Open Access Article
Open Access ArticleExploration of novel cationic amino acid-enriched short peptides: design, SPPS, biological evaluation and in silico study†
Prashant K. Chandolea,
Tushar Janardan Pawar b,
José Luis Olivares-Romero
b,
José Luis Olivares-Romero b,
Sunil R. Tivaria,
Bianney Garcia Larac,
Harun Pateld,
Iqrar Ahmadd,
Enrique Delgado-Alvarado
b,
Sunil R. Tivaria,
Bianney Garcia Larac,
Harun Pateld,
Iqrar Ahmadd,
Enrique Delgado-Alvarado e,
Siddhant V. Kokate*f and
Yashwantsinh Jadeja
e,
Siddhant V. Kokate*f and
Yashwantsinh Jadeja *a
*a
aDepartment of Chemistry, Marwadi University, Rajkot-360003, Gujarat, India. E-mail: drysjadeja@gmail.com
bRed de Estudios Moleculares Avanzados, Instituto de Ecología, A.C. Carretera Antigua a Coatepec 351, Xalapa, 91073, Veracruz, Mexico
cDepartamento de Química, Universidad de Guanajuato, Noria Alta S/N, Guanajuato-36050, Guanajuato, Mexico
dDepartment of Pharmaceutical Chemistry, R. C. Patel Institute of Pharmaceutical Education and Research, Shirpur District Dhule-425405, Maharashtra, India
eMicro and Nanotechnology Research Center, Universidad Veracruzana, Blvd. Av. Ruiz Cortines No. 455 Fracc. Costa Verde, Boca del Río 94294, Mexico
fDepartment of Chemistry, S. S. C. College, Junnar, Pune-410502, Maharashtra, India. E-mail: siddhantkokate@gmail.com
First published on 3rd June 2024
Abstract
Antimicrobial resistance (AMR) represents a critical challenge worldwide, necessitating the pursuit of novel approaches to counteract bacterial and fungal pathogens. In this context, we explored the potential of cationic amino acid-enriched short peptides, synthesized via solid-phase methods, as innovative antimicrobial candidates. Our comprehensive evaluation assessed the antibacterial and antifungal efficacy of these peptides against a panel of significant pathogens, including Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes, Candida albicans, and Aspergillus niger. Utilizing molecular docking techniques, we delved into the molecular interactions underpinning the peptides' action against these microorganisms. The results revealed a spectrum of inhibitory activities, with certain peptide sequences displaying pronounced effectiveness across various pathogens. These findings underscore the peptides' potential as promising antimicrobial agents, with molecular docking offering valuable insights into their mechanisms of action. This study enriches antimicrobial peptide (AMP) research by identifying promising candidates for further refinement and development toward therapeutic application, highlighting their significance in addressing the urgent issue of AMR.
Introduction
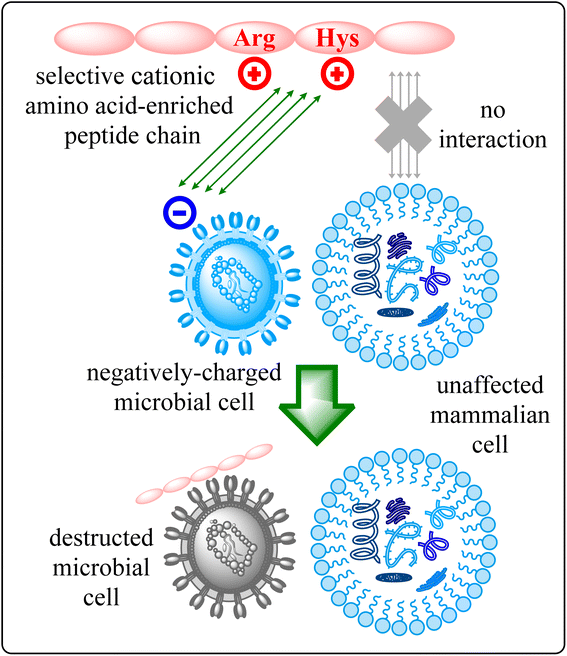
The escalating challenge of antimicrobial resistance (AMR) has become a critical concern for global health, necessitating innovative and multifaceted strategies to combat bacterial and fungal infections.1 As traditional antibiotics increasingly fall short against multidrug-resistant pathogens, the scientific community is urgently seeking innovative approaches to address this crisis.2 In this context, the development of short peptides, particularly those enriched with cationic amino acids, has emerged as a promising avenue for novel antimicrobial agents.3 Antimicrobial Peptides (AMPs) represent an essential class of molecules within the innate immune defences of various organisms, including humans, plants, and insects.4 Characterized by their broad-spectrum activity against bacteria, fungi, viruses, and certain parasites, AMPs offer a versatile approach to combating infections. Their effectiveness is primarily attributed to their ability to disrupt microbial membranes, leading to cell lysis and pathogen death.5Among AMPs, short cationic amino acid-enriched peptides have drawn significant attention due to their potent antimicrobial properties and lower risk of resistance development.6 The foundation of their potency is firmly anchored in their cationic residues that bestow a positive charge upon these peptides.6 This pivotal characteristic instigates a symphony of electrostatic interactions with microbial cell membranes adorned with negative charges, thereby triggering a nuanced molecular dialogue.6c As the peptides engage in binding, a transformative narrative unfolds, underscored by the disruption of membrane integrity, propelling a cascade of events culminating in the demise of microorganisms.6d,e Beyond their immediate disruptive potential, the positive charge holds the promise of selectivity, with intriguing indications that certain short cationic peptides exhibit a predilection for microbial cells over their mammalian counterparts.7 This intricate mode of action is visually encapsulated in Fig. 1, where the schematic representation elucidates the journey of these short cationic AA-enriched peptides—from their initial electrostatic interaction to the culmination of the microorganism's downfall. This figure serves as a beacon, illuminating the mechanistic essence that underpins the potential of these peptides to revolutionize the landscape of antimicrobial strategies.
The smaller size and higher solubility of short cationic AA-enriched peptides compared to monoclonal antibodies provide better pharmacokinetics, higher cellular uptake in the target tissue, and rapid clearance from the non-target tissues.8 Such peptides that are effectively taken up by cells are called Cell Penetrating Peptides (CPPs).9 Such short cationic AA-enriched peptides are a group of small peptides consisting of 2–30 AAs that can pass the lipid bilayer of cells without disturbing their structural and functional integrity and carry bioactive molecules, like nucleic acids,10 proteins,11 bioimaging agents12 or nanoparticles13 across the cell membrane. Thus, they are also known as protein transduction domains (PTDs). They are primarily small cationic peptides, histidine (His),14 arginine (Arg)15 or lysine (Lys)16 rich peptides. The His-rich CPPs are particularly utilized for the transduction of small interfering RNA (siRNA) or DNA derivatives inside cells.17 In contrast, Arg-rich CPPs have the capability of efficient internalization and deliver biologically active cargoes.18
In recent years, the exploration of short peptides has grown due to their inherent advantages over traditional antibiotics.19 Their adaptability for synthesis and modification, facilitated by established techniques like solid-phase peptide synthesis (SPPS),20 positions them as a platform for innovative antimicrobial design. Against this backdrop, the current study embarks on an expedition into the design, synthesis, and comprehensive evaluation of cationic AA-enriched short peptides. Rigorous and careful assessments encompass antibacterial and antifungal activities, encompassing a spectrum of clinically relevant strains. This is synergistically complemented by molecular docking analyses, which penetrate the subtleties of peptide-microbe interactions and furnish insights to guide rational design strategies. Recent studies employing molecular docking and simulation have shed light on the structural determinants of peptide efficacy and offered guidelines for the rational design of new antimicrobial candidates.19,21
Remarkably, the strength of such cationic AA-enriched short peptides can be further bolstered by their cystine-containing composition—a structural feature that imparts robust stability and endows them with the resilience to thwart enzymatic degradation.22 Moreover, Cys residue can provide a site for the conjugation of the peptide to another molecule via the –SH group. Additionally, N-terminal Cys improves the helical propensity of the molecule along with its cell penetration power, which in turn aids to improve its biological activity. Furthermore, hydrophobic AAs consisting non-polar side chains, like Leu, Val, Ala, Ile, Gly, Pro, etc. can increase the hydrophobicity and helical content of peptides, which in turn can increase their biological activity.23 It has been very well explored that the higher overall hydrophobicity of peptides could favour their penetration into the lipid bilayer of the cell membrane.24 Considering this, we have designed 7a–e which consists hydrophobic AAs with non-polar side chains, viz., Gly (7a), Ala (7b), Val (7c), Leu (7d), and Pro (7e). The hydrophobicity of each derivative will be calculated and it is anticipated that higher the hydrophobicity higher is the biological activity of the peptide derivative. Additionally, AAs with polar and hydrophobic side chains, like Ser, Thr, Tyr, Asn, and Gln enable peptide derivatives to have an amphipathic character which is a really important feature for peptide therapeutics to exhibit significant biological activities.25 Thus, in this study, we have designed one derivative, 7f, consisting Tyr which has a polar side chain with the anticipation of showing good biological activity.
