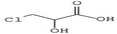
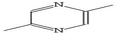
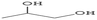Open Access Article
Open Access ArticleIn silico evaluation of potential breast cancer receptor antagonists from GC-MS and HPLC identified compounds in Pleurotus ostreatus extracts†
Magdalene Eno Effiong *ab,
Mercy Bella-Omunagbeab,
Israel Sunmola Afolabi
*ab,
Mercy Bella-Omunagbeab,
Israel Sunmola Afolabi ac and
Shalom Nwodo Chineduac
ac and
Shalom Nwodo Chineduac
aDepartment of Biochemistry, College of Science and Technology, Covenant University, Canaanland, PMB 1023, Ota, Ogun State, Nigeria. E-mail: magdalene.effiongpgs@stu.cu.edu.ng
bCovenant Applied Informatics and Communication Africa Centre of Excellence (CApIC-ACE), Nigeria
cCovenant University Public Health and Wellbeing Research Cluster (CUPHWERC), Covenant University, Canaanland, PMB 1023, Ota, Ogun State, Nigeria
First published on 9th August 2024
Abstract
Introduction: Pharmacotherapeutic targets for breast cancer include the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor (EGFR). Inhibitors of these receptors could be interesting therapeutic candidates for the treatment and management of breast cancer (BC). Aim: This study used GC-MS and HPLC to identify bioactive compounds in Pleurotus ostreatus (P. ostreatus) extracts and applied in silico methods to identify potent EGFR, ER, and PR inhibitors from the compounds as potential drug candidates. Method: GC-MS and HPLC were used to identify bioactive chemicals in P. ostreatus extracts of aqueous (PO-A), methanol (PO-M), ethanol (PO-E), chloroform (PO-C), and n-hexane (PO-H). The ER, PR, and EGFR model optimization and molecular docking of compounds/control inhibitors in the binding pocket were simulated using AutoDock Vina in PyRx. The drug-likeness, pharmacokinetic, and pharmacodynamic features of prospective docking leads were all anticipated. Result: The results indicated the existence of 29 compounds in PO-A, 36 compounds in PO-M and PO-E, 42 compounds in PO-C, and 22 compounds in PO-H extracts. With ER, only o-tolylamino-acetic acid (4-nitro-benzylidene)-hydrazide (−7.5 kcal mol−1) from the ethanolic extract could bind to the receptor. PR and EGFR, on the other hand, identified several compounds with higher binding affinities than the control. Ergotaman-3′,6′,18-trione (−8.1 kcal mol−1), 5,10-diethoxy-2,3,7,8-tetrahydro-1H,6H-dipyrrolo[1,2-a:1′,2′-d]pyrazine (−7.8 kcal mol−1) from the aqueous extract; o-tolylamino-acetic acid (4-nitro-benzylidene)-hydrazide (−8.4 kcal mol−1) from the ethanolic extract had better binding affinity compared to progesterone (−7.7 kcal mol−1). Likewise, ergotaman-3′,6′,18-trione (−9.7 kcal mol−1) from the aqueous extract and phenol, 2,4-bis(1,1-dimethyl ethyl) (−8.2 kcal mol−1) from the chloroform extract had better binding affinities compared to the control, gefitinib (−7.9 kcal mol−1) with regards to EGFR. None of the PO-H or PO-M extracts outperformed the control for any of the proteins. Phenols and flavonoids such as quercetin, luteolin, rutin, chrysin, apigenin, ellagic acid, and naringenin had better binding affinity to PR and EGFR compared to their control. Conclusion: The identified compounds in the class of phenols and flavonoids were better lead molecules due to their ability to strongly bind to the proteins' receptors. These compounds showed promising drug-like properties; they could be safe and new leads for creating anticancer medicines.
1 Introduction
Mushrooms are high in nutrients such as protein, dietary fiber, vitamins, and minerals, all of which benefit general health.1,2 They contain antioxidants, which help to neutralize damaging free radicals, potentially lowering the risk of chronic illnesses like as cancer.3,4 Mushrooms have anti-inflammatory properties that may help prevent certain cancers. They also contain immunomodulatory properties, which can help boost the body's natural defenses against cancer cells.5,6 Studies have yielded positive findings in terms of decreasing cancer cell proliferation, inducing apoptosis, and lowering tumor development.7–10 Mushrooms are frequently used as a supplement or adjuvant therapy to conventional cancer medications, with the potential to reduce treatment-related side effects and improve general well-being.11,12Breast cancer is a major public health issue worldwide due to its high prevalence, mortality rates, disparities in access to care, and emotional impact on individuals, families, communities, and healthcare systems.13–15 The disease is the most often diagnosed cancer among women worldwide, with a high mortality rate.16–18 It has a significant economic impact, including costs for diagnosis, treatment, care and psychosocial support.17–19 Tackling breast cancer involves a multifaceted strategy, including prevention, early detection, diagnosis, and treatment such as surgery, chemotherapy, radiation therapy, hormone therapy, targeted therapy, and immunotherapy.20,21 However, these methods can have side effects and do not always result in complete remission or prevent recurrence due to drug resistance and tumoral heterogeneity.22
Breast cancer has several molecular subgroups based on hormone receptor expression and gene profiles. The five primary subtypes are luminal A, luminal B, HER2-enriched, triple-negative breast cancer (TNBC), and basal-like.23,24 The disease's heterogeneity is continually changing, demanding personalized and focused treatments.25 Receptors associated with these subtypes, such as estrogen receptors (ER), progesterone receptors (PR), human epidermal growth factor receptor 2 (HER2), are important in cancer initiation, development, and treatment response.26,27 Targeting these receptors with specific drugs has resulted in significantly improved outcomes, emphasizing the importance of the discovery of natural inhibitors and drug candidates.
Oyster mushrooms, also known as Pleurotus ostreatus, have been studied for their potential anticancer properties,28 including interactions with ER+, PR+ and HER2+ breast cancer receptors.29,30 Bioactive compounds in Pleurotus ostreatus may interact with these breast cancer receptors, either independently or synergistically leading to decreased breast carcinogenesis, survival and proliferation. Therefore this study aims at utilizing gas chromatography mass spectrophotometer (GC-MS) and high performance liquid chromatography (HPLC) techniques to identify and quantify the bioactive compounds in Pleurotus ostreatus, and apply in silico techniques to evaluate their inhibitory effects on breast cancer receptors.
2 Materials and methods
2.1 Sample collection and preparation
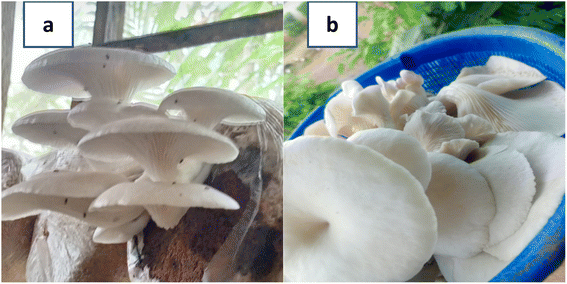 | ||
| Fig. 1 (a and b): P. ostreatus mushroom.31 (a): P. ostreatus growing on its substrate. (b): P. ostreatus freshly harvested. | ||
2.2 Gas chromatography-mass spectrometry (GC-MS) analysis of the extracts of Pleurotus ostreatus
PO extracts were analyzed by GC-MS on a GCMS-QP2010SE SHIMADZU JAPAN using a fused Optima-5MS capillary column of 30 m length, 0.25 mm diameter, and 0.25 μm film thickness. The GC conditions were pure helium (1.56 mL min−1 flow rate and 37 cm s−1 linear velocity), injector temperature (200 °C), column oven temperature (60 °C initially, then increased to 160 °C and later to 250 °C at 10 °C min−1 with 2 min/increment hold time), and injection volume and split ratio (0.5 μL and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively). The MS conditions were: ion source of 230 °C and an interface temperature of 250 °C, a solvent delay of 4.5 minutes recorded in a scan range 50 to 700 amu. Unknown constituents were detected by comparing the retention duration, mass spectrum data and fragmentation pattern of the extracts with established libraries (National Institute of Standards and Technology (NIST) and Wiley libraries).35,36
1, respectively). The MS conditions were: ion source of 230 °C and an interface temperature of 250 °C, a solvent delay of 4.5 minutes recorded in a scan range 50 to 700 amu. Unknown constituents were detected by comparing the retention duration, mass spectrum data and fragmentation pattern of the extracts with established libraries (National Institute of Standards and Technology (NIST) and Wiley libraries).35,36
2.3 HPLC analysis of the extracts of Pleurotus ostreatus
The HPLC identification and characterization of flavonoids, phenols and saponins in the five extracts of P. ostreatus was carried out using the method described by ref. 37.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) mobile phase, 205 nm wavelength, a 14 minutes run duration, sorbent type is C18 silica packed column, 5 μm, 4.6 × 2.50 mm. HPLC analysis was performed using N2000 chromatography software (4.0 version) with a high-sensitivity LC-8518 UV detector, column (150 mm × 4.6 mm) set at 40 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve to determine flavonoids content and amount in the extracts.37
30) mobile phase, 205 nm wavelength, a 14 minutes run duration, sorbent type is C18 silica packed column, 5 μm, 4.6 × 2.50 mm. HPLC analysis was performed using N2000 chromatography software (4.0 version) with a high-sensitivity LC-8518 UV detector, column (150 mm × 4.6 mm) set at 40 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve to determine flavonoids content and amount in the extracts.37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80
80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mobile phase, at 272 nm wavelength, and a run time of 25 minutes. To analyze flavonoids in extracts, N2000 chromatography software was used with a high-sensitivity LC-8518 diode array (DA) detector, a column (150 mm × 4.6 mm) set at 35 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve.37
1) mobile phase, at 272 nm wavelength, and a run time of 25 minutes. To analyze flavonoids in extracts, N2000 chromatography software was used with a high-sensitivity LC-8518 diode array (DA) detector, a column (150 mm × 4.6 mm) set at 35 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve.37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74
74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mobile phase, 210 nm wavelength, and a run period of 25 minutes. To analyze flavonoids in extracts, N2000 chromatography software was used with a high-sensitivity LC-8518 diode array (DA) detector, a column (150 mm × 4.6 mm) set at 40 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve.37
1) mobile phase, 210 nm wavelength, and a run period of 25 minutes. To analyze flavonoids in extracts, N2000 chromatography software was used with a high-sensitivity LC-8518 diode array (DA) detector, a column (150 mm × 4.6 mm) set at 40 °C, and a low-pressure gradient and solvent delivery LC-8518 pump with a high-pressure switching valve.372.4 In silico studies
3 Results
3.1 Gas chromatography-mass spectroscopy (GC-MS) results
The gas chromatography of Pleurotus ostreatus aqueous, methanol, ethanol, chloroform and n-hexane extracts revealed the presence of 29, 36, 36, 42 and 22 peaks respectively.3.2 High performance liquid chromatography (HPLC) profile of P. ostreatus extracts
The HPLC profile of P. ostreatus extracts revealed the presence of flavonoids, phenols and saponins. The extracts possessed a wider array of flavonoid compounds, followed by phenolic compounds and least amount of saponin compounds. Majority of the saponin compounds were unidentified compared to the flavonoid and phenolic compounds (Table 6).| S/N | Compounds (mg kg−1) | PO-A | PO-M | PO-E | PO-C | PO-H |
|---|---|---|---|---|---|---|
| A | FLAVONOIDS | |||||
| 1 | 2.5-Dihydroxybenzoic acid | 0.0014 | — | 0.0107* | — | 0.0009 |
| 2 | Caffeic acid | 0.0013 | — | 0.0329* | — | 0.0165 |
| 3 | Phenylacetic acid | — | — | — | 8.4189* | 0.0022 |
| 4 | Trans-cinnamic acid | 66.6939* | — | — | 27.7456 | 0.0018 |
| 5 | Quercetin | — | — | — | 35.3818 | 95.5490* |
| 6 | Naringenin | — | — | — | — | 4.4120 |
| 7 | Chrysin | — | — | — | — | 0.0161 |
| 8 | 3,4-Dimethoxybenzoic acid | — | 79.6578* | — | — | — |
| 9 | Gallic acid | — | 0.0944 | 0.0066 | — | — |
| 10 | 4-Hydroxybenzoic acid | — | 0.2174 | — | — | — |
| 11 | Salicylic acid | — | 0.1216 | — | — | — |
| 12 | p-Coumaric acid | — | 19.9073 | — | — | — |
| 13 | Rutin hydrate | 0.0009 | — | 0.0308 | — | — |
| 14 | O-Coumaric acid | 0.0011 | — | 0.0231 | — | — |
| 15 | Benzoic acid | — | — | 76.1210 | — | — |
| 16 | Chlorogenic acid | 33.2979 | — | 23.6395 | — | — |
| 17 | Luteolin | — | — | 0.0594 | — | — |
| 18 | 4-Methoxycinnamic acid | 0.0012 | — | — | — | — |
| 19 | Unidentified | 0.0012 | — | — | — | — |
| 20 | Unidentified | 0.0011 | — | — | — | — |
| 21 | Unidentified | — | — | — | 0.0185 | — |
| 22 | Unidentified | — | — | — | — | 0.0015 |
| 23 | Unidentified | — | — | 0.0291 | — | — |
| 24 | Unidentified | — | — | 0.0280 | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| B | PHENOLS | |||||
| 1 | Gallic acid | — | 0.0932 | 0.0010 | — | 0.0128 |
| 2 | Syringic acid | 59.1016 | 76.2015 | 12.0117 | 36.6614 | 49.0412 |
| 3 | Ferulic acid | — | — | — | 24.8694 | 21.2843 |
| 4 | Quercetin | 40.8968 | 22.7721 | — | 38.4585 | 17.9785 |
| 5 | Apigenin | — | 0.9332 | — | — | 11.3508 |
| 6 | Caffeic acid | 0.0016 | — | — | — | — |
| 7 | Vanillic acid | — | — | 69.9143 | — | — |
| 8 | Ellagic acid | — | — | 18.0513 | — | — |
| 9 | Gallic acid | — | 0.0932 | 0.0010 | — | 0.0128 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| C | SAPONINS | |||||
| 1 | Saponin | 168.9560* | 59.0371 | 17.5690 | 34.3375 | 16.5521 |
| 2 | Unidentified | 30.8881 | 0.0257 | 0.0019 | 1.0807 | 0.0073 |
| 3 | Unidentified | 0.1474 | 18.1823 | 0.0285 | 1.0833 | 0.2756 |
| 4 | Unidentified | 0.0009 | 22.7383 | 44.9442 | 13.4500 | 4.1655 |
| 5 | Unidentified | 0.0003 | — | 17.6892 | 0.0130 | 8.8328 |
| 6 | Unidentified | 0.0012 | — | 1.1126 | 0.0032 | 0.0092 |
| 7 | Unidentified | 0.0003 | — | 18.6404 | 0.0023 | 0.0180 |
| 8 | Unidentified | 0.0003 | — | — | 0.0020 | 0.0339 |
| 9 | Unidentified | 0.0007 | — | — | 0.0018 | 0.0198 |
| 10 | Unidentified | 0.0005 | — | — | 0.0040 | 0.0152 |
| 11 | Unidentified | 0.0017 | — | — | 0.0020 | 0.0207 |
| 12 | Unidentified | — | — | — | 0.0008 | 0.0256a |
| 13 | Unidentified | — | — | — | 0.0034 | 0.0067 |
| 14 | Unidentified | — | — | — | 0.0016 | 0.0064 |
| 15 | Unidentified | — | — | — | 0.0028 | 0.0034 |
3.3 Molecular docking results
| Compound ID | Compound name | Binding affinity (kcal mol−1) | |
|---|---|---|---|
| Aqueous extract | 10531 | Ergotaman-3′,6′,18-trione | −8.1 |
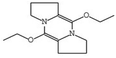
| 565346 | 5,10-Diethoxy-2,3,7,8-tetrahydro-1H,6H-dipyrrolo[1,2-a:1′,2′-d]pyrazine | −7.8 | |
| Control: 5994 | Progesterone | −7.7 | |
| Chloroform extract | Control: 5994 | Progesterone | −11.4 |
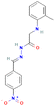
| Ethanolic extract | 6876597 | o-Tolylamino-acetic acid (4-nitro-benzylidene)-hydrazide | −8.4 |
| Control: 5994 | Progesterone | −6.9 | |
| Flavonoids | 5280445 | Luteolin | −9.6 |
| 5280343 | Apigenin | −9.5 | |
| 439246 | Naringenin | −9.2 | |
| Control: 5994 | Progesterone | −7.7 | |
| Hexane extract | Control: 5994 | Progesterone | −7.7 |
| Methanolic extract | Control: 5994 | Progesterone | −11.4 |
| Phenols | 5280343 | Apigenin | −9.5 |
| 5280443 | Quercetin | −8.1 | |
| 5281855 | Ellagic acid | −7.2 | |
| Control: 5994 | Progesterone | −7.2 |
| Compound ID | Compound name | Binding affinity (kcal mol−1) | |
|---|---|---|---|
| Aqueous extract | Control: 5757 | Estradiol | −10.9 |
| Chloroform extract | Control: 5757 | Estradiol | −10.9 |
| Ethanolic extract | 6876597 | o-Tolylamino-acetic acid (4-nitro-benzylidene)-hydrazide | −7.5 |
| Control: 5757 | Estradiol | −7 | |
| Flavonoids | Control: 5757 | Estradiol | −10.9 |
| Hexane extract | Control: 5757 | Estradiol | −8.8 |
| Methanolic extract | Control: 5757 | Estradiol | −8.6 |
| Phenols | Control: 5757 | Estradiol | −10.9 |
| Compound ID | Compound name | Binding affinity (kcal mol−1) | |
|---|---|---|---|
| Aqueous extract | 10531 | Ergotaman-3′,6′,18-trione | −9.7 |
| Control: 123631 | Gefitinib | −7.9 | |
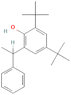
| Chloroform extract | 93344 | Phenol, 2,4-bis(1,1-dimethyl ethyl) | −8.2 |
| Control: 123631 | Gefitinib | −8 | |
| Ethanolic extract | Control: 123631 | Gefitinib | −7.9 |
| Flavonoids | 5280343 | Quercetin | −8.5 |
| 5280445 | Luteolin | −8.4 | |
| 5280805 | Rutin | −8.4 | |
| 5281607 | Chrysin | −7.8 | |
| Control: 123631 | Gefitinib | −7.8 | |
| Hexane extract | Control: 123631 | Gefitinib | −7.7 |
| Methanolic extract | Control: 123631 | Gefitinib | −7.7 |
| Phenols | 5281855 | Ellagic acid | −8.8 |
| 5280343 | Apigenin | −8.5 | |
| 5280443 | Quercetin | −7.8 | |
| Control: 123631 | Gefitinib | −7.8 |
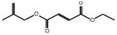
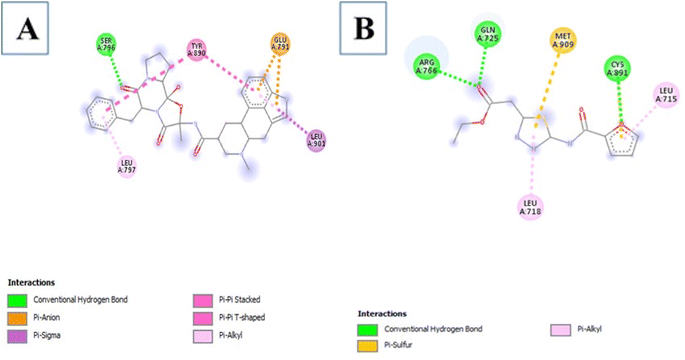
3.3.3.1 Aqueous extract. Among the 29 compounds, 2 compounds, 10531 and 565346 exhibited a low binding affinity lower than the control, (compound 5994) which exhibited a low binding affinity of −7.7 kcal mol−1. Compound 10531 exhibited the lowest binding affinity of −8.1 kcal mol−1 while compound 565346 exhibited a low binding affinity of −7.8 kcal mol−1. The interactions formed by compound 10531 with PR include; Conventional Hydrogen Bonds with SER796, Pi–sigma with LEU901, Pi–alkyl with LEU797, Pi–anion with GLU791, and Pi–Pi stacked/Pi–Pi T-shaped with TYR890 (Fig. 2A). The interactions formed by compound 565346 include Conventional Hydrogen Bonds with ARG766, GLN725, and CYS891, Pi–sulfur with MET909, and Pi–alkyl with LEU715 and LEU718 (Fig. 2B).
3.3.3.2 Chloroform extract. Among the 42 compounds, none exhibited a lower binding affinity than that of the control, 5994 at −11.4 kcal mol−1.
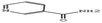
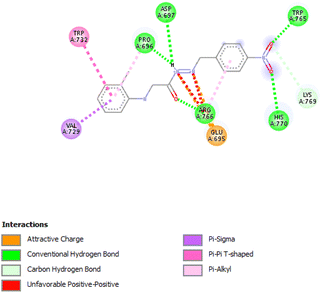
3.3.3.3 Ethanolic extract. Among the 36 compounds, compound 6876597 exhibited a binding affinity of −8.4 kcal mol−1 which was lower than that of the control 5994 at −6.9 kcal mol−1. Compound 6876597 formed interactions such as Conventional Hydrogen Bonds with PRO696, ASP697, ARG766, TRP765, and HIS770, Pi–sigma with VAL729, Pi–alkyl with PRO696, Pi–Pi T-shaped with TRP732, Attractive Charge with GLU695 and Carbon Hydrogen Bond with LYS769 (Fig. 3).
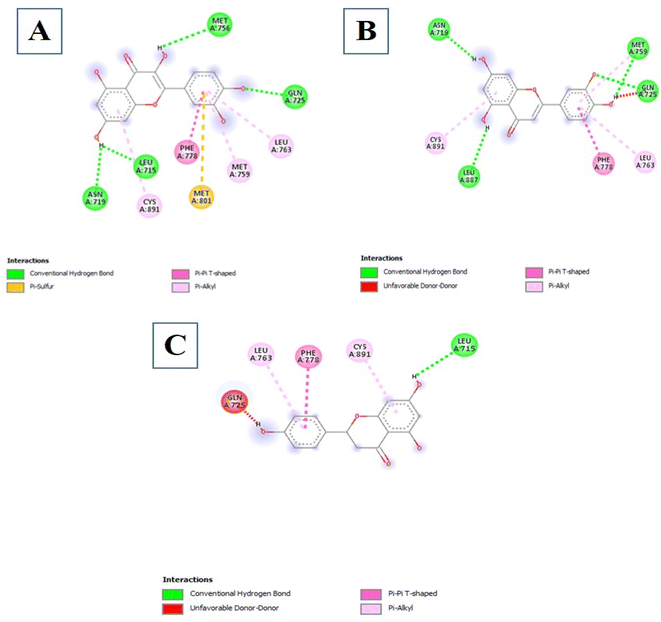
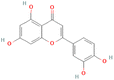
3.3.3.4 Flavonoids. Among the 18 Flavonoids studied, three of them possessed binding affinities lower than the control at −7.7 kcal mol−1. The first three are 5280445, 5280343, and 439246 at binding affinities of −9.6, −9.5, and −9.2 kcal mol−1 respectively. The interactions formed by Compound 5280445 include Conventional Hydrogen Bonds with ASN719, LEU887, MET759, and GLN725, Pi–Pi T-shaped with PHE778 and Pi–alkyl with CYS891 and LEU763 (Fig. 4B). Compound 5280343 formed interactions which include Conventional Hydrogen Bonds with ASN719, LEU715, MET756, and GLN725, Pi–sulfur with MET801, Pi–Pi T-shaped with PHE778 and Pi–alkyl with CYS891, MET759, and LEU763 (Fig. 4A). Compound 439246 formed interactions such as Conventional Hydrogen Bond with LEU715, Pi–Pi T-shaped with PHE778, and Pi–alkyl with CYS891 and LEU763 (Fig. 4C).
3.3.3.5 Hexane extract. Among the 22 compounds, none exhibited a lower binding affinity than that of the control, 5994 at −7.7 kcal mol−1.
3.3.3.6 Methanolic extract. Among the 36 compounds, none exhibited a lower binding affinity than that of the control, 5994 at −11.4 kcal mol−1.
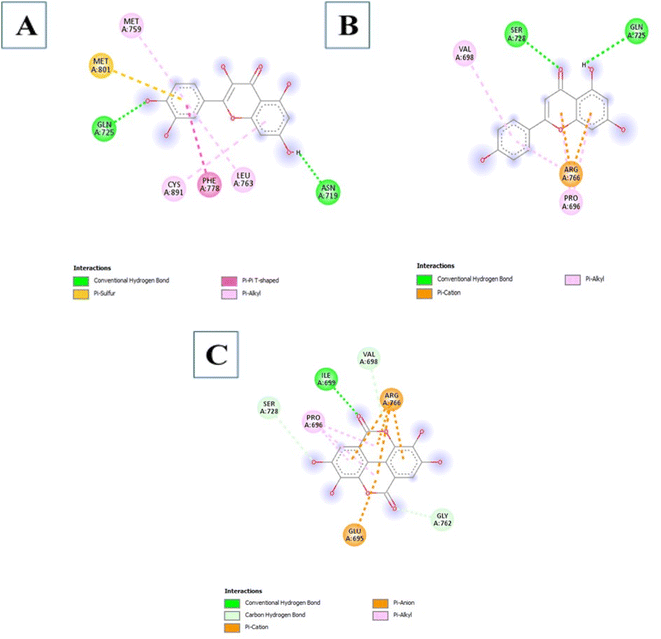
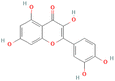
3.3.3.7 Phenols. The Phenols studied showed a low binding affinity with PR. Two compounds namely 5280343 and 5280443 showed a low binding affinity with −9.5 kcal mol−1 and −8.1 kcal mol−1 respectively which are considerably much lower than the control at −7.2 kcal mol−1. A third compound 5281855 also exhibited low binding affinity at −7.2 kcal mol−1. The interactions formed by compound 5280343 include Conventional Hydrogen Bonds with GLN725 and ASN719, Pi–sulfur with MET801, Pi–Pi T-shaped with PHE778 and Pi–alkyl with MET759, CYS891 and LEU763 (Fig. 5A). The interactions formed by compound 5280443 include Conventional Hydrogen Bonds with SER728 and GLN725, Pi–cation with ARG766, and Pi–alkyl with PRO696 and VAL698 (Fig. 5B). The interactions formed by Compound 5281855 include Conventional Hydrogen Bonds with ILE699, Carbon Hydrogen Bonds with VAL698, GLY762, and SER728, Pi–anion/Pi–cation with ARG766 and GLU695a and Pi–alkyl with PRO696 (Fig. 5C).
3.3.4.1 Aqueous extract. Among the 26 compounds, none exhibited a lower binding affinity than the control, 5757 at −10.9 kcal mol−1. Compound 10531 exhibited the next lowest binding affinity of −8.8 kcal mol−1.
3.3.4.2 Chloroform extract. Among the 42 compounds, none exhibited a lower binding affinity than that of the control, 5757 at −10.9 kcal mol−1. In another pose, the control also exhibited a low binding affinity at −8.6 kcal mol−1. Compound 88693 showed a binding affinity of −6.8 kcal mol−1 which is considerably higher than that of the control.
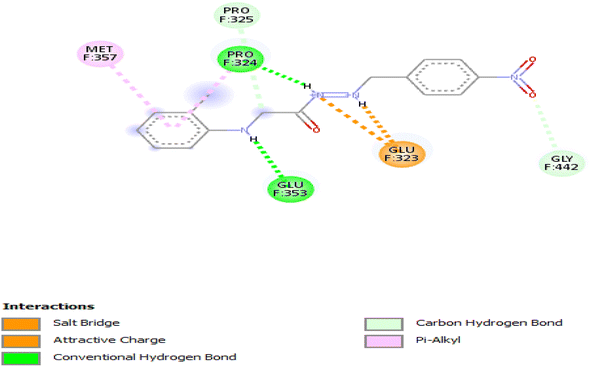
3.3.4.3 Ethanolic extract. Compound 6876597 exhibited a binding affinity of −7.5 kcal mol−1 which is lower than that of the control at −7 kcal mol−1. No other compound showed a lower binding affinity. The interactions formed by compound 6876597 are Conventional Hydrogen Bonds with PRO324 and GLU353, attractive charge/salt bridge with GLU323, Pi–alkyl with MET357 and PRO324, and Carbon Hydrogen Bond with PRO325 and GLY442 (Fig. 6).
3.3.4.4 Flavonoids. The control compound, 5757 showed better binding ability than all other compounds studied at −10.9 kcal mol−1. The next compounds with a low binding affinity were 439246 and 5281607, both at −8.2 kcal.
3.3.4.5 Hexane extract. The control compound exhibited the lowest binding affinity at −8.8 kcal mol−1. The next hit with a low binding affinity was compound 610065 at −7.5 kcal mol−1.
3.3.4.6 Methanolic extract. None of the compounds under study exhibited a lower binding affinity than 5757, the control at −8.6 kcal mol−1.
3.3.4.7 Phenols. The control, 5757 exhibited the lowest binding affinity when compared to the phenols studied. It exhibited a low binding affinity of −10.9 kcal mol−1. Compound 5281855 exhibited the next low binding affinity at −9.1 kcal mol−1.
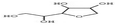
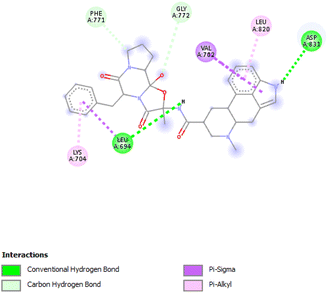
3.3.5.1 Aqueous extract. Compound 10531 exhibited the lowest binding affinity of −9.7 kcal mol−1 which was considerably lower than that of the control, 123631 at −7.9 kcal mol−1. The interactions formed are Conventional Hydrogen Bonds with LEU694 and ASP831, Pi–sigma with VAL702 and LEU694, Pi–alkyl with LYS704 and LEU820, and Carbon Hydrogen Bonds with PHE771 and GLY772 (Fig. 7).
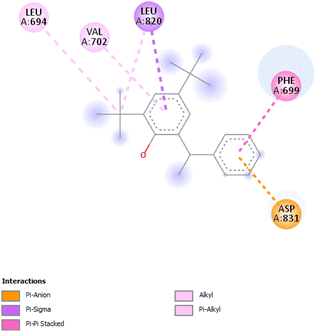
3.3.5.2 Chloroform extract. Compound 93344 exhibited a binding affinity of −8.2 kcal mol−1 which is considerably lower than the binding affinity of the control at −8 kcal mol−1. The interactions formed by compound 93344 are Pi–anion with ASP831, Pi–sigma with LEU820, Pi–Pi stacked with PHE699, and alkyl/Pi–alkyl with LEU694 and VAL702 (Fig. 8).
3.3.5.3 Ethanolic extract. No compound exhibited a lower binding affinity than that of the control, 123631 at −7.9 kcal mol−1.
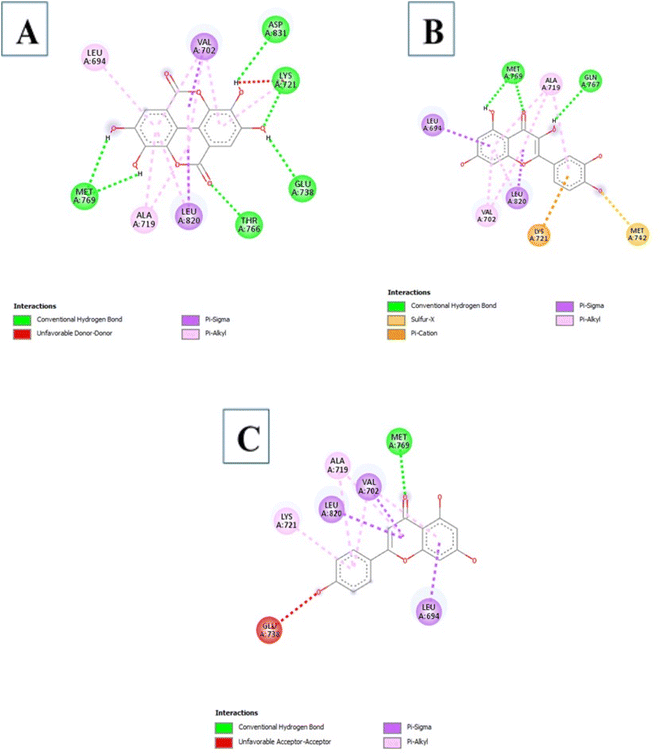
3.3.5.4 Flavonoids. Four compounds showed a better binding affinity than that of the control at −7.8 kcal mol−1. These compounds include 5280343, 5280445, 5280805, and 5281607 at best binding affinities of −8.5, −8.4, −8.4, and −7.8 kcal mol−1 respectively. Compound 5280343 formed interactions which include Conventional Hydrogen Bond with MET769, GLN767, GLU738, MET742 and ASP831, Pi–cation with LYS721, Pi–sigma with LEU820 and LEU694, sulfur–X with MET742 and Pi–alkyl with VAL702, LEU820 and ALA719 (Fig. 9A). Compound 5280445 formed interactions which include MET742 and MET769, Pi–sigma with VAL702, LEU820 and LEU694, and Pi–alkyl with LYS721, ALA719, and VAL702 (Fig. 9B). Compound 5280805 formed interactions which include Conventional Hydrogen Bonds with CYS773, LYS721, and LEU764, Pi–Pi stacked with PHE699, Pi–anion with ASP831 and Pi–alkyl with VAL702 and ALA719 (Fig. 9C). Compound 5281607 formed interactions which include Pi–cation with LYS721, Pi–sulfur with MET742, Pi–sigma with VAL702 and LEU820, and Pi–alkyl with LEU694, ALA719 and VAL702 (Fig. 9D).
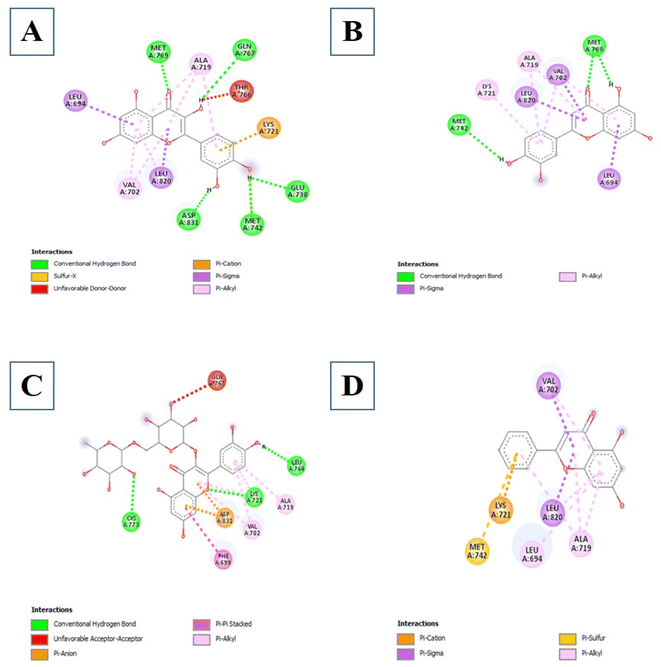 | ||
| Fig. 9 The intermolecular interactions between 5280343 (A); 5280445 (B); 5280805 (C); 5281607 (D) and EGFR. | ||
3.3.5.5 Hexane extract. Among the 22 compounds, none exhibited a lower binding affinity than that of the control, 123631 at −7.7 kcal mol−1.
3.3.5.6 Methanolic extract. Among the 36 Compounds, none exhibited a lower binding affinity than that of the control, 123631 at −7.7 kcal mol−1.
3.3.5.7 Phenols. The control compound, 123631 exhibited a low binding affinity at −7.8 kcal mol−1. Three compounds showed a better binding affinity than that of the control. These compounds are 5281855, 5280343, and 5280443 at binding affinity of −8.8, −8.5, and −7.8 kcal mol−1. Compound 5281855 formed interactions which include Conventional Hydrogen Bonds with ASP831, LYS721, GLU738, THR766 and MET769, Pi–sigma with VAL702 and LEU820, and Pi–alkyl with LEU694, ALA719, VAL702, LEU820 and LYS721 (Fig. 10A). Compound 5280343 formed interactions which include Conventional Hydrogen Bond GLN767 and MET769, sulfur–X with MET7422, Pi–cation with LYS721, Pi–sigma with LEU694 and LEU820 and Pi–alkyl with VAL702, LEU820 and ALA719 (Fig. 10B). Compound 5280443 formed interactions which include Conventional Hydrogen Bond with MET769, Pi–sigma with VAL702, LEU820, and LEU694, and Pi–alkyl with LYS721, VAL702 and ALA719 (Fig. 10C).
| Compounds | Water solubility (ESOL) | Lipophilicity (iLOGP) | GI absorption | BBB permeation | P-glycoprotein substrate | |
|---|---|---|---|---|---|---|
| Aqueous extract | 10531 | −4.88 (moderately soluble) | 3.31 | High | No | Yes |
| 565346 | −2.22 (soluble) | 1.62 | High | No | No | |
| Chloroform extract | 93344 | −6.40 (poorly soluble) | 4.01 | High | No | Yes |
| Ethanolic extract | 6876597 | −3.64 (soluble) | 2.03 | High | No | No |
| Flavonoids | 5280445 | −3.71 (soluble) | 1.86 | High | No | No |
| 5280343 | −3.16 (soluble) | 1.63 | High | No | No | |
| 439246 | −3.49 (soluble) | 1.75 | High | No | Yes | |
| 5280805 | −3.30 (soluble) | 1.58 | Low | No | Yes | |
| 5281607 | −4.19 (moderately soluble) | 2.27 | High | Yes | No | |
| Phenols | 5280343 | −3.16 (soluble) | 1.63 | High | No | No |
| 5280443 | −3.94 (soluble) | 1.89 | High | No | No | |
| 5281855 | −2.94 (soluble) | 0.79 | High | No | No |
| Compounds | Lipinski | Ghose | Veber | Egan | Muegge | |
|---|---|---|---|---|---|---|
| a Compound* – compounds with no violation of the drug likeness rules. | ||||||
| Aqueous extract | 10531 | No; 1 violation | No; 3 violation | Yes | Yes | No; 1 violation |
| 565346* | Yes | Yes | Yes | Yes | Yes | |
| Chloroform extract | 93344 | Yes | No; 1 violation | Yes | No; 1 violation | No; 2 violations |
| Ethanolic extract | 6876597* | Yes | Yes | Yes | Yes | Yes |
| Flavonoids | 5280445* | Yes | Yes | Yes | Yes | Yes |
| 5280343* | Yes | Yes | Yes | Yes | Yes | |
| 439246* | Yes | Yes | Yes | Yes | Yes | |
| 5280805 | No; 3 violations | No; 4 violations | No; 1 violation | No; 1 violation | No; 4 violations | |
| 5281607* | Yes | Yes | Yes | Yes | Yes | |
| Phenols | 5280343* | Yes | Yes | Yes | Yes | Yes |
| 5280443* | Yes | Yes | Yes | Yes | Yes | |
| 5281855 | Yes | Yes | No; 1 violation | No; 1 violation | Yes | |
| Compounds | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | |
|---|---|---|---|---|---|---|
| a Compound* – compounds with no inhibition on all the CYP isoforms. | ||||||
| Aqueous extract | 10531 | No | No | No | Yes | Yes |
| 565346* | No | No | No | No | No | |
| Chloroform extract | 93344 | No | Yes | No | Yes | No |
| Ethanolic extract | 6876597 | Yes | No | Yes | No | No |
| Flavonoids | 5280445 | Yes | No | No | Yes | Yes |
| 5280343 | Yes | No | No | Yes | Yes | |
| 439246 | Yes | No | No | No | Yes | |
| 5280805* | No | No | No | No | No | |
| 5281607 | Yes | No | No | Yes | Yes | |
| Phenols | 5280343 | Yes | No | No | Yes | Yes |
| 5280443 | Yes | No | No | Yes | Yes | |
| 5281855 | Yes | No | No | No | No | |
4 Discussion
P. ostreatus is highly nutritious and medicinal mushroom that contains a wide variety of bioactive compounds which may possess anti-cancer properties.28 The isolation and identification of these bioactive compounds using a combination of GC-MS and HPLC provides an opportunity to explore their anti-cancer potential and mechanisms of actions. As a result, this study was carried out to identify the bioactive compounds in P. ostreatus using GC-MS and HPLC and evaluate their inhibition potential on breast cancer receptors in silico.4.1 GC-MS profile of P. ostreatus bioactive compounds and their biological activities
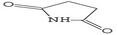
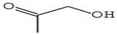
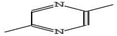
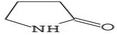
Gas chromatography-mass spectrometry (GC-MS) is a valuable analytical tool for profiling and studying secondary metabolites in a variety of biological samples.41 The GC-MS profile of the phytocompounds from the five extracts of P. ostreatus revealed that chloroform solvent extracted more versatile compounds compared to other solvents. This is similar to the reports by Effiong et al.42 Overall the GC-MS identified bioactive compounds in P. ostreatus have been found to possess numerous medicinal and biological significance ranging from anti-tumor, antioxidant, anti-cancer, anti-diabetic, anti-microbial, anti-inflammatory, immunomodulatory, anti-malarial, probiotic, neuroprotective, anti-tuberculosis, etc.Antioxidant compounds perform the role of removing free radicals that cause oxidative damage to cells and tissues. They can combat oxidative stress, protect cells from injury, maintain cellular integrity, regulate growth, and aid in immune function which plays a significant role in preventing and managing breast cancer development.43 The identified antioxidant compounds in Pleurotus ostreatus extracts were 4H-pyran-4-one, furanones,44,45 cyclohexanecarboxylic acid, pentadecanoic acid, hexadecanoic acid, dodecanoic acid, heptacosanoic acid,46,47 2-pyrrolidinone;48 niacinamide; niacin;49 1,2,4-triazol-3-amine;50 acetamide;51 propanamide;52 pyrrolizine derivatives,53 piracetam;54 D-limonene55 and eicosane.56
Anti-inflammatory compounds were also identified in the extracts of P. ostreatus. These compounds includes 4H-pyran-4-one, furanones,44 pyrazine, thiadiazole;57,58 acetamide;59 succinamide60 and pyrrolizine,61 D-limonene62 and eicosane.63 These compounds functioning as anti-inflammatory chemicals help to prevent breast cancer by modulating inflammatory pathways. They reduce inflammation, prevent pro-inflammatory cytokine production, and scavenge free radicals. They also influence the hormonal pathways and cellular mechanisms that lead to the progression of breast cancer.64
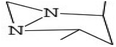
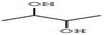
Compounds with anti-proliferative, anti-cancer and anti-tumor properties were identified among the GC-MS profile of P. ostreatus. These compounds include 2,3-butanediol,65 1-dodecanol,66 4H-pyran-4-one, furanones, succinimide,67 propanamide,68 pyrrolizine,69 acetamide,51 butyrolactone;70 piperazine,71 heptane,72 pyrazine,73 pyrrole;74 thiadiazole,75 fumaric acid;76 linoleic acid ethyl ester,77 methoxyacetic acid,78 phenol;79 D-limonene;80 eicosane81 and 1,2,4-triazol-3-amine.82 These compounds can combat cancer initiation and progression by preventing cancer cells from proliferating, dividing, and spreading uncontrolled. They can induce apoptosis and inhibit angiogenesis, preventing breast cancer initiation, progression, and metastasis.83
Immunomodulatory compounds were identified among the GC-MS profile of P. ostreatus extracts. These compounds includes succinimide,84 propanamide,85 D-limonene,86,87 fumaric acid and its derivatives.87 These compounds enhance the immune system's response to cancer cells, aid in tumor detection and eradication, hence lowering breast cancer growth and progression. Compounds with anti-diabetic properties were also identified. These compounds includes pyrazine;88 pyrrolizine;53 pyrrole;74 phenol;79 dodecanoic acid;89 D-limonene,90 acetamide91 and eicosane.56 These compounds functions in breast cancer prevention and management by addressing diabetes and obesity, which are risk factors of breast cancer. They target insulin resistance, hyperglycemia, and inflammation leading to a lower risk of breast cancer.92
Antimicrobial compounds have been demonstrated to lower breast cancer risk by altering the microbiome and immune response.93 The GC-MS profile of P. ostreatus revealed the presence of various compounds with antimicrobial properties. These compounds includes fumaric,94 dodecanoic acid, nonadecanoic acid, ethyl ester,95 heptacosanoic acid, methyl ester,96 acetic acid,97 heptane,98 nonane,99 hexanal,100 propanol,101 furanol,102 phenol,79 2-pyrrolidinone,69 furanone,103 propanamide,104 pyrazine,73 piperazine,105 pyrrolizine,106 pyrrole,74 D-limonene107 and eicosane.108 These compounds indirectly aid in breast cancer prevention by lowering chronic inflammation, promoting hormonal balance, and strengthening the body's natural defenses against malignant growth.83
4.2 HPLC profile of P. ostreatus
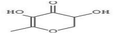
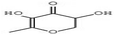
HPLC has high sensitivity, resolution, and reproducibility, making it possible to identify and quantify various classes of bioactive compounds such as phenols, flavonoids, saponins, among others.109 The HPLC profile of P. ostreatus revealed the presence of numerous unidentified saponins. The aqueous extract had the highest saponin content, followed by the methanol, chloroform, ethanol and hexane extracts. The presence of saponins suggests the ability of these extracts to prevent cancer cell proliferation by interrupting the cell cycle and triggering apoptosis. Although the specific mechanisms of action is not clearly understood with little research into their effects on breast cancer.Phenols are a class of bioactive molecules that have the potential to prevent and treat breast cancer. These bioactive compounds contain antioxidant and anti-inflammatory properties, which reduce oxidative stress, protect DNA from damage, and prevent cancer cell proliferation.83 They also influence hormone signaling pathways, potentially lowering the risk of hormone receptor-positive breast cancer.110,111 The HPLC profile of P. ostreatus extracts revealed the presence of the following phenolic compounds: gallic acid, syringic acid, ferulic acid, quercetin, apigenin, caffeic acid, vanillic acid and ellagic acid. Syringic acid was found to be the most abundant phenolic compound present in all the extracts of P. ostreatus. It targets distinct pathways in cancer cells, increasing DNA repair, apoptosis and reducing angiogenesis.112 It was highest in the methanolic extract, followed by the aqueous, n-hexane, chloroform and ethanolic extract. Ferulic acid was also identified in the non-polar extracts of P. ostreatus, with the chloroform extract having higher ferulic acid content compared to the n-hexane extract. The presence of ferulic acid shows the ability of these extracts to inhibit enzymes involved in proliferation and reduce NF-kB activity.113 Caffeic acid interferes with estrogen signaling, potentially impacting hormone-receptor positive breast cancers.114 It was only found to be present in the aqueous extract of P. ostreatus. Ellagic acid is an ellagitannin that inhibits various signaling pathways involved in cancer cell proliferation and survival. It also improves DNA repair and inhibits enzymes that enable cancer cells to evade cell death.115 Ellagic and vanillic acids was found to be present only in the ethanol extract. Quercetin and apigenin functions as antioxidants, protecting cells from free radical damage, which can result in cancer growth. They also inhibit enzymes that promote cell proliferation and induce apoptosis.116 Angenin was found to be present in the hexane and methanol extracts, with higher amounts in the hexane extract compared to the methanol extract. However, quercetin was present in all extracts except the ethanol extract, following the trend of aqueous > chloroform > methanol and hexane extracts.
Flavonoids have antioxidant qualities that neutralize free radicals, which can damage cells and promote cancer growth.117 They regulate inflammation, cell cycle progression, inhibit cell division enzymes, and modulate estrogen signaling. They also have anti-angiogenic properties, which prevent the formation of new blood vessels necessary for tumor growth.83 The HPLC profile of P. ostreatus extracts revealed the presence of 2,5-dihydroxybenzoic acid, caffeic acid, phenylacetic acid, trans-cinnamic acid, quercetin, naringenin, chrysin, 3,4-dimethoxybenzoic acid, gallic acid, 4-hydroxybenzoic acid, salicylic acid, p-coumaric acid, rutin hydrate, o-coumaric acid, benzoic acid, chlorogenic acid, luteolin, 4-methoxycinnamic acid and other six unidentified flavonoid compounds. Quercetin was found to be the most abundant flavonoid in the n-hexane extract, followed by naringenin and other flavonoids in minute quantities. Trans-cinnamic and chlorogenic acid were the most abundant flavonoids in the aqueous extract. 3,4-Dimethoxybenzoic and p-coumaric acid were the most abundant flavonoids in the methanolic extract, whereas, benzoic and chlorogenic acid were the most abundant flavonoid in the ethanolic acid. In the chloroform extract, quercetin and trans-cinnamic acid were the most abundant, phenylacetic acid was also found to be present in small quantities.
4.3 In silico evaluation of the anti-cancer effects of P. ostreatus on breast cancer receptors
Breast cancer receptors play a crucial role in breast cancer development and progression. They aid in identifying the molecular subtype of breast cancer, tumor behavior and therapy response.118 Hormone receptor-positive breast cancers, ER+ and PR+, respond well to hormonal therapy, while HER2-positive breast cancers benefit from targeted therapies combined with chemotherapy. HER2-positive breast cancers necessitate HER2-targeted therapies, whereas triple-negative breast cancers lack these receptors.119 Understanding the roles of these receptors has led to the development of personalized therapies and exploration of natural phytochemicals that have improved survival rates and quality of life for many patients.1205 Conclusion
This study focused on the discovery of potent inhibitors of EGFR, ER and PR from Pleurotus ostreatus extracts as a potential therapy for breast cancer using in silico methods. When compared with the already established drugs as well as their natural substrates, the binding energy of some of the compounds was better. With ER, only one compound out of all compounds studied exhibited a better binding affinity than estradiol, its substrate. PR and EGFR on the other hand recorded many compounds with better binding affinity than that of the control. Most of these compounds were in the class of phenols and flavonoids demonstrating the proven anticancer potential of compounds in this class. None of the hexane and methanolic extracts performed better than the control for each of the proteins. These compounds possess good ADMET properties indicating that these compounds can be considered safe for further development to anticancer drugs. Further in vitro and in vivo studies should be undertaken to validate these docking results.Data availability
Data is available upon request from the corresponding author.Author contributions
All authors conceptualized this research. Experiments, data analysis and writing of original draft was carried out by Effiong Magdalene Eno and Mercy Bella-Omunagbe. Methodology validation and writing (review and editing) was carried out by Israel Sunmola Afolabi and Shalom Nwodo Chinedu.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to Covenant Applied Informatics and Communication – Africa Centre of Excellence (CApIC-ACE) and Covenant University Centre for Research Innovation and Discovery (CUCRID) for the funding.References
- A. Hamza, A. Mylarapu, K. V. Krishna and D. S. Kumar, An insight into the nutritional and medicinal value of edible mushrooms: A natural treasury for human health, J. Biotechnol., 2024, 381, 86–99, DOI:10.1016/j.jbiotec.2023.12.014.
- M. E. Effiong, C. P. Umeokwochi, I. S. Afolabi and S. N. Chinedu, Assessing the nutritional quality of Pleurotus ostreatus (oyster mushroom), Front. Nutr., 2024, 10(January), 1–13, DOI:10.3389/fnut.2023.1279208.
- G. M. Liuzzi, T. Petraglia, T. Latronico, A. Crescenzi and R. Rossano, Antioxidant Compounds from Edible Mushrooms as Potential Candidates for Treating Age-Related Neurodegenerative Diseases, Nutrients, 2023, 15(8), 23, DOI:10.3390/nu15081913.
- K. Arunachalam, P. S. Sreeja and X. Yang, The Antioxidant Properties of Mushroom Polysaccharides can Potentially Mitigate Oxidative Stress, Beta-Cell Dysfunction and Insulin Resistance, Front. Pharmacol, 2022, 13(May), 1–23, DOI:10.3389/fphar.2022.874474.
- H. J. Park, Current Uses of Mushrooms in Cancer Treatment and Their Anticancer Mechanisms, Int. J. Mol. Sci., 2022, 23(18), 10502–10516, DOI:10.3390/ijms231810502.
- M. B. Gariboldi, E. Marras and N. Ferrario, et al., Anti-Cancer Potential of Edible/Medicinal Mushrooms in Breast Cancer, Int. J. Mol. Sci., 2023, 24, 10120–10150, DOI:10.3390/ijms241210120.
- S. Nandi, R. Sikder, S. Rapior, S. Arnould, J. Simal-Gandara and K. Acharya, A review for cancer treatment with mushroom metabolites through targeting mitochondrial signaling pathway: In vitro and in vivo evaluations, clinical studies and future prospects for mycomedicine, Fitoterapia, 2024, 172(January 2023), 105681–106703, DOI:10.1016/j.fitote.2023.105681.
- M. P. Pathak, K. Pathak and R. Saikia, et al., Immunomodulatory effect of mushrooms and their bioactive compounds in cancer: A comprehensive review, Biomed. Pharmacother., 2022, 149(April), 112901, DOI:10.1016/j.biopha.2022.112901.
- J. M. Macharia, L. Zhang and R. W. Mwangi, et al., Are chemical compounds in medical mushrooms potent against colorectal cancer carcinogenesis and antimicrobial growth?, Cancer Cell Int., 2022, 22(1), 1–13, DOI:10.1186/s12935-022-02798-2.
- P. Twardowski, N. Kanaya and P. Frankel, et al., A phase i trial of mushroom powder in patients with biochemically recurrent prostate cancer: Roles of cytokines and myeloid-derived suppressor cells for Agaricus bisporus-induced prostate-specific antigen responses, Cancer, 2015, 121(17), 2942–2950, DOI:10.1002/cncr.29421.
- J. Garcia, F. Rodrigues, M. J. Saavedra, F. M. Nunes and G. Marques, Bioactive polysaccharides from medicinal mushrooms: A review on their isolation, structural characteristics and antitumor activity, Food Biosci., 2022, 49, 101955, DOI:10.1016/j.fbio.2022.101955.
- L. Figueiredo and W. C. B. Régis, Medicinal mushrooms in adjuvant cancer therapies: an approach to anticancer effects and presumed mechanisms of action, Nutrire, 2017, 42(1), 1–10, DOI:10.1186/s41110-017-0050-1.
- A. Midlenko, K. Mussina and G. Zhakhina, et al., Prevalence, incidence, and mortality rates of breast cancer in Kazakhstan: data from the Unified National Electronic Health System, 2014–2019, Front. Public Health, 2023, 11, 1132742–1132752, DOI:10.3389/fpubh.2023.1132742.
- S. Gehlert, D. Hudson and T. Sacks, A Critical Theoretical Approach to Cancer Disparities: Breast Cancer and the Social Determinants of Health, Front. Public Health, 2021, 9(May), 1–8, DOI:10.3389/fpubh.2021.674736.
- P. Adewale Adeoye, Epidemiology of Breast Cancer in Sub-Saharan Africa, in Breast Cancer Updates, IntechOpen, 2023, pp. 1–20, DOI:10.5772/intechopen.109361.
- M. E. Effiong, I. S. Afolabi and S. N. Chinedu, Influence of age and education on breast cancer awareness and knowledge among women in South Western Nigeria Methods The participants were randomly selected from six, Afr. J. Reprod. Health, 2023, 88(March), 87–107, DOI:10.29063/ajrh2023/v27i3.11.
- A. Mohammed, D. Melak and F. Demeke Bayou, et al., Prevalence and associated factors of depression among breast cancer patients in Sub-Saharan Africa: A systematic review and meta-analysis, SAGE Open Med., 2024, 12(January), 1–9, DOI:10.1177/20503121241226897.
- A. Alzehr, C. Hulme, A. Spencer and S. Morgan-Trimmer, The economic impact of cancer diagnosis to individuals and their families: a systematic review, Support. Care Cancer, 2022, 30(8), 6385–6404, DOI:10.1007/s00520-022-06913-x.
- T. Nwankwo, A. O. Ogunyemi and B. A. Maduafokwa, et al., Psychosocial Support and Cost Burden of Cancer Among Patients Attending Tertiary Oncology Clinics in Lagos State, Nigeria, Asian Pac. J. Cancer Prev., 2023, 24(7), 2313–2319, DOI:10.31557/APJCP.2023.24.7.2313.
- D. Nagpal, R. Verma, V. Mittal, P. Jeandet and D. Kaushik, Targeted therapies against breast cancer: Clinical perspectives, obstacles and new opportunities, J. Drug Delivery Sci. Technol., 2023, 89, 105049, DOI:10.1016/j.jddst.2023.105049.
- F. Ye, S. Dewanjee and Y. Li, et al., Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer, Mol. Cancer, 2023, 22(1), 1–40, DOI:10.1186/s12943-023-01805-y.
- S. Ramón y Cajal, M. Sesé and C. Capdevila, et al., Clinical implications of intratumor heterogeneity: challenges and opportunities, J. Mol. Med., 2020, 98(2), 161–177, DOI:10.1007/s00109-020-01874-2.
- N. M. Atallah, M. Haque and C. Quinn, et al., Characterisation of luminal and triple-negative breast cancer with HER2 Low protein expression, Eur. J. Cancer, 2023, 195(July), 113371, DOI:10.1016/j.ejca.2023.113371.
- E. B. Belachew, A. F. Desta and T. Y. Gebremariam, et al., Immunohistochemistry-derived subtypes of breast cancer distribution in four regions of Ethiopia, Front. Endocrinol., 2023, 14(November), 1–11, DOI:10.3389/fendo.2023.1250189.
- K. Andrade de Oliveira, S. Sengupta, A. K. Yadav and R. Clarke, The complex nature of heterogeneity and its roles in breast cancer biology and therapeutic responsiveness, Front. Endocrinol., 2023, 14(February), 1–13, DOI:10.3389/fendo.2023.1083048.
- L. Guo, D. Kong and J. Liu, et al., Breast cancer heterogeneity and its implication in personalized precision therapy, Exp. Hematol. Oncol., 2023, 12(1), 1–27, DOI:10.1186/s40164-022-00363-1.
- A. F. Onyia, T. A. Nana and E. A. Adewale, et al., Breast Cancer Phenotypes in Africa: A Scoping Review and Meta-Analysis, JCO Global Oncology, 2023,(9), 1–10, DOI:10.1200/go.23.00135.
- M. E. Effiong, C. P. Umeokwochi, I. S. Afolabi and S. N. Chinedu, Comparative antioxidant activity and phytochemical content of five extracts of Pleurotus ostreatus (oyster mushroom), Sci. Rep., 2024, 14(1), 3794, DOI:10.1038/s41598-024-54201-x.
- N. Shafiq, B. Shakoor and N. Yaqoob, et al., A virtual insight into mushroom secondary metabolites: 3D-QSAR, docking, pharmacophore-based analysis and molecular modeling to analyze their anti-breast cancer potential, J. Biomol. Struct. Dyn., 2024, 1, 1–22, DOI:10.1080/07391102.2024.2304137.
- A. M. H. Al-Rajhi and T. M. A. Ghany, In vitro repress of breast cancer by bio-product of edible Pleurotus ostreatus loaded with chitosan nanoparticles, Appl. Biol. Chem., 2023, 66(1), 33–47, DOI:10.1186/s13765-023-00788-0.
- M. E. Effiong, I. S. Afolabi and S. N. Chinedu, Assessing the nutritional quality of Pleurotus ostreatus (oyster mushroom), Front. Nutr., 2024, 10, 1–13, DOI:10.3389/fnut.2023.1279208.
- R. Roghini and K. Vijayalakshmi, Phytochemical screening, quantitative analysis of flavonoids and minerals in ethanolic extract of Citrus paradisi, Int. J. Pharma Sci. Res., 2018, 9(11), 4859, DOI:10.13040/IJPSR.0975-8232.9(11).4859-64.
- X. Zhang, Y. Bai and Y. Wang, et al., Anticancer Properties of Different Solvent Extracts of Cucumis melo L. Seeds and Whole Fruit and Their Metabolite Profiling Using HPLC and GC-MS, BioMed Res. Int., 2020, 2020, 1–9, DOI:10.1155/2020/5282949.
- V. Mishra, S. Tomar, P. Yadav, S. Vishwakarma and M. P. Singh, Elemental Analysis, Phytochemical Screening and Evaluation of Antioxidant, Antibacterial and Anticancer Activity of Pleurotus ostreatus through In Vitro and In Silico Approaches, Metabolites, 2022, 12(9), 1–25, DOI:10.3390/metabo12090821.
- F. N. Iheagwam, E. N. Israel, K. O. Kayode, O. C. De Campos, O. O. Ogunlana and S. N. Chinedu, GC-MS Analysis and Inhibitory Evaluation of Terminalia catappa Leaf Extracts on Major Enzymes Linked to Diabetes, J. Evidence-Based Complementary Altern. Med., 2019, 2019, 1–14, DOI:10.1155/2019/6316231.
- F. N. Iheagwam, O. O. Ogunlana and S. N. Chinedu, Model Optimization and In Silico Analysis of Potential Dipeptidyl Peptidase IV Antagonists from GC- MS Identified Compounds in Nauclea latifolia Leaf Extracts, Int. J. Mol. Sci., 2019, 20(5913), 1–28, DOI:10.3390/ijms20235913.
- I. S. Afolabi, E. F. Ahuekwe, P. A. Garuba and A. J. Adigun, Enterococcus faecalis -Induced Biochemical Transformation during Fermentation of Underutilized Solenostemon monostachyus Leaves Enterococcus faecalis -Induced Biochemical Transformation during Fermentation of Underutilized Solenostemon monostachyus Leaves, Fermentation, 2023, 9(33), 1–20, DOI:10.3390/fermentation9010033.
- S. Dallakyan and A. J. Olson, Small-molecule library screening by docking with PyRx, Methods Mol. Biol., 2015, 1263, 243–250, DOI:10.1007/978-1-4939-2269-7_19.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31(2), 455–461, DOI:10.1002/JCC.21334.
- G. Xiong, Z. Wu and J. Yi, et al., ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties, Nucleic Acids Res., 2021, 49(W1), W5–W14, DOI:10.1093/nar/gkab255.
- A. Paul and S. Das, Gas Chromatography Mass Spectrometry based metabolomic investigation on five different rice cutivars (Oryza sativa L.) under different induced Cadmium stress, Food Chem. Adv., 2023, 2, 100175, DOI:10.1016/j.focha.2022.100175.
- M. E. Effiong, C. P. Umeokwochi, I. S. Afolabi and S. N. Chinedu, Comparative antioxidant activity and phytochemical content of five extracts of Pleurotus ostreatus ( oyster mushroom ), Sci. Rep., 2024,(0123456789), 1–15, DOI:10.1038/s41598-024-54201-x.
- P. Chaudhary, P. Janmeda and A. O. Docea, et al., Oxidative stress, free radicals and antioxidants: potential crosstalk in the pathophysiology of human diseases, Front. Chem., 2023, 11(May), 1–24, DOI:10.3389/fchem.2023.1158198.
- B. Borah, K. D. Dwivedi and L. R. Chowhan, Review on Synthesis and Medicinal Application of Dihydropyrano[3,2-b]Pyrans and Spiro-Pyrano[3,2-b]Pyrans by Employing the Reactivity of 5-Hydroxy-2-(Hydroxymethyl)-4H-Pyran-4-One, Polycyclic Aromat. Compd., 2022, 42(9), 5893–5937, DOI:10.1080/10406638.2021.1962923.
- Y. M. Youssef, M. E. Azab and G. A. Elsayed, et al., Synthesis and antioxidant, antimicrobial, and antiviral activity of some pyrazole-based heterocycles using a 2(3H)-furanone derivative, J. Iran. Chem. Soc., 2023, 20(9), 2203–2216, DOI:10.1007/s13738-023-02814-w.
- K. R. Renugadevi, C. V. Nachiyar and M. Zaveri, Bioactivity of Dodecanoic Acid Extracted from Geitlerinema sp. TRV57, Indian J. Pharm. Educ. Res., 2021, 55(1), 224–231, DOI:10.5530/ijper.55.1.25.
- J. Mett and U. Müller, The medium-chain fatty acid decanoic acid reduces oxidative stress levels in neuroblastoma cells, Sci. Rep., 2021, 11(1), 1–13, DOI:10.1038/s41598-021-85523-9.
- P. G. Li, M. V. Raimondi, V. Spanò, R. Holl, P. Barraja and A. Montalbano, Pyrrolidine in Drug Discovery: A Versatile Scaffold for Novel Biologically Active Compounds, Springer International Publishing, 2021, vol. 379, DOI:10.1007/s41061-021-00347-5.
- J. S. Sohn and J. S. Choi, Development and evaluation of niacinamide transdermal formulation by artificial membrane permeability, Saudi Pharm. J., 2023, 31(7), 1229–1236, DOI:10.1016/j.jsps.2023.05.018.
- T. Ihnatova, A. Kaplaushenko, Y. Frolova and E. Pryhlo, Synthesis and antioxidant properties of some new 5-phenethyl-3-thio-1,2,4-triazoles, Pharmacia, 2021, 68, 129–133, DOI:10.3897/pharmacia.68.e53320.
- R. Chauhan, P. Saini, R. Choudhary and S. Rani, A Review on Pharmacological Profile of Ethanamide and their Derivatives, Int. J. Pharm. Sci. Rev. Res., 2020, 64(2), 162–170, DOI:10.47583/ijpsrr.2020.v64i02.027.
- G. Oloyede, O. Oluwakayode, O. Bamkole and W. Akanmu, Hispanamide: An Antioxidant Phenolic Propanamide from the Leaves of Acalypha Hispida (Burn.F), J. Nat. Sci. Res., 2020, 11(14), 11–16, DOI:10.7176/JNSR/11-14-02.
- S. Poyraz, H. A. Döndaş, N. Y. Döndaş and J. M. Sansano, Recent insights about pyrrolidine core skeletons in pharmacology, Front. Pharmacol, 2023, 14(September), 1–16, DOI:10.3389/fphar.2023.1239658.
- S. A. Navarro, K. G. G. Serafim, S. S. Mizokami, M. S. N. Hohmann, R. Casagrande and W. A. Verri, Analgesic activity of piracetam: Effect on cytokine production and oxidative stress, Pharmacol., Biochem. Behav., 2013, 105, 183–192, DOI:10.1016/j.pbb.2013.02.018.
- P. Anandakumar and M. K. V. Sattu Kamaraj, D-limonene: A multifunctional compound with potent therapeutic effects, J. Food Biochem., 2020, 45(1), e13566–e13576, DOI:10.1111/jfbc.13566.
- A. Balachandran, S. B. Choi and M. M. Beata, et al., Antioxidant, Wound Healing Potential and In Silico Assessment of Naringin, Eicosane and Octacosane, Molecules, 2023, 28(3), 1043–1073, DOI:10.3390/molecules28031043.
- G. Q. Chen, H. Y. Guo, Z. S. Quan, Q. K. Shen, X. Li and T. Luan, Natural Products–Pyrazine Hybrids: A Review of Developments in Medicinal Chemistry, Molecules, 2023, 28(21), 7740–7499, DOI:10.3390/molecules28217440.
- R. B. Vinayak Singh, M. R. Pradeep Kumar, M. Stephy Grace, J. G. Babu and R. D. Nnada, An insight to the synthetic and medicinal aspects of thiadiazole scaffold, Eur. Chem. Bull., 2023, 12(4), 14530–14553, DOI:10.48047/ecb/2023.12.si4.13152023.08/06/2023.
- P. Rani, D. Pal, R. R. Hegde and S. R. Hashim, Anticancer, anti-inflammatory, and analgesic activities of synthesized 2-(substituted phenoxy) acetamide derivatives, BioMed Res. Int., 2014, 2014, 1–9, DOI:10.1155/2014/386473.
- Z. Zhao, J. Yue and X. Ji, et al., Research progress in biological activities of succinimide derivatives, Bioorg. Chem., 2021, 108, 104557, DOI:10.1016/j.bioorg.2020.104557.
- R. W. Sabnis, Pyrrolidinone Derivatives as NIK Inhibitors for Treating Inflammatory and Autoimmune Diseases, ACS Med. Chem. Lett., 2024, 15(3), 322–323, DOI:10.1021/acsmedchemlett.4c00044.
- L. Yu, J. Yan and Z. Sun, D-limonene exhibits anti - inflammatory and antioxidant properties in an ulcerative colitis rat model via regulation of iNOS , COX-2 , PGE2 and ERK signaling pathways, Mol. Med. Rep., 2017, 15(4), 2339–2346, DOI:10.3892/mmr.2017.6241.
- P. N. Okechukwu, Evaluation of Anti-Inflammatory, Analgesic, Antipyretic Effect of Eicosane, Pentadecane, Octacosane, and Heneicosane, Asian J. Pharm. Clin. Res., 2020, 13(4), 29–35, DOI:10.22159/ajpcr.2020.v13i4.36196.
- J. H. Jang and T. J. Lee, Mechanisms of Phytochemicals in Anti-Inflammatory and Anti-Cancer, Int. J. Mol. Sci., 2023, 24(9), 5–8, DOI:10.3390/ijms24097863.
- N. F. Sianipar, K. Assidqi, Y. E. Hadisaputri, S. Salam, R. Tarigan and R. Purnamaningsih, Determination of Bioactive Compounds of Superior Mutant Rodent Tuber (Typhoniumflagelliforme) in Various Fractions Using GC-MS, IOP Conf. Ser.: Earth Environ. Sci, 2021, 794(1), 10, DOI:10.1088/1755-1315/794/1/012144.
- T. Diesinger, A. Lautwein and S. Bergler, et al., A New CYP2E1 Inhibitor, 12-Imidazolyl-1-dodecanol, Represents a Potential Treatment for Hepatocellular Carcinoma, Can. J. Gastroenterol. Hepatol., 2021, 2021, 1–16, DOI:10.1155/2021/8854432.
- M. Cieślak, M. Napiórkowska and J. Kaźmierczak-barańska, et al., New succinimides with potent anticancer activity: Synthesis, activation of stress signaling pathways and characterization of apoptosis in leukemia and cervical cancer cells, Int. J. Mol. Sci., 2021, 22(9), 4318–4341, DOI:10.3390/ijms22094318.
- J. Zhang, J. Zhang and G. Hao, et al., Design, Synthesis, and Structure–Activity Relationship of 7-Propanamide Benzoxaboroles as Potent Anticancer Agents, J. Med. Chem., 2019, 62(14), 6765–6784, DOI:10.1021/acs.jmedchem.9b00736.
- D. T. Ngoc, T. H. M. Vu and X. L. Ngo, et al., A New Pyrrolidone Alkaloid and Other Constituents from Rourea oligophlebia Stems, J. Chem., 2021, 2021(1), 1–5, DOI:10.1155/2021/6659106.
- J. Hur, J. Jang and J. Sim, A Review of the Pharmacological Activities and Recent Synthetic Advances of γ-Butyrolactones, Int. J. Mol. Sci., 2021, 22, 2769–2817, DOI:10.3390/ijms22052769.
- M. Al-Ghorbani, M. A. Gouda, M. Baashen, O. Alharbi, F. A. Almalki and L. V. Ranganatha, Piperazine Heterocycles as Potential Anticancer Agents: A Review, Pharm. Chem. J., 2022, 56(1), 29–37, DOI:10.1007/s11094-022-02597-z.
- D. H. Kim, M. H. Park and Y. J. Choi, et al., Molecular Study of Dietary Heptadecane for the Anti-Inflammatory Modulation of NF-kB in the Aged Kidney, PLoS One, 2013, 8(3), e59316, DOI:10.1371/journal.pone.0059316.
- W. Hou, W. Dai and H. Huang, et al., Pharmacological activity and mechanism of pyrazines, Eur. J. Med. Chem., 2023, 258, 115544, DOI:10.1016/j.ejmech.2023.115544.
- B. H. Ganesh, A. G. Raj, B. Aruchamy, P. Nanjan and C. D. P. R. Pyrrole, A Decisive Scaffold for the Development of Therapeutic Agents and Structure-Activity Relationship, ChemMedChem, 2024, 19, e202300447, DOI:10.1002/cmdc.202300447.
- U. A. Çevik, D. Osmaniye and S. Levent, et al., Synthesis and characterization of a new series of thiadiazole derivatives as potential anticancer agents, Heterocycl. Commun., 2020, 26(1), 6–13, DOI:10.1515/hc-2020-0002.
- K. Heelan and T. Markham, Fumaric acid esters as a suitable first-line treatment for severe psoriasis: an Irish experience, Clin. Exp. Dermatol., 2012, 37(7), 793–795, DOI:10.1111/j.1365-2230.2012.04351.x.
- S. Basak and A. K. Duttaroy, Conjugated linoleic acid and its beneficial effects in obesity, cardiovascular disease, and cancer, Nutrients, 2020, 12(7), 1–7, DOI:10.3390/nu12071913.
- K. R. Parajuli, Q. Zhang and S. Liu, et al., Methoxyacetic acid suppresses prostate cancer cell growth by inducing growth arrest and apoptosis, Am. J. Clin. Exp. Urol., 2014, 2(4), 300–312 Search PubMed.
- H. H. Al Mamari, in Phenolic Compounds: Classification, Chemistry, and Updated Techniques of Analysis and Synthesis, Intech, 2022, DOI:10.5772/intechopen.98958.
- J. J. Chebet, J. E. Ehiri, D. J. Mcclelland, D. Taren and I. A. Hakim, Effect of d-limonene and its derivatives on breast cancer in human trials : a scoping review and narrative synthesis, BMC Cancer, 2021, 21(1), 1–11 CrossRef PubMed.
- X. Chuah, P. Okechukwu, F. Amini and S. Teo, Eicosane, pentadecane and palmitic acid: The effects in in vitro wound healing studies, Asian Pac. J. Trop. Biomed., 2018, 8(10), 490–499, DOI:10.4103/2221-1691.244158.
- K. Bozorov, J. Zhao and H. A. Aisa, Since January 2020 Elsevier Has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID- 19. The COVID-19 Resource Centre Is Hosted on Elsevier Connect, the Company's Public News and Information, 2020, January Search PubMed.
- Y. Olayiwola and L. Gollahon, Natural Compounds and Breast Cancer: Chemo-Preventive and Therapeutic Capabilities of Chlorogenic Acid and Cinnamaldehyde, Pharmaceuticals, 2024, 17(3), 361, DOI:10.3390/ph17030361.
- M. Cieślak, M. Napiórkowska and J. Kaźmierczak-barańska, et al., New succinimides with potent anticancer activity: Synthesis, activation of stress signaling pathways and characterization of apoptosis in leukemia and cervical cancer cells, Int. J. Mol. Sci., 2021, 22(9), 4318, DOI:10.3390/ijms22094318.
- F. Pourrajab, S. K. Forouzannia and S. A. Tabatabaee, Novel immunomodulatory function of 1,3,4-thiadiazole derivatives with leishmanicidal activity, J. Antimicrob. Chemother., 2012, 67(8), 1968–1978, DOI:10.1093/jac/dks144.
- M. F. N. Meeran, A. Seenipandi and H. Javed, et al., Heliyon Can limonene be a possible candidate for evaluation as an agent or adjuvant against infection , immunity , and in fl ammation in COVID-19, Heliyon, 2021, 7(September 2020), e05703, DOI:10.1016/j.heliyon.2020.e05703.
- K. Heelan and T. Markham, Fumaric acid esters as a suitable first-line treatment for severe psoriasis: an Irish experience, Clin. Exp. Dermatol., 2012, 37(7), 793–795, DOI:10.1111/j.1365-2230.2012.04351.x.
- D. Choudhary, S. Garg and M. Kaur, et al., Advances in the Synthesis and Bio-Applications of Pyrazine Derivatives: A Review, Polycyclic Aromat. Compd., 2023, 43(5), 4512–4578, DOI:10.1080/10406638.2022.2092873.
- K. R. Renugadevi, C. V. Nachiyar and M. Zaveri, Bioactivity of Dodecanoic Acid Extracted from Geitlerinema sp. TRV57, Indian J. Pharm. Educ. Res., 2021, 55(1), 224–231, DOI:10.5530/ijper.55.1.25.
- A. J. Vieira, F. P. Beserra, M. C. Souza, B. M. Totti and A. L. Rozza, Chem.-Biol. Interact., 2018, 1(283), 97–06, DOI:10.1016/j.cbi.2018.02.007.
- F. Hezam, A. Ostoot, Z. Salma, S. Shaukath and A. Khanum. Recent Investigations into Synthesis and Pharmacological Activities of Phenoxy Acetamide and its Derivatives (Chalcone, Indole and Quinoline) as Possible Therapeutic Candidates, Springer, Berlin Heidelberg, 2021, vol. 18, DOI:10.1007/s13738-021-02172-5.
- W. J. He, C. H. Lv and Z. Chen, et al., The Regulatory Effect of Phytochemicals on Chronic Diseases by Targeting Nrf2-ARE Signaling Pathway, Antioxidants, 2023, 12(2), 22, DOI:10.3390/antiox12020236.
- S. Parida and D. Sharma, Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights, Cells, 2020, 9(5), 1091, DOI:10.3390/cells9051091.
- S. C. V. Das Neves, S. M. B. C. da Silva and G. K. A. Costa, et al., Dietary supplementation with fumaric acid improves growth performance in nile tilapia juveniles, Animals, 2022, 12(1), 10, DOI:10.3390/ani12010008.
- F. M. Dayrit, The Properties of Lauric Acid and Their Significance in Coconut Oil, J. Am. Oil Chem. Soc., 2015, 92(1), 1–15, DOI:10.1007/s11746-014-2562-7.
- R. R. Naik, A. K. Shakya and G. A. Oriquat, et al., Fatty acid analysis, chemical constituents, biological activity and pesticide residues screening in jordanian propolis, Molecules, 2021, 26(16), 1–12, DOI:10.3390/molecules26165076.
- A. del Olmo, J. Calzada and M. Nuñez, Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy, Crit. Rev. Food Sci. Nutr., 2017, 57(14), 3084–3103, DOI:10.1080/10408398.2015.1087964.
- A. Kessenikh, E. Gnuchikh and S. Bazhenov, et al., Genotoxic effect of 2,2’-bis(bicyclo[2.2.1] heptane) on bacterial cells, PLoS One, 2020, 15(8 August 2020), 1–11, DOI:10.1371/journal.pone.0228525.
- P. Sharma and D. Thakur, Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India, Sci. Rep., 2020, 10(1), 1–18, DOI:10.1038/s41598-020-60968-6.
- M. Lehtonen, S. Kekäläinen, I. Nikkilä, P. Kilpeläinen, M. Tenkanen and K. S. Mikkonen, Active food packaging through controlled in situ production and release of hexanal, Food Chem.: X, 2020, 5(December 2019), 100074, DOI:10.1016/j.fochx.2019.100074.
- R. Jain and Y. Yan, Dehydratase mediated 1-propanol production in metabolically engineered Escherichia coli, Microb. Cell Fact., 2011, 10(1), 1–10 CrossRef PubMed.
- R. M. Rajendran and B. D. Parthiban, Identification, characterization, and antibacterial studies of furobenzopyrans from Ammi visnaga, J. Appl. Pharm. Sci., 2023, 13(9), 84–94, DOI:10.7324/JAPS.2023.138123.
- I. S. Sharafutdinov, E. Y. Trizna and D. R. Baidamshina, et al., Antimicrobial effects of sulfonyl derivative of 2(5H)-furanone against planktonic and biofilm associated methicillin-resistant and -susceptible Staphylococcus aureus, Front. Microbiol., 2017, 8(NOV), 1–12, DOI:10.3389/fmicb.2017.02246.
- A. E. Evren, L. Yurttas and M. Yılmaz-Cankilic, Synthesis of novel N -(naphthalen-1-yl)propanamide derivatives and evaluation their antimicrobial activity, Phosphorus, Sulfur Silicon Relat. Elem., 2020, 195(2), 158–164, DOI:10.1080/10426507.2019.1657428.
- G. Hussain, M. A. Abbasi and A. Aziz-Ur-Rehman, et al., Synthesis of 3-[4-(2-furoyl)-1-piperazinyl]-N-(substituted)propanamides as promising antibacterial agents with mild cytotoxicity, Trop. J. Pharm. Res., 2018, 17(7), 1397–1406, DOI:10.4314/tjpr.v17i7.25.
- M. Bodhaguru, P. Santhiyagu and M. Lakshmanan, et al., In vitro biomedicinal properties of Pyrrolidine-2,4-Dione derived from a novel actinobacterium Streptomyces rochei, a green approach, Biocatal. Agric. Biotechnol., 2019, 20(1), 101244, DOI:10.1016/j.bcab.2019.101244.
- M. D. Ibáñez, N. M. Sanchez-ballester and M. A. Blázquez, Encapsulated Limonene : A Pleasant Lemon-Like Aroma with Promising Application in the Agri- Food Industry . A Review, Molecules, 2020, 25(11), 2598–2618, DOI:10.3390/molecules25112598.
- T. Ahsan, J. Chen, X. Zhao, M. Irfan and Y. Wu, Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf, AMB Express, 2017, 7(1), 54–63, DOI:10.1186/s13568-017-0351-z.
- Y. Wang, Y. Ma and L. Tao, et al., Recent Advances in Separation and Analysis of Saponins in Natural Products, Separations, 2022, 9(7), 163–193, DOI:10.3390/separations9070163.
- F. Rispo, G. De Negri Atanasio and I. Demori, et al., An extensive review on phenolic compounds and their potential estrogenic properties on skin physiology, Front. Cell Dev. Biol., 2024, 11, 1–16, DOI:10.3389/fcell.2023.1305835.
- A. Mazurakova, L. Koklesova and M. Samec, et al., Anti-breast cancer effects of phytochemicals: primary, secondary, and tertiary care, EPMA J., 2022, 13(2), 315–334, DOI:10.1007/s13167-022-00277-2.
- A. Mihanfar, S. G. Darband and S. Sadighparvar, et al., In vitro and in vivo anticancer effects of syringic acid on colorectal cancer: Possible mechanistic view, Chem.-Biol. Interact., 2021, 337, 109337, DOI:10.1016/j.cbi.2020.109337.
- Y. Zhai, T. Wang, Y. Fu, T. Yu, Y. Ding and H. Nie, Ferulic Acid: A Review of Pharmacology, Toxicology, and Therapeutic Effects on Pulmonary Diseases, Int. J. Mol. Sci., 2023, 24(9), 8011, DOI:10.3390/ijms24098011.
- A. H. Rosendahl, C. M. Perks and L. Zeng, et al., Caffeine and Caffeic Acid Inhibit Growth and Modify Estrogen Receptor and Insulin-like Growth Factor I Receptor Levels in Human Breast Cancer, Clin. Cancer Res., 2015, 21(8), 1877–1887, DOI:10.1158/1078-0432.CCR-14-1748.
- M. Čižmáriková, R. Michalková and L. Mirossay, et al., Ellagic Acid and Cancer Hallmarks: Insights from Experimental Evidence, Biomolecules, 2023, 13(11), 1653, DOI:10.3390/biom13111653.
- P. Biswas, D. Dey and P. K. Biswas, et al., A Comprehensive Analysis and Anti-Cancer Activities of Quercetin in ROS-Mediated Cancer and Cancer Stem Cells, Int. J. Mol. Sci., 2022, 23(19), 11746, DOI:10.3390/ijms231911746.
- G. M. Adinew, E. Taka, P. Mendonca, S. S. Messeha and K. F. A. Soliman, The anticancer effects of flavonoids through miRNAs modulations in triple-negative breast cancer, Nutrients, 2021, 13(4), 1212–1249, DOI:10.3390/nu13041212.
- S. Satpathi, S. S. Gaurkar, A. Potdukhe and M. B. Wanjari, Unveiling the Role of Hormonal Imbalance in Breast Cancer Development: A Comprehensive Review, Cureus, 2023, 15(7), 1–10, DOI:10.7759/cureus.41737.
- Y. Feng, M. Spezia and S. Huang, et al., ScienceDirect Breast cancer development and progression : Risk factors , cancer stem cells , signaling pathways , genomics , and molecular pathogenesis, Genes Dis., 2018, 5(2), 77–106, DOI:10.1016/j.gendis.2018.05.001.
- R. Velaga, S. Tanaka and M. Toi, Molecular vulnerabilities and therapeutic resistance in hormone receptor positive and HER2 dependent breast cancer tumours, Cancer Drug Resist., 2022, 5(2), 487–497, DOI:10.20517/cdr.2022.10.
- V. P. Androutsopoulos, A. M. Tsatsakis and D. A. Spandidos, Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention, BMC Cancer, 2009, 9, 1–17, DOI:10.1186/1471-2407-9-187.
- R. D. Bruno and V. C. O. Njar, Targeting cytochrome P450 enzymes: A new approach in anti-cancer drug development, Bioorg. Med. Chem., 2007, 15(15), 5047–5060, DOI:10.1016/j.bmc.2007.05.046.
- M. Stielow, A. Witczyńska, N. Kubryń, Ł. Fijałkowski, J. Nowaczyk and A. Nowaczyk, The Bioavailability of Drugs—The Current State of Knowledge, Molecules, 2023, 28(24), 8038, DOI:10.3390/molecules28248038.
- Z. Li, H. Wei, S. Li, P. Wu and X. Mao, The Role of Progesterone Receptors in Breast Cancer, Drug Des., Dev. Ther., 2022, 16(January), 305–314, DOI:10.2147/DDDT.S336643.
- S. M. Swain, M. Shastry and E. Hamilton, Targeting HER2-positive breast cancer: advances and future directions, Nat. Rev. Drug Discovery, 2023, 22(2), 101–126, DOI:10.1038/s41573-022-00579-0.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03832k |
| This journal is © The Royal Society of Chemistry 2024 |