Open Access Article
Open Access ArticleMorpholine-modified Ru-based agents with multiple antibacterial mechanisms as metalloantibiotic candidates against Staphylococcus aureus infection†
Shijie Lin‡
a,
Yun Song‡b,
Yajuan Sun‡b,
Wenjing Linb,
Guangying Yub,
Xiangwen Liao b and
Qiang Yang
b and
Qiang Yang *c
*c
aDepartment of Pharmacy, Hainan General Hospital (Hainan Affiliated Hospital of Hainan Medical University), Haikou, 570311, China
bJiangxi Provincial Key Laboratory of Drug Design and Evaluation, School of Pharmacy, Jiangxi Science & Technology Normal University, Nanchang, 330013, China
cDepartment of Clinical Pharmacy, Hainan Cancer Hospital, Haikou, 570100, China. E-mail: 18789551643@163.com
First published on 24th June 2024
Abstract
Multidrug-resistant bacteria resulting from the abuse and overuse of antibiotics have become a huge crisis in global public health security. Therefore, it is urgently needed to develop new antibacterial drugs with unique mechanisms of action. As a versatile moiety, morpholine has been widely employed to enhance the potency of numerous bioactive molecules. In this study, a series of ruthenium-based antibacterial agents modified with the morpholine moiety were designed and characterized, aiming to obtain a promising metalloantibiotic with a multitarget mechanism. Antibacterial activity screening demonstrated that the most active complex Ru(II)-3 exhibited the strongest potency against Staphylococcus aureus (S. aureus) with an MIC value of only 0.78 μg mL−1, which is better than most clinically used antibiotics. Notably, Ru(II)-3 not only possessed excellent bactericidal efficacy, but could also overcome bacterial resistance. Importantly, Ru(II)-3 very efficiently removed biofilms produced by bacteria, inhibited the secretion of bacterial exotoxins, and enhanced the activity of many existing antibiotics. The results of mechanism studies confirmed that Ru(II)-3 could destroy the bacterial membrane and induce ROS production in bacteria. Furthermore, animal infection models confirmed that Ru(II)-3 showed significant anti-infective activity in vivo. Overall, this work demonstrated that a morpholine-modified ruthenium-based agent is a promising antibiotic candidate in tackling the crisis of drug-resistant bacteria.
1. Introduction
Nowadays, multidrug-resistant bacteria pose a huge threat to global health, with an increasing number of people losing their lives every year. Among them, Staphylococcus aureus (S. aureus) is one of the most common Gram-positive bacteria, causing a variety of infections, such as joint infection, skin infection, and bacteremia, ranging from sub-acute superficial skin infection to life-threatening septicemia.1–4 It has been reported that the notorious “superbug”, methicillin-resistant S. aureus (MRSA), resulted in more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths and 3.5 million disabilities in 2019.5,6 Moreover, the outbreak of COVID-19 (coronavirus disease) has triggered a massive use of antibiotics, further increasing antimicrobial resistance and worsening the problem.7,8 Therefore, it is urgently needed to develop novel antibacterial drugs, especially those with unique mechanisms of action, to combat antibiotic-resistant bacteria.
000 deaths and 3.5 million disabilities in 2019.5,6 Moreover, the outbreak of COVID-19 (coronavirus disease) has triggered a massive use of antibiotics, further increasing antimicrobial resistance and worsening the problem.7,8 Therefore, it is urgently needed to develop novel antibacterial drugs, especially those with unique mechanisms of action, to combat antibiotic-resistant bacteria.
As a representative transition metal, ruthenium (Ru) has aroused great interest because of its excellent biological activity. Its rich photophysical and chemical properties are very suitable for drug development. Moreover, the similarity in the action of ruthenium to that of iron not only reduces the toxicity of Ru-based drugs, but also endows it with different oxidation states in different physiological states.9–11 These advantages make Ru-based agents modified with a bioactive moiety exhibit robust antibacterial efficacy and effectively prevent bacteria from developing resistance.12–16
Morpholine is a natural six-membered ring, which has been widely exploited to enhance the potency of numerous bioactive molecules because of its superior physicochemical, biochemical, and metabolic properties.17 It was reported that the weakly basic N in morpholine could enhance solubility, whereas the oxygen atom forms hydrogen bonding, which enhances its potency against the target protein by establishing hydrophobic interactions.18 Notably, morpholine-modified agents exhibited a wide range of biological properties, including anti-inflammatory, antioxidant, antibacterial, and antitumor activities.19,20 In various antibiotics, the morpholine moiety also has an important effect on their antibacterial activity (Scheme 1A).17
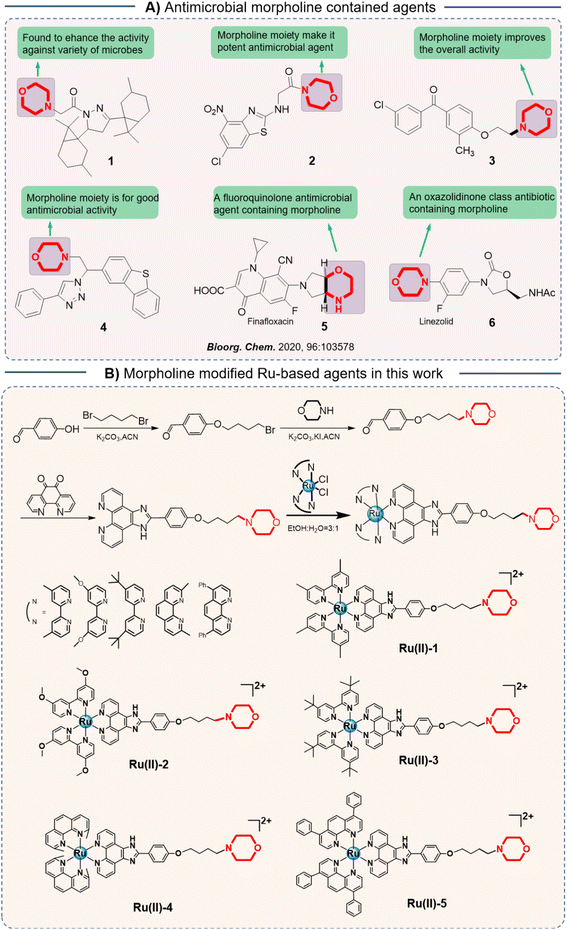 | ||
| Scheme 1 (A) Antimicrobial morpholine contained agtents reported before; (B) the design of five Ru-based agents modified with morpholine and their synthesis routes. | ||
Herein, in order to screen a promising metalloantibiotic with a multitarget mechanism and robust potency, we introduced a morpholine moiety into an Ru-based structure and then obtained a series of novel Ru-based antibacterial agents (Scheme 1B). Subsequently, their antibacterial activity against S. aureus was studied in vitro. Then, the most effective agent, Ru(II)-3, was selected for the antibacterial mechanism study. Furthermore, the ability of Ru(II)-3 to clear bacterial biofilms was confirmed, which may help overcome bacterial resistance. Notably, the anti-toxin efficacy and synergistic activity of Ru(II)-3 with existing antibiotics were explored, too. At last, its toxicity and anti-infective potency in vivo were verified using animal infection models.
