Open Access Article
Open Access ArticleFluorescent probe for imaging intercellular tension: molecular force approach†
Xiao-Hong Wang *a,
Ming Wangb,
Jian-bin Panc,
Jin-miao Zhua,
Hu Chenga,
Hua-ze Donga,
Wen-jie Bia,
Shi-wei Yanga,
Yuan-yuan chena,
Fan Xua and
Xiao-jing Duana
*a,
Ming Wangb,
Jian-bin Panc,
Jin-miao Zhua,
Hu Chenga,
Hua-ze Donga,
Wen-jie Bia,
Shi-wei Yanga,
Yuan-yuan chena,
Fan Xua and
Xiao-jing Duana
aSchool of Chemical and Pharmaceutical Engineering, Hefei Normal University, 230061, Hefei, Anhui, China
bSchool of Energy Materials and Chemical Engineering, Hefei University, Hefei 230601, China
cState Key Laboratory of Analytical Chemistry for Life Science and Collaborative Innovation Center of Chemistry for Life Sciences, School of Chemistry and Chemical Engineering, Nanjing University, 210023, China
First published on 19th July 2024
Abstract
Cellular mechanical force plays a crucial role in numerous biological processes, including wound healing, cell development, and metastasis. To enable imaging of intercellular tension, molecular tension probes were designed, which offer a simple and efficient method for preparing Au-DNA intercellular tension probes with universal applicability. The proposed approach utilizes gold nanoparticles linked to DNA hairpins, enabling sensitive visualization of cellular force in vitro. Specifically, the designed Au-DNA intercellular tension probe includes a molecular spring flanked by a fluorophore–quencher pair, which is anchored between cells. As intercellular forces open the hairpin, the fluorophore is de-quenched, allowing for visualization of cellular force. The effectiveness of this approach was demonstrated by imaging the cellular force in living cells using the designed Au-DNA intercellular tension probe.
1 Introduction
Cellular force is a fundamental biological process1 that is involved in numerous cellular processes, including migration, proliferation and differentiation. Cells regulate their behaviour through mechanical feedback and interactions with their microenvironment.2 Specifically, receptors on the cell surface bind to ligands, triggering mechanical stimuli that allow cells to sense and convert physical signals from the external environment.3 However, imaging intercellular forces is a challenge, as it typically requires specialized conditions that are not readily available in most laboratories. To address this limitation, there is a need for an efficient molecular force fluorescent probe that is suitable for most laboratory conditions and can be used with a fluorescence microscope or fluorescence confocal microscope.4,5 Mechanical cues are critical for a wide range of cellular processes, including cell adhesion, immune recognition, metastasis, and coagulation.6–8 Specifically, cells sense the mechanical properties of Extracellular Matrix (ECM) through integrin receptors on the cytoskeleton, which transmit forces bidirectionally between the cytoskeleton and ECM.9Multiple approaches are available for studying integrin-mediated cellular forces. Single molecule force spectroscopy is a technology used to detect interactions between cell receptors and their ligands. This method includes atomic force spectroscopy,10 magnetic tweezers, and optical tweezers.11 Although single-molecule force spectroscopy can capture the physicochemistry of ligand–receptor interactions, it may not recapitulate biological processes accurately because many membrane receptors function as oligomers instead of isolated molecules. Traction Force Microscopy (TFM) is another method for studying cellular mechanics, which measures the extent of deformation of the polymer substrate caused by cellular traction.12 TFM provides micron-level spatial resolution and nN-level sensitivity, but it cannot measure the force of a single receptor and the force measured by TFM is relatively rough. However, TFM and single-molecule force spectroscopy provide a foundation for understanding the impact of environmental forces on various cell functions. In contrast, the mechanical interactions between individual cells remain largely unexplored due to the difficulty of modifying and controlling functional probes at complex cell–cell interfaces. To bridge this gap, Khalid Salaita13 developed Molecular Tension-based Fluorescence Microscopy (MTFM). MTFM enables imaging cellular traction at pico-Nu (pN) resolution, allowing for the visualization of mechanical interactions between individual cells.
The molecular reporter is the technical core of MTFM and consists of an extensible “spring” unit with fluorophores and quenchers on both sides that is fixed to the substrate.14 The tension delivered to the probe causes the spring structure to extend or open, separating the fluorophore from the quencher and resulting in a sharp increase of fluorescence intensity.15,16 The spring structure comprises polyethylene glycol, protein, and DNA.14 DNA-based MTFM probes offer several advantages, including ease of synthesis, modular design, and optimal signal-to-noise ratios for imaging. Typically, DNA probes are composed of three oligonucleotide strands. The first strand anchors the probe to the underlying substrate, the second strand is a stem-loop hairpin structure with a well-characterized force–stretch relationship, and the third strand ligands that bind to cell surface receptors.17,18
MTFM molecular tension fluorescent probe is fixed at one end on a glass substrate, which requires a complex and time-consuming modification. To overcome this limitation, Zhao Bin19 developed an intercellular tension probe in 2017, which can be directly incubating the probe with cells without the need for substrate modification. Building on this approach, the design is improved by connecting gold nanoparticles (AuNPs) between DNA hairpin probes. In addition to the quenching between fluorophores and quenchers on hairpins. In addition to the quenching between the fluorophores and quenchers on hairpins, there is also quenching between AuNPs and fluorophores. The double quenching approach helps to overcome background noise, resulting in an gold nanoparticles DNA (Au-DNA) tension fluorescent probe with Nanometal Surface Energy Transfer (NSET)20 and Fluorescence Energy Resonance Transfer (FRET) effects.21–23 Khalid Salaita16 verified that the quenching efficiency of the Au-DNA molecular force probe can reach 100%, indicating very sensitive.
In this study, a novel strategy is proposed for constructing Au-DNA intercellular tension probe to visualize cellular tension. The Au-DNA intercellular tension probe are compatible with standard fluorescence microscope and can be widely used to measure intercellular tension of many systems. Due to this method is intrinsically non-invasive in principle and cannot affect the activity of the cells, which can offer promising avenue for the use of Au-DNA intercellular tension probe in other cellular mechanoconduction applications.
2 Experimental
2.1 Reagents
| Name | Sequences (5′ to 3′) |
|---|---|
| LS | HS-TTTGCTGGGCTACGTGGCGCTCTT-6-Cy3 |
| LS-C | Cholesterol- TTTGCTGGGCTACGTGGCGCTCTT-6-Cy3 |
| HS | CCCGTGAAATACCGCACAGATGCGTTTGTATAA ATGTTTTTTTCATTTATACTTTAAGAGCGCCACGTAGCCCAGC |
| AS | BHQ2-CGCATCTGTGCGGTATTTCACCCC-SH |
| nq-AS | BHQ2-CGCATCTGTGCGGTATTTCACCCC |
All DNA oligonucleotides (including circular strands, anchored AS strands, and LS-RGD strands) were placed in a 90–95 °C water bath and annealed for 5 min at a concentration of 10 μM. The synthesized hairpins are not used up and stored in a refrigerator at 4 °C.
There are three types of DNA hairpin strands, which are synthesized as follows.
Cy3-BHQ2-1 hairpin chain, containing RGD: LS-RGD, HS, AS.
Cy3-BHQ2-2 hairpin chain with cholesterol: LS-C, HS, AS.
Cy3-BHQ2-nq hairpin chain with fluorophore without RGD and cholesterol: LS, HS, nq-AS.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After 30 min, the resulting hybridized DNA solution was added to 1 mL 10 nM Au nanoparticle solution and incubated on an orbital shaker for 1 h.
1. After 30 min, the resulting hybridized DNA solution was added to 1 mL 10 nM Au nanoparticle solution and incubated on an orbital shaker for 1 h.Subsequently, phosphate buffer (0.1 M, pH = 7.4) and 10% sodium dodecyl sulfate (SDS) solution (w/v) were added to the mixture to make their concentrations 10 mM and 0.1%, respectively. The resulting solution was gradually “salted” with 6 aliquots of 2 M NaCl solution (0.05 M each time) at 2 h (20 min interval) to obtain a final NaCl solution of 0.3 M. Note that after each salting out, the Au nanoparticle solution was sonicated for 10 s to maximize DNA packaging. The particles were stirred overnight and photoprotected. Then, 30 μM passivated PEG (SH-PEG) was added to the AuNP-DNA solution and allowed to incubate for 4 h. The reaction mixture was centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 20 min) three times and resuspended in PBS solution.
000 rpm, 20 min) three times and resuspended in PBS solution.
