 Open Access Article
Open Access ArticleTherapeutic applications of RNA nanostructures
Theresa Yip ab,
Xiaodong Qibc,
Hao Yan*bc and
Yung Chang*ab
ab,
Xiaodong Qibc,
Hao Yan*bc and
Yung Chang*ab
aSchool of Life Sciences, Arizona State University, Tempe, AZ 85281, USA. E-mail: Hao.Yan@asu.edu; Yung.Chang@asu.edu
bBiodesign Center for Molecular Design and Biomimetics, Biodesign Institute, Arizona State University, Tempe, AZ 85281, USA
cSchool of Molecular Sciences, Arizona State University, Tempe, AZ 85281, USA
First published on 11th September 2024
Abstract
RNA-based therapeutics have gained wide public interest in recent years. RNA is a versatile molecule that exists in many forms including mRNA, siRNA, miRNA, ribozymes, and other non-coding RNAs and is primarily applied for gene therapy. RNA is also used as a modular building block to construct RNA nanostructures. The programmable nature of RNA nanostructures enables the generation of simple, modulable, and multi-functional RNA-based therapeutics. Although the therapeutic application of RNA may be limited due to its structural instability, advances in RNA nanotechnology have improved the stability of RNA nanostructures for greater application. Various strategies have been developed to enhance the stability of RNA nanostructures enabling their application in vivo. In this review, we examine the therapeutic applications of RNA nanostructures. Non-immunogenic RNA nanostructures can be rationally designed with functional RNA molecules to modulate gene expression for gene therapy. On the other hand, nucleic acids can be sensed by cellular receptors to elicit an innate immune response, for which certain DNA and RNA motifs can function as adjuvants. Taking advantage of this adjuvant potential, RNA nanostructures can be used for immunotherapy and be designed for cancer vaccines. Thus, we examine the therapeutic application of immunogenic RNA nanostructures for cancer immunotherapy. RNA nanostructures represent promising platforms to design new nanodrugs, gene therapeutics, immunotherapeutic adjuvants, and cancer vaccines. Ongoing research in the field of RNA nanotechnology will continue to empower the development of RNA nanostructure-based therapeutics with high efficacy and limited toxicity.
Introduction
RNA was thought of primarily as an intermediary molecule, relaying genetic information from DNA for protein translation, or as operational components to assist with protein translation (tRNA and rRNA). However, with the discovery of its catalytic ability,1 RNA began to be examined as its own functional molecule. Since then, the functionality of RNA has expanded to include the ability to edit genes, regulate gene expression, assist in translation, and more. Additionally, due to its physical properties as a modular molecule, RNA has been explored as a unique and independent building block for constructing RNA-based nanostructures for functional applications.2,3The field of RNA nanotechnology evolved alongside its more established DNA counterpart. In the 1980s Seeman conducted studies to design and characterize DNA junctions with 3 or more nucleic acid strands4–6 launching the field of DNA nanotechnology. Inspired by this research, Jaeger, Westhof, and Leontis explored the secondary and tertiary structures of RNA, using its mosaic structure to construct some of the early tectoRNAs.7–9 Building off this work, various methods have been employed to rationally design RNA nanostructures, including nanocubes from multiple RNA scaffolds (Fig. 1A),10 RNA origami tiles from single-stranded RNA (ssRNA) (Fig. 1B),11 large RNA origami from a single ssRNA folding over itself (Fig. 1C),12 and tetrameric nanocages using branched kissing loop design (Fig. 1D).13 Recently, various RNA origami structures were designed using a simplified RNA Origami Automated Design (ROAD) software16 and constructed via in vivo synthesis and assembly in Escherichia coli (E. coli).17 In addition, Guo's group developed a natural packaging RNA (pRNA) strategy from phi29 bacteriophage to generate various functionalized RNA nanostructures and explored their application for drug delivery and gene therapy (Fig. 1E).14,15 While the focus of this review is to examine the therapeutic applications of RNA nanostructures, we refer the readers to these articles detailing the design and construction of RNA nanostructures.11,12,16,18–20
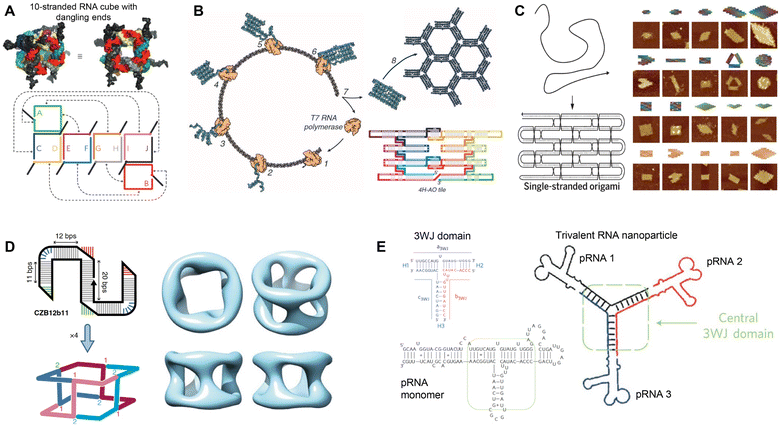 | ||
| Fig. 1 Rational assembly of RNA nanostructures. (A) Two- and three-dimensional model of RNA cube with dangling ends formed from 10 strands of RNA. Reproduced with permission from ref. 10. Copyright 2010 Springer Nature Limited. (B) Co-transcriptional assembly of RNA origami tiles from 4-helix antiparallel-odd (4H-AO) crossovers. Reproduced with permission from ref. 11. Copyright 2014 American Association for the Advancement of Science. (C) Single-stranded origami formed from parallel crossover cohesion (left). AFM images of single-stranded DNA and RNA origami (right). DNA (top 3 rows) and RNA (bottom row). Reproduced with permission from ref. 12. Copyright 2017 American Association for the Advancement of Science. (D) Tetrameric RNA nanocage formed from 4 Z-tiles using branched kissing loop interactions. Reproduced with permission from ref. 13. Copyright 2020 Springer Nature Limited. (E) Trivalent RNA nanoparticle formed from 3 pRNA sequences bound together by the 3WJ motif. Reproduced with permission from ref. 14 and 15. Copyright 2011 Springer Nature Limited. | ||
Developing successful therapeutics for in vivo application relies on many factors including stability and effective delivery. Nanoparticles, such as lipids and polymers, are commonly employed as carriers for therapeutics because of their optimal size for cellular delivery.21 In addition, lipids and polymer-based nanoparticles may provide biological payloads some protection from the hostile extracellular environment that contains nucleases and proteases. However, these nanoparticles can also be toxic and difficult to assemble in a controllable way, leading to inconsistencies in cargo payload and size between batches.22 On the other hand, nucleic acid nanostructures can be assembled into pre-defined shapes and sizes with good precision and reproducibility.12 Additionally, they can function as carriers to attach small molecules at precise locations and controllable stoichiometries, which enables the generation of uniform, precise, and functional nanostructures.12,23,24 Yet from an application point of view, DNA nanostructures have been used primarily for bioimaging and as biomedical sensors for diagnostics,25 but they showed limited therapeutic function, due to their incompatibility with in vivo physiological conditions. While DNA molecules tend to be more stable than RNA, RNA nanostructures designed by RNA assembling technologies (as detailed below) have been shown to have high stability that enables their application in vivo.26 RNA nanostructures also display some additional advantage over DNA nanostructures, including greater structural versatility and diverse functionality, attributed to many functions of RNA molecules, ranging from expression of genes to regulation of gene expression.27 Thus, RNA nanostructures can be readily programmed with a variety of RNA molecules, such as mRNA, small interfering RNA (siRNA), and ribozymes. The diversity of functional RNA molecules provides great potential for developing a range of multi-functional therapeutics. Thus, RNA nanostructures represent unique and rational platforms to design functional RNA-based therapeutics.
In this review, we examine the therapeutic applications of RNA nanostructures, highlighting their diversity and multifunctionality. We first discuss the improved stability of RNA nanostructures that enables their utilization in vivo. With translational applications in mind, we review the functionalization of RNA nanostructures as carriers for gene editing RNAs, catalytic ribozymes, and small molecule drugs. We also examine the use of RNA nanostructures for cancer immunotherapy, discussing their applications as immunomodulators and cancer vaccine platforms. Thus, the versatile nature of RNA nanostructures empowers the design of multifunctional RNA therapeutics.
Enhancing the stability of RNA nanostructures
Although RNA exhibits higher thermodynamic stability compared to DNA molecules and RNA nanostructures have great therapeutic potential, their application in vivo is currently limited due to their intrinsic instability. Aside from nuclease degradation, RNA nanostructures are also susceptible to unfolding or misfolding under in vivo conditions. Thus, several strategies have been explored to enhance the stability of RNA nanostructures. Below, we discuss the use of RNA backbone modifications, nanoparticle carriers, and compact nanostructure designs to stabilize RNA nanostructures.RNA is highly susceptible to nucleases, more so than DNA, because of the 2′-OH group present in RNA that can interact with the phosphate group, resulting in RNA degradation.28,29 Thus, chemical modifications to the ribose sugar,30,31 substituting the 2′-OH with fluorine (–F), amine (–NH2), or O-methyl (–OCH3), or RNA backbone, replacing the phosphodiester (–O−) with phosphorothioate (–S−) or boranophosphate (–BH3−)32,33 have been demonstrated to increase the stability of RNA to enzymatic degradation. Likewise, the incorporation of 2′–5′ linkages, as opposed to the typical 3′–5′ linkage, has been used to improve the stability to nucleases.34 For further information detailing modifications of RNA molecules for increased stability we refer the readers to this review.2
On the other hand, RNA therapeutics can also be packaged in lipid or polymer-based nanoparticles for enhanced stability. Lipid-based systems, including liposomes, micelles, and solid lipid nanoparticles, encapsulate RNA in nanoparticles to protect them from extracellular nucleases, enabling their application in vivo, such as mRNA vaccines made against COVID-19 and even cancers.35–37 Similarly, polymers, such as polyethyleneimine (PEI), chitosan, or dendrimers, have been employed to package RNA, increasing their stability.22 These nanoparticles can be easily modified and functionalized with targeting ligands for multifunctional therapeutics.38 However, these approaches have their own challenges, including inconsistent RNA loading, potential toxicity, and non-specific cellular internalization.22
RNA nanostructures have also been explored as carriers to assemble mRNA, siRNA, and ribozymes, owing to their simple extension of nucleotides for incorporating these functional moieties. However, many RNA nanostructures are still susceptible to nuclease degradation. In addition, similar to DNA nanostructures, the integrity of RNA nanostructures is dependent on the presence of divalent cations, such as Mg2+, to overcome the negatively charged backbone repulsions for folding into their tertiary structures.39 However, the concentration of divalent cation required for the formation of RNA nanostructure is much higher than the physiological level in the body. Thus, RNA nanostructures formed in vitro are not compatible with in vivo conditions. To overcome these obstacles, nucleic acid nanostructures were constructed with modified nucleotides or coated with inorganic molecules or peptides to enhance their stability.40,41
Alternatively, RNA nanostructures were designed in a unique way such that their tertiary structure is less dependent on cations and more resistant to nucleases than conventional RNA. For example, the Guo group used the bacteriophage phi 29 DNA packaging RNA (pRNA) that has diverse self-association. These pRNA naturally assemble into a 3-way junction (3WJ), an RNA motif formed from 3 intertwining RNA strands (see Fig. 1E), forming RNA nanostructures in the absence of divalent cations.14 Although unmodified pRNA-3WJ were still sensitive to serum nuclease, their stability could be improved by incorporating UTPs and CTPs modified with 2′-fluorine (2′-F).14,30 Thus, the modified pRNA-3WJ was demonstrated to have high stability.
