Open Access Article
Open Access ArticleCarboxymethylcellulose encapsulated fingolimod, siRNA@ZnO hybrid nanocomposite as a new anti-Alzheimer's material†
Nuha B. Aljohaniab,
Safaa Y. Qustia,
Madeeha Alsinya,
Fadwa Aljoudc,
Norah Bakheet Aljohanid,
Eman S. Alsolamie,
Khalid A. Alamry e and
Mahmoud A. Hussein
e and
Mahmoud A. Hussein *ef
*ef
aBiochemistry Department, Faculty of Science, King Abdul Aziz University, Jeddah 21589, Kingdom of Saudi Arabia
bBiochemistry Department, Faculty of Science, University of Tabuk, Tabuk, Kingdom of Saudi Arabia
cRegenerative Medicine Unit, King Fahd Medical Research Centre, King Abdul Aziz University, Jeddah 21589, Saudi Arabia
dKing Abdul Aziz University Hospital, Jeddah, Kingdom of Saudi Arabia
eChemistry Department, Faculty of Science, King Abdulaziz University, P.O. Box 80203, Jeddah 21589, Saudi Arabia. E-mail: maabdo@kau.edu.sa; mahussein74@yahoo.com
fChemistry Department, Faculty of Science, Assiut University, Assiut 71516, Egypt
First published on 12th July 2024
Abstract
Alzheimer's disease (AD) is a fatal neurological disorder that causes cognitive and memory function to deteriorate. A critical pathogenic event that speeds up the development of AD is the interaction between dysfunctional microglia and amyloid-β (Aβ). We have developed a hybrid nanocomposite material to treat AD by normalizing the dysfunctional microglia. The material is based on carboxymethylcellulose (CMC) encapsulated fingolimod, siRNA, and zinc oxide (ZnO) with variable loading (CMC-Fi-siRNA@ZnOa–d). The material was characterized using different techniques including FTIR, XRD, thermal analysis, SEM with EDX, and TEM micrographs. The chemical structure was confirmed by FTIR and XRD analyses, which indicated the successful integration of ZnO nanoparticles (NPs) into the polymer matrix, signifying a well-formed composite structure. The thermal stability order at 10% weight loss was CMC-Fi-siRNA@ZnOc > CMC-Fi-siRNA@ZnOb > CMC-Fi-siRNA@ZnOd > CMC-Fi-siRNA@ZnOa. The CMC-Fi-siRNA@ZnOd dramatically alleviates the priming of microglia by lowering the level of proinflammatory mediators and increasing the secretion of BDNF. This considerably improves the phagocytosis of Aβ. In the cell viability test in immortalized microglia cells (IMG), the hybrid nanocomposite (NP) exhibited no significant effect on cell survival after 48 hours of incubation. The NP also decreased the cytotoxicity caused by Aβ. Therefore, the CMC-hybrid NP has high potential as a drug delivery system in the development of therapeutic strategies for AD.
1. Introduction
Alzheimer's disease (AD) is the most prevalent neurodegenerative disorder that affects elderly people, causing a gradual decline in cognitive and memory functions. Researchers have primarily focused on inhibiting the aberrant generation and aggregation of amyloid-β (Aβ) to combat AD. However, several clinical trial failures to date suggest that focusing on Aβ alone is more than enough.1 Therefore, it is necessary to explore alternative strategies to treat AD. One critical pathogenic event that accelerates the progression of AD is the aberrant crosstalk between dysfunctional microglia and Aβ. Microglia are immune cells in the central nervous system that are responsible for constantly scanning the environment around the brain for cellular debris and pathogens2,3 and can phagocytose and degrade extracellular Aβ in the AD brain.4 However, abnormal Aβ aggregation impairs microglia phagocytic function and causes the production of proinflammatory mediators such as tumor necrosis factor (TNF-), interleukin 6 (IL-6), interleukin 1 (IL-1) and reactive oxygen species (ROS).5,6 These proinflammatory mediators contribute to Aβ accumulation,7 resulting in a negative feedback loop between Aβ and microglial dysfunction that accelerates AD progression.8It is, therefore, useful to normalize microglia priming and reduce Aβ to treat AD effectively.9 Fingolimod has been proven to decrease the production of inflammatory mediators in microglia, which has the effect of modulating the dysfunctional microglia.10 Unfortunately, its poor aqueous solubility restricts its application. Furthermore, the secretion of inflammatory cytokines is mediated by a gene called signal transducer and activator of transcription 3 (STAT3),11 which is expressed at higher levels in dysfunctional microglia. To control inflammation effectively, the STAT3 protein expression must be downregulated.12 Small interfering RNA (siRNA) is considered a highly effective therapeutic agent for RNA interference-mediated downregulation of gene expression.13 Additionally, it has been found that zinc oxide nanoparticles (ZnO NPs) can stimulate the Th2 response, which is effective in normalizing the microglia's polarization.14 As a result, fingolimod, siSTAT3, and ZnO NPs delivery to normalize dysfunctional microglia will have synergistic effects on the normalization of microglia.
However, the therapeutic effects of these medications are limited by their short blood half-life and poor blood–brain barrier penetration.15 Natural polymers are a promising choice for creating nanocomposites because their chemical composition is similar to biomolecules already present in extracellular matrices, making them more biocompatible compared to artificial polymers such as poly(ethylene glycol) (PEG).16,17 The field of nanoparticle drug delivery is studying a variety of these natural polymers as PEG substitutes. While very little work has been done with other polysaccharides, most of this work has concentrated on chitosan-based materials.18,19 There is an indication that polysaccharides prolong plasmatic life and decrease the adsorption of non-specific proteins.20 Furthermore, the biodegradability of these materials can be employed to induce drug release and activation using specific enzymes to promote controlled degradation of the coating, in addition to facilitating the nanocomposite's eventual clearance.21,22 These characteristics make polysaccharides very intriguing materials for the synthesis of brain-targeted nanocomposites, with their capability to interact with specific protein/cell surfaces and decrease particle aggregation.23 Hybrid polymer nanosystems are used in biomedicine and other industries. Recently studied hybrid materials include polymers and inorganic (metal oxide NPs, carbon nanotubes, graphene) or organic (lipids, proteins, and phospholipids) systems. The results show tremendous medication targeting improvements. Hybrid polymer materials generate innovative hybrid nanosystems and eliminate the need to manufacture new molecules, which is costly and time-consuming.24 Hybrid nanosystems have various benefits over non-hybrid systems, including increased circulation duration, high stability, structural disintegration, premature release, low encapsulation rate, and regulated drug release. Polymers are used to produce hybrid drug delivery materials to stabilize and improve single medication therapeutic activity due to the urgent demand for an efficient carrier system. Two or more materials can be combined to create hybrid materials with unique qualities. Due to the large superficial surface area between phases, polymers can transfer their chemical and physical characteristics to inorganic or biological polymers, resulting in synergistic and hybrid qualities. This improves system stability by changing solubility and biodistribution. The materials' conjugation extends blood circulation while retaining biological activity.25 Under basic circumstances, cellulose and chloroacetic acid form carboxymethyl cellulose (CMC).26,27 CMC has several uses, but nanocomposite biological applications are uncommon. According to brain delivery system studies, nanocomposite surface charge affects cellular uptake and composite–cell interactions.28 The negative charge of CMC improves nanocomposite colloidal stability, reduces plasma protein adsorption, and extends plasma circulation.29 These features make CMC suitable for brain-targeted nanocomposite research in vivo and in vitro.30,31 CMC has regulated drug release, biocompatibility, bio-adhesion, site-specific drug delivery, and hydrophilicity due to carboxylate groups.32
In the present work, hybrid nanocomposite frameworks, abbreviated as CMC-Fi-siRNA@ZnOa–d, have been effectively fabricated utilizing a casting approach. The products underwent comprehensive analysis using widely-used techniques such as FTIR, XRD, TGA, SEM with EDX, and TEM micrographs. The combination of CMC-encapsulated fingolimod and siRNA, together with varying amounts of ZnO hybrid nanocomposite, was identified as effective materials for preventing Alzheimer's disease. The new fabricated composites were classified as a drug delivery carrier to enhance therapeutic strategies in the treatment of AD.
