Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
5-Aminoisophthalate-based kojic acid-appended bis-1,2,3-triazole: a fluorescent chemosensor for Cu2+ sensing and in silico study†
Sachin Kumar ab,
Bajrang Lala,
Gurleen Singh
ab,
Bajrang Lala,
Gurleen Singh c,
Muskand,
Ram Kumar Tittal*a,
Jandeep Singh
c,
Muskand,
Ram Kumar Tittal*a,
Jandeep Singh *c,
Ghule Vikas D.
*c,
Ghule Vikas D. a and
Renu Sharmab
a and
Renu Sharmab
aDepartment of Chemistry, National Institute of Technology, Kurukshetra, Haryana 136119, India. E-mail: rktittaliitd@nitkkr.ac.in; Tel: +91-1744-233-542
bSchool of Applied Sciences, Om Sterling Global University, Hisar, Haryana 125001, India
cSchool of Chemical Engineering and Physical Sciences, Lovely Professional University, Phagwara, Punjab 144411, India. E-mail: singhjandeep@gmail.com
dDepartment of Chemistry, University of Delhi, Delhi 110007, India
First published on 3rd July 2024
Abstract
A new, easy-to-prepare, and highly selective fluorescent chemosensor, i.e., 5-aminoisophthalate-based kojic acid-appended bis-1,2,3-triazole, was synthesized from an alkyne of 5-aminoisophthalic acid and azido-kojic acid using Cu(I)-catalyzed click chemistry and then successfully characterized. The alkyne structure of 5-aminoisophthalic acid, 1, was supported by the single-crystal X-ray crystallographic data. The fluorescent probe 3 was found to be highly selective for Cu2+ ions supported by the Job's plot with a stoichiometric ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) metal ratio of 2
metal ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, exhibiting almost a two-fold enhancement in the emission intensity upon the addition of Cu2+ ions (0–25 μM) with a detection limit of 8.82 μM. A comparison with LODs from previously developed chemosensors for Cu2+ ions was also conducted. Reversibility analysis indicated that probe 3 could be used as both a reusable sensor and as a scavenger of copper ions. DFT calculations with the basis sets B3LYP/6-311G(d,p) and LanL2DZ were employed for geometrical optimizations of structures of the alkyne 1, azide 2, probe 3, and complex 3.Cu2+. Hirshfeld surface analysis revealed significant intermolecular interactions in compound 1. Additionally, molecular docking for the antimicrobial activity showed the better antibacterial efficacy of probe 3.
1, exhibiting almost a two-fold enhancement in the emission intensity upon the addition of Cu2+ ions (0–25 μM) with a detection limit of 8.82 μM. A comparison with LODs from previously developed chemosensors for Cu2+ ions was also conducted. Reversibility analysis indicated that probe 3 could be used as both a reusable sensor and as a scavenger of copper ions. DFT calculations with the basis sets B3LYP/6-311G(d,p) and LanL2DZ were employed for geometrical optimizations of structures of the alkyne 1, azide 2, probe 3, and complex 3.Cu2+. Hirshfeld surface analysis revealed significant intermolecular interactions in compound 1. Additionally, molecular docking for the antimicrobial activity showed the better antibacterial efficacy of probe 3.
1. Introduction
In the realm of chemical and biological research, the development of innovative fluorescent probes for the selective detection of metal ions and exploration of potential antimicrobial agents with desired characteristics have become paramount. Among these, exploring diverse heterocyclic scaffolds has attracted significant attention owing to their versatile applications in various fields including medicinal chemistry and materials science.1,2 Due to their unique electronic and structural properties, triazoles, a class of five-membered heterocyclic compounds, have emerged as intriguing candidates. Amongst the various fluorescent chemosensors available, triazoles, obtained via CuAAC through a ‘click’ methodology,3–6 which is considered the ideal synthetic route to form 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole derived from natural compounds, offer unique advantages due to their biocompatibility and eco-friendly nature. Kojic acid, a natural product obtained from fungi, has attracted significant attention for its diverse applications in medicine and materials science.7–10 The application of the synthesized 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole as a sensing platform for heavy and transition metal ions is of specific interest after prior research in related fluorescent and biologically significant hybrid compounds.11,12The significant increase in the accumulation of metal ions beyond acceptable levels has underscored the urgent need to investigate effective and robust research substitutes for their immediate recognition, even at low concentrations.13,14 The focus on metal cation detection arises from their extensively documented harmful effects on terrestrial and aquatic ecosystems,15,16 making their qualitative and quantitative identification imperative.17 As the third-most-common heavy metal ion in the human body, bivalent copper (Cu2+) is an essential ion that plays several roles in biological processes. Functioning as a catalytic cofactor for metalloenzymes,18 it plays a pivotal role in many critical physiological functions. However, an intricate balance of metal ions is crucial, as the dysregulation of Cu2+ ions can lead to oxidative stress and associated health complications. Copper deficiency in the human body is implicated in several serious conditions, such as Alzheimer's disease,19,20 and Wilson's disease.21
On the other hand, elevated concentrations of copper pose risks to vital organs, particularly the liver and kidneys.22 Therefore, it is crucial to maintain copper homeostasis, which calls for sophisticated regulatory mechanisms. Environmental pollution resulting from the accumulation of Cu2+ ions through industrial discharge is also a matter of concern.23
The research community is compelled to explore cutting-edge approaches for the efficient and selective detection of this metal, driven by various factors. Moreover, the significance of optical technologies has grown substantially owing to their sensitive responses, practical feasibility, economic viability, and abundant biocompatibility. These attributes collectively enable their extension to real-time applications, facilitating the monitoring of copper levels in biological and environmental contexts.
This research study explored the synthesis of 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole, and investigated its promising potential, particularly in metal ion sensing and in silico exploration of the synthesized probe's potential antimicrobial properties. The focus of this study revolved around the strategic incorporation of 5-amino isophthalic acid and kojic acid as crucial precursors in the synthesis process. This choice is not arbitrary but stems from their distinctive reactivity, e.g., using 5-amino isophthalic acid, with its amino functionality and aromatic character, is anticipated to introduce desirable electronic properties to the triazole backbone. Concurrently, the inclusion of kojic acid, renowned for its chelating capabilities and excellent coordination properties,24,25 can add an element of selectivity and sensitivity to the synthesized chemosensor, particularly in the context of metal ion recognition. Molecular docking, a powerful computational technique, was used to investigate the interaction between the probe and selected microbial targets. This dual-purpose investigation sheds light on the probe's ability to selectively detect Cu2+ ions and its potential as an antimicrobial agent. The outcomes of this research hold promise for contributing to the advancement of fluorescence-based sensing technologies and the discovery of novel antimicrobial agents with potential therapeutic applications.
Considering our experience in the synthesis of hybrid compounds linked with 1,2,3-triazole/fluorophoric groups for designing pharmacologically important compounds or chemosensors for selectively detecting metal ions/biothiols,26,27 we herewith present for the first time a 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole as a chemosensor for the selective detection of Cu2+ ions in the presence of other metal ions, such as Pb2+, Cr3+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Hg2+, Na+, K+, Mg2+, Ca2+, and Ba2+.
