi, H. E. Okda, B. Elgendy, T. S. Ibrahim, M. M. Abd-Alhaseeb, P. Gratteri, C. T. Supuran, A. A. Al-Karmalawy and M. Elagawany, Eur. J. Med. Chem., 2023, 250, 115180 CrossRef PubMed
.
- A. M. El-Naggar, A. M. A. Hassan, E. B. Elkaeed, M. S. Alesawy and A. A. Al-Karmalawy, Bioorg. Chem., 2022, 123, 105770 CrossRef PubMed
.
- M. M. Hammoud, M. Khattab, M. Abdel-Motaal, J. Van der Eycken, R. Alnajjar, H. Abulkhair and A. A. Al-Karmalawy, J. Biomol. Struct. Dyn., 2022, 41, 5199–5216 Search PubMed
.
- D. T. Ayele, M. L. Akele and A. T. Melese, BMC Chem., 2022, 16, 30 CrossRef PubMed
.
- A. M. Khalil, O. M. Sabry, H. I. El-Askary, S. M. El Zalabani, B. M. Eltanany, L. Pont, F. Benavente, A. Elshewy and N. M. Fayek, Phytochem. Anal., 2024, 35, 1–18 CrossRef PubMed
.
- T. Cheng, J. Ye, H. Li, H. Dong, N. Xie, N. Mi, Z. Zhang, J. Zou, H. Jin and W. Zhang, RSC Adv., 2019, 9, 8714–8727 RSC
.
- S. Yaermaimaiti, T. Wu and H. A. Aisa, Ind. Crops Prod., 2021, 172, 113977 CrossRef
.
- N. P. Araujo, H. S. Arruda, F. N. dos Santos, D. R. de Morais, G. A. Pereira and G. M. Pastore, Food Res. Int., 2020, 137, 109556 CrossRef
.
- J. A. B. Peixoto, G. Álvarez-Rivera, A. S. G. Costa, S. Machado, A. Cifuentes, E. Ibáñez, M. B. P. P. Oliveira and R. C. Alves, Antioxidants, 2023, 12, 251 CrossRef
.
- L. Zhu, S. Ma, K. Li, P. Xiong and W. Cai, Molecules, 2022, 27, 2631 CrossRef PubMed
.
- M. Kim, J. Y. Kim, H. S. Yang, J. S. Choe and I. G. Hwang, Antioxidants, 2021, 10, 1208 CrossRef
.
- J. Dai, A. Sorribas, W. Y. Yoshida and P. G. Williams, Phytochemistry, 2010, 71, 2168–2173 CrossRef PubMed
.
- K. Kramberger, D. Barlič-Maganja, D. Bandelj, A. Baruca Arbeiter, K. Peeters, A. Miklavčič Višnjevec and Z. J. Pražnikar, Metabolites, 2020, 10, 403 CrossRef PubMed
.
- K. Niaz, M. A. Nawaz, S. Pervez, U. Younas, I. Shah and F. Khan, S. Afr. J. Bot., 2022, 144, 437–447 CrossRef
.
- B. S. Zhumakanova, I. Korona-Głowniak, K. Skalicka-Woźniak, A. Ludwiczuk, T. Baj, K. K. Wojtanowski, A. Józefczyk, K. A. Zhaparkulova, Z. B. Sakipova and A. Malm, Molecules, 2021, 26, 3193 CrossRef PubMed
.
- P. López-Rojas, Á. Amesty, M. Guerra-Rodríguez, Y. Brito-Casillas, B. Guerra, L. Fernández-Pérez and A. Estévez-Braun, Pharmaceuticals, 2022, 15, 1–30 CrossRef
.
- J. Simayi, A. Abulizi, M. Nuermaimaiti, N. Khan, S. Hailati, M. Han, Z. Talihati, K. Abudurousuli, N. Maihemuti, M. Nuer, W. Zhou and A. Wumaier, Biomed Res. Int., 2022, 2022, 4176235 Search PubMed
.
- L. R. Fernández, A. Cirigliano, M. P. Fabani, B. Lima, S. Alberti, F. Kramer, A. A. Tapia, G. Cabrera, J. A. Palermo and M. Sánchez, Planta Med., 2013, 79, 1724–1729 CrossRef PubMed
.
- Y. H. Lee, B. Kim, S. Kim, M. S. Kim, H. Kim, S. R. Hwang, K. Kim and J. H. Lee, J. Food Drug Anal., 2017, 25, 776–788 CrossRef
.
- S. A. S. da Silva, A. L. Souto, M. de F. Agra, E. V. L. Da-Cunha, J. M. Barbosa-Filho, M. S. da Silva and R. Braz-Filho, Arkivoc, 2004, 54–58 Search PubMed
.
- A. Trifan, E. Wolfram, N. Esslinger, A. Grubelnik, K. Skalicka-Woźniak, M. Minceva and S. V. Luca, Phytochem. Anal., 2021, 32, 482–494 CrossRef PubMed
.
- K. N. Venugopala, V. Rashmi and B. Odhav, Biomed Res. Int., 2013, 2013, 963248 Search PubMed
.
- K. Wang, J. Tian, Y. Li, M. Liu, Y. Chao, Y. Cai, G. Zheng and Y. Fang, ACS Omega, 2021, 6, 17045–17057 CrossRef PubMed
.
- M. J. Oza and Y. A. Kulkarni, J. Pharm. Pharmacol., 2017, 69, 755–789 CrossRef
.
- N. Guo, S. Zhang, M. Gu and G. Xu, Crop J., 2021, 9, 530–542 CrossRef
.
- Z. N. Ling, Y. F. Jiang, J. N. Ru, J. H. Lu, B. Ding and J. Wu, Signal Transduct. Target. Ther., 2023, 8, 345 CrossRef PubMed
.
- F. Zhang, B. Li, Y. Wen, Y. Liu, R. Liu, J. Liu, S. Liu and Y. Jiang, Pharm. Biol., 2022, 60, 1349–1364 CrossRef PubMed
.
- Z. Li, Z. Tu, H. Wang and L. Zhang, Molecules, 2020, 25, 4507 CrossRef
.
- I. M. Abu-Reidah, M. S. Ali-Shtayeh, R. M. Jamous, D. Arráez-Román and A. Segura-Carretero, Food Chem., 2015, 166, 179–191 CrossRef
.
- Z. T. Abdel Shakour, R. H. El-Akad, A. I. Elshamy, A. E.-N. G. N. G. El Gendy, L. A. Wessjohann and M. A. Farag, Food Chem., 2022, 399, 133948 CrossRef PubMed
.
- R. Tanvir, A. Javeed and Y. Rehman, FEMS Microbiol. Lett., 2018, 365, fny114 CrossRef PubMed
.
- R. Gevrenova, G. Zengin, K. I. Sinan, E. Yıldıztugay, D. Zheleva-Dimitrova, C. Picot-Allain, M. F. Mahomoodally, M. Imran, S. Dall’acqua and S. Dall’acqua, Antioxidants, 2021, 10, 1180 CrossRef PubMed
.
- A. Masyita, R. Mustika Sari, A. Dwi Astuti, B. Yasir, N. Rahma Rumata, T. Bin Emran, F. Nainu and J. Simal-Gandara, Food Chem. X, 2022, 13, 100217 CrossRef CAS
.
- F. Han, Y. Li, X. Mao, R. Xu and R. Yin, MASS Spectrom., 2016, 51, 363–368 CrossRef CAS PubMed
.
- E. A. Mahrous, A. H. Elosaily, A. A. A. Salama, A. M. Salama and S. M. El-Zalabani, Plants, 2022, 11, 218 CrossRef CAS PubMed
.
- B. Lin, S. Guo, X. Hong, X. Jiang, H. Li, J. Li, L. Guo, M. Li, J. Chen, B. Huang, Y. Xu and M. G. Ferraro, Evid. Based Complement. Altern. Med., 2022, 2022, 1322751 Search PubMed
.
- M. E. Hussein, O. G. Mohamed, A. M. El-Fishawy, H. I. El-Askary, A. A. Hamed, M. M. Abdel-Aziz, R. Alnajjar, A. Belal, A. M. Naglah, A. A. Almehizia, A. A. Al-Karmalawy, A. Tripathi and A. S. El Senousy, Plants, 2022, 11, 3286 CrossRef PubMed
.
- S. Bhuvanendran, Y. Kumari, I. Othman and M. F. Shaikh, Front. Pharmacol., 2018, 9, 665 CrossRef PubMed
.
- G. Caruso, J. Godos, A. Privitera, G. Lanza, S. Castellano, A. Chillemi, O. Bruni, R. Ferri, F. Caraci and G. Grosso, Nutrients, 2022, 14, 819 CrossRef PubMed
.
- S. Habtemariam, Int. J. Mol. Sci., 2018, 19, 458 CrossRef PubMed
.
- H. Suzuki, D. Yamashiro, S. Ogawa, M. Kobayashi, D. Cho, A. Iizuka, M. Tsukamoto-Yasui, M. Takada, M. Isokawa, K. Nagao and Y. Fujiwara, Front. Nutr., 2020, 7, 586166 CrossRef PubMed
.
- R. Tao, S. Huang, J. Zhou, L. Ye, X. Shen, J. Wu and L. Qian, J. Nutr., 2022, 152, 889–898 CrossRef PubMed
.
- L. Taoro-González, D. Pereda, C. Valdés-Baizabal, M. González-Gómez, J. A. Pérez, F. Mesa-Herrera, A. Canerina-Amaro, H. Pérez-González, C. Rodríguez, M. Díaz and R. Marin, Int. J. Mol. Sci., 2022, 23, 7430 CrossRef
.
- Z. Bao, P. Zhang, J. Chen, J. Gao, S. Lin and N. Sun, J. Funct. Foods, 2020, 69, 103948 CrossRef
.
- P. Świt, A. Pollap and J. Orzeł, Top. Curr. Chem., 2023, 381, 16 CrossRef
.
- S. C. Jee, K. M. Lee, M. Kim, Y. J. Lee, S. Kim, J. O. Park and J. S. Sung, Int. J. Mol. Sci., 2020, 21, 9202 CrossRef CAS
.
- S. Ahmed, S. T. Khan, M. K. Zargaham, A. U. Khan, S. Khan, A. Hussain, J. Uddin, A. Khan and A. Al-Harrasi, Biomed. Pharmacother., 2021, 139, 111609 CrossRef CAS PubMed
.
- R. Joshi, ChemistrySelect, 2023, 8, 202303861 CrossRef
.
- Z. Kovarik, Z. Radić, B. Grgas, M. Škrinjarić-Špoljar, E. Reiner and V. Simeon-Rudolf, Biochim. Biophys. Acta, 1999, 1433, 261–271 CrossRef CAS PubMed
.
- L. M. Willis, B. Shukitt-Hale and J. A. Joseph, Genes Nutr., 2009, 4, 309–314 CrossRef CAS PubMed
.
- A. M. Wiedeman, S. I. Barr, T. J. Green, Z. Xu, S. M. Innis and D. D. Kitts, Nutrients, 2018, 10, 1513 CrossRef PubMed
.
- I. L. Gutiérrez, C. Dello Russo, F. Novellino, J. R. Caso, B. García-Bueno, J. C. Leza and J. L. M. Madrigal, Int. J. Mol. Sci., 2022, 23, 6134 CrossRef PubMed
.
- M. Tavan, P. Hanachi, M. de la Luz Cádiz-Gurrea, A. Segura Carretero and M. H. Mirjalili, Neurochem. Res., 2024, 49, 306–326 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06991a |
| This journal is © The Royal Society of Chemistry 2024 |