2. Results and discussion
2.1 Antibacterial activity of morpholine-modified Ru-based agents
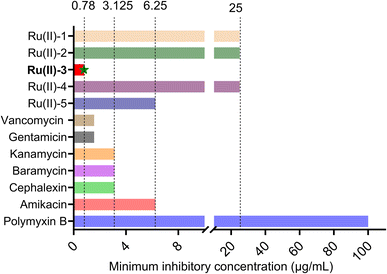
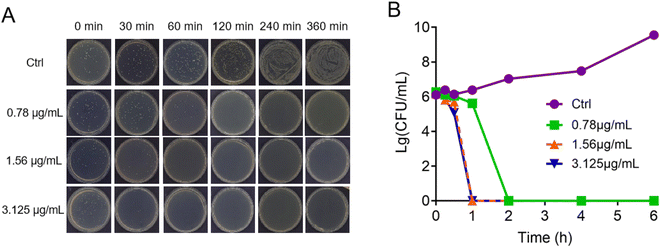
The morpholine moiety was linked to the Ru-based structure via long-chain alkyl groups and the detailed synthesis routes are shown in Scheme 1B. All the complexes were characterized by 1H-NMR, 13C-NMR and HRMS spectroscopy. In addition, their purity was confirmed by HPLC (ESI†). The characterization data are presented in the ESI.† To begin, the minimum inhibitory concentration (MIC) values of five compounds against S. aureus were explored and the data are shown in Fig. 1. Five morpholine-modified ruthenium complexes showed good antibacterial activity against S. aureus, with the MIC values ranging from 0.78 to 25 μg mL−1. Importantly, the best candidate agent, namely, Ru(II)-3, exhibited the strongest antibacterial effect, and has an MIC value of only 0.78 μg mL−1. This potency is much better than most clinically common antibiotics, too (Fig. 1). In addition, Ru(II)-3 also exhibited robust effectiveness against drug-resistant MRSA isolated from the clinic (Table S1†). Furthermore, the rapid bactericidal efficacy, a feature that helps in efficiently controlling acute bacterial infections, of Ru(II)-3 was investigated through a time–kill curve analysis. Notably, Ru(II)-3 killed more than 99% of bacteria in just 2 h at 0.78 μg mL−1. When the concentration was increased to 1.56 μg mL−1, the time required was only 1 h (Fig. 2). At last, the antibacterial effect of five agents on S. aureus was directly monitored through a bacterial growth curve. As exhibited in Fig. 3, the growth of S. aureus was completely inhibited by the five tested agents at the MIC. Collectively, these data suggest that these morpholine-modified Ru-based agents possess robust antibacterial potency in vitro.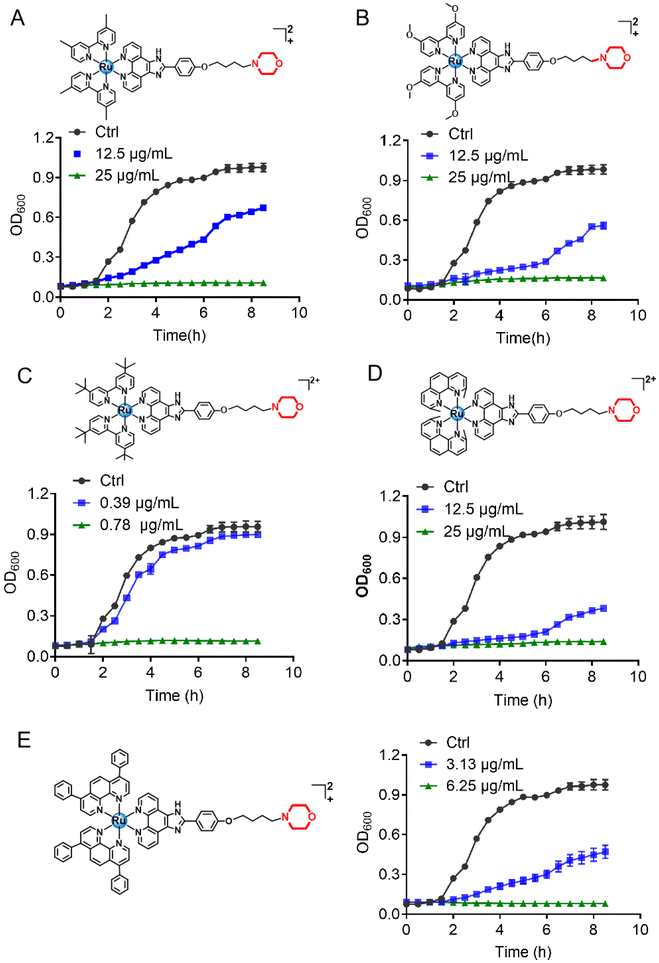 | ||
| Fig. 3 Growth curve of S. aureus in the presence of Ru-based compounds. (A) Ru(II)-1; (B) Ru(II)-2; (C) Ru(II)-3; (D) Ru(II)-4; (E) Ru(II)-5. | ||
2.2 Evaluating the frequencies of drug resistance
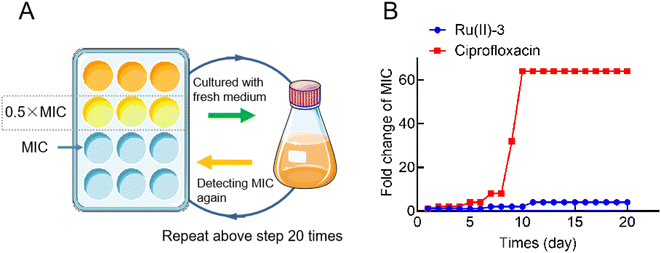
The main reason why bacterial infections have become increasingly difficult to deal with is that bacteria can easily develop resistance to antibiotics because of which clinicians are left with few weapons to deal with such multidrug-resistant bacteria. Hence, novel antibacterial drugs with unique mechanisms of action, for which it is difficult for bacteria to develop resistance, are urgently needed. Therefore, in order to verify whether complex Ru(II)-3 can overcome bacterial resistance, a resistance development assay was employed.21 As shown in Fig. 4, after continuously exposing to ciprofloxacin (0.5 MIC) for 20 generations, S. aureus developed significant resistance since the MIC of ciprofloxacin against it increased by 64 times. However, under the same condition, the MIC values of Ru(II)-3 against S. aureus only increased by 4 times, suggesting that Ru(II)-3 could not easily produce drug resistance in S. aureus.2.3 Inhibition of hemolytic toxin secretion
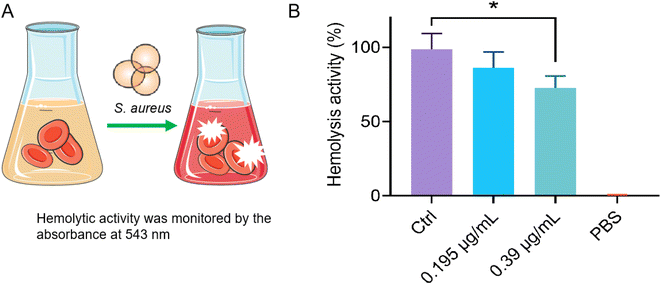
