Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design, synthesis, and antiproliferative screening of new quinoline derivatives bearing a cis-vinyl triamide motif as apoptosis activators and EGFR-TK inhibitors†
Hany M. Abd El-Lateef*ab,
Ahmed Gaafar Ahmed Gaafarc,
Arwa Sultan Alqahtanid,
Aamal A. Al-Mutairid,
Dalal Sulaiman Alshayae,
Fahmy Gad Elsaidf,
Eman Fayadg and
N. A. Farouk *h
*h
aDepartment of Chemistry, College of Science, King Faisal University, Al-Ahsa 31982, Saudi Arabia. E-mail: hmahmed@kfu.edu.sa
bDepartment of Chemistry, Faculty of Science, Sohag University, Sohag 82524, Egypt
cDepartment of Pharmacology and Toxicology, Faculty of Pharmacy, Port Said University, Port Said, Egypt
dDepartment of Chemistry, College of Science, Imam Mohammad Ibn Saud Islamic University(IMSIU), P.O. Box 90950, Riyadh 11623, Saudi Arabia
eDepartment of Biology, College of Science, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh 11671, Saudi Arabia
fDepartment of Biology, College of Science, King Khalid University, PO Box 960, Abha, 61421, Asir, Saudi Arabia
gDepartment of Biotechnology, College of Sciences, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia
hDepartment of Chemistry, Faculty of Science, Port Said University, Port Said, Egypt. E-mail: nesmafarouk566@gmail.com
First published on 7th August 2024
Abstract
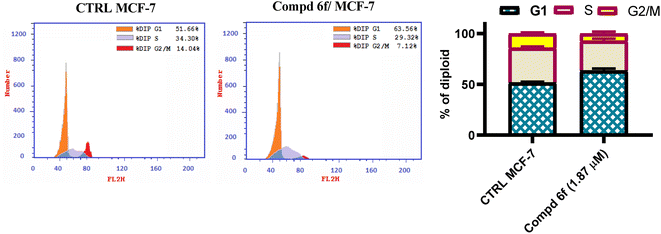
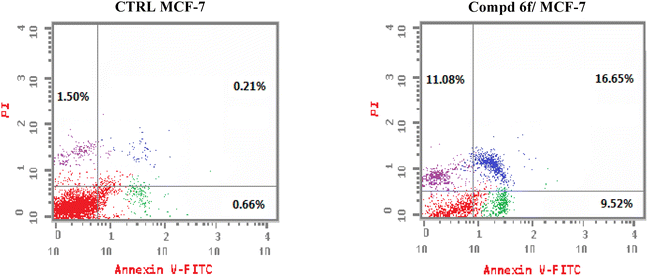
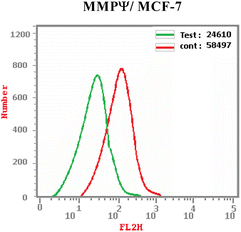
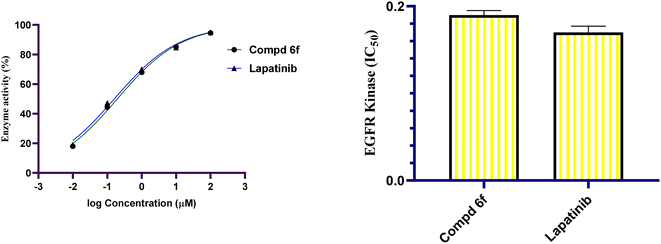
In this work, a congeneric set of quinoline-tethered cis-vinyl triamide hybrids was prepared and evaluated as EGFR tyrosine kinase inhibitors for the management of breast cancer. All of the prepared hybrids were evaluated for their antiproliferative effect against the breast MCF-7 cell line. Among the tested hybrids, compound 6f displayed the most potent antiproliferative activity with an IC50 value of 1.87 μM compared to STU (IC50 = 13.71 μM) as the standard reference. The most promising hybrid, 6f, was found to induce cellular cycle arrest at the G1 phase. Furthermore, the molecular mechanism of this hybrid revealed its ability to induce cellular apoptosis via the mitochondrial-dependent apoptotic pathway. Compound 6f decreased MCF-7 cells' MMP compared to the controls (percentage change value of 57.93%). Further investigation of the selective compound 6f showed that it can inhibit EGFR tyrosine kinase.
1. Introduction
Cancer remains the leading cause of death in the world, second only to heart disease.1,2 Cancer itself is a dynamic, complex cellular network of uncontrolled growth.3 Of all cancer subtypes, breast cancer is one of the most deadly, and also consistently beckons the largest number of new diagnoses among women.4,5 Clearly, there is a serious unmet demand for therapies targeting this cancer subtype.6,7 Effective chemotherapeutic treatments with minimal side effects are urgently needed, particularly in the face of the increasing prevalence of drug-resistant tumors.8 Therapies based on targeted biological target are intended to be less toxic than conventional chemotherapy.9,10The epidermal growth factor receptor (EGFR), being one of the most prominent protein kinases, plays a vital role in a series of cellular processes during the life cycle of the cell, such as the regulation of cell migration and cell division.11,12 Multiple prospective clinical trials were initiated to test the efficacy of EGFR-targeted therapy versus standard chemotherapy.13 Clinical trials revealed definite evidence of the superiority of EGFR tyrosine kinase inhibitors over standard chemotherapeutic regimens.14 Targeted inhibitors of EGFR signalling inhibited ligand-induced EGFR auto-phosphorylation and attenuated downstream signalling responsible for proliferation and survival of the cells.15,16 The inhibition of EGFR signalling leads, in most cases, to cell cycle arrest and/or drug-induced apoptosis.17,18 Therefore, EGFR has been regarded as an attractive target for the establishment of chemotherapeutic drugs for many cancers.19 Accordingly, the discovery of new EGFR has attracted a great deal of attention in recent years.20
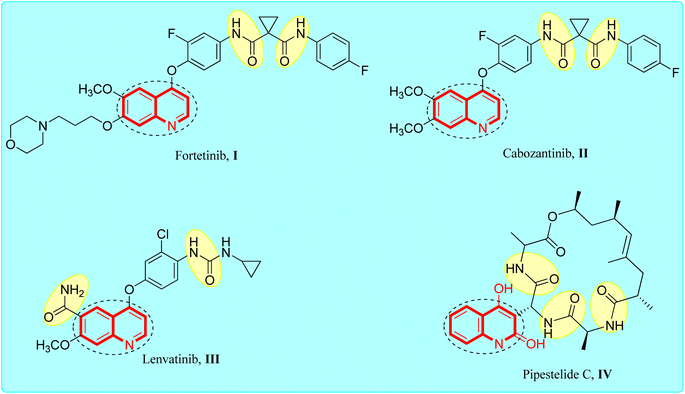
Quinolines are a distinct class of fused bicyclic hetero-aromatic core compounds that have developed into a very popular research topic.21–23 Several quinoline-based compounds are known to have chemical, biological and therapeutic applications.24,25 They were found to exhibit anticancer, antiviral and antimicrobial effects.26–28 The quinoline nucleus is also an integral component of several anticancer drugs, which have revolutionized the therapy of cancer disease.29–31 The anti-cancer activity seems to be due to a variety of mechanisms include inhibition of cellular promoting factors such as tubulin polymerization and topoisomerase.32,33 Recently, several research studies have described quinoline derivatives with anticancer efficacy via inhibition of kinases and inhibition of the anti-death Bcl-2 family of proteins34,35 (Fig. 1).