2. Experimental section
2.1 Materials and methods
All chemicals utilized in this study were procured from reputable suppliers. Specifically, 5-aminoisophthalic acid, K2CO3, NaN3, and various metal chlorides (Cr3+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Hg2+, Na+, K+, Mg2+, Ca2+, and Ba2+) and lead acetate: Pb2+ were sourced from Sigma Aldrich. Kojic acid was acquired from TCI Chemicals, while propargyl bromide was obtained from Avra. Quinine sulfate served as the reference standard for determining the fluorescence quantum yields. All solvents were purchased from Loba Chemie. NMR spectra (1H and 13C) were recorded using a Bruker Avance (II) 500 MHz instrument in CDCl3 and DMSO-d6. High-resolution mass spectrometry (HRMS) analyses were performed using an electrospray mass spectrometer. The Fourier transform infrared (FTIR) spectra were recorded using a Shimadzu IR Affinity-1S spectrophotometer. Sensing studies were conducted using SHIMADZU UV-1900 and PerkinElmer FL 6500 spectrophotometers.2.2 Synthesis of di(prop-2-yn-1-yl) 5-aminoisophthalate, 1
The alkyne synthesis was commenced by treating 5 mmol of 5-aminoisophthalic acid with 11 mmol propargyl bromide in 10 mL of DMF, utilizing 25 mmol anhydrous potassium carbonate as a base. The reaction, monitored by TLC, proceeded over 10 h. After the reaction was finished, ice-cold water was used to quench the mixture, yielding a white solid. The precipitate was thoroughly washed with chilled water and subsequently dried in a vacuum. Purification of the alkyne involved passing it through a short silica gel column using ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexane (2
hexane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v) as the eluent. This purification process isolated alkyne 1 as a white solid with an impressive 88% yield.
8, v/v) as the eluent. This purification process isolated alkyne 1 as a white solid with an impressive 88% yield.
Appearance: white solid; yield: 88%; mpt: 90–92 °C; FTIR (νmax cm−1): 3421, 3350, 3305, 3269, 2129, 1716, 1697, 1600, 1444, 1330, 1234, 1132, 1016, 906, 758, 553; 1H NMR (500 MHz, DMSO-d6) δ = 7.64(t, J = 1.55, 1H), 7.44 (d, J = 1.50, 2H), 5.83 (s, 2H), 4.95 (d, J = 2.35, 4H), 3.62 (t, J = 2.45, 2H); 13C NMR (126 MHz, CDCl3) δ = 165.27 (C), 147.02 (C), 130.99 (C), 121.11 (CH), 120.31 (CH), 77.67 (C), 75.35 (CH), 52.82 (CH2); HRMS (ESI+): [M + H]+ calculated for C14H11NO4+ is 258.0761, found 258.0767.
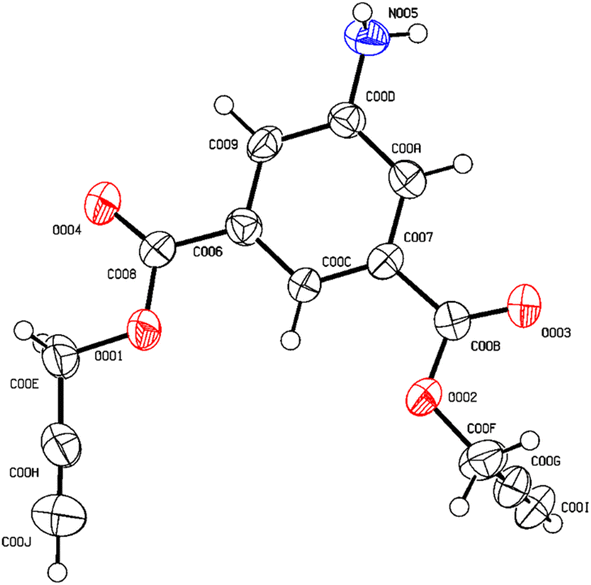
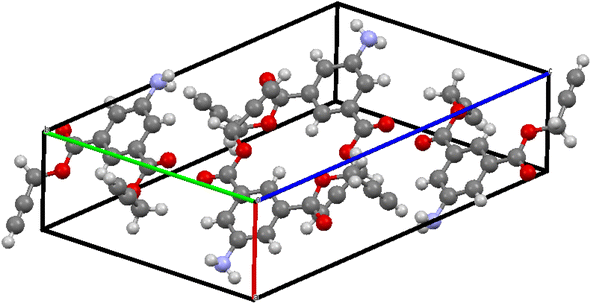
After undergoing recrystallization with a solution containing acetonitrile and n-hexane, the initially formed solid crystalline material successfully transformed into a singular crystal of compound 1, identified by CCDC no. 2300460 and depicted in Fig. 1. The complete crystal data and its structure refinement are presented in Table 1 and Fig. 2.
| a R = ∑(‖Fo| − |Fc‖)/∑|Fo|.b Rw = {∑[w(Fo2 − Fc2)2]/∑[w(Fo2)2]}1/2. | ||
|---|---|---|
| Identification code | 1 | |
| Empirical formula | C13.75H10.75NO4 | |
| Formula weight | 253.90 | |
| Temperature | 216 K | |
| Wavelength | 0.71073 Å | |
| Crystal system | Monoclinic | |
| Space group | P21/c | |
| Unit cell dimensions | a = 4.7711(6) | α = 90 |
| b = 13.430(5) | β = 94.722(10) | |
| c = 19.544(2) | γ = 90 | |
| Volume | 1248.1(5) Å3 | |
| Z | 4 | |
| Density (calculated) | 1.352 g cm−3 | |
| Absorption coefficient | 0.101 mm−1 | |
| F(000) | 529.0 | |
| Index ranges | h = 5, k = 11, l = 22 | |
| Reflections collected | 0.0567(807) | |
| Completeness to theta = 26.37° | 99.7% | |
| Final R indices [I > 2 sigma(I)]a,b | R1 = 0.0567, wR2 = 0.1805 | |
2.3 Synthesis of 2-(azidomethyl)-5-hydroxy-4H-pyran-4-one,28 2
The chloro derivative of kojic acid was synthesized by following the published literature with a few extra tweaks.29 In a 50 mL round-bottom flask, 1 mmol of kojic acid was dissolved in 2 equivalents of thionyl chloride at room temperature. The reaction mixture was stirred for 2 h. Upon completion of the reaction, work-up was performed by evaporating the reaction mixture using a rotary evaporator. After drying, the resulting solid was washed with ample water and utilized directly for azidation. A solution of sodium azide (1.5 equivalents) was heated to 85–90 °C for a minimum of 30 min. The obtained chlorokojic solid was then added to the heated solution, and at room temperature, it was stirred overnight. After the reaction, ice-cold water was used to quench the mixture, and the residue was filtered, washed with cold water, and vacuum-dried. The resulting brown solid was recrystallized in THF to obtain a pure compound.Appearance: brown solid; yield: 85%; mpt: 129–130 °C; FTIR (νmax cm−1): 3331, 3236, 3059, 3035, 2119, 1658, 1629, 1450, 1379, 1269, 1145, 929, 790, 663, 530; 1H NMR (500 MHz, CDCl3) δ = 8.02 (s, 1H), 7.86 (s, 1H), 6.49 (s, 1H), 4.21 (s, 2H); 13C NMR (126 MHz, CDCl3) δ = 173.74 (C), 162.66 (C), 145.73 (C), 137.81 (CH), 111.18 (CH), 51.13 (CH2); HRMS (ESI+): [M + H]+ cal. for C6H6N3O3+ is 168.0404, found 168.0413.
2.4 General procedure for synthesizing 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole, 3
First, 12 mL of DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8
H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v) mixture was added to a 100 mL round-bottom flask. The reactants, including 1 mmol of alkyne, 2.1 mmol of organic azide, and CuSO4·5H2O (10 mol%) along with sodium ascorbate (20 mol%) as a catalyst, were introduced into the flask with slight modifications based on literature procedures. The resulting reaction mixture was stirred at room temperature, and TLC analysis was performed and confirmed the completion of the reaction within 30 min. After the reaction was finished, the mixture was cautiously transferred into a beaker filled with ice-cold water. After filtering, extensively washing with cooled water, and vacuum-drying the resultant solid product, the pure chemical was obtained with an amazing 93% yield.
2 v/v) mixture was added to a 100 mL round-bottom flask. The reactants, including 1 mmol of alkyne, 2.1 mmol of organic azide, and CuSO4·5H2O (10 mol%) along with sodium ascorbate (20 mol%) as a catalyst, were introduced into the flask with slight modifications based on literature procedures. The resulting reaction mixture was stirred at room temperature, and TLC analysis was performed and confirmed the completion of the reaction within 30 min. After the reaction was finished, the mixture was cautiously transferred into a beaker filled with ice-cold water. After filtering, extensively washing with cooled water, and vacuum-drying the resultant solid product, the pure chemical was obtained with an amazing 93% yield.
Appearance: brownish yellow solid; yield: 93%; mpt: 189–190 °C; FTIR (νmax cm−1): 3442, 3361, 3234, 3140, 3088, 2964, 1722, 1625, 1453, 1350, 1222, 1122, 970, 756; 1H NMR (500 MHz, DMSO-d6) δ = 9.32 (s, 2H), 8.36 (s, 2H), 8.06 (s, 2H), 7.61 (s, 1H), 7.38 (s, 2H), 6.43 (s, 2H), 5.62 (s, 4H), 5.39 (s, 4H), 4.94 (s, 2H); 13C NMR (126 MHz, DMSO-d6) δ = 173.51 (C), 165.08 (C), 160.35 (C), 149.50 (C), 145.94 (C), 142.11 (C), 139.94 (C), 130.29 (CH), 125.80 (CH), 118.17 (CH), 116.64 (CH), 113.06 (CH), 57.67 (CH2), 49.89 (CH2); HRMS (ESI+): [M + H]+ cal. for C26H21N7O10+ is 592.1423, found 592.1431.
2.5 Spectrophotometric analysis
The sensing behavior of probe 3 was thoroughly investigated using UV-visible absorption spectroscopy. Metal ion recognition was explored with a diverse range of metal ions, including Cr3+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Hg2+, Pb2+, Na+, K+, Mg2+, Ca2+, and Ba2+. The sensitivity of probe 3 toward Cu2+ ions was specifically analyzed using fluorescence spectroscopy. In-depth studies on the time and temperature dependence of the probe, as well as competing metal ion titrations, were conducted to provide a comprehensive understanding of its behavior. The determination of the association constant (Ka) was achieved through the application of a Benesi–Hildebrand (B–H) plot. Ka was calculated using the formula Ka = 1/slope, where the slope was obtained by plotting 1/concentration of the analyte vs. 1/change in absorbance. Calculations of the limit of detection (LOD) and the limit of quantification (LOQ)30 were done using 3σ/S and 10σ/S, respectively. In these calculations, σ stands for the standard deviation of the blank, and S for the slope of the calibration curve. This methodology ensured a rigorous evaluation of the probe's sensing capabilities and provides valuable insights into its potential applications.2.6 Computational studies
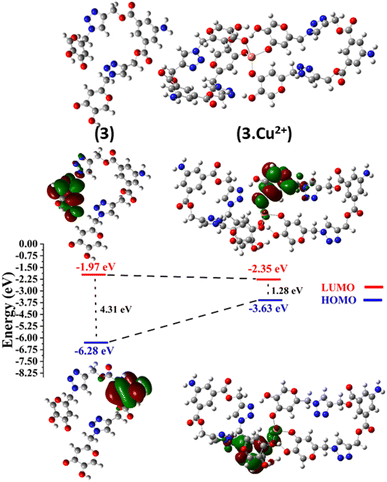
Utilizing the Gaussian09 program suite,31 the stability assessments and geometrical optimizations of 1, 2, and probe 3 were conducted employing the B3LYP32/6-311G (d,p) basis set. For complex 3.Cu2+, the LanL2DZ basis set33 within the DFT/B3LYP method was employed to ascertain the most probable stable molecular configurations. This versatile approach enabled the determination of the molecular structures of the metal complexes, particularly emphasizing their stability. All the optimized geometries were validated by confirming the absence of negative frequencies, ensuring minimum energy on the potential energy surface.
Further analysis involved the calculation of the frontier molecular orbital (FMO) and HOMO–LUMO energy gaps of probe 3 subsequent to its complexation with Cu2+ ions, shedding light on the electronic properties. These findings were pivotal in understanding the intricate interplay between the molecular structures and electronic behaviors, offering insights into the reactivity and possible applications of these compounds.
Moreover, Hirshfeld surface (HS) and 2D-fingerprint plots were generated to understand the packing arrangements and intermolecular interactions of the synthesized compounds by using the Crystal Explorer 17.5 program.34
3. Results and discussion
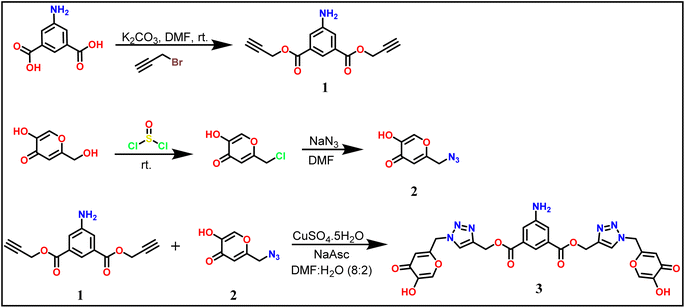
As mentioned earlier, the terminal bis-alkyne 1 with 5-amino isophthalte was synthesized using the propargylation reaction. The azidation of chlorokojic acid was performed to synthesize the organic azide 2. The proposed probe 3, named bis((1-((5-hydroxy-4-oxo-4H-pyran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)methyl) 5-aminoisophthalate was synthesized by the reaction between 1 and 2 via the CuAAC “Click reaction” using CuSO4·5H2O and NaAsc as a catalyst, as illustrated in Scheme 1. Successful characterizations were obtained for the 5-aminoisophthalte-linked terminal bis-alkyne (1), azide (2), and probe 3 (details are given in the ESI file).†3.1 Spectroscopic analysis
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–H and C
C–H and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C bonds, respectively. Likewise, the existence of the azide group (–N3) in 2 was affirmed by a sharp peak with high intensity at 2119 cm−1 in its spectrum. Upon the addition of 1 and 2, leading to the formation of the 1,2,3-triazole ring, the disappearance of the peaks at 3269, 2129, and 2119 cm−1 in the spectrum of 3 indicated the synthesis of a novel compound. Furthermore, a new peak at 3140 cm−1 validated the existence of the –C
C bonds, respectively. Likewise, the existence of the azide group (–N3) in 2 was affirmed by a sharp peak with high intensity at 2119 cm−1 in its spectrum. Upon the addition of 1 and 2, leading to the formation of the 1,2,3-triazole ring, the disappearance of the peaks at 3269, 2129, and 2119 cm−1 in the spectrum of 3 indicated the synthesis of a novel compound. Furthermore, a new peak at 3140 cm−1 validated the existence of the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H within the triazole ring.
C–H within the triazole ring.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C bonds in the 13C NMR spectrum of 1, were conspicuously absent in the spectrum of 3. This absence verified the successful synthesis of probe 3.
C bonds in the 13C NMR spectrum of 1, were conspicuously absent in the spectrum of 3. This absence verified the successful synthesis of probe 3.The observed molar mass at m/z = 592.14 further reinforced the confirmation, providing additional support for the synthesis of the desired compound.
3.2 UV-Vis analysis
The expected fine-quality UV-Vis absorption characteristics of the synthesized 5-aminoisophthalate-based kojic acid-appended 1,4-disubstituted bis-1,2,3-triazole as probe 3 were attributed to its rich aromatic and conjugated moieties embedded within its molecular structure. Subsequent absorbance analysis confirmed these expectations, revealing that probe 3 displayed a prominent absorption peak at λmax = 296 nm. Following this, the potential of the probe for metal ion recognition was explored, wherein 0.5 mM solutions of both probe 3 and various metal ions, including Cr3+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, Zn2+, Cd2+, Hg2+, Pb2+, Na+, K+, Mg2+, Ca2+, and Ba2+ were employed. The UV-Vis analyses to assess the probe's ion recognition capabilities were carried out in a solvent medium consisting of a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of DMSO/H2O. The absorption of the chemosensor remained largely unaffected by the majority of the tested metal ions. Notably, only Cu2+ ions induced a noticeable shift in absorption, thereby establishing the sensor's selective responsiveness to Cu2+ ions, as depicted in Fig. 3, where the relative absorption change of the probe for different metal ions is presented. The relative absorption change was determined by subtracting the absorbance of the reference solution from the absorbance of the solution containing the metal ion, which gave the change in absorbance due to the presence of that specific metal ion and could be then expressed as the relative change compared to the reference absorbance.
1 ratio of DMSO/H2O. The absorption of the chemosensor remained largely unaffected by the majority of the tested metal ions. Notably, only Cu2+ ions induced a noticeable shift in absorption, thereby establishing the sensor's selective responsiveness to Cu2+ ions, as depicted in Fig. 3, where the relative absorption change of the probe for different metal ions is presented. The relative absorption change was determined by subtracting the absorbance of the reference solution from the absorbance of the solution containing the metal ion, which gave the change in absorbance due to the presence of that specific metal ion and could be then expressed as the relative change compared to the reference absorbance.
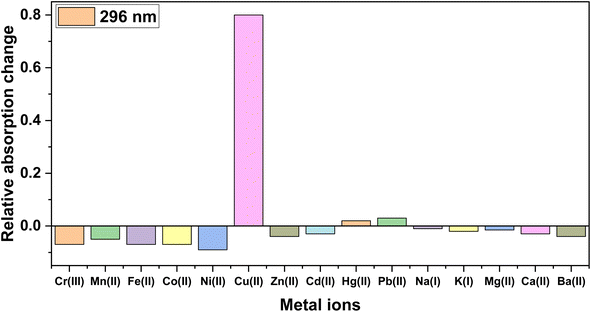 | ||
Fig. 3 Relative absorption intensity changes of probe 3 on adding different metal ions in DMSO/H2O (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1). | ||
3.3 Chemosensing potential of probe 3 for Cu2+
The selective recognition behavior of probe 3 for Cu2+ ions was investigated through UV-Vis spectroscopic analysis, wherein a 0.5 mM solution of the probe in a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
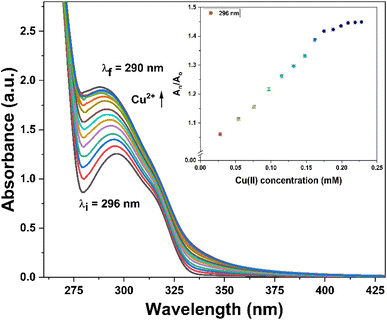
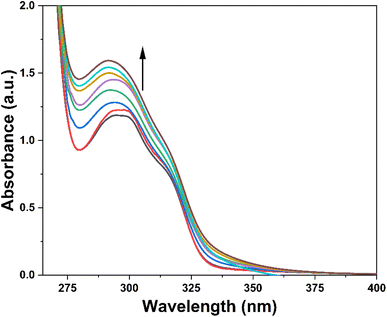
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of DMSO and H2O was sequentially subjected to gradual additions of Cu2+ ions (0–0.25 mM). The absorbance of the solution was measured after each successive addition, enabling the assessment of the probe's recognition potential. Fig. 4 illustrates the alterations in the absorption spectra of probe 3 as Cu2+ ions were progressively introduced. Initially, probe 3 exhibited a well-defined absorption peak at λmax = 296 nm. However, with the introduction of the metal ions, there was a gradual hyperchromic response, accompanied by a shift toward a shorter wavelength (blue shift) of approximately 6 nm, ultimately leading to a shift in λmax to 290 nm upon completion of the titration. Furthermore, Ka for the interaction of Cu2+ ions with 3 determined through the B–H eqn (1) was 4.89 × 103 M−1, and the B–H plot for the same is illustrated in Fig. SI-14.†
1 mixture of DMSO and H2O was sequentially subjected to gradual additions of Cu2+ ions (0–0.25 mM). The absorbance of the solution was measured after each successive addition, enabling the assessment of the probe's recognition potential. Fig. 4 illustrates the alterations in the absorption spectra of probe 3 as Cu2+ ions were progressively introduced. Initially, probe 3 exhibited a well-defined absorption peak at λmax = 296 nm. However, with the introduction of the metal ions, there was a gradual hyperchromic response, accompanied by a shift toward a shorter wavelength (blue shift) of approximately 6 nm, ultimately leading to a shift in λmax to 290 nm upon completion of the titration. Furthermore, Ka for the interaction of Cu2+ ions with 3 determined through the B–H eqn (1) was 4.89 × 103 M−1, and the B–H plot for the same is illustrated in Fig. SI-14.†
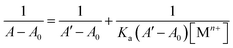 | (1) |
3.4 Competitive metal ion titration analysis
The competitive analysis of metal ions holds a crucial role in analytical chemistry, particularly in assessing the practical utility of 1,2,3-triazole-based chemosensors in addressing environmental concerns and optimizing industrial processes. To evaluate the specificity of probe 3, a competitive metal ion titration was conducted in a solvent system consisting of DMSO/H2O (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) using an equimolar mixture of various metal ions (Fig. 5). The absorption spectra observed after the titration suggested that the probe could selectively detect Cu2+ ions, regardless of the presence of other ions. This was evident from the resemblance of the recorded absorption spectra to those obtained when probe 3 was subjected to Cu2+ ions only.
1 v/v) using an equimolar mixture of various metal ions (Fig. 5). The absorption spectra observed after the titration suggested that the probe could selectively detect Cu2+ ions, regardless of the presence of other ions. This was evident from the resemblance of the recorded absorption spectra to those obtained when probe 3 was subjected to Cu2+ ions only.
 | ||
| Fig. 5 Absorption spectra of probe 3 indicating the selective recognition of Cu2+ ions among different metal ions in equimolar concentrations. | ||
3.5 Time and temperature-dependent analysis
The assessment of a chemosensor's sensitivity and stability in selectively binding metal ions requires a thorough examination of its response to changes in time and temperature. Analyzing the influence of these variables on the sensor's performance is essential for optimizing its sensitivity and selectivity, ensuring a precise and dependable detection of target analytes. In pursuit of this objective, an experiment was conducted to investigate how time and temperature impact the binding affinity of probe 3 for Cu2+ ions. The results are presented in Fig. SI-15† regarding the effect of time on the binding of Cu2+ ions with probe 3. It was observed that the absorption intensity of the probe 3.Cu2+ complex solutions remained constant even as the interaction duration was extended. This observation indicated that the probe's binding affinity for Cu2+ ions was time-independent.In the context of temperature-dependent analysis, probe 3.Cu2+ solution was subjected to a temperature range from 20 °C to 50 °C, and the absorption spectra were collected at 2 °C intervals. Fig. SI-16† illustrate the temperature effects on Cu2+ binding to probe 3, revealing that variations within this temperature range had no significant impact on the stability of probe 3.Cu2+ complexation. Thus, the complexation was found to be stable across the examined temperature range.
3.6 Reversibility analysis
The reversibility of a chemosensor is a crucial aspect in the field of analytical chemistry, providing insights into the dynamic interactions between the chemosensor and the target analyte. The significance of this analysis lies in the ability to discern the reversibility of the chemosensor-analyte complex formation, which has implications for its practical utility in various sensing applications. In this context, the 3.Cu2+ complex solution was subjected to the incremental addition of ethylenediaminetetraacetic acid (EDTA), which resulted in a decrease in absorbance (Fig. SI-17†), attributable to a competitive binding scenario, wherein EDTA competed with the chemosensor for Cu2+ binding sites. This competition was due to the higher affinity of EDTA for Cu2+ ions, leading to the displacement of the chemosensor and subsequent alteration in the optical properties of the solution. However, these preliminary results indicated that probe 3 could be used as both a reusable sensor and as a scavenger of copper ions depending on the concentration of both Cu2+ and EDTA, which may help address the toxicity concerns of Cu2+ ions.3.7 Fluorescence investigation
The photophysical attributes of probe 3 were examined through fluorescence spectroscopy analysis in conjunction with UV-Vis absorption spectroscopy analysis. The emission of a 10 μM probe 3 solution excited at 340 nm (λex) was monitored, revealing a prominent emission (λem) at approximately 436 nm accompanied by a shoulder peak at 414 nm (ϕ = 0.27). Upon the successive addition of Cu2+ ions (0–25 μM), a significant enhancement of the fluorescence emission of the probe was observed, as depicted in Fig. 6, exhibiting almost a two-fold enhancement in the fluorescence intensity (ϕ = 0.96), which confirmed the recognition of the Cu2+ ions by 3. The LOD and LOQ were calculated by analyzing the correlation plot (Fig. SI-18†) derived from the fluorescence titrations and determined to be 8.82 μM and 29.39 μM, respectively.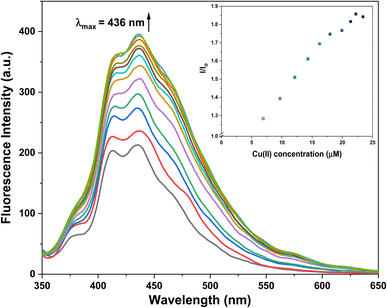 | ||
| Fig. 6 Fluorescence titration of probe 3 with the successive addition of Cu2+ ions; inset exhibits the relative emission of 3 (I/Io) as a function of Cu2+ concentration (μM). | ||
Moreover, Table 2 compares the LODs between previously synthesized chemosensors and the present study, employing 1,4-disubstituted 1,2,3-triazoles for Cu2+ ion detection. The LOD assumes particular significance in environmental monitoring, especially concerning the metal ion concentration limits established by the Environmental Protection Agency (EPA). It signifies the minimum concentration at which a specific metal ion can be consistently identified and quantified, confirming the precision and accuracy of analytical methodologies. Adhering to or exceeding the EPA's stipulated detection limit is imperative for protecting public health and the environment. This compliance allows for recognizing and regulating potentially hazardous metal impurities in air, water, and soil, thereby facilitating the implementation of operative pollution control and risk management strategies.36
| S. no. | Chemosensor | Structure | Limit of detection | Reference |
|---|---|---|---|---|
| a CBT = chalcone-based 1,2,3-triazole; APT1 = antipyrine based bis-1,2,3-triazole; APT2 = antipyrine linked bis-1,2,3-triazole, CFT = ferrocene–chalcone–triazole. | ||||
| 1 | CBT |  |
110 μM | 37 |
| 2 | APT1 |  |
63 μM | 30 |
| 3 | APT2 |  (a) X = Cl (a) X = Cl |
0.14 mM | 11 |
| (b) X = CN | 0.20 mM | |||
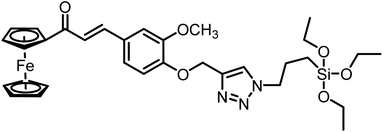
| 4 | CFT |  |
13.4 μM | 38 |
 |
9.42 μM | |||
| 5 | Carbohydrate based sensors | 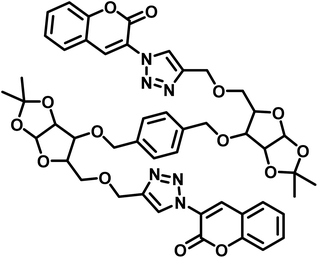 |
6.99 μM | 39 |
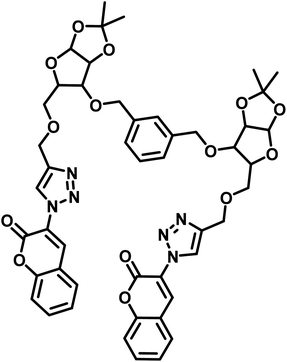 |
7.30 μM | |||
| 6 | Probe 3 | 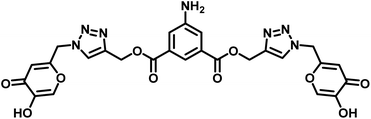 |
8.82 μM | This work |
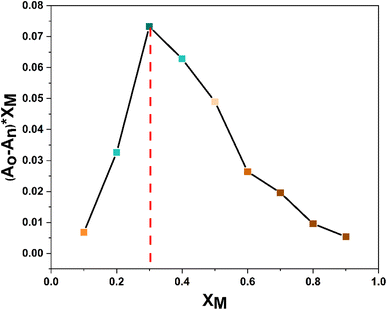
3.8 Job plot analysis
In order to ascertain the stoichiometry between probe 3 and Cu2+ ions, a Job's plot analysis was conducted, which involved preparing a range of solutions of probe 3 and Cu2+ ions with equal concentrations while systematically altering the molar ratio of the probe (or metal ion) from 0.1 to 0.9. The absorbance of each of these solutions was measured, and the plot of the relative absorption change versus the molar ratio of the metal ions was analyzed. The positions of the maximum absorbance peaks in the graph represent the ratio of ligand to metal in the complex solution. For instance, a maximum absorbance corresponding to a mole fraction ratio of 0.5 on the metal mole ratio scale suggests a complex with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand-to-metal (L
1 ligand-to-metal (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M) composition. Similarly, peaks at 0.33 and 0.67 indicate complexes with L
M) composition. Similarly, peaks at 0.33 and 0.67 indicate complexes with L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratios of 2
M ratios of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, respectively. As illustrated in Fig. 7, the maximum absorbance was at the 0.3 molar ratio, indicating a stoichiometric ratio of 2
2, respectively. As illustrated in Fig. 7, the maximum absorbance was at the 0.3 molar ratio, indicating a stoichiometric ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (L
1 (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M) between the probe and the metal ions.
M) between the probe and the metal ions.
3.9 Computational studies
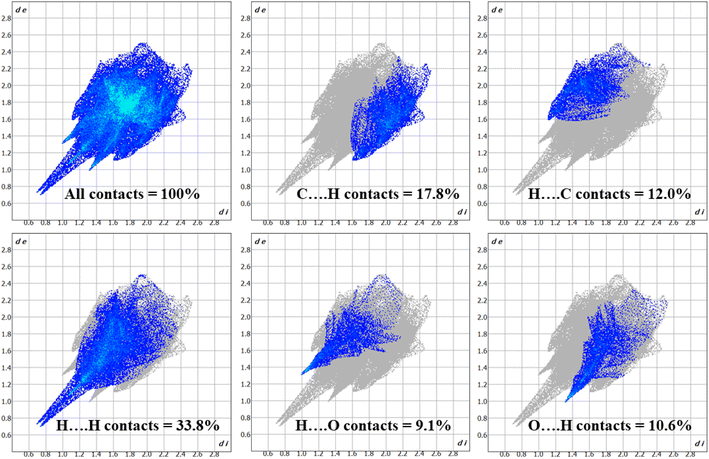
Whenever the distance between atoms equals the total of their van der Waals radii, a white spot appears on the HS. Each point on the HS signifies two distances: (i) the distance from the nearest outer nucleus to the surface (denoted as de), and (ii) the distance from the surface to the closest inner nucleus (referred to as di). The HS is shown to be a transparent surface through which the structure of the molecules can be seen. The HS plots over dnorm, di, de, shape index, curvedness, and fragment patches with a selected network of interactions for the title compound 1 are shown in Fig. 9.
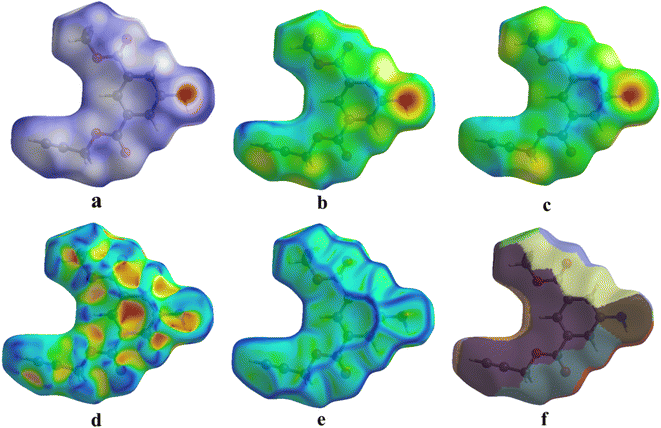 | ||
| Fig. 9 (a) dnorm, (b) di, (c) de, (d) shape index, (e) curvedness, and (f) fragment patches with a selected network of interactions of title compound 1. | ||
The 3D dnorm surface can be resolved into 2D fingerprint plots, which analyze all intermolecular contacts at the same time and give a quantitative summary of the nature and type of intermolecular contacts experienced by the molecules in a crystal. Furthermore, we may get a concise understanding of the relative% contribution and individual contact separation from various contact types in the entire HS area by 2D-fingerprint plots (de vs. di).41 The 2D fingerprint plot provides a detailed two-dimensional graphical illustration of the intermolecular contacts in a crystal system. Fig. 10 represents the 2D fingerprint plots for the title compound 1, which illustrate distinct intermolecular interactions concerning the HS area. H⋯H was the most significant contact in the crystal packing of 1, accounting for 33.8% of the total, while contacts such as C⋯H (17.8%), H⋯C (12.0%), H⋯O (9.1%), and O⋯H (10.6%) provided only a small contribution to the crystal packing.
Crystal Explorer 17.5 program was used to perform the energy framework calculations utilizing symmetry operations on the B3LYP/6-311G(d, p) functional basis set.
A collection of atoms with a radius of 3.8 Å was formed around a molecule. The computation process next involved figuring out the various kinds of molecular interaction energies.42 All the electrostatic energy (Eele) is related to the forces that exist between charged particles, whereas polarization energy (Epol) is focused on the distortion of an electron cloud inside a molecule as a result of other nearby charge distributions. The term ‘dispersive energy, Edis’ describes the weak attractive forces resulting from momentary variations in the electron distribution of a molecule. The amount of energy required to overcome the forces preventing two molecules from occupying the same space is known as repulsive energy (Erep). The overall interaction energy of the molecule under investigation may be obtained by summing the various kinds of interaction energies, as the formula below illustrates:
| Etot = Epol + Eele + Edis + Erep | (2) |
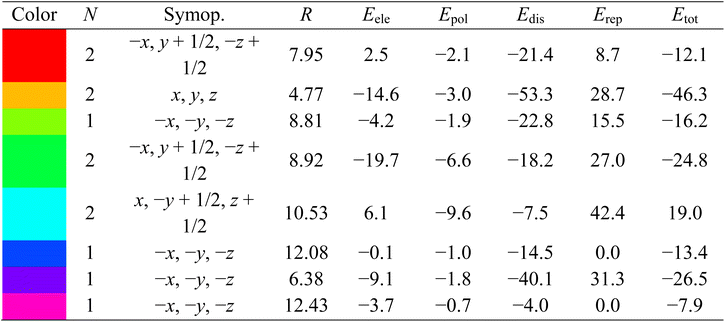
Fig. 11 illustrates the molecule used in the estimation of the interaction energies along the x, y, and z axes for compound 1. All types of interaction energies between the molecular pairs were calculated and are listed in Table 3 for compound 1. Herein, R shows the distance among the mean atomic spots (molecular centroids) and N signifies the no. of molecules at a specific distance. The scaling factors that were employed for all types of interaction energies for the B3LYP/6-311G(d, p) basis set were kele = 1.057, kpol = 0.740, kdisp = 0.871, and krep = 0.618,43 and energy values for each type of interaction are reported in kJ mol−1. A molecule exhibiting x, −y + 1/2, and z + 1/2 symmetry operations, positioned at a distance of 10.53 Å from the chosen molecule's centroid, demonstrates a maximum total interaction energy of 19.0 kJ mol−1, as Table 3 illustrates. On the other hand, the symmetry operation x, y, and z situated at 4.77 Å from the chosen molecule's centroid yields the lowest total interaction energy of −46.3 kJ mol−1. Fig. 11 depicts the Eele, Edis, and Etot for compound 1 along the x, y, and z axes, depicted in red, green, and blue, respectively.
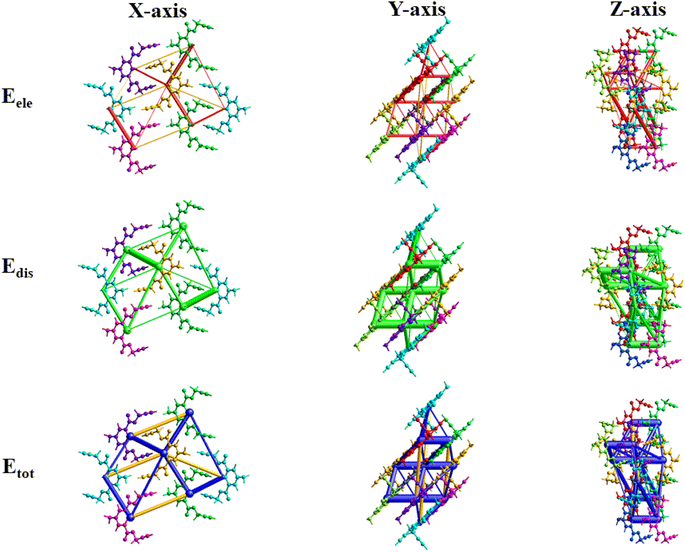 | ||
| Fig. 11 3D-interaction energy frameworks for compound 1 within the radius of 3.8 Å along the x, y, and z axes. | ||
| a R is the distance between molecular centroids (mean atomic position) in Å and N is the number of molecules at that distance, Symop = rotational symmetry operator, Eele = electrostatic energy, Epol = polarization energy, Edis = dispersion energy, Erep = repulsion energy, Etot = total interaction energy, energies are in kJ mol−1. |
|---|
 |
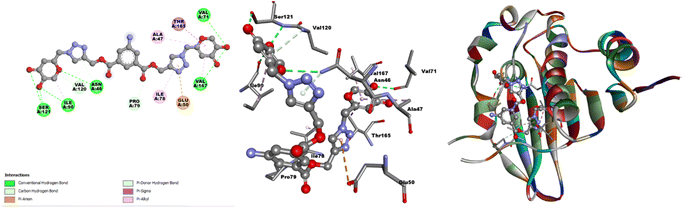 | ||
| Fig. 12 2D and 3D interactions and an illustration of probe 3 docked in the active site of DNA gyrase topoisomerase II. | ||
Similarly, when engaged with the fungal enzyme 14α-demethylase lanosterol (PDB ID 1EA1), probe 3 exhibited a docking score of −8.8 kcal mol−1, as illustrated in Table SI-3.† In particular, interactions involving six hydrogen bonds between ASP67, GLN72, PHE255, THR260, and ALA256 were identified, serving as hydrogen donors to the oxygen atom of the kojic acid group. Additionally, TYR76 acted as a hydrogen donor to the nitrogen atom of the 5-aminoisophthalate group. The benzene and triazole ring system of probe 3 showed π-orbital interactions with LEU321, ARG95, and MET79 (Fig. SI-20†). Alongside these interactions, various π-interactions, including π–π T-shaped, π–anion, and π–sigma, were observed. The smaller bond length and the more negative docking score signify enhanced the stability of the hybrid molecules. Molecular docking analyses suggested promising results, primarily for the bacterial activities. The representation of the potent molecular probe 3 docked within the active site of DNA gyrase topoisomerase II (PDB ID 1KZN) is depicted in Fig. 12. The strategic molecular docking approach aimed to augment the antimicrobial efficacy of probe 3.
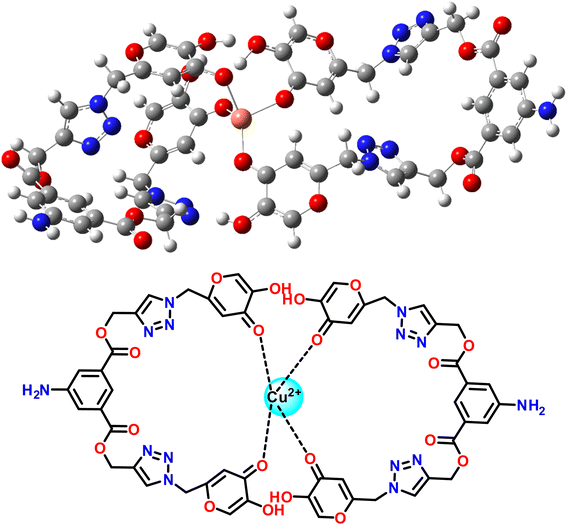
3.10 Probable binding mode
The hard and soft acids and bases (HSAB) concept categorizes Cu2+ as a borderline acid.44 In contrast, probe 3 possessed N and O atoms with lone pairs, making them electron-rich sites capable of interacting with metal ions. Moreover, the Job plot analysis of the probe with the metal ions established a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 L
1 L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratio. In light of the HSAB principles and the host–guest relationship confirmed by the Job plot analysis, a plausible binding mode is depicted in Fig. 13, illustrating the metal ion binding to the ligand through the O atoms of the probe.
M ratio. In light of the HSAB principles and the host–guest relationship confirmed by the Job plot analysis, a plausible binding mode is depicted in Fig. 13, illustrating the metal ion binding to the ligand through the O atoms of the probe.
4. Conclusion
In conclusion, we synthesized a new fluorescent chemosensor 5-aminoisophthalate and kojic acid-based 1,2,3-triazole scaffolds to recognize Cu2+ ions. Upon the addition of Cu2+ ions, an almost two-fold enhancement in the emission intensity was exhibited. From the Job's plot, the binding stoichiometries of the complex were determined to be 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (L
1 (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M). The association constant for the formation of the Cu2+ complex was determined to be 4.89 × 103 M−1. The detection limit of probe 3 was 8.82 μM, much lower than the value recommended by the EPA. The time and temperature-dependent analyses revealed that the chemosensor was stable, independent of the time and temperature upon complexation with Cu2+ ions. Reversibility analysis indicated that probe 3 could be used as both a reusable sensor and as a scavenger of copper ions. A comparison analysis of LODs from previously developed chemosensors for Cu2+ ions revealed that our probe is the most potent among them. DFT studies confirmed the complex formation mechanisms. Hirshfeld surface analysis revealed significant intermolecular interactions in compound 1. Energy framework calculations indicated favorable interactions for the title compound 1. Molecular docking was performed to investigate the interaction between the probe and selected bacterial target with a B.E. of −9.2 kcal mol−1. This dual-purpose investigation sheds light on the probe's ability to selectively detect Cu2+ ions and its potential as an antibacterial agent.
M). The association constant for the formation of the Cu2+ complex was determined to be 4.89 × 103 M−1. The detection limit of probe 3 was 8.82 μM, much lower than the value recommended by the EPA. The time and temperature-dependent analyses revealed that the chemosensor was stable, independent of the time and temperature upon complexation with Cu2+ ions. Reversibility analysis indicated that probe 3 could be used as both a reusable sensor and as a scavenger of copper ions. A comparison analysis of LODs from previously developed chemosensors for Cu2+ ions revealed that our probe is the most potent among them. DFT studies confirmed the complex formation mechanisms. Hirshfeld surface analysis revealed significant intermolecular interactions in compound 1. Energy framework calculations indicated favorable interactions for the title compound 1. Molecular docking was performed to investigate the interaction between the probe and selected bacterial target with a B.E. of −9.2 kcal mol−1. This dual-purpose investigation sheds light on the probe's ability to selectively detect Cu2+ ions and its potential as an antibacterial agent.
Author contributions
Sachin Kumar: methodology, investigation, validation, writing manuscript. Bajrang Lal: methodology, investigation, validation. Gurleen Singh: validation, writing manuscript. Muskan: crystallographer. Ram Kumar Tittal: supervision, resources, writing: review and editing. Jandeep Singh: conceptualization, resources. Vikas D. Ghule: software. Renu Sharma: supervision.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors thank the National Institute of Technology, Kurukshetra, and Lovely Professional University for providing the support and resources that allowed them to complete the present research work. The authors also acknowledge Delhi University for XRD analysis.References
- R. Kumar, A. Thakur, Sachin, D. Chandra, A. K. Dhiman, P. K. Verma and U. Sharma, Coord. Chem. Rev., 2024, 499, 215453 CrossRef CAS.