It has been reported that the exotoxin of S. aureus is essential for it to spread through tissues and survive under host immune-cell clearance. Therefore, the ability of antibacterial agents to inhibit the secretion of toxins is very beneficial for them to remove bacteria in vivo more effectively.22 Here, in order to further study the inhibitory effect of Ru(II)-3 on the secretion of α-toxin from S. aureus, a hemolysis test using rabbit blood cells was performed. In brief, S. aureus was co-incubated with a sub-inhibitory concentration of Ru(II)-3; then, the bacterial supernatant was collected and incubated with rabbit blood at 37 °C for 30 min. As shown in Fig. 5, after co-incubating with Ru(II)-3, the hemolytic effect of the bacterial supernatant on blood cells was obviously broken, indicating that Ru(II)-3 effectively inhibited α-toxin production from S. aureus. It is noteworthy that Ru(II)-3 itself did not obviously destroy red blood cells even at 250 μg mL−1 (Fig. S29†), suggesting its good biocompatibility.2.4 Biofilm inhibition by Ru(II)-3
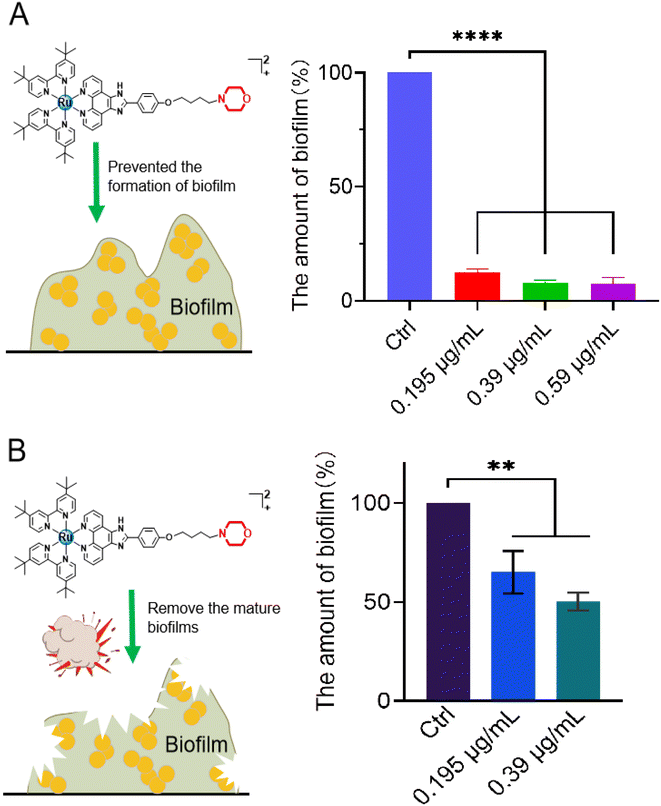
As a defense barrier, the formation of a biofilm greatly promotes the resistance of bacteria to antibiotics; therefore, the effective removal of bacterial biofilms urgently needs to be addressed. Consequently, the anti-biofilm potency of Ru(II)-3 was explored. To begin, S. aureus was co-incubated with the sub-inhibitory concentration of Ru(II)-3 at 37 °C for 48 h, and then the formation of a biofilm was monitored using a crystal violet staining assay. As shown in Fig. 6A, only at 0.195 μg mL−1 (0.25 MIC), Ru(II)-3 remarkably prevented the formation of a biofilm in S. aureus, with an inhibition rate above 90%. Next, the destructive effect of Ru(II)-3 on the already formed biofilm was investigated. The data presented in Fig. 6B clearly indicate that more than 50% of the mature biofilm was eradicated in the presence of 0.39 μg mL−1 Ru(II)-3 (0.5 MIC). Taken together, these results presented here demonstrated that Ru(II)-3 could efficiently inhibit bacterial biofilm formation. More importantly, it also has a powerful ability to remove mature biofilms. Of course, this ability of Ru(II)-3 might also explain, to some extent, why S. aureus could not easily develop resistance to it.2.5 Synergistic activity of Ru(II)-3 with antibiotics
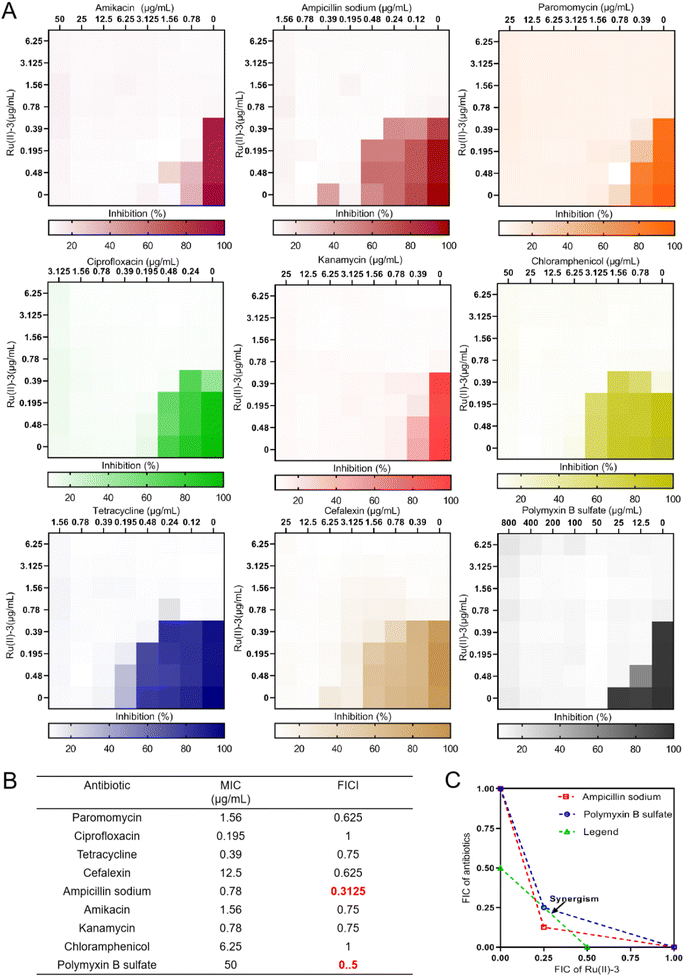
In recent years, the development of antimicrobial adjuvants—agents that can increase the antibiotic potency and even reverse drug resistance—has been an alternative approach to combat antibiotic-resistant bacteria.23 In order to investigate the synergistic activity of Ru(II)-3 with antibiotics, the checkerboard method was employed. At first, nine clinically used antibiotics from different categories were selected. Then heat-map data were drawn and the corresponding fractional inhibitory concentration index (FICI; a value of ≤0.5 suggests synergism) was calculated when a combination of these antibiotics was used with Ru(II)-3 against S. aureus. As shown in Fig. 7, Ru(II)-3 significantly enhanced the antibacterial efficacy of two antibiotics against S. aureus, namely, ampicillin (FICI, 0.31) and polymyxin B (FICI, 0.5), suggesting that Ru(II)-3 could effectively enhance the antibacterial activity of some clinically used antibiotics.2.6 Antibacterial mechanism exploration