2. Materials and methods
2.1 Materials and reagents
Immortalized microglia cells (IMG) cells were obtained from Kerafast (Boston, USA). Dulbecco's Modified Eagle Medium/Ham's F-12 (DMEM/F12), fetal bovine serum (FBS), penicillin-streptomycin solution, phosphate buffered saline (PBS), CMC, dimethyl sulfoxide (DMSO), HFIP (1,1,1,3,3,3-hexafluoro-2-propanol), SDS (sodium dodecyl sulfonate), zinc acetate (Zn(CH3CO2)·2H2O), sodium hydroxide (NaOH), ethanol (CH3CH2OH), hexane and RNeasy kit were obtained from Sigma-Aldrich (St. Louis, MO, USA). Trypsin EDTA, thiazolyl blue tetrazolium bromide, Aβ (1–42), TNF-α, IL-1β, BDNF ELISA kits, dichloro-dihydro-fluorescein diacetate (DCFH-DA), 4′,6-diamidino-2-phenylindole (DAPI), rabbit anti-phospho-STAT3 (Tyr705) antibody and goat anit-rabbit IgG/HRP antibody were from Solarbio life science (Beijing, China). Fingolimod was purchased from Medchemexpress (Monmouth junction, NJ, USA). All siRNAs were synthesized by Thermo Fisher (Waltham, MA, USA) and the sequences used were as follows:Sense (5′-3′): GAGGAGGCAUUUGGAAAGUTT.
Antisense (5′-3′): ACUUUCCAAAUGCCUCCUCTT.
2.2 Synthesis of ZnO nanoparticles
A beaker containing water was heated to 65 °C. Meanwhile, 0.10 g of Zn(CH3CO2)·2H2O was dissolved in 25 mL of isopropanol with heating in a fume hood, and 125 mL isopropanol was placed in a flask and cooled in an ice bath. Once the zinc acetate solid was completely dissolved, this solution was added to the 125 mL of chilled isopropanol. Also, 15 mL of 0.050 M NaOH was obtained in isopropanol, and then this solution was cooled as well. The chilled NaOH solution was gradually transferred to the chilled and rapidly stirred zinc acetate solution using a Pasteur pipet. The flask containing the mixed solution was then placed in the 65 °C water bath and samples were taken immediately. The UV spectra of each sample were recorded.To promote precipitation of ZnO NPs, hexane was added to the ZnO NPs solution in a volume ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The solution with hexane was then placed in separate vials and allowed to stand for 24 h. After 24 h, the ZnO NPs precipitated, and then the supernatant was removed. The precipitated ZnO NPs solution was centrifuged at 8000 rpm for 5 min, and then the solvent was removed. To remove the unreacted precursors, the ZnO NPs collected were washed using ethanol. The NPs were then redispersed in ethanol using an ultrasonic bath for 5 min. The NPs dispersed in ethanol were centrifuged, and the solvent was removed. These steps were repeated three times.11
1. The solution with hexane was then placed in separate vials and allowed to stand for 24 h. After 24 h, the ZnO NPs precipitated, and then the supernatant was removed. The precipitated ZnO NPs solution was centrifuged at 8000 rpm for 5 min, and then the solvent was removed. To remove the unreacted precursors, the ZnO NPs collected were washed using ethanol. The NPs were then redispersed in ethanol using an ultrasonic bath for 5 min. The NPs dispersed in ethanol were centrifuged, and the solvent was removed. These steps were repeated three times.11
2.3 Fabrication of CMC-Fi-siRNA@ZnOa–d hybrid NCs
To prepare CMC-Fi-siRNA@ZnOa–d hybrid NCs, a simple casting tool was utilized. First, 0.5 g of CMC was dissolved in distilled water, and then fingolimod, siRNA (20%), and ZnO NPs with varying loading (3, 6, 10, and 20%) were added to the CMC solution at room temperature. The mixture was stirred constantly for 30 min. To study the loading ability of hybrid nanocomposite to siRNA, the agarose gel retarding assay was applied in TAE buffer at 100 V for 30 min. This study was conducted using a dynamic light scattering (DLS) instrument (Malvern Zetasizer Nano ZS, UK). Table 1 illustrates the suggested symbols and compositions for pure CMC and CMC-Fi-siRNA@ZnOa–d hybrid nanocomposites. Please note for microbiological investigations, the sample preparation requires a special pre-treatment. The nanocomposite suspensions were diluted several times with their dispersion media before being treated directly onto any of the required analyses listed below.| Products | CMC% (weight, g) | Fi-siRNA% (weight, g) | ZnO% (weight, g) |
|---|---|---|---|
| Pure CMC | 100 (0.5) | — | — |
| CMC-Fi-siRNA@ZnOa | 77 (0.385) | 20 (0.1) | 3 (0.015) |
| CMC-Fi-siRNA@ZnOb | 74 (0.37) | 20 (0.1) | 6 (0.03) |
| CMC-Fi-siRNA@ZnOc | 70 (0.35) | 20 (0.1) | 10 (0.05) |
| CMC-Fi-siRNA@ZnOd | 60 (0.30) | 20 (0.1) | 20 (0.10) |
2.4 Characterization tools
Fourier transform infrared spectra (FTIR) covering a wavenumber range of 400 to 4000 cm−1 were obtained using a PerkinElmer infrared spectrophotometer. X-ray diffraction (XRD) patterns were obtained using a Bruker device at a 2θ range of 10–105° (λ = 1.54178 Å, step size of 0.02, and scan step time of 0.80 s). Thermogravimetric analysis (TGA) using a Shimadzu TGA-50 series was performed with a temperature increase rate of 10 °C min−1, holding temperature at 800 °C, in a platinum cell under air atmosphere. The nanocomposite's morphology was investigated using scanning electron microscopy (SEM) using Zeiss LEO Supra 55VP Field Emission and SEM Zeiss 1530 was used to take SEM images. Under a polished aluminum sample holder and vacuum, samples were then dried and coated in gold using an EMITECH K450X sputter coater. The high-resolution transmission electron microscope (HRTEM, JEOL TEM-2100, Japan) was used to scan the shape, size, and texture of the nanocomposite at a magnification of 20× and an accelerating voltage of 250 kV. The drug loading of fingolimod for CMC-Fi-siRNA@ZnOa–d hybrid nanocomposites was determined by high-performance liquid chromatography (HPLC) (Agilent, Japan).2.5 Preparation of amyloid-β (1–42)
The oligomeric aggregated form of Aβ is the major toxin that causes neuronal cell death.22 Herein, oligomeric Aβ was used. Minor modifications were made to previously published methods used to synthesize oligomeric Aβ.26 HFIP was used to emulsify monomeric Aβ to 1 mM using a gas-tight syringe. The samples were vortexed to produce a homogeneous solution and then centrifuged for 40 s at 1000 rpm. After that, the samples were aliquoted into microfuge tubes and kept in the biosafety cabinet overnight to allow the HFIP to evaporate. For further processing, the resulting samples were saved at −20 °C. To create oligomeric Aβ, the resulting samples were dissolved in 5 mM DMSO, vortexed for 5 s, bath sonicated for 10 min, and then centrifuged for 30 s at 1000 rpm. Next, the samples were diluted to 100 μM (monomeric Aβ concentration) by cold PBS + 0.05% SDS, and completely vortexed. To promote high-order aggregation, the Aβ aggregates were incubated at 4 °C for two weeks. When the oligomer preparations were ready after the two-week incubation, they were centrifuged to clear sediment at 4 °C and 1000 rpm for 3 min. Before the cells were treated, the oligomeric Aβ in the supernatant was diluted to 10 μM with Dulbecco's modified Eagle's medium.332.6 In vitro fingolimod release
2 mL of CMC-Fi-siRNA@ZnOa–d nanocomposite in a dialysis bag (MWCO 3500) with or without H2O2 (1 × 10−3 M) was incubated in 50 mL of 0.5% Tween-80 under horizontal shaking (150 rpm) at 37 °C. At specified time intervals, 100 μL of the medium was pulled out and replaced with an equal volume of fresh solution. The fingolimod concentration was determined using HPLC, which was supplied with a UV detector and C18 column at 35 °C (Agilent, Japan). The mobile phase for the HPLC analysis consisted of 50 mM potassium dihydrogenphosphate (pH 3.0) and acetonitrile (45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) with a flow rate of 1.0 mL min−1. At the retention time of 2.7 min and 226 nm, the fingolimod concentration was determined.11 The related HPLC chromatograms for in vitro release study were provided as illustrated in Fig. S1–S6 (see ESI file).†
50, v/v) with a flow rate of 1.0 mL min−1. At the retention time of 2.7 min and 226 nm, the fingolimod concentration was determined.11 The related HPLC chromatograms for in vitro release study were provided as illustrated in Fig. S1–S6 (see ESI file).†
2.7 Cell culture and MTT assay
IMG cells were cultured in a medium supplied with 10% fetal bovine serum and 1000 U mL−1 penicillin-streptomycin at 37 °C with 5% CO2. For the MTT assay, cells were seeded with about 5000 cells per well in 96-well plates in 100 μL of culture media for 24 h. Cells were treated for 48 h with free fingolimod, free ZnO NPs, empty CMC, and hybrid nanocomposite (CMC-Fi-siRNA@ZnOa–d). The range of tested concentration was 200–800 nM for fingolimod and 0.03–0.3 mg mL−1 for ZnO NPs. The range of CMC-based composite concentration was 0.06–0.5 mg mL−1. The cells were treated with Aβ42 (10 μM) with or without the nanocomposite and the fingolimod concentration in nanocomposite was 200 nM, while for the ZnO NPs and siRNA, the concentration was 0.03 mg mL−1 and 40 nM, respectively. Each concentration was added in triplicate. After 48 h, the vitality of the cells was determined using the MTT assay. 20 μL MTT solution (5 mg mL−1) was added to each well and incubated for 3 h. Afterward, the medium was discarded, and 100 μL DMSO was added to each well. It is important to note that, before entering the hood, we sprayed the additive bottles with 70% ethanol, making sure not to open any bottles outside the sterilized hood before adding them to the cells. Next, we filtered the additives through a 0.22 micron filter.By measuring the solution at 570 nm, the density of live cells was determined. The untreated cells were considered as a control. The percentage of absorption of treated cells compared to control cells (untreated cells) was used to determine cell viability using the following formula:
| % Cell viability = [Asample/Acontrol] × 100 |
The results are the mean and standard deviation (SD) values determined from three repeats of the experiment (n = 3).
2.8 Detection of inflammatory cytokines and Aβ-generated ROS
IMG cells were treated with 10 μM Aβ42 for 12 h in the presence or absence of a hybrid nanocomposite. The fingolimod concentration in the nanocomposite was 200 nM. After 12 h incubation, the supernatants were collected and the levels of IL-1β, IL-12, TNF-α, and BDNF were detected by performing ELISA kits following the instructions offered by the manufacturers. For ROS detection, the cells were labeled with 10 nM DCFH-DA for 20 min at 37 °C. Then, the cells were washed with PBS three times and fixed with 4% paraformaldehyde at 37 °C for 15 min. The nucleus was then counterstained for 15 min with DAPI at 37 °C. The fluorescence image was taken by Confocal Laser Scanning Microscopy (CLSM) (Zeiss Co., Germany).2.9 Polymerase chain reaction of pro- and anti-inflammatory cytokines
Quantitative real-time polymerase chain reaction (RT-qPCR) analysis was used to determine the mRNA level of gene expression. Using the RNeasy kit, total RNA was extracted from treated microglial cells following the manufacturer's instructions. Reverse transcription reagents Thermo Fisher (Waltham, MA, USA) were used for reverse transcription of RNA into cDNA. The TNF-α and TGF-β expression was determined by the RT-qPCR system. All procedures were carried out according to the manufacturer's protocols. Following normalization to the internal control (glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels), changes in mRNA levels were obtained. RT-qPCR was performed using the following primer sequences in Table 2.| Forward primer | Reverse primer | |
|---|---|---|
| GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| TGF-β | TCTGCATTGCACTTATGCTGA | AAAGGGCGATCTAGTGATGGA |
| TNF-α | TCTCAGCCTCTTCTCATTCCTGCT | AGAACTGATGAGAGGGAGGCCATT |
2.10 p-STAT3 downregulation
After 48 h of treatment with the hybrid nanocomposite, IMG cells cultured in six-well plates were incubated for 8 h with Aβ42 (10 μM). Then, PBS was used to wash the cells before they were lysed to extract the total protein. Western blot analysis was used to identify the p-STAT3 level, with β-actin as the loading control.Statistical analysis: the mean ± SD was used to express all data. GraphPad Prism version 9.5 software was used to determine the significance using Student's t-test or two-way ANOVA.
3. Results and discussion
3.1 Hybrid nanocomposite fabrication
Hybrid nanocomposites CMC-Fi-siRNA@ZnOa–d were designed and fabricated using a casting technique to encapsulate hydrophobic ZnO NPs, fingolimod, and siRNA into the CMC polymer matrix through electrostatic interaction. The presence of CMC as a hydrophilic polysaccharide helps to balance the hydrophobic and hydrophilic components of the hybrid nanocomposite. Before the fabrication process, ZnO NPs were synthesized following established procedures.11 Fig. 1a and b show the synthesized ZnO NP aqueous solutions under white light and UV light, respectively. The size distribution of 3–5 nm was measured by DLS as illustrated in Fig. 1c.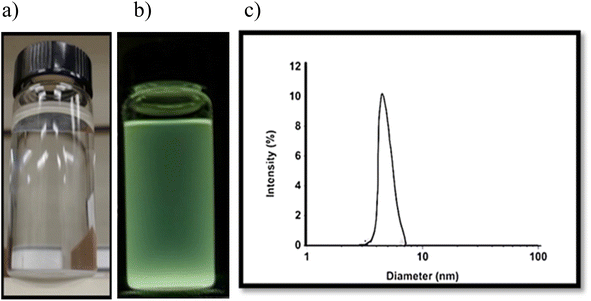 | ||
| Fig. 1 (a and b) The aqueous solution of ZnO NPs under white light and UV light; (c) the ZnO NPs size distribution measured by DLS. | ||
The hybrid nanocomposites CMC-Fi-siRNA@ZnOa–d were fabricated and their chemical structures were confirmed using various analytical techniques such as FTIR, XRD, thermal analysis (including TGA and DTG), SEM with energy-dispersive X-ray spectroscopy (EDX), and TEM micrographs. The interaction between ZnO NPs and the CMC-Fi-siRNA composite was explored using FTIR spectroscopy. In Fig. 2, the analysis of CMC-Fi-siRNA@ZnOa–d nanocomposites filled with 3, 6, 10, and 20% concentrations of ZnO NPs are depicted to investigate potential intermolecular interactions among the system's components. The presence of ZnO leads to the broadening of the hydroxyl group, extending the vibration to 3342 cm−1 and 3390 cm−1, associated with the stretching frequency of the –OH group, while another band at 2920–2869 cm−1 corresponds to C–H stretching vibration from the –CH2 group. Intense absorption bands between 1516–1592 cm−1 and 1408–1418 cm−1 indicate the presence of the CH2COO– group.34–37 Notably, the exact peaks of the CMC-Fi-siRNA@ZnOa–d blend remain unchanged after compounding, suggesting structural preservation, although some shifts are observed after the addition of zinc nanofiller. The observed favorable intermolecular interactions between these groups and NPs contribute to variations in the characteristic absorption bands.
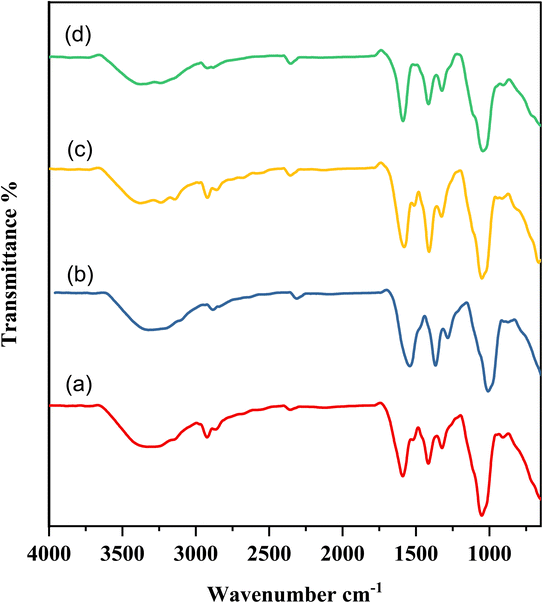 | ||
| Fig. 2 FT-IR spectra of hybrid nanocomposites (a) CMC-Fi-siRNA@ZnOa, (b) CMC-Fi-siRNA@ZnOb, (c) CMC-Fi-siRNA@ZnOc, (d) CMC-Fi-siRNA@ZnOd. | ||
XRD was used to investigate the crystal structure of the synthesized CMC-Fi-siRNA@ZnOa–d, to determine whether the materials were crystalline or amorphous. The XRD patterns, depicted in Fig. 3, were consistent with previous studies on pure CMC, which showed lower crystallinity indicated by a characteristic broad peak at 21.39°, attributed to the cleavage of hydrogen bonds.38 Additionally, the XRD pattern of CMC-Fi-siRNA@ZnOa–d displays well-defined diffraction peaks at specific angles, including 2θ = 31.97° (100), 34.26° (002), 36.39° (101), 47.38° (102), 56.38° (110), 62.81° (103), 66.46° (200), 68.13° (112), 69.21° (201), 72.55° (004), and 81.41° (202). These peaks are characteristic of ZnO NPs, as reported in a previous study.39 This observation indicates the successful integration of ZnO NPs into the polymer matrix, signifying a well-formed composite structure.
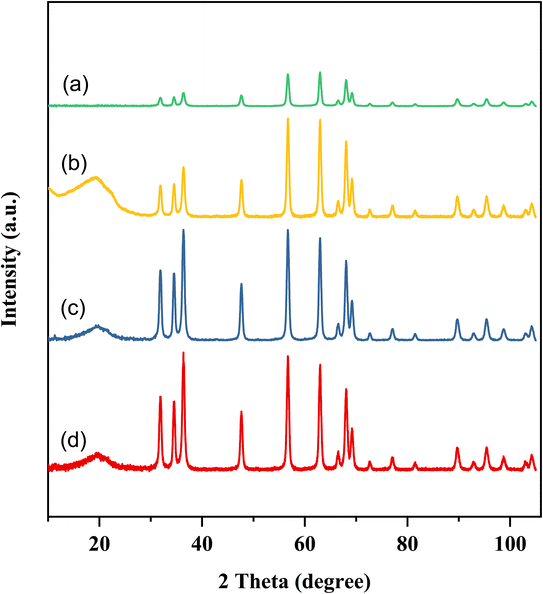 | ||
| Fig. 3 XRD diffraction patterns of hybrid nanocomposites (a) CMC-Fi-siRNA@ZnOa, (b) CMC-Fi-siRNA@ZnOb, (c) CMC-Fi-siRNA@ZnOc, (d) CMC-Fi-siRNA@ZnOd. | ||
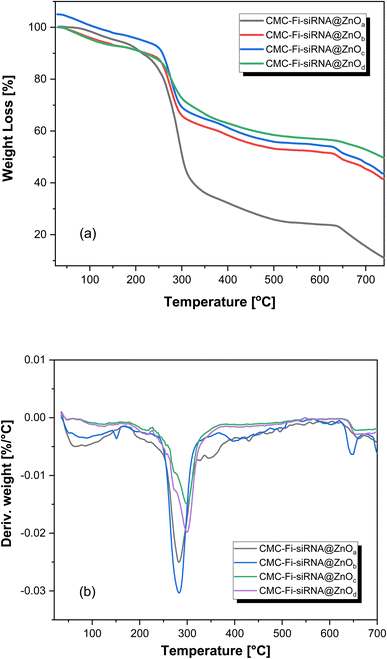
The samples undergo three consecutive decomposition steps during thermal degradation. The first step begins at temperatures ranging from 201 to 251 °C and ends at 340 to 303 °C for CMC-Fi-siRNA@ZnOa, CMC-Fi-siRNA@ZnOb, CMC-Fi-siRNA@ZnOc, and CMC-Fi-siRNA@ZnOd, respectively. The second step commences at 301 °C for all derivatives except CMC-Fi-siRNA@ZnOa, which starts at 335 °C and ends at approximately 621 °C. Notably, only CMC-Fi-siRNA@ZnOa exhibits significant separation between the three degradation steps. The initial stage of the thermogravimetric curves shows a weight reduction ranging from 1 to 5%, which is due to the loss of moisture and solvents. Table 3 provides the temperatures at which weight losses of 10%, 20%, 30%, and 40% occur (represented as 10–40% values). The 10% weight loss temperatures of the samples range from 201.71 to 271.64 °C, indicating their thermal stabilities. Among the CMC-Fi-siRNA@ZnOa–d derivatives, the thermal stability at 10% weight loss follows this order: CMC-Fi-siRNA@ZnOc > CMC-Fi-siRNA@ZnOb > CMC-Fi-siRNA@ZnOd > CMC-Fi-siRNA@ZnOa. The T40 value, which represents the temperature at 40% weight loss, plays a crucial role in determining the relative thermal stability of a polymer. The T40 values for CMC-Fi-siRNA@ZnOa–d range from 301 to 515 °C. The maximum polymer degradation temperature (PDTmax), which indicates the degree of maximum weight loss, is observed in the range of 282.32 to 298.60 °C, as shown in Fig. 4 and Table 3. At a temperature of 800 °C, the weight loss of the CMC-Fi-siRNA@ZnOa–d samples is 71.41%, 42.71%, 45.57%, and 39.57%, respectively. This indicates the high thermal stability of these polymers, particularly CMC-Fi-siRNA@ZnOa, which retains 71.41% of its weight even at a temperature of 800 °C. Comparatively, incorporating the ZnO nanofiller into CMC-Fi-siRNA@ZnOb, CMC-Fi-siRNA@ZnOc, and CMC-Fi-siRNA@ZnOd samples resulted in improved stability and reduced weight loss during degradation. This suggests that these samples exhibit enhanced thermal properties and overall improved characteristics, as indicated by a higher percentage of weight retention.
| Products | Temperature (°C) for various percentage decompositionsa | ||||
|---|---|---|---|---|---|
| PDTmaxb | 10% | 20% | 30% | 40% | |
| a Determined by TGA at a heating rate of 10 °C min−1.b The values were determined from DTG curves. | |||||
| CMC-Fi-siRNA@ZnOa | 282.32 | 201.71 | 258.98 | 288.11 | 301.64 |
| CMC-Fi-siRNA@ZnOb | 283.10 | 235.29 | 292.51 | 300.82 | 411.68 |
| CMC-Fi-siRNA@ZnOc | 299.53 | 271.64 | 287.13 | 301.64 | 419.83 |
| CMC-Fi-siRNA@ZnOd | 298.60 | 230.72 | 288.92 | 345.33 | 515.19 |
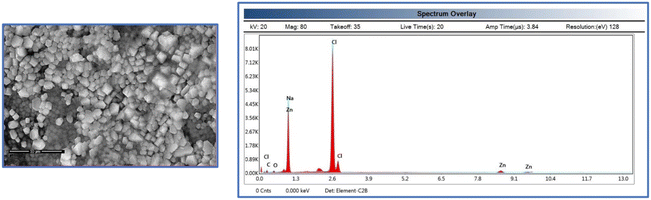
SEM and TEM were used to investigate the morphology and size distribution of the nanocomposite. The micrographs suggest that the fabricated hybrid nanocomposites are stable. The surface properties of the CMC-Fi-siRNA@ZnOa–d derivatives were assessed using SEM and EDX. The hybrid nanocomposites mostly appeared as additional aggregates, with a narrow size distribution and a spherical shape. SEM analyses were conducted at different magnifications: (a) for pure CMC at X = 2.5 K, (b and c) for CMC-Fi-siRNA@ZnOb at X = 5 K and 10 K, and (d and e) for CMC-Fi-siRNA@ZnOd at X = 15 K and 30 K, as depicted in Fig. 5. In the SEM images of CMC-Fi-siRNA@ZnOb (Fig. 5B and C) at X = 5 K and 10 K, aggregated cubic shapes indicative of loaded ZnO2 NPs were observed in certain regions on the composite surface. Meanwhile, for CMC-Fi-siRNA@ZnOd (d and e) at X = 15 K and 30 K, larger crystallites with a truncated octahedral morphology were apparent. EDX analysis of the synthesized CMC-Fi-siRNA@ZnOd nanocomposite confirmed the presence of carbon, oxygen, zinc, and sodium, providing evidence of ZnO NPs incorporation within the polymer nanocomposite, as illustrated in Fig. 6. Furthermore, TEM images (Fig. 7) of the CMC-Fi-siRNA@ZnOb and CMC-Fi-siRNA@ZnOd nanocomposites distinctly revealed the presence of immersed cubic-shaped ZnO within the nanocomposites.
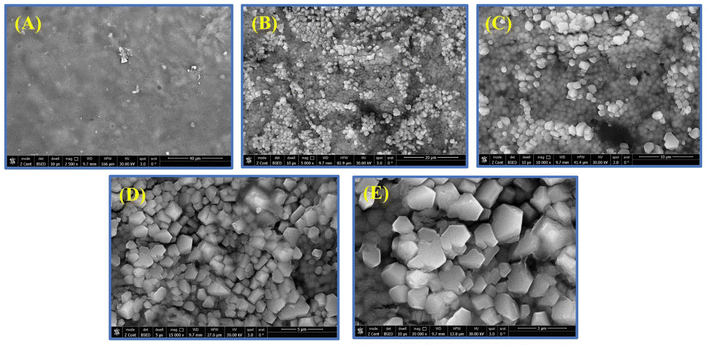 | ||
| Fig. 5 SEM images of pure CMC (A) (magnification X = 2.5 K), CMC-Fi-siRNA@ZnOb, (B and C) (magnifications X = 5 K and 10 K), CMC-Fi-siRNA@ZnOd, (D and E) (magnifications X = 15 K and 30 K). | ||
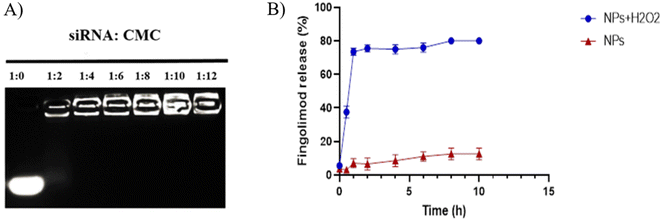
The zeta potential of drug-loaded nanocomposites has a significant impact on various properties, including the dosage, circulation release rate, and absorption of the drug. This is dependent on the surface charge of the outer layer of the hybrid nanocomposite.28 In general, nanocomposites with neutral, positively, or negatively charged low zeta potential tend to rapidly aggregate in aqueous solutions. The CMC-Fi-siRNA@ZnO nanocomposite was fabricated in the presence of a CMC polymer matrix, resulting in a highly negative charge indicating that CMC molecules were present on the surface of the hybrid nanocomposite. The potent negative charge of CMC provided the nanocomposite with colloidal stability, indicating that CMC is a suitable polymer for in vivo drug delivery.10,16 Using these values, the formulated CMC-Fi-siRNA@ZnOa–d hybrid nanocomposites were studied for their stability and biological function. Furthermore, to investigate STAT3 siRNA loading on the nanocomposite, a siRNA retarding assay was used. As shown in Fig. 8A, no siRNA band could be observed with all siRNA/CMC-Fi-siRNA@ZnO ratios, suggesting that siRNA could be fully loaded by nanocomposite through electronic complexing at all ratios. Next, to study the fingolimod release kinetics, the CMC-Fi-siRNA@ZnO was incubated with or without H2O2. After 1 h of incubation, the release of fingolimod increased to 80% in the presence of H2O2, while the release percentages were only about 10% without H2O2. These results demonstrated that the hybrid CMC-Fi-siRNA@ZnO nanocomposite has great ROS-sensitive ability as shown in Fig. 8B.
3.2 MTT assay
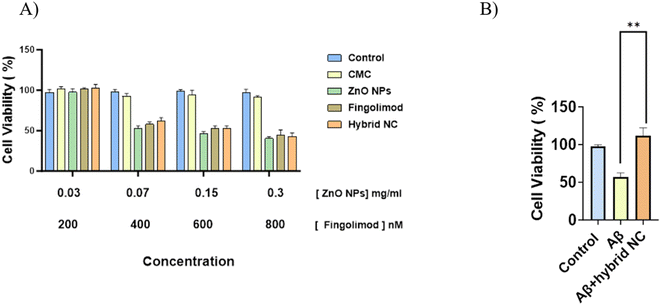
The MTT assay was used to determine the effect on IMG cell viability after 48 h of incubation with empty CMC, free ZnO NPs, free fingolimod, and the hybrid nanocomposite (CMC-Fi-siRNA@ZnO). The toxicity analysis after incubation of IMG cells with all concentrations of ZnO NPs and fingolimod alone showed induced higher toxicity in comparison to cells treated with 0.03 mg mL−1 and 200 nM for ZnO NPs and fingolimod (90–100% cell viability concerning the sample of control). Low nanocomposite toxicity is critical to be used safely and effectively in biomedical applications. No negative effect on the viability of the cells was observed when the CMC was loaded with 200 nM fingolimod, and 0.03 mg mL−1 ZnO NPs indicating that the hybrid nanocomposite at these concentrations has good biocompatibility (Fig. 9A). The potential cytotoxicity of aggregated Aβ42 was generally recognized.2 We used MTT assay also to investigate the nanocomposite effect on Aβ-induced cytotoxicity. Aβ42 was applied to IMG cells either with or without a hybrid nanocomposite. As shown in Fig. 9B, cell viability significantly decreased to 60% after exposure to Aβ42 for 48 h. The treatment with CMC-Fi-siRNA@ZnO hybrid nanocomposite significantly increased the viability of the cell to 125%, indicating that the CMC-hybrid nanocomposite could not only reduce the Aβ-induced cytotoxicity but also enhance cell proliferation.3.3 Inflammatory inhibition effect of the hybrid nanocomposite
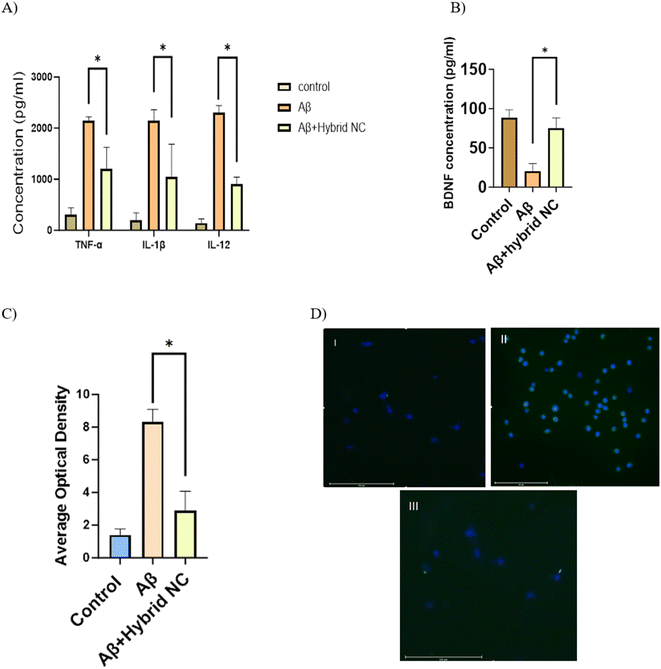
The aggregation of Aβ can activate microglia and cause the release of proinflammatory cytokines such as TNF-α and IL-1β and IL-12. This, in turn, promotes Aβ aggregation and neuronal death.12 To measure the levels of the proinflammatory cytokines, an enzyme-linked immunosorbent assay (ELISA) was used. The results, as displayed in Fig. 10A, show that when Aβ42 was incubated alone, the TNF-α supernatant level significantly increased to 2300 pg mL−1. However, when co-incubated with CMC-Fi-siRNA@ZnO hybrid nanocomposite, the production of TNF-α was considerably reduced compared to the Aβ42 alone group. The CMC-Fi-siRNA@ZnO hybrid nanocomposite also reduced IL-1β and IL-12 secretion, whereas the Aβ group consistently showed higher levels (Fig. 10A). Additionally, sandwich ELISA was used to measure the BDNF level. In contrast to the Aβ42 group, CMC-Fi-siRNA@ZnO hybrid nanocomposite increased the level of BDNF, demonstrating their neuroprotective effect (Fig. 10B). As a result, CMC-Fi-siRNA@ZnO hybrid nanocomposite was able to enhance the production of BDNF and lower the levels of pro-inflammatory cytokines, which might be attributed to their inflammatory inhibition effect. Moreover, Aβ-42 can induce microglial ROS production, which can result in a series of cellular component damages.4,31 Based on the performance of the CMC-Fi-siRNA@ZnO hybrid nanocomposite in reducing cytotoxicity of Aβ, the ROS level was assessed in microglia. The content of intracellular ROS was observed by CLSM. The green fluorescence was significantly increased by 5.15 times in the presence of Aβ42 alone compared to the control group, indicating an elevated ROS level. However, treating the cells with CMC-Fi-siRNA@ZnO hybrid nanocomposite caused a decrease in ROS level compared to the Aβ42 positive control, which was similar to that of cells of the control group (Fig. 10C and D).3.4 Pro-and anti-inflammatory cytokines' polymerase chain reaction
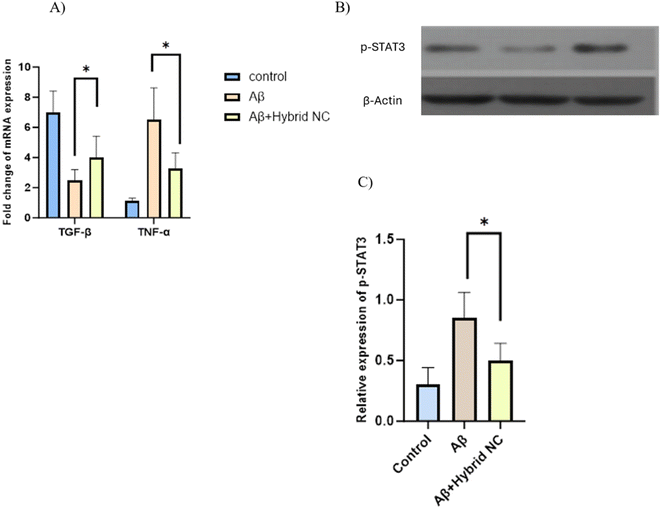
After the treatment with the hybrid nanocomposite, RT qPCR analysis revealed that the TNF-α proinflammatory M1 gene was decreased, while the TGF-β anti-inflammatory M2 gene was slightly elevated indicating that CMC-Fi-siRNA@ZnO hybrid nanocomposite have the potential to attenuate the inflammation in the activated microglia cells compared to Aβ treated group (Fig. 11A).3.5 p-STAT3 silencing
Western blot analysis was used to determine the p-STAT3 expression, the active form of STAT3, to detect the efficiency of hybrid nanocomposite silencing. Following Aβ42 treatment, IMG cells showed increased p-STAT3 expression. However, the CMC-Fi-siRNA@ZnO hybrid nanocomposite was able to reduce p-STAT3 expression (Fig. 11B and C). The results demonstrate that the CMC-based hybrid nanocomposite has efficient silencing capabilities.4. Conclusion
In this study, hybrid nanocomposite structures with the general abbreviations CMC-Fi-siRNA@ZnOa–d have been successfully fabricated using CMC biopolymer as a matrix and encapsulated with fingolimod, siRNA and in the presence of variable loading of ZnO using a casting technique. The final products were fully characterized by the most common characterization tools. The newly developed composites were categorized as a drug delivery system for the advancement of therapeutic approaches in the treatment of Alzheimer's disease (AD). The fabricated hybrid materials were able to normalize the microglia priming due to the synergistic effects of the drugs, which caused a significant increase in the phagocytosis of Aβ. The obtained MTT results indicated that the hybrid nanocomposites (CMC-Fi-siRNA@ZnO) were not cytotoxic, highly biocompatible, and could significantly reduce the Aβ-induced cytotoxicity. The pro-inflammatory cytokines and ROS are reduced because of the significant switching of the dysfunctional microglia toward an alternative activation phenotype by CMC-Fi-siRNA@ZnO hybrid nanocomposite. The synergistic strategy significantly restricted the interaction between microglia and Aβ, which reduced the AD pathology. The CMC-hybrid nanocomposite provided a new horizon of a carboxymethylcellulose-based @ ZnO of different loading material for the treatment of AD and other neurodegenerative diseases. The inclusion of ZnO NP and siRNA in the final formulations significantly improve the phagocytosis of Aβ. The CMC-Fi-siRNA@ZnOd which contains 20% ZnO loading dramatically alleviates the priming of microglia by lowering the level of proinflammatory mediators and increasing the secretion of BDNF.Data availability
All data was introduced in the text. The authors confirm that all data were included in the revised manuscript.Author contributions
N. B. Aljohani: methodology, investigation, writing – original draft preparation; S. Y. Qusti: supervision, writing – reviewing and editing; M. Alsiny: supervision, writing – reviewing and editing; F. Aljoud: methodology; N. B. Aljohani: writing – original draft preparation; E. S. Alsolami: writing – original draft preparation; K. A. Alamry: writing – reviewing and editing; M. A. Hussein: conceptualization, investigation, writing – reviewing and editing.Conflicts of interest
There are no conflicts to declare.References
- D. E. Ehrnhoefer, B. K. Y. Wong and M. R. Hayden, Nat. Rev. Drug Discovery, 2011, 10, 853 CrossRef CAS.
- M. T. Heneka, M. J. Carson, J. El Khoury, G. E. Landreth, F. Brosseron, D. L. Feinstein, A. H. Jacobs, T. Wyss-Coray, J. Vitorica, R. M. Ransohoff, K. Herrup, S. A. Frautschy, B. Finsen, G. C. Brown, A. Verkhratsky, K. Yamanaka, J. Koistinaho, E. Latz, A. Halle, G. C. Petzold, T. Town, D. Morgan, M. L. Shinohara, V. H. Perry, C. Holmes, N. G. Bazan, D. J. Brooks, S. Hunot, B. Joseph, N. Deigendesch, O. Garaschuk, E. Boddeke, C. A. Dinarello, J. C. Breitner, G. M. Cole, D. T. Golenbock and M. P. Kummer, Lancet Neurol., 2015, 14, 388–405 CrossRef CAS PubMed.
- P. Sharma, N. Y. Jang, J. W. Lee, B. C. Park, Y. K. Kim and N. H. Cho, Pharmaceutics, 2019, 11, 493, DOI:10.3390/pharmaceutics11100493.
- W. E. Bawarski, E. Chidlowsky, D. J. Bharali and S. A. Mousa, Nanomedicine, 2008, 4, 273–282, DOI:10.1016/j.nano.2008.06.002.
- D. J. Selkoe and J. Hardy, EMBO Mol. Med., 2016, 8, 595–608, DOI:10.15252/emmm.201606210.
- J. Flores, M. L. Fillion and A. C. LeBlanc, Cell Death Dis., 2022, 13, 864 CrossRef CAS.
- S. Lee, C. Altaner, J. Puls and B. Saake, Carbohydr. Polym., 2003, 54, 353–362 CrossRef CAS.
- D. Felsky, T. Roostaei, K. Nho, S. L. Risacher, E. M. Bradshaw, V. Petyuk, A. Schneider, J. A. Saykin, D. A. Bennett and P. L. De Jager, Nat. Commun., 2019, 10, 409 CrossRef CAS PubMed.
- A. K. Y. Fu, K. Huang, M. Y. F. Yuen, X. P. Zhou, D. S. Y. Mak, I. C. W. Chan, T. H. Cheung, B. R. Zhang, W. Fu, F. Y. Liew and N. Y. Ip, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2705 Search PubMed.
- B. B. Hu, F. Y. Dai, Z. M. Fan, G. H. Ma, Q. W. Tang and X. Zhang, Adv. Mater., 2015, 27, 5499 CrossRef CAS PubMed.
- M. V. Guillot-Sestier, K. R. Doty, D. Gate, J. Rodriguez Jr, B. P. Leung, K. Rezai-Zadeh and T. Town, Neuron, 2015, 85, 534–548 CrossRef CAS PubMed.
- S. F. Hunter, J. D. Bowen and A. T. Reder, CNS Drugs, 2016, 30, 135–147 CrossRef CAS.
- R. Liu, J. Yang, L. Liu, Z. Lu, Z. Shi, W. Ji, J. Shen and X. Zhang, Adv. Sci., 2019, 7, 1901555 CrossRef.
- Y. Guan, M. Li, K. Dong, N. Gao, J. Ren, Y. Zheng and X. Qu, Biomaterials, 2016, 98, 92–102 CrossRef CAS PubMed.
- W. B. Wang, J. X. Xu and A. Q. Wang, eXPRESS Polym. Lett., 2011, 5, 385–400 CrossRef CAS.
- I. Solanki, P. Parihar, M. L. Mansuri and M. S. Parihar, Adv. Nutr., 2015, 6, 64–72 CrossRef CAS.
- B. Wilson, M. K. Samanta, M. S. Muthu and G. Vinothapooshan, Ther. Delivery, 2011, 5, 599–609 CrossRef PubMed.
- L. Zhong, Y. Xu, R. Zhuo, T. Wang, K. Wang, R. Huang, D. Wang, Y. Gao, Y. Zhu, X. Sheng, K. Chen, N. Wang, L. Zhu, D. Can, Y. Marten, M. Shinohara, C. C. Liu, D. Du, H. Sun, L. Wen, H. Xu, G. Bu and X. F. Chen, Nat. Commun., 2019, 10, 1365 CrossRef PubMed.
- S. Kim, C. K. Lim, J. Na, Y. D. Lee, K. Kim, K. Choi, J. F. Leary and I. C. Kwon, Chem. Commun., 2010, 46, 1617–1619 RSC.
- J. J. Palop and L. Mucke, Nat. Neurosci., 2010, 13, 812–818 CrossRef CAS.
- M. Creixell, A. P. Herrera, M. Latorre-Esteves, V. Ayala, M. Torres-Lugo and C. Rinaldi, J. Mater. Chem., 2010, 20, 8539–8547 RSC.
- M. Prabaharan and J. F. Mano, Drug Deliv., 2005, 12, 41–57 CrossRef CAS PubMed.
- Z. Liu, Y. Jiao, Y. Wang, C. Zhou and Z. Zhang, Adv. Drug Delivery Rev., 2008, 60, 1650–1662 CrossRef CAS.
- D. C. Ferreira Soares, S. C. Domingues, D. B. Viana and M. L. Tebaldi, Biomed. Pharmacother., 2020, 131, 110695 CrossRef CAS.
- C. Boyer, M. R. Whittaker, V. Bulmus, J. Liu and T. P. Davis, NPG Asia Mater., 2010, 2, 23–30 CrossRef.
- G. Aguilera, C. C. Berry, R. M. West, E. Gonzalez-Monterrubio, A. Angulo-Molina, Ó. Arias-Carrión and M. Á. Méndez-Rojas, Nanoscale Adv., 2018, 1, 671–685 RSC.
- V. Ayala, A. P. Herrera, M. Latorre-Esteves, M. Torres-Lugo and C. Rinaldi, J. Nanopart. Res., 2013, 15, 1874 CrossRef PubMed.
- S. I. A. Cohen, P. Arosio, J. Presto, F. R. Kurudenkandy, H. Biverstal, L. Dolfe, C. Dunning, X. Yang, B. Frohm, M. Vendruscolo, J. Johansson, C. M. Dobson, A. Fisahn, T. P. J. Knowles and S. Linse, Nat. Struct. Mol. Biol., 2015, 22, 207–213 CrossRef CAS.
- C. Altaner, J. Puls and B. Saake, Cellulose, 2003, 10, 391–395 CrossRef CAS.
- W. J. Deardorff and G. T. Grossberg, Expert Rev. Neurother., 2017, 17, 17–32 CrossRef CAS.
- M. Eufemi, R. Cocchiola, D. Romaniello, V. Correani, L. Di Francesco, C. Fabrizi, B. Maras and M. E. Schininà, Neurochem. Int., 2015, 81, 48–56 CrossRef CAS.
- C. Turrina, M. Schoenen, D. Milani, A. Klassen, D. M. R. Gonzaléz, G. Cvirn, P. Mela, S. Berensmeier, I. Slabu and S. P. Schwaminger, Colloids Surf., B, 2023, 228, 113428 CrossRef CAS.
- C. Ren, D. Li, Q. Zhou and X. Hu, Biomaterials, 2020, 232, 119752 CrossRef CAS.
- A. Latif, T. Anwar and S. Noor, J. Chem. Soc. Pak., 2007, 29, 143 CAS.
- V. K. Varshney, P. K. Gupta, S. Naithani, R. Khullar, A. Bhatt and P. L. Soni, Carbohydr. Polym., 2006, 63, 40–45 CrossRef CAS.
- S. Tawfik, S. H. Abd Elsalam, H. M. El-Hennawi, I. Abd El-Thalouth and E. Adel, From the Proceeding International Conference on Applied Life Sciences, ed. F. Nejadkoorki, 2012 Search PubMed.
- V. Pushpamalar, S. J. Langford, M. Ahmad and Y. Y. Lim, Carbohydr. Polym., 2006, 64, 312–318 CrossRef CAS.
- M. El-Sakhawy, S. Kamel, A. Salama and H. A. S. Tohamy, Cellul. Chem. Technol., 2018, 52, 193–200 CAS.
- P. Vanathi, P. Rajiv, S. Narendhran, S. Rajeshwari, P. K. Rahman and R. Venckatesh, Mater. Lett., 2014, 134, 13–15 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra01965b |
| This journal is © The Royal Society of Chemistry 2024 |