Open Access Article
Open Access ArticleInclusion complex of quercetin with sulfobutylether β-cyclodextrin: preparation, characterization, antioxidant and antibacterial activities and the inclusion mechanism
Kunkun Dai†
a,
Jiayi Wu†a,
Xinyang Liua,
Suilou Wanga,
Yihang Liua,
Hehe Li*b and
Haixiang Wang *a
*a
aDepartment of Food Nutrition and Health, School of Engineering, China Pharmaceutical University, Nanjing 211198, China. E-mail: wanghaixiang@cpu.edu.cn
bBeijing Laboratory of Food Quality and Safety, Key Laboratory of Alcoholic Beverages Quality and Safety of China Light Industry, Beijing Technology and Business University, Beijing 100048, China. E-mail: xyzhehe@126.com
First published on 20th March 2024
Abstract
Quercetin (QCT) has a variety of pharmacological effects, such as antioxidant, antibacterial, anticancer, anticardiovascular and antiaging effects. However, its poor water solubility, stability and bioavailability limit its applications. The special structure of cyclodextrins and their derivatives with a hydrophobic inner cavity and hydrophilic outer wall can load a variety of hydrophobic drugs of a suitable size and shape, thereby improving the stability and solubility of these molecules. In this study, an inclusion complex of quercetin and sulfobutylether-β-cyclodextrin was prepared. It was characterized via FT-IR, UV, 1H NMR, XRD, DSC, and SEM analysis, which revealed the successful formation of the inclusion complex. In vitro biological activity estimations were carried out and the results indicated that the inclusion complex displayed higher antioxidative and antibacterial properties compared with free QCT. In addition, the mechanisms of inclusion were explored using 1H NMR analysis and docking calculations, thus providing a theoretical basis for obtaining an inclusion complex.
1. Introduction
Quercetin (QCT) is a type of bioactive substance widely existing in foods such as broccoli, blueberries and apples and has a variety of pharmacological effects, including antioxidant,1,2 antibacterial,3 anticancer,4,5 and anticardiovascular activities.6 However, the poor water solubility and stability of QCT in aqueous systems severely restricts its applications.7–10The formation of cyclodextrin (CD) inclusion complexes is a method to increase the solubility of insoluble composites. CDs are cyclic oligosaccharides obtained from the degradation of starch by glucosyltransferase and are composed of 6–12 D-(+)-glucopyranose units in a chair conformation connected by α-1,4-glucosidic bonds.11 The most common CDs possess 6, 7 and 8 glucose units (defined as α-CD, β-CD, and γ-CD, respectively), among which β-CD has a medium cavity size and low production cost and is the most common and widely used CD in food, cosmetic and pharmaceutical fields.12,13 Cyclodextrins have a special structure of a hydrophobic inner cavity and hydrophilic outer wall, which can entrap a variety of hydrophobic molecules of suitable size and shape, presenting a high inclusion rate and improving molecular stability and solubility. Presently, cyclodextrins are often used for the inclusion of water-insoluble molecules for different purposes, such as solubilization; slow release; and flavor and taste masking in food, drugs, and cosmetics.14,15 However, β-CD is poorly soluble in water (25 °C, 18.5 mg mL−1), highly hemolytic and highly nephrotoxic,16,17 and thus its application is very limited. A wide variety of water-soluble and fat-soluble cyclodextrin derivatives has been produced, such as methylated-β-CD, hydroxypropyl-β-CD and acetylated-β-CD. Sulfobutylether-β-cyclodextrin (SBE-β-CD) is obtained through a substitution reaction between 1,4-butane sulfonated lactone and the hydroxyl groups of the carbon at the 2, 3 and 6 position of the glucose unit under alkaline conditions, with an average degree of substitution between 6.2 and 6.9.18 Due to the extended butyl side chain of the hydrophobic cavity and high acid dissociation constant of SBE-β-CD, it possesses high water solubility (>500 mg mL−1), low toxicity, good molecule binding ability and good hemocompatibility.19 It remains in the same ionized state at any pH, always with a negative surface charge, and is an anionic derivative.20
In this work, a novel inclusion complex of QCT and SBE-β-CD was prepared by a stirring combined freeze-drying method, and the stoichiometry parameter and association constant of the guest/host in the QCT/SBE-β-CD inclusion complex (QCT/SBE-β-CD) were investigated by analyzing the phase-solubility of QCT in SBE-β-CD solutions at different concentrations. Subsequently, QCT/SBE-β-CD was characterized by Fourier transform infrared spectroscopy (FT-IR), ultraviolet-visible spectroscopy (UV), 1H nuclear magnetic resonance (1H NMR), X-ray diffraction (XRD), differential scanning calorimetry (DSC), and scanning electron microscopy (SEM). Lastly, the antioxidative and antibacterial properties and inclusion mechanism of QCT/SBE-β-CD were studied.