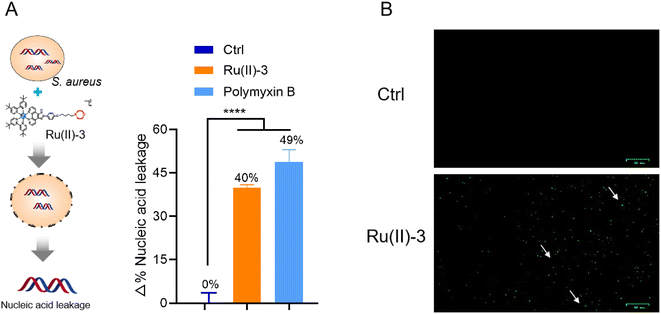
The advantage of Ru-based agents is that they exhibit multitarget actions, such as bacterial membrane destruction and reactive oxygen species (ROS) induction, an antimicrobial mechanism different from that of traditional antibiotics.16 Therefore, the destruction effect of Ru(II)-3 on the bacterial membrane was first verified by measuring the leakage of nucleic acid. As shown in Fig. 8A, after incubation with Ru(II)-3 or polymyxin B for 2 h, the nucleic acid leakage from S. aureus increased by 40% and 49%, respectively, indicating a breakdown of the bacterial cell membrane. It has been reported that the breakage of the bacterial membrane can lead to its depolarization.24 Therefore, 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)], a dye for depolarization monitoring, was employed to further explore the effect of Ru(II)-3 on the bacterial membrane potential. As shown in Fig. 8B, a strong green fluorescence appeared when S. aureus was incubated with Ru(II)-3 for 2 h, indicating the membrane depolarization. Next, PI and DAPI, two common fluorescence probes for bacterial-cell staining,25 were employed to further monitor the damage effect of Ru(II)-3 on the bacterial membrane. Normally, if some bacteria showed red fluorescence, it indicates that their cell membrane is damaged. As shown in Fig. 9, after treatment with Ru(II)-3 for 2 h, it could be clearly observed that some bacteria were stained with red fluorescence in the presence of two dyes. Notably, a similar phenomenon was also observed in the vancomycin-treated group. This result indicated that Ru(II)-3 could destroy the integrity of bacterial membranes. Finally, the membrane destruction effect of Ru(II)-3 was further visually detected by a scanning electron microscope (SEM). As shown in Fig. 10, the surface of S. aureus in the PBS-treated group was complete and smooth. However, after treatment with Ru(II)-3, some bacteria were seriously ruptured. Collectively, these results clearly suggested that Ru(II)-3 could cause the destruction of the bacterial membrane, thereby achieving the bactericidal effect.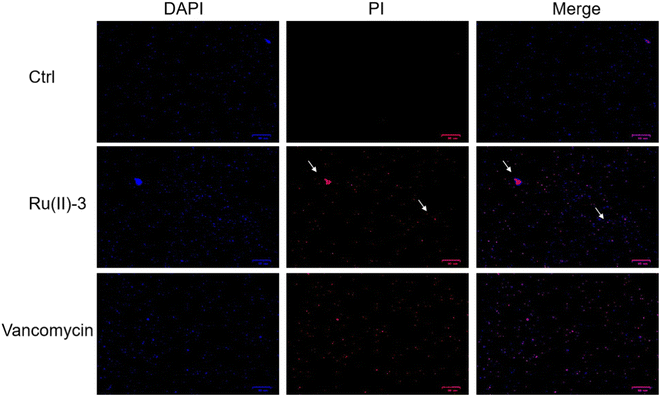 | ||
| Fig. 9 DAPI and PI staining results of S. aureus after treatment with PBS, Ru(II)-3 or vancomycin at 1 MIC. All the images are 25 μm in scale. | ||
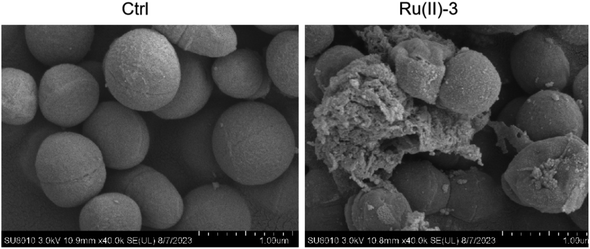 | ||
| Fig. 10 Scanning electron microscopy (SEM) images of S. aureus after treatment with 0.78 μg mL−1 Ru(II)-3. | ||
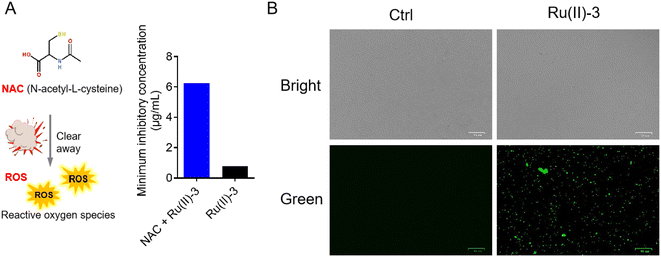