3 Results and discussion
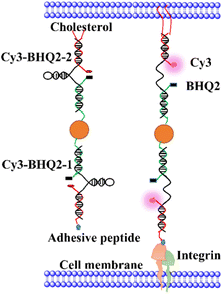
Fig. 1 illustrates the structure of Au-DNA intercellular tension probe. The DNA strand consists of three strands: (1) an anchor strand with a fluorophore, (2) a DNA strand with aquencher, and (3) a cholesterol-modified stem-loop DNA hairpin strand. After annealing, the three strands pair complementally. The Cy3-BHQ2-1 probe contains an RGD peptide that specifically binds to integrin on the cell membrane. The self-assembled Cy3-BHQ2-2 probe has a pair of cholesterol anchors at one end, allowing the DNA probe to spontaneously insert into living cell membranes through hydrophobic interactions. The core of the probe is retractable DNA hairpin, which exhibits a “spring” characteristic. A section of DNA is linked to a cRGD peptide that binds to the integrin receptor on the cell surface, while cholesterol at the other end enables stably anchoring on the cell membrane. Fluorescent dyes and quenchers effectively quench fluorescence via FRET and NSET of Au nanoparticles as DNA is folded. When DNA hairpins are opened by intercellular tension force, the fluorescent dyes restore fluorescence. If the tension applied by neighboring cells exceeds the threshold force required for unfolded DNA hairpins, the fluorophore dissociates from the quencher, resulting in an increase in fluorescence intensity.In Fig. 1, each tension probe is composed of a DNA hairpin labeled with a fluorophore quencher pair that is attached to AuNPs. The fluorescent groups are double quenched by molecular quenching agents via FRET and NSET, reducing background signals and improving the sensitivity of traditional molecular tension sensors.15,16 When the applied force exceeds the hairpin threshold unfolding force, the DNA hairpin opens, resulting in fluorescence.
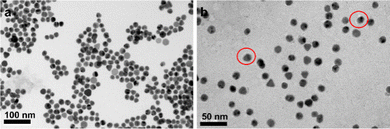
Although larger diameter AuNPs have been reported to be more efficient at quenching fluorescence,15 smaller AuNPs are selected to minimize the potential for each particle to bind multiple integrins. Based on the structural data and previous literature,16 17 nm AuNPs are chosed. As shown in Fig. 2, the diameter of Au-DNA nanoparticles is larger compared to Au-DNA particles, and certain white circles around the AuNPs can also be observed. Additionally, TEM results indicated that most AuNPs were well separated from each other, and there were no aggregated particles observed.
To verify the binding of DNA to AuNPs, Dynamic Light Scattering (DLS) measurements are conducted. As shown in Fig. 3, the hydration radius of Au-DNA is larger than that of Au alone. DLS results (Fig. 3a) indicate that the diameter of the Au-DNA structure increases from 17.4 nm to 38.12 nm, suggesting that AuNPs follow a specific pattern when bound to DNA and form complex structures with AuNPs and Au-DNA. These DLS measurements support the observation that the average diameter of the particles increases from 17 nm to 38 nm (Fig. 3a). After synthesizing AuNPs using literature procedures, their hydrodynamic diameters were determined to be approximately 17 nm by DLS. Following oligonucleotides modification, the average hydrodynamic diameter of Au-DNA increased from to around 38 nm. Among the different nanospheres, the sample AuNPs exhibited the maximum displacement (λMax ≈ 15 nm) because of their larger size, which resulted in lighter scattering. Upon addition of DNA to the AuNPs sample, the UV absorption spectrum displayed a slight red shift, as shown in Fig. 3b, indicating successful coating of the DNA layer on the surface of AuNPs.
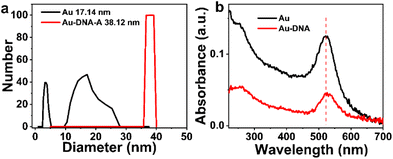 | ||
| Fig. 3 (a) Dynamic light scattering particle size distributions (b) UV-vis absorption spectra of Au and Au-DNA comparing mean diameter data. | ||
It was investigated whether Au-DNA molecular force fluorescent probes could be anchored to cellular membranes sensitive to cellular mechanics, such as DMCK cells. Au-DNA intercellular tension probe was observed anchored to the cell membrane within 30 min of incubation (Fig. 4b). The efficient membrane anchoring is due to cholesterol-mediated hydrophobic interactions and RGD-integrin interactions (Fig. 4b). Au-DNA intercellular tension probe unfolds by integrin-mediated forces, allowing Cy3 to dissociate from BHQ2. Compared to probes without RGD peptides (Fig. 4a), the fluorescence signal of the Au-DNA tension probe (Fig. 4b) on cell membrane is clearly visible. On the cell membrane, there are many signal receptors such as integrins and cadherins.13,19 These receptors bind to ligands on neighboring cells or ECM surfaces, mediating mechanical transmission. Integrins are excellent sensors for cellular force reception.13 When the receptor binds to the ligand, the cell forces generated by the cell cytoskeleton are transmitted to the surrounding environment. Focal adhesions contain thousands of integrin receptors, making the intercellular tension probe signal very stable.
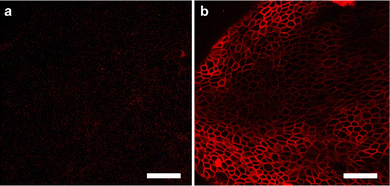 | ||
| Fig. 4 (a) Confocal fluorescence diagram of Au DNA without RGD ligand and cholesterol and (b) confocal fluorescence diagram of Au DNA containing ligand and cholesterol. Scale bar: 50 μm. | ||
To further determine whether intercellular tension was driven by contraction of actomyosin II, the Rho-associated kinase (ROCK) inhibitor Y-27632 (Fig. 5 and movie S1†) were used to inhibit the contractility of myosin. The ROCK inhibit led to a decrease in intercellular tension overtime. After treatment with the inhibitor Y-27632, the tension signal was reduced, indicating that the intercellular tension was driven by the contraction of actomyosin and confirming the feasibility of the probe Furthermore, additional experiments were conducted by incubating the cell force probe with cells and then adding the cytoskeleton inhibitor ML-7 (Fig. S1†). The experimental results showed a significant decrease in cell force signal after the inhibitor was added, indicating that the probe we synthesized is an intercellular force probe.
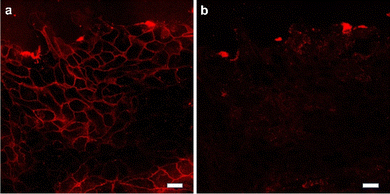 | ||
| Fig. 5 The cells are treated with ROCK inhibitor. Representative fluorescence images showed the change of Au-DNA tension probe before (a) and after (b) Y27632 treatment 30 min. Scale bar: 20 μm. | ||
4 Conclusions
In summary, the Au-DNA intercellular tension probe successfully imaged MDCK integrin-mediated tension, with clear fluorescence signal observed on the cell membrane. Compared to techniques such as traction microscopy and optical tweezers, the DNA spring probes are simpler to use, requiring only incubation and self-assembly on the cell membrane, without the need for more complex and time-consuming cloning or transfection. The Au-DNA intercellular tension probe is an effective and user-friendly method that has the potential to become a widely-used tool. By studying intercellular tension, this technique can shed light on various collective migration behaviors, including tumor invasion, wound healing, and embryonic development. These probes offer a promising approach for future tissue engineering and regenerative medicine applications.Data availability
No additional data are available.Author contributions
XHW: conceived ideas, completed experiments, and wrote the article. MW: conceived ideas, and revised the article for language issues. WJB and JBP: revised the article for language issues. SWY: collected and organized the data. JMZ and HZD: designed and funded experiments. HC: discussed the results. YYC, FX and XJD: assisted in completing experiments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The acknowledgements come at the end of an article after the conclusions and before the notes and references. This work was supported by the Science and Technology Major Project of Anhui Province of China (201903a07020003), the Science and Technology Major Project of Fuyang of Anhui Province of China (FK20208018); Anhui Engineering Laboratory Project for the Development and Utilization of Natural Resources; Derived from Medicines and Edibles (YSTY2022020); Hefei Normal University 2022 Scientific Research Launch Fund for Introducing High level Talents (2022rcjj42, 2022rcjj26); 2023 Anhui university research project (2023AH051323); University natural science research project of Anhui Department of Education (KJ2021A0921).Notes and references
- I. Muhamed, J. Wu, P. Sehgal, X. Kong, A. Tajik, N. Wang and D. E. Leckband, J. Cell Sci., 2016, 129, 1843–1854 CrossRef CAS PubMed.
- G. M. Allen, K. C. Lee, E. L. Barnhart, M. A. Tsuchida, C. A. Wilson, E. Gutierrez, A. Groisman, J. A. Theriot and A. Mogilner, Cell Syst., 2020, 11, 286–299 CrossRef CAS PubMed.
- B. Ladoux and R.-M. Mège, Nat. Rev. Mol. Cell Biol., 2017, 18, 743–757 CrossRef CAS.
- A. Colom, E. Derivery, S. Soleimanpour, C. Tomba, M. D. Molin, N. Sakai, M. González-Gaitán, S. Matile and A. Roux, Nat. Chem., 2018, 10, 1118–1125 CrossRef CAS.
- Q. Tian, F. Yang, H. Jiang, P. Bhattacharyya, T. Xie, A. A. Ali, Y. Sun and M. You, Front. Cell Dev. Biol., 2023, 11, 1220079 CrossRef.
- S. Checa, M. K. Rausch, A. Petersen, E. Kuhl and G. N. Duda, Biomech. Model. Mechanobiol., 2015, 14, 1–13 CrossRef.
- T. Mammoto and D. E. Ingber, Development, 2010, 137, 1407–1420 CrossRef CAS.
- B. G. Godard and C.-P. Heisenberg, Curr. Opin. Cell Biol., 2019, 60, 114–120 CrossRef CAS PubMed.
- D. N. Clarke and A. C. Martin, Curr. Biol., 2021, 31, R667–R680 CrossRef CAS PubMed.
- A. Hategan, R. Law, S. Kahn and D. E. Discher, Biophys. J., 2003, 85, 2746–2759 CrossRef CAS PubMed.
- M. Capitanio and F. S. Pavone, Biophys. J., 2013, 105, 1293–1303 CrossRef CAS.
- H. Colin-York, Y. Javanmardi, L. Barbieri, D. Li, K. Korobchevskaya, Y. Guo, C. Hall, A. Taylor, S. Khuon, G. K. Sheridan, T.-L. Chew, D. Li, E. Moeendarbary and M. Fritzsche, Nano Lett., 2019, 19, 4427–4434 CrossRef CAS PubMed.
- K. Galior, Y. Liu, K. Yehl, S. Vivek and K. Salaita, Nano Lett., 2016, 16, 341–348 CrossRef CAS PubMed.
- Y. Liu, K. Galior, V. P.-Y. Ma and K. Salaita, Acc. Chem. Res., 2017, 50, 2915–2924 CrossRef CAS PubMed.
- V. P.-Y. Ma, Y. Liu, L. Blanchfield, H. Su, B. D. Evavold and K. Salaita, Nano Lett., 2016, 16, 4552–4559 CrossRef CAS PubMed.
- Y. Liu, L. Blanchfield, V. P.-Y. Ma, R. Andargachew, K. Galior, Z. Liu, B. Evavold and K. Salaita, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5610–5615 CrossRef CAS.
- B. Zhao, N. Li, T. Xie, Y. Bagheri, C. Liang, P. Keshri, Y. Sun and M. You, Chem. Sci., 2020, 11, 8558–8566 RSC.
- P. Keshri, B. Zhao, T. Xie, Y. Bagheri, J. Chambers, Y. Sun and M. You, Angew. Chem., Int. Ed., 2021, 60, 15548–15555 CrossRef CAS.
- B. Zhao, C. O'Brien, A. P. K. K. K. Mudiyanselage, N. Li, Y. Bagheri, R. Wu, Y. Sun and M. You, J. Am. Chem. Soc., 2017, 139, 18182–18185 CrossRef CAS PubMed.
- H.-B. Wang, Y. Li, Y. Chen, Z.-P. Zhang, T. Gan and Y.-M. Liu, Microchim. Acta, 2018, 185, 102 CrossRef.
- H.-B. Wang, Y. Li, H.-Y. Bai and Y.-M. Liu, Sens. Actuators, B, 2018, 259, 204–210 CrossRef CAS.
- H.-B. Wang, B.-B. Tao, N.-N. Wu, H.-D. Zhang and Y.-M. Liu, Spectrochim. Acta, Part A, 2022, 271, 120948 CrossRef CAS.
- H.-B. Wang, Y. Chen, N. Li and Y.-M. Liu, Microchim. Acta, 2017, 184, 515–523 CrossRef CAS.
- X.-H. Wang, F. Yang, J.-B. Pan, B. Kang, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2020, 92, 16180–16187 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02647k |
| This journal is © The Royal Society of Chemistry 2024 |