DOI:
10.1039/D4RA01865F
(Paper)
RSC Adv., 2024,
14, 12081-12095
In vitro and in silico docking and molecular dynamic of antimicrobial activities, alpha-glucosidase, and anti-inflammatory activity of compounds from the aerial parts of Mussaenda saigonensis†
Received
11th March 2024
, Accepted 9th April 2024
First published on 16th April 2024
Abstract
Twelve compounds were isolated from Mussaenda saigonensis aerial parts through phytochemical analysis and the genus Mussaenda is the first place where the compounds 4–6 and 11–12 have been found. Based on the ability to inhibit NO production in RAW264.7 cells, compound 2 has demonstrated the strongest anti-inflammatory activity in vitro with an IC50 of 7.6 μM, as opposed to L-NMMA's IC50 of 41.3 μM. Compound 12 was found to be the most effective inhibitor of alpha-glucosidase enzyme in vitro, with an IC50 value of 42.4 μM (compared to 168 μM for acarbose). Compounds 1–12 were evaluated in vitro for antimicrobial activity using the paper dish method. Compound 11 demonstrated strong antifungal activity against M. gypseum with a MIC value of 50 μM. In silico docking for antimicrobial activity, pose 90 or compound 11 docked well to the 2VF5 enzyme, PDB, which explains why compound 11 had the highest activity in vitro. Entry 2/pose 280 demonstrated excellent anti-inflammatory activity in silico. The stability of the complex between pose 280 and the 4WCU enzyme for anti-inflammatory activity has been assessed using molecular dynamics over a simulation course ranging from 0 to 100 ns. It has been found to be stable from 60 and 100 ns. The Tyr 159 (95%, H-bond via water bridge), Asp 318 (200%, multiple contacts), Met 273 (75%, hydrophobic interaction via water bridge), and Gln 369 (75%, H-bond via water bridge) interacted well within the time range of 0 to 100 ns. It has more hydrophilic or polar pharmacokinetics.
1 Introduction
Mussaenda (Rubiaceae) is found in tropical and subtropical regions. Some of its plants are utilized in traditional Chinese medicine.1 Saponins, terpenoids, iridoids, and flavonoids are the major components found in the Mussaenda genus.2–14 Mussaenda plants have been found to have a variety of medicinal qualities, including antioxidant, anticancer, and anti-inflammatory effects.15–19 There are two native species in Vietnam: Mussaenda recurvata and Mussaenda saigonensis.20 M. recurvata is thought to be a good source of anti-inflammatory chemicals.21,22 Previous research on this Vietnamese plant found nine anti-inflammatory compounds: five saponins and four oleanane-type triterpenes.21,22 However, not much is known about Mussaenda saigonensis's chemical and biological data. The potential of M. recurvata components as alpha-glucosidase and NO inhibitors and the high potent activity of M. saigonensis extracts in these assays encouraged us to investigate the titled plant. The main objective of the research was to explore the potential of isolated compounds in suppressing alpha-glucosidase activity and NO production. In this paper, the phytochemical investigation of the aerial parts of Mussaenda saigonensis has performed resulting in the isolation of 12 compounds: shanzilactone (1), 6-acetyl shanzhiside methyl ester (2), barlerin (3), harpagoside (4), (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5), indole-3-carboxylic acid (6), ursolic acid (7), quinovic acid (8), rotundic acid (9), clethric acid (10), martynoside (11), and verbacoside (12) as seen in Fig. 1.
 |
| | Fig. 1 Structures of compounds 1–12 have been isolated from M. saigonensis material. | |
2 Materials and methods
2.1. Materials
For nuclear magnetic resonance (NMR) and high-resolution electrospray ionization mass spectroscopic (HRESIMS) spectra, the Bruker Avance III and MicrOTOF-Q mass spectrometers have been selected. The following deuterated solvents were used to record the 1H NMR (500, 600 MHz) and 13C NMR (125, 150 MHz) spectra: CDCl3, DMSO-d6, and CD3OD. Normal-phase and reverse-phase silica gel (Merck) were used for thin-layer chromatography (TLC). The Himedia silica gel 60 was utilized for the column chromatography (CC) procedure. For the alpha-glucosidase inhibitory experiment, yeast Saccharomyces cerevisiae, α-glucosidase enzyme (Sigma-Aldrich, EC3.2.1.20) and the positive control acarbose (Sigma-Aldrich, EC260-030-7) were acquired. For the NO inhibition experiment, Griess' reagent (Sigma-Aldrich, EC215-981-2) and the positive control, L-NMMA (Merck, M7033), were utilized.
2.2. Plant materials
In November 2020, plant material belonging to Mussaenda saigonensis was gathered in Tay Ninh province, Vietnam. For the study, the aerial sections were chosen. Dr Dang Van Son used the voucher specimen (VNM_Tri467) kept in the VNM Herbarium to identify the botanical name.
2.3. Extraction and isolation
After being dried and ground, 8.0 kg of M. saigonensis material was allowed to macerate in 3 × 40 L of EtOH (24 h each), and the mixture was then concentrated using the aid of an evaporator to yield 1.200 g of EtOH extract. Step-by-step liquid–liquid partitioning was carried out with solvents that became more polar: n-hexane, dichloromethane, and ethyl acetate. The resulting extracts were n-hexane (70.0 g), CH2Cl2 (300.0 g), and EtOAc (410.0 g), in that order. Eight fractions, E1–E8, were obtained by applying the (EtOAc, 410.0 g) extract to silica gel CC using the gradient system of CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (30
MeOH (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–0
1–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
Six fractions, E2.1–E2.6, were obtained by further fractionating fraction E2 (32.0 g) using silica gel CC eluted with CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (30
MeOH (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–5
1–5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Compounds 1 (8.0 mg) and 5 (12.0 mg) were obtained by rechromatographing the fraction E.2.2 (2.5 g) on silica gel CC with the n-hexane–CH2Cl2–MeOH (3
1, v/v). Compounds 1 (8.0 mg) and 5 (12.0 mg) were obtained by rechromatographing the fraction E.2.2 (2.5 g) on silica gel CC with the n-hexane–CH2Cl2–MeOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v) solvent system. Four subfractions (E2.5.1–E2.5.4) have been created from the further chromatography of fraction E2.5 (4.8 g) on silica gel CC. Combinations of n-hexane and EtOAc were used to elute the fraction (2
0.1, v/v/v) solvent system. Four subfractions (E2.5.1–E2.5.4) have been created from the further chromatography of fraction E2.5 (4.8 g) on silica gel CC. Combinations of n-hexane and EtOAc were used to elute the fraction (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and 1
2, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v). In order to collect compounds 7 (20.0 mg) and 9 (6.0 mg), subfraction E2.5.3 (690 mg) was passed over a silica gel CC with an eluent of n-hexane
5, v/v). In order to collect compounds 7 (20.0 mg) and 9 (6.0 mg), subfraction E2.5.3 (690 mg) was passed over a silica gel CC with an eluent of n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Subsequently, six fractions, E5.1–E5.6, were obtained by dissolving the 42 g of fraction E5 employing a gradient system of CH2Cl2–MeOH (10
1). Subsequently, six fractions, E5.1–E5.6, were obtained by dissolving the 42 g of fraction E5 employing a gradient system of CH2Cl2–MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–0
1–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). This was done using silica gel CC. Five fractions (E5.3.1–5.3.5) were isolated by rechromatographing fraction E5.3 (11.0 g) using a gradient of CH2Cl2
1, v/v). This was done using silica gel CC. Five fractions (E5.3.1–5.3.5) were isolated by rechromatographing fraction E5.3 (11.0 g) using a gradient of CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (10
MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → 1
1 → 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Compounds 6 (5.0 mg) and 8 (7.0 mg) were extracted by repeated the subfraction E5.3.2 (950 mg) by chromatography on a silica gel column eluting with CH2Cl2
1, v/v). Compounds 6 (5.0 mg) and 8 (7.0 mg) were extracted by repeated the subfraction E5.3.2 (950 mg) by chromatography on a silica gel column eluting with CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (10
MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). By implementing n-hexane
1, v/v). By implementing n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) as an eluent, column chromatography was used to separate subfraction E5.3.4 (860 mg), yielding compounds 10 (12.0 mg) and 4 (8.0 mg). Compounds 2 (14.0 mg) and 3 (10.0 mg) were obtained from fraction E5.4 (12 g) by two consecutive silica gel CC: a normal-phase column with a mobile phase as n-hexane
1, v/v/v) as an eluent, column chromatography was used to separate subfraction E5.3.4 (860 mg), yielding compounds 10 (12.0 mg) and 4 (8.0 mg). Compounds 2 (14.0 mg) and 3 (10.0 mg) were obtained from fraction E5.4 (12 g) by two consecutive silica gel CC: a normal-phase column with a mobile phase as n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) and a C-18 reverse-phase column using a solvent system of MeOH
1, v/v/v) and a C-18 reverse-phase column using a solvent system of MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (3
H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). A gradient system of EtOAc–MeOH was used to elute the silica gel CC used to extract fraction E7 (53 g) (10
1, v/v). A gradient system of EtOAc–MeOH was used to elute the silica gel CC used to extract fraction E7 (53 g) (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v) to give five subfractions. E7.1–E7.5. Subfraction E7.3 (14.0 g) was fractionated by silica gel CC using a solvent system consisting of n-hexane, ethyl acetate, and methanol (1
10, v/v) to give five subfractions. E7.1–E7.5. Subfraction E7.3 (14.0 g) was fractionated by silica gel CC using a solvent system consisting of n-hexane, ethyl acetate, and methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 to 1
0.1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v) to afford six fractions. E7.3.1–E7.3.6. Fraction E7.3.4 (945 mg) was applied to C-18-RP silica gel CC, eluted with MeOH
0.1, v/v/v) to afford six fractions. E7.3.1–E7.3.6. Fraction E7.3.4 (945 mg) was applied to C-18-RP silica gel CC, eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (2
H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to afford compounds 11 (8.0 mg) and 12 (11.0 mg).
1, v/v) to afford compounds 11 (8.0 mg) and 12 (11.0 mg).
2.4. Alpha-glucosidase inhibition
Isolated compounds were evaluated for the alpha-glucosidase inhibition using the same procedure reported previously.23 Each sample was dissolved in DMSO, then diluted in different concentrations. The alpha-glucosidase enzyme and p-nitrophenyl-D-glucopyranoside (pNPG) were dissolved in sodium phosphate buffer at pH 6.9. The prepared samples (each 50 μL) were preincubated with alpha-glucosidase at 37 °C for 20 min. The mixture then added by 40 μL of pNPG and continued for 20 min. Finally, sodium carbonate (130 μL, 0.2 M) was added to stop the reaction. Each sample has been evaluated in triplicate at nine different concentrations surrounding the IC50 values, and the average values were used for further analysis.
2.5. Measurement of NO production assay
NO inhibition of compounds 1–12 were determined with the same procedure reported previously.21,22 Dulbecco's Modified Eagle's Medium (DMEM) contained L-glutamine (2 mM), HEPES (10 mM), sodium pyruvate (1 mM), and 10% fetal bovine serum (FBS). RAW 264.7 cells were cultured in DMEM, seeded in 96-well culture plates (each well 2 × 105 cells), and incubated for 3 days (the tested condition: 37 °C in 5%-CO2 humidified air). NO amount was measured using the Griess reagent. L-NMMA was used as a positive control.
2.6. In vitro anti-microbial activities
The antimicrobial and antifungal activity of compounds 1–12, and control drugs such as ampicillin and fluconazole have been performed based on paper disk diffusion for quality and followed article.24 All compounds, 1–13 had performed for its inhibition activity against bacteria Escherichia coli (E.C), Pseudomonas aeruginosa (P.A), Streptococcus faecalis (S.F) and methicillin-resistant (MRSA) and fungi Candida albicans, T. mentagrophytes, and T. rubrum, and M. gypseum by the disk diffusion method. The ampicillin and fluconazole had been used as standard drugs at a concentration of 1 mg mL−1. The microbial test organisms were prepared in LB broth for 24 hours at 37 °C. Muller–Hinton agar (MHA) or Potato Dextrose Agar (PDA) media have been used to prepare gel plates for bacteria or fungi. Each strain was swabbed uniformly into the individual plate using sterile cotton swabs. The paper dishes of 6 mm diameter were put on surfaces of MHA disks. For this step, entries 1–12 at the concentrations from 3.125 to 300 μM were taken equally at 30 μL, meanwhile, each bacterium sample were added with positive drug into a paper disk. (The 30 μL of every sample of entries 1–12 and the positive drug put each of paper disk for every bacterium/fungus.) After incubation at 25 °C for 24 hours, measuring the level of inhibited zone was conducted.
2.7. In silico docking study and MD
2.7.1. Procedure docking. In silico docking study of compounds has performed based on Scheme 1, for antimicrobial activity of compounds, enzyme 2VF5: PDB are used to explain the mechanism why compound has shown good interactions with enzyme in silico docking and led compound inhibited well with enzyme.1,26 The active center on 2VF5 has determined at coordination of X, Y, Z: 26.577, 22.697, 8.113, respectively.27 The docking parameters has determined by spacing, the numbers of elements on X, Y, Z axis are 0.500 and (60, 60, 60), respectively. For docking to account for the activity inflammatory, the 4WCU enzyme has been selected to calculated docking.26 In docking of ligand or the best pose, the docking parameters has been set up as active center, spacing, and the numbers of elements on X, Y, and Z axis that are (X, Y, and Z of 29.286, −50.843, and −26.674), 0.5, and (60, 60, and 60).
 |
| | Scheme 1 General docking procedure of the best docking pose of ligand to target enzyme. | |
2.7.2. MD simulation. The complex of the best docking pose 280 (the best conformation docking among 500 conformations, run = 500 models), the best confirmation of compound (4) and the crystal structure 4WCU enzyme had been saved in complex_pose_280_4WCU.pdb file by autodock tools, and has imported the structure of this complex to Desmond Schrodinger 2018, version 4 in Linux operation system for molecular dynamic (MD).28–31 The MD simulation course focuses on utilizing thermostat and barostat to maintain constant temperature (T = constant) in a system with variables N, V, and E. The Nosé–Hoover thermostat is typically used in molecular dynamics simulations. Barostats, which can fluctuate based on cell volume in liquids or cell parameters in solids, are also commonly employed.32 When combined with the Charmm36 force field, the SPC water model is the most effective option.33,34 In order to derive the protein force field and the atomic partial charges, quantum chemical calculations were performed on the interactions that occurred between model compounds and water via Scheme 2. The parameters set up in system were 300 K, 100 atm of pressure, 100 ns of simulation time, 24 of CPU, 1000 trajectory course in molecular dynamic panel. The initial conditions had been chosen via Desmond system builder panel including solvation and ions. The MD simulation course in Desmond – Schrodinger software in Linux has 3 part need to be finished: (1) preparing protein wizard, (2) Desmond system builder, and (3) MD simulation.
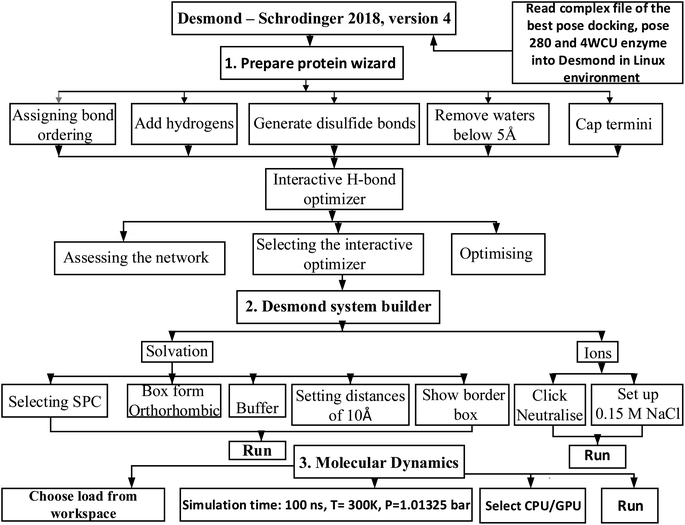 |
| | Scheme 2 General procedure for MD simulation of one complex of the best docking pose 280 and 4WCU on real environment has been conducted by Desmond in Linux. | |
3 Results and discussions
3.1. Isolation and identification of isolated compounds
12 compounds have isolated from Mussaenda saigonensis. To confirm the chemical structures, the NMR and HRESIMS spectroscopic methods were used. Shanzilactone (1), 6-acetyl shanzhiside methyl ester (2), barlerin (3), harpagoside (4), and (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5), indole-3-carboxylic acid (6), ursolic acid (7), quinovic acid (8), rotundic acid (9), clethric acid (10), martynoside (11) and verbacoside (12). Iridoids (1–4), sesquiterpenes (5), alkaloids (6), triterpenes (7–10), and phenylethanoid derivatives (11 and 12) are the five types of skeletons. All data NMR of entry 1–12 and heteronuclear multiple bond correlations (HMBC) described in Tables S1–S12 and Fig. S13.1 to S24.5.† Iridoids 1–3 were discovered in Mussaenda plants: shanzilactone (1) from M. incana,35 6-acetyl shanzhiside methyl ester (2) from M. pubescens and barlerin (3) from M. pubescens.36 Harpagoside (4), (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5), and indole-3-carboxylic acid (6) were discovered for the first time in Mussaenda. Triterpenoid and triterpenoid glycosides are well-known in Mussaenda plants, including ursolic acid (7) from M. recurvata,22 quinovic acid (8) from M. pilosissima and M. glabra,12,14 rotundic acid (9) from M. macrophylla,8 and clethric acid (10) from M. pubescens.4 This is the first time a phenylethanoid skeleton (compounds 11 and 12) has been discovered in the Mussaenda genus. Compounds 11 and 12 are new members that have contributed to the Mussaenda genus' chemical data. Our preliminary study on this species indicated the presence of six compounds: 2α,3β-dihydroxyurs-12-en-28-oic acid, 3β,23-dihydroxyurs-12-en-28-oic acid, coumaric acid, caffeic acid, aloe-emodin and 11-O-α-rhamnopyranosylaloe-emodin. They are anthraquinones, phenolic monocyclic compounds, and triterpenes that were found for the first time in Mussaenda saigonensis. However, no biological activity of compounds was reported. This paper focused on the bioguided-isolation based on antimicrobial activities, alpha-glucosidase, and anti-inflammatory activity to investigated bioactive components from Mussaenda saigonensis.37
The physicochemical properties of compounds 1–12 were described in the ESI documentation.†
3.2. Inhibition of NO production
Compounds 1–12 were assessed for their capability to hinder the production of NO in RAW264.7 cells when stimulated with LPS as seen in Fig. 2. Compound 2 is strongest inhibition NO among 12 compounds and positive control. Compounds 2, 7 and 4 reduced nitrite accumulation in LPS-stimulated RAW 264.7 cells, with IC50 values of 7.6 ± 0.98, 18.2 ± 0.70 and 25.9 ± 3.32 μM, respectively, and others were weak or inactive. The positive control used was L-NMMA, with an IC50 value of 41.3 ± 6.6 μM. Iridoids 2–4 with a glucose unit performed better than non-sugar iridoid 1, indicating the importance of this glucose moiety. Compound 7 was the most active of the ursanes 7–10, implying that substitutions at C-19, C-23, C-24, and C-26 would reduce activity. Iridoids are well-known anti-inflammatory compounds, according to a comprehensive review.38 Harpagoside (4) was found to be a significant anti-inflammatory constituent of Harpagophytum procumbens.39 Our findings were consistent with those found in the literature. Phenylethanoids 11 and 12 have also been reported to be weak NO inhibitors1 According to the authors, 11 and 12 are both weaker than 4. Other compounds that inhibit NO have previously been reported: 5,40 6,41 7,42 8,41,42 9,43,44 and 10.45
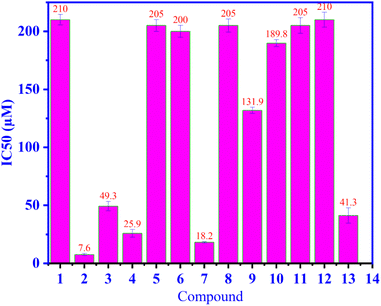 |
| | Fig. 2 The ability of NO inhibition of compound 1–12, and positive control (13), L-NMMA are indicate in the values of IC50. | |
3.3. Alpha-glucosidase inhibition
Compounds 1–12 were assessed for their ability to inhibit alpha glucosidase. The compounds all displayed moderate activity, with IC50 values covering a wide range from 42.4 to 250 μM, as seen in Fig. 3 compared to the positive control acarbose IC50 168 μM. Among 1–12, ursolic acid is believed to be a well-known inhibitor with many published reports.43,46,47 Biological data of 8–12 gave consistency with those reported previously.48–50 Compound 12 are the best candidate for alpha glucosidase enzyme inhibition among us and positive control, acarbose with the IC50 value of 42.4 μM.
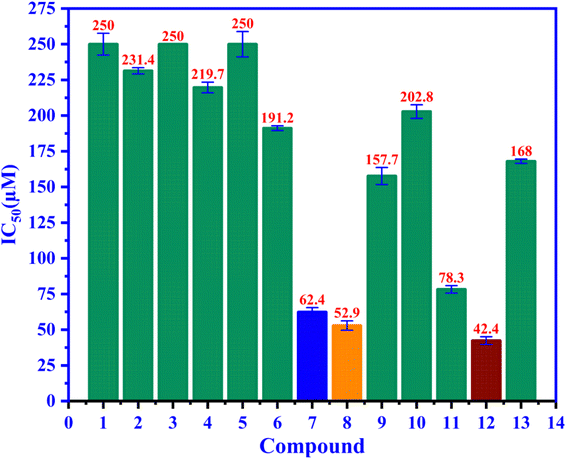 |
| | Fig. 3 The ability of alpha-glucosidase inhibition of entries 1–12 and positive control 13, acarbose have been presented in the IC50 values. | |
3.4. In vitro antimicrobial activities
3.4.1. Antibacterial activity. As shown in Fig. 4, the antibacterial activity of compound 1–12 and ampicillin drug (13) (positive control) have been exposed in the values of the minimum inhibition concentrations (MICs), as seen in Fig. 4, the MIC values of compound 1–12 are varied from 100 to 300 μM and they compared to the MICs of standard drug, ampicillin that changed from 3.125 to 6.25 μM against four bacteria such as E. coli, P. aeruginosa, MRSA, S. faecalis. All compounds 1–12 indicated the antibacterial activity ability are moderate activity against E. coli, P. aeruginosa, MRSA, S. faecalis. Among compounds exposed low or inactivity antibacterial activity, compound 2 and 5 demonstrated good inhibition against E. coli at concentration of 100 μM and compound 10 inhibited against MRSA and S. faecalis at concentration of 100 and 100 μM, respectively.
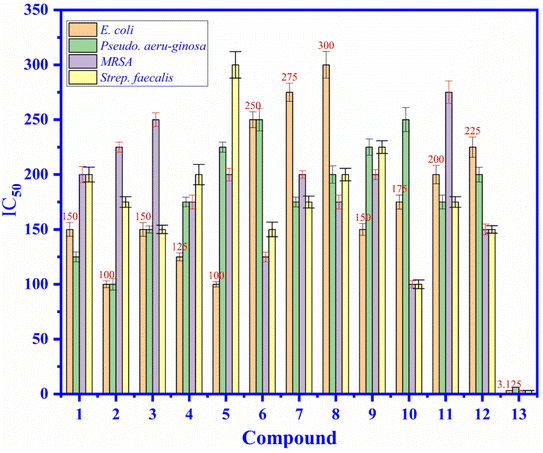 |
| | Fig. 4 The antibacterial activity of compound (1) to (12), and (13): ampicillin drug (positive control) at concentrations of 3.125 to 300 μM against E. coli, P. aeruginosa, MRSA, and S. faecalis bacteria presented in the values of MIC (μM). | |
3.4.2. Antifungal activity. As indicated in Fig. 5, the antifungal activity of compound 1–12, and positive control drug, fluconazole against C. albicans, T. mentagrophytes, T. rubrum, and M. gypseum fungus is demonstrated that compound 11 had shown the most excellent activity against M. gypseum fungi with the value of MIC of 50 μM that compare to the values of the others changed from 100 to 275 μM and the value of standard drug of 6.12 μM. Some compounds had shown good inhibition with the values of MIC of 100 μM because the compound 2 inhibited against C. albicans, and compound 6, 7, and 10 inhibited against M. gypseum. Some compounds indicated weak inhibitions or inactive such as compound 1, 2, 3, 4, 5, 8, 9, and 12 (all fungus), 6, 7, 10, 11 (E. coli, P. aeruginosa, and MRSA). As seen in Fig. 6, the antimicrobial activity of compound 11 (martynoside) has inhibited M. gypseum fungi at different concentrations of C1, C2, C3, C4, and C5 of 50, 25, 12.5, 6.25 and negative control (biochemistry DMSO solvent), respectively.
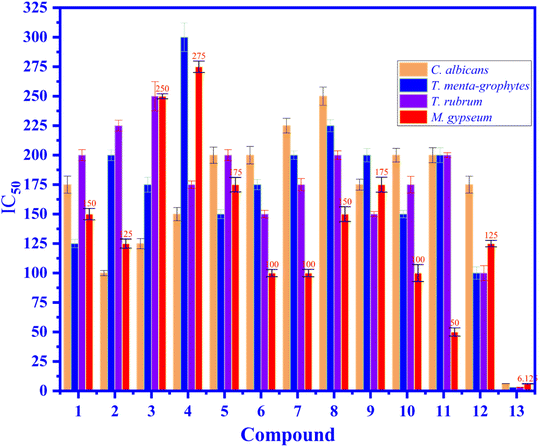 |
| | Fig. 5 The antifugal activity of compound 1–12, and 13: fluconazole, positive control drug at concentrations of 3.125 to 300 μM against fungus such as C. albicans, T. mentagrophytes, T. rubrum, and M. gypseum fungus are exposed in the values of MIC (μM). | |
 |
| | Fig. 6 The antimicrobial activity of compound 11/Martynoside has inhibited against M. gypseum fungi at different concentrations of C1 = 50 μM, C2 = 25, C3 = 12.5, C4 = 6.25 and C5: negative control biochemistry DMSO solvent. | |
3.5. In silico docking and MD
3.5.1. Antimicrobial activity. The docking results of the most activity poses of compounds 1–11 have been indicated in Table S13,† Fig. 4 to 5 (the active pose) and Fig. S25–S33† (the inactive poses). As seen in Table S13,† at thermodynamic site (free Gibbs energy or Ki), these poses had docked to enzyme 2VF5, in order to rank by rotundic acid (9) > quinovic acid (8) > clethric acid (10) > (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5) > barlerin (3) > 6-acetyl shanzhiside methyl ester (2) > harpagoside (4) > shanzilactone (1) > verbacoside (12) > martynoside (11) > indole-3-carboxylic acid (6). Base on ligand interaction model, those have been ranked by martynoside (11) > harpagoside (4) > 6-acetyl shanzhiside methyl ester (2) > rotundic acid (9) = quinovic acid (8) = clethric acid = (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5) = barlerin (3) = shanzilactone (1) = verbacoside (12) = indole-3-carboxylic acid (6).51
3.5.1.1 Pose 90. The best one stable conformation among 500 docking conformations of compound 11 were docked on 2VF5 enzyme with the values of free Gibbs energy and inhibition constant of −4.92 kcal mol−1 and 248 μM, respectively to explain general mechanism why conformation inhibits well enzyme to make inactive bacteria or fungi, as seen in Fig. 4 to 5.25 Based on analyzing the docking interaction model, Fig. 7, pose 90 interacted well with enzyme because three parts of pose 90 have exposed full interactions with 2VF5 such as functional group: Glu 325 to hydrogen atoms of hydroxyl groups, Leu 480 to hydrogen atom of hydroxyl phenolic, Tyr 312 to hydrogen atom of hydroxyl phenolic group, and Tyr 304 to oxygen and hydrogen atoms; capping group: pi–pi stacked interaction from Tyr 476 to pi electron system of phenyl group, and pi–alkyl or alkyl from Leu 480 to pi electron system of phenyl group; and linker part: Leu 480 to methyl group, and Tyr 476 to methyl of methoxy group. The pose 90 ranked strongest interaction because it has significant hydrophobic such as pi–pi stacked and pi–pi alkyl or alkyl interactions (Tyr 476), pi–pi alkyl or alkyl interactions (Ala 496 and Leu 480). Those interactions belong to be characteristic hydrophobic of pose/drug. As seen in Table S13,† those hydrogen bondings, 7 hydrogen bondings develop the attribute hydrophilic of pose 90. Among hydrogen bondings, pose 90:H – X:Glu 325:O is strongest bonding because of shortest bond length. As seen in Fig. 8, one ligand map indicated the secondary interactions forming between pose 90 and 2VF5 including hydrogen bondings (brown color lines): Tyr 312, Tyr 304, Glu 325, Val 324, and Asn 305, steric interactions (light green colors): Met 308, Tyr 304, Glu 325, and Tyr 312, and overlap interactions (violet circles) on atoms of pose 90. The bigger the diameter of violet circles, the stronger overlap interactions are. Other poses that interacted weakly with 2VF5 are explained in Table S13† (column describes).
 |
| | Fig. 7 One 2D diagram exposed the significant ligand interactions between pose 90, compound 11 and 2VF5: full interaction in ligand interaction model. | |
 |
| | Fig. 8 One ligand map shown the secondary interactions between pose 90, compound 11 and 2VF5, hydrogen bondings: brown color, light green color: steric interactions, violet circles: overlap interactions. | |
3.5.2. Anti-inflammatory.
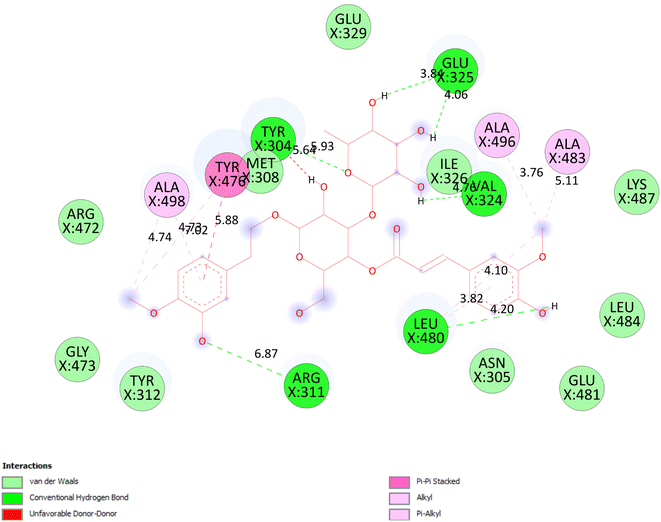
3.5.2.1 Pose 280. At thermodynamic site, the best docking pose 280 among 500 conformation of 6-acetyl shanzhiside methyl ester of compound (2) have docked to pocket site of 4WCU enzyme to explain why compound 2 or pose 280 indicated high alpha glucoside enzyme inhibition in vitro by calculation and modeling in silico docking study and MD simulations.52,53 This pose anchored to 4WCU with the values of free Gibbs energy and inhibition constant of −6.38 kcal mol−1 and 21.0 μM, respectively. It formed 5 hydrogen bonding that are listed as B:Tyr 159:O – pose 280:O (2.86 Å), pose 280:H – B:Asp 318:O (2.41 Å); pose 280:H – B:Asp 318:O (1.98 Å); pose 280:H – B:Asp 318:O (2.04 Å); pose 280:H – B:Asp 201:O (1.86 Å). Among them, hydrogen bonding, pose 280:H – B:Asp 201:O is strongest because of the shortest bond length. As seen in Fig. 9 or pose 280, at ligand interaction model site, pose 280 is proved good interactions with enzyme because 3 parts of pose have shown full interactions such as capping group or protein identification of pose 280: carbon hydrogen bond from Asp 318 to carbon atom of cyclohexene ether, linker part of pose 280: carbon hydrogen bond from Thr 271 to hydroxyl methylene carbohydrate group, and functional group of this pose: Tyr 159 to hydroxyl alcohol of cyclopentyl group; Asp 201 to hydrogen atom of carbohydrate group; Asp 318 to hydrogen atom of hydroxyl alcohol of cyclopentenyl group, hydrogen atoms of hydroxyl alcohol groups of carbohydrate, respectively. Pose 280 interacted well with 4WCU at ligand interaction model site.26 As exposed in Fig. 10, one ligand map has shown the secondary interactions such as hydrogen bonding (light brown color lines): Asp 201, Asp 318, steric interaction (light green color lines): Met 273, Phe 372, Phe 340, Leu 319, Asp 201, Tyr 159, Asp 318, Thr 271, and Glu 230, and overlap interactions that are violet circles on atoms of pose 280. The bigger diameters of violet circles, the stronger overlap interactions are.
 |
| | Fig. 9 The significant interactions between pose 280, compound 2 and 4WCU: PDB enzyme on one 2D diagram. | |
 |
| | Fig. 10 One ligand map presented the secondary interaction between pose 280, compound 2 and 4WCU, hydrogen bondings: brown color, light green color: steric interactions, violet circles: overlap interactions. | |
3.5.3. Molecular dynamic (MD) of complex of pose 280 and 4WCU in silico of the anti-inflammatory activity. The simulation interactions of complex of pose 280, one the best docking pose 280 of compound 6-acetyl shanzhiside methyl ester, compound (2) and 4WCU: enzyme to explain to be stable of complex in real environment such as 300 K, 100.102 of simulation time, the numbers of atoms of 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682, the number of water molecules of 11
682, the number of water molecules of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399, 0 charge, and 24 CPUs. For 4WCU enzyme, the enzyme information is presented by a 339 residual in chain, one chain (B), 5411 atoms, 2722 heavy atoms, and −15 charge. The ligand properties are reported by 59 total atoms including 31 heavy atoms, 0 charge, 5 fragments, 12 rotation bonds. The counter ion (salt information) is Na type, 15 atom, concentration of 23.926 mM, and 15 positive charges. As seen in Fig. 11A–J, the enzyme secondary structure is 52.62% helix, 0% strand, and 52.62% total SSE (secondary structure elements). As indicated in Fig. 11B, the curves of RMSD of 4WCU and pose 280 show the RMSDs of 4WCU (left Y-axis), the RMSD of pose 280 (Y-right axis) against simulation time of 0 to 100 ns (X-axis). The plots of the values of RMSD of 4WCU have been varied from 1.0 to 2.0 Å for C-alpha (light blue curve), 1.0 to 2.2 for backbone plot (green color curve), and 1.0 to 3.4 side chain curve are in order of 1–3 Å are perfectly acceptable for small, globular proteins. Changes much smaller than that, however, indicate that the 4WCU is undergoing a small conformational change during the simulation. It is also important that your simulation converges – the RMSD values stabilize around a fixed value. The values of curve of RMSD of pose 280 fit 4WCU (‘Lig fit Pro’) are a little higher than the values of RMSD of backbone of the reference and then the RMSD of the heavy atoms of pose 280 are measured. The values observed are a little larger than the RMSD of the 4WCU enzyme, so it is likely that the pose 280 has a little diffused away from its initial binding site of 4WCU. The curve of RMSD of ‘Lig fit Pro’, dark red color has equilibrated from 60 to 100 ns, varied a little change from 20 to 60 ns, and make larger fluctuation from 0 to 20 ns. The values of RMSD of pose 280, ‘Lig fit Lig’ – light violet color shows the RMSD of pose 280 that aligned and measured just on its reference conformation. This RMSD value measures the internal fluctuations of the atoms of pose 280. The plot of ‘Lig fit Lig’ has exposed that the values of RMSD of pose 280 are changed from 0.6 to 1.2 Å (≤2.0 Å) that proved the validation of docking model of pose 280 anchoring to 4WCU about oriental docking, docking conformation, docking method, docking parameters, and docking model for interesting pose 280 to active site on 4WCU.54 As seen in Fig. 11C, residual peaks indicated areas of the protein that fluctuated the most during the simulation time of 100 ns. On this curve, peaks indicate areas of the protein that fluctuated the most during the simulation are 125 to 140, 200 to 220, 250 to 275, and 320 to 340. Typically, the tails (N- and C-terminal) fluctuated more than any other part of 4WCU. Secondary structure elements like alpha helices and beta strands are usually more rigid than the unstructured part of the enzyme, and thus fluctuate less than the loop regions. As indicated in Fig. 11D, structure of pose 280 have 10 torsion bondings including C–OH (brown color), C–OH (violet), C–OH (orange), C–O (ether, red color) of carbohydrate part, C–O (green color), C–C of ester (light orange), C–O of ester (light violet) of cyclopentenyl ether, C–O (light green) of alcohol group, C–O (light blue) of acetyl group, and C–O (light pink) of alcohol group of cyclopentyl group. As seen in Fig. 11E, the interactions of 4WCU with the pose 280 have been presented by H bond that interacted with pose 280 via water bridges maintained over 95% simulation course from Tyr 159 on 4WCU. The residual amino acid Asp 318 on 4WCU have made multiple contacts of same subtype with the pose 280 via water bridges (200%). The Asp 318 has formed H-bonds with atoms on pose 280 via water bridge interactions. The Met 273 linked H-bonds via hydrophobic and water bridge that control 75% of simulation time. The Gln 369 has formed H-bond via water bridges maintained at 75% simulation course. The Phe 372 has created the hydrophobic interactions that control over 55% simulation course. The Asp 272 linked H-bond via water bridge that took over 60% simulation time. The Asn 321 and Tyr 329 formed water bridges that established over 40 and 55% simulation time, respectively. As indicated in Fig. 11F, the numbers of contracts appeared one more time in whole simulation course such as Tyr 159 (0 to 100 ns), His 160 (0 to less than 50 ns), Asp 318 (0 to 100 ns), Tyr 329 (10 to 100 ns), Met 273 (0 to 100 ns), Thr 333 (10 to 90 ns), Met 357 (50 to 100 ns), Gln 369 (0 to 100 ns), and Phe 372 (0 to 100 ns). As exposed in Fig. 11G, one 2D diagram has presented the atom of pose 280 interactions with the residues of 4WCU that included one brown boundary around pose 280 that proved this pose is more polar because of charged negative (brown color). The polar interactions: Asn 321 and Gln 369. The Asn 321 formed H-bond to oxygen atom of C
399, 0 charge, and 24 CPUs. For 4WCU enzyme, the enzyme information is presented by a 339 residual in chain, one chain (B), 5411 atoms, 2722 heavy atoms, and −15 charge. The ligand properties are reported by 59 total atoms including 31 heavy atoms, 0 charge, 5 fragments, 12 rotation bonds. The counter ion (salt information) is Na type, 15 atom, concentration of 23.926 mM, and 15 positive charges. As seen in Fig. 11A–J, the enzyme secondary structure is 52.62% helix, 0% strand, and 52.62% total SSE (secondary structure elements). As indicated in Fig. 11B, the curves of RMSD of 4WCU and pose 280 show the RMSDs of 4WCU (left Y-axis), the RMSD of pose 280 (Y-right axis) against simulation time of 0 to 100 ns (X-axis). The plots of the values of RMSD of 4WCU have been varied from 1.0 to 2.0 Å for C-alpha (light blue curve), 1.0 to 2.2 for backbone plot (green color curve), and 1.0 to 3.4 side chain curve are in order of 1–3 Å are perfectly acceptable for small, globular proteins. Changes much smaller than that, however, indicate that the 4WCU is undergoing a small conformational change during the simulation. It is also important that your simulation converges – the RMSD values stabilize around a fixed value. The values of curve of RMSD of pose 280 fit 4WCU (‘Lig fit Pro’) are a little higher than the values of RMSD of backbone of the reference and then the RMSD of the heavy atoms of pose 280 are measured. The values observed are a little larger than the RMSD of the 4WCU enzyme, so it is likely that the pose 280 has a little diffused away from its initial binding site of 4WCU. The curve of RMSD of ‘Lig fit Pro’, dark red color has equilibrated from 60 to 100 ns, varied a little change from 20 to 60 ns, and make larger fluctuation from 0 to 20 ns. The values of RMSD of pose 280, ‘Lig fit Lig’ – light violet color shows the RMSD of pose 280 that aligned and measured just on its reference conformation. This RMSD value measures the internal fluctuations of the atoms of pose 280. The plot of ‘Lig fit Lig’ has exposed that the values of RMSD of pose 280 are changed from 0.6 to 1.2 Å (≤2.0 Å) that proved the validation of docking model of pose 280 anchoring to 4WCU about oriental docking, docking conformation, docking method, docking parameters, and docking model for interesting pose 280 to active site on 4WCU.54 As seen in Fig. 11C, residual peaks indicated areas of the protein that fluctuated the most during the simulation time of 100 ns. On this curve, peaks indicate areas of the protein that fluctuated the most during the simulation are 125 to 140, 200 to 220, 250 to 275, and 320 to 340. Typically, the tails (N- and C-terminal) fluctuated more than any other part of 4WCU. Secondary structure elements like alpha helices and beta strands are usually more rigid than the unstructured part of the enzyme, and thus fluctuate less than the loop regions. As indicated in Fig. 11D, structure of pose 280 have 10 torsion bondings including C–OH (brown color), C–OH (violet), C–OH (orange), C–O (ether, red color) of carbohydrate part, C–O (green color), C–C of ester (light orange), C–O of ester (light violet) of cyclopentenyl ether, C–O (light green) of alcohol group, C–O (light blue) of acetyl group, and C–O (light pink) of alcohol group of cyclopentyl group. As seen in Fig. 11E, the interactions of 4WCU with the pose 280 have been presented by H bond that interacted with pose 280 via water bridges maintained over 95% simulation course from Tyr 159 on 4WCU. The residual amino acid Asp 318 on 4WCU have made multiple contacts of same subtype with the pose 280 via water bridges (200%). The Asp 318 has formed H-bonds with atoms on pose 280 via water bridge interactions. The Met 273 linked H-bonds via hydrophobic and water bridge that control 75% of simulation time. The Gln 369 has formed H-bond via water bridges maintained at 75% simulation course. The Phe 372 has created the hydrophobic interactions that control over 55% simulation course. The Asp 272 linked H-bond via water bridge that took over 60% simulation time. The Asn 321 and Tyr 329 formed water bridges that established over 40 and 55% simulation time, respectively. As indicated in Fig. 11F, the numbers of contracts appeared one more time in whole simulation course such as Tyr 159 (0 to 100 ns), His 160 (0 to less than 50 ns), Asp 318 (0 to 100 ns), Tyr 329 (10 to 100 ns), Met 273 (0 to 100 ns), Thr 333 (10 to 90 ns), Met 357 (50 to 100 ns), Gln 369 (0 to 100 ns), and Phe 372 (0 to 100 ns). As exposed in Fig. 11G, one 2D diagram has presented the atom of pose 280 interactions with the residues of 4WCU that included one brown boundary around pose 280 that proved this pose is more polar because of charged negative (brown color). The polar interactions: Asn 321 and Gln 369. The Asn 321 formed H-bond to oxygen atom of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of acetyl group on pose 280 (light pink color line) via one water bridge that controlled over 82% simulation time and Gln 369 formed H-bond linked H-bond via one water bridge that took over 70% time of simulation course. The amino acid Asp 318 (215%), brown color and charged negative amino acid created H bonds via one water bridge from oxygen atoms of hydroxyl atoms of carbohydrate and cyclopentyl group. Asp 272 formed one H bond via water bridge that controlled at 88% time of simulation course. The Tyr 159, one hydrophobic amino acid created one H bond to H atom of hydroxyl group of cyclopentanyl group that took 94% simulation time. Other hydrophobic amino acids such as Met 273 (H bond and taking 45% time), Tyr 329 (H bond via water bridge to oxygen of acetyl group taking 98%), and Met 357 (H bond, via water bridge to oxygen atom of C
O of acetyl group on pose 280 (light pink color line) via one water bridge that controlled over 82% simulation time and Gln 369 formed H-bond linked H-bond via one water bridge that took over 70% time of simulation course. The amino acid Asp 318 (215%), brown color and charged negative amino acid created H bonds via one water bridge from oxygen atoms of hydroxyl atoms of carbohydrate and cyclopentyl group. Asp 272 formed one H bond via water bridge that controlled at 88% time of simulation course. The Tyr 159, one hydrophobic amino acid created one H bond to H atom of hydroxyl group of cyclopentanyl group that took 94% simulation time. Other hydrophobic amino acids such as Met 273 (H bond and taking 45% time), Tyr 329 (H bond via water bridge to oxygen of acetyl group taking 98%), and Met 357 (H bond, via water bridge to oxygen atom of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of ester group, 72%). The morphology kinetic of this pose indicated that pose 280 is more polar and including polar and non polar interactions (hydrophobic interactions), as seen in Fig. 11G. As exposed in Fig. 11H, the properties of pose 280 has shown the RMSD, rGyr, Intra HB, Mol SA, SASA, and PSA that calculated 1.2 to 1.8 Å, 3.8 to 4.0 Å2, 0 to 1, 352 to 368 Å2, 40 to 120 Å2, and 240 to 290 Å2. As seen in Fig. 11I, The Ligand Root Mean Square Fluctuation (L-RMSF) is useful for characterizing changes in the ligand atom positions. The values of RMSF of pose 280 are varied from 0.5 to 2.0 Å and two oxygen atoms of two hydroxyl groups are strongest fluctuations (labels 26 and 27). As demonstrated in Fig. 11J, the torsion profile of bondings of pose 280 has been calculated and exposed that the most strong torsion bondings are C–O of acetyl group (light blue), C–C of ester binding to ring ether (orange), and C–O of ester (light violet color) that are belongs to torsion energies of 13.47 kcal mol−1 (−180° torsion), 10.14 kcal mol−1 (−180° torsion), and 10.00 (−180° torsion).
O of ester group, 72%). The morphology kinetic of this pose indicated that pose 280 is more polar and including polar and non polar interactions (hydrophobic interactions), as seen in Fig. 11G. As exposed in Fig. 11H, the properties of pose 280 has shown the RMSD, rGyr, Intra HB, Mol SA, SASA, and PSA that calculated 1.2 to 1.8 Å, 3.8 to 4.0 Å2, 0 to 1, 352 to 368 Å2, 40 to 120 Å2, and 240 to 290 Å2. As seen in Fig. 11I, The Ligand Root Mean Square Fluctuation (L-RMSF) is useful for characterizing changes in the ligand atom positions. The values of RMSF of pose 280 are varied from 0.5 to 2.0 Å and two oxygen atoms of two hydroxyl groups are strongest fluctuations (labels 26 and 27). As demonstrated in Fig. 11J, the torsion profile of bondings of pose 280 has been calculated and exposed that the most strong torsion bondings are C–O of acetyl group (light blue), C–C of ester binding to ring ether (orange), and C–O of ester (light violet color) that are belongs to torsion energies of 13.47 kcal mol−1 (−180° torsion), 10.14 kcal mol−1 (−180° torsion), and 10.00 (−180° torsion).
 |
| | Fig. 11 The simulation of complex of pose 280 and 4WCU enzyme over time period of 100 ns: (A) protein secondary structure are presented as 52.62 helix, 0% strand, and 52.62% total SSE (secondary structure elements), light blue: beta-strands and brown color: alpha-helices. (B) The RMSD curves of 4WCU and pose 280. (C) Residual peaks indicated areas of the protein that fluctuate the most during the simulation time of 100 ns. (D) Torsion profile of pose 280. (E) The interactions of 4WCU with the pose 280. (F) The numbers of contacts between 4WCU and pose 280 in simulation course from 0 to 100 ns. (G) One schematic of detailed atom of pose 280 interactions with the residues of 4WCU. (H) The properties of pose 280. (I) The RMSF of pose 280. (J) The ligand torsions plot summarizes. | |
4 Conclusion
Twelve compounds have been discovered from Mussaenda saigonensis including Shanzilactone (1), 6-acetyl shanzhiside methyl ester (2), barlerin (3), harpagoside (4), and (3S,5R,6R,7E,9S)-megastiman-7-ene-3,5,6,9-tetraol (5), indole-3-carboxylic acid (6), ursolic acid (7), quinovic acid (8), rotundic acid (9), clethric acid (10), martynoside (11) and verbacoside (12). They are classified into five types of skeletons: iridoids (1–4), sesquiterpene (5), alkaloid (6), triterpenes (7–10), and phenylethanoid derivatives (11 and 12). The compounds 4–6 and 11–12 have been discovered in the genus Mussaenda for the first time. Compounds 2, 7, and 4 have inhibited NO stronger than positive control, L-NMMA, and specially has compound 2 inhibited the most excellent among them in vitro. Compound 12 has exposed highest alpha-glucosidase inhibition among entries and positive control, acarbose in vitro. Antimicrobial activities, in vitro, compound 11 has inhibited against M. gypseum fungi with MIC of 50 μM and all compound 1–12 have shown moderate activity against four bacteria. In silico docking study pose 90 or compound 11 docked well to 2VF5 to explain how compound 11 has high activity against M. gypseum fungi based on enzyme inhibition mechanism 2VF5. For anti-inflammatory in vitro, the best pose docking 280 has selected among 500 conformations of compound 2 that docked to 4WCU enzyme to explain general in anti-inflammatory mechanism in vitro and complex of pose 280 and 4WCU continued performing in silico MD simulation. Results of simulation course of 0 to 100 ns indicated that complex pose 280 and 4WCU are hydrophilic interaction around pose 280 and residual amino acids appeared more than one time in whole simulation period such as Tyr 159, Asp 318, Met 273, Gln 369, Phe 372, and Asp 372.
Author contributions
Conceptualization, writing – original draft, writing – review & editing, data curation, formal analysis, funding acquisition, investigation, methodology, and project administration: Tran Thi Ngoc Mai, Mai Dinh Tri, Tran Nguyen Minh An; resources, software, supervision, validation, and visualization: Phan Nhat Minh, Nguyen Tan Phat, Mai Thanh Chi, Thuc Huy Duong, Nguyen Hong Nhi Phan, Van-Son Dang, Nguyen Van Hue, Nguyen Thi Hong Anh.
Conflicts of interest
No potential conflict of interest was reported by the authors.
Acknowledgements
The funding of this research was provided by Vietnam Academy of Science and Technology (VAST) (grant number NCVCC15.02/22-23).
References
- M. I. A. Chowdury, M. N. Alam, S. Chowdhury, M. S. Biozid, M. Faruk, M. M. U. Mazumdar and A. I. Chowdhury, Evaluation of Ex-vivo Anti-arthritic, Anti-inflammatory, Anti-cancerous and Thrombolytic Activities of Mussaenda roxburghii Leaf, Eur. J. Med. Plants, 2015 DOI:10.9734/ejmp/2015/20483.
- Y. Takeda, H. Nishimura and H. Inouye, Two new iridoid glucosides from Mussaenda parviflora and Mussaenda shikokiana, Phytochemistry, 1977, 16, 1401 CrossRef CAS.
- W. Zhao, P. Wang, R. Xu, G. Qin, S. Jiang and H. Wu, Saponins from Mussaenda pubescens, Phytochemistry, 1996 DOI:10.1016/0031-9422(95)00028-3.
- W. Zhao, J. L. Wolfender, K. Iiostettmann, K. Cheng, R. Xu and G. Qin, Triterpenes and triterpenoid saponins from Mussaenda pubescens, Phytochemistry, 1997, 45, 1073 CrossRef CAS PubMed.
- W. Zhao, J. Xu, G. Qin and R. Xu, New triterpenoid saponins from Mussaenda pubescens, J. Nat. Prod., 1994, 57, 1613 CrossRef CAS PubMed.
- W. M. Zhao, J. P. Xu, G. W. Qin and R. S. Xu, Saponins from Mussaenda pubescens, Phytochemistry, 1995 DOI:10.1016/0031-9422(94)00861-M.
- B. Dinda, S. Debnath, S. Majumder, S. Arima, N. Sato and Y. Harigaya, Chemical constituents of Mussaenda incana, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2005 DOI:10.1002/chin.200610184.
- N. C. Kim, A. E. Desjardins, C. D. Wu and A. D. Kinghorn, Activity of triterpenoid glycosides from the root bark of Mussaenda macrophylla against two oral pathogens, J. Nat. Prod., 1999 DOI:10.1021/np9901579.
- V. K. Thu, N. X. Bach, L. T. Anh, D. T. Trang, N. X. Nhiem, B. H. Tai, P. V. Kiem, C. V. Minh, S. J. Park, Y. Seo, W. Namkung and S. H. Kim, Discovery of cycloartane-type triterpene saponins from Mussaenda glabra, Phytochem. Lett., 2019 DOI:10.1016/j.phytol.2019.07.006.
- V. K. Thu, N. X. Bach, C. T. Dung, N. T. Cuong, T. H. Quang, Y. C. Kim, H. Oh and P. V. Kiem, Iridoids and cycloartane saponins from Mussaenda pilosissima valeton and their inhibitory NO production in BV2 cells, Nat. Prod. Res., 2021 DOI:10.1080/14786419.2020.1737059.
- S. M. Mohamed, E. Y. Bachkeet, S. A. Bayoumi, S. Jain, S. J. Cutler, B. L. Tekwani and S. A. Ross, Potent antitrypanosomal triterpenoid saponins from Mussaenda luteola, Fitoterapia, 2015 DOI:10.1016/j.fitote.2015.10.011.
- S. M. Mohamed, E. Y. Backheet, S. A. Bayoumi and a. Ross, New cycloartane saponin and monoterpenoid glucoindole alkaloids from Mussaenda luteola, Fitoterapia, 2016 DOI:10.1016/j.fitote.2016.03.009.
- N. X. Bach, V. K. Thu, D. T. Trang and P. V. Kiem, Quinovic acid glycosides from Mussaenda pilosissima Valeton, Vietnam J. Chem., 2019 DOI:10.1002/vjch.201960010.
- N. X. Bach, V. K. Thu, N. X. Nhiem, D. T. Trang and P. V. Kiem, Triterpenoid saponins from Mussaenda glabra Vahl, Vietnam J. Chem., 2019 DOI:10.1002/vjch.201900060.
- B. Dinda, S. Majumder, S. Arima, N. Sato and Y. Harigaya, Iridoid glucoside and sterol galactoside from Mussaenda macrophylla, J. Nat. Med., 2008 DOI:10.1007/s11418-008-0273-9.
- D. B. Menon, J. M. Sasikumar, R. Scholar and J. M. Sasikumar, Antioxidant and ant inflammatory activities of the root of Mussaenda glabrata antioxidant and anti-inflammatory activities of the root of Mussaenda glabrata, J. Pharm. Res., 2011,(10), 3320–3322 CAS.
- L. D. M. Sahadevan and D. B. Menon, Mussaenin A Isolated from Mussaenda glabrata Induces Apoptosis in the Liver Cancer
Cells Via Mitochondrial Pathway, Int. J. Pharmacogn. Phytochem. Res., 2017 DOI:10.25258/phyto.v9i09.10315.
- M. A. A. Sikder, R. B. Rashid, F. Islam, A. K. M. N. Hossian, A. B. Siddique, S. Kabir, M. R. Haque, M. S. Rahman and M. A. Rashid, Screening of ten medicinal plants of Bangladesh for analgesic activity on Swiss-albino mice, Orient. Pharm. Exp. Med., 2013 DOI:10.1007/s13596-013-0117-3.
- D. Kar, S. Rout, L. Moharana and S. Majumdar, Evaluation of anticonvulsant activity of hydroalcoholic extract of Mussaenda philippica on animals, J. Acute Dis., 2014 DOI:10.1016/s2221-6189(14)60010-x.
- A. Naiki, S. Tagane, H. Toyama, V. S. Dang and T. Yahara, Mussaenda recurvata (Rubiaceae), a new species from Southern Vietnam with observations on its heterostyly, Phytotaxa, 2017 DOI:10.11646/phytotaxa.328.2.7.
- M. D. Tri, N. T. Phat, P. N. Minh, M. T. Chi, B. M. Hao, T. N. M. An, M. Alam, N. V. Kieu, V. S. Dang, T. T. N. Mai and T. H. Duong, In vitro anti-inflammatory, in silico molecular docking and molecular dynamics simulation of oleanane-type triterpenes from aerial parts of Mussaenda recurvata, RSC Adv., 2023 10.1039/d2ra06870b.
- M. D. Tri, L. H. Ngoc, T. M. N. An, N. T. Phat, P. N. Minh, N. V. Kieu, V. S. Dang, T. T. N. Mai and T. H. Duong, Recurvataside, a new saponin from aerial parts of Mussaenda recurvata, Nat. Prod. Res., 2023 DOI:10.1080/14786419.2022.2039137.
- N. H. Nguyen, N. M. A. Tran, T. H. Duong and G. V. Vo, α-Glucosidase inhibitory activities of flavonoid derivatives isolated from Bouea macrophylla: in vitro and in silico studies, RSC Adv., 2023 10.1039/d3ra00650f.
- N. Duy, T. Ha, T. Phuong, N. V. Cuong and T. M. A. Nguyen, Novel Benzimidazol-2-Thione Derivatives: Synthesis, In Vitro Anticancer, Antimicrobial Activities, And In Silico Molecular Docking Study, ChemistrySelect, 2023, 8(17), e202300246, DOI:10.1002/slct.202300246.
- T. C. Mai, N. T. Tran, D. T. Mai, T. T. N. Mai, N. H. T. Duyen, T. N. M. An, M. Alam, C. H. Dang and T. D. Nguyen, Supercritical CO2 assisted extraction of essential oil and naringin from Citrus grandis peel: in vitro antimicrobial activity and docking study, RSC Adv., 2022 10.1039/d2ra04068a.
- F. U. Eze, U. C. Okoro, D. I. Ugwu and S. N. Okafor, Biological Activity Evaluation of Some New Benzenesulphonamide Derivatives, Front. Chem., 2019, 7 DOI:10.3389/fchem.2019.00634.
- X. Y. Meng, H. X. Zhang, M. Mezei and M. Cui, Molecular docking: a powerful approach for structure-based drug discovery. Current computer-aided drug design, Curr. Comput.-Aided Drug Des., 2011, 7(2), 146–157, DOI:10.2174/157340911795677602.
- D. E. Shaw Research, Desmond Version 3.0 Tutorial, 2011 Search PubMed.
- J. Ash and D. Fourches, Characterizing the Chemical Space of ERK2 Kinase Inhibitors Using Descriptors Computed from Molecular Dynamics Trajectories, J. Chem. Inf. Model., 2017 DOI:10.1021/acs.jcim.7b00048.
- A. Ece, Computer-aided drug design, 2023, DOI:10.1186/s13065-023-00939-w.
- J. Bhachoo and T. Beuming, Investigating protein–peptide interactions using the schrödinger computational suite, Methods Mol. Biol., 2017 DOI:10.1007/978-1-4939-6798-8_14.
- B. L. Hilian and A. F. Voter, Thermostated molecular dynamic: how to avoid the Toda demon hidden in Nosé-Hoover dynamics, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1995 DOI:10.1103/PhysRevE.52.2338.
- S. W. Chiu, S. A. Pandit, H. L. Scott and E. Jakobsson, An improved united atom force field for simulation of mixed lipid bilayers, J. Phys. Chem. B, 2009 DOI:10.1021/jp807056c.
- A. D. Mackerell, M. Feig and C. L. Brooks, Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulation, J. Comput. Chem., 2004 DOI:10.1002/jcc.20065.
- L. Zandi, M. Makungu, J. J. E. Munissi, S. Duffy, R. Puttreddy, D. V. D. Heiden, K. Rissanen, V. M. Avery, S. S. Nyandoro and M. Erdélyi, Secoiridoids and Iridoids from Morinda asteroscepa, J. Nat. Prod., 2020 DOI:10.1021/acs.jnatprod.0c00447.
- I. I. Tatli, Z. S. Akdemir, E. Yesilada and E. Küpeli, Anti-Inflammatory and Antinociceptive Potential of Major Phenolics from Verbascum salviifolium Boiss, 2008, https://www.znaturforsch.com.
- M. Phan, P. Nguyen, K. Tran, D. Bui, H. Tan, S. Dang and T. Mai, Chemical constituents from aerial parts of Mussaenda saigonensis, Sci. Technol. Dev. J.: Nat. Sci., 2023 DOI:10.32508/stdjns.v7i4.1276.
- A. Viljoen, M. Mncwangi and I. Vermaak, Anti-Inflammatory Iridoids of Botanical Origin, Curr. Med. Chem., 2012 DOI:10.2174/092986712800229005.
- V. Gyurkovska, K. Alipieva, A. Maciuk, P. Dimitrova, N. Ivanovska, C. Haas, T. Bley and M. Georgiev, Anti-inflammatory activity of Devil's claw in vitro systems and their active constituents, 2011, DOI:10.1016/j.foodchem.2010.08.056.
- L. V. Xiao-Jing, Y. Li, S. G. Ma, J. Qu, J. B. Liu, Y. H. Li, D. Zhang, L. Li and S. S. Yu, Bioactive megastigmane glucosides and monoterpenes from Lyonia ovalifolia, J. Asian Nat. Prod. Res., 2019 DOI:10.1080/10286020.2018.1509313.
- M. B. Gupta, T. N. Bhalla, G. P. Gupta, C. R. Mitra, K. P. Bhargava and K. Pbhargava, Inflammatory activity of natural products (i) triterpenoids, Eur. J. Pharmacol., 1969 DOI:10.1016/0014-2999(69)90067-3.
- C. Y. Wang, H. J. Jang, Y. K. Han, X. D. Su, S. W. Lee, M. C. Rho, S. H. Wang, S. Y. Yang and Y. H. Kim, Alkaloids from Tetrastigma hemsleyanum and their anti-inflammatory effects on LPS-induced RAW264.7 cells, Molecules, 2018 DOI:10.3390/molecules23061445.
- N. N. Tuan, H. N. Thi, C. L. T. My, T. X. Hai, H. T. Trung, A. N. T. Kim, T. N. Tan, T. L. Van, C. Q. Nguyen, Q. D. Tran, P. C. Kuo, Q. L. Dang and T. D. Thang, Inhibition of α-Glucosidase, Acetylcholinesterase, and Nitric Oxide Production by Phytochemicals Isolated from Millettia speciosa—In Vitro and Molecular Docking Studies, Plants, 2022 DOI:10.3390/plants11030388.
- Y. Han, L. Zhang, W. Li, X. Liu, J. Xiao, G. Chen and N. Li, Natural CAC chemopreventive agents from Ilex rotunda Thunb, J. Nat. Med., 2019 DOI:10.1007/s11418-019-01281-z.
- J. Y. Tao, S. J. Dai, F. Zhao, J. F. Liu, W. S. Fang and K. Liu, New ursane-type triterpene with NO production suppressing activity from Nauclea officinalis, J. Asian Nat. Prod. Res., 2012 DOI:10.1080/10286020.2011.628839.
- J. Wang, J. Zhao, Y. Yan, D. Liu, C. Wang and H. Wang, Inhibition of glycosidase by ursolic acid: in vitro, in vivo and in silico study, J. Sci. Food Agric., 2020 DOI:10.1002/jsfa.10098.
- P. P. Wu, B. J. Zhang, X. P. Cui, Y. Yang, Z. Y. Jiang, Z. H. Zhou, Y. Y. Zhong, Y. Y. Mai, Z. Ouyang, H. H. S. Chen, J. Zheng, S. Q. Zhao and K. Zhang, Synthesis and biological evaluation of novel ursolic acid analogues as potential α-glucosidase inhibitors, Sci. Rep., 2017 DOI:10.1038/srep45578.
- K. Budipramana, J. Junaidin, K. R. Wirasutisna, Y. B. Pramana and S. Sukrasno, An integrated in silico and in vitro assays of dipeptidyl peptidase-4 and α-glucosidase inhibition by stellasterol from Ganoderma australe, Sci. Pharm., 2019 DOI:10.3390/scipharm87030021.
- R. Kazemi, M. R. Delnavazi, H. P. Khankandi, S. Mojtabavi, A. Hoseinsalari and M. A. Faramarzi, α-Glucosidase Inhibitors from Marrubium astracanicum: Isolation and Molecular Docking, Rev. Bras. Farmacogn., 2022 DOI:10.1007/s43450-022-00287-1.
- J. N. Nyemb, R. T. Tchuenguem, A. Venditti, A. T. Tchinda, C. Henoumont, E. Talla, S. Laurent and J. Iqbal, Antimicrobial and α-glucosidase inhibitory activities of chemical constituents from Gardenia aqualla (Rubiaceae), Nat. Prod. Res., 2022 DOI:10.1080/14786419.2022.2031187.
- G. Giannini, W. Cabri, C. Fattorusso and M. Rodriquez, Histone deacetylase inhibitors in the treatment of cancer: Overview and perspectives, Future Med. Chem., 2012 DOI:10.4155/fmc.12.80.
- P. N. T. Trinh, T. N. M. An, T. N. Danh, N. T. H. Anh, N. V. Kieu, T. H. Duong and L. T. Dung, The In silico and In vitro Anti-inflammatory and Antibacterial Activities of Flavonoids from Artemisia vulgaris in Vietnam, Curr. Org. Chem., 2013 DOI:10.2174/1385272827666230913105836.
- T. N. T. Pham, T. T. Nguyen, T. L. T. Nguyen, T. N. M. An, N. N. Tuan, T. D. Tong and D. T. Le, Antioxidant and Anti-Inflammatory Activities of Phytochemicals from Ruellia tuberosa, J. Chem., 2022 DOI:10.1155/2022/4644641.
- N. H. Nguyen, N. M. A. Tran, T. H. Duong and G. V. Vo, α-Glucosidase inhibitory activities of flavonoid derivatives isolated from Bouea macrophylla : in vitro and in silico studies, RSC Adv., 2023 10.1039/d3ra00650f.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article bc,
Mai Thanh Chibc,
Thuc Huy Duongd,
Nguyen Hong Nhi Phand,
Tran Nguyen Minh An
bc,
Mai Thanh Chibc,
Thuc Huy Duongd,
Nguyen Hong Nhi Phand,
Tran Nguyen Minh An *e,
Van-Son Dangbf,
Nguyen Van Hueg,
Nguyen Thi Hong Anhh and
Mai Dinh Tri*bc
*e,
Van-Son Dangbf,
Nguyen Van Hueg,
Nguyen Thi Hong Anhh and
Mai Dinh Tri*bc
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (30
MeOH (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–0
1–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (30
MeOH (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–5
1–5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Compounds 1 (8.0 mg) and 5 (12.0 mg) were obtained by rechromatographing the fraction E.2.2 (2.5 g) on silica gel CC with the n-hexane–CH2Cl2–MeOH (3
1, v/v). Compounds 1 (8.0 mg) and 5 (12.0 mg) were obtained by rechromatographing the fraction E.2.2 (2.5 g) on silica gel CC with the n-hexane–CH2Cl2–MeOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v) solvent system. Four subfractions (E2.5.1–E2.5.4) have been created from the further chromatography of fraction E2.5 (4.8 g) on silica gel CC. Combinations of n-hexane and EtOAc were used to elute the fraction (2
0.1, v/v/v) solvent system. Four subfractions (E2.5.1–E2.5.4) have been created from the further chromatography of fraction E2.5 (4.8 g) on silica gel CC. Combinations of n-hexane and EtOAc were used to elute the fraction (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and 1
2, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v). In order to collect compounds 7 (20.0 mg) and 9 (6.0 mg), subfraction E2.5.3 (690 mg) was passed over a silica gel CC with an eluent of n-hexane
5, v/v). In order to collect compounds 7 (20.0 mg) and 9 (6.0 mg), subfraction E2.5.3 (690 mg) was passed over a silica gel CC with an eluent of n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Subsequently, six fractions, E5.1–E5.6, were obtained by dissolving the 42 g of fraction E5 employing a gradient system of CH2Cl2–MeOH (10
1). Subsequently, six fractions, E5.1–E5.6, were obtained by dissolving the 42 g of fraction E5 employing a gradient system of CH2Cl2–MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–0
1–0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). This was done using silica gel CC. Five fractions (E5.3.1–5.3.5) were isolated by rechromatographing fraction E5.3 (11.0 g) using a gradient of CH2Cl2
1, v/v). This was done using silica gel CC. Five fractions (E5.3.1–5.3.5) were isolated by rechromatographing fraction E5.3 (11.0 g) using a gradient of CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (10
MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → 1
1 → 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Compounds 6 (5.0 mg) and 8 (7.0 mg) were extracted by repeated the subfraction E5.3.2 (950 mg) by chromatography on a silica gel column eluting with CH2Cl2
1, v/v). Compounds 6 (5.0 mg) and 8 (7.0 mg) were extracted by repeated the subfraction E5.3.2 (950 mg) by chromatography on a silica gel column eluting with CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (10
MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). By implementing n-hexane
1, v/v). By implementing n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) as an eluent, column chromatography was used to separate subfraction E5.3.4 (860 mg), yielding compounds 10 (12.0 mg) and 4 (8.0 mg). Compounds 2 (14.0 mg) and 3 (10.0 mg) were obtained from fraction E5.4 (12 g) by two consecutive silica gel CC: a normal-phase column with a mobile phase as n-hexane
1, v/v/v) as an eluent, column chromatography was used to separate subfraction E5.3.4 (860 mg), yielding compounds 10 (12.0 mg) and 4 (8.0 mg). Compounds 2 (14.0 mg) and 3 (10.0 mg) were obtained from fraction E5.4 (12 g) by two consecutive silica gel CC: a normal-phase column with a mobile phase as n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2
CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (1
MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) and a C-18 reverse-phase column using a solvent system of MeOH
1, v/v/v) and a C-18 reverse-phase column using a solvent system of MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (3
H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). A gradient system of EtOAc–MeOH was used to elute the silica gel CC used to extract fraction E7 (53 g) (10
1, v/v). A gradient system of EtOAc–MeOH was used to elute the silica gel CC used to extract fraction E7 (53 g) (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v) to give five subfractions. E7.1–E7.5. Subfraction E7.3 (14.0 g) was fractionated by silica gel CC using a solvent system consisting of n-hexane, ethyl acetate, and methanol (1
10, v/v) to give five subfractions. E7.1–E7.5. Subfraction E7.3 (14.0 g) was fractionated by silica gel CC using a solvent system consisting of n-hexane, ethyl acetate, and methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 to 1
0.1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v) to afford six fractions. E7.3.1–E7.3.6. Fraction E7.3.4 (945 mg) was applied to C-18-RP silica gel CC, eluted with MeOH
0.1, v/v/v) to afford six fractions. E7.3.1–E7.3.6. Fraction E7.3.4 (945 mg) was applied to C-18-RP silica gel CC, eluted with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (2
H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to afford compounds 11 (8.0 mg) and 12 (11.0 mg).
1, v/v) to afford compounds 11 (8.0 mg) and 12 (11.0 mg).






![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682, the number of water molecules of 11
682, the number of water molecules of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399, 0 charge, and 24 CPUs. For 4WCU enzyme, the enzyme information is presented by a 339 residual in chain, one chain (B), 5411 atoms, 2722 heavy atoms, and −15 charge. The ligand properties are reported by 59 total atoms including 31 heavy atoms, 0 charge, 5 fragments, 12 rotation bonds. The counter ion (salt information) is Na type, 15 atom, concentration of 23.926 mM, and 15 positive charges. As seen in Fig. 11A–J, the enzyme secondary structure is 52.62% helix, 0% strand, and 52.62% total SSE (secondary structure elements). As indicated in Fig. 11B, the curves of RMSD of 4WCU and pose 280 show the RMSDs of 4WCU (left Y-axis), the RMSD of pose 280 (Y-right axis) against simulation time of 0 to 100 ns (X-axis). The plots of the values of RMSD of 4WCU have been varied from 1.0 to 2.0 Å for C-alpha (light blue curve), 1.0 to 2.2 for backbone plot (green color curve), and 1.0 to 3.4 side chain curve are in order of 1–3 Å are perfectly acceptable for small, globular proteins. Changes much smaller than that, however, indicate that the 4WCU is undergoing a small conformational change during the simulation. It is also important that your simulation converges – the RMSD values stabilize around a fixed value. The values of curve of RMSD of pose 280 fit 4WCU (‘Lig fit Pro’) are a little higher than the values of RMSD of backbone of the reference and then the RMSD of the heavy atoms of pose 280 are measured. The values observed are a little larger than the RMSD of the 4WCU enzyme, so it is likely that the pose 280 has a little diffused away from its initial binding site of 4WCU. The curve of RMSD of ‘Lig fit Pro’, dark red color has equilibrated from 60 to 100 ns, varied a little change from 20 to 60 ns, and make larger fluctuation from 0 to 20 ns. The values of RMSD of pose 280, ‘Lig fit Lig’ – light violet color shows the RMSD of pose 280 that aligned and measured just on its reference conformation. This RMSD value measures the internal fluctuations of the atoms of pose 280. The plot of ‘Lig fit Lig’ has exposed that the values of RMSD of pose 280 are changed from 0.6 to 1.2 Å (≤2.0 Å) that proved the validation of docking model of pose 280 anchoring to 4WCU about oriental docking, docking conformation, docking method, docking parameters, and docking model for interesting pose 280 to active site on 4WCU.54 As seen in Fig. 11C, residual peaks indicated areas of the protein that fluctuated the most during the simulation time of 100 ns. On this curve, peaks indicate areas of the protein that fluctuated the most during the simulation are 125 to 140, 200 to 220, 250 to 275, and 320 to 340. Typically, the tails (N- and C-terminal) fluctuated more than any other part of 4WCU. Secondary structure elements like alpha helices and beta strands are usually more rigid than the unstructured part of the enzyme, and thus fluctuate less than the loop regions. As indicated in Fig. 11D, structure of pose 280 have 10 torsion bondings including C–OH (brown color), C–OH (violet), C–OH (orange), C–O (ether, red color) of carbohydrate part, C–O (green color), C–C of ester (light orange), C–O of ester (light violet) of cyclopentenyl ether, C–O (light green) of alcohol group, C–O (light blue) of acetyl group, and C–O (light pink) of alcohol group of cyclopentyl group. As seen in Fig. 11E, the interactions of 4WCU with the pose 280 have been presented by H bond that interacted with pose 280 via water bridges maintained over 95% simulation course from Tyr 159 on 4WCU. The residual amino acid Asp 318 on 4WCU have made multiple contacts of same subtype with the pose 280 via water bridges (200%). The Asp 318 has formed H-bonds with atoms on pose 280 via water bridge interactions. The Met 273 linked H-bonds via hydrophobic and water bridge that control 75% of simulation time. The Gln 369 has formed H-bond via water bridges maintained at 75% simulation course. The Phe 372 has created the hydrophobic interactions that control over 55% simulation course. The Asp 272 linked H-bond via water bridge that took over 60% simulation time. The Asn 321 and Tyr 329 formed water bridges that established over 40 and 55% simulation time, respectively. As indicated in Fig. 11F, the numbers of contracts appeared one more time in whole simulation course such as Tyr 159 (0 to 100 ns), His 160 (0 to less than 50 ns), Asp 318 (0 to 100 ns), Tyr 329 (10 to 100 ns), Met 273 (0 to 100 ns), Thr 333 (10 to 90 ns), Met 357 (50 to 100 ns), Gln 369 (0 to 100 ns), and Phe 372 (0 to 100 ns). As exposed in Fig. 11G, one 2D diagram has presented the atom of pose 280 interactions with the residues of 4WCU that included one brown boundary around pose 280 that proved this pose is more polar because of charged negative (brown color). The polar interactions: Asn 321 and Gln 369. The Asn 321 formed H-bond to oxygen atom of C
399, 0 charge, and 24 CPUs. For 4WCU enzyme, the enzyme information is presented by a 339 residual in chain, one chain (B), 5411 atoms, 2722 heavy atoms, and −15 charge. The ligand properties are reported by 59 total atoms including 31 heavy atoms, 0 charge, 5 fragments, 12 rotation bonds. The counter ion (salt information) is Na type, 15 atom, concentration of 23.926 mM, and 15 positive charges. As seen in Fig. 11A–J, the enzyme secondary structure is 52.62% helix, 0% strand, and 52.62% total SSE (secondary structure elements). As indicated in Fig. 11B, the curves of RMSD of 4WCU and pose 280 show the RMSDs of 4WCU (left Y-axis), the RMSD of pose 280 (Y-right axis) against simulation time of 0 to 100 ns (X-axis). The plots of the values of RMSD of 4WCU have been varied from 1.0 to 2.0 Å for C-alpha (light blue curve), 1.0 to 2.2 for backbone plot (green color curve), and 1.0 to 3.4 side chain curve are in order of 1–3 Å are perfectly acceptable for small, globular proteins. Changes much smaller than that, however, indicate that the 4WCU is undergoing a small conformational change during the simulation. It is also important that your simulation converges – the RMSD values stabilize around a fixed value. The values of curve of RMSD of pose 280 fit 4WCU (‘Lig fit Pro’) are a little higher than the values of RMSD of backbone of the reference and then the RMSD of the heavy atoms of pose 280 are measured. The values observed are a little larger than the RMSD of the 4WCU enzyme, so it is likely that the pose 280 has a little diffused away from its initial binding site of 4WCU. The curve of RMSD of ‘Lig fit Pro’, dark red color has equilibrated from 60 to 100 ns, varied a little change from 20 to 60 ns, and make larger fluctuation from 0 to 20 ns. The values of RMSD of pose 280, ‘Lig fit Lig’ – light violet color shows the RMSD of pose 280 that aligned and measured just on its reference conformation. This RMSD value measures the internal fluctuations of the atoms of pose 280. The plot of ‘Lig fit Lig’ has exposed that the values of RMSD of pose 280 are changed from 0.6 to 1.2 Å (≤2.0 Å) that proved the validation of docking model of pose 280 anchoring to 4WCU about oriental docking, docking conformation, docking method, docking parameters, and docking model for interesting pose 280 to active site on 4WCU.54 As seen in Fig. 11C, residual peaks indicated areas of the protein that fluctuated the most during the simulation time of 100 ns. On this curve, peaks indicate areas of the protein that fluctuated the most during the simulation are 125 to 140, 200 to 220, 250 to 275, and 320 to 340. Typically, the tails (N- and C-terminal) fluctuated more than any other part of 4WCU. Secondary structure elements like alpha helices and beta strands are usually more rigid than the unstructured part of the enzyme, and thus fluctuate less than the loop regions. As indicated in Fig. 11D, structure of pose 280 have 10 torsion bondings including C–OH (brown color), C–OH (violet), C–OH (orange), C–O (ether, red color) of carbohydrate part, C–O (green color), C–C of ester (light orange), C–O of ester (light violet) of cyclopentenyl ether, C–O (light green) of alcohol group, C–O (light blue) of acetyl group, and C–O (light pink) of alcohol group of cyclopentyl group. As seen in Fig. 11E, the interactions of 4WCU with the pose 280 have been presented by H bond that interacted with pose 280 via water bridges maintained over 95% simulation course from Tyr 159 on 4WCU. The residual amino acid Asp 318 on 4WCU have made multiple contacts of same subtype with the pose 280 via water bridges (200%). The Asp 318 has formed H-bonds with atoms on pose 280 via water bridge interactions. The Met 273 linked H-bonds via hydrophobic and water bridge that control 75% of simulation time. The Gln 369 has formed H-bond via water bridges maintained at 75% simulation course. The Phe 372 has created the hydrophobic interactions that control over 55% simulation course. The Asp 272 linked H-bond via water bridge that took over 60% simulation time. The Asn 321 and Tyr 329 formed water bridges that established over 40 and 55% simulation time, respectively. As indicated in Fig. 11F, the numbers of contracts appeared one more time in whole simulation course such as Tyr 159 (0 to 100 ns), His 160 (0 to less than 50 ns), Asp 318 (0 to 100 ns), Tyr 329 (10 to 100 ns), Met 273 (0 to 100 ns), Thr 333 (10 to 90 ns), Met 357 (50 to 100 ns), Gln 369 (0 to 100 ns), and Phe 372 (0 to 100 ns). As exposed in Fig. 11G, one 2D diagram has presented the atom of pose 280 interactions with the residues of 4WCU that included one brown boundary around pose 280 that proved this pose is more polar because of charged negative (brown color). The polar interactions: Asn 321 and Gln 369. The Asn 321 formed H-bond to oxygen atom of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of acetyl group on pose 280 (light pink color line) via one water bridge that controlled over 82% simulation time and Gln 369 formed H-bond linked H-bond via one water bridge that took over 70% time of simulation course. The amino acid Asp 318 (215%), brown color and charged negative amino acid created H bonds via one water bridge from oxygen atoms of hydroxyl atoms of carbohydrate and cyclopentyl group. Asp 272 formed one H bond via water bridge that controlled at 88% time of simulation course. The Tyr 159, one hydrophobic amino acid created one H bond to H atom of hydroxyl group of cyclopentanyl group that took 94% simulation time. Other hydrophobic amino acids such as Met 273 (H bond and taking 45% time), Tyr 329 (H bond via water bridge to oxygen of acetyl group taking 98%), and Met 357 (H bond, via water bridge to oxygen atom of C
O of acetyl group on pose 280 (light pink color line) via one water bridge that controlled over 82% simulation time and Gln 369 formed H-bond linked H-bond via one water bridge that took over 70% time of simulation course. The amino acid Asp 318 (215%), brown color and charged negative amino acid created H bonds via one water bridge from oxygen atoms of hydroxyl atoms of carbohydrate and cyclopentyl group. Asp 272 formed one H bond via water bridge that controlled at 88% time of simulation course. The Tyr 159, one hydrophobic amino acid created one H bond to H atom of hydroxyl group of cyclopentanyl group that took 94% simulation time. Other hydrophobic amino acids such as Met 273 (H bond and taking 45% time), Tyr 329 (H bond via water bridge to oxygen of acetyl group taking 98%), and Met 357 (H bond, via water bridge to oxygen atom of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of ester group, 72%). The morphology kinetic of this pose indicated that pose 280 is more polar and including polar and non polar interactions (hydrophobic interactions), as seen in Fig. 11G. As exposed in Fig. 11H, the properties of pose 280 has shown the RMSD, rGyr, Intra HB, Mol SA, SASA, and PSA that calculated 1.2 to 1.8 Å, 3.8 to 4.0 Å2, 0 to 1, 352 to 368 Å2, 40 to 120 Å2, and 240 to 290 Å2. As seen in Fig. 11I, The Ligand Root Mean Square Fluctuation (L-RMSF) is useful for characterizing changes in the ligand atom positions. The values of RMSF of pose 280 are varied from 0.5 to 2.0 Å and two oxygen atoms of two hydroxyl groups are strongest fluctuations (labels 26 and 27). As demonstrated in Fig. 11J, the torsion profile of bondings of pose 280 has been calculated and exposed that the most strong torsion bondings are C–O of acetyl group (light blue), C–C of ester binding to ring ether (orange), and C–O of ester (light violet color) that are belongs to torsion energies of 13.47 kcal mol−1 (−180° torsion), 10.14 kcal mol−1 (−180° torsion), and 10.00 (−180° torsion).
O of ester group, 72%). The morphology kinetic of this pose indicated that pose 280 is more polar and including polar and non polar interactions (hydrophobic interactions), as seen in Fig. 11G. As exposed in Fig. 11H, the properties of pose 280 has shown the RMSD, rGyr, Intra HB, Mol SA, SASA, and PSA that calculated 1.2 to 1.8 Å, 3.8 to 4.0 Å2, 0 to 1, 352 to 368 Å2, 40 to 120 Å2, and 240 to 290 Å2. As seen in Fig. 11I, The Ligand Root Mean Square Fluctuation (L-RMSF) is useful for characterizing changes in the ligand atom positions. The values of RMSF of pose 280 are varied from 0.5 to 2.0 Å and two oxygen atoms of two hydroxyl groups are strongest fluctuations (labels 26 and 27). As demonstrated in Fig. 11J, the torsion profile of bondings of pose 280 has been calculated and exposed that the most strong torsion bondings are C–O of acetyl group (light blue), C–C of ester binding to ring ether (orange), and C–O of ester (light violet color) that are belongs to torsion energies of 13.47 kcal mol−1 (−180° torsion), 10.14 kcal mol−1 (−180° torsion), and 10.00 (−180° torsion).