- D. Quiroga and E. Coy-barrera, Organics, 2023, 4, 503–523 CrossRef CAS.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew Chem. Int. Ed. Engl., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- R. C. Gupta, S. S. Razi, R. Ali, S. K. Dwivedi, P. Srivastava, P. Singh, B. Koch, H. Mishra and A. Misra, Sens. Actuators, B, 2017, 251, 729–738 CrossRef CAS.
- T. Puthiyedath and D. Bahulayan, Sens. Actuators, B, 2018, 272, 110–117 CrossRef CAS.
- A. Kumar, P. S. Chae and S. Kumar, Dyes Pigm., 2020, 174, 108092 CrossRef CAS.
- G. Sanjotha, C. T. Shivasharana and S. I. Manawadi, J. Pure Appl. Microbiol., 2019, 13, 2261–2272 CrossRef CAS.
- M. Rosfarizan, M. S. Mohamed, N. Suhaili, M. M. Salleh and A. B. Ariff, Biotechnol. Mol. Biol. Rev., 2010, 5, 24–37 Search PubMed.
- H. B. Wang, Y. Chen, Y. Li and Y. M. Liu, Anal. Methods, 2016, 8, 8322–8328 RSC.
- B.-B. Tao, N.-N. Wu, H.-D. Zhang and H.-B. Wang, J. Electrochem. Soc., 2022, 169, 037529 CrossRef CAS.
- B. Lal, S. Kumar, R. K. Tittal, G. Singh, J. Singh, V. D. Ghule, R. S. Mathpati and J. K. Sabane, J. Mol. Struct., 2024, 1297, 136995 CrossRef CAS.
- R. K. Naveen, P. Tittal, K. Yadav, G. V. D. Lal and A. Kumar, New J. Chem., 2019, 43, 8052–8058 RSC.
- M. Ramachandran, S. Anandan and M. Ashokkumar, New J. Chem., 2019, 43, 9832–9842 RSC.
- G. Singh, J. D. Kaur, D. Pawan, S. Sushma, P. S. Shilpy and K. N. Singh, New J. Chem., 2022, 46, 7055–7069 RSC.
- Y. Wu, H. Pang, Y. Liu, X. Wang, S. Yu, D. Fu, J. Chen and X. Wang, Environ. Pollut., 2019, 246, 608–620 CrossRef CAS PubMed.
- B. K. Bansod, T. Kumar, R. Thakur, S. Rana and I. Singh, Biosens. Bioelectron., 2017, 94, 443–455 CrossRef CAS PubMed.
- Q. Zheng, Q. Li, S. Hu, X. Guo and H. Yang, J. Environ. Chem. Eng., 2023, 11, 109079 CrossRef CAS.
- J. L. Zhao, H. Tomiyasu, C. Wu, H. Cong, X. Zeng, S. Rahman, P. E. Georghiou, D. L. Hughes, C. Redshaw and T. Yamato, Tetrahedron, 2015, 71, 8521–8527 CrossRef CAS.
- S. Bagheri, R. Squitti, T. Haertlé, M. Siotto and A. A. Saboury, Front. Aging Neurosci., 2018, 9, 1–15 CrossRef PubMed.
- R. Patel and M. Aschner, Toxics, 2021, 9, 1–11 CrossRef PubMed.
- G. J. Brewer, Exp. Biol. Med., 2001, 226, 665–673 CrossRef CAS PubMed.
- J. Zhang, M. Zhu, D. Jiang, H. Zhang, L. Li, G. Zhang, Y. Wang, C. Feng and H. Zhao, New J. Chem., 2019, 43, 10176–10182 RSC.
- G. Izydorczyk, K. Mikula, D. Skrzypczak, K. Moustakas, A. Witek-Krowiak and K. Chojnacka, Environ. Res., 2021, 197, 111050 CrossRef CAS PubMed.
- J. C. Zilles, F. L. dos Santos, I. C. Kulkamp-Guerreiro and R. V. Contri, Exp. Dermatol., 2022, 31, 1500–1521 CrossRef PubMed.
- X. Liu, C. Yu, B. Su and D. Zha, RSC Med. Chem., 2023, 14, 268–276 RSC.
- G. Singh, B. Lal, R. Singh, N. George, G. Singh, Diksha, G. Kaur, H. Singh, R. K. Tittal, G. Kaur and J. Singh, Spectrochim. Acta, Part A, 2023, 302, 123163 CrossRef CAS PubMed.
- N. Nehra, V. D. Ghule and R. K. Tittal, J. Mol. Struct., 2022, 1250, 131755 CrossRef CAS.
- M. Saraei, Z. Ghasemi, G. Dehghan, M. Hormati and K. Ojaghi, Monatsh. Chem., 2017, 148, 917–923 CrossRef CAS.
- S. Emami, E. Ghafouri, M. A. Faramarzi, N. Samadi, H. Irannejad and A. Foroumadi, Eur. J. Med. Chem., 2013, 68, 185–191 CrossRef CAS PubMed.
- S. Kumar, B. Lal, R. K. Tittal, G. Singh, J. Singh, V. D. Ghule, R. Sharma and J. K. Sabane, Sens. Diagn., 2023, 2, 1267 RSC.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, and D. J. Fox, Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- S. Chiodo, N. Russo and E. Sicilia, J. Chem. Phys., 2006, 125, 104107 CrossRef CAS PubMed.
- P. R. Spackman, M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, D. Jayatilaka and M. A. Spackman, J. Appl. Crystallogr., 2021, 54, 1006–1011 CrossRef CAS PubMed.
- A. Allouche, J. Comput. Chem., 2012, 32, 174–182 CrossRef PubMed.
- M. Marzi, M. Farjam, Z. Kazeminejad, A. Shiroudi, A. Kouhpayeh and E. Zarenezhad, J. Chem., 2022, 2022, 7884316 Search PubMed.
- R. Singh, G. Singh, N. George, G. Singh, A. Devi, H. Singh, G. Kaur and J. Singh, RSC Adv., 2023, 13, 32399–32412 RSC.
- G. Singh, A. Arora, S. Rani, P. Kalra and M. Kumar, ChemistrySelect, 2017, 2, 3637–3647 CrossRef CAS.
- D. Kushwaha, R. S. Singh and V. K. Tiwari, Tetrahedron Lett., 2014, 55, 4532–4536 CrossRef CAS.
- M. Ashfaq, M. N. Tahir, S. Muhammad, K. S. Munawar, A. Ali, G. Bogdanov and S. S. Alarfaji, ACS Omega, 2021, 6, 31211–31225 CrossRef CAS PubMed.
- H. Aziz, A. Saeed, C. J. McAdam, J. Simpson, T. Hökelek, E. Jabeen, A. Khurshid, M. Saleem and H. R. El-Seedi, J. Mol. Struct., 2023, 1281, 135108 CrossRef CAS.
- K. Kumara, F. H. Al-Ostoot, Y. H. E. Mohammed, S. A. Khanum and N. K. Lokanath, Chem. Data Collect., 2019, 20, 100195 CrossRef CAS.
- D. Dey, S. Bhandary, S. P. Thomas, M. A. Spackman and D. Chopra, Phys. Chem. Chem. Phys., 2016, 18, 31811–31820 RSC.
- R. G. Pearson, J. Am. Chem. Soc., 1963, 85, 3533–3539 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2300460. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra02372b |
| This journal is © The Royal Society of Chemistry 2024 |