In this study, we investigate the potential of such cationic AA-enriched short peptides as potent antimicrobial agents. Solid-phase peptide synthesis (SPPS) technology further supports the development of these peptides by enabling precise control over their composition and structure. This study builds upon the foundation of molecular docking insights and SPPS advancements to design, synthesize, and evaluate a new series of cationic amino acid-enriched short peptides. Through a comprehensive set of antibacterial and antifungal assays, coupled with detailed molecular docking simulations, we explore the interactions between these peptides and various pathogens, including Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pyogenes, Candida albicans, and Aspergillus niger.
Material and methods
The 2-chlorotrityl chloride (2-CTC) resin was employed for the synthesis of a series of Cys rich cationic AA-enriched short peptides. Highly efficient loading estimation is obtained from using such resin. This resin is mainly used for the synthesis of short-chain peptides.26 It was purchased from Merck. Fmoc(9-fluorenylmethoxycarbonyl)-protected L-amino acids were utilized and acquired from Sichuan, China. HBTU (2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium) and triisopropylsilane (TIS) were acquired from survival chemical. HOBt·H2O (N-hydroxybenztriazole monohydrate), diisopropylethylamine (DIPEA) and trifluoroacetic acid (TFA) acquired from spectrochem. Phenol acquired from SD fine chemicals. A fully automated CSBio peptide synthesizer (CS136X) was utilized for the synthesis of derivatives.21An open capillary method was utilized to determine the melting points which were uncorrected. The Kaiser test was used for monitoring the deprotection and coupling reactions. A mixture of ethyl acetate and n-hexanes was used for the purification of the derivatives. Shimadzu liquid chromatography mass spectrometry (LC-MS) (at 70 eV) mass spectrometer (ESI) was used to determine the mass spectra. Bruker Avance 400 MHz NMR (nuclear magnetic resonance) spectrometer was used to determine the 1H and 13C NMR spectra using DMSO-d6 (deuterated dimethyl sulfoxide) solvent.21
Experimental section
Synthetic route for the formation of intermediate Fmoc-Cys(Trt)-CT resin 1
The 2-CTC resin was introduced to the peptide synthesizer with a substitution of 1.0 mmol g−1. The resin was initially rinsed with dichloromethane (DCM; 10 volumes) and then drained. After that, DCM (10 volumes) was added, and the reaction mass was stirred for 60 minutes for swelling before being drained. Following this, Fmoc-Cys(Trt)-OH (3.0 equiv.) was dissolved in DCM (8 volumes) and transferred into the reaction vessel. DIPEA (6 equiv.) was subsequently added to the reaction vessel and stirred at 28 °C for 2 hours. The peptidyl resin was filtered after the 2 hours mark and washed twice with DCM and once with dimethylformamide (DMF).27 A solution containing DIPEA, methanol (MeOH), and DCM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) was used to cap the unreacted functional groups of the resin. The loading percentage was monitored using an ultraviolet (UV) spectrophotometer.21
7) was used to cap the unreacted functional groups of the resin. The loading percentage was monitored using an ultraviolet (UV) spectrophotometer.21
Synthetic route for the formation of intermediates 6a–f
The Standard Fmoc protocol was employed to produce the desired intermediates 6a–f through the Solid Phase Peptide Synthesis (SPPS) method. In the SPPS reaction vessel, the Fmoc-Cys(Trt)-CT resin was swollen in 20 volumes of DMF for thirty minutes. The CSBio peptide synthesizer facilitated the synthesis of the intended intermediates. Post-swelling, the following steps were repeated for the synthesis of the intermediates: (a) deprotection of the Fmoc group was carried out using 20% piperidine/DMF (v/v) (10 v). The resin was washed twice (5 minutes and 10 minutes each). The Kaiser test was used to ensure complete deprotection, indicated by the appearance of a blue colour test solution and resin beads. Subsequently, the reaction mass (peptidyl resin) was filtered and washed thrice with DMF, once with iso-propyl alcohol (IPA), and thrice again with DMF. (b) For all the coupling reactions, Fmoc AA (3.0 equiv., 3 mmol concerning initial resin loading), HBTU (3.0 equiv., 3 mmol), HOBt·H2O (3.0 equiv., 3 mmol), and DIPEA (6.0 equiv., 6 mmol) in 8 volumes of DMF were utilized. All the coupling reactions required one hour of stirring. The Kaiser test was used to confirm the completion of the reaction, indicated by a colourless test solution and resin beads. The reaction mass (peptidyl resin) was then filtered and washed five times with DMF to yield fmoc-protected intermediates.28 Additionally, fmoc group deprotection was carried out using 20% piperidine/DMF (v/v) (10 v) to obtain the desired intermediates 6a–i. The Kaiser test was used to confirm the deprotection of the Fmoc group, where blue colour resin beads and test solution were observed, confirming the completion of the step. Following this, the reaction mass (peptidyl resin) was filtered and washed five times with DMF to produce 6a–i.21Synthetic route for the synthesis of desired cationic AA-enriched short peptides 7a–f
A cleavage cocktail consisting of a mixture of TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TFA (10
TFA (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80) (10 mL g−1) was utilized for the cleavage of the sidechain and CTC resin.29 The protected peptidyl resin was stirred in the cleavage cocktail for 3 hours at 27 ± 2 °C. After 3 h, the reaction mass was filtered, and the filtrate obtained was precipitated using chilled diisopropyl ether (DIPE) (50 v). Subsequently, the reaction mass was stirred at 0–5 °C for 2 hours, filtered, and washed thrice with chilled DIPE (10 v). Finally, the wet cake was dried at 30 °C under vacuum to yield the desired products.21
80) (10 mL g−1) was utilized for the cleavage of the sidechain and CTC resin.29 The protected peptidyl resin was stirred in the cleavage cocktail for 3 hours at 27 ± 2 °C. After 3 h, the reaction mass was filtered, and the filtrate obtained was precipitated using chilled diisopropyl ether (DIPE) (50 v). Subsequently, the reaction mass was stirred at 0–5 °C for 2 hours, filtered, and washed thrice with chilled DIPE (10 v). Finally, the wet cake was dried at 30 °C under vacuum to yield the desired products.21
Results and discussion
Chemistry
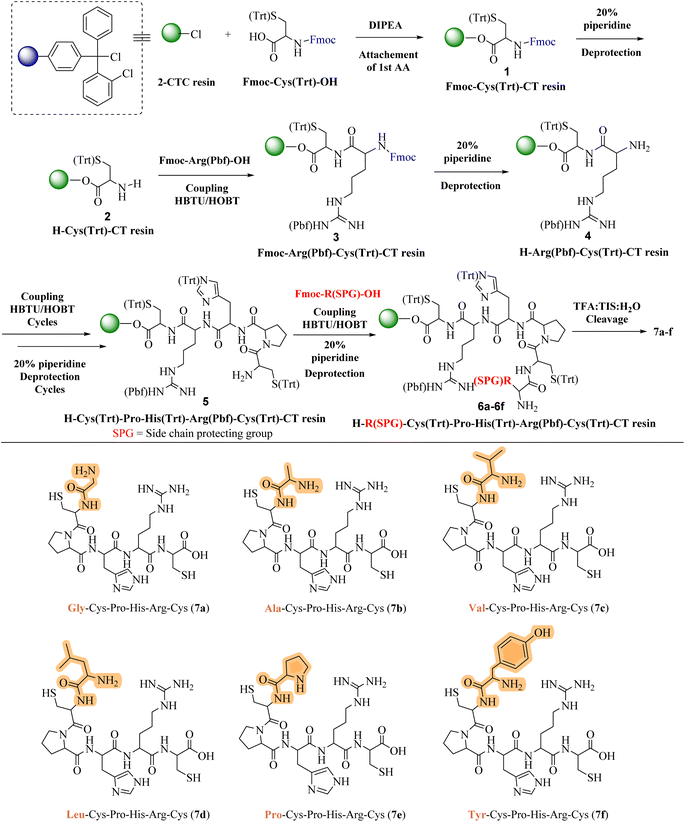
In response to the escalating challenge of antimicrobial resistance, our study embarked on the synthesis of a novel series of cationic short peptides, enriched with cysteine and denoted as compounds 7a–f. Utilizing solid-phase peptide synthesis (SPPS) with the HBTU/HOBt coupling protocol, we successfully synthesized these peptides in a stable, powdered form (Scheme 1).The synthesis commenced with the synthesis of Fmoc-Cys(Trt)-2-CT resin 1. This was achieved by attaching Fmoc-Cys(Trt)-OH to the 2-CTC resin, utilizing DIPEA and DCM as the solvent. Following this, the Fmoc protective group was removed from compound 1 using a 20% solution of piperidine in DMF. This deprotection step yielded the H-Cys(Trt)-2-CT resin 2, revealing the free amino (–NH2) group at its N-terminus. The successful removal of the Fmoc group was confirmed by a positive Kaiser test, indicated by the transformation of the test solution and resin to a dark blue color. After the initial deprotection, Fmoc-Arg(Pbf)-OH was coupled to compound 2 under conditions facilitated by HBTU/HOBt, leading to the formation of Fmoc-Arg(Pbf)-Cys(Trt)-2-CT resin 3. The completion of this coupling reaction was verified through the Kaiser test, with a colorless test solution and resin beads indicating completion of the reaction. Further removal of the Fmoc group from compound 3 resulted in the formation of H-Arg(Pbf)-Cys(Trt)-2-CT resin 4.
Continuing the synthesis, in situ coupling of the compound 4 with the next set of amino acids (AAs)—namely histidine, proline, and an additional cysteine—was conducted under the established HBTU/HOBt conditions, followed by the deprotection of the Fmoc group. This iterative process yielded the H-Cys(Trt)-Pro-His(Trt)-Arg(Pbf)-Cys(Trt)-2-CT resin intermediates 5. Subsequently, six different protected amino acids (Fmoc-R(SPG)-OH) were incorporated using the same HBTU/HOBt conditions, with subsequent deprotection of the Fmoc group. This sequence of actions produced the H–R(SPG)-Cys(Trt)-Pro-His(Trt)-Arg(Pbf)-Cys(Trt)-2-CT resin intermediates 6a–f.
The culmination of the synthesis was marked by global cleavage, performed over three hours at room temperature with a cleavage cocktail of TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TFA in a 10
TFA in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 ratio. This step precipitated the desired short peptides through DIPE, effectively separating them from the CTC resin and side-product groups (SPGs). This step is also known as “acidolysis.” Purification using ethyl acetate and n-hexane refined the crude product, resulting in the isolation of compounds 7a–f with yields ranging from 78% to 89%.
80 ratio. This step precipitated the desired short peptides through DIPE, effectively separating them from the CTC resin and side-product groups (SPGs). This step is also known as “acidolysis.” Purification using ethyl acetate and n-hexane refined the crude product, resulting in the isolation of compounds 7a–f with yields ranging from 78% to 89%.
Each peptide, depicted in Scheme 1, underwent thorough characterization via 1H, 13C NMR (ESI†), and mass spectral analysis, affirming the integrity of their structures. The series encompassed peptides with varied AA substitutions—Gly in 7a, Ala in 7b, Val in 7c, Leu in 7d, Pro in 7e, and Tyr in 7f—to explore and compare their antimicrobial activities. The inclusion of different Fmoc-protected AAs and SPGs at strategic synthesis points introduced a diversity of structural features crucial for a comprehensive biological evaluation.
Docking study
Candida albicans, commonly known as C. albicans, exists as a commensal organism, coexisting harmoniously as part of the natural human microflora in approximately 50% of the population.30 While yeast is generally benign in individuals with robust health, it has the potential to transform into a pathogen, instigating life-threatening infections in those who are immunocompromised or debilitated. The proteolytic activity stands out as a crucial virulence factor associated with C. albicans. This proteolytic activity is linked to the secreted aspartic proteinases (Saps) of C. albicans, comprising a minimum of 10 distinct members. Sap1–8 are released into the extracellular space, while Sap 9–10 manifest as glycosylphosphatidylinositol (GPI)-anchored membrane-bound proteins. Several studies have substantiated the significance of Sap5 in contributing to virulence during both local and systemic infections caused by C. albicans.31Docking protocol was cross-validated by redocking a ligand back into its binding site in the protein structure to compare the predicted binding pose with the experimentally observed pose. Both, co-crystalized ligand and docked ligand were having RMSD value of 1.82 Å. The docking study of the cationic amino acid-enriched short peptides towards the aspartic proteinase (Sap 5) from C. albicans revealed that short peptides formed multiple interactions with the Sap 5 enzyme (Fig. 2 and 3). Peptide 7e showed the highest docking score of the −10.198 kcal mol−1 towards the Sap 5 enzyme. It formed 09 hydrogen bond interactions with the 07 amino acids of Sap 5 enzyme, namely Asp 218, Thr 221, Thr 222, Arg 299, Glu 300, Gly 85, and Asp 86. Compound 7d, which was found to be a potent compound of the series against the in vitro assay against the C. albicans, was having a docking score of −8.52 kcal mol−1 and showed interaction with the 07 amino acids via the hydrogen bonding as shown in Table 1. This interaction includes the following amino acid residues: Glu 132, Asp 303, Lys 83, Gly 85, Asp 86, Gly 220, and Lys 192 (Fig. 2).32
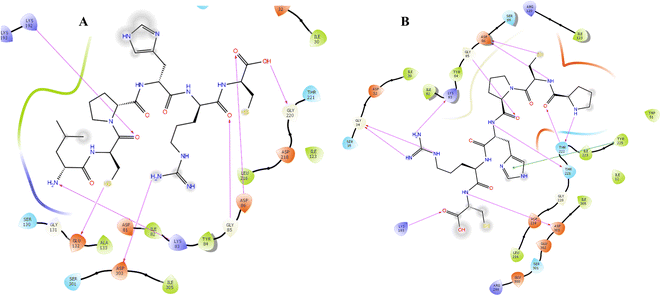 | ||
| Fig. 2 Docking interaction of compound 7d (A) and 7e (B) with the aspartic proteinase (Sap 5) from C. albicans. | ||
| Title | Docking score (kcal mol−1) | Glide emodel | Glide energy |
|---|---|---|---|
| 7e | −10.198 | −122.438 | −79.703 |
| 7d | −8.52 | −119.225 | −82.513 |
| 7a | −8.344 | −112.734 | −85.848 |
| 7b | −8.197 | −120.133 | −78.636 |
| 7c | −8.045 | −104.461 | −69.745 |
| 7f | −6.771 | −94.334 | −72.8 |
MD simulation result
Molecular dynamic simulation (MDS) is a powerful tool for studying ligand binding, protein folding, and conformational changes.33 It can reveal the positions of all the atoms at femtosecond temporal resolution and provide accurate estimates of ligand binding affinities. The MD simulation was performed for 200 ns to gain insights into the conformational changes that occur in the Sap 5 enzyme when compound 7d binds into the pocket of the enzyme. A single file of compound 7d in complex with Sap 5 enzyme was saved in the pdb file format and later subjected to the 200 ns time of simulation.To conduct an MDS, a system was constructed using the SPC solvent model. The SPC model is a widely used explicit solvent model that employs a combination of interactions, such as van der Waals and electrostatic forces, to represent the behavior of solvent molecule.34 The system was set up with an orthorhombic boundary condition and a box size of 5 × 5 × 5 Å. This boundary condition is a type of periodic boundary condition that closely resembles a cubic cell in use. To achieve system neutralization, counter ions (13 Na+ and 18 Cl−) were introduced. This process is essential to balance the system's overall charge, particularly in the presence of a charged biomolecular system, ensuring that the electrostatic energy is accurately calculated during the simulation.35 The system was then prepared for the 200 ns production phase of the MDS through a series of minimization and equilibration steps under NPT conditions. After the minimization process, a 200 ns MDS was performed to investigate the conformational stability and changes of the complex involving Sap 5 and compound 7d.