2. Experimental
2.1. Materials
Sulfobutylether-β-cyclodextrin (SBE-β-CD, >99%, substitution degree = 6.7, MW = 2187.513) was purchased from Shandong Binzhou Zhiyuan Biochemistry Co., Ltd (Shandong, China). Quercetin (QCT, 98%) was purchased from Bide Pharmatech Ltd (Shanghai, China). 2,2′-Azinobis-(3-ethylbenzothiazoline-6-sulphonate) (ABTS, 99%) and 1,1-diphenyl-2-picrylhydrazyl free radical (DPPH, 99%) was purchased from Aladdin Biochemical Technology Co., Ltd (Shanghai, China). Escherichia coli (E. coli) ATCC 25922 and Staphylococcus aureus (S. aureus) ATCC 25923 strains were obtained from China General Microbiological Culture Collection Center and kept in the College of Engineering, China Pharmaceutical University. All other reagents were of analytical grade.2.2. Phase solubility
The phase-solubility analysis was carried out according to the reported method.21 Excess amounts of QCT (5 mg) were added to SBE-β-CD solutions (0–10 mM) and sonicated for 30 min. The suspensions were shaken constantly at different temperatures (27 °C, 37 °C and 50 °C) in a thermostatic shaker for 48 h until equilibrium was reached. Then, the suspensions were filtered using 0.45 μm micropore membrane filters, and examined by UV-vis spectroscopy. All samples were characterized in triplicate. The stability constant (Ks) was calculated according to the following formula using the phase-solubility diagram:
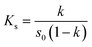 | (1) |
2.3. Preparation of the QCT/SBE-β-CD inclusion complex
The inclusion complexes of QCT with SBE-β-CD were prepared by a stirring combined freeze-drying method, as described in the literature with some modifications.22 Briefly, SBE-β-CD (0.914 g) was dissolved in 43 mL of distilled water and kept at 40 °C and QCT (50 mg) was dissolved in 7 mL anhydrous ethanol, and then slowly added into the prepared solution of SBE-β-CD. The mixture was magnetically stirred for 2 h at 40 °C, and then evaporated under vacuum condition to remove the organic solvent. The resulting solution was freeze dried for 48 h to obtain the inclusion complexes. Before analysis, the inclusion complexes were washed with ethanol solution to remove the unloaded QCT and dried under vacuum. The entrapment efficiency (EE) was calculated using eqn (2):
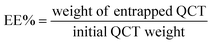 | (2) |
2.4. Preparation of physical mixture of QCT and SBE-β-CD
QCT and SBE-β-CD were simply mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 M ratio to obtain a physical mixture.
2.5 M ratio to obtain a physical mixture.
2.5. Characterization of inclusion complexes
2.6. Antioxidant activity determination
The antioxidant activity of free QCT and the inclusion complex against ABTS radicals was assessed using a modified reported method.23 In this method, an appropriate volume of 5 mmol L−1 ABTS solution was allowed to react with the same volume of 2.45 mmol L−1 potassium persulfate, and the mixture was left to stand in the dark at room temperature for 15–20 h to produce the blue-green ABTS radical cation chromophore (ABTS˙). The ABTS radical solution was diluted with PBS to obtain an absorbance value of 0.700 ± 0.002 at 734 nm. 2 mL of diluted ABTS radical solution was added to 1 mL of free QCT and inclusion complex aqueous solutions, respectively, and their absorbance values were recorded at 734 nm after mixing for 10 min. All samples were characterized in triplicate.The antioxidant activities of free QCT and the inclusion complex against DPPH radicals were also evaluated using a modified reported method.24 1 mL of free QCT and inclusion complex aqueous solution was immersed in 2 mL of DPPH solution. The samples were mixed well and placed in the dark for 30 min, and then their absorbance values were recorded at 517 nm. All samples were characterized in triplicate.
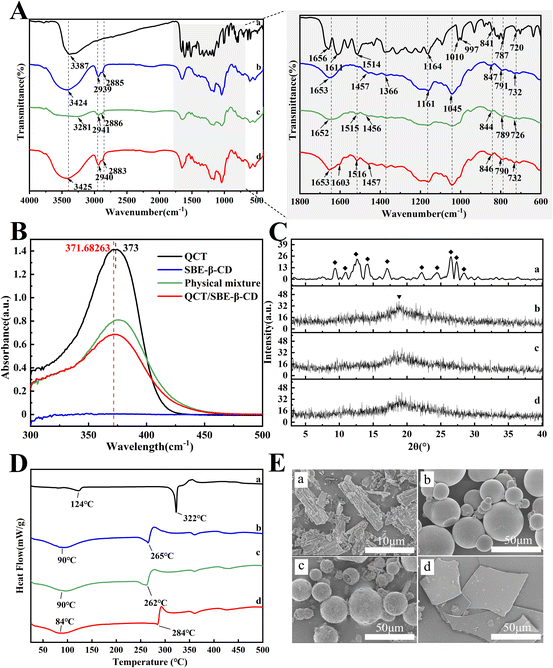
The scavenging activity (%) of ABTS radicals or DPPH radicals was computed according to the following equation:
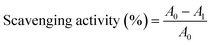 | (3) |
2.7. Antibacterial activity determination
Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) were selected to test the antibacterial activity of the samples, and the operations were conducted under sterile conditions.2.8. Molecular docking
To study the inclusion mechanism between QCT and SBE-β-CD, the structures were calculated and simulated by the molecular simulation docking method. The molecular structure of QCT was downloaded from the PubChem database and the conformation with energy-minimized configuration was obtained by processing and optimization with the Docking Suite, which was saved in “mol2” format for subsequent use. The construction of SBE-β-CD was based on β-CD by adding seven sulfobutyl groups at the wider edge of the glucopyranose unit. After the construction of the molecule, its water of crystallization was removed and it was hydrogenated. Then, the lowest energy conformation was obtained with its energy optimized, which was saved in “mol2” format for subsequent use. The SYBYL software was used to perform simulated docking calculations using QCT and SBE-β-CD as test objects. The specific operations were as follows: the SBE-β-CD structure was imported to generate the active site, and then the QCT structure was imported for docking. The combination modes were retained with three conformations, and the one with the highest score was chosen.2.9. Statistical analysis
Statistical analysis was carried out using an independent sample t-test (Origin 2021, SPSS 26). “*” represents significant (P < 0.05), “**” represents very significant (P < 0.01), and “***” represents extremely significant (P < 0.001).3. Results and discussion
3.1. Phase-solubility analysis
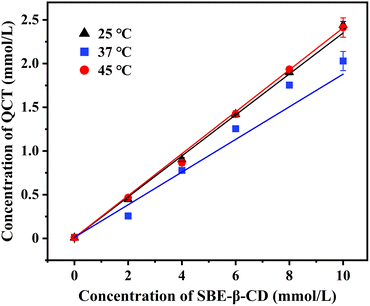
The phase-solubility diagrams of the inclusion complexes in water at different temperatures are shown in Fig. 1. The concentration of QCT and SBE-β-CD showed a good linear relationship at different temperatures, indicating that the inclusion was performed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion ratio.28 The Ks value was calculated to be 4504.69, 10
1 inclusion ratio.28 The Ks value was calculated to be 4504.69, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185.98 and 11
185.98 and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 521.81 L mol−1, respectively. The Ks values were high at different temperatures, indicating the strong binding force and good stability of the inclusion complex. Moreover, Ks increased with temperature, which was significantly temperature dependent, demonstrating that the inclusion was an endothermic process, an increase in temperature was conducive to the formation of the inclusion complex, and the stability of the inclusion complex increased accordingly. Therefore, the appropriate temperature has an important effect in the preparation of the inclusion complex. Meanwhile, the Y-axis cutoff values also increased with an increase in temperature, resulting in an increase in the solubility of QCT. These data are presented in Table 1.
