Open Access Article
Open Access ArticleDrug repositioning identifies salvinorin A and deacetylgedunin (DCG) enriched plant extracts as novel inhibitors of Mpro, RBD–ACE2 and TMPRRS2 proteins†
Mariana J. Shayo a,
Baraka Samwelb,
Daniel M. Shadrackcd,
Joel Cassel
a,
Baraka Samwelb,
Daniel M. Shadrackcd,
Joel Cassel e,
Joseph M. Salvino
e,
Joseph M. Salvino e,
Luis J. Montaner
e,
Luis J. Montaner e,
Geradius Deogratias
e,
Geradius Deogratias *f,
Ian Tietjene,
Lucy Kirurig,
Samson Hilongah and
Ester Innocenta
*f,
Ian Tietjene,
Lucy Kirurig,
Samson Hilongah and
Ester Innocenta
aDepartment of Biological and Pre-clinical Studies, Institute of Traditional Medicine, Muhimbili University of Health and Allied Sciences, P.O.Box 65001, Dar es Salaam, Tanzania
bDepartment of Natural Products, Institute of Traditional Medicines, Muhimbili University of Health and Allied Sciences, P.O.Box 65001, Dar es Salaam, Tanzania
cDepartment of Chemistry, Faculty of Natural and Applied Sciences, St. John's University of Tanzania, P.O.Box 47, Dodoma, Tanzania
dSchool of Life Science and Bio-engineering, Nelson Mandela African Institute of Science and Technology, P.O.Box 447, Arusha, Tanzania
eThe Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104, USA
fChemistry Department, College of Natural and Applied Sciences, University of Dar es Salaam, P.O.Box 35061, Dar es Salaam, Tanzania. E-mail: dgeradius@udsm.ac.tz
gKenyata University, Department of Chemistry, P.O.Box 43844-00100, Nairobi, Kenya
hDepartment of Medical Botany, Plant Breeding and Agronomy, Institute of Traditional Medicine, Muhimbili University of Health and Allied Sciences, P.O.Box 65001, Dar es Salaam, Tanzania
First published on 4th July 2024
Abstract
The coronavirus disease 2019 (COVID-19) has spread worldwide with severe health, social, and economic repercussions. Although vaccines have significantly reduced the severity of symptoms and deaths, alternative medications derived from natural products (NPs) are vital to further decrease fatalities, especially in regions with low vaccine uptake. When paired with the latest computational developments, NPs, which have been used to cure illnesses and infections for thousands of years, constitute a renewed resource for drug discovery. In the present report, a combination of computational and in vitro methods reveals the repositioning of NPs and identifies salvinorin A and deacetylgedunin (DCG) as having potential anti-SARS-CoV-2 activities. Salvinorin A was found both in silico and in vitro to inhibit both SARS-CoV-2 spike/host ACE2 protein interactions, consistent with blocking viral cell entry, and well as live virus replication. Plant extracts from Azadirachta indica and Cedrela odorata, which contain high levels of DCG, inhibited viral cell replication by targeting the main protease (Mpro) and/or inhibited viral cell entry by blocking the interaction between spike RBD–ACE2 protein at concentrations lower than salvinorin A. Our findings suggest that salvinorin A represent promising chemical starting points where further optimization may result in effective natural product-derived and potent anti-SARS-CoV-2 inhibitors to supplement vaccine efforts.
1 Introduction
The global response to the COVID-19 pandemic witnessed unprecedented efforts from scientific laboratories that rapidly advanced drug and vaccine development to confront the devastating impact of the virus.1,2 This remarkable cooperation has led to the approval of 13 vaccines by the World Health Organization (WHO) for emergency use since late 2020 and over 170 vaccines at various stages of development.3 However, despite this rapid progress, a decline in public confidence in vaccines has emerged that has contributed to lower vaccination rates and hindered full control of the virus.4,5 This hesitancy, influenced by misconceptions about vaccine safety and efficacy, as well as a general mistrust of the institutions involved, presents a substantial challenge in achieving widespread immunization.6 For example, in many high-income countries, up to 30% of the population remains unvaccinated, while in many developing nations, vaccine hesitancy surpasses 60%.2,7,8To help to address this gap, alternative approaches, including the exploration of natural product-based drugs, are necessary.9,10 For years, natural products have been demonstrated to possess antiviral, anti-inflammatory, and antioxidant properties, offering a valuable resource in the fight against viruses.11–13 As a result, they also represent a promising resource from which to identify new therapeutic leads against SARS-CoV-2 that may be more acceptable to local communities, particularly those that rely on medicinal plants as part of their healthcare.
The entry of SARS-CoV-2 into host cells initiates viral replication, with its viral RNA genome guiding the production of key proteins. This includes papain-like cysteine protease (PLpro) and 3-chymotrypsin-like cysteine protease (3CLpro), also known as main protease (Mpro). Furthermore, the virus exploits host machinery, such as TMPRSS2, facilitating its entry and subsequent replication within the host cell.14–16 Due to their role in protein processing, both PLpro and Mpro have emerged as promising targets for drug development.17–19 Another molecular target of SARS-CoV-2 is through the inhibition of viral cell entry. For example, SARS-CoV-2 cellular binding and entry is controlled in large part by the receptor binding domain (RBD) of the viral spike protein, which interacts with the host receptor ACE2. Therapeutic antibodies that disrupt this interaction are potent inhibitors of SARS-CoV-2 replication, although viral mutation in RBD can rapidly generate resistance to these antibodies as well as circumvent host immunity.
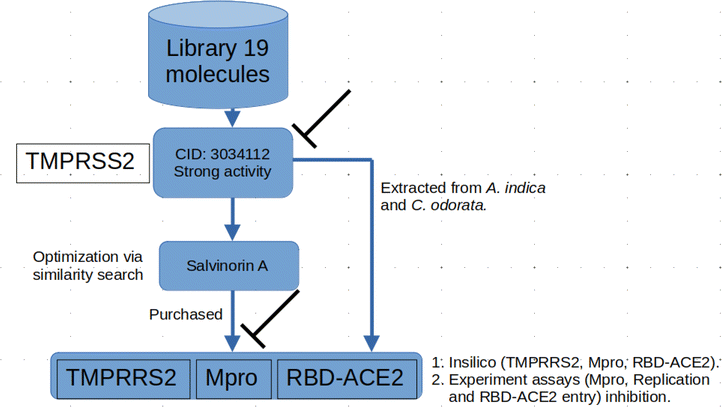
Natural products that inhibit the RBD–ACE2 interaction through mechanisms that are distinct from therapeutic antibodies might present a higher genetic barrier to drug resistance, particularly if they can also target other essential proteins like the viral proteases. In this context, this work builds upon previous efforts, aiming to offer new therapeutic leads for combating the spread of COVID-19. The findings from this present investigation reveal the potential of salvinorin-A, as well as deacetylgedunin (DCG)-containing plant extracts such as those from Cedrela odorata (CO) and the neem tree (Azadirachta indica, AI), in inhibiting specific viral processes of SARS-CoV-2.
2 Materials and methods
2.1 In silico studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(P(x)/P(y)), where F is the free energy surface, kB is the Boltzmann constant, T is temperature and P(x)/P(y) is the probability distribution along the selected reaction coordinates.
ln(P(x)/P(y)), where F is the free energy surface, kB is the Boltzmann constant, T is temperature and P(x)/P(y) is the probability distribution along the selected reaction coordinates.| ΔGbind = Gcomplex − (Gprotein + Gligand) | (1) |
| Gx = 〈EMM〉 + 〈Gsol〉 − TΔS | (2) |
| 〈EMM〉 = 〈Ebonded〉 + 〈Enon-bonded〉 = 〈Ebonded〉 + (EvdW + Eelec) | (3) |
| 〈Gsol〉 = Gpolar + Gnon-polar = Gpolar + (γ × SASA + b) | (4) |
2.2 Plant identification and crude extraction
The collection sites for A. indica plant leaves (Pwani, Tanzania, voucher no. 980) and C. odorata bark (Kilombero, Tanzania, voucher no. 981) were selected deliberately. Botanical identification and voucher specimens were deposited in the Herbarium of the Institute of Traditional Medicine, the Muhimbili University of Health and Allied Sciences. Powdered air-dried backs of C. odorata (500 g) and leaves of A. indica (500 g) were extracted separately with methanol and ethanol. The leaves were soaked for 24 hours, and the process was repeated three times to exhaust the extracts. The filtrates obtained were concentrated under decreased pressure and kept at 4 °C until needed.2.3 In vitro studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 96-well format, and compounds and extracts were then added to final concentrations in 3-fold replicates and incubated for an additional 2 hours before infection with 150× TCID50 of SARS-CoV-2 virus (USA-WA1/2020). Cells were incubated for an additional 96 hours, treated with Alamar Blue for 4 hours, fixed with paraformaldehyde to a final concentration of 4%, and incubated at room temperature for at least 30 minutes to inactivate the virus. Fluorescence intensity was then measured using a ClarioStar plate reader. Background fluorescence was subtracted from wells containing resazurin and D10+ medium but no cells.
000 cells per well in 96-well format, and compounds and extracts were then added to final concentrations in 3-fold replicates and incubated for an additional 2 hours before infection with 150× TCID50 of SARS-CoV-2 virus (USA-WA1/2020). Cells were incubated for an additional 96 hours, treated with Alamar Blue for 4 hours, fixed with paraformaldehyde to a final concentration of 4%, and incubated at room temperature for at least 30 minutes to inactivate the virus. Fluorescence intensity was then measured using a ClarioStar plate reader. Background fluorescence was subtracted from wells containing resazurin and D10+ medium but no cells.3 Results and discussion
3.1 Salvinorin A and deacetylgedunin (DCG) block SARS-CoV-2 viral cell entry through TMPRRS2 inhibition
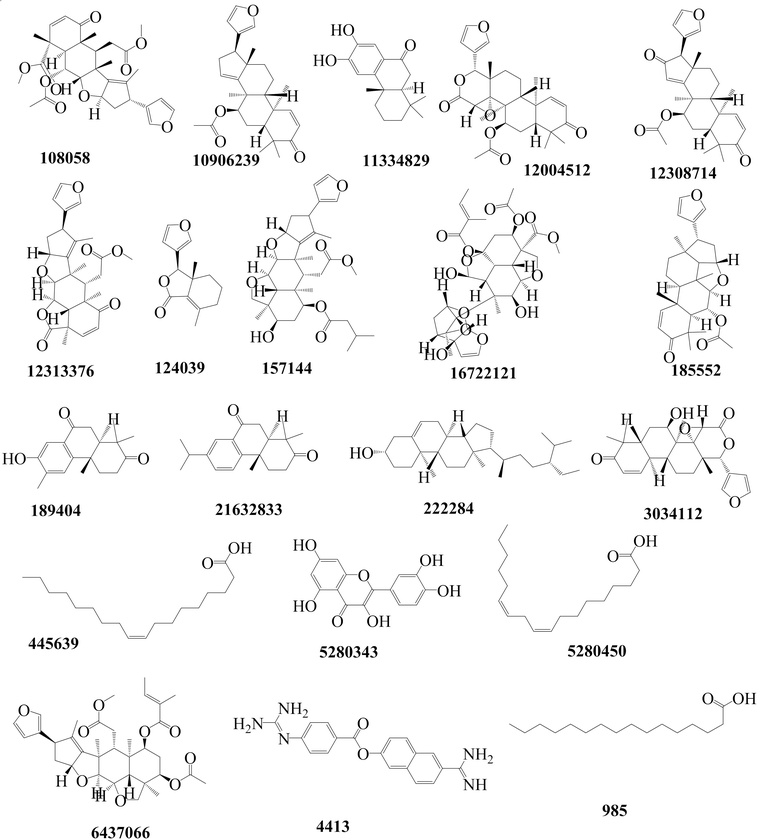
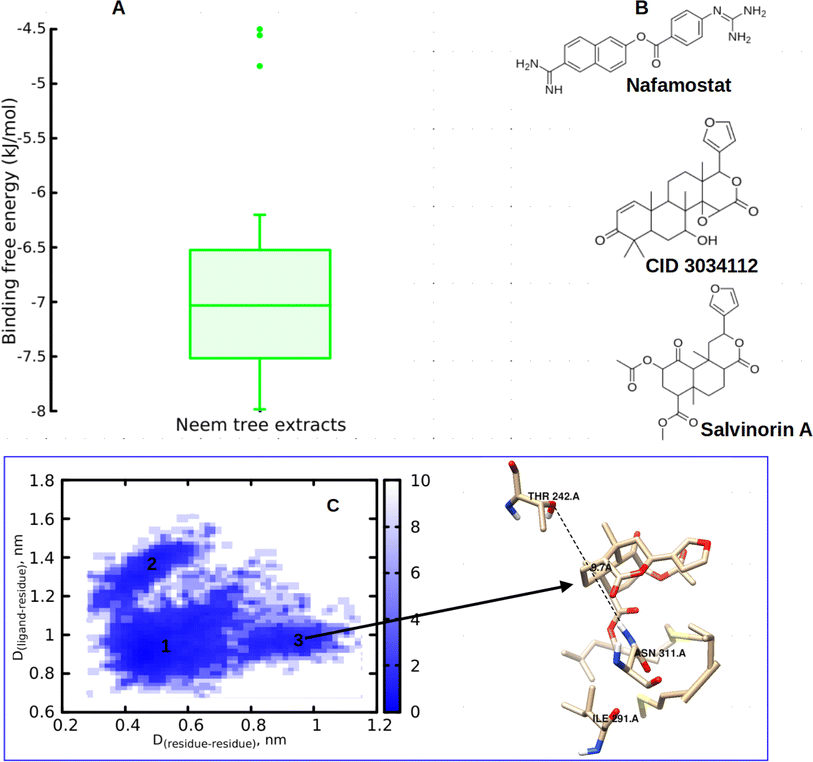
In this study, we began by investigating the potential of neem tree extracts to impede the enzymatic activity of TMPRSS2 protein, a critical player known for activating the ACE2 receptor binding domain, thus facilitating viral entry into human cells. It is worth noting that we have previously reported the ADMET profile of the neem tree extract, and DCG exhibited favourable drug-like properties.20 Conversely, salvinorin A is an approved molecule with a well-established ADMET profile. Furthermore, before commencing docking calculations, we thoroughly validated our docking protocols using an experimental dataset, as detailed in our previous works.20,23 Through molecular docking calculations (Fig. 3A), we identified compounds from the neem tree demonstrating noteworthy binding affinities, comparable to that of the reference molecule, nafamostat (−7.8 kcal mol−1). This suggested that these neem-derived compounds have the potential to obstruct viral cell entry by inhibiting TMPRSS2. In our pursuit of identifying clinically proven medications for combating COVID-19, we conducted a similarity search on the top-scoring neem tree extracts, with a focus on DCG (CID 3034112) which had an average binding affinity of −7.86 kcal mol−1. Our investigation also led to the identification of salvinorin A with a Tanimoto index of 0.9. We hypothesized that molecules with structural similarities often share similar modes of action. As anticipated, salvinorin A exhibited a comparable average binding affinity of −7.9 kcal mol−1. Since our objective was to find molecules in clinical applications for other indications that can be repurposed, our discussion will now focus on salvinorin A. To further elucidate the inhibitory effects of salvinorin A on the enzymatic catalytic activities of TMPRSS2, we conducted classical molecular dynamics simulations. The 2D free energy surface, considering residues–residues and residues–ligand distances, revealed three minima, with the deepest minimum observed at 0.5, 0.9 nm for residues–residues, residue–ligand distances. This indicates the potential of salvinorin A to block the enzymatic catalytic activities of TMPRSS2 by increasing the distances between residues. Notably, catalytic residues Thr242 and Asn311 are pivotal for the activation and catalytic activities of the protein. TMPRSS2 inhibitors, such as nafamostat, typically block the recognition of these residues, thereby interfering with catalytic activities. We performed MM-PBSA free energy calculation, the time-lapse binding free energy suggests that salvinorin A remained stable with a strong affinity until 50 ns, where it showed the unbinding and binding pattern from the protein, which considerably reduced its binding affinity (Fig. S1A–C†). Such observed fluctuations suggest that salvinorin A could exhibit inhibition to TMPRSS2 albeit with low efficacy. It is noteworthy that we extended our analysis by correlating the free energy with three key reaction coordinates: RMSD, pose RMSD, and distance (between the protein active site and ligand). Among these, only pose RMSD exhibited a consistent fluctuation pattern from 50 ns onwards, mirroring the observations from the time-dependent free energy calculation (Fig. S1D†). This suggests a significant role of pose RMSD in capturing the unbinding process, whereas distance and RMSD displayed negligible changes over time. This underscores the potential efficacy of pose RMSD as a reliable reaction coordinate in drug design for predicting in vitro or in vivo activities. Given that salvinorin A is an approved kappa opioid receptor (KOR) agonist and a drug for neurological diseases, this discovery motivated us to explore its efficacy in blocking the spike ACE2–RBD interaction and inhibiting Mpro, as detailed in the subsequent sections (Table 1).| Ligand ID | TMPRSS2 |
|---|---|
| CID 108058 | −6.88 |
| CID 10906239 | −7.20 |
| CID 11334829 | −6.88 |
| CID 12004512 | −7.50 |
| CID 12308714 | −7.60 |
| CID 12313376 | −7.60 |
| CID 124039 | −6.20 |
| CID 157144 | −6.50 |
| CID 16722121 | −7.50 |
| CID 185552 | −7.52 |
| CID 189404 | −7.00 |
| CID 21632833 | −7.06 |
| CID 222284 | −6.54 |
| CID 3034112 | −7.86 |
| CID 445639 | −4.56 |
| CID 5280343 | −7.50 |
| CID 5280450 | −4.84 |
| CID 6437066 | −6.54 |
| CID 985 | −4.50 |
| Nafamostat | −7.80 |
| Salvinorin A | −7.90 |
3.2 Inhibition of ACE2–RBD interaction by salvinorin A and DCG
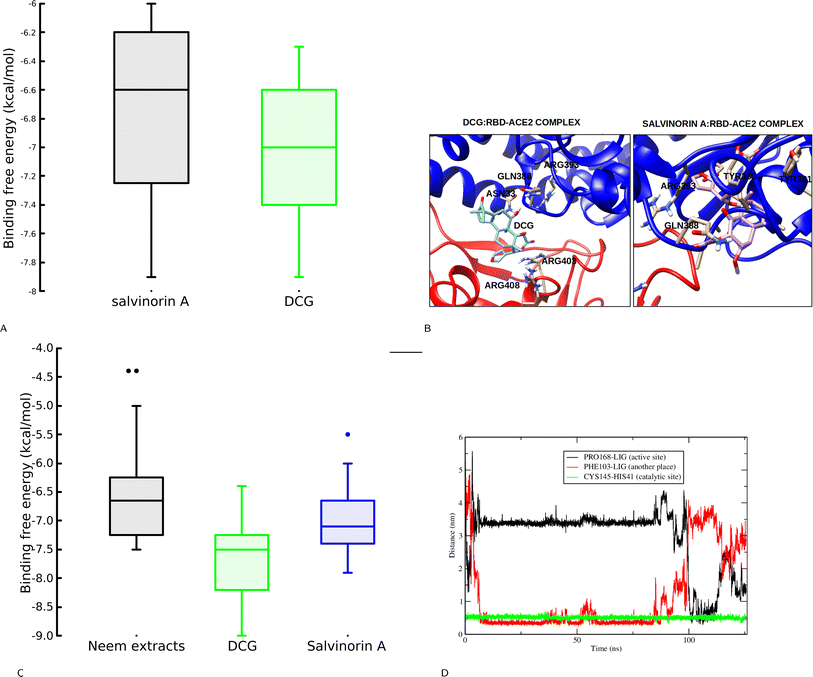
This section explores the ability of identified salvinorin A and DCG to block viral cell entry through ACE2–RBD inhibition. To deepen our understanding, both rigid and flexible docking calculations based on a relaxed complex scheme (RCS) were performed. At first glance, the rigid docking calculation resulted in a binding affinity of −6.6 and −7.4 kcal mol−1 for salvinorin A and DCG, respectively. This encouraged us to perform an RCS which has been effective in reducing biases in docking calculations.24,30 Interestingly, some snapshots from RCS showed higher binding affinity as compared to the crystal structure with an average binding affinity of −6.75 and −7.0 kcal mol−1 for salvinorin A and DCG, respectively (Fig. 4A). Binding mode analysis shows that salvinorin A bound at the interface and interacted with Tyr505, Arg403, Tyr463, and Lys417 from the RBD and residues from the human ACE2 were His42, Asn33, Glu37, and Arg393 thereby, blocking the interaction of the human ACE2 and the viral RBD hence preventing its entry into human cells (Fig. 4B). Interestingly, although DCG bound at the same place as salvinorin A, it has a different binding mode with additional interacting residues which could be attributed to its observed strong binding affinity. The furan ring in DCG is pointing towards the spike RBD while in salvinorin A, the furan points towards the human ACE2 (Fig. 4B). Both docking calculation based on rigid and ensemble structures from MD simulation suggests that salvinorin A could show activity in vitro, despite it having slightly less affinity as compared to DCG. Indeed, this observation is consistent with our in vitro assay described in subsequent subsections. Previous in silico and in vitro studies have shown that both ligands or peptides binding at the spike ACE2–RBD and interacting with Tyr505, Arg403, Lys417, His42, Glu37, Asn33 and Arg393 can potentially block viral cell entry.24,31–333.3 Inhibition of Mpro by salvinorin A and DCG
To gain a deeper understanding of the potential inhibitory effect of salvinorin A and DCG on SARS-CoV-2 Mpro, we next conducted molecular dynamics-based docking calculations to assess the stability of its binding to the target protein. The Mpro's binding pocket and structure have been extensively characterized, consisting of three distinct subunits: S1, S2, and S3 with three domains I–III (domain I, residues 8–101, domain II, residues 102–184 and domain III, residues 201–303).34,35 It is known that a co-crystallized N3 covalent inhibitor binds to the catalytic dyad cavity which can accommodate four substrate residues spanning from position P1′ to P4.34,35 Our initial docking calculations indicated that salvinorin A binds to the Mpro pocket with a binding affinity of −6.2 kcal mol−1. It forms interactions with P′ catalytic dyad cavity residues His41 and Cys145. The limitations of the current docking algorithm, such as the lack of global protein flexibility and the omission of water molecules, caution us against placing full trust in docking poses and affinities. Consequently, we conducted classical molecular dynamics (MD) simulations to gain deeper insights into binding stability. MD simulations offer the advantage of providing protein sampling and flexibility, allowing the ligand to explore various binding poses more freely. We further extracted 20 snapshots from the MD simulation and performed docking calculations. It is interesting, to note that, when neem extracts docked to single crystal structures DCG exhibited a superior binding affinity of −7.5 kcal mol−1. DCG and salvinorin A were further docked to the 20 ensemble structures, it's worth noting, that ensemble structures improved the sensitivity of docking, as a result, both DCG and salvinorin A had good scores compared to the crystal structure (Fig. 4C).The MD simulation of salvinorin A complex demonstrated a dynamic behavior by shifting from its initial docking position (P1′) within the first 5 nanoseconds, eventually accommodating the cavity P4 where the N3 inhibitor binds. After 100 nanoseconds, salvinorin A resumed its original docking pose at the cavity (P1′) interacting with the catalytic dyad residues with fluctuations (Fig. 4D). The observed dynamical changes suggest that salvinorin A may undergo different adaptations inside the cavity, potentially leading to the inhibition of viral enzymatic activities. Furthermore, this hypothesis is supported by the increased distance of the catalytic dyad residues His41–Cys145, which measured 5 Å throughout the simulation time, compared to the crystal structure distance of 3.8 Å (Fig. 4D). Notably, Mpro's enzymatic activity hinges on catalytic site reactions, and inhibiting these activities disrupts viral polymerization, ultimately curbing SARS-CoV-2 viral replication. These findings indicate that salvinorin A exhibits activity against Mpro, albeit with a moderate inhibitory potential. This observation is consistent with the results reported by Ullah et al., through docking calculations.36 Our in silico findings align well with our in vitro results, where salvinorin A displayed a moderate yet significant inhibition of Mpro and viral replication.
3.4 In vitro inhibition of SARS-CoV-2 by salvinorin A and plant extracts
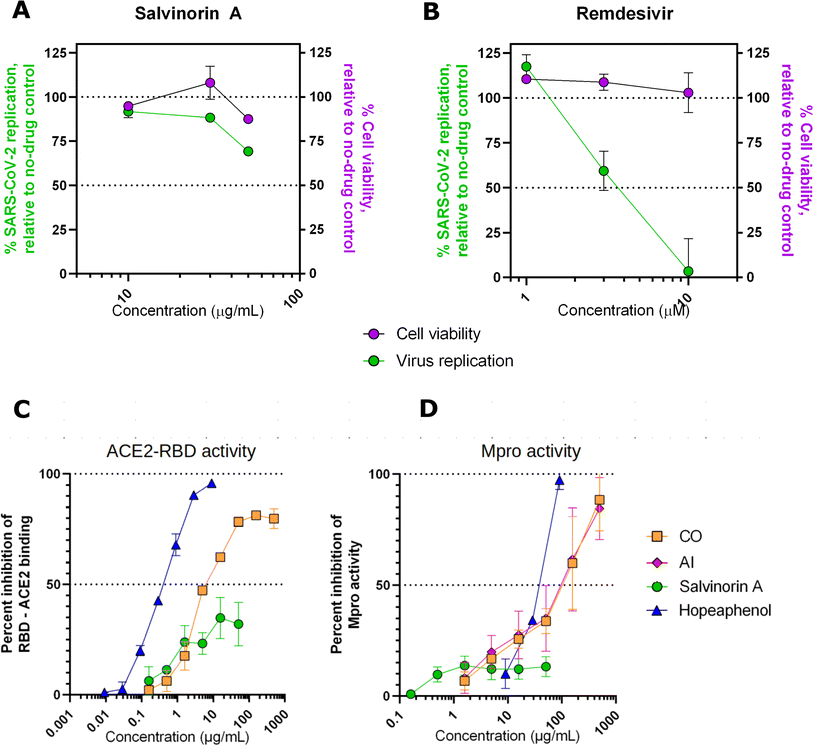
Previous in silico subsections have highlighted the potential of salvinorin A as a potential lead to inhibit SARS-CoV-2 through multiple mechanisms. While salvinorin A was readily obtained commercially, we were unable to purchase DCG due to supplier issues. To circumvent this gap, we then searched for plants with documented high levels of DCG. Literature shows that DCG, besides being found in neem trees, can be isolated from Cedrela odorata.37 This piqued our interest to investigate the potential of DCG derived from Cedrela odorata, CO, and neem tree (Azadirachta indica, AI). As a result, we extended our research to include the assessment of crude extracts from CO and AI for their anti-SARS-CoV-2 activities.To assess the effects of salvinorin A and the plant extracts on live SARS-CoV-2 replication, we performed cell viability restoration studies in the presence of infectious SARS-CoV-2 as previously described.29 Briefly, Vero-E6 cells were incubated with the compound for 2 hours before infection with SARS-CoV-2 (USA-WA1/2020, “wild-type” variant). After 4 days viability of infected cells was measured with Alamar Blue stain. In the absence of the drug, we observed an average 47.4 ± 21.1% (mean ± SD) reduction in viability consistent with cytopathic effects due to virus infection. In this assay, salvinorin A was observed to have moderate but reproducible antiviral activity, with 30.7 ± 1.8% and 11.7 ± 1.2% restoration of cell viability at 50 and 30 μg mL−1, respectively (Fig. 5A). Due to limits of salvinorin A solubility in this assay, we were unable to test higher concentrations; as a result, the full dose–response curve for this compound could not be achieved. In contrast, over 95% and 40.6 ± 11.0% restoration of cell viability was observed in cells treated with 10 or 3 μM, respectively, of control SARS-CoV-2 inhibitor remdesivir (Fig. 5B). These results indicate that the antiviral activity of salvinorin A, at least on its own, does not match the activity of licensed drugs like remdesivir. However, improved activity may be isolated from either compound combinations obtained from parental plants and/or future synthetic derivatives of salvinorin A. Our observation shows no major effects (i.e. >50%) on cell viability in the presence of salvinorin A in uninfected cells (Fig. 5A, purple), indicating that salvinorin A did not induce major cytotoxicity at antiviral concentrations.
Interestingly, as observed in our in silico calculations, salvinorin A demonstrated an appreciable viral cell entry inhibition at 10 μg mL−1 by inhibiting at least 40%, while CO crude extract showed more activity at 7 μg mL−1 by inhibiting 50% of RBD–ACE2 binding (AI could not be assessed in this assay due to autofluorescence issues). This suggests that both salvinorin A and the crude extracts possess spike RBD–ACE2 viral entry inhibitory activities. We noted that both crude extracts from Azadirachta indica (neem tree) and Cedrela odorata also exhibited strong activities compared to the pure salvinorin A. Both extracts were also able to inhibit 50% of the Mpro activities at 100 μg mL−1 concentration, while salvinorin A only inhibited ≈20% of Mpro activities at the same concentration. Based on our current findings, we hypothesize that the crude extracts have other compounds that work synergistically or additively toward inhibiting viral cell entry and the Mpro activities. The re-isolation and testing of individual compounds from the crude extracts of Cedrela odorata is highly recommended.
4 Conclusion
The discovery and development of new therapeutic leads to combat COVID-19 is an ongoing initiative in order to supplement vaccine efforts. Given the rapid advances in computational algorithms and hardware, it has become possible to screen large libraries and identify potential compounds. The work started by screening neem tree extracts and tested them for their potential to inhibit SARS-COV-2 by targeting TMPRRS2, ACE2–RBD, Mpro and viral replication. Salvinorin A obtained through similarity search demonstrated activity both in silico and in vitro by moderately inhibiting viral cell entry, Mpro and replication. Both crude extracts (CO and AI) showed more potent activity than salvinorin A. Further isolation and purification and in vitro testing of individual compounds from CO is therefore recommended. Further in vivo validation is also suggested towards developing more effective therapies for SARS-COV-2.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Software, writing-review & editing: Mariana J. Shayo, Daniel M. Shadrack, Geradius Deogratias, Ester Innocent, Ian Tietjen, Baraka Samwel, Samson Hilonga. Methodology, data analysis, visualization: Ian Tietjen, Daniel M. Shadrack, Joel Cassel, Joseph M. Salvino, Luis J. Montaner, Daniel M. Shadrack, Geradius Deogratias. Funding acquisition: Ester Innocent, Ian Tietjen, Mariana J. Shayo, Daniel M. Shadrack, Geradius Deogratias. Computational resources: Lucy Kiruri, Daniel M. Shadrack.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors, MJS, BS, DMS, GD, and EI, acknowledge the funding received from the Swedish International Development Cooperation Agency (Sida seed grant) for Junior Faculty Scheme through Muhimbili University of Health and Allied Sciences. LJM and JS received support from the Wistar Science Discovery Fund, while IT received assistance from the Canadian Institutes for Health Research (CIHR PJT-153057). Additionally, LJM expresses gratitude for the support provided by the Robert I. Jacobs Fund of the Philadelphia Foundation and the Herbert Kean, M. D., Family Professorship. LK and DMS recognize the support for accessing the Lengau high-performance computing facility in South Africa. DMS acknowledges the AGNES junior research grant award.Notes and references
- J. S. Tregoning, K. E. Flight, S. L. Higham, Z. Wang and B. F. Pierce, Nat. Rev. Immunol., 2021, 21, 626–636 CrossRef CAS PubMed.
- T. Hartonen, B. Jermy, H. Sõnajalg, P. Vartiainen, K. Krebs, A. Vabalas, FinnGen, Estonian Biobank Research Team, T. Leino, H. Nohynek, J. Sivelä, R. Mägi, M. Daly, H. M. Ollila, L. Milani, M. Perola, S. Ripatti and A. Ganna, et al., Nat. Human Behav., 2023, 1–15 Search PubMed.
- Z. Huang, S. Xu, J. Liu, L. Wu, J. Qiu, N. Wang, J. Ren, Z. Li, X. Guo and F. Tao, et al., Nat. Commun., 2023, 14, 2009 CrossRef CAS PubMed.
- J. V. Lazarus, K. Wyka, T. M. White, C. A. Picchio, L. O. Gostin, H. J. Larson, K. Rabin, S. C. Ratzan, A. Kamarulzaman and A. El-Mohandes, Nat. Med., 2023, 1–10 Search PubMed.
- R. J. D. Vergara, P. J. D. Sarmiento and J. D. N. Lagman, J. Publ. Health, 2021, 43, e291–e292 CrossRef PubMed.
- J. V. Lazarus, K. Wyka, T. M. White, C. A. Picchio, K. Rabin, S. C. Ratzan, J. P. Leigh, J. Hu and A. El-Mohandes, Nat. Commun., 2022, 13, 3801 CrossRef CAS PubMed.
- A. I. Mohamud, S. A. Mohamed and K. Abdullahi, IOSR J. Dent. Med. Sci., 2021, 20, 1–4 Search PubMed.
- M. D. H. Hawlader, M. L. Rahman, A. Nazir, T. Ara, M. M. A. Haque, S. Saha, S. Y. Barsha, M. Hossian, K. F. Matin and S. R. Siddiquea, et al., Int. J. Infect. Dis., 2022, 114, 1–10 CrossRef CAS PubMed.
- J. Shawon, Z. Akter, M. M. Hossen, Y. Akter, A. Sayeed, M. Junaid, S. S. Afrose and M. A. Khan, Curr. Pharm. Des., 2020, 26, 5241–5260 CrossRef CAS PubMed.
- A. Hensel, R. Bauer, M. Heinrich, V. Spiegler, O. Kayser, G. Hempel and K. Kraft, Planta Med., 2020, 86, 659–664 CrossRef CAS PubMed.
- H. Zhang, J. M. Penninger, Y. Li, N. Zhong and A. S. Slutsky, Intensive Care Med., 2020, 46, 586–590 CrossRef CAS PubMed.
- S. Jo, S. Kim, D. H. Shin and M.-S. Kim, J. Enzym. Inhib. Med. Chem., 2020, 35, 145–151 CrossRef CAS PubMed.
- A. d. S. Antonio, L. S. M. Wiedemann and V. F. Veiga-Junior, RSC Adv., 2020, 10, 23379–23393 RSC.
- Y. Chen, Q. Liu and D. Guo, J. Med. Virol., 2020, 92, 418–423 CrossRef CAS PubMed.
- Y. W. Chen, C.-P. B. Yiu and K.-Y. Wong, F1000Research, 2020, 9, 1–19 Search PubMed.
- B. Krichel, S. Falke, R. Hilgenfeld, L. Redecke and C. Uetrecht, Biochem. J., 2020, 477, 1009–1019 CrossRef CAS PubMed.
- K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Science, 2003, 300(5626), 1763–1767 CrossRef CAS PubMed.
- V. Mody, J. Ho, S. Wills, A. Mawri, L. Lawson, M. C. Ebert, G. M. Fortin, S. Rayalam and S. Taval, Commun. Biol., 2021, 4, 93 CrossRef CAS PubMed.
- J. Zhu, H. Zhang, Q. Lin, J. Lyu, L. Lu, H. Chen, X. Zhang, Y. Zhang and K. Chen, Drug Des., Dev. Ther., 2022, 1067–1082 CrossRef CAS PubMed.
- D. M. Shadrack, S. A. Vuai, M. G. Sahini and I. Onoka, RSC Adv., 2021, 11, 26524–26533 RSC.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, J. Cheminf., 2011, 3, 1–14 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- D. M. Shadrack, H. S. Swai and A. Hassanali, J. Mol. Graph. Model., 2020, 96, 107510 CrossRef CAS PubMed.
- D. M. Shadrack, G. Deogratias, L. W. Kiruri, I. Onoka, J.-M. Vianney, H. Swai and S. S. Nyandoro, J. Mol. Model., 2021, 27, 1–15 CrossRef PubMed.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- R. Kumari, R. Kumar, O. S. D. D. Consortium and A. Lynn, J. Chem. Inf. Model., 2014, 54, 1951–1962 CrossRef CAS PubMed.
- I. Tietjen, J. Cassel, E. T. Register, X. Y. Zhou, T. E. Messick, F. Keeney, L. D. Lu, K. D. Beattie, T. Rali and P. Tebas, et al., Antimicrob. Agents Chemother., 2021, 65, 10–1128 CrossRef PubMed.
- J.-H. Lin, A. L. Perryman, J. R. Schames and J. A. McCammon, J. Am. Chem. Soc., 2002, 124, 5632–5633 CrossRef CAS PubMed.
- M. Quagliata, M. A. Stincarelli, A. M. Papini, S. Giannecchini and P. Rovero, ACS Omega, 2023, 8(25), 22665–22672 CrossRef CAS PubMed.
- S. Pourmand, S. Zareei, M. Shahlaei and S. Moradi, Comput. Biol. Med., 2022, 146, 105625 CrossRef CAS PubMed.
- G. Jaiswal and V. Kumar, PLoS One, 2020, 15, e0240004 CrossRef CAS PubMed.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang and C. Peng, et al., Nature, 2020, 582, 289–293 CrossRef CAS PubMed.
- D. W. Kneller, G. Phillips, H. M. O'Neill, R. Jedrzejczak, L. Stols, P. Langan, A. Joachimiak, L. Coates and A. Kovalevsky, Nat. Commun., 2020, 11, 3202 CrossRef CAS PubMed.
- S. Ullah, B. Munir, A. G. Al-Sehemi, S. Muhammad, I.-u. Haq, A. Aziz, B. Ahmed and A. Ghaffar, Saudi J. Biol. Sci., 2022, 29, 103274 CrossRef CAS PubMed.
- T. S. R. Nogueira, M. d. S. Passos, L. P. S. Nascimento, M. B. d. S. Arantes, N. O. Monteiro, S. I. d. S. Boeno, A. d. C. Junior, O. d. A. Azevedo, W. d. S. Terra and M. G. C. Vieira, et al., Molecules, 2020, 25, 5401 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra02593h |
| This journal is © The Royal Society of Chemistry 2024 |