The amide pharmacophore is an important framework for developing drugs and discovery.36 According to medicinal chemistry sources, one or more amide bonds serve as the basic component in more than 25% of natural and synthetic drugs available in the market.37 The amide functional group enhances the anticancer activity by making molecules more polar as well as forming a hydrogen bond acceptor–donor domain with the target receptor.38 Therefore, the introduction of an amide motif bearing a cis-vinyl group into the quinoline core is likely to significantly influence biological activity.39
The aforementioned intriguing findings, combined with our ongoing quest for more potent anticancer agents, led to the molecular hybridization of the quinoline core and the bioactive triamide motif bearing a cis-vinyl group in order to integrate them into a single molecular framework and achieve a new hybrid that could have potential antiproliferative activity (Fig. 2). The study's goal is to look into the benefits of such hybridization in terms of predicted biological activity and to see whether this resulted in better biological activity for the produced hybrids. Lastly, we used in vitro tests to assess the antiproliferative action of the prepared quinoline-cis-vinyl triamide hybrids, as well as FACS and ELISA methods to determine the mechanism of cellular action.
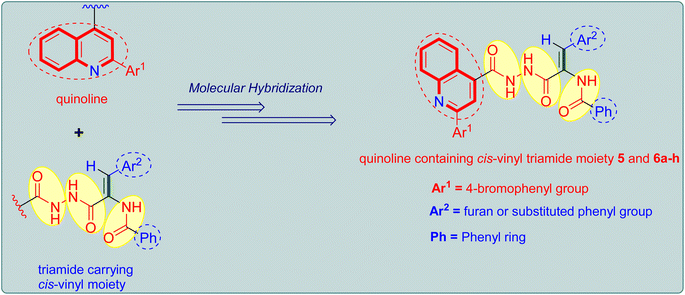 | ||
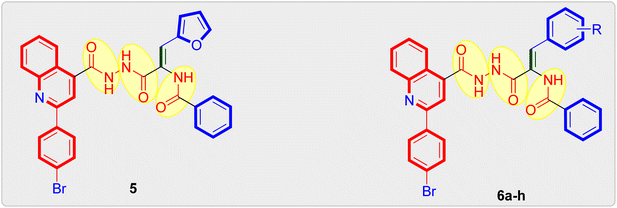
| Fig. 2 Design strategy adopted for the design of target quinoline tethered cis-vinyl triamides 5 and 6a–h. | ||
2. Results and discussion
2.1. Chemistry
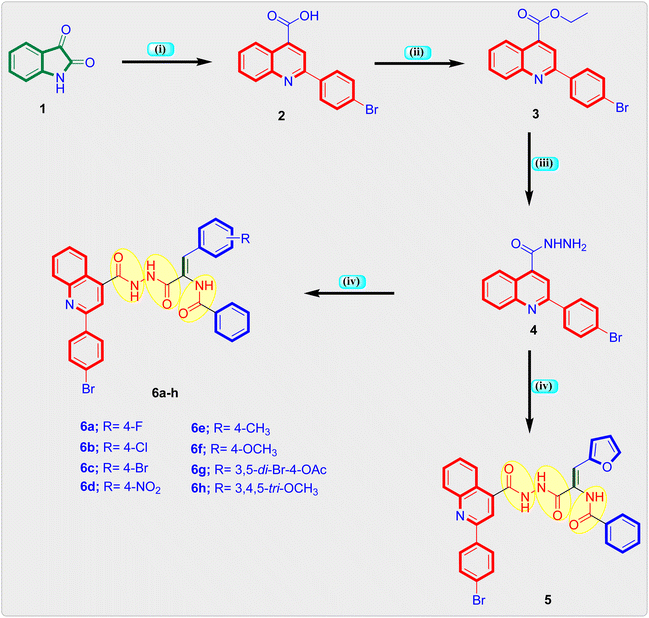
The synthetic route used in the preparation of the intermediate and final quinoline based triamide compounds is depicted in Scheme 1. In the initial step appropriate 2-(4-bromoquinoline)-4-carboxylic acid 2 was obtained from reaction of isatin 1 and 4-bromoacetophenone in the presence of 33% KOH in pure ethanol by refluxing which was then heated to reflux in pure ethanol in the presence of concentrated sulphuric acid to obtain ethyl quinoline-4-carboxylate 3.26 This is followed by hydrazinolysis with pure hydrazine hydrate in boiling ethanol to furnish the key intermediate quinoline-4-carbohydrazide compound 4.40,41 Lastly, final quinolone-linked triamide derivatives 5 and 6a–h were obtained by the reaction of quinoline-4-carbohydrazide 4 and respective methyl 3-aryl-2-(benzamido)-2-propenoate derivative in freshly molten sodium acetate and glacial acetic acid by refluxing and yielding 64–78%. The formation of the quinoline linked triamide compounds 5 and 6a–h in general were authenticated by 1H-NMR, 13C-NMR and elemental analysis. 1H-NMR spectra of the final quinoline-like triamide molecules displayed three broad singlets at range δ 10.89–9.99 ppm ascribed to the three amide protons and singlet peak at δ 7.48–7.37 ppm range assigned to olefinic (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) proton. The characteristic doublet signal at about δ 8.46–8.42 ppm is attributed to the C8–H of the quinoline motif. In addition, the protons of C7–H and C6–H of quinoline motif appeared as two triplet signals with one proton each in the range at δ 7.88–7.72 and 7.72–7.63 ppm, respectively.
CH) proton. The characteristic doublet signal at about δ 8.46–8.42 ppm is attributed to the C8–H of the quinoline motif. In addition, the protons of C7–H and C6–H of quinoline motif appeared as two triplet signals with one proton each in the range at δ 7.88–7.72 and 7.72–7.63 ppm, respectively.
The 13C-NMR spectra of the final compounds displayed signals for aromatic and olefinic carbon atoms were observed at around δ 153.09–107.71 ppm, whereas signals for aliphatic carbon atoms in compounds 6f, 6e, 6g and 6h were observed at around δ 60.54–20.65. The characteristic C2 carbon of quinoline moiety was identified in the range δ 155.03–155.00 ppm. Furthermore, the carbonyl carbons of the three amide functions were seen at about δ 168.85–164.45 ppm providing a substantial argument in support of the compounds' ascribed structures. Each of the target compounds was characterized using the melting point technique, and their purity was verified using the TLC method. The newly prepared quinoline-triamide molecules' analytical and spectral data (1H-NMR and 13C-NMR spectra) agreed fully with the suggested structures.
2.2. Biology
3. Conclusions
A new set of quinoline-tethered cis-vinyl triamide hybrids 5 and 6a–h was designed and constructed as inhibitors of EGFR tyrosine kinase for the treatment of breast cancer. All the prepared quinoline-tethered cis-vinyl triamide hybrids were screened for their in vitro antiproliferative activity and revealed moderate to potent activity. Among them, quinoline compounds 6e bearing [2-(4-methylphenyl)-cis-vinyl] triamide and 6f bearing [2-(4-methoxyphenyl)-cis-vinyl] triamide were found to be the most potent hybrids against the MCF-7 breast cancer cell line, with IC50 values of 1.87 and 1.88 μM, respectively, compared to STU (IC50 = 13.77 μM). Additional mechanistic studies demonstrated that quinoline-[2-(4-methoxyphenyl)-cis-vinyl] triamide 6f effectively blocked the G1 phase of the cell cycle and was found to promote cellular apoptosis. It is notable that quinoline-[2-(4-methoxyphenyl)-cis-vinyl] triamide 6f increased the percentage population at G1 phase by 1.2-fold more than controls. In addition, it boosted the level of both early and late apoptosis by almost 14.4- and 79.3-fold, respectively, compared to controls. Further mechanistic apoptotic studies confirmed that treatment in MCF-7 cells with the most active member; quinoline-[2-(4-methoxyphenyl)-cis-vinyl] triamide 6f, decreased the level of MMP of MCF-7 cells (57.93% percent change value) less than controls. Notably, quinoline-[2-(4-methoxyphenyl)-cis-vinyl] triamide 6f also demonstrated potent EGFR tyrosine kinase inhibition with an IC50 value of 0.19 μM, which was equipotent to reference standard Lapatinib (IC50 = 0.17 μM). The previous findings indicated that quinoline-[2-(4-methoxyphenyl)-cis-vinyl] triamide 6f is a potent apoptosis-active anticancer molecule that exerted its action via EGFR tyrosine kinase inhibition as a promising structure that could serve as a novel template for developing powerful and selective agents in cancer therapy.4. Experimental
4.1. Chemistry
4.1.1.1 (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-1-(furan-2-yl)-3-oxoprop-1-en-2-yl)benzamide (5). Yield 76%; mp: 188–190 °C. 1H-NMR (400 MHz, DMSO-d6) δ: 10.80 (s, 1H, NH), 10.49 (s, 1H, NH), 9.99 (s, 1H, NH), 8.42 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.26 (d, J = 8.3 Hz, 2H), 8.19–8.14 (m, 2H), 8.13–8.07 (m, 2H), 7.88 (t, J = 7.6 Hz, C7–H quinoline), 7.84–7.78 (m, 3H), 7.71 (t, J = 7.7 Hz, 2H, C6–H quinoline and Ar–H), 7.63 (dd, J = 8.3, 6.2 Hz, 1H), 7.56 (t, J = 7.5 Hz, 2H), 7.35 (s, 1H), 6.82 (d, J = 3.5 Hz, 1H), 6.63 (dd, J = 3.5, 1.8 Hz, 1H). 13C-NMR (101 MHz, DMSO) δ 166.32, 166.11, 164.45, 155.01, 150.03, 148.30, 145.42, 141.99, 137.75, 134.30, 132.45, 132.14, 131.06, 129.97, 129.74, 128.75, 128.47, 127.98, 126.18, 126.01, 124.22, 124.05, 119.42, 117.14, 115.14, 112.88. C30H21BrN4O4 (581.42): calcd: C, 61.97; H, 3.64; N, 9.64. Found: C, 62.09; H, 3.78; N, 9.51.
4.1.1.2. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-1-(4-fluorophenyl)-3-oxoprop-1-en-2-yl)benzamide (6a). Yield 68%; mp: 177–179 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.68 (d, J = 90.2 Hz, 2H, 2NH), 10.11 (s, 1H, NH), 8.46 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.31–8.23 (m, 2H, Ar–H), 8.16 (t, J = 4.3 Hz, 2H, Ar–H), 8.06 (d, J = 7.6 Hz, 2H, Ar–H), 7.87 (t, J = 7.6 Hz, 1H, C7–H quinoline), 7.83–7.77 (m, 2H, Ar–H), 7.71 (td, J = 5.6, 2.9 Hz, 3H, C6–H quinoline and Ar–H), 7.65–7.59 (m, 1H, Ar–H), 7.55 (t, J = 7.4 Hz, 2H, Ar–H), 7.43 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.26 (td, J = 8.9, 1.6 Hz, 2H, Ar–H). 13C-NMR (101 MHz, DMSO) δ 168.85, 166.50, 166.05, 156.50, 155.00, 148.32, 142.34, 142.19, 137.80, 134.03, 132.45, 132.25, 132.21, 132.13, 131.06, 131.00, 129.96, 129.74, 128.79, 128.47, 127.91, 126.12, 124.22, 117.14, 116.19, 115.98. C32H22BrFN4O3 (609.44): calcd: C, 63.06; H, 3.64; N, 9.19. Found: C, 62.95; H, 3.55; N, 9.33.
CH), 7.26 (td, J = 8.9, 1.6 Hz, 2H, Ar–H). 13C-NMR (101 MHz, DMSO) δ 168.85, 166.50, 166.05, 156.50, 155.00, 148.32, 142.34, 142.19, 137.80, 134.03, 132.45, 132.25, 132.21, 132.13, 131.06, 131.00, 129.96, 129.74, 128.79, 128.47, 127.91, 126.12, 124.22, 117.14, 116.19, 115.98. C32H22BrFN4O3 (609.44): calcd: C, 63.06; H, 3.64; N, 9.19. Found: C, 62.95; H, 3.55; N, 9.33.
4.1.1.3. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-1-(4-chlorophenyl)-3-oxoprop-1-en-2-yl)benzamide (6b). Yield 73%; mp: 185–187 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.83 (s, 1H, NH), 10.58 (s, 1H, NH), 10.13 (s, 1H, NH), 8.43 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.27 (d, J = 8.2 Hz, 2H, Ar–H), 8.16 (s, 2H, Ar–H), 8.08–8.02 (m, 2H, Ar–H), 7.88 (t, J = 7.6 Hz, 1H, C7–H quinoline), 7.81 (d, J = 8.2 Hz, 2H, Ar–H), 7.72 (t, J = 7.7 Hz, 2H, C6–H quinoline), 7.67 (d, J = 8.3 Hz, 2H, Ar–H), 7.62 (d, J = 7.2 Hz, 1H, Ar–H), 7.55 (t, J = 7.5 Hz, 2H, Ar–H), 7.48 (d, J = 8.3 Hz, 2H, Ar–H), 7.40 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH). 13C-NMR (101 MHz, DMSO) δ 166.49, 166.15, 164.96, 155.02, 148.30, 141.98, 137.74, 133.95, 133.84, 133.43, 132.46, 132.29, 131.61, 131.07, 129.98, 129.80, 129.75, 129.53, 129.13, 128.79, 128.48, 128.00, 125.98, 124.23, 124.05, 117.15. C32H22BrClN4O3 (625.90): calcd: C, 61.41; H, 3.54; N, 8.95. Found: C, 61.59; H, 3.39; N, 9.09.
CH). 13C-NMR (101 MHz, DMSO) δ 166.49, 166.15, 164.96, 155.02, 148.30, 141.98, 137.74, 133.95, 133.84, 133.43, 132.46, 132.29, 131.61, 131.07, 129.98, 129.80, 129.75, 129.53, 129.13, 128.79, 128.48, 128.00, 125.98, 124.23, 124.05, 117.15. C32H22BrClN4O3 (625.90): calcd: C, 61.41; H, 3.54; N, 8.95. Found: C, 61.59; H, 3.39; N, 9.09.
4.1.1.4. (Z)-N-(1-(4-Bromophenyl)-3-(2-(2-(4-bromophenyl)quinoline-4-carbonyl)hydrazinyl)-3-oxoprop-1-en-2-yl)benzamide (6c). Yield 78%; mp: 191–193 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.83 (s, 1H, NH), 10.58 (s, 1H, NH), 10.13 (s, 1H, NH), 8.43 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.26 (d, J = 8.2 Hz, 2H, Ar–H), 8.17 (d, J = 7.7 Hz, 2H, Ar–H), 8.05 (d, J = 7.7 Hz, 2H, Ar–H), 7.87 (t, J = 7.8 Hz, 1H, C7–H quinoline), 7.80 (d, J = 8.1 Hz, 2H, Ar–H), 7.71 (t, J = 7.7 Hz, 2H, C6–H quinoline), 7.60 (h, J = 8.0, 7.3 Hz, 6H, Ar–H), 7.37 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH). 13C-NMR (101 MHz, DMSO) δ 166.46, 166.14, 164.95, 155.02, 148.31, 141.99, 137.75, 133.96, 133.80, 132.46, 132.29, 132.05, 131.84, 131.07, 129.99, 129.91, 129.75, 129.55, 128.79, 128.49, 127.99, 126.00, 124.23, 124.06, 122.59, 117.15. C32H22Br2N4O3 (670.35): calcd: C, 57.33; H, 3.31; N, 8.36. Found: C, 57.42; H, 3.36; N, 8.29.
CH). 13C-NMR (101 MHz, DMSO) δ 166.46, 166.14, 164.95, 155.02, 148.31, 141.99, 137.75, 133.96, 133.80, 132.46, 132.29, 132.05, 131.84, 131.07, 129.99, 129.91, 129.75, 129.55, 128.79, 128.49, 127.99, 126.00, 124.23, 124.06, 122.59, 117.15. C32H22Br2N4O3 (670.35): calcd: C, 57.33; H, 3.31; N, 8.36. Found: C, 57.42; H, 3.36; N, 8.29.
4.1.1.5. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-1-(4-nitrophenyl)-3-oxoprop-1-en-2-yl)benzamide (6d). Yield 69%; mp: 207–209 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.89 (s, 1H, NH), 10.72 (s, 1H, NH), 10.30 (s, 1H, NH), 8.44 (d, J = 8.3 Hz, 1H, C8–H quinoline), 8.27 (dt, J = 9.0, 2.9 Hz, 4H, Ar–H), 8.18 (t, J = 4.2 Hz, 2H, Ar–H), 8.05 (d, J = 7.7 Hz, 2H, Ar–H), 7.90 (d, J = 8.7 Hz, 3H, Ar–H), 7.81 (d, J = 8.1 Hz, 2H, Ar–H), 7.72 (t, J = 7.7 Hz, 1H, C7–H quinoline), 7.63 (t, J = 7.4 Hz, 1H, C6–H quinoline), 7.56 (t, J = 7.5 Hz, 2H, Ar–H), 7.43 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH). 13C-NMR (101 MHz, DMSO) δ 166.53, 166.14, 164.73, 155.03, 148.31, 147.25, 142.33, 141.88, 141.54, 137.74, 132.45, 132.37, 131.07, 130.83, 129.99, 129.76, 128.82, 128.55, 128.01, 127.79, 127.62, 125.96, 125.90, 124.17, 124.05, 117.18. C32H22BrN5O5 (636.45): calcd: C, 60.39; H, 3.48, N; 11.00. Found: C, 60.24; H, 3.57; N, 11.12.
CH). 13C-NMR (101 MHz, DMSO) δ 166.53, 166.14, 164.73, 155.03, 148.31, 147.25, 142.33, 141.88, 141.54, 137.74, 132.45, 132.37, 131.07, 130.83, 129.99, 129.76, 128.82, 128.55, 128.01, 127.79, 127.62, 125.96, 125.90, 124.17, 124.05, 117.18. C32H22BrN5O5 (636.45): calcd: C, 60.39; H, 3.48, N; 11.00. Found: C, 60.24; H, 3.57; N, 11.12.
4.1.1.6. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-3-oxo-1-p-tolylprop-1-en-2-yl)benzamide (6e). Yield 70%; mp: 211–213 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.79 (s, 1H, NH), 10.49 (s, 1H, NH), 10.07 (s, 1H, NH), 8.44 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.27 (d, J = 8.3 Hz, 2H, Ar–H), 8.17 (d, J = 9.0 Hz, 2H, Ar–H), 8.07 (d, J = 7.6 Hz, 2H, Ar–H), 7.88 (t, J = 7.5 Hz, 1H, C7–H quinoline), 7.81 (d, J = 8.2 Hz, 2H, Ar–H), 7.72 (t, J = 7.8 Hz, 1H, C6–H quinoline), 7.58 (dt, J = 15.1, 7.2 Hz, 5H, Ar–H), 7.42 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.22 (d, J = 7.9 Hz, 2H, Ar–H), 2.32 (s, 3H, Ar–CH3). 13C-NMR (101 MHz, DMSO) δ 167.67, 166.78, 166.10, 164.42, 155.03, 148.30, 145.77, 141.91, 137.74, 135.63, 133.91, 133.45, 132.42, 131.43, 131.08, 130.00, 129.75, 128.82, 128.38, 128.02, 127.40, 125.96, 124.24, 124.04, 117.64, 117.15, 20.65. C33H25BrN4O3 (605.48): calcd: C, 65.46; H, 4.16; N, 9.25. Found: C, 65.60; H, 4.28; N, 9.11.
CH), 7.22 (d, J = 7.9 Hz, 2H, Ar–H), 2.32 (s, 3H, Ar–CH3). 13C-NMR (101 MHz, DMSO) δ 167.67, 166.78, 166.10, 164.42, 155.03, 148.30, 145.77, 141.91, 137.74, 135.63, 133.91, 133.45, 132.42, 131.43, 131.08, 130.00, 129.75, 128.82, 128.38, 128.02, 127.40, 125.96, 124.24, 124.04, 117.64, 117.15, 20.65. C33H25BrN4O3 (605.48): calcd: C, 65.46; H, 4.16; N, 9.25. Found: C, 65.60; H, 4.28; N, 9.11.
4.1.1.7. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-1-(4-methoxyphenyl)-3-oxoprop-1-en-2-yl)benzamide (6f). Yield 75%; mp: 206–208 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.76 (s, 1H, NH), 10.43 (s, 1H, NH), 10.04 (s, 1H, NH), 8.44 (d, J = 8.4 Hz, 1H, C8–H quinoline), 8.30–8.23 (m, 2H, Ar–H), 8.19–8.13 (m, 2H, Ar–H), 8.12–8.05 (m, 2H, Ar–H), 7.87 (t, J = 7.5 Hz, 1H, C7–H quinoline), 7.84–7.77 (m, 2H, Ar–H), 7.71 (t, J = 7.8 Hz, 1H, C6–H quinoline), 7.63 (d, J = 8.5 Hz, 3H, Ar–H), 7.55 (t, J = 7.4 Hz, 2H, Ar–H), 7.43 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.98 (d, J = 8.8 Hz, 2H, Ar–H), 3.78 (s, 3H, Ar–OCH3). 13C-NMR (101 MHz, DMSO) δ 166.44, 166.16, 165.27, 160.36, 155.01, 142.10, 140.27, 137.76, 134.18, 132.46, 132.17, 131.83, 131.42, 130.83, 129.97, 129.74, 128.77, 128.46, 127.97, 126.85, 126.75, 126.04, 124.22, 124.07, 117.13, 114.60, 55.72. C33H25BrN4O4 (621.48): calcd; C, 63.78; H, 4.05; N, 9.02. Found: C, 63.86; H, 3.96; N, 8.95.
CH), 6.98 (d, J = 8.8 Hz, 2H, Ar–H), 3.78 (s, 3H, Ar–OCH3). 13C-NMR (101 MHz, DMSO) δ 166.44, 166.16, 165.27, 160.36, 155.01, 142.10, 140.27, 137.76, 134.18, 132.46, 132.17, 131.83, 131.42, 130.83, 129.97, 129.74, 128.77, 128.46, 127.97, 126.85, 126.75, 126.04, 124.22, 124.07, 117.13, 114.60, 55.72. C33H25BrN4O4 (621.48): calcd; C, 63.78; H, 4.05; N, 9.02. Found: C, 63.86; H, 3.96; N, 8.95.
4.1.1.8. (Z)-4-(2-Benzamido-3-(2-(2-(4-bromophenyl)quinoline-4-carbonyl)hydrazinyl)-3-oxoprop-1-enyl)-2,6-dibromophenyl acetate (6g). Yield 64%; mp: 181–183 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.86 (s, 1H, NH), 10.65 (s, 1H, NH), 10.22 (s, 1H, NH), 8.43 (d, J = 8.5 Hz, 1H, C8–H quinoline), 8.27 (d, J = 8.4 Hz, 2H, Ar–H), 8.17 (d, J = 8.8 Hz, 2H, Ar–H), 8.03 (d, J = 10.7 Hz, 4H, Ar–H), 7.88 (t, J = 7.6 Hz, C7–H quinoline), 7.81 (d, J = 8.3 Hz, 2H, Ar–H), 7.71 (t, J = 7.8 Hz, 1H, C6–H quinoline), 7.63 (t, J = 7.2 Hz, 1H, Ar–H), 7.56 (t, J = 7.5 Hz, 2H, Ar–H), 7.40 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 2.40 (s, 3H, Ar–OCOCH3). 13C-NMR (101 MHz, DMSO) δ 166.48, 166.18, 165.19, 165.17, 155.02, 148.31, 142.06, 139.30, 137.76, 134.15, 132.47, 132.20, 131.61, 131.39, 131.08, 130.04, 129.75, 129.69, 128.77, 128.45, 128.23, 128.00, 126.05, 126.01, 124.24, 124.07, 117.13, 21.41. C34H23Br3N4O5 (807.28): calcd: C, 50.59; H, 2.87; N, 6.94. Found: C, 50.44; H, 2.79; N, 7.07.
CH), 2.40 (s, 3H, Ar–OCOCH3). 13C-NMR (101 MHz, DMSO) δ 166.48, 166.18, 165.19, 165.17, 155.02, 148.31, 142.06, 139.30, 137.76, 134.15, 132.47, 132.20, 131.61, 131.39, 131.08, 130.04, 129.75, 129.69, 128.77, 128.45, 128.23, 128.00, 126.05, 126.01, 124.24, 124.07, 117.13, 21.41. C34H23Br3N4O5 (807.28): calcd: C, 50.59; H, 2.87; N, 6.94. Found: C, 50.44; H, 2.79; N, 7.07.
4.1.1.9. (Z)-N-(3-(2-(2-(4-Bromophenyl)quinoline-4-carbonyl)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (6h). Yield 67%; mp: 169–171 °C. 1H-NMR (400 MHz, DMSO-d6) δ 10.80 (s, 1H, NH), 10.51 (s, 1H, NH), 10.10 (s, 1H, NH), 8.45 (dd, J = 8.4, 1.4 Hz, 1H, C8–H quinoline), 8.31–8.23 (m, 2H, Ar–H), 8.17 (d, J = 10.0 Hz, 2H, Ar–H), 8.12 (dd, J = 7.1, 2.0 Hz, 2H, Ar–H), 7.88 (t, J = 7.5 Hz, C7–H quinoline), 7.84–7.78 (m, 2H, Ar–H), 7.72 (t, J = 7.7 Hz, 1H, C6–H quinoline), 7.62 (t, J = 7.4 Hz, 1H, Ar–H), 7.55 (t, J = 7.5 Hz, 2H, Ar–H), 7.48 (s, 1H,
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.03 (s, 2H, Ar–H), 3.68 (s, 3H, OCH3), 3.63 (s, 6H, 2OCH3). 13C-NMR (101 MHz, DMSO) δ 166.48, 166.13, 164.95, 155.02, 153.09, 152.91, 148.31, 142.04, 138.72, 137.76, 134.00, 132.46, 132.28, 131.90, 131.07, 129.98, 129.74, 128.71, 128.40, 128.14, 127.97, 126.03, 124.23, 124.07, 117.13, 107.71, 60.54, 56.08. C35H29BrN4O6 (681.53): calcd: C, 61.68; H, 4.29; N, 8.22. Found: C, 61.81; H, 4.36; N, 8.10.
CH), 7.03 (s, 2H, Ar–H), 3.68 (s, 3H, OCH3), 3.63 (s, 6H, 2OCH3). 13C-NMR (101 MHz, DMSO) δ 166.48, 166.13, 164.95, 155.02, 153.09, 152.91, 148.31, 142.04, 138.72, 137.76, 134.00, 132.46, 132.28, 131.90, 131.07, 129.98, 129.74, 128.71, 128.40, 128.14, 127.97, 126.03, 124.23, 124.07, 117.13, 107.71, 60.54, 56.08. C35H29BrN4O6 (681.53): calcd: C, 61.68; H, 4.29; N, 8.22. Found: C, 61.81; H, 4.36; N, 8.10.
4.2. Biological studies
All the experimental procedure used in the biological studies were shown in the ESI.†Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its ESI.†Conflicts of interest
No potential conflict of interest was reported by the author(s).Acknowledgements
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. KFU241438). The authors extend their appreciation to Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R465), Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia, and the authors extend their appreciation to the Deanship of Scientific Research and Graduate Studies at King Khalid University for funding this work through the Large Research Project under grant number RGP2/61/45.References
- C. Santucci, S. Mignozzi, M. Malvezzi, P. Boffetta, G. Collatuzzo, F. Levi, C. La Vecchia and E. Negri, European cancer mortality predictions for the year 2024 with focus on colorectal cancer, Ann. Oncol., 2024, 35, 308–316 CrossRef CAS PubMed.
- R. L. Siegel, A. N. Giaquinto and A. Jemal, Cancer statistics, Ca-Cancer J. Clin., 2024, 74, 12–49 CrossRef PubMed.
- P. Nenclares and K. J. Harrington, The biology of cancer, Medicine, 2020, 48, 67–72 CrossRef.
- K. M. Cuthrell and N. Tzenios, Breast Cancer: Updated and Deep Insights, Int. Res. J. Oncol., 2023, 6, 104–118 Search PubMed.
- M. Ensenyat-Mendez, P. Llinàs-Arias, J. I. J. Orozco, S. Íñiguez-Muñoz, M. P. Salomon, B. Sesé, M. L. DiNome and D. M. Marzese, Current Triple-Negative Breast Cancer Subtypes: Dissecting the Most Aggressive Form of Breast Cancer, Front. Oncol., 2021, 11, 681476–681483 CrossRef CAS PubMed.
- J. Wang and S.-G. Wu, Breast Cancer: An Overview of Current Therapeutic Strategies, Challenge, and Perspectives, Breast Cancer: Targets Ther., 2023, 15, 721–730 CAS.
- J. G. Elmore and C. I. Lee, Toward More Equitable Breast Cancer Outcomes, JAMA, 2024, 331, 1896–1897 CrossRef PubMed.
- Brianna and S. H. Lee, Chemotherapy: how to reduce its adverse effects while maintaining the potency?, Med. Oncol., 2023, 40, 88 CrossRef CAS PubMed.
- M. Puzzo, M. De Santo, C. Morelli, A. Leggio and L. Pasqua, The Advent of Molecular Targeted Therapies Against Cancer. Toward Multi-Targeting Drugs Through Materials Engineering: A Possible Future Scenario, Small Sci., 2024, 2400113 CrossRef.
- H. Jin, L. Wang and R. Bernards, Rational combinations of targeted cancer therapies: background, advances and challenges, Nat. Rev. Drug Discovery, 2023, 22, 213–234 CrossRef CAS PubMed.
- M. L. Uribe, I. Marrocco and Y. Yarden, EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance, Cancers, 2021, 13, 2748 CrossRef CAS PubMed.
- A. W. Burgess, Regulation of Signaling from the Epidermal Growth Factor Family, J. Phys. Chem. B, 2022, 126, 7475–7485 CrossRef CAS PubMed.
- A. Dang, S. Dang and B. Vallish, Efficacy and Safety of EGFR Inhibitors in the Treatment of EGFRPositive NSCLC Patients: A Meta-analysis, Rev. Recent Clin. Trials, 2021, 16, 193–201 CrossRef CAS PubMed.
- S.-Y. Liu, S.-Y. M. Liu, W.-Z. Zhong and Y.-L. Wu, Targeted Therapy in Early Stage Non-small Cell Lung Cancer, Curr. Treat. Options Oncol., 2022, 23, 1169–1184 CrossRef PubMed.
- S. Halder, S. Basu, S. P. Lall, A. K. Ganti, S. K. Batra and P. Seshacharyulu, Targeting the EGFR signaling pathway in cancer therapy: What's new in 2023?, Expert Opin. Ther. Targets, 2023, 27, 305–324 CrossRef CAS PubMed.
- K. S. Alharbi, M. A. Javed Shaikh, O. Afzal, A. S. Alfawaz Altamimi, W. H. Almalki, S. I. Alzarea, I. Kazmi, F. A. Al-Abbasi, S. K. Singh, K. Dua and G. Gupta, An overview of epithelial growth factor receptor (EGFR) inhibitors in cancer therapy, Chem. Biol. Interact., 2022, 366, 110108 CrossRef CAS.
- F. Volta, S. La Monica, A. Leonetti, L. Gnetti, M. Bonelli, A. Cavazzoni, C. Fumarola, M. Galetti, K. Eltayeb, R. Minari, P. G. Petronini, M. Tiseo and R. Alfieri, Intrinsic Resistance to Osimertinib in EGFR Mutated NSCLC Cell Lines Induced by Alteration in Cell-Cycle Regulators, Targeted Oncol., 2023, 18, 953–964 CrossRef PubMed.
- D. Nasser Binjawhar, F. A. Al-Salmi, M. A. Alghamdi, A. s. Alqahtani, E. Fayad, R. M. Saleem, I. Zaki and A. M. Youssef Moustafa, Design, Synthesis, and Biological Evaluation of Newly Synthesized Cinnamide-Fluorinated Containing Compounds as Bioactive Anticancer Agents, ACS Omega, 2024, 9, 18505–18515 CrossRef CAS.
- R. Roskoski, Properties of FDA-approved small molecule protein kinase inhibitors: A 2024 update, Pharmacol. Res., 2024, 200, 107059 CrossRef CAS PubMed.
- N. Ryad, A. A. Elmaaty, I. M Ibrahim, A. H. Ahmed Maghrabi, M. A. Yahya Alahdal, R. M. Saleem, I. Zaki and L. M. A. A. Ghany, Harnessing molecular hybridization approach to discover novel quinoline EGFR-TK inhibitors for cancer treatment, Future Med. Chem., 2024, 16, 1087–1107 CrossRef PubMed.
- V. Yadav, J. Reang, V. Sharma, J. Majeed, P. C. Sharma, K. Sharma, N. Giri, A. Kumar and R. K. Tonk, Quinoline-derivatives as privileged scaffolds for medicinal and pharmaceutical chemists: A comprehensive review, Chem. Biol. Drug Des., 2022, 100, 389–418 CrossRef CAS PubMed.
- O. O. Ajani, K. T. Iyaye and O. T. Ademosun, Recent advances in chemistry and therapeutic potential of functionalized quinoline motifs–a review, RSC Adv., 2022, 12, 18594–18614 RSC.
- D. N. Binjawhar, F. A. Al-Salmi, O. A. A. Ali, M. A. Alghamdi, E. Fayad, R. M. Saleem, I. Zaki and N. Farouk, Design, synthesis and cytotoxic activity of molecular hybrids based on quinolin-8-yloxy and cinnamide hybrids and their apoptosis inducing property, RSC Adv., 2024, 14, 11443–11451 RSC.
- C. dos Santos Chagas, F. L. A. Fonseca and I. A. Bagatin, Quinoline-derivative coordination compounds as potential applications to antibacterial and antineoplasic drugs, Mater. Sci. Eng. C, 2019, 98, 1043–1052 CrossRef CAS PubMed.
- M. Mohasin, M. Zafer Alam, Q. Ullah, A. Ahmad, P. F. Rahaman and S. A. Khan, A Review on Synthesis and Biological Applications of Quinoline Derivative as Fused Aromatic Compounds, Polycyclic Aromat. Compd., 2024, 1–30 Search PubMed.
- M. Ilakiyalakshmi and A. Arumugam Napoleon, Review on recent development of quinoline for anticancer activities, Arabian J. Chem., 2022, 15, 104168 CrossRef CAS.
- S. Hu, J. Chen, J.-X. Cao, S.-S. Zhang, S.-X. Gu and F.-E. Chen, Quinolines and isoquinolines as HIV-1 inhibitors: Chemical structures, action targets, and biological activities, Bioorg. Chem., 2023, 136, 106549 CrossRef CAS PubMed.
- H. M. Abd El-Lateef, A. A. Elmaaty, L. M. A. Abdel Ghany, M. S. Abdel-Aziz, I. Zaki and N. Ryad, Design and Synthesis of 2-(4-Bromophenyl)Quinoline-4-Carbohydrazide Derivatives via Molecular Hybridization as Novel Microbial DNA-Gyrase Inhibitors, ACS Omega, 2023, 8, 17948–17965 CrossRef CAS PubMed.
- I. A. Bala, O. F. Al Sharif, A. M. Asiri and R. M. El-Shishtawy, Quinoline: A versatile bioactive scaffold and its molecular hybridization, Results Chem., 2024, 7, 101529 CrossRef CAS.
- V. Snehi, H. Verma, S. Saha, S. Kumar and D. Pathak, An extensive review on biological interest of quinoline and its analogues, Int. J. Sci. Healthcare Res., 2023, 8, 45–66 CAS.
- Y. Chauhan, K. Neha, S. Wakode, M. Shahfaiz, R. B. Bodla and K. Sharma, Progression and Expansion of Quinoline as Bioactive Moiety: A Patent Review, Pharm. Pat. Anal., 2023, 12, 287–314 CrossRef CAS PubMed.
- Y. Ren, Y. Ruan, B. Cheng, L. Li, J. Liu, Y. Fang and J. Chen, Design, synthesis and biological evaluation of novel acridine and quinoline derivatives as tubulin polymerization inhibitors with anticancer activities, Bioorg. Med. Chem., 2021, 46, 116376 CrossRef CAS PubMed.
- H. K. Swedan, A. E. Kassab, E. M. Gedawy and S. E. Elmeligie, Topoisomerase II inhibitors design: Early studies and new perspectives, Bioorg. Chem., 2023, 136, 106548 CrossRef CAS PubMed.
- T. Fan, Y. Ji, D. Chen, X. Peng, J. Ai and B. Xiong, Design, synthesis and biological evaluation of 4-aminoquinoline derivatives as receptor-interacting protein kinase 2 (RIPK2) inhibitors, J. Enzyme Inhib. Med. Chem., 2023, 38, 282–293 CrossRef CAS PubMed.
- R. Hamdy, S. A. Elseginy, N. I. Ziedan, A. T. Jones and A. D. Westwell, New Quinoline-Based Heterocycles as Anticancer Agents Targeting Bcl-2, Molecules, 2019, 24, 1274 CrossRef PubMed.
- H. M. Abd El-Lateef, E. E. M. Toson, A. H. Abu Almaaty, R. M. Saleem, A. H. A. Maghrabi, E. H. El-Sayed, I. Zaki and M. M. Youssef, Synthesis, Characterization and Biological Evaluation of New Enamide Fluorinated-Schiff Base Derivatives as Potential Cytotoxic and Apoptosis-Inducing Agents, ChemistrySelect, 2023, 8, e202303070 CrossRef CAS.
- C. Zhao, Y. Wang, Q. Pham, C. Dai, A. Chatterjee and M. Wasa, Chemical Tagging of Bioactive Amides by Cooperative Catalysis: Applications in the Syntheses of Drug Conjugates, J. Am. Chem. Soc., 2023, 145, 14233–14250 CrossRef CAS PubMed.
- Z. S. El-Fakharany, Y. M. Nissan, N. K. Sedky, R. K. Arafa and S. M. Abou-Seri, New proapoptotic chemotherapeutic agents based on the quinolone-3-carboxamide scaffold acting by VEGFR-2 inhibition, Sci. Rep., 2023, 13, 11346 CrossRef CAS.
- K. O. Mohamed, I. Zaki, I. M. El-Deen and M. K. Abdelhameid, A new class of diamide scaffold: Design, synthesis and biological evaluation as potent antimitotic agents, tubulin polymerization inhibition and apoptosis inducing activity studies, Bioorg. Chem., 2019, 84, 399–409 CrossRef CAS.
- H. M. Abd El-Lateef, L. M. A. Ghany, R. M. Saleem, A. H. A. Maghrabi, M. A. Y. Alahdal, E. H. K. Ali, B. Y. Beshay, I. Zaki and R. E. Masoud, Design, synthesis and antiproliferative screening of newly synthesized coumarin-acrylamide hybrids as potential cytotoxic and apoptosis inducing agents, RSC Adv., 2023, 13, 32547–32557 RSC.
- L. M. A. A. Ghany, N. Ryad, M. S. Abdel-Aziz, H. M. A. El-Lateef, I. Zaki and B. Y. Beshay, Design, synthesis, antimicrobial evaluation, and molecular modeling of new sulfamethoxazole and trimethoprim analogs as potential DHPS/DHFR inhibitors, J. Mol. Struct., 2024, 1309, 138170–138183 CrossRef CAS.
- K. Gousias, T. Theocharous and M. Simon, Mechanisms of Cell Cycle Arrest and Apoptosis in Glioblastoma, Biomed, 2022, 10, 564 CAS.
- X. Guo and L. Chen, From G1 to M: a comparative study of methods for identifying cell cycle phases, Briefings Bioinf., 2024, 25, 517–526 CrossRef PubMed.
- D. Bertheloot, E. Latz and B. S. Franklin, Necroptosis, pyroptosis and apoptosis: an intricate game of cell death, Cell. Mol. Immunol., 2021, 18, 1106–1121 CrossRef CAS PubMed.
- E. Ozyerli-Goknar and T. Bagci-Onder, Epigenetic Deregulation of Apoptosis in Cancers, Cancers, 2021, 13, 3210 CrossRef CAS PubMed.
- S. Dadsena, C. Zollo and A. J. García-Sáez, Mechanisms of mitochondrial cell death, Biochem. Soc. Trans., 2021, 49, 663–674 CrossRef CAS PubMed.
- M. Abate, A. Festa, M. Falco, A. Lombardi, A. Luce, A. Grimaldi, S. Zappavigna, P. Sperlongano, C. Irace, M. Caraglia and G. Misso, Mitochondria as playmakers of apoptosis, autophagy and senescence, Semin. Cell Dev. Biol., 2020, 98, 139–153 CrossRef CAS PubMed.
- K. Takács-Vellai, Apoptosis and Autophagy, Different Modes of Cell Death: How to Utilize Them to Fight Diseases?, Int. J. Mol. Sci., 2023, 24, 11609 CrossRef PubMed.
- H. Liu, B. Zhang and Z. Sun, Spectrum of EGFR aberrations and potential clinical implications: insights from integrative pan-cancer analysis, Cancer Commun., 2020, 40, 43–59 CrossRef CAS PubMed.
- E. N. Alsahafi, S. Thavaraj, N. Sarvestani, O. Novoplansky, M. Elkabets, B. Ayaz, M. Tavassoli and M. F. Legends, EGFR overexpression increases radiotherapy response in HPV-positive head and neck cancer through inhibition of DNA damage repair and HPV E6 downregulation, Cancer Lett., 2021, 498, 80–97 CrossRef CAS PubMed.
- A. E. Maennling, M. K. Tur, M. Niebert, T. Klockenbring, F. Zeppernick, S. Gattenlöhner, I. Meinhold-Heerlein and A. F. Hussain, Molecular Targeting Therapy against EGFR Family in Breast Cancer: Progress and Future Potentials, Cancers, 2019, 11, 1826 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04915b |
| This journal is © The Royal Society of Chemistry 2024 |