521.81 L mol−1, respectively. The Ks values were high at different temperatures, indicating the strong binding force and good stability of the inclusion complex. Moreover, Ks increased with temperature, which was significantly temperature dependent, demonstrating that the inclusion was an endothermic process, an increase in temperature was conducive to the formation of the inclusion complex, and the stability of the inclusion complex increased accordingly. Therefore, the appropriate temperature has an important effect in the preparation of the inclusion complex. Meanwhile, the Y-axis cutoff values also increased with an increase in temperature, resulting in an increase in the solubility of QCT. These data are presented in Table 1.
| T (° C) | Linear equation | R2 | Ks (L mol−1) |
|---|---|---|---|
| 25 | y = 0.215x − 0.0608 | 0.9903 | 4504.69 |
| 37 | y = 0.2435x − 0.0316 | 0.9988 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185.98 185.98 |
| 45 | y = 0.2426x − 0.0278 | 0.9981 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 521.81 521.81 |
3.2. Entrapment efficiency (EE) analysis
The EE of the inclusion complexes was 86.06 ± 1.43% in this study. In previous reports, some cyclodextrin inclusion complexes prepared by co-grinding, spray-drying, microwave and co-precipitation methods had encapsulation efficiencies of 31% to 80%,29–32 which are lower than the encapsulation efficiency in this study. The inclusion complex prepared by stirring combined with freeze-drying method was beneficial for improving its encapsulation efficiency, and other reports also supported similar conclusions.33,34 The reason for this phenomenon may be due to the stirring process increasing the contact area between the host and guest, reducing the repulsion between them, and the freeze-drying process minimizing the possibility of chemical decomposition and the loss of bioactivity.35 This study also investigated the effects of some factors (such as host and guest inclusion ratio, inclusion time, inclusion temperature and solvent composition) on the EE during stirring combined with freeze-drying process to obtain the optimal preparation conditions.3.3. FT-IR analysis
The FTIR spectra are shown in Fig. 2A. The broad absorption peak at 3387 cm−1 in the spectrogram of QCT (Fig. 2A(a)) represents the phenolic hydroxyl group in the benzene ring. The absorption peak at 1656 cm−1 represents the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond and that at 1611 cm−1 represents the C
C bond and that at 1611 cm−1 represents the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond in the benzene ring. The peak at 1514 cm−1 corresponds to the C
C bond in the benzene ring. The peak at 1514 cm−1 corresponds to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration. The peaks at 1465–1340 cm−1 and 1000–675 cm−1 correspond to the C–H stretching vibration and bending vibration, respectively. The peak at 1164 cm−1 is ascribed to the C–O bond. In the spectrum of SBE-β-CD (Fig. 2A(b)), the peak at 3424 cm−1 is ascribed to the hydroxyl group bond. The peaks at 2939 at 2885 cm−1 is ascribed to the –CH2 bond. The peaks at 1653, 1366, 1161, and 1045 cm−1 correspond to the adsorbed water, S
O stretching vibration. The peaks at 1465–1340 cm−1 and 1000–675 cm−1 correspond to the C–H stretching vibration and bending vibration, respectively. The peak at 1164 cm−1 is ascribed to the C–O bond. In the spectrum of SBE-β-CD (Fig. 2A(b)), the peak at 3424 cm−1 is ascribed to the hydroxyl group bond. The peaks at 2939 at 2885 cm−1 is ascribed to the –CH2 bond. The peaks at 1653, 1366, 1161, and 1045 cm−1 correspond to the adsorbed water, S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, glycosidic bond, and S–O bond, respectively. The physical mixture (Fig. 2A(c)) was simply a combination of the QCT characteristic peak and the SBE-β-CD characteristic peak, indicating no other interactions. Alternatively, the intensity of the absorption peak of the inclusion complex (Fig. 2A(d)) at 1603 cm−1 changed, indicating that QCT may be partially embedded (ring A or ring B) in the cavity to form a new compound, and no new absorption peak was found, indicating that no new chemical bond was generated.