Another strategy is to construct compact RNA origami nanostructures, which presumably render less accessibility to nucleases. Building off previous work done by Afonin, et al.,42 Han, et al. developed a parallel crossover cohesion design to form compact double-stranded RNA (dsRNA) nanostructures from a single RNA strand.12 Qi, et al. demonstrated that this ssRNA molecule could self-assemble efficiently into its dsRNA rectangular conformation in the absence of divalent cations, eliminating the dependence on Mg2+ for folding.26 Additionally, without nucleotide modifications or packaging materials, this RNA origami was found to be highly stable in saline for more than 10 months and resistant to RNase and serum nucleases, demonstrating its great thermostability and enzymatic stability for in vivo application.
Furthermore, several computational simulation software have been developed to assess structural stability and construct stable RNA nanostructures,16,18,43,44 greatly facilitating rational design of stable RNA nanostructures.
Therapeutic application of RNA nanostructures
RNA-based therapeutics have gained public attention since the COVID-19 pandemic leading to new and renewed interest in RNA as therapeutics. In addition to being a template for protein translation, RNA can also silence gene expression via siRNA and confer enzymatic activity as ribozymes. Designing functional RNA therapeutics relies on effective assembly and delivery. To this end, RNA can also function as a carrier to assemble these RNA-based therapeutics as well as small drug molecules for delivery. Although lipid- and polymer-based nanoparticles have been widely investigated as carriers for similar payloads, RNA nanostructures represent a new platform for rationally designing RNA-based therapeutics.22,36Due to their programmability, RNA nanostructures can assemble small functional molecules at pre-defined positions with stoichiometric control.18 In addition, because of the simplicity of assembling or extending functional RNA strands on RNA nanostructures, the modularity in altering and switching RNA strands, and the versatility of attaching multifunctional molecules, RNA nanostructures demonstrate great potential for designing RNA-based therapeutics by displaying molecules that confer specific targeting, gene regulation, immune-stimulation, and even toxicity.2 Thus, RNA nanostructures offer consistency, precision, and functionality for designing therapeutics.
Targeted therapeutics can enhance drug efficacy, reduce drug dosage, and limit toxicity associated with off-target effects, especially during systemic application. RNA nanotechnology has been used to assemble targeting molecules such as RNA/DNA aptamers, small 20–100 nucleotide sequences that, like antibodies, bind to specific molecules via their 3D conformation.45 Aptamers have several advantages including small size, rapid generation, versatile RNA modification, and lack of immunogenicity.46 Furthermore, RNA aptamers can be readily incorporated into RNA nanostructures by extending strands of RNA nanostructures to include the aptamer sequence or by linking the RNA aptamer to the RNA nanostructure via complementary base pairing of nucleotides.
In addition to aptamers, a library of other functional RNA and non-RNA molecules such as gene regulation sequences, small molecule drugs, vaccine adjuvants, antigens, and targeting ligands, could be incorporated to develop diverse functional RNA nanostructures. Here, we describe RNA nanostructures that have been developed for therapeutics.
RNA interference (RNAi)
RNA nanostructures are most widely functionalized for application in RNAi therapy. RNAi, a highly conserved biological process in response to dsRNA, silences the expression of specific genetic sequences, functioning as a defense mechanism to viruses and other foreign nucleic acids.47 In the cell, dsRNA or pre-miRNA are cleaved into 20–27 nucleotide sequenced siRNA and miRNA, which then serve as templates for binding and silencing specific mRNA sequences. The ability to control expression of targeted genes has broad translational application, and siRNA and miRNA mimics, as gene silencers, have been extensively studied to treat a wide range of diseases where certain genes are overexpressed, while anti-miRNA can silence overproduced miRNAs to increase gene expression.22,48 Because of their versatile nature, RNA nanostructures represent an excellent platform for carrying siRNA and miRNA mimics to develop RNAi-based therapies.Unlike lipid- or polymer-based nanoparticle delivery systems which rely on the encapsulation of siRNA and miRNA mimics, RNA-based systems can directly incorporate the RNAi sequence into their overall nanostructure design. For example, Lee, et al. used the rolling circle transcription (RCT) method49 to design RNA microsponges containing half a million copies of a single or multiple siRNAs that were directly transcribed and then self-assembled into microsponges (Fig. 2A).50,55 After coating with PEI, they were able to condense microsponge RNA into a nanometer range for greater cellular uptake. This simple strategy demonstrated the ability to incorporate many functional siRNA sequences within RNA nanoparticles.
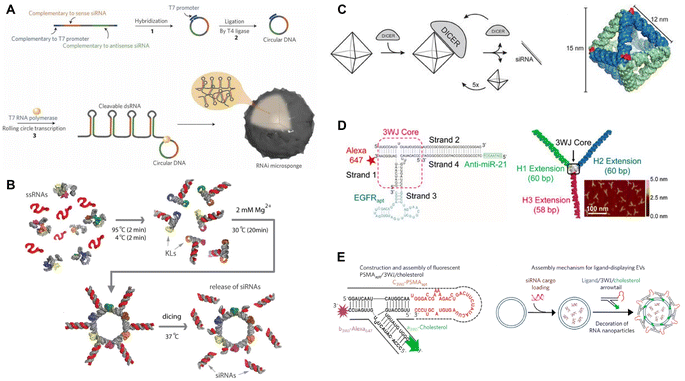 | ||
| Fig. 2 RNA nanostructures functionalized for RNAi or anti-miRNA. (A) RNAi microsponge formation with siRNA. Reproduced with permission from ref. 50. Copyright 2012 Springer Nature Limited. (B) RNA nanorings functionalized with dicer substrates. Reproduced from ref. 51. Copyright 2014 American Chemical Society. (C) RNA octahedron formed with intrinsic siRNAs. Reproduced with permission from ref. 52. Copyright 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (D) pRNA-3WJ functionalized with anti-miRNA and RNA aptamer. Reproduced from ref. 53. Copyright 2015 American Chemical Society. (E) pRNA-3WJ functionalized with cholesterol and RNA aptamer to assemble onto exosomes. Reproduced with permission from ref. 54. Copyright 2017 Springer Nature Limited. | ||
To attach siRNA to RNA nanostructures at predefined positions, Afonin, et al. designed a method for self-assembling RNA strands into a nanoring with 6 siRNAs or tetrahedrons with up to 12 siRNAs.56,57 Their one-pot co-transcriptional assembly method streamlined the production of these functionalized nanostructures by eliminating the need for separate transcription and assembly steps. To demonstrate functionality, the Shapiro group used the RNA nanorings to assemble 6 short hairpin RNAs (shRNAs) targeting different regions of the human immunodeficiency virus (HIV)-1, which reduced the replication of HIV in transfected cells (Fig. 2B).51
Instead of combining the siRNAs as additional elements, Høiberg, et al. incorporated the siRNA into the wire-frame structure of an octahedron RNA origami, using the unstructured siRNA as staple strands in the RNA octahedron (Fig. 2C).52 This incorporation provides the RNA origami nanostructure with RNAi activity and also opens opportunities for further functionalization of the nanostructures, e.g., by attaching targeting molecules, like target-specific aptamers, to enable targeted RNAi therapeutics. Guo's group and others applied this approach in their RNA nanostructures. They designed a tumor targeting RNAi therapeutic using the pRNA-3WJ motif to assemble different RNA aptamers, siRNAs, anti-miRNA, and locked nucleic acids (LNA) (Fig. 2D).14,53,58–61 These multi-functional RNA nanostructures were demonstrated to decrease respective gene expression or silence specific miRNAs in targeted tumor cells, leading to tumor regression in the mouse models. For further application, cholesterol-conjugated pRNA-3WJ were assembled onto exosomes that were then loaded with siRNAs, which had the potential to be used more broadly for other functional small molecules (Fig. 2E).54,62 These studies demonstrate effective tumor targeting and tumor reduction in vivo models and reflect the programmable nature of RNA nanostructures for designing RNAi-based targeted tumor therapeutics. At the moment, RNA nanostructure-based RNAi therapeutics are tested in pre-clinical stages, with promising outcomes in reducing early tumor growth in mouse tumor models.53,59,60 In particular, some RNA nanostructures display high stability that enables systemic delivery in mice, giving this platform potential as an RNAi therapeutic.53,60
mRNA
Eukaryotic mRNAs are composed of a 5′-untranslated region (UTR), open reading frame that codes for the gene, 3′-UTR, and poly(A) tail.63 These are commonly delivered with lipid nanoparticles that enable direct release of mRNA into the cytoplasm for translation, providing a synthetic approach to direct protein delivery.63 Traditionally, the difficulty with developing mRNA therapeutics is their low stability, need for cytoplasmic delivery, and potential innate immunity.64 Due to improvements in RNA nanotechnology that have enhanced RNA stability and modified nanostructures for reduced innate immunity, RNA nanostructures present an excellent opportunity to construct functional mRNA delivery platforms.Recently, Hu, et al. assembled an RNA origami lantern-shaped nanostructure containing Smad4 mRNA scaffolded between 2 staple strands with a tumor targeting peptide, which was designed to restore Smad4 expression in colorectal tumor cells (Fig. 3).65 Observing different levels of expression with different lantern designs, they suggested that the flexible lantern structure enabled mRNA functionality, proposing a new design strategy for functionalizing RNA nanostructures. While there are very few studies that employ RNA nanostructures to deliver mRNA, the success of this platform paves the way for more mRNA nanostructure designs with even greater stability to be explored for therapeutic applications.
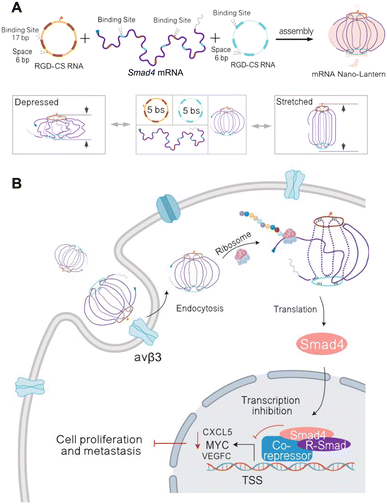 | ||
| Fig. 3 RNA nanostructure functionalized with mRNA. (A) Assembly of mRNA nano-lantern from Smad4 mRNA and two circular staple RNAs (csRNA) containing complementary mRNA binding sites (bs) and RGD (Arg-Gly-Asp) for cell targeting (top). Flexible conformation of mRNA nano-lantern (bottom). (B) Cellular delivery of lantern-shaped flexible RNA origami for Smad4 mRNA translation. Reproduced from ref. 65. Copyright 2023 Springer Nature Limited. Originally created in http://Biorender.com. | ||
Ribozymes
Ribozymes are enzymatic RNA molecules that have various catalytic functions, including cleavage and splicing of nucleic acids. Cleaving ribozymes can be divided into large ribozymes, such as ribonuclease P (RNase P), and small, self-cleaving ribozymes, such as hairpin, hammerhead, hepatitis delta virus, Varkud Satellite (VS), pistol, twister, twister sister, and hatchet ribozymes, named for their specific RNA motifs.66 The ability of these ribozymes to self-cleave and splice is dependent on their folding into appropriate functional conformations. Thus, identifying the conditions at which native ribozymes fold in vitro helps optimize the folding conditions of other RNA nanostructures.67,68 Ribozymes have been proposed and studied as therapeutic treatments, particularly for treating viral infections.69In 2003, a hepatitis B virus (HBV) hammerhead ribozyme was fused to pRNA-3WJ, conferring catalytic activity to this RNA nanostructure.70 Since then, the Guo group has used this HBV ribozyme to functionalize RNA triangles, squares, and tetrahedrons, enabling these nanostructures to cleave HBV genomic RNA (Fig. 4A).14,24,71,73 While a handful of ribozymes have been developed, very few have made it to clinical trials due to the difficulty in design and low stability in vivo.74 However, a better understanding of ribozyme structure and functional motifs may improve their efficacy and therapeutic application.75
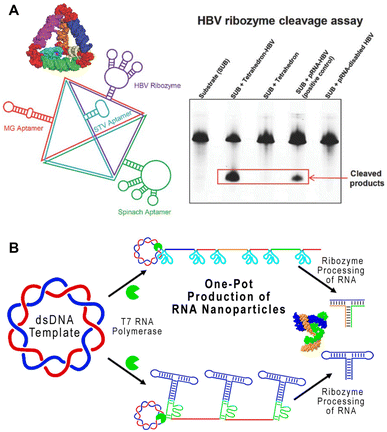 | ||
| Fig. 4 RNA nanostructures functionalized with ribozymes. (A) RNA tetrahedron functionalized with HBV ribozyme and various aptamers (left). HBV cleavage assay demonstrating the ability of tetrahedron assembled HBV ribozymes to cleave the RNA substrate (right). Reproduced with permission from ref. 71. Copyright 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) One-pot production of pRNA-3WJ nanostructures using self-cleaving ribozymes. Reproduced with permission from ref. 72. Copyright 2019 American Chemical Society. | ||
On the other hand, some ribozymes have been incorporated successfully into RNA nanostructures. Similar to the previously described method of utilizing ribozymes for cleaving RCT products,76 a one-pot RCT approach containing self-cleaving hammerhead ribozymes was used to transcribe and cleave RNA sequences used to generate the pRNA-3WJ motif (Fig. 4B).72 This streamlined the overall construction of pRNA-3WJ, combining transcription and assembly, which enables further study and exploration of ribozyme-containing RNA nanostructures for in vivo applications.
CRISPR
CRISPR is a gene editing tool that has been utilized to modify targeted genomic DNA sequences. Recently, RNA origami was used as a scaffold for carrying functional motifs for the CRISPR/dCas9 gene regulatory system (Fig. 5).77 This sgRNAO was designed by fusing a single-guide RNA (sgRNA) to RNA origami and incorporating transcription factors bound to coat protein to enhance transcription. Pothoulakis, et al.77 explored the stoichiometry and positioning of these functional components and designed a sgRNAO system to increase protein production in Saccharomyces cerevisiae yeast. This platform opens another avenue for exploring RNA nanostructures functionalized for CRISPR gene regulation.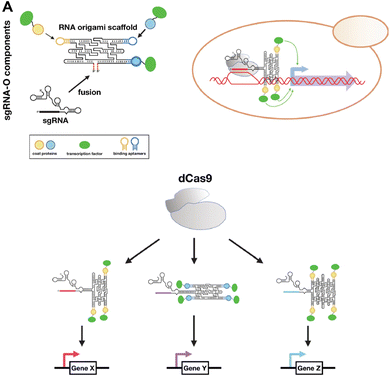 | ||
| Fig. 5 RNA nanostructures functionalized with CRISPR. (A) sgRNA-O for regulating transcriptional in S. cerevisiae. sgRNA-O is formed by fusing sgRNA with RNA origami scaffold containing coat protein targeting aptamers for binding coat protein linked transcription factors (top left). sgRNA-O utilizes dCas9 to regulate gene expression (top right). Multiple sgRNA-O can be constructed to control the transcription of various genes via dCas9 (bottom). Reproduced from ref. 77. Copyright 2022 Pothoulakis, et al. Published by Oxford University Press on behalf of Nucleic Acids Research. | ||
Small molecule drugs
In addition to carrying functional RNA molecules, RNA nanostructures have also been examined as carriers for small molecules drugs. In 2019, Piao, et al. used pRNA-3WJ for carrying camptothecin (CPT),15 a potent antitumor drug that targets topoisomerase I but is not widely used because of its low solubility in aqueous solutions.78 Using click reactions to assemble CPT to RNA along with a tumor targeting ligand, the Guo group reported enhanced solubility of CPT in water and drug-induced KB cell (an epithelial carcinoma cell line) killing leading to reduced tumor growth (Fig. 6A). The same group also designed a four-way junction (4WJ) RNA, an RNA motif formed from 4 intertwining RNA strands, which demonstrated higher stability and greater loading capacity compared to the 3WJ-RNA, to carry paclitaxel (PTX) or SN38, chemotherapy drugs, and tumor targeting aptamers.79,80 Treatment with the drug-loaded 4WJ-RNA nanostructures promoted tumor-targeted cell death and reduced tumor growth in mice (Fig. 6B). Thus, stable 4WJ-RNA nanostructures represent an effective method for controlled incorporation of small molecule drugs.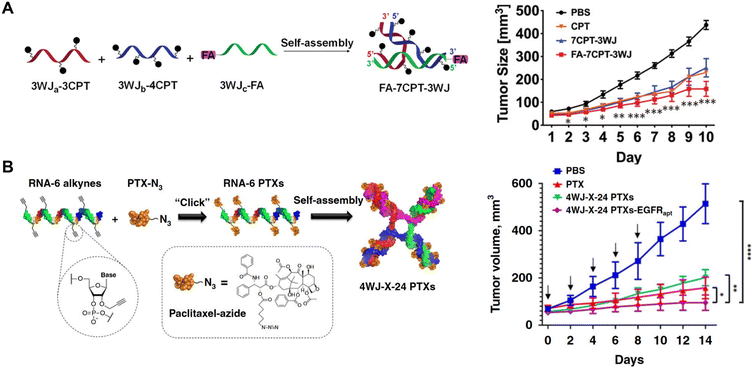 | ||
| Fig. 6 RNA nanostructures functionalized with small molecule drugs. (A) Assembly of a tumor targeting RNA nanostructure drug using pRNA-3WJ strands containing camptothecin (CPT) or folic acid (FA) for targeting (left). Reduction of KB tumor growth in mice treated with the assembled RNA nanostructure drug (FA-7CPT-3WJ) (right). Reproduced from ref. 15. Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) Assembly of 4WJ RNA nanostructure drug using 4 paclitaxel (PTX)-linked RNA strands (left). Reduction of MDA-MB-231 tumor growth in mice treated with 4WJ RNA assembled with PTX and EGFR aptamer (4WJ-X-24 PTXs-EGFRapt) (right). Reproduced from ref. 79. Copyright 2020 Springer Nature Limited. | ||
These studies demonstrate the ability of RNA nanostructures to function as tumor-targeting drug carriers, effectively delivering the drugs to tumors for targeted cell death. The strategy of using simple click reactions to stoichiometrically control drug assembly highlights the potential of these RNA nanostructures for assembling any other functional small molecules for cancer therapy. Currently, RNA nanostructure-conjugated cytotoxic drug therapeutics have been explored in mouse tumor models and were shown to have some benefit in promoting targeted drug delivery and reducing early tumor growth.15,79,80 As drug carriers these RNA nanostructures demonstrated stability for systemic administration and further studies are warranted demonstrating greater survival in mice.
Therapeutic applications of RNA nanostructures in cancer immunotherapy
The field of immunotherapy research has gained momentum over the last decade, largely due to the success of treatments such as immune checkpoint blockades (ICB) and chimeric antigen receptor (CAR)-T cell transfer.81 Immunotherapies aim to enhance the immune response against tumor cells and/or prevent tumor growth. Aside from ICB and CAR-T cell therapy, other common immunotherapy strategies include cytokine therapy that can increase the number or functionality of immune cells, antibody therapy that can offer targeted elimination of tumor cells, and cancer vaccines that can elicit tumor-specific immunity.82–84 More recently, with the increase of genomic sequencing data, there has been an opportunity to develop more personalized therapeutics. With a toolkit of functional RNA molecules, such as mRNA, siRNA, and RNA aptamers, RNA-based therapeutics, along with RNA nanotechnology, offers the potential to develop both general and personalized immunotherapies. Below we discuss new approaches for applying RNA nanostructures to modulate immune responses and design vaccines for cancer immunotherapy.Immune modulation
Immunotherapy adopts different strategies to modulate the immune response. One widely used strategy is employing adjuvants for immunostimulation. Adjuvants range from synthetic products, like aluminum salts and oil-in-water emulsions, to natural motifs, such as pathogen associated molecular patterns (PAMPs). Upon their binding to pattern recognition receptors (PRRs), i.e., toll-like receptors (TLRs), NOD-like receptors (NLR), RIG-I-like receptors (RLR), PAMPs can quickly activate many cells, especially immune cells to generate a robust immunostimulatory response.85 Thus, resembling viral infections, nucleic acid-based adjuvants, such as unmethylated CpG-oligodeoxynucleotides (ODN) DNA (TLR9 agonist), polyinosinic:polycytidylic acid (PolyIC) dsRNA (TLR3 agonist), Resiquimod or ssRNA (TLR7/8 agonists), and cytoplasmic dsDNA (cyclic GMP–AMP synthase (cGAS) stimulator of interferon genes (STING) agonist), provoke strong innate immunity and therefore are widely explored for cancer immunotherapies.Given the ability to incorporate DNA and RNA motifs, RNA nanostructures represent a rational platform for developing cancer immunotherapeutics. Indeed in 2014, Khisamutdinov, et al. used the pRNA-3WJ motif for assembling up to 5 and 6 CpG-ODN on RNA polygon and RNA nanoprism nanostructures, respectively.86,87 Treatment with these RNA polygon-CpG (containing 2 mg kg−1 CpG) induced systemic levels of pro-inflammatory TNF-α and IL-6 cytokines. This group demonstrated that the pro-inflammatory response was dose dependent and could be controlled by incorporating fewer CpG molecules (Fig. 7A).
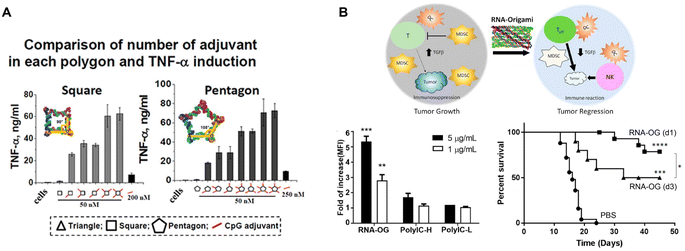 | ||
| Fig. 7 RNA nanostructures as adjuvants. (A) CpG-dependent production of TNF-α by RAW 264.7 macrophages upon treatment with square and pentagon RNA nanostructures functionalized with various numbers of the CpG DNA adjuvant. Reproduced from ref. 86. Copyright 2014 Khisamutdinov, et al. Published by Oxford University Press on behalf of Nucleic Acids Research, (B) schematic of an RNA origami nanostructure with intrinsic adjuvant activity eliciting anti-tumor immunity (top). RNA origami (RNA-OG), PolyIC H (high molecular weight), PolyIC L (low molecular weight) stimulates RAW 264.7 macrophages (bottom left). Increased CT26 tumor-free survival of mice treated with the assembled RNA-OG administered on day 1 or 3 (d1 or d3) (bottom right). Reproduced with permission from ref. 26. Copyright 2020 American Chemical Society. | ||
More recently, Qi, et al. developed an RNA origami nanostructure with intrinsic TLR3 activity, due to its folding into dsRNA, for cancer immunotherapy (Fig. 7B).26 This RNA origami was demonstrated to be a potent, but selective TLR3 agonist that is more stable and safer than PolyIC, the conventional TLR3 agonist. Treatment with RNA origami (0.8 mg kg−1) induced a strong innate immune response that subsequently activated adaptive tumor-specific immunity and reduced immunosuppression in the tumor microenvironment, resulting in tumor regression of a colorectal tumor mouse model without systemic toxicity.26
Further studies by Dai, et al. assessing the intrinsic stimulatory ability of this RNA origami, revealed that the edge length of this nanostructure is critical to its stimulatory activity, presumably due to its ability to engage TLR3 dimerization for cellular activation.88 These strategies highlight the potential of RNA nanostructures for designing immunotherapies either by assembling with adjuvants or as intrinsic adjuvants to enhance the host immunity. On the other hand, Dai, et al. also generated RNA origami nanostructures with functional epigenetic nucleoside analogues that enabled RNA origami nanostructures to evade immune recognition, demonstrating the ability to epigenetically control immunogenicity,89 which could potentially be applied in the construction of functional RNA therapeutics that require no or low immunogenicity (as discussed in Therapeutic application of RNA nanostructures). Thus, different methods can be used to regulate immune stimulation. The immunostimulatory ability of RNA nanostructures has been applied as adjuvants for cancer immunotherapy in mouse tumor models, which showed success in reducing overall tumor growth and increasing mouse survival. Additional studies combining these adjuvants with other cancer therapeutics will help further enhance the anti-tumor efficacy, as discussed in the next section.
Cancer vaccine platform
RNA nanostructures can also be designed as assembly platforms for cancer vaccines. Unlike many anti-viral vaccines that primarily focus on B cell immunity, cancer vaccines are dependent on the induction of T cell immunity, which involves the stimulation and antigen presentation, especially antigen cross-presentation, of antigen presenting cells (APCs) to activate CD8+ T cells.90 These vaccines typically consist of adjuvants, such as TLR ligands that stimulate APCs, and tumor antigens, including peptides or proteins, for antigen cross-presentation.85 Different nanostructures have been explored to bring these components together, including lipid- or polymer-based nanoparticles, as well as RNA nanostructures. While other reviews discuss the generation of cancer vaccines via lipid and polymer packaging,91,92 we will focus on RNA nanotechnology directed design and construction of cancer vaccines.Zhu, et al. used RCT and rolling circle replication (RCR) followed by PEG-grafted polypeptide (PPT-g-PEG) condensation to generate an intertwining DNA–RNA nanocapsule (iDR-NC) vaccine consisting of CpG DNA, Stat3 shRNA, and short hydrophobic peptides (Fig. 8A).93 This functionalized RNA nanovaccine was demonstrated to stimulate dendritic cells (DCs), reduce stat3 expression, and deliver peptides to APCs. Vaccination with iDR-NC induced a neoantigen-specific memory T cell response, ultimately leading to the reduction of systemic MC38 colorectal tumor growth in mice. This study demonstrates the feasibility of assembling peptide vaccines using RNA nanotechnology.
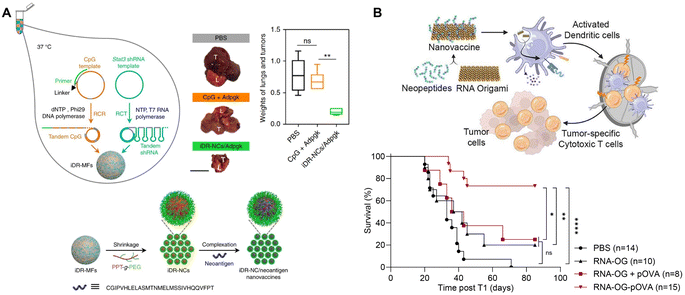 | ||
| Fig. 8 RNA nanostructures functionalized as cancer vaccines. (A) Schematic of intertwining DNA RNA nanocapsule (iDR/NC) containing CpG, stat3, and neoantigen peptide cancer vaccine. iDR/NC is formed using RCT and RCR followed by shrinkage with PEG-grafted polypeptide (PPT-g-PEG) linker and then addition of neoantigen peptide (top left and bottom). Reduction of MC38 tumor size in the lungs of mice treated with iDR-NC/Adpgk peptide (top right). L (lung), T (tumor); scale bar = 1 cm. Reproduced from ref. 93. Copyright 2017 Springer Nature Limited. (B) Schematic of an RNA origami (RNA-OG) peptide nanovaccine formed from an RNA-OG nanostructure with intrinsic adjuvant activity and neopeptides. RNA-OG nanovaccine induces DC activation and stimulation which mobilizes cytotoxic T cells to eliminate tumor cells (top). Originally created in http://Biorender.com. Greater B16-OVA tumor-free survival of mice treated with the assembled RNA-OG-pOVA peptide vaccine. Reproduced with permission from ref. 94. Copyright 2024 American Chemical Society. | ||
Wang, et al. proposed several methodologies to further functionalize RCT generated RNA nanostructures, termed hybrid RNA nanoflowers (hRNF).95 Using DBCO- or biotin-modified UTP during RCT, they generated DBCO-hRNF and biotin-hRNF, which could be covalently bound to azide- or avidin-linked molecules, respectively, post-transcription. This strategy offers a way to design versatile functionalized RNA nanostructures.
Recently, Yip, et al., used an RNA origami nanostructure with intrinsic TLR3 activity to develop a nanovaccine platform to assemble neoantigenic peptides by attaching short azide-linked peptides to DBCO-containing RNA origami through DBCO/azide click reactions (Fig. 8B).94 This RNA origami peptide (RNA-OG-peptide) nanovaccine was found to stimulate and promote cross-presentation in DCs and elicit a strong tumor-specific cytotoxic T cell response. Vaccination with RNA-OG-peptide led to tumor regression and greater survival in mice with B16-OVA melanoma tumors. This protective immunity was further demonstrated to be long-term, because of the generation of tumor-specific memory T cells that rendered the tumor-bearing animals resistant to the second challenge of tumor cells. Without the addition of adjuvants or carriers, this RNA origami nanostructure represents a simple and modulable platform for designing RNA origami-based peptide vaccines.
While only a few studies have applied RNA nanostructures for cancer immunotherapy, with the success of these platforms in mouse studies, the improved stability of RNA nanostructures, and the growing toolkit of nanotechnology, more effort will be directed toward development of RNA nanostructure-based immunotherapeutics.
Challenges & future perspectives
The use of RNA nanostructures for therapeutics has moved this technology towards translational application. With the abundance of functional RNA molecules in addition to its potential adjuvant activity, RNA nanostructures can be employed for various purposes (see summary in Table 1). Here, we categorize the therapeutic applications into two main functions – (1) rational design of functional RNA molecules without immunogenicity, (2) vaccine design using RNA nanostructures with intrinsic adjuvant activity. Below, we highlight some of the benefits and challenges going forward with each of these platforms.| Applications of RNA nanostructures | Description | Methods of attachment | Examples |
|---|---|---|---|
| Targeting | RNA nanostructures attached to RNA aptamers or cell-targeting ligands | Incorporation into RNA nanostructure sequence; complementary base paired to RNA nanostructure | pRNA-3WJ (3-way junction) -aptamer or -ligand (e.g. folate)14,58 (Fig. 2D) |
| RNAi | RNA nanostructures assembled with siRNA to silence gene expression | Incorporation into RNA nanostructure sequence; complementary base paired to RNA nanostructure | RNAi microsponge,50 RNA nanoring,56,57 RNA octahedron origami,52 pRNA-3WJ siRNA14,58,60 (Fig. 2) |
| mRNA | RNA nanostructures assembled with mRNA to express specific genes | Complementary base paired to RNA nanostructure | mRNA nano-lantern65 (Fig. 3) |
| Ribozymes | RNA nanostructures attached to ribozymes to confer enzymatic activity | Incorporation into RNA nanostructure sequence | RNA tetrahedron HBV (hepatitis B virus) ribozyme,14,73 self-cleaved pRNA-3WJ72 (Fig. 4) |
| CRISPR | RNA nanostructures assembled with single-guide RNA (sgRNA) to regulate gene expression | Incorporation into RNA nanostructure sequence; complementary base paired to RNA nanostructure | sgRNAO (sgRNA origami)77 (Fig. 5) |
| Small molecule drugs | RNA nanostructures attached to small molecule drugs to induce tumor cell killing | Incorporation of chemically modified nucleotides in RNA nanostructure followed by click chemistry | pRNA-3WJ-CPT (camptothecin),15 4WJ RNA-PTX (paclitaxel) or SN38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 79,80 (Fig. 6) 79,80 (Fig. 6) |
| Immune modulation | RNA nanostructures with intrinsic adjuvant activity or attached to pathogen associated molecular patterns (PAMPs) | Complementary base paired to RNA nanostructure | RNA origami (dsRNA),26 pRNA-3WJ-CpG-ODN86 (Fig. 7) |
| Cancer vaccines | RNA nanostructures assembled with tumor peptides (and adjuvants) | Addition of PEG-polypeptide linker followed by peptide addition; incorporation of chemically modified nucleotides in RNA nanostructure followed by click chemistry | Intertwining DNA–RNA nanocapsule (iDR-NC),93 RNA origami-peptide vaccine94 (Fig. 8) |
RNA nanostructures as carriers for functional RNA
RNA nanostructures have been studied extensively as carriers for functional RNA molecules. Due to the simplicity of extending RNA strands, mRNAs, siRNAs, anti-miRNAs, ribozymes, and sgRNAs have been rationally incorporated into RNA nanostructures. In addition, their ability to carry multiple RNA enables the design of multi-functional RNA nanostructures, including targeted cancer therapeutics.Nonetheless, there are some considerations for these functional RNA nanostructures. Utilized primarily as cancer therapeutics, these nanostructures require delivery directly into tumor cells, however, tumor cell targeting and cellular uptake of functional RNA nanostructures are challenging. To overcome this, some RNA therapeutics were designed for receptor-mediated endocytosis;96 as such, ligands or aptamers targeting tumor cell receptors have successfully been used to target these cells and promote internalization.53,59 Immunogenicity is another consideration. Due to the presence of various RNA sensors in a cell, RNA nanostructures may trigger innate immune responses, which, while beneficial for vaccine design, may be harmful for siRNA or mRNA delivery. Some studies have reported that small, negatively charged RNA nanostructures lack immunogenicity,97 but other reports showed that larger RNA origami nanostructures have intrinsic adjuvant activity,26,88,94 indicating that the immunogenicity varies from structure to structure, probably owing to the presence or absence of motifs recognized by cellular RNA sensors. Ultimately, RNA nanostructures without immunogenicity, or modified to reduce immunogenicity,89 provide an excellent platform for assembling RNA-based therapeutics. These have predominantly been used to design tumor-targeting therapeutics for controlling gene and protein expression.
RNA nanostructures with intrinsic adjuvant activity as vaccine platforms
Given that immune cells have receptors to detect foreign nucleic acids, e.g. TLR3, TLR7, TLR8, and RLRs, that recognize ssRNA or dsRNA,98 RNA nanostructures can function as intrinsic adjuvants to induce an immune response.26 This feature can be utilized for vaccine design, i.e., RNA nanostructures functioning as self-adjuvanted platform to carry antigens of interest.94 Nanostructures (particularly between 10–100 nm) are ideal for drainage to and retention in lymph nodes (LNs), because they are less likely to diffuse to blood circulation for rapid elimination.99,100 Due to their ability to interact with and engulf antigens via various routes, DCs can take up nanoparticles and process the antigens carried by the nanoparticles for antigen cross-presentation.101,102However, there are some considerations for this vaccine platform. For example, due to the small size of RNA nanostructures (typically <50 nm),12 the payload capacity, especially for large molecules, such as proteins or synthetic long peptides, may be limited; however, some strategies, such as RCT, have been used to form larger RNA nanoparticles (250 nm) with a peptide linker to incorporate neopeptides.93 Additionally, payloads may require some protection for their in vivo delivery. To address this, RNA nanostructure design may need to be optimized to encapsulate antigens, such that the nanostructure not only increases the payload, but also protects it from the environment, as was demonstrated with DNA origami.103 Thus, RNA nanostructures with intrinsic adjuvant activity offer a robust platform to stimulate the immune system and design cancer vaccines. While previous strategies required the incorporation of immunostimulatory elements such as CpG DNA,86,93 the approach utilizing intrinsically immunogenic RNA origami nanostructures provides a rational and simpler platform to design nanovaccines against cancer, which can be extended to the construction of vaccines against infectious diseases or other targets.
Conclusion
RNA nanostructures represent a new and robust platform to construct RNA-based therapeutics. Their design enables precise assembly with functional RNAs, including mRNA, siRNA, ribozymes, aptamers, as well as small molecules drugs to create multi-functional RNA therapeutics that are stable for in vivo application. On the other hand, taking advantage of the intrinsic adjuvant activity of our RNA origami nanostructures can greatly simplify the design and construction of effective and personalized anti-cancer vaccines. Taken together, RNA nanostructures are promising platforms to design multifunctional therapeutics for a wide range of applications. Continuing research in RNA nanotechnology may lead to translational success for RNA nanostructures in the future.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by Arizona State University Women and Philanthropy Fund and Skysong Catalyst Seed Fund.References
- K. Kruger, P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling and T. R. Cech, Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena, Cell, 1982, 31(1), 147–157, DOI:10.1016/0092-8674(82)90414-7.
- D. Jasinski, F. Haque, D. W. Binzel and P. Guo, Advancement of the Emerging Field of RNA Nanotechnology, ACS Nano, 2017, 11(2), 1142–1164, DOI:10.1021/acsnano.6b05737.
- P. Guo, The Emerging Field of RNA Nanotechnology, Nat. Nanotechnol., 2010, 5(12), 833–842, DOI:10.1038/nnano.2010.231.
- N. R. Kallenbach, R. I. Ma and N. C. Seeman, An Immobile Nucleic Acid Junction Constructed from Oligonucleotides, Nature, 1983, 305(5937), 829–831, DOI:10.1038/305829a0.
- N. C. Seeman and N. R. Kallenbach, Design of Immobile Nucleic Acid Junctions, Biophys. J., 1983, 44(2), 201–209, DOI:10.1016/S0006-3495(83)84292-1.
- N. C. Seeman, Nucleic Acid Junctions and Lattices, J. Theor. Biol., 1982, 99(2), 237–247, DOI:10.1016/0022-5193(82)90002-9.
- E. Westhof, B. Masquida and L. Jaeger, RNA Tectonics: Towards RNA Design, Folding Des., 1996, 1(4), 78–88, DOI:10.1016/S1359-0278(96)00037-5.
- L. Jaeger, E. Westhof and N. B. Leontis, TectoRNA: Modular Assembly Units for the Construction of RNA Nano-Objects, Nucleic Acids Res., 2001, 29(2), 455–463, DOI:10.1093/NAR/29.2.455.
- A. Chworos, I. Severcan, A. Y. Koyfman, P. Weinkam, E. Oroudjev, H. G. Hansma and L. Jaeger, Building Programmable Jigsaw Puzzles with RNA, Science, 2004, 306(5704), 2068–2072, DOI:10.1126/SCIENCE.1104686/SUPPL_FILE/CHWOROS.SOM.SMALLER.PDF.
- K. A. Afonin, E. Bindewald, A. J. Yaghoubian, N. Voss, E. Jacovetty, B. A. Shapiro and L. Jaeger, In Vitro Assembly of Cubic RNA-Based Scaffolds Designed in Silico, Nat. Nanotechnol., 2010, 5(9), 676–682, DOI:10.1038/nnano.2010.160.
- C. Geary, P. W. K. Rothemund and E. S. Andersen, A Single-Stranded Architecture for Cotranscriptional Folding of RNA Nanostructures, Science, 2014, 345(6198), 799–804, DOI:10.1126/SCIENCE.1253920/SUPPL_FILE/GEARY.SM.REVISION.1.PDF.
- D. Han, X. Qi, C. Myhrvold, B. Wang, M. Dai, S. Jiang, M. Bates, Y. Liu, B. An, F. Zhang, H. Yan and P. Yin, Single-Stranded DNA and RNA Origami, Science, 2017, 358(6369), eaao2648, DOI:10.1126/SCIENCE.AAO2648.
- D. Liu, C. W. Geary, G. Chen, Y. Shao, M. Li, C. Mao, E. S. Andersen, J. A. Piccirilli, P. W. K. Rothemund and Y. Weizmann, Branched Kissing Loops for the Construction of Diverse RNA Homooligomeric Nanostructures, Nat. Chem., 2020, 12(3), 249–259, DOI:10.1038/s41557-019-0406-7.
- D. Shu, Y. Shu, F. Haque, S. Abdelmawla and P. Guo, Thermodynamically Stable RNA Three-Way Junction for Constructing Multifunctional Nanoparticles for Delivery of Therapeutics, Nat. Nanotechnol., 2011, 6(10), 658–667, DOI:10.1038/nnano.2011.105.
- X. Piao, H. Yin, S. Guo, H. Wang and P. Guo, RNA Nanotechnology to Solubilize Hydrophobic Antitumor Drug for Targeted Delivery, Adv. Sci., 2019, 6(22), 1900951, DOI:10.1002/ADVS.201900951.
- C. Geary, G. Grossi, E. K. S. McRae, P. W. K. Rothemund and E. S. Andersen, RNA Origami Design Tools Enable Cotranscriptional Folding of Kilobase-Sized Nanoscaffolds, Nat. Chem., 2021, 13(6), 549–558, DOI:10.1038/s41557-021-00679-1.
- M. Li, M. Zheng, S. Wu, C. Tian, D. Liu, Y. Weizmann, W. Jiang, G. Wang and C. Mao, In Vivo Production of RNA Nanostructures via Programmed Folding of Single-Stranded RNAs, Nat. Commun., 2018, 9(1), 1–9, DOI:10.1038/s41467-018-04652-4.
- E. Poppleton, N. Urbanek, T. Chakraborty, A. Griffo, L. Monari and K. Göpfrich, RNA Origami: Design, Simulation and Application, RNA Biol., 2023, 20(1), 510–524, DOI:10.1080/15476286.2023.2237719.
- L. Jaeger and A. Chworos, The Architectonics of Programmable RNA and DNA Nanostructures, Curr. Opin. Struct. Biol., 2006, 16(4), 531–543, DOI:10.1016/j.sbi.2006.07.001.
- E. K. S. McRae, H. Ø. Rasmussen, J. Liu, A. Bøggild, M. T. A. Nguyen, N. Sampedro Vallina, T. Boesen, J. S. Pedersen, G. Ren, C. Geary and E. S. Andersen, Structure, Folding and Flexibility of Co-Transcriptional RNA Origami, Nat. Nanotechnol., 2023, 18(7), 808–817, DOI:10.1038/s41565-023-01321-6.
- A. A. Yetisgin, S. Cetinel, M. Zuvin, A. Kosar and O. Kutlu, Therapeutic Nanoparticles and Their Targeted Delivery Applications, Molecules, 2020, 25(9), 2193, DOI:10.3390/MOLECULES25092193.
- G. Kara, G. A. Calin and B. Ozpolat, RNAi-Based Therapeutics and Tumor Targeted Delivery in Cancer, Adv. Drug Delivery Rev., 2022, 182, 114113, DOI:10.1016/J.ADDR.2022.114113.
- P. W. K. Rothemund, Folding DNA to Create Nanoscale Shapes and Patterns, Nature, 2006, 440(7082), 297–302, DOI:10.1038/nature04586.
- E. F. Khisamutdinov, D. L. Jasinski and P. Guo, RNA as a Boiling-Resistant Anionic Polymer Material to Build Robust Structures with Defined Shape and Stoichiometry, ACS Nano, 2014, 8(5), 4771–4781, DOI:10.1021/NN5006254/SUPPL_FILE/NN5006254_SI_001.PDF.
- D. Mathur and I. L. Medintz, The Growing Development of DNA Nanostructures for Potential Healthcare-Related Applications, Adv. Healthcare Mater., 2019, 8(9), 1801546, DOI:10.1002/ADHM.201801546.
- X. Qi, X. Liu, L. Matiski, R. D. V. Rodriguez, T. Yip, F. Zhang, S. Sokalingam, S. Jiang, L. Liu, H. Yan and Y. Chang, RNA Origami Nanostructures for Potent and Safe Anticancer Immunotherapy, ACS Nano, 2020, 14, 4740, DOI:10.1021/acsnano.0c00602.
- M. Zhao, R. Wang, K. Yang, Y. Jiang, Y. Peng, Y. Li, Z. Zhang, J. Ding and S. Shi, Nucleic Acid Nanoassembly-Enhanced RNA Therapeutics and Diagnosis, Acta Pharm. Sin. B, 2023, 13(3), 941, DOI:10.1016/J.APSB.2022.10.019.
- Y. Li and R. R. Breaker, Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2′-Hydroxyl Group, J. Am. Chem. Soc., 1999, 121(23), 5364–5372, DOI:10.1021/JA990592P/ASSET/IMAGES/MEDIUM/JA990592PE00015.GIF.
- L. Fang, L. Xiao, Y. W. Jun, Y. Onishi and E. T. Kool, Reversible 2′-OH Acylation Enhances RNA Stability, Nat. Chem., 2023, 15(9), 1296–1305, DOI:10.1038/s41557-023-01246-6.
- J. Liu, S. Guo, M. Cinier, L. S. Shlyakhtenko, Y. Shu, C. Chen, G. Shen and P. Guo, Fabrication of Stable and RNase-Resistant RNA Nanoparticles Active in Gearing the Nanomotors for Viral DNA Packaging, ACS Nano, 2011, 5(1), 237–246, DOI:10.1021/NN1024658/SUPPL_FILE/NN1024658_SI_001.PDF.
- S. M. Freier and K. H. Altmann, The Ups and Downs of Nucleic Acid Duplex Stability: Structure-Stability Studies on Chemically-Modified DNA:RNA Duplexes, Nucleic Acids Res., 1997, 25(22), 4429–4443, DOI:10.1093/NAR/25.22.4429.
- F. Eckstein, Phosphorothioates, Essential Components of Therapeutic Oligonucleotides, Nucleic Acid Ther., 2014, 24(6), 374–387, DOI:10.1089/nat.2014.0506.
- A. H. S. Hall, J. Wan, E. E. Shaughnessy, B. R. Shaw and K. A. Alexander, RNA Interference Using Boranophosphate SiRNAs: Structure–Activity Relationships, Nucleic Acids Res., 2004, 32(20), 5991–6000, DOI:10.1093/NAR/GKH936.
- A. R. Chandrasekaran, J. Mathivanan, P. Ebrahimi, J. Vilcapoma, A. A. Chen, K. Halvorsen and J. Sheng, Hybrid DNA/RNA Nanostructures with 2′-5′ Linkages, Nanoscale, 2020, 12(42), 21583–21590, 10.1039/D0NR05846G.
- H. N. Jung, S. Y. Lee, S. Lee, H. Youn and H. J. Im, Lipid Nanoparticles for Delivery of RNA Therapeutics: Current Status and the Role of in Vivo Imaging, Theranostics, 2022, 12(17), 7509–7531, DOI:10.7150/THNO.77259.
- Y. X. Lin, Y. Wang, S. Blake, M. Yu, L. Mei, H. Wang and J. Shi, RNA Nanotechnology-Mediated Cancer Immunotherapy, Theranostics, 2020, 10(1), 299, DOI:10.7150/THNO.35568.
- R. Kesharwani, P. Jaiswal, D. K. Patel and P. K. Yadav, Lipid-Based Drug Delivery System (LBDDS): An Emerging Paradigm to Enhance Oral Bioavailability of Poorly Soluble Drugs, J. Biomed. Mater. Dev., 2022, 1(2), 648–663, DOI:10.1007/s44174-022-00041-0.
- B. Begines, T. Ortiz, M. Pérez-Aranda, G. Martínez, M. Merinero, F. Argüelles-Arias and A. Alcudia, Polymeric Nanoparticles for Drug Delivery: Recent Developments and Future Prospects, Nanomaterials, 2020, 10(7), 1–41, DOI:10.3390/NANO10071403.
- S. J. Chen, RNA Folding: Conformational Statistics, Folding Kinetics, and Ion Electrostatics, Annu. Rev. Biophys., 2008, 37, 197–214, DOI:10.1146/ANNUREV.BIOPHYS.37.032807.125957.
- G. Zhu, Y. Liu, X. Yang, Y. H. Kim, H. Zhang, R. Jia, H. S. Liao, A. Jin, J. Lin, M. Aronova, R. Leapman, Z. Nie, G. Niu and X. Chen, DNA–Inorganic Hybrid Nanovaccine for Cancer Immunotherapy, Nanoscale, 2016, 8(12), 6684–6692, 10.1039/C5NR08821F.
- N. Ponnuswamy, M. M. C. Bastings, B. Nathwani, J. H. Ryu, L. Y. T. Chou, M. Vinther, W. A. Li, F. M. Anastassacos, D. J. Mooney and W. M. Shih, Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation, Nat. Commun., 2017, 8(1), 1–9, DOI:10.1038/ncomms15654.
- K. A. Afonin, D. J. Cieply and N. B. Leontis, Specific RNA Self-Assembly with Minimal Paranemic Motifs, J. Am. Chem. Soc., 2008, 130(1), 93–102, DOI:10.1021/JA071516M/SUPPL_FILE/JA071516MSI20071018_043644.PDF.
- M. Paliy, R. Melnik and B. A. Shapiro, Molecular Dynamics Study of the RNA Ring Nanostructure: A Phenomenon of Self-Stabilization, Phys. Biol., 2009, 6(4), 046003, DOI:10.1088/1478-3975/6/4/046003.
- J. D. Yesselman, D. Eiler, E. D. Carlson, M. R. Gotrik, A. E. d'Aquino, A. N. Ooms, W. Kladwang, P. D. Carlson, X. Shi, D. A. Costantino, D. Herschlag, J. B. Lucks, M. C. Jewett, J. S. Kieft and R. Das, Computational Design of Three-Dimensional RNA Structure and Function, Nat. Nanotechnol., 2019, 14(9), 866–873, DOI:10.1038/s41565-019-0517-8.
- L. Gold, Oligonucleotides as Research, Diagnostic, and Therapeutic Agents, J. Biol. Chem., 1995, 270(23), 13581–13584, DOI:10.1074/jbc.270.23.13581.
- J. Zhou and J. Rossi, Aptamers as Targeted Therapeutics: Current Potential and Challenges, Nat. Rev. Drug Discov., 2017, 16(3), 202, DOI:10.1038/NRD.2016.199.
- R. W. Carthew and E. J. Sontheimer, Origins and Mechanisms of MiRNAs and SiRNAs, Cell, 2009, 136(4), 641–655, DOI:10.1016/J.CELL.2009.01.035.
- B. Hu, L. Zhong, Y. Weng, L. Peng, Y. Huang, Y. Zhao and X. J. Liang, Therapeutic SiRNA: State of the Art, Signal Transduction Targeted Ther., 2020, 5(1), 1–25, DOI:10.1038/s41392-020-0207-x.
- S. L. Daubendiek, K. Ryan and E. T. Kool, Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase, J. Am. Chem. Soc., 1995, 117(29), 7818–7819 CrossRef CAS PubMed.
- J. B. Lee, J. Hong, D. K. Bonner, Z. Poon and P. T. Hammond, Self-Assembled RNA Interference Microsponges for Efficient SiRNA Delivery, Nat. Mater., 2012, 11(4), 316–322, DOI:10.1038/nmat3253.
- K. A. Afonin, M. Viard, A. Y. Koyfman, A. N. Martins, W. K. Kasprzak, M. Panigaj, R. Desai, A. Santhanam, W. W. Grabow, L. Jaeger, E. Heldman, J. Reiser, W. Chiu, E. O. Freed and B. A. Shapiro, Multifunctional RNA Nanoparticles, Nano Lett., 2014, 14(10), 5662–5671, DOI:10.1021/NL502385K/SUPPL_FILE/NL502385K_SI_001.PDF.
- H. C. Høiberg, S. M. Sparvath, V. L. Andersen, J. Kjems and E. S. Andersen, An RNA Origami Octahedron with Intrinsic SiRNAs for Potent Gene Knockdown, Biotechnol. J., 2019, 14(1), 1700634, DOI:10.1002/BIOT.201700634.
- D. Shu, H. Li, Y. Shu, G. Xiong, W. E. Carson, F. Haque, R. Xu and P. Guo, Systemic Delivery of Anti-MiRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology, ACS Nano, 2015, 9(10), 9731–9740, DOI:10.1021/ACSNANO.5B02471/ASSET/IMAGES/LARGE/NN-2015-024717_0005.JPEG.
- F. Pi, D. W. Binzel, T. J. Lee, Z. Li, M. Sun, P. Rychahou, H. Li, F. Haque, S. Wang, C. M. Croce, B. Guo, B. M. Evers and P. Guo, Nanoparticle Orientation to Control RNA Loading and Ligand Display on Extracellular Vesicles for Cancer Regression, Nat. Nanotechnol., 2017, 13(1), 82–89, DOI:10.1038/s41565-017-0012-z.
- Y. Hoon Roh, J. Z. Deng, E. C. Dreaden, J. H. Park, D. Soo Yun, K. E. Shopsowitz and P. T. Hammond, A Multi-RNAi Microsponge Platform for Simultaneous Controlled Delivery of Multiple Small Interfering RNAs, Angew. Chem., Int. Ed., 2016, 55(10), 3347–3351, DOI:10.1002/ANIE.201508978.
- K. A. Afonin, M. Kireeva, W. W. Grabow, M. Kashlev, L. Jaeger and B. A. Shapiro, Co-Transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized with SiRNAs, Nano Lett., 2012, 12(10), 5192–5195, DOI:10.1021/NL302302E/SUPPL_FILE/NL302302E_SI_001.PDF.
- P. Zakrevsky, W. K. Kasprzak, W. F. Heinz, W. Wu, H. Khant, E. Bindewald, N. Dorjsuren, E. A. Fields, N. De Val, L. Jaeger and B. A. Shapiro, Truncated Tetrahedral RNA Nanostructures Exhibit Enhanced Features for Delivery of RNAi Substrates, Nanoscale, 2020, 12(4), 2555–2568, 10.1039/C9NR08197F.
- D. Cui, C. Zhang, B. Liu, Y. Shu, T. Du, D. Shu, K. Wang, F. Dai, Y. Liu, C. Li, F. Pan, Y. Yang, J. Ni, H. Li, B. Brand-Saberi and P. Guo, Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying Both Ligand and SiRNA, Sci. Rep., 2015, 5(1), 1–14, DOI:10.1038/srep10726.
- Y. Xu, L. Pang, H. Wang, C. Xu, H. Shah, P. Guo, D. Shu and S. Y. Qian, Specific Delivery of Delta-5-Desaturase SiRNA via RNA Nanoparticles Supplemented with Dihomo-γ-Linolenic Acid for Colon Cancer Suppression, Redox Biol., 2019, 21, 101085, DOI:10.1016/J.REDOX.2018.101085.
- L. Zhang, C. Mu, T. Zhang, Y. Wang, Y. Wang, L. Fan, C. Liu, H. Chen, J. Shen, K. Wei and H. Li, Systemic Delivery of Aptamer-Conjugated XBP1 SiRNA Nanoparticles for Efficient Suppression of HER2+ Breast Cancer, ACS Appl. Mater. Interfaces, 2020, 12(29), 32360–32371, DOI:10.1021/ACSAMI.0C07353/ASSET/IMAGES/LARGE/AM0C07353_0005.JPEG.
- S. Obad, C. O. Dos Santos, A. Petri, M. Heidenblad, O. Broom, C. Ruse, C. Fu, M. Lindow, J. Stenvang, E. M. Straarup, H. F. Hansen, T. Koch, D. Pappin, G. J. Hannon and S. Kauppinen, Silencing of MicroRNA Families by Seed-Targeting Tiny LNAs, Nat. Genet., 2011, 43(4), 371–378, DOI:10.1038/ng.786.
- Z. Zheng, Z. Li, C. Xu, B. Guo and P. Guo, Folate-Displaying Exosome Mediated Cytosolic Delivery of SiRNA Avoiding Endosome Trapping, J. Controlled Release, 2019, 311–312, 43–49, DOI:10.1016/J.JCONREL.2019.08.021.
- P. S. Kowalski, A. Rudra, L. Miao and D. G. Anderson, Delivering the Messenger: Advances in Technologies for Therapeutic MRNA Delivery, Mol. Ther., 2019, 27(4), 710–728, DOI:10.1016/J.YMTHE.2019.02.012.
- K. A. Hajj and K. A. Whitehead, Tools for Translation: Non-Viral Materials for Therapeutic MRNA Delivery, Nat. Rev. Mater., 2017, 2(10), 1–17, DOI:10.1038/natrevmats.2017.56.
- M. Hu, C. Feng, Q. Yuan, C. Liu, B. Ge, F. Sun and X. Zhu, Lantern-Shaped Flexible RNA Origami for Smad4 MRNA Delivery and Growth Suppression of Colorectal Cancer, Nat. Commun., 2023, 14(1), 1–15, DOI:10.1038/s41467-023-37020-y.
- R. Micura and C. Höbartner, Fundamental Studies of Functional Nucleic Acids:Aptamers, Riboswitches, Ribozymes and DNAzymes, Chem. Soc. Rev., 2020, 49, 7331–7353, 10.1039/d0cs00617c.
- S. L. Heilman-Miller and S. A. Woodson, Effect of Transcription on Folding of the Tetrahymena Ribozyme, RNA, 2003, 9(6), 722–733, DOI:10.1261/RNA.5200903.
- Y. Li, A. Arce, T. Lucci, R. A. Rasmussen and J. B. Lucks, Dynamic RNA Synthetic Biology: New Principles, Practices and Potential, RNA Biol., 2023, 20(1), 817–829, DOI:10.1080/15476286.2023.2269508.
- B. Dönmüş, S. Ünal, F. C. Kirmizitaş and N. Türkoğlu Laçin, Virus-Associated Ribozymes and Nano Carriers against COVID-19, Artif. Cells, Nanomed., Biotechnol., 2021, 49(1), 204–218, DOI:10.1080/21691401.2021.1890103.
- S. Hoeprich, Q. Zhou, S. Guo, D. Shu, G. Qi, Y. Wang and P. Guo, Bacterial Virus Phi29 PRNA as a Hammerhead Ribozyme Escort to Destroy Hepatitis B Virus, Gene Ther., 2003, 10(15), 1258–1267, DOI:10.1038/sj.gt.3302002.
- H. Li, K. Zhang, F. Pi, S. Guo, L. Shlyakhtenko, W. Chiu, D. Shu and P. Guo, Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting, Adv. Mater., 2016, 28(34), 7501–7507, DOI:10.1002/ADMA.201601976.
- D. L. Jasinski, D. W. Binzel and P. Guo, One-Pot Production of RNA Nanoparticles via Automated Processing and Self-Assembly, ACS Nano, 2019, 13(4), 4603–4612, DOI:10.1021/ACSNANO.9B00649/ASSET/IMAGES/LARGE/NN-2019-00649A_0007.JPEG.
- D. L. Jasinski, E. F. Khisamutdinov, Y. L. Lyubchenko and P. Guo, Physicochemically Tunable Polyfunctionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties, ACS Nano, 2014, 8(8), 7620–7629, DOI:10.1021/NN502160S/SUPPL_FILE/NN502160S_SI_001.PDF.
- B. C. Wong, U. Shahid, H. S. Tan, B. C. Wong, U. Shahid and H. S. Tan, Ribozymes as Therapeutic Agents against Infectious Diseases, RNA Therapeutics - History, Design, Manufacturing, and Applications, 2022, DOI:10.5772/INTECHOPEN.107141.
- A. J. Trujillo, J. M. Myers, Z. S. Fayazi, M. C. Butler and J. M. Sullivan, A Discovery with Potential to Revitalize Hammerhead Ribozyme for Treatment of Inherited Retinal Degenerations, Adv. Exp. Med. Biol., 2019, 1185, 124, DOI:10.1007/978-3-030-27378-1_20.
- X. Wang, C. Li, X. Gao, J. Wang and X. Liang, Preparation of Small RNAs Using Rolling Circle Transcription and Site-Specific RNA Disconnection, Mol. Ther. Nucleic Acids, 2015, 4(1), e215, DOI:10.1038/mtna.2014.66.
- G. Pothoulakis, M. T. A. Nguyen and E. S. Andersen, Utilizing RNA Origami Scaffolds in Saccharomyces Cerevisiae for DCas9-Mediated Transcriptional Control, Nucleic Acids Res., 2022, 50(12), 7176–7187, DOI:10.1093/NAR/GKAC470.
- E. Martino, S. Della Volpe, E. Terribile, E. Benetti, M. Sakaj, A. Centamore, A. Sala and S. Collina, The Long Story of Camptothecin: From Traditional Medicine to Drugs, Bioorg. Med. Chem. Lett., 2017, 27(4), 701–707, DOI:10.1016/J.BMCL.2016.12.085.
- S. Guo, M. Vieweger, K. Zhang, H. Yin, H. Wang, X. Li, S. Li, S. Hu, A. Sparreboom, B. M. Evers, Y. Dong, W. Chiu and P. Guo, Ultra-Thermostable RNA Nanoparticles for Solubilizing and High-Yield Loading of Paclitaxel for Breast Cancer Therapy, Nat. Commun., 2020, 11(1), 1–11, DOI:10.1038/s41467-020-14780-5.
- X. Li, K. Jin, T.-C. Cheng, Y.-C. Liao, W.-J. Lee, A. S. Bhullar, L.-C. Chen, P. Rychahou, M. A. Phelps, Y. S. Ho and P. Guo, RNA Four-Way Junction (4WJ) for Spontaneous Cancer-Targeting, Effective Tumor-Regression, Metastasis Suppression, Fast Renal Excretion and Undetectable Toxicity, Biomaterials, 2024, 305, 122432, DOI:10.1016/J.BIOMATERIALS.2023.122432.
- S. Kruger, M. Ilmer, S. Kobold, B. L. Cadilha, S. Endres, S. Ormanns, G. Schuebbe, B. W. Renz, J. G. D'Haese, H. Schloesser, V. Heinemann, M. Subklewe, S. Boeck, J. Werner and M. Von Bergwelt-Baildon, Advances in Cancer Immunotherapy 2019 - Latest Trends, J. Exp. Clin. Cancer Res., 2019, 38(1), 1–11, DOI:10.1186/S13046-019-1266-0/FIGURES/2.
- R. E. Hollingsworth and K. Jansen, Turning the Corner on Therapeutic Cancer Vaccines, Vaccines, 2019, 4(1), 1–10, DOI:10.1038/s41541-019-0103-y.
- T. A. Waldmann, Cytokines in Cancer Immunotherapy, Cold Spring Harbor Perspect. Biol., 2018, 10(12), a028472, DOI:10.1101/CSHPERSPECT.A028472.
- L. Liu and J. Chen, Therapeutic Antibodies for Precise Cancer Immunotherapy: Current and Future Perspectives, Med. Rev., 2022, 2(6), 555, DOI:10.1515/MR-2022-0033.
- T. Zhao, Y. Cai, Y. Jiang, X. He, Y. Wei, Y. Yu and X. Tian, Vaccine Adjuvants: Mechanisms and Platforms, Signal Transduction Targeted Ther., 2023, 8(1), 1–24, DOI:10.1038/s41392-023-01557-7.
- E. F. Khisamutdinov, H. Li, D. L. Jasinski, J. Chen, J. Fu and P. Guo, Enhancing Immunomodulation on Innate Immunity by Shape Transition among RNA Triangle, Square and Pentagon Nanovehicles, Nucleic Acids Res., 2014, 42(15), 9996–10004, DOI:10.1093/NAR/GKU516.
- E. F. Khisamutdinov, D. L. Jasinski, H. Li, K. Zhang, W. Chiu and P. Guo, Fabrication of RNA 3D Nanoprism for Loading and Protection of Small RNAs and Model Drugs, Adv. Mater., 2016, 28(45), 10087, DOI:10.1002/ADMA.201603180.
- K. Dai, Y. Xu, Y. Yang, J. Shen, X. Liu, X. Tu, L. Yu, X. Qi, J. Li, L. Wang, X. Zuo, Y. Liu, H. Yan, C. Fan and G. Yao, Edge Length-Programmed Single-Stranded RNA Origami for Predictive Innate Immune Activation and Therapy, J. Am. Chem. Soc., 2023, 145(31), 17112–17124, DOI:10.1021/JACS.3C03477/ASSET/IMAGES/LARGE/JA3C03477_0007.JPEG.
- K. Dai, C. Gong, Y. Xu, F. Ding, X. Qi, X. Tu, L. Yu, X. Liu, J. Li, C. Fan, H. Yan and G. Yao, Single-Stranded RNA Origami-Based Epigenetic Immunomodulation, Nano Lett., 2023, 23(15), 7188–7196, DOI:10.1021/ACS.NANOLETT.3C02185/ASSET/IMAGES/LARGE/NL3C02185_0004.JPEG.
- L. Buonaguro and M. Tagliamonte, Peptide-Based Vaccine for Cancer Therapies, Front. Immunol., 2023, 14, 1210044, DOI:10.3389/FIMMU.2023.1210044/BIBTEX.
- D. Chatzikleanthous, D. T. O'Hagan and R. Adamo, Lipid-Based Nanoparticles for Delivery of Vaccine Adjuvants and Antigens: Toward Multicomponent Vaccines, Mol. Pharm., 2021, 18(8), 2867–2888, DOI:10.1021/ACS.MOLPHARMACEUT.1C00447/ASSET/IMAGES/LARGE/MP1C00447_0004.JPEG.
- R. J. Nevagi, M. Skwarczynski and I. Toth, Polymers for Subunit Vaccine Delivery, Eur. Polym. J., 2019, 114, 397–410, DOI:10.1016/j.eurpolymj.2019.03.009.
- G. Zhu, L. Mei, H. D. Vishwasrao, O. Jacobson, Z. Wang, Y. Liu, B. C. Yung, X. Fu, A. Jin, G. Niu, Q. Wang, F. Zhang, H. Shroff and X. Chen, Intertwining DNA-RNA Nanocapsules Loaded with Tumor Neoantigens as Synergistic Nanovaccines for Cancer Immunotherapy, Nat. Commun., 2017, 8(1), 1–13, DOI:10.1038/s41467-017-01386-7.
- T. Yip, X. Qi, H. Yan and Y. Chang, RNA Origami Functions as a Self-Adjuvanted Nanovaccine Platform for Cancer Immunotherapy, ACS Nano, 2024, 18(5), 4056–4067, DOI:10.1021/ACSNANO.3C07284.
- Y. Wang, E. Kim, Y. Lin, N. Kim, W. Kit-Anan, S. Gopal, S. Agarwal, P. D. Howes and M. M. Stevens, Rolling Circle Transcription-Amplified Hierarchically Structured Organic-Inorganic Hybrid RNA Flowers for Enzyme Immobilization, ACS Appl. Mater. Interfaces, 2019, 11(26), 22932–22940, DOI:10.1021/ACSAMI.9B04663/ASSET/IMAGES/LARGE/AM-2019-04663X_0006.JPEG.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Cellular Uptake of Nanoparticles: Journey inside the Cell, Chem. Soc. Rev., 2017, 46(14), 4218–4244, 10.1039/C6CS00636A.
- S. Guo, H. Li, M. Ma, J. Fu, Y. Dong and P. Guo, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles, Mol. Ther. Nucleic Acids, 2017, 9, 399–408, DOI:10.1016/j.omtn.2017.10.010.
- S. G. Reed, M. Tomai and M. J. Gale, New Horizons in Adjuvants for Vaccine Development, Curr. Opin. Immunol., 2020, 65, 97–101, DOI:10.1016/J.COI.2020.08.008.
- P. Yousefpour, K. Ni and D. J. Irvine, Targeted Modulation of Immune Cells and Tissues Using Engineered Biomaterials, Nat. Rev. Bioeng., 2023, 1(2), 107–124, DOI:10.1038/s44222-022-00016-2.
- G. P. Howard, G. Verma, X. Ke, W. M. Thayer, T. Hamerly, V. K. Baxter, J. E. Lee, R. R. Dinglasan and H. Q. Mao, Critical Size Limit of Biodegradable Nanoparticles for Enhanced Lymph Node Trafficking and Paracortex Penetration, Nano Res., 2019, 12(4), 837–844, DOI:10.1007/S12274-019-2301-3/METRICS.
- J. P. Lim and P. A. Gleeson, Macropinocytosis: An Endocytic Pathway for Internalising Large Gulps, Immunol. Cell Biol., 2011, 89(8), 836–843, DOI:10.1038/ICB.2011.20.
- F. M. Cruz, A. Chan and K. L. Rock, Pathways of MHC I Cross-Presentation of Exogenous Antigens, Semin. Immunol., 2023, 66, 101729, DOI:10.1016/J.SMIM.2023.101729.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, A DNA Nanodevice-Based Vaccine for Cancer Immunotherapy, Nat. Mater., 2020, 20(3), 421–430, DOI:10.1038/s41563-020-0793-6.
| This journal is © The Royal Society of Chemistry 2024 |