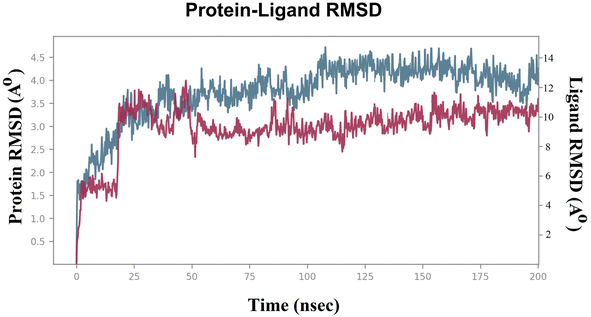
To evaluate the stability of the system during the MDS, parameters such as the “root mean square deviation (RMSD)” and “root mean square fluctuation (RMSF)” were calculated for both the Sap 5 enzyme and compound 7d.36,37 The RMSD measures the deviation of the protein structure from its initial conformation, while the RMSF measures the fluctuation of each atom in the protein over time.22,23 These parameters are commonly used to assess the stability of the system during the simulation and to identify regions of the protein that undergo significant conformational changes upon ligand binding.22,23 Lower RMSD values indicate a more stable system.22,23 In the case of the compound 7d-Sap 5 complex, the ligand's RMSD initially exhibited fluctuations ranging from 4.5 Å to 6.0 Å (net RMSD 1.5 Å) during the first 5 ns, primarily due to equilibration (Fig. 3). The fluctuations in ligand RMSD observed around the 25 ns were likely due to the flexibility of the peptide (Fig. 3). From 50 ns to 200 ns, the RMSD of the compound 7d-Sap 5 complex remained relatively constant, fluctuating between 8.5 Å and 10.5 Å (net RMSD 2.0 Å) (Fig. 3).
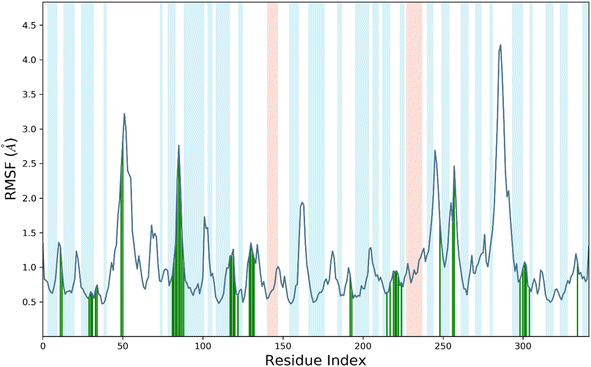
RMSF, or root mean square fluctuation, serves as an indicator of protein structure flexibility, which is crucial for assessing system stability during MDS.22,23 It measures the fluctuations in backbone and ligand atoms throughout the 200 ns MDS of the compound 7d-Sap 5 complex. This analysis provides insights into the dynamic behavior of the complex, allowing for the assessment of its stability and the identification of conformational changes over the course of the simulation.22,23 The analysis in Fig. 4 indicates that there were no significant fluctuations in the amino acid residues upon the binding of compound 7d to the active site of Sap 5. This suggests that the binding of compound 7d did not induce substantial conformational changes in the protein structure, and the complex remained relatively stable throughout the simulation. The RMSF values for the protein's backbone residues within the catalytic domain ranged from 0.5 Å to 1.8 Å (Fig. 4). Slightly higher fluctuations were observed around the 50 residue index and 80–90 residue index, where the RMSF reached 3 Å. Compound 7d interacted with 42 amino acids of the Sap 5, including Ala-11, Thr-13, Val-29, Asp-32, Gly-34, Ser-35, Lys-50, Trp-51, Asp-81, Lys-83, Tyr-84, Gly-85, Asp-86, Gly-87, Ser-88, Tyr-89, Thr-117, Ala-119, Arg-120, Lys-121, Ile-12, Ser-130, Gly-131, Glu-132, Lys-192, Lys-193, Leu-216, Asp-218, Ser-219, Gly-220, Thr-221, Thr-222, Tyr-225, Asn-249, Lys-257, Thr-258, Ile-298, Ser-301, Gly-302, Asp-303, Ile-305 and Glu-335 (Fig. 4). The RMSF values for all of these interacting residues were below 1.8 Å, except for Lys-50, Trp-51, Asp-81, Lys-83, Tyr-84, Gly-85, Asp-86, Gly-87, Ser-88, and Tyr-89, which exhibited an RMSF from 2.8–3.2 Å, as indicated by the green vertical bars as shown in Fig. 4.
We conducted a detailed analysis of the interaction between compound 7d and Sap 5 throughout the 200 ns simulation (Fig. 5). This analysis aimed to provide insights into the dynamic behaviour of the complex and the specific regions that contribute most to its flexibility during the simulation. The Sap 5 enzyme exhibited multiple interactions with compound 7d, with the most significant interaction observed between the Gly-85, Ser-88, and Lys-50 residues and the peptide backbone of compound 7d. The imidazole ring of compound 7d formed a hydrogen bond interaction with the Asp-218 residue of Sap-5 (Fig. 5). We have also measured the radius of gyration of the compound 7d during 200 ns time of simulation. Radius of gyration indicates the overall fluctuation of ligand and on an average, it was found to be 0.8 Å (5.6 to 6.4 Å) (Fig. 5).
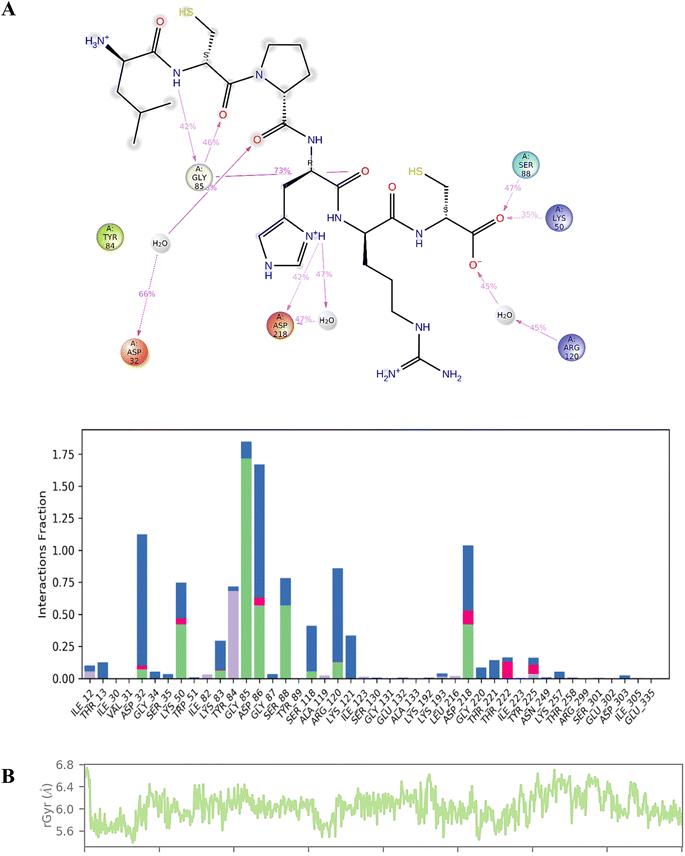 | ||
| Fig. 5 (A) Interaction profile of the compound 7d with Sap 5 enzyme during 200 ns time of simulation; (B) radius of gyration of compound 7d during 200 ns time of simulation. | ||
Docking and MD simulation procedure
The Glide SP docking molecular docking tool of Schrödinger Inc., USA was employed for molecular docking study. Protein preparation was conducted using the ‘protein preparation wizard’ within Maestro 8.0 to optimize the protein structure of Sap 5 enzyme (2QZX) using the OPLS 2005 force field. The grid was meticulously prepared by centering the ligand within the crystal structure using default box dimensions. The ligands were built using the “build panel of LigPrep 2.2” module, which generates the low energy conformer employing the OPLS 2005 force field. The low-energy conformers were meticulously docked into the generated grid using the standard precision (SP) method.32To conduct MDS, a solitary file containing the compound 7d in complex with the Sap5 enzyme was saved in the pdb file format. Subsequently, it underwent a 200 ns simulation period using the SPC solvent model. The system was set up with an orthorhombic boundary condition and a box size of 5 × 5 × 5 Å. The system was neutralized by 13 Na+ and 18 Cl− counter ions. The system was then prepared for the 200 ns production phase of the MDS through a series of minimization and equilibration steps under NPT conditions. After the minimization process, a 200 ns MDS was performed to investigate the conformational stability and changes of the ligand–receptor complex.33–37
Determination of peptide hydrophobicity
The hydrophobicity values of designed cationic AA enriched short peptide 7a–f were theoretically calculated using the relevant equations. The Eisenberg method38 was used to calculate the mean hydrophobicity value for each cationic AA enriched short peptide by employing the following eqn (1):
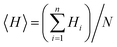 | (1) |
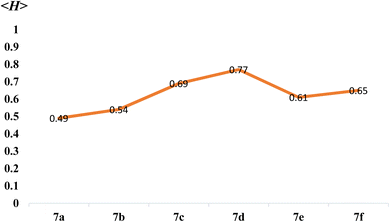
As mentioned earlier in the article, higher overall hydrophobicity of peptides could favour their penetration into the lipid bilayer of the cell membrane and thus increase their biological activity. As shown in Table 2, theoretically, compound 7d showed the highest mean hydrophobicity value of 0.77 among all synthesized cationic AA enriched short peptides 7a–f and is expected to show the highest biological activity. Whereas, theoretically, peptide derivative 7a showed the least mean hydrophobicity value of 0.49 (Fig. 6) and is expected to show the least biological activity among all the peptide derivatives. It is well known fact that higher the <H> value, greater is the hydrophobicity of the peptide sequence.
Biological evaluation
The microorganisms, including Escherichia coli MTCC 443, Pseudomonas aeruginosa MTCC 1688, Streptococcus pyogenes MTCC 442, Staphylococcus aureus MTCC 96, Aspergillus niger MTCC 282, and Candida albicans MTCC 227, were obtained from King Abdullah University Hospital (KAUH), Irbid, Jordan. They were stored at −70 °C in trypticase-soy broth with 20% glycerol from BBL Microbiology Systems (Cockeysville, Md, USA) until required for batch susceptibility testing. To ensure purity and viability, the organisms were thawed and subcultured three times. The in vitro antimicrobial activities of the synthesized compounds 7a–f were studied using the conventional Mueller Hinton Broth-microdilution method.40 This method is used for quantitative antimicrobial susceptibility testing and to determine the minimum inhibitory concentration (MIC) of antimicrobial agents. The test involved screening microorganisms for visible growth in broth, consisting of different dilutions of the antimicrobial agents. The MIC value indicates the lowest concentration of an antimicrobial agent that inhibits visible growth within a specified time frame. The microbroth dilution method offers several advantages over the macrobroth dilution method, including miniaturization and automation through the use of disposable small plastic “microdilution” trays. Its practicality and popularity in research are attributed to advantages such as reproducibility, prepared panels, cost-effective reagents, and space optimization resulting from its miniaturization.41,42The procedure for antibacterial activity assay
The strains employed in the study were recently acquired and stored under optimal conditions. The compounds underwent screening for their antibacterial activity in triplicate experiments against the bacteria, employing different concentrations of 1000, 500, 250, and 200 μg mL−1 substances exhibiting effective inhibition were subsequently subjected to dilution and further testing. The antibacterial assay employed the Mueller Hinton Broth dilution method. A suspension of the respective bacteria at a concentration of 10 μg mL−1 was inoculated on suitable media, and growth was monitored following incubation at 37 °C for one to two days. The test mixture was intended to contain 108 cells per mL. Ampicillin served as the standard drug for assessing antibacterial activity in this study.40The procedure for antifungal activity assay
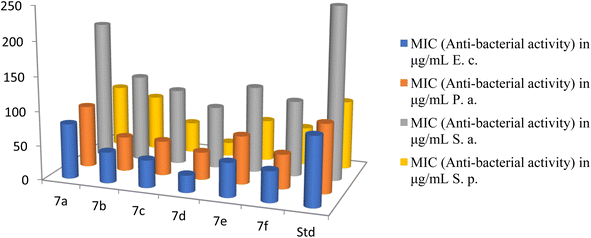
The strains employed in this study were recently acquired and appropriately stored. The substances underwent screening for their anti-fungal properties in triplicate experiments against the fungi at varying concentrations of 1000, 500, 250, and 200 μg mL−1. The active compounds were further diluted and subjected to additional testing. The antifungal assay utilized the Muller Hinton Broth dilution method. Fungal growth was monitored using Sabouraud's dextrose broth at 28 °C under aerobic conditions for a period of 48 hours. Nystatin served as the standard for comparative analysis.40The antibacterial activities of novel cationic AA enriched short peptide conjugate 7a–f against all bacterial strains are shown in Table 3. The cationic peptide conjugate 7d containing Leu was found to be 4 times more potent against E. coli and showed an excellent activity with the highest MIC value of 25 μg mL−1 among all the synthesized peptide conjugates as compared with the standard drug Ampicillin which had a MIC of 100 μg mL−1 against the same bacterial strain E. coli (Fig. 7). The compound 7c with Val showed an excellent activity with MIC value of 40 μg mL−1, 2.5 times more potency against E. coli as compared with standard Ampicillin. The cationic peptide conjugates 7b, 7e, and 7f also exhibited good activities with MIC values (in μg mL−1) of 45, 50 and 45, respectively, and were around 2 times more potent than the control drug. Against P. aeruginosa, 7d exhibited the highest activity among all the peptide derivatives with MIC value of 40 μg mL−1, 2.5 times more potency as compared with the standard drug Ampicillin which had a MIC of 100 μg mL−1 against the same. Moreover, compounds 7b, 7c and 7f also exhibited good activity with MIC values of 50 μg mL−1 and were 2 times more potent than the control drug. Furthermore, among all the synthesized cationic AA enriched short peptides, 7d showed the best antibacterial activity against Staphylococcus aureus with MIC value of 90 μg mL−1 as compared to the standard drug Ampicillin (250 μg mL−1). The other peptide derivatives, 7b, 7c, 7e, and 7f, were almost twice potent as the control drug against Staphylococcus aureus with MIC values (in μg mL−1) of 125, 110, 125, and 110, respectively. For the activity against Streptococcus pyogenes, the derivative 7d showed an excellent activity with a MIC value of 20 μg mL−1 and was five time more potent as compared with a standard drug which had a MIC of 100 μg mL−1. The compound 7c also showed good activity with MIC value of 45 μg mL−1, 2 times more potency as compared with standard Ampicillin, whereas derivatives 7b, 7e and 7f exhibited moderate activities (MIC of 80, 60 and 55 μg mL−1, respectively) as compared to the control drug.
| Compound | MIC (anti-bacterial activity) in μg mL−1 | MIC (anti-fungicidal activity) in μg mL−1 | ||||
|---|---|---|---|---|---|---|
| E. c. | P. a. | S. a. | S. p. | C. a. | A. n. | |
| a E. c.: Escherichia coli, P. a.: Pseudomonas aeruginosa, S. a.: Staphylococcus aureus, S. p.: Streptococcus pyogenes, C. a.: Candida albicans, A. n.: Aspergillus niger. | ||||||
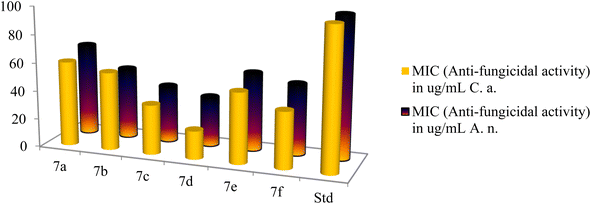
| 7a | 80 | 90 | 200 | 90 | 60 | 65 |
| 7b | 45 | 50 | 125 | 80 | 55 | 50 |
| 7c | 40 | 50 | 110 | 45 | 35 | 40 |
| 7d | 25 | 40 | 90 | 20 | 20 | 35 |
| 7e | 50 | 70 | 125 | 60 | 50 | 55 |
| 7f | 45 | 50 | 110 | 55 | 40 | 50 |
| Ampicillin | 100 | 100 | 250 | 100 | — | — |
| Nystatin | — | — | — | — | 100 | 100 |
From the antibacterial study data of peptide derivatives 7a–f against all four bacterial strains, it was seen that the peptide conjugate 7a showed almost same activities as the standard drug Ampicillin and was least active among all the synthesized derivatives, whereas the peptide derivative 7d was the most potent among them. Cationic AA enriched short peptides containing hydrophobic AAs which has non-polar sidechains, composed mostly of carbon and hydrogen, like Leu, Val, Ala, Pro, are known for their excellent cell-penetrating ability and biological activities.43–45 This is well supported by the theoretical calculations of the mean hydrophobicity of synthesized peptides 7a–f (Table 2), in which derivative 7d exhibited the highest mean hydrophobicity value of 0.77 and was expected to be the most potent among all the synthesized peptides, whereas the peptide derivative 7a showed the lowest mean hydrophobicity value of 0.49 and was expected to show the least potency against all bacterial strains. Similar to compound 7d, compounds 7c and 7f showed higher mean hydrophobicity 0.69 and 0.65, respectively, due to the presence of higher number of hydrocarbons in their side chains and thus, showed better cell penetrating ability and in turn showed better antibacterial activity as compared to 7a, 7b and 7e, which has lesser number of hydrocarbons in their sidechains and thus, their ability to penetrate cell membrane is weaker, which explains their weaker activity against bacterial strains.
The antifungal activities of cationic AA enriched short peptide conjugates 7a–f against all the fungal strains are shown in Table 3. Against Candida albicans, the peptide conjugate 7d was the most active with an excellent MIC value of 20 μg mL−1 and was five times more potent as compared to Nystatin which was active at 100 μg mL−1 (Fig. 8). Peptide conjugates 7c, 7e, and 7f also exhibited good activities against Candida albicans with MIC values of 35, 50 and 40 μg mL−1, respectively. Moreover, peptide derivatives 7a and 7b showed moderate activities with MIC values of 60 and 55 μg mL−1, respectively, as compared to the standard drug Nystatin. Against the second fungal strain Aspergillus niger, the peptide conjugate 7d was the most potent with MIC value of 35 μg mL−1 and was almost three times more potent as compared to Nystatin which was active at 100 μg mL−1. Compounds 7b, 7c and 7f also displayed good inhibition against Aspergillus niger with a MIC value of 50, 40 and 50 μg mL−1, respectively, as compared to the control drug. However, synthesized derivatives 7a and 7e showed moderate activities with MIC values of 65 and 55 μg mL−1 against Aspergillus niger.
From the antifungal study results of peptide derivatives 7a–f against both fungal strains, it was seen that the peptide conjugate 7a was least active among all the synthesized derivatives, whereas the peptide derivative 7d was the most potent among them. For this aspect, the antifungal study results showed the same trend as the antibacterial study. The antifungal study results support the theoretical prediction draw by the hydrophobicity data of all cationic AA enriched short peptide derivatives, where 7d was expected to be the most potent against both fungal strains among all the synthesized peptides due the highest mean hydrophobicity value of 0.77, whereas the peptide derivative 7a showed the least potency against both fungal strains due to the lowest mean hydrophobicity value of 0.49. Similar to compound 7d, compounds 7c and 7f showed higher mean hydrophobicity 0.69 and 0.65, respectively, due to the presence of higher number of hydrocarbons in their side chains and thus, showed better cell penetrating ability and in turn showed better antifungal activity as compared to 7a, 7b and 7e, which has lesser number of hydrocarbons in their sidechains and thus, their ability to penetrate cell membrane is weaker, which explains their weaker activity against fungal strains.
Conclusions
In conclusion, our research underscores the significant potential of cationic amino acid-enriched short peptides in combating antimicrobial resistance, leveraging solid-phase peptide synthesis for its adaptability and scalability. Extensive biological evaluations against key clinical pathogens have unveiled a spectrum of antimicrobial activity, highlighting sequences with outstanding inhibitory effectiveness. Analyses of peptide hydrophobicity have offered crucial insights, clarifying the role of hydrophobic amino acids in enhancing antimicrobial efficacy. Furthermore, molecular docking studies have provided essential mechanistic understanding, connecting empirical outcomes to theoretical perspectives and underscoring the impact of structural attributes on peptide functionality. As we advance in the antimicrobial research domain, these peptides emerge as innovative assets with broad implications for medicinal chemistry and microbiology. In the face of growing antibiotic resistance challenges, this study contributes vital insights and opens new pathways for the development of effective antimicrobial strategies, positioning cationic amino acid-enriched short peptides as pivotal in reshaping future infection control and therapeutic approaches, and igniting renewed hope for more efficacious antimicrobial interventions.Author contributions
All the authors are responsible for the following: study conception and design, data collection, analysis, interpretation of results and manuscript preparation. All the authors reviewed the results and approved the final version of the manuscript.Conflicts of interest
The authors declare no conflicts of interest regarding the publication of this paper.Acknowledgements
The authors thank University Grants Commission, New Delhi, and Marwadi University, for technical and academic support. Also, TJP acknowledges CONAHCyT for a postdoctoral fellowship (I1200/320/2022-MOD.ORD./09/2022). We are grateful to Dr Juan Luis Monribot-Villanueva for his expertise in HRMS-QTOF analysis.References
- For selected reviews on antimicrobial resistance, see: (a) P. Butaye, E. V. Duijkeren, J. Prescott and S. Schwarz, Antimicrobial resistance in bacteria from animals and the environment, Vet. Microbiol., 2014, 171, 269–272 CrossRef PubMed; (b) R. Patini, G. Mangino, L. Martellacci, G. Quaranta, L. Masucci and P. Gallenzi, The Effect of Different Antibiotic Regimens on Bacterial Resistance: A Systematic Review, Antibiotics, 2020, 9, 22 CrossRef CAS PubMed; (c) R. Urban-Chmiel, A. Marek, D. Stępień-Pyśniak, K. Wieczorek, M. Dec, A. Nowaczek and J. Osek, Antibiotic Resistance in Bacteria-A Review, Antibiotics, 2022, 11, 1079 CrossRef CAS PubMed; (d) M. Chatzopoulou and L. Reynolds, The Role of Antimicrobial Restrictions in Bacterial Resistance Control: A Systematic Literature Review, J. Hosp. Infect., 2020, 104, 125–136 CrossRef CAS PubMed.
- (a) R. M. V. Harten, R. Willems, N. Martin and A. Hendrickx, Multidrug-Resistant Enterococcal Infections: New Compounds, Novel Antimicrobial Therapies?, Trends Microbiol., 2017, 25, 467–479 CrossRef PubMed; (b) G. Cheng, M. Dai, S. Ahmed, H. Hao, X. Wang and Z. Yuan, Antimicrobial Drugs in Fighting against Antimicrobial Resistance, Front. Microbiol., 2016, 7, 470 Search PubMed; (c) E. Kamali, A. Jamali, A. Ardebili, F. Ezadi and A. Mohebbi, Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa, BMC Res. Notes, 2020, 13, 27 CrossRef CAS PubMed.
- For selected reviews on short peptides and their antimicrobial activities, see: (a) N. H. Ishak and N. M. Sarbon, A Review of Protein Hydrolysates and Bioactive Peptides Deriving from Wastes Generated by Fish Processing, Food Bioprocess Technol., 2017, 11, 2–16 CrossRef; (b) W. Zhang, Y. Zhang, R. Wang, P. Zhang, Y. Zhang, E. Randell, M. Zhang and Q. Jia, A review: Development and application of surface molecularly imprinted polymers toward amino acids, peptides, and proteins, Anal. Chim. Acta, 2022, 1234, 340319 CrossRef CAS PubMed; (c) S. Yang, M. Wang, T. Wang, M. Sun, H. Huang, X. Shi, S. Duan, Y. Wu, J. Zhu and F. Liu, Self-assembled short peptides: Recent advances and strategies for potential pharmaceutical applications, Mater. Today Bio, 2023, 20, 100644 CrossRef CAS PubMed; (d) X. Hu, M. Liao, H. Gong, L. Zhang, H. Cox, T. A. Waigh and J. R. Lu, Recent advances in short peptide self-assembly: from rational design to novel applications, Curr. Opin. Colloid Interface Sci., 2020, 45, 1–13 CrossRef CAS; (e) R. Perlikowska, Whether short peptides are good candidates for future neuroprotective therapeutics?, Peptides, 2021, 140, 170528 CrossRef CAS PubMed.
- For selected reviews on antimicrobial peptides (AMP's), see: (a) H. Luong, T. T. Thanh and T. H. Tran, Antimicrobial peptides – Advances in development of therapeutic applications, Life Sci., 2020, 260, 118407 CrossRef CAS PubMed; (b) B. Bechinger and S. Gorr, Antimicrobial Peptides: Mechanisms of Action and Resistance, J. Dent. Res., 2017, 96, 254–260 CrossRef CAS PubMed; (c) D. Fry, Antimicrobial Peptides, Surg. Infect., 2018, 19, 804–811 CrossRef PubMed; (d) D. K. Govindarajan and K. Kandaswamy, Antimicrobial peptides: A small molecule for sustainable healthcare applications. Medicine in Microecology, 2023, 100090 (e) K. Nogrado, P. Adisakwattana and O. Reamtong, Antimicrobial peptides: On future antiprotozoal and anthelminthic applications, Acta Trop., 2022, 235, 106665 CrossRef PubMed.
- For selected articles on mode of action of antimicrobial peptides, see: (a) S. Li, Y. Wang, Z. Xue, Y. Jia, R. Li, C. He and H. Chen, The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review, Trends Food Sci. Technol., 2021, 109, 103–115 CrossRef CAS; (b) J. E. Nielsen, V. A. Bjørnestad, V. Pipich, H. Jenssen and R. Lund, Beyond structural models for the mode of action: How natural antimicrobial peptides affect lipid transport, J. Colloid Interface Sci., 2021, 582, 793–802 CrossRef CAS PubMed; (c) M. Wenzel, P. Schriek, P. Prochnow, H. B. Albada, N. Metzler-Nolte and J. E. Bandow, Influence of lipidation on the mode of action of a small RW-rich antimicrobial peptide, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 1004–1011 CrossRef CAS PubMed.
- For selected reviews on cationic peptides, see: (a) U. L. Urmi, A. K. Vijay, R. Kuppusamy, S. Islam and M. D. P. Willcox, A review of the antiviral activity of cationic antimicrobial peptides, Peptides, 2023, 166, 171024 CrossRef CAS PubMed; (b) M. I. Sajid, M. Moazzam, R. Stueber, S. E. Park, Y. Cho, N. U. A. Malik and R. K. Tiwari, Applications of amphipathic and cationic cyclic cell-penetrating peptides: Significant therapeutic delivery tool, Peptides, 2021, 141, 170542 CrossRef CAS PubMed; (c) A. Vedadghavami, C. Zhang and A. G. Bajpayee, Nanotoday, 2020, 34, 100898 CrossRef CAS PubMed; (d) D. Ciumac, H. Gong, X. Hu and J. R. Lu, Membrane targeting cationic antimicrobial peptides, J. Colloid Interface Sci., 2019, 537, 163–185 CrossRef CAS PubMed; (e) S. Matamouros, S. I. Miller and S. Typhimurium, strategies to resist killing by cationic antimicrobial peptides, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 3021–3025 CrossRef CAS PubMed.
- (a) R. González-Castro, M. Gómez-Lim and F. Plisson, Cysteine-Rich Peptides: Hyperstable Scaffolds for Protein Engineering, ChemBioChem, 2020, 22, 961–973 CrossRef PubMed; (b) C. Navo, A. Asín, E. Gómez-Orte, M. Gutiérrez-Jiménez, I. Compañón, B. Ezcurra, A. Avenoza, J. Busto, F. Corzana, M. Zurbano, G. Jiménez-Osés, J. Cabello and J. Peregrina, Cell-Penetrating Peptides Containing Fluorescent D-Cysteines, Chemistry, 2018, 24, 7991–8000 CrossRef CAS PubMed.
- D. Wu, Y. Gao, Y. Qi, L. Chen, Y. Ma and Y. Li, Cancer Lett., 2014, 351(1), 13–22 CrossRef CAS PubMed.
- S. R. MacEwan and A. Chilkoti, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5(1), 31–48 CAS.
- E. Belnoue, A. A. Leystra, S. Carboni, H. S. Cooper, R. T. Macedo, K. N. Harvey, K. N. Colby, K. S. Campbell, L. A. Vanderveer, M. L. Clapper and M. Derouazi, Cancer, 2021, 13(4), 845 CrossRef CAS PubMed.
- M. Lichtenstein, S. Zabit, N. Hauser, S. Farouz, O. Melloul, J. Hirbawi and H. Lorberboum-Galski, Life, 2021, 11(9), 924 CrossRef CAS PubMed.
- E. Kondo, H. Iioka and K. Saito, Cancer Sci., 2021, 112, 2118–2121 CrossRef CAS PubMed.
- Y. Wang, C.-H. Liu, T. Ji, M. Mehta, W. Wang, E. Marino, J. Chen and D. S. Kohane, Nat. Commun., 2019, 10(1), 804 CrossRef CAS PubMed.
- S. Alizadeh, S. Irani, A. Bolhassani, S. M. Sadat and J. Avicenna, Med. Biotechnol., 2020, 12(1), 8 Search PubMed.
- S. Futaki, T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda and Y. Sugiura, J. Biol. Chem., 2001, 276(8), 5836–5840 CrossRef CAS PubMed.
- J. C. Mai, H. Shen, S. C. Watkins, T. Cheng and P. D. Robbins, J. Biol. Chem., 2002, 277(33), 30208–30218 CrossRef CAS PubMed.
- L. Porosk, P. Arukuusk, K. Põhako, K. Kurrikoff, K. Kiisholts, K. Padari, M. Pooga and U. Langel, Biomater. Sci., 2019, 7(10), 4363–4374 RSC.
- G. Tünnemann, G. Ter-Avetisyan, R. M. Martin, M. Stöckl, A. Herrmann and M. C. Cardoso, J. Pept. Sci., 2008, 14(4), 469–476 CrossRef PubMed.
- (a) V. Apostolopoulos, J. Bojarska, T. Chai, S. Elnagdy, K. Kaczmarek, J. Matsoukas, R. New, K. Parang, O. Lopez, H. Parhiz, C. Perera, M. Pickholz, M. Remko, M. Saviano, M. Skwarczynski, Y. Tang, W. Wolf, T. Yoshiya, J. Zabrocki, P. Zielenkiewicz, M. AlKhazindar, V. Barriga, K. Kelaidonis, E. Sarasia and I. Toth, A Global Review on Short Peptides: Frontiers and Perspectives, Molecules, 2021, 26, 430 CrossRef CAS PubMed; (b) A. Cerrato, S. Aita, A. Capriotti, C. Cavaliere, C. Montone, A. Laganà and S. Piovesana, A new opening for the tricky untargeted investigation of natural and modified short peptides, Talanta, 2020, 219, 121262 CrossRef CAS PubMed; (c) S. R. Tivari, S. V. Kokate, U. B. Shelar and Y. Jadeja, The design, synthesis and biological evaluation of the peptide derivatives containing guanidine moiety with 5-chloro-thiophene-2-carboxylic acid conjugates, Rasayan J. Chem., 2022, 15, 875–884 CrossRef CAS; (d) S. R. Tivari, S. V. Kokate, M. S. Gayke, I. Ahmad, H. Patel, S. G. Kumar and Y. Jadeja, A Series of Dipeptide Derivatives Containing (S)-5-Oxo-pyrrolidine-2-carboxylic acid Conjugates: Design, Solid-Phase Peptide Synthesis, in vitro Biological Evaluation, and Molecular Docking Studies, ChemistrySelect, 2022, 7, e202203462 CrossRef CAS; (e) S. R. Tivari, S. V. Kokate, E. M. Sobhia, S. G. Kumar, U. B. Shelar and Y. Jadeja, A Series of Novel Bioactive Cyclic Peptides: Synthesis by Head-to-Tail Cyclization Approach, Antimicrobial Activity and Molecular Docking Studies, ChemistrySelect, 2022, 7, e202201481 CrossRef CAS.
- H. Yan and F.-E. Chen, Recent Progress in Solid-Phase Total Synthesis of Naturally Occurring Small Peptides, Adv. Synth. Catal., 2022, 364, 1934–1961 CrossRef CAS.
- S. R. Tivari, S. V. Kokate, E. Delgado-Alvarado, M. S. Gayke, A. Kotmale, H. Patel, I. Ahmad, E. M. Sobhia, S. G. Kumar, B. G. Lara, V. D. Jain and Y. Jadeja, A novel series of dipeptide derivatives containing indole-3-carboxylic acid conjugates as potential antimicrobial agents: the design, solid phase peptide synthesis, in vitro biological evaluation, and molecular docking study, RSC Adv., 2023, 13, 24250–24263 RSC.
- C. T. Walsh, Tailoring enzyme strategies and functional groups in biosynthetic pathways, Nat. Prod. Rep., 2023, 40, 326–386 RSC.
- C. Chen, J. Hu, S. Zhang, P. Zhou, X. Zhao, H. Xu, X. Zhao, M. Yaseen and J. R. Lu, Biomaterials, 2012, 33(2), 592–603 CrossRef CAS PubMed.
- K.-Y. Huang, Y.-J. Tseng, H.-J. Kao, C.-H. Chen, H.-H. Yang and S.-L. Weng, Sci. Rep., 2021, 11, 13594 CrossRef CAS PubMed.
- (a) A. Elliott, J. Huang, S. Neve, J. Zuegg, I. Edwards, A. Cain, C. Boinett, L. Barquist, C. Lundberg, J. Steen, M. Butler, M. Mobli, K. Porter, M. Blaskovich, S. Lociuro, M. Strandh and M. Cooper, Nat. Commun., 2020, 11, 3184 CrossRef CAS PubMed; (b) F. Qiu, Y. Chen, C. Tang and X. Zhao, Int. J. Nanomed., 2018, 13, 5003–5022 CrossRef CAS PubMed.
- D. M. M. Jaradat, Thirteen decades of peptide synthesis: key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation, Amino Acids, 2017, 50, 39–68 CrossRef PubMed.
- V. Made, S. Els-Heindl and A. Beck-Sickinger, Automated solid-phase peptide synthesis to obtain therapeutic peptides, Beilstein J. Org. Chem., 2014, 10, 1197–1212 CrossRef PubMed.
- E. Kaiser, R. L. Colescott, C. D. Bossinger and P. I. Cook, Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides, Anal. Biochem., 1970, 34, 595–598 CrossRef CAS PubMed.
- R. Behrendt, P. White and J. Offer, Advances in Fmoc solid-phase peptide synthesis, J. Pept. Sci., 2016, 22, 4–27 CrossRef CAS PubMed.
- (a) J. R. Naglik, S. J. Challacombe and B. Hube, Candida albicans secreted aspartyl proteinases in virulence and pathogenesis, Adv. Synth. Catal., 2003, 67, 400–428 CAS; (b) M. Schaller, C. Borelli, H. C. Korting and B. Hube, Hydrolytic enzymes as virulence factors of Candida albicans, Mycoses, 2005, 48, 365–377 CrossRef CAS PubMed.
- C. Borelli, E. Ruge, J. H. Lee, M. Schaller, A. Vogelsang, M. Monod, H. C. Korting, R. Huber and K. Maskos, X-ray structures of Sap1 and Sap5: structural comparison of the secreted aspartic proteinases from Candida albicans, Proteins, 2008, 72, 1308–1319 CrossRef CAS PubMed.
- (a) H. M. Patel, M. Palkar and R. Karpoormath, Exploring MDR-TB inhibitory potential of 4-aminoquinazolines as mycobacterium tuberculosis N-acetylglucosamine-1-phosphate uridyltransferase (GlmUMTB) inhibitors, Chem. Biodiversity, 2020, 17, e2000237 CrossRef CAS PubMed; (b) H. Patel, K. Dhangar, Y. Sonawane, S. Surana, R. Karpoormath, N. Thapliyal, M. Shaikh, M. Noolvi and R. Jagtap, In search of selective 11 β-HSD type 1 inhibitors without nephrotoxicity: an approach to resolve the metabolic syndrome by virtual based screening, Arabian J. Chem., 2018, 11, 221–232 CrossRef CAS.
- I. Ahmad, D. Kumar and H. Patel, Computational investigation of phytochemicals from Withania somnifera (Indian ginseng/ashwagandha) as plausible inhibitors of GluN2B-containing NMDA receptors, J. Biomol. Struct. Dyn., 2022, 40(17), 7991–8003 CrossRef CAS PubMed.
- I. Ahmad, M. Shaikh, S. Surana, A. Ghosh and H. Patel, p38α MAP kinase inhibitors to overcome EGFR tertiary C797S point mutation associated with osimertinib in non-small cell lung cancer (NSCLC): emergence of fourth-generation EGFR inhibitor, J. Biomol. Struct. Dyn., 2022, 40(7), 3046–3059 CrossRef CAS PubMed.
- A. R. Zala, H. N. Naik, I. Ahmad, H. Patel, S. Jauhari and P. Kumari, Design and synthesis of novel 1, 2, 3-triazole linked hybrids: Molecular docking, MD simulation, and their antidiabetic efficacy as α-Amylase inhibitors, J. Mol. Struct., 2023, 1285, 135493 CrossRef CAS.
- Y. O. Ayipo, I. Ahmad, W. Alananzeh, A. Lawal, H. Patel and M. N. Mordi, Computational modelling of potential Zn-sensitive non-β-lactam inhibitors of imipenemase-1 (IMP-1), J. Biomol. Struct. Dyn., 2022, 29, 1–21 Search PubMed.
- N. C. Desai, A. S. Maheta, A. M. Jethawa, U. P. Pandit, I. Ahmad and H. Patel, Zeolite (Y-H)-based green synthesis, antimicrobial activity, and molecular docking studies of imidazole bearing oxydibenzene hybrid molecules, J. Heterocycl. Chem., 2022, 59(5), 879–889 CrossRef CAS.
- (a) D. Eisenberg, R. M. Weiss and T. C. Terwilliger, The helical hydrophobic moment: a measure of the amphiphilicity of a helix, Nature, 1982, 299(5881), 371–374 CrossRef CAS PubMed; (b) D. Eisenberg, E. Schwarz, M. Komaromy and R. Wall, Analysis of membrane and surface protein sequences with the hydrophobic moment plot, J. Mol. Biol., 1984, 179(1), 125–142 CrossRef CAS PubMed.
- (a) V. Pliška, M. Schmidt and J.-L. Fauchère, Partition coefficients of amino acids and hydrophobic parameters π of their sidechains as measured by thin-layer chromatography, J. Chromatogr. A, 1981, 216, 79–92 CrossRef; (b) J.-L. Fauchère and V. Pliska, Hydrophobic parameters π of amino-acid side chains from the partitioning of N-acetyl-amino-acid amides, Eur. J. Med. Chem., 1983, 18(4), 7 Search PubMed.
- N. C. Desai, K. A. Jadeja, D. J. Jadeja, V. M. Khedkar, P. C. Jha and P. C. Design, synthesis, antimicrobial evaluation, and molecular docking study of some 4-thiazolidinone derivatives containing pyridine and quinazoline moiety, Synth. Commun., 2021, 51, 952–963 CAS.
- B. Kowalska-Krochmal and R. Dudek-Wicher, The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance, Pathogens, 2021, 10, 165 CrossRef CAS PubMed.
- J. H. Jorgensen and M. J. Ferraro, Antimicrobial susceptibility testing: a review of general principles and contemporary practices, Clin. Infect. Dis., 2009, 49, 1749–1755 CrossRef CAS PubMed.
- R. Hadianamrei, M. Tomeh, S. Brown, J. Wang and X. Zhao, Rationally designed short cationic α-helical peptides with selective anticancer activity, J. Colloid Interface Sci., 2021, 607, 488–501 CrossRef PubMed.
- Y. Huang, L. He, H. Jiang and Y. Chen, Role of Helicity on the Anticancer Mechanism of Action of Cationic-Helical Peptides, Int. J. Mol. Sci., 2012, 13, 6849–6862 CrossRef CAS PubMed.
- J. Hu, C. Chen, S. Zhang, X. Zhao, H. Xu, X. Zhao and J. Lu, Designed antimicrobial and antitumor peptides with high selectivity, Biomacromolecules, 2011, 12, 3839–3843 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The spectral data of the study is provided. It includes 1H and 13C NMR, and mass spectral data. See DOI: https://doi.org/10.1039/d3ra08313f |
| This journal is © The Royal Society of Chemistry 2024 |