O bond, glycosidic bond, and S–O bond, respectively. The physical mixture (Fig. 2A(c)) was simply a combination of the QCT characteristic peak and the SBE-β-CD characteristic peak, indicating no other interactions. Alternatively, the intensity of the absorption peak of the inclusion complex (Fig. 2A(d)) at 1603 cm−1 changed, indicating that QCT may be partially embedded (ring A or ring B) in the cavity to form a new compound, and no new absorption peak was found, indicating that no new chemical bond was generated.
 | ||
| Fig. 2 FT-IR spectrum (A), UV spectrum (B), powder X-ray diffraction patterns (C), DSC curves (D), and SEM images (E) of QCT (a), SBE-β-CD (b), physical mixture (c) and inclusion complex (d). | ||
3.4. UV analysis
The UV absorption spectra are shown in Fig. 2B. QCT had a strong absorption peak at 373 nm, while SBE-β-CD had a very weak UV absorption and no characteristic peak due to the lack of unsaturated bonds. The absorption peak position of the inclusion complex showed a blue shift. This may be due to the reduced conjugation effect of SBE-β-CD on QCT and the solvent effect of the different samples (QCT was in ethanol and others were in water). This demonstrated that no new unsaturated bond was formed and the formation of the inclusion complex did not alter the UV absorption characteristics of QCT.363.5. XRD analysis
The powder X-ray diffractograms are shown in Fig. 2C. QCT (Fig. 2C(a)) displayed obvious sharp crystalline diffraction peaks at 2θ = 9.3°, 10.8°, 12.6°, 14.2°, 17°, 22.1°, 24.4°, 26.5°, 27.3° and 28.4°, indicating its crystal structure. SBE-β-CD (Fig. 2C(b)) exhibited a broadened and weak amorphous diffraction peak near 18.8°, indicating its amorphous form with an A-type diffraction pattern. The crystallinity peak was still detectable in the physical mixture (Fig. 2C(c)) and was a simple overlap of the diffraction peaks of QCT and SBE-β-CD. On the contrary, the diffraction spectrum of the inclusion complex (Fig. 2C(d)) was equal to that of SBE-β-CD, and the disappearance of the crystallinity diffraction peaks of QCT at 12.6° and 17° implied the formation of the amorphous inclusion complex.3.6. DSC thermograms
The differential scanning calorimetry curves are shown in Fig. 2D. QCT (Fig. 2D(a)) showed phase change heat absorption peaks (dissolution peak) at 124 °C and 322 °C. The peak at 124 °C was related to the dehydration of QCT, while the sharp heat absorption peak at 322 °C was generated by the heat absorption of QCT melting (melting point 317 °C). SBE-β-CD (Fig. 2D(b)) had a broad heat absorption peak between 60 °C and 120 °C, reaching the maximum at 90 °C, which resulted from the evaporation of water, and subsequently its breakdown peak appeared at 265 °C. In the physical mixture (Fig. 2D(c)), it could be seen that it was more like a superposition of endothermic peaks of QCT and SBE-β-CD. The absorption peak of SBE-β-CD at 265 °C became sharp and moved to 284 °C, indicating that the host and guest interacted during the successful formation of the inclusion and the inclusion complex (Fig. 2D(d)). The endothermic peak at 84 °C can be ascribed to the evaporation of residual ethanol during the preparation of the inclusion complex.3.7. SEM analysis
The SEM images are shown in Fig. 2E. Free QCT exhibited a needle-like morphology (Fig. 2E(a)), while SBE-β-CD presented a smooth spherical structure with a size of approximately 50 μm (Fig. 2E(b)). The physical mixture showed both crystalline components, with QCT attached to the exterior of SBE-β-CD (Fig. 2E(c)). Conversely, the structure of the inclusion complex (Fig. 2E(d)) appeared as an irregular, amorphous piece, in which the original shape of the two elements vanished, confirming the formation of the inclusion complex. The results indicate that by only mixing, the two compounds still maintained their original form, but when the inclusion complex was formed and underwent close association, QCT no longer existed in the crystalline state.3.8. 1H NMR analysis
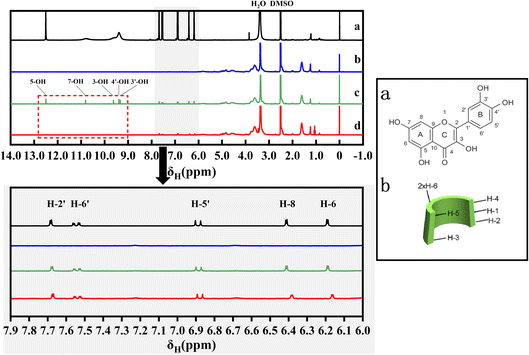
The 1H NMR spectra are shown in Fig. 3. The peaks at δ2.508 (DMSO) and δ3.388 (H2O) are the solvent peaks, and the chemical shifts of QCT are mainly in the range of 6.0 to 8.0 ppm, which are different from that of the SBE-β-CD protons in the range of 3.5 to 4.0 ppm. As shown in the red dashed box, the chemical shifts of QCT at 9.27, 9.33, 9.56, 10.76, and 12.48 ppm (3′-OH, 4′-OH, 3-OH, 7-OH, and 5-OH, respectively) disappeared in the inclusion complex, indicating that these positions of QCT were encapsulated in the cavity or formed hydrogen bonds. In the case of QCT, SBE-β-CD caused significant changes in the field shift of its H-8 and H-6 protons, while the chemical shifts of H-2′, H-6′ and H-5′ were small, indicating that the interaction between ring A and the cavity was greater than that of ring B. In the case of SBE-β-CD, H-1 (Δδ0.004 ppm, upper field) and H-3 (Δδ0.004 ppm, lower field) exhibited relatively significant chemical shifts compared to H-5 (Δδ0.003 ppm, lower field) and H-4 after the formation of the inclusion complex, indicating that QCT entered from the inner surface and the wider side of the cavity (H-3 was near to the wide side of the cavity and H-5 was near to the narrow side of the cavity), but it did not penetrate deeply and influence the chemical environment of H-5. Therefore, H-5 demonstrated a weaker chemical shift. These data are depicted in Table 2.| Substance | Proton | δ(free) | δ(complex) | Δδ |
|---|---|---|---|---|
| QCT | H-2′ | 7.683 | 7.673 | 0.01 |
| H-6′ | 7.524 | 7.523 | 0.001 | |
| H-5′ | 6.87 | 6.864 | 0.006 | |
| H-8 | 6.412 | 6.384 | 0.028 | |
| H-6 | 6.191 | 6.164 | 0.027 | |
| SBE-β-CD | H-1 | 5.057 | 5.053 | 0.004 |
| H-3 | 3.767 | 3.771 | −0.004 | |
| H-5 | 3.624 | 3.627 | −0.003 | |
| H-2′/H-3′ | 1.617 | 1.615 | 0.002 |
3.9. Antioxidant activity
The antioxidant activity of QCT and the inclusion complex against ABTS radicals and DPPH radicals is shown in Fig. 4. The scavenging ability of all the samples was enhanced with an increase in the concentration of QCT, and there was a dose-effect relation between scavenging ability and concentration.36 In the case of ABTS radicals, the scavenging ability of the inclusion complex greatly increased compared to QCT, the scavenging rate was essentially twice that of QCT. The scavenging rate of the inclusion complex was close to 100% at the concentration of 0.05 mg mL−1. In the case of the DPPH radicals, the scavenging ability of the inclusion complex was stronger than that of QCT at the concentration of 0.01 mg mL−1, which was 6.6-times higher than that of QCT. There was also a dose-dependent relation between scavenging ability and concentration. The results showed that the inclusion complex could significantly increase the antioxidant activity of QCT at certain concentrations, and its scavenging effect on ABTS radicals was greater than the two free radicals.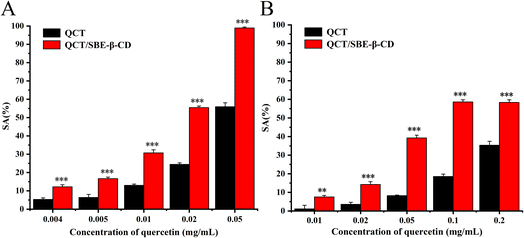 | ||
| Fig. 4 Antioxidant activity of quercetin and the inclusion complex against ABTS radicals (A) and DPPH radicals (B). | ||
The scavenging effect of antioxidants on free radicals is associated with their hydrogen supply capacity and the antioxidant performance of phenolic compounds (e.g. QCT) is determined by their structure. One hydroxyl in the ortho hydroxyl groups was hydrogenated to become a carbonyl group and the other formed an intramolecular hydrogen bond, which further resonated to form o-benzoquinone, thus stabilizing the compound. Thus, the ortho hydroxyl group in ring B of QCT was the crucial structure that determined its scavenging ability for free radicals. The antioxidant performance of the inclusion complex was closely related to its binding mode. During the inclusion process, the hydroxyl of QCT was close enough to the secondary hydroxyl of SBE-β-CD, and one or more intermolecular hydrogen bonds were formed between the host and guest. This strong intermolecular binding weakened the intramolecular hydrogen bond of QCT and the covalent bond between the oxygen and hydrogen in the hydroxyl improved the hydrogen supply capacity of the QCT hydroxyl, and thus improved the antioxidant activity of inclusion complex.
3.10. MIC analysis
It was found that the inhibition effect of either QCT or the inclusion complex on S. aureus was stronger than that of E. coli, where the inhibition effect on E. coli was very poor in all experimental concentration ranges (when the highest concentration reached 900 mg mL−1, there was still no inhibition circle. The MIC of E. coli was at least 42-times higher than that of S. aureus, and the solution was already approached to the viscous state, and thus there was no research significance anymore). The reason for this may be related to the structure of the bacteria, where Gram-negative bacteria have a rigid cell wall and an outer membrane that create resistance, whereas although Gram-positive bacteria have a thick peptidoglycan layer, they lack an outer membrane, and thus their cellular contents can easy to leak, and eventually the bacteria die.37 Therefore, subsequent experiments were conducted only on S. aureus.The control group could not inhibit the growth of bacteria, and the experimental groups showed a significant concentration dependence with an increase in the concentration of QCT. The reason for this was that the initial amount of the inclusion complex used was low, and it was not possible to release enough QCT to inhibit the growth of the bacteria. As the dosage gradually increased, the antibacterial activity increased, and the inhibitory area also increased. The amount of QCT released by the inclusion complex could have a certain degree of inhibition on the growth of S. aureus. The inhibition diameters of the inclusion complex against S. aureus were 15.9 ± 0.1 mm, 12.75 ± 0.25 mm, 12.1 ± 0.1 mm and 10.65 ± 0.15 mm at different concentrations (170 mg mL−1, 85 mg mL−1, 42.5 mg mL−1 and 21.25 mg mL−1), respectively. The MIC of the inclusion complex against S. aureus was 21.25 mg mL−1. The inhibition effect of the inclusion complex was better than that of QCT, and the main reason for this was that although SBE-β-CD itself did not have antibacterial properties, the inhibition activity of QCT itself was not affected after the inclusion.38 However, its solubility, dissolution rate and stability in water were significantly improved, and its utilization rate improved, and thus the bacterial inhibitory effect was enhanced, which helped it fully contact with the pathogens in solution. These data are presented in Table 3.
| Compounds | Concentration (mg mL−1) | Diameter of inhibition zone (mm) |
|---|---|---|
| QCT | 170 | 15.0 ± 0.1 |
| 85 | — | |
| 42.5 | — | |
| 21.25 | — | |
| 10.625 | — | |
| 5.3125 | — | |
| QCT/SBE-β-CD | 200 | 15.9 ± 0.1 |
| 100 | 12.75 ± 0.25 | |
| 50 | 12.1 ± 0.1 | |
| 25 | 10.65 ± 0.15 | |
| 12.5 | — | |
| 6.25 | — |
3.11. MBC analysis
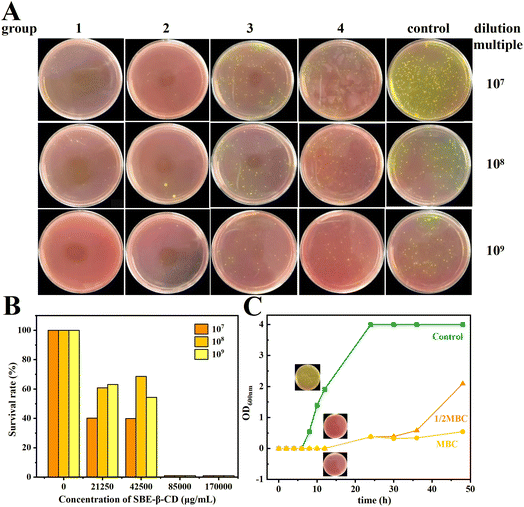
The MBC and bacterial survival of S. aureus by the inclusion complex inhibition are displayed in Fig. 5A and B, respectively. It can be seen that the growth state of S. aureus was significantly inhibited with different concentrations of inclusion complexes. Taking the bacterial solution diluted by 109 times as an example, at the concentration of 21.25 mg mL−1 (MIC), the survival rate of S. aureus was only 63.2% of that of the control group, indicating that the bacteria were only inhibited in growth but could not be completely killed under the effect of MIC. As the concentration of the inclusion complex increased, the survival rate of the bacteria gradually decreased. When increasing the concentration to 42.5 mg mL−1 (2 MIC), the median lethal rate of S. aureus was reached (54.4%). When the concentration further increased to 85 mg mL−1 (4 MIC), there were few viable bacterial colonies surviving on the plates, with a bacterial survival rate of 1%, indicating that the inclusion complex could kill 99% of S. aureus at this concentration, and thus the MBC of the inclusion complex was 85 mg mL−1.3.12. Growth curve
The growth inhibition curves of S. aureus by the inclusion complex are shown in Fig. 5C. The bacterial growth state of the control group was good, growing slowly in 0–6 h and starting to grow rapidly after 8 h. The bacterial growth was significantly inhibited by both 42.5 mg mL−1 (1/2 MBC) and 85 mg mL−1 (MBC) of inclusion complex, and the OD600 nm values hardly changed during the initial 12 h, indicating that inclusion complex could effectively inhibit the growth and reproduction of S. aureus, especially during the logarithmic phase of bacterial growth. At 12 h, the bacteria concentration of blank group reached to (6.02 ± 0.36) ×1011 CFU mL−1, while the bacteria concentration of the 1/2 MBC and MBC groups was still not observed at that time.3.13. Molecular docking
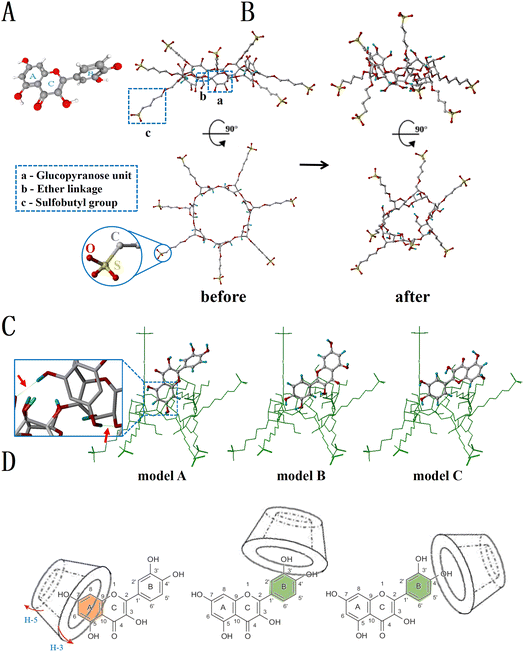
The three-dimensional stereo structures of QCT and SBE-β-CD are shown in Fig. 6A and B, respectively. QCT (Fig. 6A) is a linear molecule containing a polar group (carbonyl group) and a nonpolar aliphatic chain, and made up of two benzene rings (ring A and ring B) and a pyran ring (ring C), with ring A and ring C forming a 2-phenylacrylic acid structure with an m-phenol hydroxyl group on ring A and o-phenol hydroxyl group on ring B. The surface of SBE-β-CD (Fig. 6B) became highly hydrophilic due to the exposed hydroxyl groups in C-2 and C-3, sulfobutyl groups, and the α-1,4 glycosidic bonds. The three best conformations of the inclusion complexes obtained by molecular docking are shown in Fig. 6C. Mode A scored the highest (2.912) compared to mode B (2.625) and mode C (2.501), indicating that it was most likely that the end of ring A and ring C of QCT entered the cavity along the direction of the broad side of SBE-β-CD, while ring B was at the edge of the broad side of SBE-β-CD. The green dotted line indicated that the hydroxyl groups at H-5 and H-7 of ring A and the secondary hydroxyl groups of SBE-β-CD form hydrogen bond interactions, which connected the host and guest with intermolecular hydrogen bonds. Previous reports observed that reserpine (RSP) also approached SBE-β-CD from the broad side of the cavity and attached to its inner surface, with the formation of hydrogen bonds and only one hydrogen bond formed in the inclusion of protocatechuic acid and vanillic acid with SBE-β-CD.39,40Combining the phase-solubility, 1H NMR and molecular docking results, we concluded that the positions of 3′-OH, 4′-OH, 3-OH, 7-OH and 5-OH of QCT inserted into the cavity of SBE-β-CD or formed hydrogen bonds with SBE-β-CD. In addition, 5-OH and 7-OH could also form hydrogen bonds. Ring A interacted with the cavity but not deep, only entering from the inner surface of the cavity, while ring B could only partially penetrate the cavity due to the spatial site resistance and was located at the wide side. Therefore, several probable inclusion modes of the inclusion complex were deduced, as shown in Fig. 6D.
4. Conclusion
In our study, we successfully prepared an inclusion complex of QCT and SBE-β-CD via a stirring combined freeze-drying method and analyzed their structural characteristics by FT-IR, UV, XRD, DSC and SEM. According to the phase-solubility experiments, SBE-β-CD could theoretically form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complex with QCT, maintaining strong inclusion bonding and good stability, and the Ks values increased with an increase in temperature, which had a good solubilization effect on quercetin. The antioxidant studies showed that the inclusion complex significantly improved the scavenging ability of QCT for ABTS radicals and DPPH radicals. The inhibition activity study showed that the inhibition effects of the inclusion complex and QCT on S. aureus were stronger than that for E. coli and the inhibition effect of the inclusion complex was greatly better than that of QCT. The inclusion complex could effectively inhibit the growth of S. aureus. Besides, the mechanism of inclusion was also explored, providing a theoretical basis for obtaining an inclusion complex. Ring A of QCT was embedded in the cavity of SBE-β-CD, while ring B could only partially penetrate the cavity, which was located at the wide side. Future studies should focus on the application of inclusion complexes in the food industry and their bioavailability in vivo.
1 inclusion complex with QCT, maintaining strong inclusion bonding and good stability, and the Ks values increased with an increase in temperature, which had a good solubilization effect on quercetin. The antioxidant studies showed that the inclusion complex significantly improved the scavenging ability of QCT for ABTS radicals and DPPH radicals. The inhibition activity study showed that the inhibition effects of the inclusion complex and QCT on S. aureus were stronger than that for E. coli and the inhibition effect of the inclusion complex was greatly better than that of QCT. The inclusion complex could effectively inhibit the growth of S. aureus. Besides, the mechanism of inclusion was also explored, providing a theoretical basis for obtaining an inclusion complex. Ring A of QCT was embedded in the cavity of SBE-β-CD, while ring B could only partially penetrate the cavity, which was located at the wide side. Future studies should focus on the application of inclusion complexes in the food industry and their bioavailability in vivo.
Author contributions
Kunkun Dai and Jiayi Wu contributed equally to this work. Kunkun Dai: conceptualization, software and writing – review & editing; Jiayi Wu: methodology, investigation and writing – original draft; Xinyang Liu: validation and data curation; Suilou Wang: supervision; Yihang Liu: visualization; Hehe Li: conceptualization and resources; Haixiang Wang: project administration, supervision, funding acquisition and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the open project funds of Beijing Laboratory of Food Quality and Safety/Key Laboratory of Alcoholic Beverages Quality and Safety of China Light Industry (grant number FQS-202203), as well as the National Innovation and Entrepreneurship Training Program for Undergraduates (grant number 2023103161106).Notes and references
- M. J. Vallier, C. L. Bourvellec and O. Dangles, Food Funct., 2020, 11, 9144–9156 RSC.
- S. S. Pekkarinen, I. M. Heinonen and A. I. Hopia, J. Sci. Food Agric., 1999, 79(4), 499–506 CrossRef CAS.
- S. Anandam and S. Selvamuthukumar, J. Mater. Sci., 2014, 49, 8140–8153 CrossRef CAS.
- T. A. Dias, C. L. Duarte, C. F. Lima, M. F. Proença and C. Pereira-Wilson, Eur. J. Med. Chem., 2013, 65, 500–510 CrossRef CAS.
- J. N. Lockhart, D. M. Stevens, D. B. Beezer, A. Kravitz and E. Harth, J. Controlled Release, 2015, 220, 751–757 CrossRef CAS PubMed.
- R. V. Patel, B. M. Mistry, S. K. Shinde, R. Syed, V. Singh and H. S. Shin, Eur. J. Med. Chem., 2018, 155, 889–904 CrossRef CAS PubMed.
- T. V. Ilyich, T. A. Kovalenia, E. A. Lapshina, A. Stępniak, B. Palecz and I. B. Zavodnik, J. Mol. Liq., 2021, 325, 115184 CrossRef CAS.
- L. Gao, G. Liu, X. Wang, F. Liu, Y. Xu and J. Ma, Int. J. Pharm., 2011, 404(1–2), 231–237 CrossRef CAS PubMed.
- M. Igual, E. Garcia-Martinez, M. M. Camacho and N. Martinez-Navarrete, Innovative Food Sci. Emerging Technol., 2011, 12(2), 153–162 CrossRef CAS.
- L. G. Ranilla, M. I. Genovese and F. M. Lajolo, J. Agric. Food Chem., 2009, 57(13), 5734–5742 CrossRef CAS.
- S. G. Frank, J. Pharm. Sci., 1975, 64(10), 1585–1604 CrossRef CAS PubMed.
- V. J. Stella and R. A. Rajewski, Int. J. Pharm., 2020, 583, 119396 CrossRef CAS PubMed.
- S. Saffarionpour and L. L. Diosady, Food Funct., 2023, 14, 5062–5077 RSC.
- A. P. Sherj, B. R. Dravyakar, D. Kadam and M. Jadhav, Carbohydr. Polym., 2017, 173, 37–49 CrossRef PubMed.
- F. Odeh, H. Nsairat, W. Alshaer, S. Alsotari, R. Buqaien, S. Ismail, A. Awidi and A. A. Bawab, RSC Adv., 2019, 9, 37148 RSC.
- H. Wang, X. Xie, F. Zhang, Q. Zhou, Q. Tao, Y. Zou, C. Chen, C. Zhou and S. Yu, Food Chem. Toxicol., 2011, 49, 1387–1393 CrossRef CAS.
- L. Zhang, Z. Zhang, N. Li, N. Wang, Y. Wang, S. Tang, L. Xu and Y. Ren, Int. J. Biol. Macromol., 2013, 61, 494–500 CrossRef CAS.
- V. J. Stella and R. A. Rajewski, Int. J. Pharm., 2020, 583, 119396 CrossRef CAS PubMed.
- D. Xu, X. Li, Y. Huang, Z. Tang and G. Shu, J. Mol. Struct., 2020, 1223(2), 128969 Search PubMed.
- L. Samuelsen, R. Holm and C. SchÖnbek, J. Pharm. Sci., 2021, 110(7), 2661–2668 CrossRef CAS.
- F. S. d. M. Nathalie, G. Renato, H. R. Andre and F. Leonardo, J. Pharm. Biomed. Anal., 2008, 47, 865–869 CrossRef.
- N. Devasari, C. P. Dora, C. Singh, S. R. Paidi, V. Kumar, M. E. Sobhia and S. Suresh, Carbohydr. Polym., 2015, 134, 547–556 CrossRef CAS PubMed.
- R. Re, N. Pellegrini, A. Proteggente, A. Pannala, M. Yang and C. Rice-Evans, Free Radical Biol. Med., 1999, 26, 1231–1237 CrossRef CAS PubMed.
- Q. Liu, Y. Jing, C. Han, H. Zhang and Y. Tian, Food Hydrocolloids, 2019, 93, 432–442 CrossRef CAS.
- J. Gómez-Estaca, A. M. López de Lacey, M. E. López-Caballero, M. d. C. Gómez-Guillén and P. Montero, Food Microbiol., 2010, 27(7), 889–896 CrossRef.
- A. R. Chowdhuri, S. Tripathy, C. Haldar, S. Chandra, B. Das, S. Royb and S. K. Sahu, RSC Adv., 2015, 5, 21515 RSC.
- S. Hayet, M. Ghrayeb, D. N. Azulay, Z. Shpilt, E. Y. Tshuva and L. Chai, RSC Med. Chem., 2023, 14, 983–991 RSC.
- T. W. J. Nicol, N. Matubayasibc and S. Shimizu, Phys. Chem. Chem. Phys., 2016, 18, 15205–15217 RSC.
- A. Cid-Samamed, J. Rakmai, J. C. Mejuto, J. Simal-Gandara and G. Astray, Food Chem., 2022, 384, 132467 CrossRef CAS PubMed.
- C. Saldanha do Carmo, C. Maia, J. Poejo, I. Lychko, P. Gamito, I. Nogueira, M. R. Bronze, A. T. Serra and C. M. M. Duarte, RSC Adv., 2017, 7, 32065–32075 RSC.
- C. D. Nicolettia, M. d. S. H. Queiroza, C. G. d. S. Limab, F. d. C. da Silvab, D. O. Futuroa and V. F. Ferreiraa, J. Drug Delivery Sci. Technol., 2020, 58, 101777 CrossRef.
- M. I. Rodríguez-López, M. T. Mercader-Ros, S. López-Miranda, J. A. Pellicer, A. Pérez-Garrido, H. Pérez-Sánchez, E. Núñez-Delicado and J. A. Gabaldón, J. Sci. Food Agric., 2019, 99(3), 1322–1333 CrossRef.
- L. E. Hill, C. Gomes and T. M. Taylor, LWT--Food Sci. Technol., 2013, 51(1), 86–93 CrossRef CAS.
- N. Wangsawangrung, C. Choipang, S. Chaiarwut, P. Ekabutr, O. Suwantong, P. Chuysinuan, S. Techasakul and P. Supaphol, Gels, 2022, 8(9), 573 CrossRef CAS PubMed.
- E. M. Del Valle, Process Biochem., 2004, 39(9), 1033–1046 CrossRef CAS.
- M. Zhao, H. Wang, B. Yang and H. Tao, Food Chem., 2010, 120(4), 1138–1142 CrossRef CAS.
- I. Rasooli, M. B. Rezaei and A. Allameh, Food Control, 2006, 17(5), 359–364 CrossRef CAS.
- L. Szente and J. Szejtli, Trends Food Sci. Technol., 2004, 15(3–4), 137–142 CrossRef CAS.
- X. Xiong, M. Wu, X. Zhao and Z. Song, Luminescence, 2014, 29(6), 621–625 CrossRef CAS.
- X. Xiong, X. Zhao and Z. Song, Anal. Biochem., 2014, 460, 54–60 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |