Open Access Article
Open Access ArticleA enzyme-free fluorescence quenching sensor for amplified detection of kanamycin in milk based on competitive triggering strategies†
Yangyinchun Bao a,
Yidan Sanga,
Xuemei Yana,
Mengyang Hu
a,
Yidan Sanga,
Xuemei Yana,
Mengyang Hu a,
Na Wang
a,
Na Wang a,
Yafei Dong*ab and
Luhui Wang
a,
Yafei Dong*ab and
Luhui Wang *a
*a
aCollege of Life Science, Shaanxi Normal University, Xi'an, Shaanxi 710119, PR China. E-mail: dongyf@snnu.edu.cn
bCollege of Computer Sciences, Shaanxi Normal University, Xi'an, Shaanxi 710119, PR China
First published on 13th June 2024
Abstract
In this work, we constructed a FAM fluorescence quenching biosensor based on an aptamer competition recognition and enzyme-free amplification strategy. We design a competing unit consisting of an aptamer chain and a complementary chain, and a catalytic hairpin self-assembly (CHA) unit consisting of two hairpins in which the complementary chain can trigger the catalytic hairpin self-assembly. In the initial state, the aptamer chain is combined with the complementary chain, the catalytic hairpin self-assembly unit is inhibited, the FAM fluorescence group was far away from the BHQ1 quenching group, and the fluorescence is turn-on. In the presence of kanamycin, the aptamer chain recognizes kanamycin and doesn't form double chains, resulting in the free complementary chain triggering hairpin 1 (H1), and then H1 triggering hairpin 2 (H2), FAM fluorophore is close to the BHQ1 quenching group, and the fluorescence is off–on. When H1 and H2 form a cyclic reaction, enzyme-free amplification is achieved and there is significant output of the fluorescence signal. Therefore, the biosensor has good performance in detecting kanamycin, the detection line is 54 nM, the linear range is 54 nM–0.9 μM, and it can achieve highly selective detection of kanamycin. Kanamycin residue may cause serious harm to human health. The high sensitivity detection of kanamycin is urgent, so this project has a great application potential for food detection.
Introduction
Kanamycin is a very important glucoside antibiotic, which is widely used in the treatment of infections caused by Gramella.1,2 However, uncontrolled use of kanamycin may lead to its residual and accumulation in animals, and its entry into the human body through the food chain, which may lead to ototoxicity, nephrotoxicity and antibiotic resistance.3 In order to protect the life and health of consumers, different countries and regions have formulated relevant regulations and clarified the maximum content of kanamycin in milk, which is 200 μg kg−1 in China and 150 μg kg−1 in the European Union.4 Animal-derived foods, especially milk, have a large global consumer market. In order to protect the safety of consumers and reduce the risk of serious adverse reactions caused by kanamycin, it is urgent to establish an effective, rapid and reliable antibiotic detection platform.At present, there are many detection methods for kanamycin, including high performance liquid chromatography,5 liquid chromatography-mass spectrometry,6 immunochromatography,7 chemiluminescence,8 electrochemiluminescence9 and enzyme-linked immunization10 and other traditional methods. Due to the high cost, high operation requirements and expensive equipment, these methods are difficult to widely used, so a simple detection scheme is urgently needed.
The aptamer can recognize and bind to the target molecule with high specificity, and its chemical nature is single-stranded oligonucleotide.11 Compared with antibodies, aptamers are cheap, low transportation and storage costs, and have strong resistance to environmental interferences. Due to these advantages, aptamers have received extensive attention and are used in various fields such as antibiotics,12 toxins,13,14 disease diagnosis and treatment15,16 and so on. Using these properties of aptamers, kanamycin biosensors such as gold nanoparticle,17 silver nanoparticle18 and graphene oxide19 have been designed. However, the target will be non-specific adsorbed by nanomaterials, leading to leakage. Due to the maturity of the kanamycin aptamer strategy, many biosensors dependent on exonuclidenase have been established in recent years.20–22 Although these sensors dependent on enzyme amplification improve the detection performance, the extraction and purification of enzymes are expensive, and the enzyme activity is easily affected by the environment, making it difficult to cope with the complex detection environment.
Chain replacement is a thermodynamically-driven DNA hybridization reaction that can cascade.23 The substrate and the output chain are partially hybridized, the exposed overhang fulcrum on the substrate chain guides the hybridization with the initiator chain, and the output chain is thermodynamically driven to migrate in the substrate chain, at the same time the output chain is released. Catalytic hairpin self-assembly is a kind of enzyme-free thermostatic amplification, which has the advantages of strong specificity, high sensitivity, good repeatability and simple operation. High signal amplification sensors based on DNA self-assembly have been widely used in miRNA detection.24 In addition, the detection platform of toxins,25 antibiotics26 and bacteria,27 which combines catalytic hairpin self-assembly with aptamer technology, has also been widely reported.
Here, we designed a FAM fluorescence quenching biosensor based on aptamer recognition competition triggering enzyme-free amplification signal amplification to detect kanamycin in milk. First, we design a competing unit that includes an aptamer chain and a complementary chain and two hairpin units H1 and H2 for catalytic self-assembly. In the initial state, the aptamer chain is combined with the complementary chain, the catalytic hairpin self-assembly unit is inhibited, and the sensor is in the off state. At this time, the FAM fluorophore of H1 is turn-on. The aptamer chain recognizes kanamycin and forms a polymer, thus preventing the formation of a double chain, and the sensor turns on. At this time, the free complementary chain triggers H1 and starts the catalytic hairpin self-assembly, while hairpin 1 and hairpin 2 form a cyclic reaction, the FAM fluorophore is close to the BHQ1 quenching group, the fluorescence is quenched, and FAM acts as a signal molecule to realize signal output. The biosensor has a good detection performance of kanamycin, the detection line is 54 nM, and the linear range is 54 nM–0.9 μM. Therefore, this project has great application potential in the field of food detection.
Materials and methods
Reagents and instruments
The aptamer sequence used to identify kanamycin in this study was 5′-TGG GGG TTG AGG CTA AGC CGA-3′.28 All probes in this study (Table S1†) were obtained by Shanghai Shenggong Biotechnology Co., LTD, China. H1 (TCGGCTGCCATATAGGCTTATGGCAGCCGATATGGC – FAM) and H2 (BHQ1 – GCCATATCGGCTGACATAAGCCGATATGGCAGCCGA) using HPLC purification, the remaining oligonucleotide sequences were purified by polyacrylamide gel electrophoresis. Kanamycin sulfate was supplied by Shanghai Aladdin Biochemical Technology Co., LTD. Amoxicillin, tetracycline hydrochloride, ofloxacin by Beijing Century Aoke Biotechnology Co., LTD. The milk (3.2 g protein, 3.6 g fat, 4.9 g carbohydrates, 55 mg sodium and 100 mg calcium per 100 ml) was purchased from a local supermarket. Other reagents involved in this work are analytical grade and have not been treated. Ultrapure water (18.25 MΩ cm) is required for solution preparation. The fluorescence signal was detected using the BioTek Synergy LX multifunctional microplate detector (BioTek, USA).Experimental operation detection
First, the aptamer probe and two hairpin probes (H1 and H2) were annealed at 95 °C for 10 min, then gradually cooled to room temperature and held for 2 h to stabilize the hairpin structure. Secondly, the 500 nM aptamer probe was mixed with different concentrations of kanamycin in the buffer solution (500 nM Tris-HCl, 2.5 mM NaCl, 500 mm KCl, pH = 7.5) and incubated at 37 °C for 15 min to complete the specific recognition of the aptamer sequence and kanamycin. Subsequently, a 500 nM aptamer complementary probe was added to the system and reacted for 15 min to form a double strand. Finally, 500 nM H1 and hairpin H2 were added and cultured at 37 °C for 60 min to complete the catalytic hairpin assembly reaction and detect the fluorescence signal.Selectivity of kanamycin
Using different antibiotics (amoxicillin, tetracycline hydrochloride and ofloxacin) to study the selective detection of kanamycin by this method. We added 500 nM kanamycin, 10 times the amount of antibiotic analogues and mixtures to the aptamer sensor for detection. Determine the selectivity of the aptamer sensor for kanamycin based on the fluorescence intensity detected in the presence of different antibiotics.Milk sample preparation and testing
Through the detection of kanamycin in milk, the prospect of practical application of this method was investigated. In order to facilitate testing, milk samples need to be processed.29 First, dilute 10 ml of milk sample with 10 ml of ultra-pure water. The protein was precipitated with 20% acetic acid and the pH of the diluted milk sample was adjusted to 4.6. Centrifuge at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 25 min, remove the precipitated protein and retain the supernatant. Subsequently, the pH is adjusted to 7.0. The sample was then filtered with a 0.22 μm filter membrane. Finally, different concentrations of kanamycin were added to milk samples and detected by aptamer sensor.
000 rpm for 25 min, remove the precipitated protein and retain the supernatant. Subsequently, the pH is adjusted to 7.0. The sample was then filtered with a 0.22 μm filter membrane. Finally, different concentrations of kanamycin were added to milk samples and detected by aptamer sensor.
Results and discussion
Principle of the sensor
In Scheme 1, we described the aptamer recognition principle of a competitive FAM fluorescence quenched biosensor, which triggers amplification without enzyme involvement. First, we designed a kanamycin aptamer probe (APT) for recognizing kanamycin, and a complementary chain (C-APT) partially complementary to the kanamycin aptamer probe, which has a trigger region for opening the H1 hairpin structure. Then, we also designed two highly complementary hairpins (H1 and H2) to form catalytic hairpin self-assembly. The 5′end of H1 had a 6 base overhanging region toehold to start catalytic hairpin self-assembly, and 3′ modified FAM fluorophore. The 3′end of H2 has a 6 base overhanging region toehold to trigger an enzyme-free amplification cycle reaction, and the 5′ end modifies the fluorescence quenching group of BHQ1. In the initial state, the complementary chain forms a double-chain complex with the kanamycin aptamer probe, the complementary chain cannot be released, the trigger region is blocked and deactivated, the toehold of H1 cannot be recognized, the cyclic reaction between H1 and H2 cannot be triggered, and the fluorescence signal is high. When the sensor is turned on, kanamycin and kanamycin aptamer form complex. Kanamycin-aptamer complexe can be stabilized because of structural changes. And due to competition inhibition, complementary chains are free in the solution. The trigger region in the complementary chain specifically recognizes the toehold at the 3′end of H1. The triggering region hybridizes with the hanging region chain of H1, and then triggers an entropy driven chain displacement reaction, which opens the stem structure domain of H1, thereby opening the hairpin H1. Subsequently, chain hybridization occurs between the open stem domain at the H1 5′end of the hairpin and the overhanging region of H2, and then triggers the overhanging domain mediated chain displacement reaction, which completely opens the hairpin H2 and forms the H1–H2 complex. The FAM group at the 3′ end of H1 is close to the BHQ1 quenching group at the 5 'end of H2 in space, and the fluorescence signal is low. And again releases the complementary chain that contains the trigger region. The released trigger chain again triggers the hybridization reaction between H1 and H2. After several cycles, a large number of H1–H2 complexes are formed, FAM fluorescence signal is quenched, the fluorescence is realized from on to off, and the sensitive detection of kanamycin is realized. | ||
| Scheme 1 Principle of an enzyme-free fluorescence quenching sensor based on a competing trigger strategy. | ||
Feasibility of kanamycin detection
First, we used Autoduck Vina to perform molecular docking between the adaptation sequence and kanamycin molecules, as shown in Fig. S1.† The simulation results showed that the recognition strategy was feasible.We chose the relationship of fluorescence intensity with time to explore whether our proposed fluorescent aptamer sensor is suitable for the detection of kanamycin. As can be seen from curve a (purple curve) in Fig. 1, hairpin H1 modified with FAM fluorophores exhibited very high fluorescence. It can be seen from curve b (green curve) that the fluorescence intensity of H1 slightly decreases, which is due to the spontaneous binding of H1 and H2 leading to partial leakage. After adding kanamycin aptamer probe and complementary chain on the basis of the above, compared with H1 and H2 mixture, curve c shows that the fluorescence intensity after adding kanamycin probe and complementary chain is lower (blue curve), because the reaction of kanamycin probe and complementary chain to form double-chain complex is incomplete in the initial state, resulting in partial dissociation of complementary chain. This triggers a subsequent cyclic hybridization reaction that catalyzes the self-assembly of the hairpin, leading to the occurrence of partial leakage. After the addition of kanamycin, kanamycin induced conformational change of aptamer probe, which led to the dissociation of complementary chains that triggered the assembly of H1 and H2 cycles, forming a large number of H1–H2 complexes, and significantly quenching of fluorescence intensity (curve d, red curve). Curve e (black curve) shows the theoretical minimum value of the fluorescence intensity of the sensor we designed when it is fully opened, and its fluorescence intensity is slightly lower than that in curve d, which is in line with expectations. All of the above controlled experiments confirm that significant fluorescence signal attenuation is specifically dependent on the fluorescence quenching induced by target-activated chain displacement reactions.
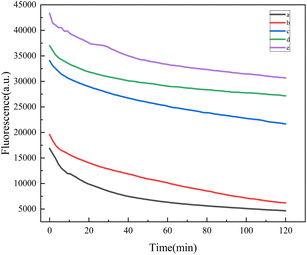 | ||
| Fig. 1 Response of fluorescence intensity over time. (a) C-Apt + H1 + H2; (b) kanamycin + Apt + C-Apt + C-Apt + H1 + H2; (c) Apt + C-Apt + H1 + H2; (d) H1 + H2; (e) H1. | ||
Polyacrylamide gel electrophoresis (PAGE) analysis also directly confirmed the reaction mechanism of the biosensor in homogeneous phase, as shown in Fig. 2. Lane 1, lane 2, lane 3, lane 4, lane 5 and lane 6 were corresponding bands of aptamer, complementary chain, H1, H2, positive control and negative control, respectively. After kanamycin was incubated with the reaction system (positive sample), the bands at H1 and H2 positions of the hairpin were weakened, the bands at double-strand positions formed by aptamer probes and complementation were weakened, and the bands of the H1–H2 complex were strengthened, because kanamycin and aptamer formed complexes simultaneously released complementary chains and formed H1–H2 complexes through cascade chain replacement reaction. Thus, these results suggest that only the addition of the target kanamycin can trigger the catalytic hairpin self-assembly reaction to amplify the signal output. The results show that the proposed fluorescent aptamer sensor is feasible for the detection of kanamycin. In order to improve the efficiency of the use of hairpins, we also made a base mismatch treatment for H1 and H2. As shown in Fig. S2,† the results show that there is a pair of mismatched bases with lower fluorescence leakage and higher fluorescence efficiency, which may be due to the high repetition of the fully complementary hairpin oligonucleotide sequence, the uncertainty of the structure formed by the hairpin, and the possibility of two hairpins in the system, which is also supported by the results of the NUPACK simulation in Fig. S3.†
 | ||
| Fig. 2 Analysis of polyacrylamide gel electrophoresis. The concentrations of Apt, C-Apt, H1 and H2 were 5 μM. The concentration of kanamycin is 10 μM. | ||
Optimization of reaction conditions
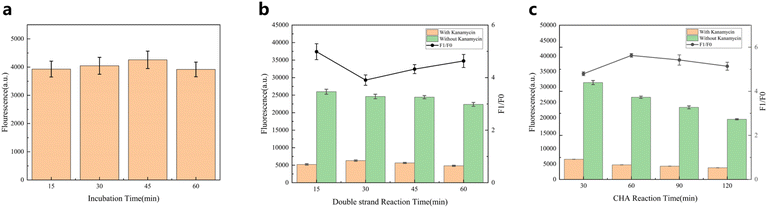
We need to optimize the experimental conditions and then perform quantitative analysis of kanamycin under the optimized conditions. The optimum conditions included incubation time of kanamycin and aptamer, binding time of aptamer and double strand, reaction time of catalytic hairpin assembly, reaction temperature and ionic strength.Incubation time of kanamycin and aptamer affects the binding efficiency of subsequent trigger chain and complementary chain. As shown in Fig. 3(a), when incubation time is 15 min, the binding of kanamycin and aptamer reaches the plateau stage, and we choose 15 min for follow-up experiments. In addition, the time of double-chain combination will also have an impact on the complementary chain triggering the self-assembly of the hairpin, as shown in Fig. 3(b), we finally chose 15 min. The trigger region of the complementary chain of the sensor hybridizes with H1, initiating the cyclic hybridization reaction of H1 and H2, inducing fluorescence quenching. Therefore, it is necessary to optimize the reaction time of catalytic hairpin assembly to obtain strong signal output. In Fig. 3(c), F/F0 increases as the catalytic hairpin assembly reaction time increases from 30 minutes to 60 minutes, which is due to the increase in reaction time for the reaction to fully occur. When the reaction time continued to increase, F/F0 began to decrease slowly, which was due to the slight attenuation of FAM fluorescence with the increase of reaction time. Finally, we chose 60 min as the time of hairpin self-assembly reaction. In addition, the effects of reaction temperature and ionic strength on probe stability and reaction rate were optimized. As shown in Fig. S4,† when the reaction temperature is 37 °C, F/F0 reaches the maximum value. In Fig. S5,† the ionic strength was selected as 500 nM Tris-HCl, 2.5 mM NaCl, 500 mM KCl. As shown in the Fig. S6,† we performed a significance analysis of the reaction conditions to determine that our choice was justified.
Detection performance of fluorescent sensor
Under the above optimal reaction conditions, the performance of our proposed fluorescent aptamer sensor for the detection of kanamycin was studied. As it is shown in Fig. 4, the results showed that the concentration of kanamycin increased from 0 to 0.9 μM, and the fluorescence intensity decreased with the increase of concentration. Kanamycin induces the conformational change of aptamer probe through specific recognition, competitively inhibits the binding of complementary chains, triggers the catalytic hairpin assembly reaction, and then the spatial proximity between the quenching group and the fluorescent group caused attenuation of the fluorescence signal. The more kanamycin is added, the more fluorescence is quenched correspondingly, and the signal output is significant. The concentration of kanamycin was linearly related to the fluorescence intensity (F) in the linear range of 54 nM ∼0.9 μM, and the linear fitting equation was F = −2876x +26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 679 (R2 = 0.9956). After that, the detection limit of the sensor is calculated to be 54 nM based on the triple standard deviation. The previous working methods had the disadvantages of being expensive, taking too long, and relying heavily on specialized equipment and professional operators. Due to the catalytic amplification reaction of the hairpin self-assembly, the sensor in this scheme has obtained good detection performance, and can realize simple, fast and low-cost detection. The calculated results are equivalent to or lower than the detection limits of existing reports, and the specific comparison results are shown in Table 1. This means that the biosensors designed will have potential applications in chemical, biomedical and environmental monitoring.
679 (R2 = 0.9956). After that, the detection limit of the sensor is calculated to be 54 nM based on the triple standard deviation. The previous working methods had the disadvantages of being expensive, taking too long, and relying heavily on specialized equipment and professional operators. Due to the catalytic amplification reaction of the hairpin self-assembly, the sensor in this scheme has obtained good detection performance, and can realize simple, fast and low-cost detection. The calculated results are equivalent to or lower than the detection limits of existing reports, and the specific comparison results are shown in Table 1. This means that the biosensors designed will have potential applications in chemical, biomedical and environmental monitoring.
| Analytical method | Strategy | Linear range (pg mL−1) | LOD (pg mL−1) | References |
|---|---|---|---|---|
| Fluorescence | MIP | 5 × 104–1.0 × 107 | 1.3 × 104 | 30 |
| Fluorescence | FRET | 5.83 × 102−4.66 × 104 | 1.69 × 102 | 31 |
| Fluorescence | Enzyme and AuNPs | 2.91 × 102−1.17 × 104 | 1.87 × 102 | 32 |
| Electrochemistry | AuNPs | 2−105 | 0.88 | 33 |
| Electrochemistry | MoS2 nanosheets | 58.3–5.83 × 104 | 16.89 | 34 |
| Electrochemistry | AgNPs | 5 × 102–1.0 × 105 | 60 | 35 |
| Colorimetric | AuNPs | Not mentioned | 5.82 | 36 |
| Fluorescence | ZIF-8@TPE | 7.3 × 103–106 | 7.3 × 103 | 37 |
| Fluorescence | Enzyme-free | 0.03–5.27 × 103 | 0.03 | This work |
We studied the selectivity of the fluorescent aptamer sensor to kanamycin using different analogues of kanamycin. As can be seen from Fig. S7,† although the reduction of fluorescence intensity caused by analogues is not similar to that of blank samples, it is still significantly compared with negative control. In contrast, the reduction in fluorescence intensity caused by kanamycin was significantly more intense, which was almost the same as that caused by kanamycin and its analogues when they were present together. The reason is that kanamycin specifically recognizes the aptamer probe, which then triggers the catalytic self-assembly of the hairpin, which outputs a strong signal. These results indicate that the fluorescence ligand sensor has good selectivity to kanamycin.
Milk sample detection
The potential application of fluorescent ligand sensor was investigated by the detection of kanamycin plus scalar in milk. See Table 2. The recoveries of different concentrations of kanamycin (100 nM, 200 nM and 300 nM) in milk were 96.89–106.75%, and the relative standard deviations (RSD) were 1.41–2.89%. These acceptable results confirm the potential application of the fluorescent aptamer sensor.| Sample number | Spiked (nM) | Mean found (nM) | Recovery (%) | RSD (%, n = 3) |
|---|---|---|---|---|
| Sample 1 | 100 | 106.75 | 106.75 | 1.41 |
| Sample 2 | 200 | 193.79 | 96.89 | 2.89 |
| Sample 3 | 300 | 303.89 | 101.30 | 2.77 |
Conclusions
In summary, we constructed a FAM fluorescence quenching biosensor based on aptamer recognition competition triggering enzyme-free amplification signal amplification for quantitative detection of kanamycin in milk. Kanamycin aptamer specifically recognizes kanamycin, the conformation of the aptamer probe is induced to change, the trigger unit is released, the catalytic hairpin self-assembly is started, and then the FAM fluorescence is quench to achieve signal output. The method has good sensitivity, which is far below the maximum allowable level in China and the European Union. In order to verify the practical application of the sensor, objective experiments were carried out on milk samples, and the recovery rate was satisfactory. In addition, the fluorescent aptamer sensor has good specificity and can distinguish kanamycin and its analogues. Therefore, it has great application prospects in the field of food safety. However, in order to fully bind the target molecule to the aptamer, the required DNA strands were added step by step in the experiment, which increased the experimental steps and complicated the experimental process to a certain extent. In addition, the detection of milk samples requires processing of the samples first, which limits the practical use of biosensors to some extent.Author contributions
Yangyinchun Bao: conceptualization, methodology, formal analysis, writing – original draft, writing – review & editing. Luhui Wang: conceptualization, methodology. Yidan Sang: conceptualization, software. Xumei Yan: methodology. Na Wang: conceptualization, software. Mengyang Hu: methodology. Yafei Dong: writing – review & editing, supervision, project administration.Conflicts of interest
There are no conflicts to resolve.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 62073207).Notes and references
- R. Y. Robati, A. Arab, M. Ramezani, F. A. Langroodi, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2016, 82, 162–172 CrossRef CAS PubMed.
- H. S. Hurd and S. Malladi, Risk Anal., 2008, 28, 695–710 CrossRef PubMed.
- R. Oertel, V. Neumeister and W. Kirch, J. Chromatogr. A, 2004, 1058, 197–201 CrossRef CAS PubMed.
- M. Bacanlı and N. Başaran, Food Chem. Toxicol., 2019, 125, 462–466 CrossRef PubMed.
- A. Tsiasioti and P. D. Tzanavaras, Food Chem., 2024, 443, 138577 CrossRef CAS PubMed.
- A. Cafaro, S. Barco, F. Pigliasco, C. Russo, M. Mariani, A. Mesini, C. Saffioti, E. Castagnola and G. Cangemi, J. Mass Spectrom. Adv. Clin. Lab, 2024, 31, 33–39 CrossRef CAS PubMed.
- M. Q. Zou, J. F. Li, F. Zhang and Y. Jin, Anal. Lett., 2010, 43, 867–875 CrossRef CAS.
- X. Chen and J. Du, Chin. J. Anal. Lab., 2010, 29, 75–78 CAS.
- C. Sun, T. Kang, L. Lu and S. Cheng, Phys. Test. Chem. Anal. Part B Chem. Anal., 2023, 59, 249–256 CAS.
- F. Xu, J. Y. Li, J. Zhou, M. L. Liu, Y. M. Liu, J. F. Wang, S. Y. Ding and X. B. Li, Chin. J. Anal. Chem., 2015, 43, 881–885 CAS.
- X. Xiao, H. Li, L. Zhao, Y. Zhang and Z. Liu, Biomed. Pharmacother., 2021, 143, 112232 CrossRef CAS PubMed.
- Y. Huang, X. Yan, L. Zhao, X. Qi, S. Wang and X. Liang, Microchem. J., 2019, 150, 104179 CrossRef CAS.
- L. Li, Y. Zhao, X. Yan, X. Qi, L. Wang, R. Ma, S. Wang and X. Mao, Sensor. Actuator. B Chem., 2021, 344, 130320 CrossRef CAS.
- J. Liu, M. Zheng, L. Wang, H. Qu and L. Zheng, Microchem. J., 2024, 110257, DOI:10.1016/j.microc.2024.110257.
- S. K. Choi, C. Lee, K. S. Lee, S.-Y. Choe, I. P. Mo, R. H. Seong, S. Hong and S. H. Jeon, Mol. Cells, 2011, 32, 527–534 CrossRef CAS PubMed.
- M. Ajamgard, J. J. Sardroodi, A. R. Ebrahimzadeh and M. R. Kamelabad, Comput. Biol. Chem., 2021, 95, 107595 CrossRef CAS PubMed.
- T. K. Sharma, R. Ramanathan, P. Weerathunge, M. Mohammadtaheri, H. K. Daima, R. Shukla and V. Bansal, Chem. Commun., 2014, 50, 15856–15859 RSC.
- R. K. Singh, B. Panigrahi, S. Mishra, B. Das, R. Jayabalan, P. K. Parhi and D. Mandal, J. Mol. Liq., 2018, 269, 269–277 CrossRef CAS.
- Y. Q. Duan, Y. T. Yang, J. X. Wang, H. M. Li and K. P. Liu, Anal. Methods, 2020, 12, 132–140 RSC.
- M. Liu, Z. Yang, B. Li and J. Du, Food Chem., 2021, 339, 128059 CrossRef CAS PubMed.
- M. Li, Y. Xie, J. Zhang, L. Lei and X. Su, Chem. Eng. J., 2023, 471, 144184 CrossRef CAS.
- Q. Zhao, S. Shi, Y. Zhang and X. Yu, Sci. Technol. Food Ind., 2015, 36(86–89), 100 Search PubMed.
- S. Y. Xiao and H. J. Liang, Acta Phys. Sin., 2016, 65(17), 178106 CrossRef.
- N. D. Chen, J. Y. Li, X. Z. Feng, Y. P. Yang, L. Zhu, X. M. Chen, X. Liu, Y. Li, C. C. Wang and L. G. Xia, Microchim. Acta, 2020, 187(8), 432 CrossRef CAS PubMed.
- G. Shi, C. Yan and J. Chen, J. Agric. Food Chem., 2022, 70, 16446–16452 CrossRef CAS PubMed.
- J. Wang, H. Li, C. Du, Y. Li, X. Ma, C. Yang, W. Xu and C. Sun, Talanta, 2022, 243, 123318 CrossRef CAS PubMed.
- R. Cai, F. Yin, H. Chen, Y. Tian and N. Zhou, Microchim. Acta, 2020, 187, 304 CrossRef CAS PubMed.
- K.-M. Song, M. Cho, H. Jo, K. Min, S. H. Jeon, T. Kim, M. S. Han, J. K. Ku and C. Ban, Anal. Biochem., 2011, 415, 175–181 CrossRef CAS PubMed.
- W. Cui, G. Hu, E. Lv, C. Li, Z. Wang, Q. Li, Z. Qian, J. Wang, S. Xu and R. Wang, Food Control, 2022, 133, 108654 CrossRef CAS.
- Y. Geng, M. Guo, J. Tan, S. Huang, Y. Tang, L. Tan and Y. Liang, Sensor. Actuator. B Chem., 2018, 268, 47–54 CrossRef CAS.
- J. Deng, Y. Liu, X. Lin, Y. Lyu, P. Qian and S. Wang, Sensor. Actuator. B Chem., 2018, 273, 1495–1500 CrossRef CAS.
- M. Ramezani, N. M. Danesh, P. Lavaee, K. Abnous and S. M. Taghdisi, Sensor. Actuator. B Chem., 2016, 222, 1–7 CrossRef CAS.
- Z. Chen, G. Lai, S. Liu and A. Yu, Sensor. Actuator. B Chem., 2018, 273, 1762–1767 CrossRef CAS.
- Y. Zhou, F. Li, H. Wu, Y. Chen, H. Yin, S. Ai and J. Wang, Sensor. Actuator. B Chem., 2019, 296, 126664 CrossRef CAS.
- S. Cheng, H. Liu, H. Zhang, G. Chu, Y. Guo and X. Sun, Sensor. Actuator. B Chem., 2020, 304, 127367 CrossRef CAS.
- L. Zou, X. Li and Y. Lai, Microchem. J., 2021, 162, 105858 CrossRef CAS.
- S. Liu, Y. Chen, Z. Ruan, J. Lin and W. Kong, Environ. Res., 2022, 206, 112617 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra01703j |
| This journal is © The Royal Society of Chemistry 2024 |