Apart from membrane destruction, ROS production is also one of the most important ways for some Ru-based antibacterial agents to kill bacteria.16 Next, to further confirm the involvement of ROS in the bactericidal process of Ru(II)-3, we evaluated the ROS production in S. aureus using two probes, namely, N-acetyl-cysteine (NAC) and 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA). Notably, NAC is an ROS scavenger, whereas DCFH-DA is a green fluorescent probe for intracellular ROS.26 First, the effect of NAC on the antibacterial efficacy of Ru(II)-3 against S. aureus was investigated. As shown in Fig. 11A, the MIC value of Ru(II)-3 increased by eight times in the presence of ROS scavenger (NAC), suggesting that the clearance of ROS reduced its bactericidal efficacy. Second, the generation of ROS in bacteria was visualized using DCFH-DA.27 As shown in Fig. 11B, no green fluorescent signals were detected in the PBS-treated group. However, after co-incubation with Ru(II)-3, lots of green fluorescent signals were observed in S. aureus, indicating the generation of ROS. Taken together, it could be concluded that Ru(II)-3 mainly killed bacteria in two ways: induce ROS formation and damage the bacterial membrane.
2.7 Antibacterial efficacy in vivo
At last, given that Ru(II)-3 showed excellent antibacterial potency in vitro, we further explored its ant-infective efficacy in vivo. First, the toxicity of Ru(II)-3 was verified using Galleria mellonella (G. mellonella) larva. In brief, G. mellonella was injected with a high dose of Ru(II)-3 and their survival rate was recorded within 7 days. The results demonstrated that complex Ru(II)-3 has pretty low toxicity in vivo, similar to two clinically used antibiotics, vancomycin and polymyxin B. As shown in Fig. 12, even though the injected dose of Ru(II)-3 reached 200 mg kg−1, above 90% of G. mellonella survived.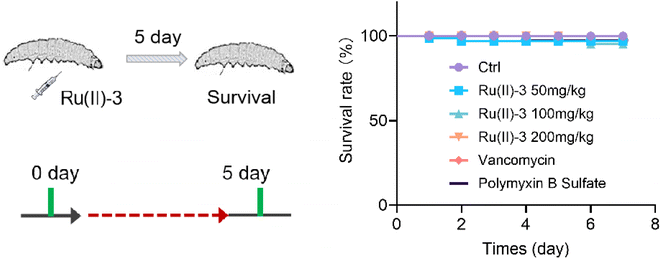 | ||
| Fig. 12 Toxicity of Ru(ii)-3 in vivo. The survival rate of G. mellonella wax worms treated with a high dose of Ru(II)-3, vancomycin, or polymyxin B. | ||
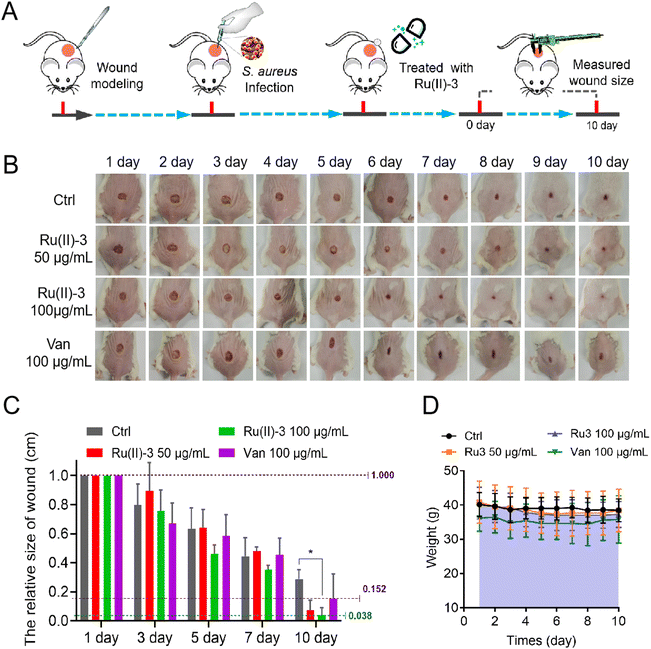
Next, two animal infection models using G. mellonella larva and mouse were employed to investigate the anti-infective potency of Ru(II)-3. For the G. mellonella infection model,28 S. aureus was injected into them and then treated with Ru(II)-3 or vancomycin after 1 h. As shown in Fig. 13, all the S. aureus-infected G. mellonella died on the second day. However, the survival rate of the infected G. mellonella after treatment with Ru(II)-3 was significantly elevated. At a dose of 50 mg kg−1, above 50% G. mellonella survived, suggesting the high anti-infective efficacy of Ru(II)-3. Next, a mouse wound infection model29 was established to further verify its antibacterial activity in vivo. The schematic of the wound infection model is exhibited in Fig. 14A. The photograph of the mouse wound and changes in size clearly demonstrated that a sterile cream containing Ru(II)-3 has a good therapeutic effect. At a dose of 100 mg kg−1, Ru(II)-3 showed stronger anti-infective potency than vancomycin, resulting in a faster wound-healing rate. The size of the mouse wounds in the Ru(II)-3-treated group on the 10th day was significantly smaller than that of the vancomycin-treated group (Fig. 14B and C). Importantly, the weight of the mouse did not change significantly throughout the treatment, indicating the low toxicity of Ru(II)-3 to mice (Fig. 14D). Collectively, these results confirm that Ru(II)-3 not only exhibited low toxicity but also had a strong anti-infection effect on S. aureus in vivo.
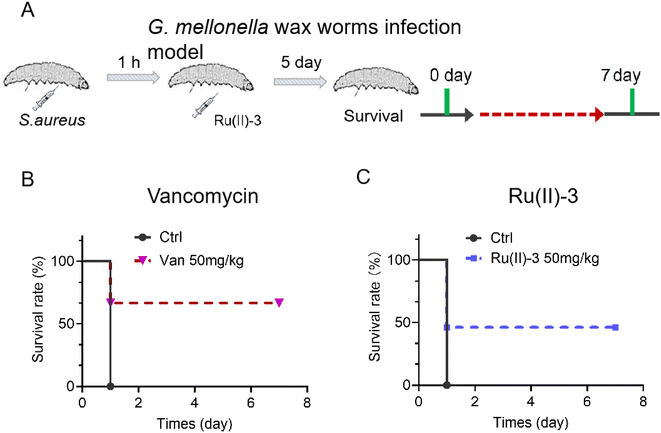 | ||
| Fig. 13 (A) Scheme of the G. mellonella larva infection model. The survival rate of S. aureus-infected G. mellonella wax worms after treatment with Ru(II)-3 (C) or vancomycin (B). | ||
3. Conclusion
In summary, a versatile morpholine moiety was linked into an Ru-based skeleton and then successfully developed into five novel Ru-based antibacterial agents with multiple antibacterial mechanisms in this work. The in vitro antibacterial potency evaluation confirmed that all the five agents showed robust bactericidal activity against S. aureus. In particular, the most-active agent, Ru(II)-3, could kill more than 99% S. aureus in just 2 h at 0.78 μg mL−1. In addition, it could effectively inhibit the α-toxin secretion from S. aureus. More importantly, Ru(II)-3 could not only efficiently inhibit bacterial biofilm formation but also had a powerful ability to remove mature biofilms. These properties will help it to remove bacteria more effectively in vivo. In fact, Ru(II)-3 could effectively enhance the antibacterial activity of most traditional antibiotics when a combination was employed. Antibacterial mechanism studies further demonstrated that Ru(II)-3 exhibited multitarget action: it killed bacteria in mainly two ways: induce ROS formation and damage the bacterial membrane. Due to this series of advantages, Ru(II)-3 not only showed much better bactericidal potency than most clinically common antibiotics but also efficiently prevented S. aureus from achieving drug resistance. At last, two animal infection models also demonstrated that Ru(II)-3 showed significant efficacy against S. aureus in vivo and with low toxicity. Taken together, the results presented here clearly demonstrated that a morpholine-modified ruthenium-based agent is a promising antibiotics candidate in tackling the crisis of drug-resistant bacteria.4. Experimental section
4.1 Synthesis and characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). Dark red product. Yield: 196.02 mg, 54.0%. 1H NMR (400 MHz, DMSO-d6) δ 14.16 (s, 1H), 9.05 (d, J = 11.2 Hz, 2H), 8.71 (d, J = 16.2 Hz, 4H), 8.24 (d, J = 6.8 Hz, 2H), 8.05 (s, 2H), 7.91 (s, 2H), 7.66 (s, 2H), 7.42 (s, 4H), 7.29–7.12 (m, 4H), 4.12 (s, 2H), 3.57 (s, 4H), 2.55 (s, 8H), 2.45 (s, 4H), 2.38 (s, 6H), 1.81 (s, 2H), 1.63 (s, 2H). 13C NMR (101 MHz, DMSO-d6) δ 160.95 (s), 156.74 (d, J = 14.5 Hz), 153.35 (s), 150.94 (d, J = 15.6 Hz), 149.98 (d, J = 14.3 Hz), 137.36 (s), 130.49 (s), 128.98–128.11 (m), 127.73 (s), 126.71 (s), 126.48 (s), 125.96 (s), 125.46 (d, J = 6.1 Hz), 122.34 (s), 121.60 (s), 115.65 (s), 80.06–74.15 (m), 80.06–70.32 (m), 69.17 (d, J = 211.6 Hz), 66.21 (dd, J = 595.2, 425.0 Hz), 58.22 (s), 55.80 (d, J = 450.6 Hz), 29.45 (s), 26.91 (s), 21.15 (d, J = 9.5 Hz). IR (KBr): ν = 3383, 2932, 2860, 2324, 2167, 1978, 1742, 1618, 1553, 1501, 1455, 1383, 1265, 1102, 1030, 841, 553 cm−1. HRMS (ESI): m/z: calcd for [M − 2PF6]2+ C51H51N9O2Ru, 461.6611; found: 461.6613.
1 v/v). Dark red product. Yield: 196.02 mg, 54.0%. 1H NMR (400 MHz, DMSO-d6) δ 14.16 (s, 1H), 9.05 (d, J = 11.2 Hz, 2H), 8.71 (d, J = 16.2 Hz, 4H), 8.24 (d, J = 6.8 Hz, 2H), 8.05 (s, 2H), 7.91 (s, 2H), 7.66 (s, 2H), 7.42 (s, 4H), 7.29–7.12 (m, 4H), 4.12 (s, 2H), 3.57 (s, 4H), 2.55 (s, 8H), 2.45 (s, 4H), 2.38 (s, 6H), 1.81 (s, 2H), 1.63 (s, 2H). 13C NMR (101 MHz, DMSO-d6) δ 160.95 (s), 156.74 (d, J = 14.5 Hz), 153.35 (s), 150.94 (d, J = 15.6 Hz), 149.98 (d, J = 14.3 Hz), 137.36 (s), 130.49 (s), 128.98–128.11 (m), 127.73 (s), 126.71 (s), 126.48 (s), 125.96 (s), 125.46 (d, J = 6.1 Hz), 122.34 (s), 121.60 (s), 115.65 (s), 80.06–74.15 (m), 80.06–70.32 (m), 69.17 (d, J = 211.6 Hz), 66.21 (dd, J = 595.2, 425.0 Hz), 58.22 (s), 55.80 (d, J = 450.6 Hz), 29.45 (s), 26.91 (s), 21.15 (d, J = 9.5 Hz). IR (KBr): ν = 3383, 2932, 2860, 2324, 2167, 1978, 1742, 1618, 1553, 1501, 1455, 1383, 1265, 1102, 1030, 841, 553 cm−1. HRMS (ESI): m/z: calcd for [M − 2PF6]2+ C51H51N9O2Ru, 461.6611; found: 461.6613.4.2 Biological activity test
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in fresh TSB and cultured to the logarithmic phase. Then, the supernatant was removed by centrifugation, and the bacteria were resuspended in PBS to OD600 = 1. After co-incubating with Ru(II)-3 or polymyxin B sulfate for 2 h, the ultraviolet absorption (260 nm) of the bacterial supernatant was measured.
1000 in fresh TSB and cultured to the logarithmic phase. Then, the supernatant was removed by centrifugation, and the bacteria were resuspended in PBS to OD600 = 1. After co-incubating with Ru(II)-3 or polymyxin B sulfate for 2 h, the ultraviolet absorption (260 nm) of the bacterial supernatant was measured.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added and the absorbance at 595 nm was measured. For the biofilm eradication assay, S. aureus was cultured in 24-well plates at 37 °C for 48 h to induce the formation of a biofilm; then, the mature biofilm was co-incubated with Ru(II)-3 for 24 h. Afterward, the biofilm was washed and fresh TSB containing MTT was added. At last, DMSO was added and the UV absorption at 595 nm was measured.
1) was added and the absorbance at 595 nm was measured. For the biofilm eradication assay, S. aureus was cultured in 24-well plates at 37 °C for 48 h to induce the formation of a biofilm; then, the mature biofilm was co-incubated with Ru(II)-3 for 24 h. Afterward, the biofilm was washed and fresh TSB containing MTT was added. At last, DMSO was added and the UV absorption at 595 nm was measured.Ethical statement
All mouse were purchased from Nanjing Keruisi animal Co., LTD. The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Jiangxi Science & Technology Normal University, and complied with the academic guidelines for the care and use of laboratory animals of China.Conflicts of interest
The authors have declared that there is no conflict of interest.Acknowledgements
We gratefully acknowledge the generous support provided by Health Research Project of Hainan Province (Grant No. 22A200010) and Jiangxi Science & Technology Normal University (2021QNBJRC001).References
- G. Maddalena, R. Matteo and V. Pierluigi, Antimicrobial resistance in organ transplant recipients, Infect. Dis. Clin. N. Am., 2023, 37(3), 515–537 CrossRef PubMed.
- X. Jiaqi, F. Weiguo, W. Jiaye, W. Ruichen, Z. Bowen, B. Letao, C. Zhesheng, Y. Hui and S. Leming, Antimicrobial peptides for combating drug-resistant bacterial infections, Drug Resist. Updates, 2023, 68, 100954 CrossRef PubMed.
- W. Haibo, W. Minji, X. Xiaohan, G. Peng, X. Zeling, Z. Qi, L. Hongyan, Y. Aixin, Y. T. K. Richard and S. Hongzhe, Multi-target mode of action of silver against Staphylococcus aureus endows it with capability to combat antibiotic resistance, Nat. Commun., 2021, 12(1), 3331 CrossRef PubMed.
- C. Gordon, B. Justin and O. Michael, Pathogenicity and virulence of Staphylococcus aureus, Virulence, 2021, 12(1), 547–569 CrossRef PubMed.
- A. A. Bandar, A. J. Intisar, M. A. Jawhra, M. A. Hussein, G. Bandar, A. Kholod and Y. Y. Nik, Antimicrobial resistance in methicillin-resistant Staphylococcus aureus, Saudi J. Biol. Sci., 2023, 30(4), 103604 CrossRef PubMed.
- C. J. Murray, et al., Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399(10325), 629–655 CrossRef CAS PubMed.
- Antimicrobial resistance in the age of COVID-19, Nat. Microbiol., 2020, 5, p. 779.
- T. M. Rawson, D. Ming, R. Ahmad, L. S. P. Moore and A. H. Holmes, Antimicrobial use, drug-resistant infections, and COVID-19, Nat. Rev. Microbiol., 2020, 18, 409–410 CrossRef CAS PubMed.
- M. A. Tiburcio, A. R. Rocha, R. A. Romano, N. M. Inada, V. S. Bagnato, R. M. Carlos and H. H. Buzzá, In vitro evaluation of the cis-[Ru(phen)2(pPDIp)]2+ complex for antimicrobial photodynamic therapy against Sporothrix brasiliensis and Candida albicans, J. Photochem. Photobiol., B, 2022, 229, 112414 CrossRef CAS PubMed.
- F. Angelo, R. Riccardo, T. Solmaz, B. Olivier, A. Philipp, F. Ariane, M. Tim, S. Leone and G. Gilles, Synthesis, characterization, and biological evaluation of new Ru(II) polypyridyl photosensitizers for photodynamic therapy, J. Med. Chem., 2014, 57(17), 7280–7292 CrossRef PubMed.
- P. Sudhindra, R. Nilmadhab, K. Binoy and P. Priyankar, Construction of homo and heteronuclear Ru(II), Ir(III) and Re(I) complexes for target specific cancer therapy, Coord. Chem. Rev., 2022, 460, 214462 CrossRef.
- R. B. Wang, M. Wei, X. R. Wang, Y. S. Chen, Y. S. Xiong, J. X. Cheng, Y. H. Tan, X. W. Liao and J. T. Wang, Synthesis of ruthenium polypyridine complexes with benzyloxyl groups and their antibacterial activities against Staphylococcus aureus, J. Inorg. Biochem., 2022, 236, 111954 CrossRef CAS PubMed.
- L. Jiang, Y. Y. Ma, Y. S. Xiong, Y. H. Tan, X. M. Duan, X. W. Liao and J. T. Wang, Ruthenium polypyridine complexes with triphenylamine groups as antibacterial agents against Staphylococcus aureus with membrane-disruptive mechanism, Front. Chem., 2022, 10, 1035741 CrossRef CAS PubMed.
- C. Y. Zhang, R. J. Yu, L. Q. Wang, H. Y. Huang, M. Q. Xiao, X. M. Duan, J. T. Wang, X. W. Liao and Y. S. Xiong, Synthesis and evaluation of sulfonyl-substituted ruthenium complex as potential antibacterial activity against Staphylococcus aureus, New J. Chem., 2022, 46, 14805–14815 RSC.
- R. B. Wang, X. M. Zhou, J. J. Chen, Y. S. Chen, Y. S. Xiong, X. M. Duan, X. W. Liao and J. T. Wang, Ruthenium polypyridine complexes containing prenyl groups as antibacterial agents against Staphylococcus aureus through a membrane-disruption mechanism, Arch. Pharm., 2023, 356, 2300175 CrossRef CAS PubMed.
- C. Y. Zhang, R. J. Yu, L. Q. Wang, H. Y. Huang, J. T. Wang, X. W. Liao, X. M. Duan and Y. S. Xiong, Design, synthesis, and evaluation of aryl-thioether ruthenium polypyridine complexes: a multi-target antimicrobial agents against gram-positive bacteria, Eur. J. Med. Chem., 2022, 240, 114562 CrossRef CAS PubMed.
- K. Archana and K. S. Rajesh, Morpholine as ubiquitous pharmacophore in medicinal chemistry: deep insight into the structure-activity relationship (SAR), Bioorg. Chem., 2020, 96, 103578 CrossRef PubMed.
- L. Elena, C. Lorenzo and T. Andrea, Occurrence of morpholine in central nervous system drug discovery, ACS Chem. Neurosci., 2021, 12(3), 378–390 CrossRef PubMed.
- C. Rishav, B. Indira, R. Souryadip, P. Kallol, S. K. Tuhin, G. Arnab and M. Arindam, Synthesis, characterization, and cytotoxicity of morpholine-containing ruthenium(II) p-cymene complexes, Inorg. Chem., 2021, 60(16), 12172–12185 CrossRef PubMed.
- T. Ariadni, X. Dimitrios and P. K. Angeliki, Morpholine as a scaffold in medicinal chemistry: an update on synthetic strategies, ChemMedChem, 2020, 15(5), 392–403 CrossRef PubMed.
- S. Bu, G. Jiang, G. Jiang, J. Liu, X. Lin, J. Shen, Y. Xiong, X. Duan, J. Wang and X. Liao, Antibacterial activity of ruthenium polypyridyl complexes against Staphylococcus aureus and biofilms, J. Biol. Inorg. Chem., 2020, 25(5), 747–757 CrossRef CAS PubMed.
- M. E. Ivarsson, D. Estelle, H. Corina, H. Samuel, H. Chrismita, F. Jillian, A. Fernando, L. Jun, F. V. Elena, B. Premysl, M. Susan, C. Wang, L. Jean-Christophe and C. Bastien, Small-Molecule allosteric triggers of Clostridium difficile toxin B auto-proteolysis as a therapeutic strategy, Cell Chem. Biol., 2019, 26, 17–26 CrossRef CAS PubMed.
- D. Hana, A. Véronique, P. Otto and M. B. Jean, Antibiotic adjuvants: make antibiotics great again, J. Med. Chem., 2019, 62(19), 8665–8681 CrossRef PubMed.
- F. E. Raquel, E. P. Jake, O. W. Jonathan, B. S. Paul and M. E. Richard, Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins, Antimicrob. Agents Chemother., 2010, 54(9), 3708–3713 CrossRef PubMed.
- G. Yong, H. Enhua, W. Tingyu, Y. Xiaoting, H. Meiyue, B. Li-Ping, F. Xiangjing, L. Jifeng and Q. Shangshang, Development of Membrane-Active Honokiol/Magnolol Amphiphiles as Potent Antibacterial Agents against Methicillin-Resistant Staphylococcus aureus (MRSA), J. Med. Chem., 2021, 64, 12903–12916 CrossRef PubMed.
- L. Guo, C. Zhang, G. Chen, M. Wu, W. Liu, C. Ding, Q. Dong, E. Fan and Q. Liu, Reactive oxygen species inhibit biofilm formation of Listeria monocytogenes, Microb. Pathog., 2018, 127, 183–189 CrossRef PubMed.
- E. Evgeniy and K. Sergei, Identification of ROS using oxidized DCFDA and flow-cytometry, Methods Mol. Biol., 2010, 594, 57–72 CrossRef PubMed.
- K. M. James, P. S. Joseph, P. B. Benjamin, M. M. Gabriel, M. Andre, H. L. Sophia, K. Hahn, D. R. Joshua, T. Athanasios, M. S. Mikhail, Z. W. Maxwell and G. Zemer, A dual-mechanism antibiotic kills Gram-negative bacteria and avoids drug resistance, Cell, 2020, 181, 1–15 CrossRef PubMed.
- J. J. Hu, N. Liu, Q. Q. Fan, Y. Q. Gu, S. J. Chen, F. Zhu and Y. Y. Cheng, A fluorous peptide amphiphile with potent antimicrobial activity for the treatment of MRSA-induced sepsis and chronic wound infection, Angew. Chem., Int. Ed., 2024, e202403140 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02667e |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |