Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A review of typical biological activities of glycyrrhetinic acid and its derivatives
Liang Chen
 a,
Jingwen Gong
a,
Xu Yong
*b,
Youbin Li
a and
Shuojin Wang
a,
Jingwen Gong
a,
Xu Yong
*b,
Youbin Li
a and
Shuojin Wang
 *a
*a
aHainan Provincial Key Laboratory for Research and Development of Tropical Herbs, Key Laboratory of Tropical Translational Medicine of Ministry of Education, School of Pharmacy Hainan Medical University, No. 3, XueYuan Road, LongHua District, Haikou City, Hainan Province 571199, China. E-mail: wang.shuojin@hainmc.edu.cn
bDepartment of Thoracic Surgery, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, Shanghai 200433, China. E-mail: xuyong@tongji.edu.cn
First published on 22nd February 2024
Abstract
Glycyrrhetinic acid, a triterpenoid compound primarily sourced from licorice root, exhibits noteworthy biological attributes, including anti-inflammatory, anti-tumor, antibacterial, antiviral, and antioxidant effects. Despite these commendable effects, its further advancement and application, especially in clinical use, have been hindered by its limited druggability, including challenges such as low solubility and bioavailability. To enhance its biological activity and pharmaceutical efficacy, numerous research studies focus on the structural modification, associated biological activity data, and underlying mechanisms of glycyrrhetinic acid and its derivatives. This review endeavors to systematically compile and organize glycyrrhetinic acid derivatives that have demonstrated outstanding biological activities over the preceding decade, delineating their molecular structures, biological effects, underlying mechanisms, and future prospects for assisting researchers in finding and designing novel glycyrrhetinic acid derivatives, foster the exploration of structure–activity relationships, and aid in the screening of potential candidate compounds.
Introduction
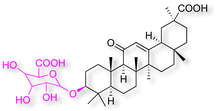
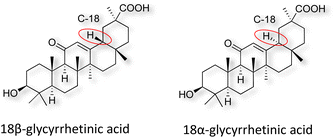
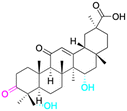
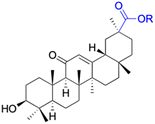
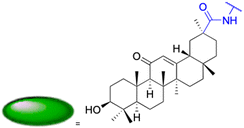
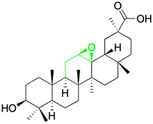
Natural products play a crucial role in the exploration of new drugs as they possess broad-spectrum activity against bacteria, fungi, viruses, cancer, and other diseases, and they exhibit a vast array of chemically diverse structures, which hold the potential to serve as lead compounds in drug discovery. In particular, numerous compounds derived from natural products have already exhibited substantial therapeutic potential in the treatment of specific ailments.1–5 Among these natural products, glycyrrhetinic acid is the triterpenoid aglycone constituent of glycyrrhizinic acid (Fig. 1), derived from the roots of the licorice plant (Glycyrrhiza glabra).6,7 There are two isomers of glycyrrhetinic acid (GA), one is (3β,18β)-3-hydroxy-11-oxoolean-12-en-30-oic acid, often called 18β-glycyrrhetinic acid or enoxolone, denoted by 18β-GA. Another one is (3β,18α)-3-hydroxy-11-oxoolean-12-en-29-oic acid, known as 18α-glycyrrhetinic acid, denoted by 18α-GA, as shown in Fig. 2. 18β-GA is the major bioactive constituent of Glycyrrhiza glabra and has been investigated to possess a wide range of biological activities, including anti-inflammatory, antitumor, antibacterial, antiviral, and antioxidant. Apart from these characteristic activities, glycyrrhetinic acid has been observed to exhibit additional properties, such as anti-diabetic, anticoagulant, immunoregulatory, anti-cholinesterase, antiarrhythmic, and anti-tetanus toxin actions.8However, 18β-GA's poor druggability, including low solubility and bioavailability, limits its clinical use.9–12 To improve the pharmacokinetic properties and enhance the bioactivity, various structural modifications of glycyrrhetinic acid have been carried out to develop novel derivatives for making them attractive candidates for further development as potential drug leads; in the process, extensive studies on the structure–activity relationship (SAR) of 18β-GA and its derivatives have been extensively investigated.13 Furthermore, these modifications focused on altering the chemical structure, including the introduction of functional groups, changes in stereochemistry, and modifications of the aglycone skeleton. Studies on the pharmacological activities of 18β-GA derivatives have shown their potential as therapeutics for various diseases, such as inflammatory diseases, cancer, bacterial and viral infections, diabetes, and liver diseases, especially in the past two years.
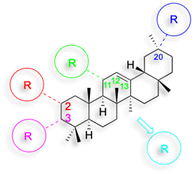
The references incorporated in this review were exclusively sourced from the databases of Google Scholar, PubMed, and Web of Science. The compilation focusing on 18β-GA and its derivatives was based in works published within the temporal span of 2000 to 2023. Significantly, the majority of these citations were published within the most recent half-decade, highlighting the contemporaneity of our curated selection. In addition, we meticulously scrutinized 266 compounds with significant biological activity from a pool of over 500 derivatives sourced from these cited references. To provide a more comprehensive and organized overview, we have compiled tables summarizing the chemical structures and effects or mechanisms of the typical biological activities of 18β-GA and its derivatives, including anti-inflammatory, anti-tumor, antibacterial, antiviral and antioxidant effects. The labeling scheme for the modification sites of all 18β-GA derivatives is described in the form of a diagram. Please refer to Fig. 3 for a visual representation of the labeling scheme.
Anti-inflammatory activity
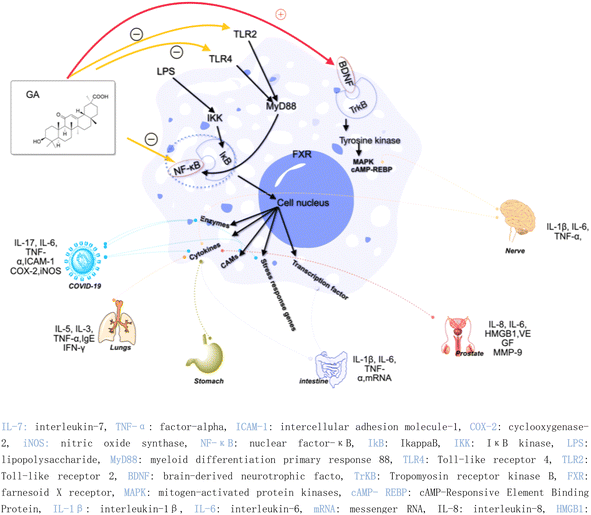
Inflammation is considered to be a driver of many diseases, including arteriosclerosis, cancer, autoimmunity, and chronic infections.14 The inflammatory process involves multiple cell types, signaling pathways, and molecular mechanisms, leading to adverse reactions such as immunosuppression and gastrointestinal problems.15–21 Therefore, the design and optimization of drugs become more complicated. The presence of active ingredients in natural products opens up new opportunities for the development of anti-inflammatory drugs. Extensive research has shown that 18β-GA demonstrates anti-inflammatory effects and holds significant potential as a therapeutic agent for various ailments.22 For instance, 18β-GA inhibits the expression of various inflammatory mediators, such as intercellular adhesion molecule-1 (ICAM-1), tumor necrosis factor-alpha (TNF-α), cyclooxygenase-2 (Cox-2), and inducible nitric oxide synthase (iNOS), by inhibiting the activity of the nuclear factor-κB (NF-κB) pathway.23 Additionally, 18β-GA has been found to reduce the production of inflammatory cytokines by inhibiting the activity of NF-κB and phosphoinositide 3-kinase (PI3K) and inhibiting the production of NO, prostaglandin E2 (PGE2), and reactive oxygen species (ROS) under lipopolysaccharide (LPS) stimulation.24 However, in an Ana-1 mouse macrophage model, 18β-GA induced the expression of Toll-like receptor 4 and activated the TLR-4 signaling pathway via the myeloid differentiation primary response 88 (MYD88) pathway.25In recent years, the research of 18β-GA on anti-inflammation has been deepened. 18β-GA (40 mg kg−1 day−1) has been found to effectively improve lung function in ovalbumin (OVA)-induced asthma mouse model, reduce lung inflammation and inflammatory cell infiltration, and inhibit the phosphorylation of NF-κB in the treatment of airway allergic inflammation. These effects are achieved through a decrease in the levels of interleukin-5 (IL-5) by approximately 40%, interleukin-13 (IL-13) by approximately 30%, and TNF-α by approximately 70%. Additionally, there is an increase in the levels of nuclear factor erythroid 2-related factor2 (Nrf2) by approximately 50% and heme oxygenase 1 (HO-1) by approximately 50%.26 Gupta et al. found that 18β-GA has potential therapeutic effects in treating depression. Specifically, it can improve symptoms caused by chronic unpredictable mild stress by activating the brain-derived neurotrophic factor (BDNF)/Tropomyosin receptor kinase B (TrkB) signaling pathway in the prefrontal cortex (PFC) and hippocampus. This activation leads to a reduction in neuroinflammation, liver biomarkers, and stress hormones while increasing the body weight and brain neurotransmitter concentrations.27
Additionally, the complex of 18β-GA also exhibits remarkable anti-inflammatory activity. Ishida et al. demonstrated that the complex of 18β-GA and hydroxypropyl-β-cyclodextrin can mitigate indomethacin-induced small intestinal injury by reducing TNF-α expression by 27.5%, interleukin-6 (IL-6) by 16.2%, and interleukin-1β (IL-1β) by 17.9% compared to indomethacin-treated tissue.28 The salt of 18β-GA and L-arginine can be formed through a co-solvent evaporation reaction, and a solid dispersion called 18β-GA-SD can be created by adding a polymer solvent, Soluplus®, with a hydrophilic-hydrophobic chemical structure. 18β-GA-SD has higher solubility, cell utilization rate, and bioavailability than 18β-GA itself. Following treatment with 18β-GA-SD, enzyme-linked immunosorbent assay (ELISA) analysis revealed an increase in LPS-induced secretion levels of cytokines such as IL-1β, IL-6, macrophage inflammatory protein-1 (MCP-1), TNF-α, interleukin-23 (IL-23), and interleukin-17A (IL-17A) in RAW 264.7 cells; meanwhile, there was a decrease in the levels of interleukins-4 (IL-4) and -10 (IL-10).11
In the context of COVID-19, 18β-GA has been found to affect the disease by inhibiting the interleukin-17 (IL-17), IL-6, and TNF-α signaling pathways, thereby holding potential as a treatment strategy.29 Another study found that a combination of 18β-GA and vitamin C (VC) treatment for COVID-19 was associated with an increase in immunity and a decrease in inflammatory stress, as well as activation of the T cell receptor signaling pathway, regulation of Fc gamma R-mediated phagocytosis, ErbB signaling pathway, and vascular endothelial growth factor signaling pathway.30 Furthermore, highly biocompatible 18β-GA nanoparticles have been synthesized and have shown promise as a treatment strategy for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections.31 Zhou et al. demonstrated that 18β-GA inhibited the expression of intercellular adhesion molecule-1 (ICAM-1), TNF-α, COX-2, and iNOS, which was attributed to the inhibition of NF-κB expression and the attenuation of NF-κB nuclear translocation.32
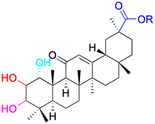
Moreover, another study discovered that 18α-GA suppressed the invasion on Matrigel-coated transwells of DU145 prostate cancer cells by regulating the expression of nu NF-κB (p65), vascular endothelial growth factor (VEGF), and metalloproteinase-9 (MMP-9). 18α-GA also augmented the expression of non-steroidal anti-inflammatory gene-1 (NAG-1) in DU-145 cells, thereby indicating its capacity for anti-inflammatory activity against prostate cancer cells.33 The mechanisms underlying the anti-inflammatory effects of GA discussed above are graphically depicted in Fig. 4. In the realm of hepatoprotective activity, 18β-GA has been shown to mitigate hepatic inflammatory injury caused by hepatitis virus infection by blocking the release of the high mobility group box 1 (HMGB1) cytokine and inhibiting its activity.34,35 Furthermore, 18β-GA has potential as a hepatoprotective agent through activating of Nuclear factor erythroid 2-related factor 2 (Nrf2) and peroxisome proliferator-activated receptor gamma (PPAR-γ), and subsequent suppression of NF-κB, and 18β-GA has been shown to protect the liver from cholestatic liver injury induced by lithocholic acid (LCA) by inhibiting the TLR2/NF-κB pathway and upregulating hepatic farnesoid X receptor (FXR) expression, while reducing inflammation and promoting bile excretion. 18β-GA significantly increased the protein levels of the tubular bile acid (BA) efflux transporter bile salt export pump (BSEP) and the basolateral BA efflux transporters multidrug resistance-associated proteins 3 and 4 (MRP3 and MRP4) but decreased the expression of the BA uptake transporter OATP2A1.23,36–39 Since the hepatic protection effect of 18β-GA is not only realized through the anti-inflammatory mechanism but could also through the antioxidant mechanism, the review about hepatic protection discussion is in the antioxidant part; Fig. 6 depicts all relevant studies.
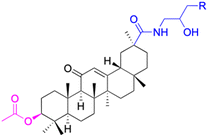
In other investigations, various compounds derived from 18β-GA, such as 1–15 (Table 1), have exhibited anti-inflammatory effects. For instance, Ma et al. identified three major metabolites (compounds 1–3) produced by the microbial transformation of 18β-GA. These metabolites exhibited potent anti-inflammatory activity by inhibiting LPS-induced NO production in mouse microglia BV2 cells.40 The structure and inhibitory activity are shown in Table 1. Another investigation found that compound 4 showed improved pharmacokinetic properties and reduced toxicity in a similar way to fungal metabolism and LPS-induced mouse models.41 Li et al. found that compound 5 decreased the expression of iNOS, COX-2, and mitogen-activated protein kinases (MAPKs) as well as the activation of NF-κB in LPS-stimulated RAW 264.7 cells.42 More recently, Yang et al. investigated the anti-inflammatory effects of compound 6 on ear edema in mice and LPS-stimulated RAW 264.7 macrophages, respectively.43 Compound 6 was shown to decrease approximately 59.69% of 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced ear edema with a gavage treatment of 40.0 mg mL−1, and immunohistochemistry results revealed that this effect was related to the inhibition of TPA-induced upregulation of TNF-α. Compound 7 effectively inhibited the protein and mRNA expression of iNOS and the mRNA expression of TNF-α, IL-6, and IL-1β in LPS-stimulated RAW 264.7 macrophages. Bian et al. investigated the anti-inflammatory effects of compound 8 on LPS-induced RAW 264.7 cells and found that it suppressed the expression of pro-inflammatory cytokines including IL-6, TNF-α, and NO.44 Compounds 9–12 showed significant inhibition activity against NO and IL-6.45–47 Among these compounds, compound 12 was identified as the most potent anti-inflammatory agent, exhibiting a significant reduction in inflammatory cytokine levels in the mouse model of AKI by inhibiting TNF-α and IL-6 in a dose-dependent manner. Compound 13 also has anti-inflammatory activity, and studies have shown that it interacts with proteins in the inflammatory process, such as matrix metalloproteinase MMP9, neutrophil elastase, and thrombin.48 Tu et al. focus on the anti-inflammatory activity of novel 18β-GA derivatives. The study evaluated the derivatives' activity in mouse models of acute inflammation induced by carrageenan. The results showed that several compounds demonstrated significant inhibition of paw edema and leukocyte infiltration.49 The results obtained from both in vitro and in vivo experiments indicate that compound 14 and compound 15 exhibit anti-inflammatory effects by reducing the expression of NO, pro-inflammatory cytokines, and chemokines, such as IL-1β, IL-6, IL-12, TNF-α, MCP-1, and macrophage inflammatory protein-1 alpha (MIP-1α) while increasing the expression of anti-inflammatory cytokine IL-10. Wang et al. introduced Soluplus®-glycyrrhetinic acid solid dispersion, which significantly improves the bioavailability and anti-inflammatory activity of 18β-GA. The solubility of 18β-GA increased with the addition of Soluplus®, and the bioavailability was enhanced 2.61-fold. The anti-inflammatory activity of 18β-GA was also improved by 32.3%.11 Compounds 16–21 have been structurally modified at the C-2 and C-30 carboxyl positions of 18β-GA. These derivatives of 18β-GA have previously demonstrated outstanding anti-inflammatory activity, as seen in Table 1.50–52
| Compounds | 18β-GA | 1 | 2 | 3 |
|---|---|---|---|---|
| Structure | 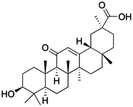 |
 |
 |
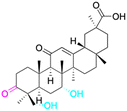 |
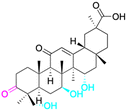
| Effects or mechanisms | 11 β-HSD1: IC50 = 0.778 μM | 1: | 2: | 3: |
| 11 β-HSD2: IC50 = 0.257 μM | NO inhibitory assay in microglia BV2 cells: IC50 = 760 μM | NO inhibitory assay in microglia BV2 cells: IC50 = 940 μM | NO inhibitory assay in microglia BV2 cells: IC50 = 160 μM | |
| Reference | 51 | 40 | 40 | 40 |
| Compounds | 4 | 5 | ||
| Structure |  |
 |
||
| Effects or mechanisms | 4: | 5: | ||
| NO inhibitory assay in RAW 264.7: IC50 = 10.13 μM | Inhibited iNOS, COX-2, MAPKs, and NF-κB in the LPS-stimulated RAW 264.7 cells | |||
| Reference | 46 | 42 | ||
| Compounds | 6 | 7 | 8 | |
| Structure | 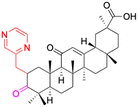 |
 |
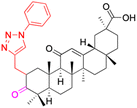 |
|
| Effects or mechanisms | 6: | 7: | 8: | |
| Delayed TPA-induced (20 mg kg−1) overexpression of TNF-α was better than the ibuprofen (40 mg kg−1). For IL-1β, at 40 mg kg−1 was preferable to ibuprofen at 40 mg kg−1 | Inhibited LPS-induced NO production. Inhibited iNOS, TNF-α, IL-6, and IL-1β in LPS-stimulated RAW 264.7 macrophages | Inhibited TPA-induced up regulation of the pro-inflammatory cytokines TNF-α and IL-1β and decreased the expression level of p65 in the NF-κB signaling pathway | ||
| Inhibition at 50 μM: 99.08% | ||||
| Reference | 43 | 53 | 44 | |
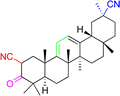
| Compounds | 9 | 10 | ||
| Structure |  |
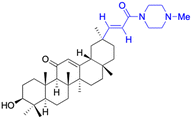 |
||
| Effects or mechanisms | 9: | 10: | ||
| NO inhibitory assay in RAW 264.7: IC50 = 18.5 μM | NO and IL-6 inhibitory activity in RAW 264.7: IC50 = 13.3 μM | |||
| Reference | 45 | 46 | ||
| Compounds | 11 | 12 | ||
| Structure | 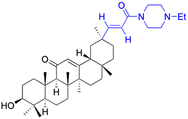 |
 |
||
| Effects or mechanisms | 11: | 12: | ||
| NO and IL-6 inhibitory activity in RAW 264.7: IC50 = 15.5 μM | NO inhibitory assay in RAW 264.7: IC50 = 2.04 μM | |||
| Reference | 46 | 47 | ||
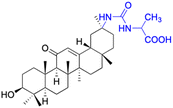
| Compounds | 13 | 14–15 | ||
| Structure |  |
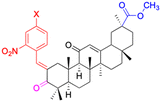 |
||
| Effects or mechanisms | 13: | 14: | ||
| Inhibit inflammatory response (10–50 μM) induced by IFNγ in macrophages in vitro and carrageenan in murine models in vivo, probably by primary interactions with active sites of MMP9, neutrophil elastase, and thrombin | X = Cl, IC50 = 53.0 μM | |||
| 15: | ||||
| X = F, IC50 = 55.4 μM | ||||
| Anti-inflammatory activities through the downregulation of NO, pro-inflammatory cytokines and chemokines (IL-1β, IL-6, IL-12, TNF-α, MCP-1, and MIP-1α) and upregulation of anti-inflammatory cytokines (IL-10). IC50 of NO inhibitory assay in microglia BV2 cells | ||||
| Reference | 48 and 52 | 49 | ||
| Compounds | 16 | 17 | ||
| Structure |  |
 |
||
| Effects or mechanisms | 16: | 17: | ||
| 11β-HSD2: IC50 = 0.004 nM | 11β-HSD1: IC50 = 0.14 μM | |||
| 11β-HSD2: IC50 = 0.011 μM | ||||
| Reference | 50 | 51 | ||
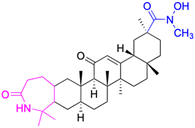
| Compounds | 18 | 19 | ||
| Structure | 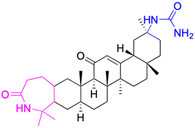 |
 |
||
| Effects or mechanisms | 18: | 19: | ||
| 11β-HSD1: IC50 = 45 μM | 11β-HSD1: IC50 > 40 μM | |||
| 11β-HSD2: IC50 = 0.033 μM | 11β-HSD2: IC50 = 0.011 μM | |||
| Reference | 54 | 54 | ||
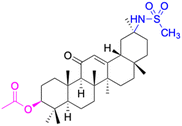
| Compounds | 20 | 21 | ||
| Structure | 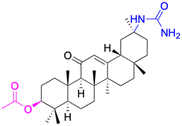 |
 |
||
| Effects or mechanisms | 20: | 21: | ||
| 11β-HSD1: IC50 = 8.3 μM | 11β-HSD1: IC50 > 40 μM | |||
| 11β-HSD2: IC50 = 0.104 μM | 11β-HSD2: IC50 = 0.0069 μM | |||
| Reference | 52 | 52 | ||
| Abbreviations | IL-6: the Interleukin-6. NF-κB: nuclear factor kappa-light-chain-enhancer of activated b cells. TNF-α: the tumor necrosis factor. COX-2: cyclooxygenase-2. MAPKs: mitogen-activated protein kinases. MIP-1α: macrophage inflammatory protein-1 alpha. 11β-HSD: 11β-hydroxysteroid dehydrogenase | |||
In conclusion, 18β-GA has potential therapeutic applications for various conditions due to its anti-inflammatory effects. Although more research is required, the use of 18β-GA and its derivatives may provide new avenues for treating inflammation-related diseases.
Antitumor activity
Cancer ravages and cripples the earth's inhabitants, ranking among the foremost destroyers of life.55 For countless years, scholars have been devoting themselves to the quest for a cure for tumors. Presently, the globe is awash with more than 80 conventional anti-tumor medications, ranging from cytotoxic drugs and hormones, to biological response modifiers (BRMs) and monoclonal antibodies.56 The majority of anticancer medications exhibit notable toxicity and necessitate administration in periodic cycles to mitigate adverse effects and impede the emergence of drug resistance. However, the excellent vitality of natural compounds adds new impetus to the research and development of anticancer drugs.57 And within this pantheon of treatment options stands the 18β-GA compound—a veritable powerhouse in its ability to vanquish cancerous cells from any part of the human body with unrivaled efficacy. Scores of meticulous studies attest to the fact that this drug is a game-changer in the fight against various forms of cancer. The sterling performance against malignant cells has been proven time and time again, and it holds immense potential as an agent in the battle against cancer. Wang et al. demonstrated that 18β-GA has potent inhibitory effects on colorectal cancer cell proliferation in vitro and in vivo. This study showed that 18β-GA treatment resulted in a significant reduction in cell migration, invasion, and wound healing capability, accompanied by the downregulation of matrix metalloproteinase (MMP) expression. Moreover, 18β-GA decreased the protein levels of phosphorylated PI3K, protein kinase B (AKT), Signal Transducer and Activator of Transcription 3 (STAT3), c-Jun N-terminal Kinase (JNK), p38 mitogen-activated protein kinase (p38), and NF-κB p65, where the phosphorylation of PI3K and STAT3 decreased as early as 2 h after 18β-GA treatment.58 Luo et al. found that 18β-GA-induced apoptosis and G2/M cell cycle arrest and inhibited migration via the ROS/MAPK/STAT3/NF-κB signaling pathways in A549 lung cancer cells. They also found that 18β-GA could reduce tumor growth in a mouse xenograft model. In breast cancer treatment,59 Shi et al. found that a combination of 18β-GA and doxorubicin enhanced cytotoxicity, apoptosis, and loss of mitochondrial membrane potential via the upregulation of a mitochondrial-dependent apoptosis pathway against MCF-7 (breast adenocarcinoma cell line) cells.60 In recent years, 18β-GA has also been found to have potential in liver cancer-targeted therapy. Speciale et al. provided a comprehensive review of the topic.61The derivatives of 18β-GA have been unearthed to harbor even more potent cancer properties in comparison to the progenitor compound. One of the most remarkable advantages of 18β-GA lies in its all-encompassing efficacy in targeting a myriad of cancer types. It has conspicuously showcased outstanding effectiveness against cancers of the digestive tract, liver, nervous system, reproductive system, immune system, thyroid, and other organ-related cancers. This renders it an invaluable weapon in the war against cancer.62,63 The 18β-GA's anti-cancer effects are believed to stem from its capacity to incite apoptosis, a process of purposeful cell death, in cancer cells. Additionally, it also exhibits anti-inflammatory and antioxidant properties that can shield cells from harm and amplify the growth of healthy cells. As demonstrated in Table 2, we have amassed an extensive collection of 18β-GA derivatives with extraordinary anticancer activity.
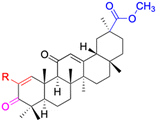
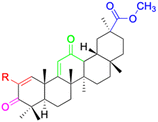
| Compounds | 18β-GA | 22–25 | 26–27 | 28–33 |
| Structure | 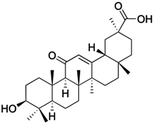 |
 |
 |
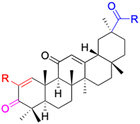 |
| Effects or mechanisms | 518A2: IC50 = 83.92 μM | 22: R = SO2CH3 | 26: R = I | 28: R = OCH3 |
| 8505C: IC50 = 86.50 μM | KU7: IC50 = 3.3 μM | 253JB-V: C50 = 3.6 μM | 253JB-V: IC50 = 0.25 μM | |
| A253: IC50 = 80.78 μM | Panc-1: IC50 = 7.6 μM | KU7I: C50 = 2.6 μM | KU7: IC50 = 1.59 pM | |
| A2780: IC50 = 74.57 μM | Panc-28: IC50 = 9.7 μM | Panc-1: IC50 = 4.4 μM | Panc-1: IC50 = 1.22 μM | |
| A431: IC50 = 79.58 μM | 23: R = I | Panc-28: IC50 = 3.6 μM | Panc-28: IC50 = 1.80 μM | |
| A549: IC50 = 82.76 μM | 253JB-V: IC50 = 2.6 μM | 27: R = CF3 | 29: R = H | |
| DLD-1: IC50 = 81.21 μM | KU7: IC50 = 3.0 μM | 253JB-V: IC50 = 0.3 μM | 253JB-V: IC50 = 6.10 μM | |
| FADU: IC50 = 84.55 μM | Panc-1: IC50 = 4.0 μM | KU7: IC50 = 1.3 μM | KU7: IC50 = 5.88 μM | |
| HCT-8: IC50 = 78.85 μM | 24: R = P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O(OCH3)2 O(OCH3)2 |
Panc-1: IC50 = 0.68 μM | Panc-1: IC50 = 3.81 μM | |
| HT-29: IC50 = 80.09 μM | 253JB-V: IC50 = 7.9 μM | Panc-28: IC50 = 1.1 μM | Panc-28: IC50 = 7.32 μM | |
| LIPO: IC50 = 81.44 μM | KU7: IC50 = 3.7 μM | 30: R = piperidinyl | ||
| MCF-7: IC50 = 84.70 μM | Panc-1: IC50 = 6.1 μM | HL-60: IC50 = 1.4 μM | ||
| SW480: IC50 = 86.80 μM | Panc-28: IC50 = 8.1 μM | 31: R= 1,4-bipiperidinyl | ||
| SW1736: IC50 = 76.93 μM | 25: R = CF3 | HL-60: IC50 = 0.8 μM | ||
| NIH 3T3: IC50 = 18.52 μM | 253JB-V: IC50 = 0.67 μM | 32: R = 4-methylpiperazinyl | ||
| HCT-11: IC50 = 78.83 μM | KU7: IC50 = 0.38 μM | HL-60: IC50 = 1.2 μM | ||
| HCT-116:IC50 = 78.83 μM | Panc-1: IC50 = 0.82 μM | 33: R = piperazinyl | ||
| Panc-28: IC50 = 1.1 μM | HL-60: IC50 = 1.7 μM | |||
| Reference | 59–61 and 88 | 82 and 83 | 82 and 83 | 82 and 83 |
| Compounds | 34 | 35–37 | 38–41 | 42–43 |
| Structure | 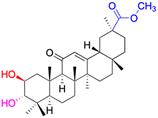 |
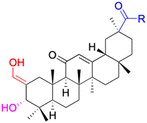 |
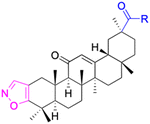 |
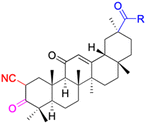 |
| Effects or mechanisms | 34: | 35: R = piperidinyl | 38: R = piperidinyl | 42: R = piperidinyl |
| HepG-2: | HL-60: IC50 = 5.5 μM | HL-60: IC50 = 1.7 μM | HL-60: IC50 = 8.6 μM | |
| IC50 = 0.22 μM | 36: R = 1,4′-bipiperidinyl | 39: R = 1,4′-bipiperidinyl | 43: R = 1,4′-bipiperidinyl | |
| HL-60: IC50 = 3.3 μM | HL-60: IC50 = 7.7 μM | HL-60: IC50 = 7.5 μM | ||
| 37: R = 4-methylpiperazinyl | 40: R = 4-methylpiperazinyl | |||
| HL-60: IC50 = 6.1 μM | HL-60: IC50 = 7.9 μM | |||
| 41: R = piperazinyl | ||||
| HL-60: IC50 = 8.2 μM | ||||
| Reference | 65 | 83 | 83 | 83 |
| Compounds | 44 | 45 | ||
| Structure | 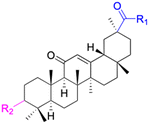 |
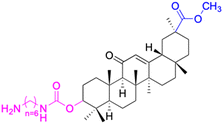 |
||
| Effects or mechanisms | 44: | 45: | ||
| R1 = O-i-Pr or OEt or OCH3 or OBn | 518A2: IC50 = 1.0 μM, 8505C: IC50 = 1.6 μM, A253: IC50 = 1.1 μM | |||
| R2 = O-β-alanine or O-L-alanine or O-glycine | A2780: IC50 = 1.3 μM, A549: IC50 = 1.5 μM, DLD-1: IC50 = 0.91 μM | |||
| 8505C: IC50 = 1.9–7.4 μM, A253: IC50 = 2.2–6.2 μM, A2780: IC50 = 1.3–5.9 μM | FADU: IC50 = 1.7 μM, HCT-116: IC50 = 1.1 μM, HCT-8: IC50 = 0.6 μM | |||
| A549: IC50 = 1.7–6.4 μM, DLD-1: IC50 = 2.5–8.5 μM, LIPO: IC50 = 2.3–7.5 μM | HT-29: IC50 = 0.5 μM, LIPO: IC50 = 1.5 μM, MCF-7: IC50 = 1.1 μM | |||
| Average: | SW1736: IC50 = 1.6 μM, SW480: IC50 = 2.2 μM | |||
| IC50 = 2.3–7.0 μM | ||||
| Reference | 89 | 80 | ||
| Compounds | 46–51 | 52–60 | ||
| Structure | 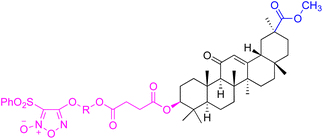 |
 |
||
| Effects or mechanisms | 46: R = (CH2)2O | 52: R1 = (CH2)2, R2 = H | ||
| BEL7402: IC50 = 7.8 μM | HepG2: IC50 = 9.0 μM, BEL7402: IC50 = 1.3 μM | |||
| 47: R = (CH2)3O | 53: R1 = (CH2)3, R2 = H | |||
| BEL7402: IC50 = 9.2 μM | HepG2: IC50 = 3.7 μM, BEL7402: IC50 = 0.43 μM | |||
| 48: R = (CH2)2CH(CH3)O | 54: R1 = (CH2)2CH(CH3), R2 = H | |||
| BEL7402: IC50 = 6.0 μM | HepG2: IC50 = 3.0 μM, BEL7402: IC50 = 1.1 μM | |||
| 49: R = (CH2)4O | 55: R1 = (CH2)4, R2 = H | |||
| BEL7402: IC50 = 8.2 μM | HepG2: IC50 = 6.7 μM, BEL7402: IC50 = 0.25 μM | |||
50: R = CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2O CHCH2O |
56: R1 = (CH2)2O(CH2)2, R2 = H | |||
| HepG2: IC50 = 7.9 μM, BEL7402: IC50 = 7.3 μM | HepG2: IC50 = 5.1 μM, BEL7402: IC50 = 3.7 μM | |||
| 51: R = CH2CH2NH | 57: R1 = CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2, R2 = H CHCH2, R2 = H |
|||
| HepG2: IC50 = 2.9 μM, BEL7402: IC50 = 2.9 μM | HepG2: IC50 = 1.3 μM, BEL7402: IC50 = 0.32 μM | |||
58: R1 = CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2, R2 = H CHCH2, R2 = H |
||||
| HepG2: IC50 = 3.3 μM, BEL7402: IC50 = 0.84 μM | ||||
| 59: R1 = (CH2)4, R2 = Ac | ||||
| HepG2: IC50 = 8.3 μM, BEL7402: IC50 = 4.8 μM | ||||
| 60: R1 = (CH2)2O(CH2)2, R2 = H | ||||
| HepG2: IC50 = 6.4 μM, BEL7402: IC50 = 9.4 μM | ||||
| Reference | 64 | 64 | ||
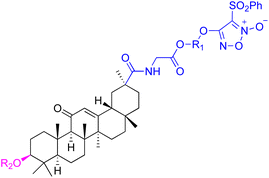
| Compounds | 61–62 | 63 | ||
| Structure | 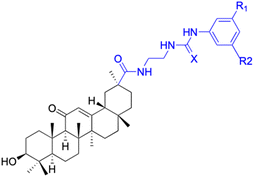 |
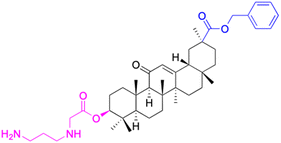 |
||
| Effects or mechanisms | 61: R1 = R2 = CF3, X = O | 63: | ||
| A549: IC50 = 7 μM, SKMEL: IC50 = 9 μM | 518A2: IC50 = 5.1 μM, 8505C: IC50 = 2.0 μM, A253: IC50 = 1.9 μM | |||
| HS683: IC50 = 6 μM, U373: IC50 = 6 μM | A549: IC50 = 4.7 μM, DLD-1: IC50 = 4.9 μM, Lipo: IC50 = 2.9 μM | |||
| PC3: IC50 = 8 μM, MCF7: IC50 = 4 μM | ||||
| 816F10: IC50 = 4 μM | ||||
| 62: R1 = R2 = H, X = S | ||||
| HS683: IC50 = 8 μM, PC3: IC50 = 9 μM | ||||
| Reference | 74 | 90 | ||
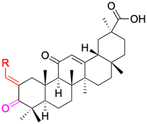
| Compounds | 64 | 65–66 | ||
| Structure | 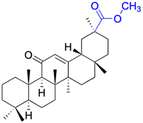 |
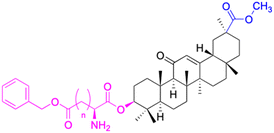 |
||
| Effects or mechanisms | 64: | 65: n = 1 | ||
| 518A2: IC50 = 23.69 μM, 8505C: IC50 = 24.30 μM, A2780: IC50 = 10.39 μM | A253: IC50 = 7.9 μM, A2780: IC50 = 8.8 μM, MCF-7: IC50 = 7.3 μM | |||
| LIPO: IC50 = 25.52 μM, SW1736: IC50 = 16.98 μM | 66: n = 2 | |||
| 518A2: IC50 = 1.7 μM, 8505C: IC50 = 1.7 μM, A253: IC50 = 1.2 μM | ||||
| A2780: IC50 = 1.6 μM, A549: IC50 = 1.7 μM, LIPO: IC50 = 1.7 μM | ||||
| MCF-7: IC50 = 1.2 μM, SW1736: IC50 = 2.3 μM | ||||
| Reference | 90 | 91 | ||
| Compounds | 67–71 | 72 | ||
| Structure | 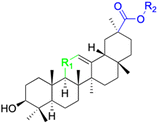 |
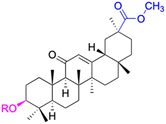 |
||
| Effects or mechanisms | 67: R1 = CH2, R2 = H | 72: | ||
| 518A2: IC50 = 71.49 μM, 8505C: IC50 = 78.52 μM, A2780: IC50 = 62.78 μM | R = L-2,4-diaminobutanoyl or D-alanyl or sacrosyl or L-prolyl or L-phenylalanyl or L-methionyl or L-ornithyl or L-lysyl | |||
| A431: IC50 = 86.13 μM, A549: IC50 = 79.13 μM, DLD-1: IC50 = 90.50 μM | 8505C: IC50 = 2.4–9.6 μM, A253: IC50 = 2.2–7.4 μM, A2780: IC50 = 1.5–5.5 μM | |||
| HCT-116: IC50 = 87.70 μM, HCT-8: IC50 = 88.76 μM, HT-29: IC50 = 90.30 μM | A549: IC50 = 2.1–9.9 μM, DLD-1: IC50 = 1.4–8.7 μM, LIPO: IC50 = 0.8–7.9 μM | |||
| LIPO: IC50 = 73.88 μM, MCF-7: IC50 = 90.19 μM, SW1736: IC50 = 72.47 μM | MCF-7: IC50 = 2.2–6.0 μM | |||
| NIH 3T3: IC50 = 68.70 μM | ||||
68: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = CH3 O, R2 = CH3 |
||||
| 518A2: IC50 = 27.54 μM, 8505C: IC50 = 26.07 μM, A2780: IC50 = 25.54 μM | ||||
| A431: IC50 = 25.28 μM, A549: IC50 = 23.50 μM, DLD-1: IC50 = 26.12 μM | ||||
| HCT-116: IC50 = 22.10 μM, HCT-8: IC50 = 24.36 μM, HT-29: IC50 = 27.54 μM | ||||
| LIPO: IC50 = 20.47 μM, MCF-7: IC50 = 22.14 μM, SW1736: IC50 = 34.87 μM | ||||
| NIH 3T3: IC50 = 22.81 μM | ||||
| 69: R1 =CH2, R2 = CH3 | ||||
| 518A2: IC50 = 34.54 μM, 8505C: IC50 = 33.88 μM, A2780: IC50 = 23.58 μM | ||||
| A431: IC50 = 33.55 μM, A549: IC50 = 31.59 μM, DLD-1: IC50 = 31.73 μM | ||||
| HCT-116: IC50 = 31.82 μM, HCT-8: IC50 = 31.34 μM, HT-29: IC50 = 23.89 μM | ||||
| LIPO: IC50 = 34.81 μM, MCF-7: IC50 = 34.37 μM, SW1736: IC50 = 32.35 μM | ||||
| NIH 3T3: IC50 = 42.22 μM | ||||
70: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = Et O, R2 = Et |
||||
| 518A2: IC50 = 25.23 μM, 8505C: IC50 = 24.58 μM, A2780: IC50 = 26.96 μM | ||||
| A431: IC50 = 23.45 μM, A549: IC50 = 22.74 μM, DLD-1: IC50 = 28.14 μM | ||||
| HCT-116: IC50 = 21.58 μM, HCT-8: IC50 = 43.42 μM, HT-29: IC50 = 22.14 μM | ||||
| LIPO: IC50 = 27.66 μM, MCF-7: IC50 = 18.61 μM, SW1736: IC50 = 13.37 μM | ||||
| NIH 3T3: IC50 = 23.66 μM | ||||
| 71: R1 = CH–OH, R2 = Et | ||||
| 518A2: IC50 = 51.52 μM, 8505C: IC50 = 52.80 μM, A2780: IC50 = 57.01 μM | ||||
| A431: IC50 = 46.55 μM, A549: IC50 = 48.97 μM, DLD-1: IC50 = 52.80 μM | ||||
| HCT-116: IC50 = 47.78 μM, HCT-8: IC50 = 44.32 μM, HT-29: IC50 = 44.32 μM | ||||
| LIPO: IC50 = 52.80 μM, MCF-7: IC50 = 48.97 μM, SW1736: IC50 = 45.48 μM | ||||
| NIH 3T3: IC50 = 43.16 μM | ||||
| Reference | 91 | 92 | ||
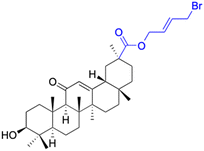
| Compounds | 73 | 74 | ||
| Structure | 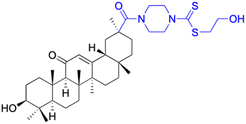 |
 |
||
| Effects or mechanisms | 73: | 74: | ||
| SMMC-7721 (after 72 h): IC50 = 14.42 μg mL−1 | 8505C: IC50 = 8.8 μM, SW1736: IC50 = 1.8 μM | |||
| Reference | 73–76 | 93 | ||
| Compounds | 75 | 76 | ||
| Structure | 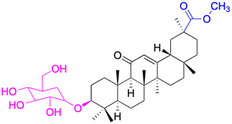 |
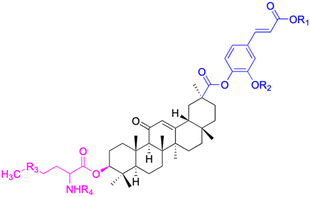 |
||
| Effects or mechanisms | 75: | 76: | ||
| MCF-7: IC50 = 1.8–8.6 μM | R1 = CH3 or Et, R2 = CH3 or H, R3 = S or Se, R4 = CO2tBu or H | |||
| MDA-MB-231: IC50 = 1.3–6.4 μM | MCF-7: IC50 = 1.8–8.6 μM | |||
| MDA-MB-231: IC50 = 1.3–6.4 μM | ||||
| Reference | 75 | 94 | ||
| Compounds | 80–90 | |||
| Structure | 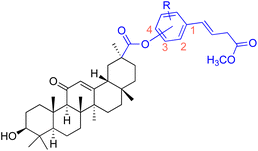 |
|||
| Effects or mechanisms | 80: 18β-GAO-4 | 87: 18β-GAO-3 | ||
| R = H | R = 4-OCH3 | |||
| A549: IC50 = 2.0 μM, SKMEL: IC50 = 3.0 μM, T98G: IC50 = 3.0 μM | KB: ED50 = 0.9 μM, KB-VIN: ED50 = 1.9 μM, A549: ED50 = 2.8 μM | |||
| HS683: IC50 = 3.0 μM, U373: IC50 = 2.0 μM, PC3: IC50 = 2.0 μM | 1A9: ED50 = 1.6 μM, HCT-8: ED50 = 2.0 μM, ZR-751: ED50 = 1.9 μM | |||
| MCF7: IC50 = 3.0 μM, 816F10: IC50 = 3.0 μM | PC-3: ED50 = 2.8 μM, DU-145: ED50 = 9.9 μM, LN-Cap: ED50 = 6.5 μM | |||
| 81: 18β-GAO-4 | 88: 18β-GAO-3 | |||
| R = 3-OCH3 | R = 3-OEt | |||
| MDA-MB-231: IC50 = 5.0 μM | KB: ED50 = 1.8 μM, KB-VIN: ED50 = 1.7 μM, A549: ED50 = 1.7 μM | |||
| 82: 18β-GAO-4 | 1A9: ED50 = 1.1 μM, HCT-8: ED50 = 2.7 μM, ZR-751: ED50 = 5.2 μM | |||
| R = 3-OEt | PC-3: ED50 = 3.3 μM, DU-145: ED50 = 5.8 μM, LN-Cap: ED50 = 1.1 μM | |||
| MDA-MB-231: IC50 = 8.1 μM | 89: 18β-GAO-2 | |||
| 83: 18β-GAO-3 | R = 3-OCH3 | |||
| R = 4-OCH3 | KB: ED50 = 0.8 μM, KB-VIN: ED50 = 2.8 μM, A549: ED50 = 2.2 μM | |||
| MCF-7: IC50 = 8.5 μM, MDA-MB-231: IC50 = 7.3 μM | 1A9: ED50 = 0.8 μM, HCT-8: ED50 = 1.9 μM, ZR-751: ED50 = 3.0 μM | |||
| 84: 18β-GAO-3 | PC-3: ED50 = 1.1 μM, DU-145: ED50 = 3.6 μM, LN-Cap: ED50 = 2.8 μM | |||
| R = 4-OEt | 90: 18β-GAO-2 | |||
| MDA-MB-231: IC50 = 9.4 μM | R = 3-F | |||
| 85: 18β-GAO-4 | KB: ED50 = 3.0 μM, KB-VIN: ED50 = 8.7 μM, A549: ED50 = 3.2 μM | |||
| R = 3-OCH3 | 1A9: ED50 = 1.3 μM, HCT-8: ED50 = 2.2 μM, ZR-751: ED50 = 2.7 μM | |||
| KB: ED50 = 1.6 μM, KB-VIN: ED50 = 2.5 μM, A549: ED50 = 2.0 μM | PC-3: ED50 = 1.6 μM, DU-145: ED50 = 2.7 μM, LN-Cap: ED50 = 4.4 μM | |||
| 1A9: ED50 = 0.9 μM, HCT-8: ED50 = 1.7 μM, ZR-751: ED50 = 2.8 μM | ||||
| PC-3: ED50 = 1.4 μM, DU-145: ED50 = 3.1 μM, LN-Cap: ED50 = 0.6 μM | ||||
| 86: 18β-GAO-2 | ||||
| R = 3-OEt | ||||
| KB: ED50 = 2.9 μM, A549: ED50 = 3.0 μM, 1A9: ED50 = 1.8 μM | ||||
| HCT-8: ED50 = 4.9 μM, ZR-751: ED50 = 8.8 μM, PC-3: ED50 = 3.5 μM | ||||
| LN-Cap: ED50 = 6.8 μM | ||||
| Reference | 75 and 76 | |||
| Compounds | 91–93 | 94–99 | ||
| Structure | 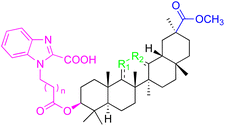 |
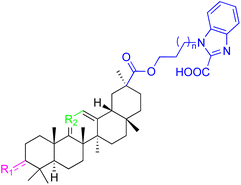 |
||
| Effects or mechanisms | 91: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = CH, n = 0 O, R2 = CH, n = 0 |
94: R1 = B-OAc, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, n = 1 O, n = 1 |
||
| Pin1 inhibition: IC50 = 1.0 μM | Pin1 inhibition: IC50 = 1.3 μM | |||
| PC-3: IC50 = 7.80 μM | 95: R1 = B-OAc, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, n = 0 O, n = 0 |
|||
| 92: R1 = CH, R2 = CH, n = 0 | Pin1 inhibition: IC50 = 1.0 μM | |||
| Pin1 inhibition: IC50 = 2.3 μM | 96: R1 = B–OH, R2 = CH2, n = 1 | |||
93: R1 = CH2, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, n= 1 O, n= 1 |
Pin1 inhibition: IC50 = 2.8 μM | |||
| Pin1 inhibition: IC50 = 2.3 μM | 97: R1 = B-OAc, R2 = CH2, n = 0 | |||
| Pin1 inhibition: IC50 = 2.1 μM | ||||
| PC-3: IC50 = 3.52 μM, LNCaP: IC50 = 7.92 μM | ||||
| 98: R1 = B-OAc, R2 = CH2, n = 0 | ||||
| Pin1 inhibition: IC50 = 4.7 μM | ||||
| 99: R1 = O, R2 = CH2, n = 1 | ||||
| Pin1 inhibition: IC50 = 3.8 μM | ||||
| Reference | 95 | 95 | ||
| Compounds | 100 | 101 | ||
| Structure | 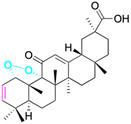 |
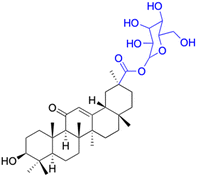 |
||
| Effects or mechanisms | 100: | 101: | ||
| A375: EC50 = 1.5 μM, A2780: EC50 = 1.0 μM, HT29: EC50 = 1.7 μM | HepG2: IC50 = 7.2 μM, MCF-7: IC50 = 7.7 μM | |||
| MCF7: EC50 = 2.9 μM, 518A2: EC50 = 1.2 μM | ||||
| Reference | 81 | 69 | ||
| Compounds | 102 | 103–108 | ||
| Structure | 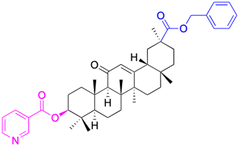 |
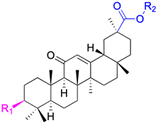 |
||
| Effects or mechanisms | 102: | 103: R1 = (E)-3-(4-acetoxyphenyl)acryl, R2 = Bn | ||
| SGC-7901: IC50 = 7.57 μM, MCF-7: IC50 = 5.51 μM, Eca-109: IC50 = 5.03 μM | HeLa: IC50 = 4.3 μM | |||
| HeLa: IC50 = 20.21 μM, Hep-G2: IC50 = 4.11 μM, HSF: IC50 = 23.18 μM | 104: R1 = nicotinyl, R2 = Bn | |||
| SGC-7901: IC50 = 7.5 μM, MCF-7: IC50 = 5.5 μM | ||||
| Eca-109: IC50 = 5.0 μM, Hep-G2: IC50 = 4.1 μM | ||||
| 105: R1 = isonicotinyl, R2 = Bn | ||||
| MCF-7: IC50 = 8.6 μM, Hep-G2: IC50 = 8.7 μM | ||||
| 106: R1 = 3-acetoxybenzyl, R2 = Bn | ||||
| HeLa: IC50 = 7.8 μM | ||||
| 107: R1 = 2-ethoxy-2-oxoacetyl, R2 = H | ||||
| A-549: IC50 = 1.0 μM | ||||
| 108: R1 = dodecanyl, R2 = H | ||||
| A-549: IC50 = 1.2 μM | ||||
| Reference | 70 and 71 | 70 and 71 | ||
| Compounds | 109–114 | 115 | ||
| Structure | 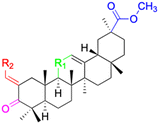 |
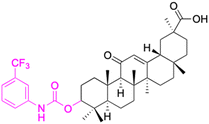 |
||
| Effects or mechanisms | 109: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = 1-imidazolyl O, R2 = 1-imidazolyl |
115: | ||
| MCF7: IC50 = 6.4 μM, SH-SY5Y: IC50 = 6.0 μM, Jurkat: IC50 = 3.2 μM | A549: IC50 = 2.81 μM, HT29: IC50 = 3.19 μM, HepG2: IC50 = 5.55 μM | |||
| 110: R1 = CH2, R2 = 1-imidazolyl | MCF-7: IC50 = 5.26 μM, PC-3: IC50 = 5.96 μM, Karpas299: IC50 = 5.59 μM | |||
| HT-29: IC50 = 3.3 μM, A549: IC50 = 2.8 μM, MIAPaca2: IC50 = 3.3 μM | ||||
| HeLa: IC50 = 2.2 μM, A375: IC50 = 2.0 μM, MCF7: IC50 = 3.0 μM | ||||
| HepG2: IC50 = 3.1 μM, SH-SY5Y: IC50 = 1.7 μM, Jurkat: IC50 = 1.1 μM | ||||
| BJ: IC50 = 6.9 μM | ||||
111: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = 2-methyl-1-imidazolyl O, R2 = 2-methyl-1-imidazolyl |
||||
| HT-29: IC50 = 9.4 μM, A375: IC50 = 7.1 μM, MCF7: IC50 = 5.6 μM | ||||
| SH-SY5Y: IC50 = 5.6 μM, Jurkat: IC50 = 2.4 μM | ||||
| 112: R1 = CH2, R2 = 2-methyl-1-imidazolyl | ||||
| HT-29: IC50 = 3.6 μM, A549: IC50 = 3.1 μM, MIAPaca2: IC50 = 3.3 μM | ||||
| HeLa: IC50 = 2.6 μM, A375: IC50 = 2.3 μM, MCF7: IC50 = 3.2 μM | ||||
| HepG2: IC50 = 3.5 μM, SH-SY5Y: IC50 = 2.2 μM, Jurkat: IC50 = 1.3 μM | ||||
113: R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R2 = 1,2,3-triazolyl-4-methyl carboxylate O, R2 = 1,2,3-triazolyl-4-methyl carboxylate |
||||
| A375: IC50 = 7.2 μM, MCF7: IC50 = 6.0 μM, SH-SY5Y: IC50 = 3.7 μM | ||||
| Jurkat: IC50 = 1.7 μM | ||||
| 114: R1 = CH2, R2 = 1,2,3-triazolyl-4-methyl carboxylate | ||||
| HT-29: IC50 = 8.9 μM, A549: IC50 = 7.9 μM, MIAPaca2: IC50 = 6.9 μM | ||||
| HeLa: IC50 = 5.4 μM, A375: IC50 = 4.9 μM, MCF7: IC50 = 5.2 μM | ||||
| HepG2: IC50 = 9.0 μM, SH-SY5Y: IC50 = 3.2 μM, Jurkat: IC50 = 1.5 μM | ||||
| Reference | 66 | 67 | ||
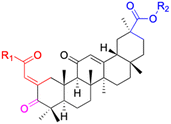
| Compounds | 116–122 | 123–127 | 128–143 | |
| Structure | 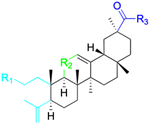 |
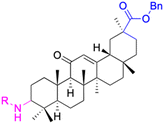 |
 |
|
| Effects or mechanisms | 116: R1 = CO2H, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R3 = OBn O, R3 = OBn |
123: R = L-ala | 128: R1 = OH, R2 = Bn | |
| NTUB1: IC50 = 2.3 μM | A549: IC50 = 2.109 μM | MCF-7: IC50 = 3.8 μM, PC-3: IC50 = 1.6 μM | ||
117: R1 = CO2 CH3, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R3 = OBn O, R3 = OBn |
MCF-7: IC50 = 2.135 μM | 129: R1 = OCH3, R2 = Bn | ||
| NTUB1: IC50 = 9.4 μM | HepG2: IC50 = 2.439 μM | MCF-7: IC50 = 1.1 μM, PC-3: IC50 = 1.2 μM | ||
118: R1 = CO2H, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R3 = NHC6H5 O, R3 = NHC6H5 |
HeLa: IC50 = 2.39 μM | 130: R1 = NHCH3, R2 = Bn | ||
| NTUB1: IC50 = 3.3 μM | MDCK: IC50 = 4.645 μM | MCF-7: IC50 = 1.1 μM, PC-3: IC50 = 0.40 μM | ||
119: R1 = CO2H, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R3 = NHCH(CH3)2 O, R3 = NHCH(CH3)2 |
124: R = L-gly | 131: R1 = NHEt, R2 = Bn | ||
| NTUB1: IC50 = 4.7 μM | A549: IC50 = 2.442 μM | MCF-7: IC50 = 0.59 μM, PC-3: IC50 = 0.27 μM | ||
| 120: R1 = CO2CH3, R2 = H2, R3 = NHCH(CH3)CO2Me | MCF-7: IC50 = 2.853 μM | 132: R1 = NH-nPr, R2 = Bn | ||
| Jurkat: IC50 = 9.6 μM | HepG2: IC50 = 3.472 μM | MCF-7: IC50 = 1.4 μM, PC-3: IC50 = 0.46 μM | ||
| 121: R1 = CO2 CH3, R2 = CH2, R3 = NHCH(CH3)CO2CH3 | HeLa: IC50 = 3.01 μM | 133; R1 = pyrrolidinyl, R2 = Bn | ||
| Jurkat: IC50 = 6.1 μM | MDCK: IC50 = 3.749 μM | MCF-7: IC50 = 3.0 μM, PC-3: IC50 = 3.4 μM | ||
122: R1 = CO2Et, R2 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, R3 = OEt O, R3 = OEt |
125: R = L-Boc-gly | 134: R1 = morpholinyl, R2 = Bn | ||
| 518A2: IC50 = 9.2 μM, A2780: IC50 = 5.8 μM | A549: IC50 = 2.751 μM | MCF-7: IC50 = 4.9 μM, PC-3: IC50 = 5.2 μM | ||
| MCF-7: IC50 = 3.811 μM | 135: R1 = 1,4-bipiperidinyl, R2 = Bn | |||
| HepG2: IC50 = 3.306 μM | MCF-7: IC50 = 2.1 μM, PC-3: IC50 = 3.0 μM | |||
| HeLa: IC50 = 3.296 μM | 136: R1 = piperazinyl, R2 = Bn | |||
| MDCK: IC50 = 4.431 μM | MCF-7: IC50 = 3.1 μM, PC-3: IC50 = 2.7 μM | |||
| 126: R = L-phe | 137: R1 = 1-methylpiperazinyl, R2 = Bn | |||
| A549: IC50 = 3.006 μM | MCF-7: IC50 = 3.3 μM, PC-3: IC50 = 3.1 μM | |||
| MCF-7: IC50 = 3.281 μM | 138: R1 = 1-Boc-piperazinyl, R2 = Bn | |||
| HepG2: IC50 = 5.048 μM | MCF-7: IC50 = 0.44 μM, PC-3: IC50 = 0.23 μM | |||
| HeLa: IC50 = 3.296 μM | 139: R1 = anilinyl, R2 = Bn | |||
| MDCK: IC50 = 5.024 μM | MCF-7: IC50 = 0.73 μM, PC-3: IC50 = 0.45 μM | |||
| 127: R = L-pro | 140: R1 = 4-nitroanilinyl, R2 = Bn | |||
| A549: IC50 = 3.261 μM | MCF-7: IC50 = 5.8 μM, PC-3: IC50 = 2.0 μM | |||
| MCF-7: IC50 = 7.623 μM | 141: R1 = 4-chloroanilinyl, R2 = Bn | |||
| HepG2: IC50 = 2.143 μM | MCF-7: IC50 = 8.9 μM, PC-3: IC50 = 0.85 μM | |||
| HeLa: IC50 = 2.209 μM | 142: R1 = 4-aminoperidinyl, R2 = Bn | |||
| MDCK: IC50 = 2.528 μM | MCF-7: IC50 = 0.98 μM, PC-3: IC50 = 0.69 μM | |||
| 143: R1 = 1-Boc-piperazinyl, R2 = CH3 | ||||
| MCF-7: IC50 = 1.0 μM, PC-3: IC50 = 0.68 μM | ||||
| Reference | 66 and 96 | 68 | 97 | |
| Compounds | 144 | 145–146 | ||
| Structure | 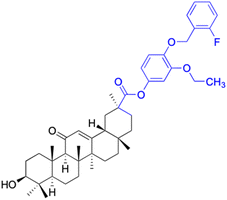 |
 |
||
| Effects or mechanisms | 144: HeLa: IC50 = 1.1 μM | 145: R = 2,4-diCl, R1 = C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
||
| MDA-MB-231: IC50 = 9.6 μM | ||||
| 146: R= 3-OEt, 5-F, 4-(methoxymethyl)benzene, R1 = OH | ||||
| HeLa: IC50 = 1.1 μM | ||||
| Reference | 98 | 98 | ||
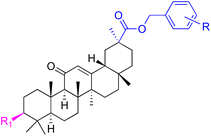
| Compounds | 147 | 148 | 149 | |
| Structure |  |
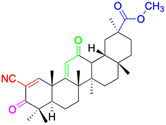 |
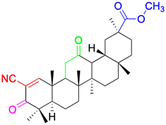 |
|
| Effects or mechanisms | 147: | 148: | 149: KB-3-1: IC50 = 5.5 μM | |
| Karpas299: IC50 = 6.51 μM, A549: IC50 > 40 μM | 253JB-V: IC50 = 0.11 μM | |||
| HepG2: IC50 = 6.93 μM, MCF-7: IC50 = 18.85 μM | KU7: IC50 = 0.12 μM | |||
| PC-3: IC50 = 18.18 μM | Panc-1: IC50 = 0.07 μM | |||
| Panc-28: IC50 = 0.05 μM | ||||
| KB-3-1: IC50 = 0.3 μM | ||||
| KB-8-5: IC50 = 1.2 μM | ||||
| HeLa: IC50 = 1.3 μM | ||||
| MCF-7: IC50 = 5 μM | ||||
| SK-N-MC: IC50 = 0.8 μM | ||||
| MDA-MB-231: IC50 = 5.97 μM | ||||
| Reference | 72 | 99 | 99 | |
| Compounds | 150 | 151 | ||
| Structure | 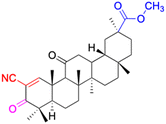 |
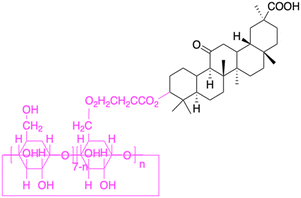 |
||
| Effects or mechanisms | 150: KB-3-1: IC50 = 5.5 μM | 151: Hep3B: cytotoxicity (28% cell viability) | ||
| Reference | 84–87 | 77 | ||
| Compounds | 152 | 153 | ||
| Structure | 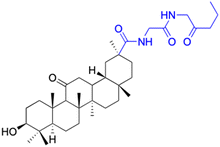 |
 |
||
| Effects or mechanisms | 152: | 153: | ||
| MCF-7: IC50/ = 5.1 μM, HCT-116: IC50 = 7.40 μM | MCF-7: IC50 = 5.0 μM, HCT-116: IC50 = 5.2 μM | |||
| Reference | 79 | 100 | ||
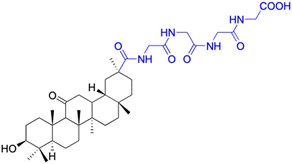
| Compounds | 154 | 155–156 | ||
| Structure | 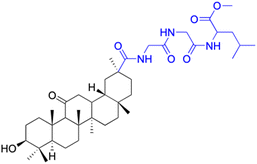 |
 |
||
| Effects or mechanisms | 154: | 155: R = CH3 | ||
| MCF-7: IC50 = 3.70 μM, HCT-116: IC50 = 3.0 μM, HepG-2: IC50 = 3.30 μM | MCF-7: IC50 = 6.9 μM, HepG2: IC50 = 9.9 μM | |||
| 156: R = 4-(trifluoromethyl)benzene | ||||
| MCF-7: IC50 = 9.5 μM, HepG2: IC50 = 25.6 μM | ||||
| Reference | 79 | 101 | ||
| Abbreviations | HepG-2: hepatocellular carcinoma cell line. HCT-116: colorectal carcinoma cell line. MCF-7: breast adenocarcinoma cell line. MDCK: Madin–Darby canine kidney cell line. HeLa: cervical cancer cell line. A549: lung adenocarcinoma cell line. Hep3B: hepatocellular carcinoma cell line. SW1736: thyroid carcinoma cell line. LIPO: liposarcoma cell line. A2780: ovarian cancer cell line. 8505C: thyroid cancer cell line. 518A2: melanoma cell line. HSF: fibroblast cell line. Eca-109: esophageal carcinoma cell line. SGC-7901: gastric cancer cell line. average: unknown cell line. DU-145: prostate cancer cell line. Karpas299: lymphoma cell line. DLD-1: colorectal adenocarcinoma cell line. NIH 3T3: mouse embryonic fibroblast cell line. BEL7402: hepatocellular carcinoma cell line. SMMC-7721: hepatocellular carcinoma cell line. NTUB1: bladder cancer cell line. LN-Cap: prostate adenocarcinoma cell line. Jurkat: T-cell leukemia cell line. ZR-751: breast cancer cell line. KB: oral epidermoid carcinoma cell line. KB-VIN: multidrug-resistant oral epidermoid carcinoma cell line. A549: lung adenocarcinoma cell line. 1A9: human lymphoblastoid cell line. HCT-8: colorectal adenocarcinoma cell line. HT-29: colorectal adenocarcinoma cell line. CT-26: colorectal carcinoma cell line. PC-3: prostate cancer cell line. SKMEL: melanoma cell line. T98G: glioblastoma cell line. HS683: glioma cell line. U373: glioblastoma cell line. 816F10: melanoma cell line. Pin1: peptidyl–prolyl cis–trans isomerase NIMA-interacting 1. FADU: hypopharyngeal carcinoma cell line. Panc-28: pancreatic carcinoma-28 cell line. Panc-1: pancreatic carcinoma-1 cell line. 253JB-V: bladder carcinoma cell line. KU7: a cell line derived from human bladder cancer. SK-N-MC: human neuroblastoma cell line. KB-8-5: human epidermoid carcinoma cell line. KB-3-1: human epidermoid carcinoma cell line. SH-SY5Y: human neuroblastoma cell line. MIAPaca2: pancreatic carcinoma cell line. HL-60: human promyelocytic leukemia cell line | |||
In the realm of liver cancer treatment, researchers have discovered that 18β-GA holds significant potential due to its ability to exhibit toxicity against multiple liver cancer cell lines. A study conducted by Lai et al. found that 18β-GA derivatives 46–60 demonstrated selective cell toxicity against human hepatocellular carcinoma, hepG2 (hepatocellular carcinoma cell line) cells, and BEL-7402 (hepatocellular carcinoma cell line) cells.64 Similarly, derivatives 34, 101–102, 109–115, 123–127, and 147 displayed excellent cell toxicity against hepG2.65–72 Moreover, derivatives 73 and 74, which were modified at position C30, exhibited noteworthy cell toxicity against SMMC-7721 (hepatocellular carcinoma cell line).73–76 Researchers also discovered the complex of 18β-GA-conjugated-β-cyclodextrin and emodin's superior cell toxicity against hep3B (hepatocellular carcinoma cell line) cells when compared to emodin alone.77
In the domain of gastrointestinal cancers, encompassing those that affect the mouth, esophagus, colon, and stomach, the extraordinary cytotoxicity of 18β-GA and its derivatives has been strikingly demonstrated, particularly against colon cancer cell lines. The literature is replete with evidence of 18β-GA's potent effects on HCT-116 (colorectal carcinoma cell line), HCT-8 (colorectal adenocarcinoma cell line), DLD-1 (colorectal adenocarcinoma cell line), and HT-29 (colorectal adenocarcinoma cell line) cells. For instance, derivatives 152–154 and 45 exhibit toxicity towards HCT-116, with derivative 45 also affecting HCT-8 cells and DLD-1. Likewise, derivatives 109–125 display remarkable cytotoxicity towards HT-29 cells.78–80 Moreover, Seribian et al.'s study unveiled the high cytotoxicity of 18β-GA 1,9-peroxide on numerous human tumor cell lines, including HT-29 cells.81 Compounds 22–29 manifest substantial activity against Panc-1 (pancreatic carcinoma-1 cell line) and Panc-28 (pancreatic carcinoma-28 cell line) cells, and compounds 109–114 have been established as inhibitors of MIAPaca2 (pancreatic carcinoma cell line) cells.66,67,82,83 As for human oral epidermoid cancer cell lines, such as KB-3-1, KB-8-5, KB, and KB-VIN, compounds 85–90 and 148–150 have displayed their significant prowess.74,76,84–87
In the context of prostate cancer cell lines such as PC-3 (androgen-independent) and LN-Cap, compounds 61–62, 86–90, and 128–143 have demonstrated significant inhibitory effects.75,76,97 In ovarian cancer cell lines like A2780, compounds 64–71 exhibited inhibitory activity up to 1.5 μM.90,91 Notably, compounds 109–114, 103, 106,102, 144, and 146 displayed notable inhibitory activity against HeLa cells (cervical cancer cell line).70,71,81,98 Additionally, compounds 152–156 showed strong inhibitory activity against MCF-9 breast cancer cell line.79,101
Beyond these realms, GA and its derivatives have also exhibited their anticancer activity in other areas. Prior research has established that GA and its derivatives have the ability to inhibit Neurosystem-associated cancer cell lines, such as SH-SY5Y (human neuroblastoma cell line) and SK-N-MC (human neuroblastoma cell line).66,84 In the investigation conducted by Csuk et al. conducted an investigation, which found that GA and its derivatives displayed robust activity against thyroid cancer.91 Li et al. found that 18β-GA exert anticancer effects as pin1 inhibitors.95 Furthermore, GA and its derivatives have demonstrated significant inhibitory activity against various types of cancer cells including those associated with lung cancer, lymphoma, melanoma, and breast cancer.66–68,74–76,80,82,83,89,91–94,96
In conclusion, 18β-GA and its derivatives have shown promising anti-tumor properties in various types of cancer, including colorectal, breast, lung, and liver. The cytotoxic effects of 18β-GA have been attributed to its ability to induce apoptosis, cell cycle arrest, inhibit migration, and downregulate various signaling pathways involved in cancer progression. In addition, 18β-GA has been shown to enhance the cytotoxicity of conventional chemotherapeutic agents, making it a potential adjuvant therapy for cancer treatment. Although 18β-GA and its derivatives have shown potential as anti-tumor agents, further studies are needed to fully understand their mechanisms of action and to optimize their pharmacological properties for clinical applications.
Antibacterial activity
The emergence and spread of drug-resistant bacteria pose a significant threat to global health. Conventional antibiotics are often rendered ineffective against these resistant strains, leading to prolonged and complicated treatment regimens, as well as increased morbidity and mortality rates. Consequently, there is a critical need to identify novel antibiotics that can effectively target and eliminate these drug-resistant bacteria.102 Researchers have turned their attention to natural compounds as potential sources of new antibiotics. Natural compounds have long been recognized for their diverse chemical structures and biological activities. By studying and modifying these compounds, scientists hope to develop more potent and effective antibiotics. Among the natural compounds explored for their antibacterial properties, 18β-GA and related compounds have shown promise. These compounds have exhibited antibacterial effects against various bacterial strains, suggesting their potential as therapeutic agents. Further investigations are underway to elucidate the mechanisms of action and optimize the activity of these compounds.103The antimicrobial properties of 18β-GA, a compound extracted from the licorice plant, have been extensively studied by various researchers. Kim et al. discovered that 18β-GA has the ability to disrupt bacterial cell membranes, leading to the eradication of these microorganisms. This finding has generated significant interest in the potential of 18β-GA as a novel antibacterial agent.104 Salari et al. further supported the antibacterial activity of 18β-GA against periodontopathogenic and capnophilic bacteria, while another investigation found that this natural compound can inhibit the growth of Helicobacter pylori.105,106 In a comprehensive study, Schrader et al. explored the antibacterial properties of various natural plant compounds, including 18β-GA and 18α-GA, and evaluated their efficacy against common pathogens found in pond-cultured channel catfish.107 It has been demonstrated that 18β-GA can effectively combat antibiotic-resistant bacterial strains, such as methicillin-resistant Staphylococcus aureus (MRSA), by inhibiting their survival and virulence gene expression.108 Furthermore, this compound has shown potential in preventing the growth and formation of supragingival plaque bacteria and treating H. pylori infections.109,110 In the fight against opportunistic nosocomial P. aeruginosa, 18β-GA has proven to be a valuable ally.111 Additionally, 18β-GA has been investigated for its ability to enhance the activity of tobramycin and polymyxin B against MRSA.112 In the quest to combat opportunistic nosocomial P. aeruginosa, 18β-GA has been found to be a valuable ally.113 Moreover, 18β-GA has been used in combination with nanoparticles and hydrogels to combat bacterial infections. Darvishi et al. developed and evaluated the antibacterial activity of 18β-GA-loaded PL18β-GA nanoparticles, which demonstrated significant antibacterial activity against both Gram-positive and Gram-negative bacteria.114 Similarly, Zhao et al. engineered an injectable moldable hydrogel assembled from natural glycyrrhizic acid, which exhibited remarkable antibacterial activity against both types of bacteria.115 Recently, the remarkable antibacterial capabilities of 18β-GA derivatives have come to light. These derivatives have shown promising inhibitory effects against various bacterial strains, making them potential candidates for combating bacterial infections.116 In this review, our objective is to classify and elucidate the antibacterial activities of different 18β-GA derivatives against specific bacterial species. 18β-GA and its derivatives, as shown in Table 3, have demonstrated significant potential in inhibiting pathogens.
| Compounds | 18β-GA | 157–163 | 164–166 |
| Structure | 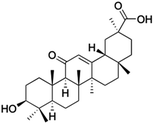 |
 |
 |
| Effects or mechanisms | Bacillus subtilis: MIC = 7.6 μg mL−1 | 157: R = CH2CH3 | 164: R = |
| Staphylococcus: MIC = 12.5 μg mL−1 | B. subtilis: MIC = 16.9 μg mL−1 |  |
|
| A. actinomycetemcomitans: | S. scabies: MIC = 2.1 μg mL−1 | Xoo: EC50 = 2.28 μg mL−1 | |
| MIC = 8 μg mL−1 | S. aureus: MIC = 4.2 μg mL−1 | Xac: EC50 = 1.42 μg mL−1 | |
| E. corrodens: MIC = 16 μg mL−1 | MRSA: MIC = 4.0 μg mL−1 | 165: R = | |
| C. sputigena: MIC = 8 μg mL−1 | 158: R = (CH2)2CH3 |  |
|
| Edwardsiella ictaluri: | B. subtilis: MIC = >34.8 μg mL−1 | Xoo: EC50 = 3.57 μg mL−1 | |
| MIC > 470.7 μg mL−1 | S. scabies: MIC = 4.3 μg mL−1 | Xac: EC50 = 0.93 μg mL−1 | |
| H. pylori: MIC = 20.8 μg mL−1 | S. aureus: MIC = 4.3 μg mL−1 | 166: R = | |
| P. aeruginosa: MIC = 160 μg mL−1 | MRSA: MIC = 2.0 μg mL−1 | 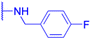 |
|
| P. gingivalis ATCC 33277: | 159: R = (CH2)3CH3 | Xoo: EC50 = 2.63 μg mL−1 | |
| MIC = 64 μg mL−1 | B. subtilis: MIC = >34.8 μg mL−1 | Xac: EC50 = 2.31 μg mL−1 | |
| S. gordonii: MIC = 64 μg mL−1 | S. scabies: MIC = 4.3 μg mL−1 | ||
| N. gonorrhoeae: | S. aureus: MIC = 4.3 μg mL−1 MRSA: MIC = 2.0 μg mL−1 | ||
| MIC = 3.9–62.5 μg mL−1 | 160: R = CH3 | ||
| B. subtilis: MIC = 4.0 μg mL−1 | |||
| S. scabies: MIC = 1.0 μg mL−1 | |||
| S. aureus: MIC = 2.0 μg mL−1 | |||
| 161: R = CH2CH3 | |||
| B. subtilis: MIC = 2.0 μg mL−1 | |||
| S. scabies: MIC = 4.1 μg mL−1 | |||
| S. aureus: MIC = 1.0 μg mL−1 | |||
| MRSA: MIC = 1.0 μg mL−1 | |||
| 162: R = CH(CH3)2 | |||
| B. subtilis: MIC = >33.9 μg mL−1 | |||
| S. scabies: MIC = 4.2 μg mL−1 | |||
| S. aureus: MIC = 8.4 μg mL−1 | |||
| MRSA: MIC = 2.0 μg mL−1 | |||
| 163: R = (CH2)3CH3 | |||
| B. subtilis: MIC = >34.8 μg mL−1 | |||
| S. scabies: MIC = 4.3 μg mL−1 | |||
| S. aureus: MIC = >34.8 μg mL−1 | |||
| MRSA: MIC = >32.0 μg mL−1 | |||
| Reference | 104, 105, 107, 110, 111, 113 and 125 | 117 | 118 |
| Compounds | 167–171 | 172 | |
| Structure | 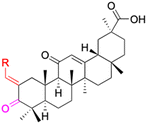 |
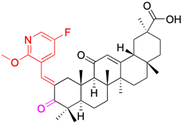 |
|
| Effects or mechanisms | 167: R = | 172: | |
 |
Staphylococcus aureus (ATCC 6538): | ||
| Staphylococcus aureus (ATCC 6538): MIC = 54.88 μg mL−1 | MIC = 6.25 μmol L−1 | ||
| Staphylococcus aureus (ATCC 29213): MIC = 6.86 μg mL−1 | Staphylococcus aureus subsp. aureus (ATCC 29213): | ||
| Staphylococcus epidermidis (ATCC 12228): MIC = 27.44 μg mL−1 | MIC = 6.25 μmol L−1 | ||
| 168: R = | Staphylococcus epidermidis (ATCC 12228): | ||
 |
MIC = 6.25 μmol L−1 | ||
| Staphylococcus aureus (ATCC 6538): MIC = 3.39 μg mL−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 6.79 μg mL−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 3.39 μg mL−1 | |||
| 169: R = | |||
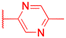 |
|||
| Staphylococcus aureus (ATCC 6538): MIC = 2.72 μg mL−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 2.72 μg mL−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 2.72 μg mL−1 | |||
| 170: R = | |||
 |
|||
| Staphylococcus aureus (ATCC 6538): MIC = 6.83 μg mL−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 13.67 μg mL−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 6.83 μg mL−1 | |||
| 171: R = | |||
 |
|||
| Staphylococcus aureus (ATCC 6538): MIC = 27.34 μg mL−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 54.68 μg mL−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 27.34 μg mL−1 | |||
| Reference | 43 | 123 | |
| Compounds | 173 | 174 | |
| Structure | 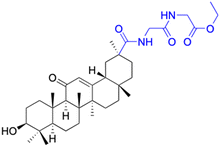 |
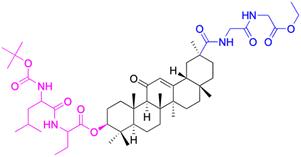 |
|
| Effects or mechanisms | 173: | 174: | |
| Streptococcus pneumonia RCMB 010010: | Streptococcus pneumonia RCMB 010010: | ||
| Diameter of inhibition zone = 15 mm | Diameter of inhibition zone = 12 mm | ||
| Staphylococcus aureus ATCC25923: | Staphylococcus aureus ATCC25923: | ||
| Diameter of inhibition zone = 15 mm | Diameter of inhibition zone = 17 mm | ||
| Micrococcus luteus: | Micrococcus luteus: | ||
| Diameter of inhibition zone = 30 mm | Diameter of inhibition zone = 30 mm | ||
| Escherichia coli ATCC25922: | Escherichia coli ATCC25922: | ||
| Diameter of inhibition zone = 20 mm | Diameter of inhibition zone = 18 mm | ||
| Pseudomonas aeruginosa ATCC7853: | Pseudomonas aeruginosa ATCC7853: | ||
| Diameter of inhibition zone = 18 mm | Diameter of inhibition zone = 15 mm | ||
| Reference | 79 | 79 | |
| Compounds | 175 | 176 | |
| Structure |  |
 |
|
| Effects or mechanisms | 175: | 176: | |
| Streptococcus pneumonia RCMB 010010: | Streptococcus pneumonia RCMB 010010: | ||
| Diameter of inhibition zone = 17 mm | Diameter of inhibition zone = 11 mm | ||
| Staphylococcus aureus ATCC25923: | Staphylococcus aureus ATCC25923: | ||
| Diameter of inhibition zone = 17 mm | Diameter of inhibition zone = 10 mm | ||
| Micrococcus luteus: | Micrococcus luteus: | ||
| Diameter of inhibition zone = 30 mm | Diameter of inhibition zone = 29 mm | ||
| Escherichia coli ATCC25922: | Escherichia coli ATCC25922: | ||
| Diameter of inhibition zone = 16 mm | Diameter of inhibition zone = 13 mm | ||
| Pseudomonas aeruginosa ATCC7853: | Pseudomonas aeruginosa ATCC7853: | ||
| Diameter of inhibition zone = 15 mm | Diameter of inhibition zone = 13 mm | ||
| Proteus vulgaris RCMB 010085: | Proteus vulgaris RCMB 010085: | ||
| Diameter of inhibition zone = 17 mm | Diameter of inhibition zone = 12 mm | ||
| Candida albicans: | Candida albicans: | ||
| Diameter of inhibition zone = 12 mm | Diameter of inhibition zone = 15 mm | ||
| Reference | 119 | 124 | |
| Compounds | 177 | 178 | |
| Structure |  |
 |
|
| Effects or mechanisms | 177: | 178: | |
| Xoo: EC50 = 5.89 μg mL−1 | Xoo: EC50 = 36.5 μg mL−1 | ||
| Psa: EC50 = 16.1 μg mL−1 | Psa: EC50 = 114 μg mL−1 | ||
| Xac: EC50 = 3.64 μg mL−1 | Xac: EC50 = 29.1 μg mL−1 | ||
| Reference | 119 | 119 | |
| Compounds | 179–184 | 185–187 | |
| Structure | 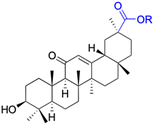 |
 |
|
| Effects or mechanisms | 179: | 185: R = | |
| R = Bn |  |
||
| Theileria annulata (T339): GI50 = 7.431 μmol L−1 | Xoo: EC50 = 4.69 μg mL−1, Xac: EC50 = 6.29 μg mL−1 | ||
| Theileria annulata (T5815): GI50 = 7.595 μmol L−1 | 186: R = | ||
| 180: |  |
||
| R = CH2CH3 | Xoo: EC50 = 3.64 μg mL−1, Xac: EC50 = 20.5 μg mL−1 | ||
| Theileria annulata (T339): GI50 = 5.638 μmol L−1 | 187: R = | ||
| Theileria annulata (T5815): GI50 = 7.557 μmol L−1 |  |
||
| 181: | Xoo: EC50 = 5.56 μg mL−1, Xac: EC50 = 8.83 μg mL−1 | ||
| R = CH3 | |||
| Theileria annulata (T5815): GI50 = 5.977 μmol L−1 | |||
| 182: | |||
| R = CH2CH2CH3 | |||
| Theileria annulata (T339): GI50 = 5.549 μmol L−1 | |||
| Theileria annulata (T5815): GI50 = 3.55 μmol L−1 | |||
| 183: | |||
| R = CH(CH3)2 | |||
| Theileria annulata (T339): GI50 = 1.638 μmol L−1 | |||
| Theileria annulata (T5815): GI50 = 1.499 μmol L−1 | |||
| 184: | |||
| R = (CH2)3CH3 | |||
| Theileria annulata (T5815): GI50 = 9.946 μmol L−1 | |||
| Reference | 124 | 120 | |
| Compounds | 188–189 | 190–195 | |
| Structure | 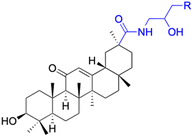 |
 |
|
| Effects or mechanisms | 188: R = | 190: R = 5-fluoro-2-nitro-benzene | |
 |
Staphylococcus aureus (ATCC 6538): MIC = 10 μmol L−1 | ||
| Xoo: EC50 = 10.2 μg mL−1 | Staphylococcus aureus (ATCC 29213): MIC = 10 μmol L−1 | ||
| Xac: EC50 = 4.16 μg mL−1 | Staphylococcus epidermidis (ATCC 12228): MIC = 10 μmol L−1 | ||
| 189: R = | MRSA: MIC = 16 μmol L−1 | ||
 |
191: R = 4-chloro-2-nitro-benzene | ||
| Xoo: EC50 = 10.9 μg mL−1 | Staphylococcus aureus (ATCC 6538): MIC = 10 μmol L−1 | ||
| Xac: EC50 = 5.16 μg mL−1 | Staphylococcus aureus (ATCC 29213): MIC = 5 μmol L−1 | ||
| Staphylococcus epidermidis (ATCC 12228): MIC = 5 μmol L−1 | |||
| MRSA: MIC = 8 μmol L−1 | |||
| 192: R = 4-methoxy-2-nitro-benzene | |||
| Staphylococcus aureus (ATCC 6538): MIC = 5 μmol L−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 5 μmol L−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 5 μmol L−1 | |||
| MRSA: MIC = 4 μmol L−1 | |||
| 193: R = 5-bromo-2-nitro-benzene | |||
| Staphylococcus aureus (ATCC 6538): MIC = 2.5 μmol L−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 2.5 μmol L−1 | |||
| Staphylococcus epidermidis (ATCC 12228):MIC = 2.5 μmol L−1 | |||
| MRSA: MIC = 16 μmol L−1 | |||
| 194: R = 4-bromo-2-nitro-benzene | |||
| Staphylococcus aureus (ATCC 6538): MIC = 12.5 μmol L−1 | |||
| Staphylococcus aureus (ATCC 29213):MIC = 12.5 μmol L−1 | |||
| Staphylococcus epidermidis (ATCC 12228):MIC = 12.5 μmol L−1 | |||
| MRSA: MIC = 16 μmol L−1 | |||
| 195: R = 4-fluoro-2-nitro-benzene | |||
| Staphylococcus aureus (ATCC 6538): MIC = 5 μmol L−1 | |||
| Staphylococcus aureus (ATCC 29213): MIC = 5 μmol L−1 | |||
| Staphylococcus epidermidis (ATCC 12228): MIC = 5 μmol L−1 | |||
| MRSA: MIC = 8 μmol L−1 | |||
| Reference | 120 | 49 | |
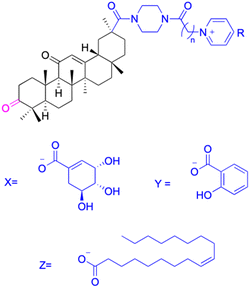
| Compounds | 196–201 | 202–225 | |
| Structure | 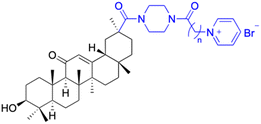 |
 |
|
| Effects or mechanisms | 196: | 202: | |
| n = 5 | n = 5; R = Br− | ||
| Xoo: EC50 = 8.57 μg mL−1, Xac: EC50 = 7.67 μg mL−1 | Xoo: EC50 = 9.47 μg mL−1, Xac: EC50 = 11.8 μg mL−1 | ||
| 195: | 203: | ||
| n = 6 | n = 6; R = Br− | ||
| Xoo: EC50 = 5.24 μg mL−1, Xac: EC50 = 9.55 μg mL−1 | Xoo: EC50 = 9.18 μg mL−1, Xac: EC50 = 34.5 μg mL−1 | ||
| 197: | 204: | ||
| n = 7 | n = 7; R = Br− | ||
| Xoo: EC50 = 5.06 μg mL−1, Xac: EC50 = 8.16 μg mL−1 | Xoo: EC50 = 7.12 μg mL−1, Xac: EC50 = 9.53 μg mL−1 | ||
| 198: | 205: | ||
| n = 8 | n = 8; R = Br− | ||
| Xoo: EC50 = 3.54 μg mL−1, Xac: EC50 = 10.3 μg mL−1 | Xoo: EC50 = 3.38 μg mL−1, Xac: EC50 = 18.7 μg mL−1 | ||
| 199: | 206: | ||
| n = 9 | n = 9; R = Br− | ||
| Xoo: EC50 = 3.47 μg mL−1, Xac: EC50 = 34.1 μg mL−1 | Xoo: EC50 = 2.29 μg mL−1, Xac: EC50 = 25.6 μg mL−1 | ||
| 200: | 207: | ||
| n = 10 | n = 10; R = Br− | ||
| Xoo: EC50 = 6.60 μg mL−1, Xac: EC50 = 17.4 μg mL−1 | Xoo: EC50 = 1.37 μg mL−1, Xac: EC50 = 37.4 μg mL−1 | ||
| 208: | |||
| n = 5; R = X | |||
| Xoo: EC50 = 14.08 μg mL−1, Xac: EC50 = 14.76 μg mL−1 | |||
| 209: | |||
| n = 5; R = Y | |||
| Xoo: EC50 = 19.53 μg mL−1, Xac: EC50 = 6.8 μg mL−1 | |||
| 210: | |||
| n = 5; R = Z | |||
| Xoo: EC50 = 19.06 μg mL−1, Xac: EC50 = 4.59 μg mL−1 | |||
| 211: | |||
| n = 6; R = X | |||
| Xoo: EC50 = 12.11 μg mL−1, Xac: EC50 = 6.88 μg mL−1 | |||
| 212: | |||
| n = 6; R = Y | |||
| Xoo: EC50 = 12.9 μg mL−1, Xac: EC50 = 25.03 μg mL−1 | |||
| 213: | |||
| n = 6; R = Z | |||
| Xoo: EC50 = 20.59 μg mL−1, Xac: EC50 = 14.81 μg mL−1 | |||
| 214: | |||
| n = 7; R = X | |||
| Xoo: EC50 = 6.5 μg mL−1, Xac: EC50 = 14.81 μg mL−1 | |||
| 215: | |||
| n = 7; R = Y | |||
| Xoo: EC50 = 6.17 μg mL−1, Xac: EC50 = 11.69 μg mL−1 | |||
| 216: | |||
| n = 7; R = Z | |||
| Xoo: EC50 = 17.25 μg mL−1, Xac: EC50 = 14.39 μg mL−1 | |||
| 217: | |||
| n = 8; R = X | |||
| Xoo: EC50 = 5.17 μg mL−1, Xac: EC50 = 7.16 μg mL−1 | |||
| 218: | |||
| n = 9; R = X | |||
| Xoo: EC50 = 4.18 μg mL−1, Xac: EC50 = 10.32 μg mL−1 | |||
| 219: | |||
| n = 10; R = X | |||
| Xoo: EC50 = 1.6 μg mL−1, Xac: EC50 = 8.48 μg mL−1 | |||
| 220: | |||
| n = 8; R = Y | |||
| Xoo: EC50 = 4.93 μg mL−1, Xac: EC50 = 3.82 μg mL−1 | |||
| 221: | |||
| n = 9; R = Y | |||
| Xoo: EC50 = 7.56 μg mL−1, Xac: EC50 = 4.38 μg mL−1 | |||
| 222: | |||
| n = 10; R = Y | |||
| Xoo: EC50 = 4.14 μg mL−1, Xac: EC50 = 10.15 μg mL−1 | |||
| 223: | |||
| n = 8; R = Z | |||
| Xoo: EC50 = 13.77 μg mL−1, Xac: EC50 = 22.17 μg mL−1 | |||
| 224: | |||
| n = 9; R = Z | |||
| Xoo: EC50 = 12.46 μg mL−1, Xac: EC50 = 2.07 μg mL−1 | |||
| 225: | |||
| n = 10; R = Z | |||
| Xoo: EC50 = 2.98 μg mL−1, Xac: EC50 = 6.08 μg mL−1 | |||
| Reference | 121 | 121 | |
| Compounds | 226 | ||
| Structure | 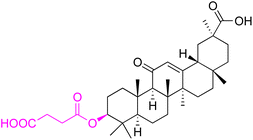 |
||
| Effects or mechanisms | 226: | ||
| MRSA SA5002: MIC = 16 mg L−1 | |||
| MRSA SA5053: MIC = 16 mg L−1 | |||
| MSSA SA5028: MIC = 16 mg L−1 | |||
| Reference | 122 | ||
| Abbreviations | Xac: Xanthomonas citri subsp. citri. Xoo: Xanthomonas oryzae pv. oryzae. Psa: Pseudomonas syringae pv. actinidiae. MRSA: methicillin-resistant Staphylococcus aureus | ||
Compounds 157–163 have emerged as potent inhibitors of Streptomyces scabies, a notorious plant pathogen. These derivatives have exhibited remarkable inhibitory activity, suggesting their potential application in managing plant bacterial diseases.117 Compound 161 has demonstrated superior inhibitory activity against Bacillus subtilis, Staphylococcus aureus, and MRAS compared to conventional antibiotics such as ampicillin, streptomycin, and vancomycin. This finding highlights the potential of 18β-GA derivatives as effective alternatives for combating drug-resistant bacterial strains.
Furthermore, compounds 164–166, compounds 177–178, compounds 183–187, and compounds 196–225 have displayed robust inhibitory activity against Xanthomonas oryzae pv. oryzae (Xoo) and X. axonopodis pv. citri (Xac).118–121 Xiang et al. particularly emphasized the potency of compounds 164 and 165. In vivo trials have further confirmed the potential of these compounds in managing rice bacterial blight disease, with control efficacy ranging between 50.57% and 53.70% at 200 μg mL−1.118
Moreover, Yang et al. discovered that derivatives of 18β-GA (compounds 167–176, 190–195, and 226) exhibit potent antibacterial activity against Staphylococcus aureus, Staphylococcus epidermidis, and MRAS.43,122 Compound 172, as identified by Guo et al., has demonstrated robust antibacterial properties and has been used to prepare supramolecular self-assembly hydrogels with exceptional thermodynamic stability and high melting temperatures.123 Additionally, compounds 173–176 have exhibited high activity against various bacteria, particularly showing enhanced antibacterial effects against Micrococcus luteus compared to gentamicins.79
Tropical bovine theileriosis (TBT) is one of the progressive and lymphoproliferative tick-borne diseases caused by Theileria annulata. Buvanesvaragurunathan et al. investigated the effect of 18β-GA esters (compounds 179–184) on the growth of Theileria annulata and found that they induced apoptosis in parasite cells. Among these esters, the isopropyl ester of 18β-GA (compound 183) showed improved anti-theileriosis efficacy than other 18β-GA derivatives.124
In conclusion, the rise of drug-resistant bacteria necessitates the discovery of novel antibiotics that can effectively combat these resilient strains mentioned above. Natural compounds, such as 18β-GA and its derivatives, offer a promising avenue for antibiotic development. Future research efforts should focus on understanding the mode of action of these compounds and optimizing their efficacy against drug-resistant bacteria.
Antiviral activity
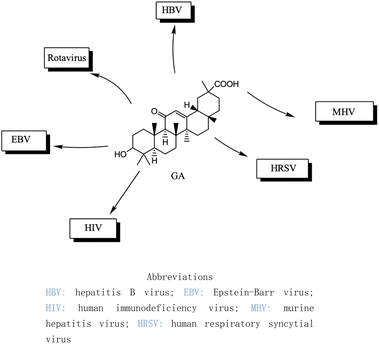
Over the past two decades, the potencies have been extensively investigated for pentacyclic triterpenoids, such as asiatic acid, betulinic acid, boswellic acid, glycyrrhizin, 18β-GA, lupeol, oleanolic acid, and ursolic acid, and their analogs and derivatives, as potent antitumor and antiviral agents. These triterpenoids have displayed remarkable cytotoxic activity against various tumor cell lines and exhibit antiviral properties, in particular, anti-HIV activity.126 The main active constituents of licorice are triterpenoids, which have shown inhibitory effects on several viruses, including SARS-CoV-2.127 It has been revealed that these compounds achieve their antiviral effects through various mechanisms such as inhibiting virus replication, directly inactivating viruses, halting inflammation mediated by HMGB1/TLR4, preventing β-chemokines, reducing the binding of HMGB1 to DNA to weaken virus activity, and inhibiting reactive oxygen species formation.128,129 While these natural products offer great potential as anti-viral and anti-microbial agents, they comprise complex mixtures of organic molecules, making it difficult to determine their exact effectiveness. Hence, further research is required to gain an intricate understanding of their mechanisms of action and their potential for use as food or herbal medicine. Additionally, it is vital to carefully consider the pleiotropic effects of these compounds to avoid potential negative consequences.Several studies have shown that 18β-GA inhibit several viruses (Fig. 5), for example, Sato et al. reported that 18β-GA inhibits hepatitis B virus (HBV) by suppressing surface antigens,130 while Hardy et al. showed that 18β-GA exhibits significant antiviral activity against rotavirus replication in vitro.131 Other investigations demonstrated that 18β-GA inhibited rotavirus SA11 via the Fas/FasL pathway, inhibits Epstein–Barr virus (EBV) in superinfected Raji cells, showed significant antiviral activity against human immunodeficiency virus (HIV), inhibits infection of human respiratory syncytial virus (HRSV), and significantly protects against murine hepatitis virus (MHV)-induced severe hepatic injury by suppressing HMGB1 release.35,132–135
In recent years, researchers have also worked on the antiviral properties of 18β-GA derivatives (Table 4). Baltina et al. synthesized a series of 18β-GA derivatives. They found that compounds 227–230 exert the most significant antiviral activity (IC50 = 0.13 μM) against ZIKV, with compound 227 demonstrating promising potential as an antiviral agent against ZIKV infection.136 Similarly, Zígolo et al. reported that compound 231 exhibited significant antiviral activity against TK+ and TK− strains of herpes simplex virus type 1 (HSV-1).137 Liang et al. found that water-soluble β-cyclodextrin-18β-GA (compounds 232–237) showed promising antiviral activity against the influenza A/WSN/33 (H1N1) virus.138,139 More recently, Ding et al. suggested that 18β-GA and its derivatives (compounds 238–241) could alleviate the symptoms of COVID-19 patients.140 Additionally, Wang et al. synthesized several compounds and observed that compounds 242–243 exhibited significant inhibitory activities against HBV DNA replication.73 These findings highlight the potential of 18β-GA and its derivatives as potent antiviral agents with remarkable antiviral activity against numerous viral infections.
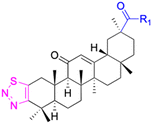
| Compounds | 227–228 | 229–230 | 231 |
| Structure | 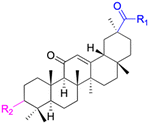 |
 |
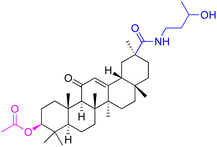 |
| Effects or mechanisms | 227: | 229: | 231: |
| R2 = OAc | R1 = COOBu | HSV-1 virus: CC50 = 190.2 μM, EC50 = 4.95 μM, CC50/EC50 = 38.38 | |
| R2 = | ZIKA virus: CC50 > 50 μM, IC50 = 0.29 μM, CC50/IC50 > 172.4 | ||
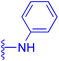 |
230: | ||
| ZIKA virus: CC50 > 50 μM | R1 = COOCH3 | ||
| IC50 = 0.13 μM, CC50/IC50 > 384 | ZIKA virus: CC50 > 50 μM, IC50 = 0.56 μM | ||
| 228: | CC50/IC50 > 89.3 | ||
| R2 = | |||
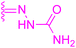 |
|||
| R1 = COOBu | |||
| ZIKA virus: CC50 > 50 μM | |||
| IC50 = 0.55 μM, CC50/IC50 > 90.9 | |||
| Reference | 136 | 136 | 137 |
| Compounds | 232–237 | 238–241 | |
| Structure |  |
 |
|
 |
|||
| Effects or mechanisms |  |
238: | |
| 232: | R = | ||
| R = Ac |  |
||
| Influenza A/WSN/33 (H1N1) virus: IC50 = 12.1 μM, CC50 > 100 μM, SI > 8.3 | HBV: CC50 > 985.68 μM, IC50 = 5.71 μM, SI > 172.6 | ||
| 233: | 239: | ||
| R = H | R = | ||
| Influenza A/WSN/33 (H1N1) virus: IC50 = 9.03 μM, CC50 > 100 μM, SI > 11.1 |  |
||
 |
HBV: CC50 > 1373.13 μM, IC50 = 5.36 μM, SI > 255.9 | ||
| 234: | 240: | ||
| R = Ac | R = | ||
| Influenza A/WSN/33 (H1N1) virus: IC50 = 20.7 μM, CC50 > 100 μM, SI > 4.8 | 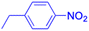 |
||
| 235: | HBV: CC50 > 1327.92 μM, IC50: 8.90 μM, SI > 149.2 | ||
| R = H | 241: | ||
| Influenza A/WSN/33 (H1N1) virus: IC50 = 11.0 μM, CC50 > 100 μM, SI > 9.1 | R = | ||
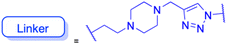 |
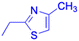 |
||
| 236: | HBV: CC50 = 37.17 μM, IC50 = 9.08 μM, SI = 4.1 | ||
| R = Ac | |||
| Influenza A/WSN/33 (H1N1) virus:IC50 = 20.7 μM, CC50 > 100 μM, SI > 4.8 | |||
| 237: | |||
| R = H | |||
| Influenza A/WSN/33 (H1N1) virus: IC50 = 11.0 μM, CC50 > 100 μM, SI > 9.1 | |||
| Reference | 138 and 139 | 140 | |
| Compounds | 242 | 243 | |
| Structure | 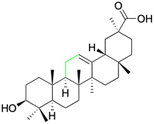 |
 |
|
| Effects or mechanisms | 242: | 243: | |
| HBV: | HBV: | ||
| CC50 = 161.68 μM | CC50 = 35.71 μM | ||
| IC50 = 47.00 μM | IC50 = 18.37 μM | ||
| SI = 3.4 | SI = 1.9 | ||
| Reference | 73 | 73 |
In summary, the research on pentacyclic triterpenoids, including 18β-GA and its derivatives, suggests their immense potential as effective and safe antiviral agents. These compounds have demonstrated varying degrees of antiviral activity against numerous viral infections, making them a promising area of ongoing research. However, further studies are necessary to comprehensively investigate their mechanisms of action and how they can be effectively used as food or herbal medicine while considering the possible negative consequences of their pleiotropic effects.
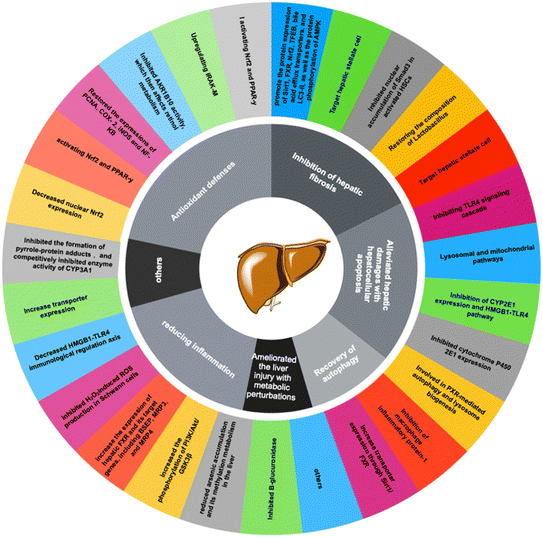
Antioxidant activity
18β-GA has been found to exhibit significant antioxidant activity, which makes it of great interest in the research of antioxidants. Alanazi et al. found that the serum concentrations of final glucose, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) in mice treated with 20 mg per kg acrylamide (Acr) increased to 131 ± 12.2 mg dL−1, 76.5 ± 12.0 μ U−1, 47.7 ± 9.17 μ L−1, and 82.5 ± 10.3 μ L−1, which is much higher than the normal concentrations (serum final glucose, AST, ALT, and alkaline ALP concentrations of 87.7 ± 5.93 mg dL−1, 21.1 ± 2.60 μ U−1, 10.7 ± 1.16 μ L−1, and 24.1 ± 3.97 μ L−1), respectively, compared to these serums in the 18β-GA-Acr (50 mg per kg 18β-GA) group. The biochemical variables of rats return to normal. The findings provide sufficient evidence to demonstrate that 18β-GA possesses the capability to suppress the production of oxygen species and reinstate the antioxidant mechanisms in diabetic rats afflicted with acrylamide-induced liver and kidney cytotoxicity.141 Similarly, Melekoglu et al. discovered that the antioxidant defense system parameters, encompassing malondialdehyde (MDA), reduced glutathione (GSH), superoxide dismutase (SOD), and catalase (CAT), were significantly higher in the ovarian tissues of rats treated with 18β-GA (100 mg kg−1 day−1) compared to those subjected to ischemia-reperfusion (I/R) alone.142 These findings suggest that 18β-GA may have protective effects against oxidative stress in a variety of tissues and systems. In addition to its potential antioxidant properties, recent research has also explored the potential therapeutic applications of 18β-GA in the context of viral infections. For example, Rehman et al. found that 18β-GA exhibited a solid binding affinity for several SARS-CoV-2 protein targets, including main protease (binding energy = −9.46 kcal mol−1), helicase (binding energy = −9.91 kcal mol−1), spike glycoprotein (S) (binding energy = −8.08 kcal mol−1), and E-channel proteins (binding energy = −9.72 kcal mol−1), through ligand–protein interactions. This finding suggests that 18β-GA may have the potential as a therapeutic agent in the fight against COVID-19.143We have discovered that a significant number of studies on the antioxidant properties of 18β-GA focus on its hepatoprotective function. In the mouse model of carbon tetrachloride (CCl4)-induced chronic liver fibrosis, it was observed that CCl4 inhibited the expression of Nrf2 regulatory genes, including CAT, glutathione peroxidase 2 (GPX2), and superoxide dismutase 3 (SOD3). However, 18β-GA was found to protect the mouse liver from oxidative stress by potentially activating the nuclear trans of Nrf2, enhancing the expression of its target genes, and increasing the activity of antioxidant enzymes.37 Furthermore, 18β-GA was also found to have the ability to inhibit the activity of xanthine oxidase (XO) significantly. XO is responsible for reducing O2 to superoxide anionic radical O2, leading to oxidative stress.144 In a mouse model of methotrexate (MTX)-induced liver injury, Mahmoud et al. discovered that 18β-GA was able to reverse the significant manifestations of Nrf2, hemooxygenase-1, and PPARg induced by MTX, thus restoring antioxidant defense.38 Another study demonstrated that 18β-GA significantly reduced alpha-naphthylisothiocyanate (ANIT)-induced liver damage primarily by increasing the expression of nuclear factors (such as Sirt1, FXR, and Nrf2) and their targeted excretion transporters in the liver, which play a crucial role in maintaining bile acidosis in hepatocytes. The plasma levels of ALT, AST, ALP, γ-glutamyl transpeptidase (GGT), and total bilirubin (TBIL) were significantly elevated by 31.2-, 33.4-, 5.1-, 5.0-, and 91.3-fold, respectively, in rats induced with ANIT (P < 0.0001). However, for 18β-GA (60 mg kg−1 for 7 days treatment), all of these levels showed a significant reduction of 62.0%, 38.5%, 45.7%, 51.6%, and 39.7%, respectively (P < 0.05).145 Moreover, the study also revealed that 18β-GA exerts its hepatoprotective effects against RTS-induced liver damage through the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway and enhanced glycogen synthase kinase 3 beta (GSK3β) pathway, which promotes the Nrf2-mediated antioxidant system.146 Fig. 6 briefly illustrates the hepatoprotective effect of 18β-GA based on anti-inflammatory and antioxidant mechanisms. Additionally, other hepatoprotective mechanisms are also discussed, such as the inhibition of the TLR/NF-κB pathway and upregulation of hepatic FXR to facilitate bile acid synthesis, transport, and detoxification, competitive inhibition of cyto P450 (CYP) enzymes responsible for the activation of pyrrolizidine alkaloid (PA) metabolism, particularly C3A1, which protects against liver damage, activation of PXR to regulate autophagy and lysosomal biogenesis, thereby alleviating acute liver injury, inhibition of hepatic stellate cell activation, and direct transcriptional inhibition of α2 (I) collagen gene (COL1A2), as observed in transgenic reporter mice, and other mechanisms.147–150
18β-GA derivatives (Table 5) also demonstrated significant antioxidant activity. It was discovered that compounds 244–247 exhibited robust antioxidant activity and inhibited ROS activity by up to 41%.151 Maitraie et al. observed that compounds 249–258 displayed both anti-inflammatory and antioxidant properties, with compound 254 specifically exerting inhibitory effects on NO and superoxide anions in RAW 246.7 cells.152 Moreover, Zhang et al. found that compounds 259–263 hindered the proliferation of activated hepatic stellate cells (HSC)-T6 cells by inducing apoptosis and arresting them in the G0/G1 phase. They used rat hepatic stellate cell line T6 cells activated by transforming growth factor-β-1 as the cell model and as the 18β-GA control. The IC50 value of the compound on cell proliferation was determined by tetrazolium salt colorimetry. It was found that the inhibitory effect of compounds 259–263 on activated HSC-T6 was stronger than that of GA (IC50 = 78.4 ± 2.3 μM).153 Numerous studies have demonstrated a strong association between COX-2 and the activation of hepatic stellate cells (HSCs), thereby facilitating the initiation and progression of hepatic fibrosis. Among them, compounds 262 and 265 strongly inhibit the activation of HSC-T6 cells by downregulating the expression of alpha-smooth muscle actin (α-SMA) and type I collagen (Col1) proteins, which are biomarkers of liver fibrosis. After treatment with compound activated HSC-T6, the expression levels of the two biomarkers were down-regulated. Second, both compounds downregulated the expression levels of COX-2 and transforming growth factor beta1 (TGF-β1) and reduced ROS levels in a concentration-dependent manner. This suggests that they inhibit HSC-T6 activation and may also be due to downregulation of COX-2 levels, inhibition of the TGF-β1 signaling pathway, and reduction of ROS levels.
| Compounds | 244 | 245 | 246 | 247 |
| Structure | 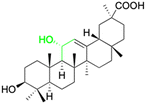 |
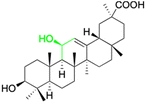 |
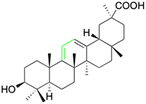 |
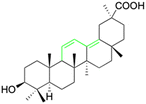 |
| Effects or mechanisms | 244: | 245: | 246: | 247: |
| ROS: | ROS: | ROS: | ROS: | |
| Inhibition of 50% activity (1.0 mg mL−1) | Inhibition of 51% activity (1.0 mg mL−1) | Inhibition of 44% activity (1.0 mg mL−1) | Inhibition of 41% activity (1.0 mg mL−1) | |
| Reference | 151 | 151 | 151 | 151 |
| Compounds | 248 | 249–258 | ||
| Structure |  |
 |
||
| Effects or mechanisms | 248: R = CH3 | 249: R1 = H, R2 = H | ||
| PMA: IC50 = 12.9 μM | fMLP/CB: IC50 = 7.0 μM | |||
| 250: R1 = CH3, R2 = H | ||||
| RAW 264.7 (B): IC50 = 26.1 μM | ||||
| 251: R1 = CH3, R2 = CH3 | ||||
| RAW 264.7 (A): IC50 = 44.3 μM | ||||
| 252: R1 = CH3, R2 = CH(CH3)2 | ||||
| RAW 264.7 (A): IC50 = 43.0 μM | ||||
| 253: R1 = CH3, R2 = Bn | ||||
| PMA: IC50 = 17.0 ± 1.5 μM | ||||
| RAW 264.7 (A): IC50 = 44.5 μM | ||||
| RAW 264.7 (B): IC50 = 13.7 μM | ||||
| 254: R1 = CH3, R2 = CH(CH3)2 | ||||
| PMA: IC50 = 15.6 μM | ||||
| RAW 264.7 (A): IC50 = 13.1 μM | ||||
| 255: R1 = CH3, R2 = C6H5 | ||||
| RAW 264.7 (B): IC50 = 15.5 μM | ||||
| 256: R1 = H, R2 = Bn | ||||
| RAW 264.7 (B): IC50 = 2.3 μM | ||||
| 257: R1 = H, R2 = CH(CH3)2 | ||||
| fMLP/CB: IC50 = 9.8 μM | ||||
| 258: R1 = H, R2 = C6H5 | ||||
| RAW 264.7 (B): IC50 = 27.7 μM | ||||
| Reference | 152 | 152 | ||
| Compounds | 259 | 260 | ||
| Structure |  |
 |
||
| Effects or mechanisms | 259: | 260: | ||
| HSC-T6*: IC50 = 17.6 μM | HSC-T6*: IC50 = 63.8 μM | |||
| Reference | 153 | 153 | ||
| Compounds | 261 | 262 | ||
| Structure |  |
 |
||
| Effects or mechanisms | 261: | 262: | ||
| HSC-T6*: IC50 = 54.5 μM | HSC-T6*: IC50 = 30.3 μM | |||
| Reference | 153 | 153 | ||
| Compounds | 263 | |||
| Structure |  |
|||
| Effects or mechanisms | 263: | |||
| HSC-T6*: IC50 = 59.8 μM | ||||
| Reference | 153 | |||
| Abbreviations | ROS: reactive oxygen species. PMA: superoxide anion formation fromrat neutrophils stimulated with PMA. fMLP/CB: superoxide anion formation from rat neutrophils stimulated with fMLP/CB. RAW 264.7 (A): the accumulation of NO2 inRAW 264.7 cells stimulated with LPS. RAW 264.7 (B): TNF-a formation from RAW 264.7 cellsstimulated with LPS. HSC-T6*: HSCs activated by TGF-β1 (10 ng mL−1) | |||
Overall, while the study of oxidative stress and its effects on the body is complex, recent research has shed light on the potential benefits of compounds like 18β-GA in combatting this process. By exploring the mechanisms of these compounds and their effects on various tissues and systems, we can better understand how to combat oxidative stress and its associated health risks.
Discussion
Experience has imparted the understanding that when a compound manifests a biological activity characterized by an IC50 value lower than 10 μM, it may be classified as potential biological efficacy. Additionally, in the process of scrutinizing lead and candidate compounds, it is importance to consider both cost-effectiveness and the intricacy of synthetic routes. Keeping these pivotal factors in consideration, the investigation unveiled that compounds 16–21 exhibited noteworthy inhibitory activities against 11β-HSD2 within the sub-micromolar (nM) range. Particularly remarkable is compound 16, which boasts an exceptionally modest synthetic complexity, necessitating a single-step reaction initiated from 18β-GA. The incorporation of amide and hydroxyl groups at the C-30 position has substantially augmented the solubility of 18β-GA. Compounds of this kind exhibit tremendous promise for further in-depth exploration. Moreover, numerous studies have demonstrated that the majority of structural alterations to 18β-GA revolve around rigid five-ring skeleton structure, encompassing the addition, removal, and replacement of functional groups. Comparatively, few studies explore the strategy, such as scaffold hopping and changes in the skeleton itself to the biological activity. Reports about compounds 38–41, 116–122, 227–230, and 248–259 have discernible indicated that brought about a substantial augmentation in the anti-tumor, antiviral, and antioxidant properties of 18β-GA through the processes of ring opening and ring expansion. The modifications in 18β-GA from the complexity of the derivative structure is mainly due to addition rather than subtraction. It may be connected with that there are few reaction methods for removing carbon atoms in the rigid alkyl skeleton.It is particularly noteworthy that compounds 227–230 demonstrate an inhibitory activity against the ZIKA virus within the sub-micromolar (nM) range. Perhaps designing modifications that involve adding or reducing rings could provide excellent solutions for enhancing the target binding strength, selectivity, bioavailability, selective tissue distribution, and metabolic stability of 18β-GA derivatives. However, further studies are necessary to comprehensively reveal their mechanisms or the target protein to further guide the modification of compounds. Moreover, 18β-GA derivatives that self-assemble, including gels, micelles, nanoparticles, and liposomes, hold potential for application in food additives and intelligent drug delivery due to availability, biocompatibility, and controllable degradability.154 Additionally, while the mainstream research direction focuses on the aforementioned topics, shifting the focus to other biologically active research areas such as anti-diabetes, anti-coagulation, and neuroprotection, could prove worthwhile, as the studies in these areas are still relatively scarce. This could further broaden the development prospects of 18β-GA derivatives and increase their role in various fields.
Conclusions
In conclusion, the past decade has yielded promising research on the therapeutic potential of 18β-GA and its derivatives for various diseases, including cancer, inflammation, bacterial infection, hepatic diseases, and viral infections. Pharmacological effects have been observed through a variety of pathways, including inflammation-related signaling, immune response modulation, and gene expression regulation. However, it is unfortunate that no derivatives have entered clinical trials (from https://www.clinicaltrials.gov) due to their poor pharmacological properties, low bioavailability, significant toxic side effects, and other factors.The review of over 200 chemical structures and key activity data in this review article serves as a valuable data resource for pharmaceutical chemists and also provides future research directions. Future research, except self-assembling derivatives, as well as exploring other related fields should more focus on revealing the mechanisms of action or the target protein and the relationship with the SAR of derivates and to further guide the structural modifications. With further research and optimization, 18β-GA derivatives will address the above crucial issues that hold great promise as potential therapeutic agents for various diseases.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author thanks Hainan Provincial Natural Science Foundation of China (No. 2019RC229 and No. 20152023) and Hainan Provincial Graduate Students Scientific Innovative Foundation (Qhys2021-368) for financial support. Sincerely thanks Professor Wang, Professor Li and Professor Xu for their selfless help and guidance in the process of writing.Notes and references
- J. Padmavathy and S. Devarajan, Bangladesh J. Pharmacol., 2017, 12, 151–161 CrossRef.
- C. Sabbadin, L. Bordin, G. DonÃ, J. Manso, G. Avruscio and D. Armanini, Front. Endocrinol., 2019, 10, 484 CrossRef PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- S. E. Osagie-Eweka, N. E. J. Orhue, E. K. I. Omogbai and F. C. Amaechina, Toxicol Rep, 2021, 8, 239–247 CrossRef CAS.
- H. B. Wang, Y. He, M. L. Jian, X. G. Fu, Y. H. Cheng, Y. J. He, J. Fang, L. Li and D. Zhang, Molecules, 2022, 27, 21 Search PubMed.
- M. Ahmad, M. Jalaluddin and B. P. Panda, Ann. Microbiol., 2013, 64, 683–688 CrossRef.
- Q. Ni, Y. Gao, X. Yang, Q. Zhang, B. Guo, J. Han and S. Chen, Front. Pharmacol, 2022, 13, 1001018 CrossRef CAS PubMed.
- C. S. Graebin, The Pharmacological Activities of Glycyrrhizinic Acid (“Glycyrrhizin”) and Glycyrrhetinic Acid, Springer International Publishing, Cham, 2018 Search PubMed.
- A. Kowalska and U. Kalinowska-Lis, Int. J. Cosmet. Sci., 2019, 41(4), 325–331 CrossRef CAS PubMed.
- L. Jin, L. Dai, M. Ji and H. Wang, Bioorg. Chem., 2019, 85, 179–190 CrossRef CAS PubMed.
- H. Wang, R. W. Li, Y. Rao, S. X. Liu, C. H. Hu, Y. Zhang, L. C. Meng, Q. L. Wu, Q. H. Ouyang, H. Liang and M. Qin, Pharmaceutics, 2022, 14, 1797 CrossRef CAS PubMed.
- A. Mittal, M. Nagpal and V. K. Vashistha, Rev. Bras. Farmacogn., 2023, 33, 1154–1169 CrossRef.
- R. Guo, Y. Liu, R. Sheng and J. Fan, Mini-Rev. Med. Chem., 2022, 22, 2024–2066 CrossRef PubMed.
- M. G. Netea, F. Balkwill, M. Chonchol, F. Cominelli, M. Y. Donath, E. J. Giamarellos-Bourboulis, D. Golenbock, M. S. Gresnigt, M. T. Heneka, H. M. Hoffman, R. Hotchkiss, L. A. B. Joosten, D. L. Kastner, M. Korte, E. Latz, P. Libby, T. Mandrup-Poulsen, A. Mantovani, K. H. G. Mills, K. L. Nowak, L. A. O'Neill, P. Pickkers, T. van der Poll, P. M. Ridker, J. Schalkwijk, D. A. Schwartz, B. Siegmund, C. J. Steer, H. Tilg, J. W. M. van der Meer, F. L. van de Veerdonk and C. A. Dinarello, Nat. Immunol., 2017, 18, 826–831 CrossRef CAS PubMed.
- R. Medzhitov, Cell, 2010, 140, 771–776 CrossRef CAS PubMed.
- T. Lawrence, Cold Spring Harbor Perspect. Biol., 2009, 1, a001651 Search PubMed.
- M. Groslambert and B. F. Py, J. Inflammation Res., 2018, 11, 359–374 CrossRef CAS PubMed.
- A. Quintás-Cardama and S. Verstovsek, Clin. Cancer Res., 2013, 19, 1933–1940 CrossRef PubMed.
- L. A. O'neill, D. Golenbock and A. G. Bowie, Nat. Rev. Immunol., 2013, 13, 453–460 CrossRef PubMed.
- J.-M. Cavaillon, Clin. Rev. Allergy Immunol., 2023, 65, 183–187 CrossRef CAS PubMed.
- V. Thiruchenthooran, E. Sánchez-López and A. Gliszczyńska, Cancers, 2023, 15, 475 CrossRef CAS PubMed.
- S. A. Richard, Mediators Inflammation, 2021, 2021, 1–15 CrossRef PubMed.
- J.-X. Zhou and M. Wink, Medicines, 2019, 6, 55 CrossRef CAS PubMed.
- C.-Y. Wang, T.-C. Kao, W.-H. Lo and G.-C. Yen, J. Agric. Food Chem., 2011, 59, 7726–7733 CrossRef CAS PubMed.
- L.-N. Peng, L. Li, Y.-F. Qiu, J.-H. Miao, X.-Q. Gao, Y. Zhou, Z.-X. Shi, Y.-L. Xu, D.-H. Shao, J.-C. Wei and Z.-Y. Ma, J. Asian Nat. Prod. Res., 2011, 13, 942–950 CrossRef CAS PubMed.
- J. Liu, Y. Xu, M. Yan, Y. Yu and Y. Guo, Sci. Rep., 2022, 12, 3121 CrossRef CAS PubMed.
- G. L. Gupta, L. Sharma and M. Sharma, Neurochem. Res., 2023, 48, 551–569 CrossRef CAS PubMed.
- T. Ishida, I. Miki, T. Tanahashi, S. Yagi, Y. Kondo, J. Inoue, S. Kawauchi, S. Nishiumi, M. Yoshida, H. Maeda, C. Tode, A. Takeuchi, H. Nakayama, T. Azuma and S. Mizuno, Eur. J. Pharmacol., 2013, 714, 125–131 CrossRef CAS PubMed.
- W. Zheng, X. Huang, Y. Lai, X. Liu, Y. Jiang and S. Zhan, Front. Pharmacol, 2021, 12, 631206 CrossRef CAS PubMed.
- R. Li, K. Wu, Y. Li, X. Liang, K. P. Lai and J. Chen, Briefings Bioinf., 2021, 22, 1161–1174 CrossRef CAS PubMed.
- Z. Zhao, Y. Xiao, L. Xu, Y. Liu, G. Jiang, W. Wang, B. Li, T. Zhu, Q. Tan and L. Tang, ACS Appl. Mater. Interfaces, 2021, 13, 20995–21006 CrossRef CAS PubMed.
- J.-X. Zhou and M. Wink, Medicines, 2019, 6, 55 CrossRef CAS PubMed.
- A. V. Shetty, S. Thirugnanam, G. Dakshinamoorthy, A. Samykutty, G. Zheng, A. Chen, M. C. Bosland, A. Kajdacsy-Balla and M. Gnanasekar, Int. J. Oncol., 2011, 39, 635–640 CAS.
- Y. Xiao, J. Xu, C. Mao, M. Jin, Q. Wu, J. Zou, Q. Gu, Y. Zhang and Y. Zhang, J. Biol. Chem., 2010, 285, 1128–1137 CrossRef CAS PubMed.
- X. Shi, L. Yu, Y. Zhang, Z. Liu, H. Zhang, Y. Zhang, P. Liu and P. Du, Int. Immunopharmacol., 2020, 84, 106578 CrossRef CAS PubMed.
- A. M. Mahmoud and H. S. Al Dera, Genes Nutr., 2015, 10, 1–13 CrossRef CAS PubMed.
- S. Chen, L. Zou, L. Li and T. Wu, PLoS One, 2013, 8, e53662 CrossRef CAS PubMed.
- A. M. Mahmoud, O. E. Hussein, W. G. Hozayen and S. M. Abd El-Twab, Chem.-Biol. Interact., 2017, 270, 59–72 CrossRef CAS PubMed.
- Z. Wang, J. Ma, Y. He, K. K. Miu, S. Yao, C. Tang, Y. Ye and G. Lin, Phytomedicine, 2022, 102, 154162 CrossRef CAS PubMed.
- Y. Ma, J. M. Liu, R. D. Chen, X. Q. An and J. G. Dai, Chin. Chem. Lett., 2017, 28, 1200–1204 CrossRef CAS.
- B. Y. Fan, B. C. Jiang, S. S. Yan, B. H. Xu, H. L. Huang and G. T. Chen, Planta Med., 2019, 85, 56–61 CrossRef CAS PubMed.
- B. Li, Y. Yang, L. Chen, S. Chen, J. Zhang and W. Tang, MedChemComm, 2017, 8, 1498–1504 RSC.
- Y. Yang, Q. Zhu, Y. Zhong, X. Cui, Z. Jiang, P. Wu, X. Zheng, K. Zhang and S. Zhao, Bioorg. Chem., 2020, 101, 103985 CrossRef CAS PubMed.
- M. Bian, D. Zhen, Q.-K. Shen, H.-H. Du, Q.-Q. Ma and Z.-S. Quan, Bioorg. Chem., 2021, 107, 104598 CrossRef CAS PubMed.
- Q. P. Zhang, Y. N. Wang, Z. Y. Wang, E. A. H. Mohammed, Q. Y. Zhao, D. He and Z. Wang, Bioorg. Chem., 2022, 119, 105542 CrossRef CAS PubMed.
- B. Li, S. Cai, Y. A. Yang, S. C. Chen, R. Chen, J. B. Shi, X. H. Liu and W. J. Tang, Eur. J. Med. Chem., 2017, 139, 337–348 CrossRef CAS PubMed.
- H. B. Wang, J. W. Zuo, L. Zha, X. Jiang, C. X. Wu, Y. A. Yang, W. J. Tang and T. L. Shi, Bioorg. Chem., 2021, 110, 104755 CrossRef CAS PubMed.
- A. V. Markov, A. V. Sen’kova, I. I. Popadyuk, O. V. Salomatina, E. B. Logashenko, N. I. Komarova, A. A. Ilyina, N. F. Salakhutdinov and M. A. Zenkova, Int. J. Mol. Sci., 2020, 21, 3511 CrossRef CAS PubMed.
- B. Tu, J. Liang, Y. Ou, X. Zhang, W. Zheng, R. Wu, L. Gan, D. Li, Y. Lu, J. Wu, W. David Hong, K. Zhang, P. Wu, J. Jin and W.-L. Wong, Bioorg. Chem., 2022, 122, 105714 CrossRef CAS PubMed.
- X. Su, H. Lawrence, D. Ganeshapillai, A. Cruttenden, A. Purohit, M. J. Reed, N. Vicker and B. V. L. Potter, Bioorg. Med. Chem., 2004, 12, 4439–4457 CrossRef CAS PubMed.
- R. Gaware, R. Khunt, L. Czollner, C. Stanetty, T. D. Cunha, D. V. Kratschmar, A. Odermatt, P. Kosma, U. Jordis and D. Claßen-Houben, Bioorg. Med. Chem., 2011, 19, 1866–1880 CrossRef CAS PubMed.
- D. V. Kratschmar, A. Vuorinen, T. D. Cunha, G. Wolber, D. Classen-Houben, O. Doblhoff, D. Schuster and A. Odermatt, J. Steroid Biochem. Mol. Biol., 2011, 125, 129–142 CrossRef CAS PubMed.
- R. You, W. Long, Z. Lai, L. Sha, K. Wu, X. Yu, Y. Lai, H. Ji, Z. Huang and Y. Zhang, J. Med. Chem., 2013, 56, 1984–1995 CrossRef CAS PubMed.
- B. Xu, G.-R. Wu, X.-Y. Zhang, M.-M. Yan, R. Zhao, N.-N. Xue, K. Fang, H. Wang, M. Chen, W.-B. Guo, P.-L. Wang and H.-M. Lei, Molecules, 2017, 22, 924 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- S. Tauro, B. Dhokchawle, P. Mohite, D. Nahar, S. Nadar and E. Coutinho, Curr. Med. Chem., 2023, 7, 848–870 Search PubMed.
- S. Wang, Y. Shen, R. Qiu, Z. Chen, Z. Chen and W. Chen, Int. J. Oncol., 2017, 51, 615–624 CrossRef CAS PubMed.
- Y.-H. Luo, C. Wang, W.-T. Xu, Y. Zhang, T. Zhang, H. Xue, Y.-N. Li, Z.-R. Fu, Y. Wang and C.-H. Jin, OncoTargets Ther., 2021, 14, 5131 CrossRef CAS PubMed.
- J. Shi, J. Li, J. Li, R. Li, X. Wu, F. Gao, L. Zou, W. W. S. Mak, C. Fu and J. Zhang, Phytomedicine, 2021, 81, 153408 CrossRef CAS PubMed.
- A. Speciale, C. Muscarà, M. S. Molonia, M. Cristani, F. Cimino and A. Saija, Molecules, 2022, 27, 1775 CrossRef CAS PubMed.
- C. Liu, Q. Ma, G. Gong and F. Su, Molecules, 2023, 28, 5855 CrossRef CAS PubMed.
- H. Hussain, I. Ali, D. Wang, F. L. Hakkim, B. Westermann, I. Ahmed, A. M. Ashour, A. Khan, A. Hussain, I. R. Green and S. T. A. Shah, Expert Opin. Drug Discovery, 2021, 16, 1497–1516 CrossRef CAS PubMed.
- Y. Lai, L. Shen, Z. Zhang, W. Liu, Y. Zhang, H. Ji and J. Tian, Bioorg. Med. Chem. Lett., 2010, 20, 6416–6420 CrossRef CAS PubMed.
- J. Hu, Y. Wu, C. Zhao, Y. Ju and Y. Ju, Chem. J. Chin. Univ., 2010, 31, 1762–1768 CAS.
- D. P. Alho, J. A. Salvador, M. Cascante and S. Marin, Molecules, 2019, 24, 766 CrossRef PubMed.
- D. Cai, Z. Zhang, Y. Chen, Y. Zhang, Y. Sun and Y. Gong, Molecules, 2019, 24, 3631 CrossRef PubMed.
- F. Zhou, G.-R. Wu, D.-S. Cai, B. Xu, M.-M. Yan, T. Ma, W.-B. Guo, W.-X. Zhang, X.-M. Huang, X.-h. Jia, Y.-Q. Yang, F. Gao, P.-L. Wang and H.-M. Lei, Eur. J. Med. Chem., 2019, 178, 623–635 CrossRef CAS PubMed.
- L. Dai, J. Li, J. Yang, Y. Men, Y. Zeng, Y. Cai and Y. Sun, Catalysts, 2018, 8, 615 CrossRef.
- R. Wang, Y. Li, X. Huai, Q. Zheng, W. Wang, H.-J. Li and Q. Huai, Drug Des., Dev. Ther., 2018, 12, 1321–1336 CrossRef CAS PubMed.
- D. K. Yadav, A. Meena, A. Srivastava, D. Chanda, F. Khan and S. Chattopadhyay, Drug Des., Dev. Ther., 2010, 4, 173–186 CAS.
- D. Cai, Z. hua Zhang, Y. Chen, C. Ruan, S. qiang Li, S. qin Chen and L. shan Chen, RSC Adv., 2020, 10, 11694–11706 RSC.
- L.-J. Wang, C.-A. Geng, Y.-B. Ma, X.-Y. Huang, J. Luo, H. Chen, X.-M. Zhang and J.-J. Chen, Bioorg. Med. Chem. Lett., 2012, 22, 3473–3479 CrossRef CAS PubMed.
- B. Lallemand, M. Gelbcke, J. Dubois, M. Prévost, I. Jabin and R. Kiss, Mini-Rev. Med. Chem., 2011, 11, 881–887 CrossRef CAS PubMed.
- Y. Li, L. Feng, Z.-F. Song, H.-B. Li and Q.-Y. Huai, Molecules, 2016, 21, 199 CrossRef PubMed.
- J. Tatsuzaki, M. Taniguchi, K. F. Bastow, K. Nakagawa-Goto, S. L. Morris-Natschke, H. Itokawa, K. Baba and K.-H. Lee, Bioorg. Med. Chem., 2007, 15, 6193–6199 CrossRef CAS PubMed.
- S.-C. Yu, Y.-T. Hou, C.-M. Hsu, F.-J. Tsai and Y. Tsai, J. Inclusion Phenom. Macrocyclic Chem., 2022, 102, 339–346 CrossRef CAS.
- R. Csuk, S. Schwarz, R. Kluge and D. Ströhl, Phytomedicine, 2010, 45, 5718–5723 CAS.
- G. O. Moustafa, A. Shalaby, A. M. Naglah, M. M. Mounier, H. El-Sayed, M. M. Anwar and E. S. Nossier, Molecules, 2021, 26, 4573 CrossRef CAS PubMed.
- R. Csuk, S. Schwarz, R. Kluge and D. Ströhl, Eur. J. Med. Chem., 2010, 45, 5718–5723 CrossRef CAS PubMed.
- I. Serbian, R. K. Wolfram, L. Fischer, A. Al-Harrasi and R. Csuk, Mediterr. J. Chem., 2018, 7, 286–293 CrossRef CAS.
- G. Chadalapaka, I. Jutooru, A. McAlees, T. Stefanac and S. Safe, Bioorg. Med. Chem. Lett., 2008, 18, 2633–2639 CrossRef CAS PubMed.
- Y. Gao, X. Guo, X. Li, D. Liu, D. Song, Y. Xu, M. Sun, Y. Jing and L. Zhao, Molecules, 2010, 15, 4439–4449 CrossRef CAS PubMed.
- E. B. Logashenko, O. V. Salomatina, A. V. Markov, D. V. Korchagina, N. F. Salakhutdinov, G. A. Tolstikov, V. V. Vlassov and M. A. Zenkova, ChemBioChem, 2011, 12, 784–794 CrossRef CAS PubMed.
- O. V. Salomatina, A. V. Markov, E. B. Logashenko, D. V. Korchagina, M. A. Zenkova, N. F. Salakhutdinov, V. V. Vlassov and G. A. Tolstikov, Bioorg. Med. Chem., 2014, 22, 585–593 CrossRef CAS PubMed.
- L.-F. Yang, Y. Xing, J.-X. Xiao, J. Xie, W. Gao, J. Xie, L.-T. Wang, J. Wang, M. Liu and Z. Yi, ACS Med. Chem. Lett., 2018, 9, 1105–1110 CrossRef CAS PubMed.
- P. Alper, O. V. Salomatina, N. F. Salakhutdinov, E. Ulukaya and F. Ari, Bioorg. Med. Chem., 2021, 30, 115963 CrossRef CAS PubMed.
- S. Wang, Y. Shen, R. Qiu, Z. Chen, Z. Chen and W. Chen, Int. J. Oncol., 2017, 51, 615–624 CrossRef CAS PubMed.
- S. Schwarz and R. Csuk, Bioorg. Med. Chem., 2010, 18, 7458–7474 CrossRef CAS PubMed.
- R. Csuk, S. Schwarz, B. Siewert, R. Kluge and D. Ströhl, Eur. J. Med. Chem., 2011, 46, 5356–5369 CrossRef CAS PubMed.
- R. Csuk, S. Schwarz, R. Kluge and D. Ströhl, Arch. Pharm., 2012, 345, 28–32 CrossRef CAS PubMed.
- R. Csuk, S. Schwarz, B. Siewert, R. Kluge and D. Ströhl, Z. Naturforsch. B Chem. Sci., 2012, 67, 731–746 CrossRef CAS.
- R. Csuk, S. Schwarz, B. Siewert, R. Kluge and D. Ströhl, Arch. Pharm., 2012, 345, 223–230 CrossRef CAS PubMed.
- W. Guo, M. Yan, B. Xu, F. Chu, W. Wang, C. Zhang, X. Jia, Y. Han, H. Xiang and Y. Zhang, Chem. Cent. J., 2016, 10, 1–11 CrossRef PubMed.
- K. Li, T. Ma, J. Cai, M. Huang, H. Guo, D. Zhou, S. Luan, J. Yang, D. Liu and Y. Jing, Bioorg. Med. Chem., 2017, 25, 5441–5451 CrossRef CAS PubMed.
- K.-W. Lin, A.-M. Huang, T.-C. Hour, S.-C. Yang, Y.-S. Pu and C.-N. Lin, Bioorg. Med. Chem., 2011, 19, 4274–4285 CrossRef CAS PubMed.
- M. Huang, P. Gong, Y. Wang, X. Xie, Z. Ma, Q. Xu, D. Liu, Y. Jing and L. Zhao, Bioorg. Chem., 2020, 103, 104187 CrossRef CAS PubMed.
- Q.-X. Zheng, R. Wang, Y. Xu, C.-X. He, C.-Y. Zhao, Z.-F. Wang, R. Zhang, W. Dehaen, H.-J. Li and Q.-Y. Huai, Biol. Pharm. Bull., 2020, 43, 102–109 CrossRef CAS PubMed.
- J. Sun, H.-Y. Liu, C.-Z. Lv, J. Qin and Y.-F. Wu, J. Agric. Food Chem., 2019, 67, 9643–9651 CrossRef CAS PubMed.
- N. Dheman, N. Mahoney, E. M. Cox, J. J. Farley, T. Amini and M. L. Lanthier, Clin. Infect. Dis., 2020, 73, e4444–e4450 CrossRef PubMed.
- J. Sun, H.-Y. Liu, C.-Z. Lv, J. Qin and Y.-F. Wu, J. Agric. Food Chem., 2019, 67, 9643–9651 CrossRef CAS PubMed.
- N. Dheman, N. Mahoney, E. M. Cox, J. J. Farley, T. Amini and M. L. Lanthier, Clin. Infect. Dis., 2020, 73, ciaa859 Search PubMed.
- D. Langer, B. Czarczynska-Goslinska and T. Goslinski, Curr. Issues Pharm. Med. Sci., 2016, 29, 118–123 CrossRef CAS.
- H. K. Kim, Y. Park, H. N. Kim, B. H. Choi, H. G. Jeong, D. G. Lee and K.-S. Hahm, Biotechnol. Lett., 2002, 24, 1899–1902 CrossRef CAS.
- M. H. Salari and Z. Kadkhoda, Clin. Microbiol. Infect., 2003, 9, 987–988 CrossRef CAS PubMed.
- J. Bielenberg and R. Krausse, Phytother. Res., 2004, 2, 37–39 Search PubMed.
- K. K. Schrader, Toxins, 2010, 2, 1676–1689 CrossRef CAS PubMed.
- R. Long Danyelle, J. Mead, M. Hendricks Jay, E. Hardy Michele and M. Voyich Jovanka, Antimicrob. Agents Chemother., 2013, 57, 241–247 CrossRef CAS PubMed.
- N. Dewake, X. Ma, K. Sato, S. Nakatsu, K. Yoshimura, Y. Eshita, H. Fujinaka, Y. Yano, N. Yoshinari and A. Yoshida, Microbiol. Immunol., 2021, 65, 343–351 CrossRef CAS PubMed.
- M. M. Celik and N. Duran, Revista Romana de Medicina de Laborator, 2019, 27, 63–71 CrossRef.
- S. Kannan, G. Sathasivam and M. Marudhamuthu, Microb. Pathog., 2019, 126, 332–342 CrossRef CAS PubMed.
- A. d. Breij, T. G. Karnaoukh, J. Schrumpf, P. S. Hiemstra, P. H. Nibbering, J. T. v. Dissel and P. C. d. Visser, Eur. J. Clin. Microbiol. Infect. Dis., 2016, 35, 555–562 CrossRef PubMed.
- Y. Zhao and X. Su, BB Rep., 2023, 33, 101427 CAS.
- B. Darvishi, S. Manoochehri, M. Esfandyari-Manesh, N. Samadi, M. Amini, F. Atyabi and R. Dinarvand, Drug Res., 2015, 65, 617–623 CrossRef CAS PubMed.
- X. Zhao, H. Zhang, Y. Gao, Y. Lin and J. Hu, ACS Appl. Bio Mater., 2020, 3, 648–653 CrossRef CAS PubMed.
- E. A. H. Mohammed, Y. Peng, Z. Wang, X. Qiang and Q. Zhao, Russ. J. Bioorg. Chem., 2022, 48, 906–918 CrossRef CAS PubMed.
- L.-R. Huang, X.-J. Hao, Q.-J. Li, D.-P. Wang, J.-X. Zhang, H. Luo and X.-S. Yang, J. Nat. Prod., 2016, 79, 721–731 CrossRef CAS PubMed.
- M. Xiang, Y.-L. Song, J. Ji, X. Zhou, L.-W. Liu, P.-Y. Wang, Z.-B. Wu, Z. Li and S. Yang, Pest Manage. Sci., 2020, 76, 2959–2971 CrossRef CAS PubMed.
- L. Zhang, Y. Fu, Y. Ding, J. Meng, Z. Wang and P. Wang, Chem. Res. Chin. Univ., 2021, 37, 662–667 CrossRef CAS.
- Y.-l. Song, H.-w. Liu, Y.-h. Yang, J.-j. He, B.-x. Yang, L.-l. Yang, X. Zhou, L.-w. Liu, P.-y. Wang and S. Yang, J. Integr. Agric., 2022, 22, 2759–2771 CrossRef.
- J.-J. He, T. Li, H.-W. Liu, L.-L. Yang, Y.-H. Yang, Q.-Q. Tao, X. Zhou, P.-Y. Wang and S. Yang, Arabian J. Chem., 2023, 16, 104771 CrossRef CAS.
- K. Oyama, M. Kawada-Matsuo, Y. Oogai, T. Hayashi, N. Nakamura and H. Komatsuzawa, PLoS One, 2016, 11, e0165831 CrossRef PubMed.
- S. Guo, S. Chen, N. Cao, W. Zheng, D. Li, Z. Sheng, X. Xu, Q. Zhang, X. Zheng, K. Wu, P. Wu, K. Zhang and W. D. Hong, J. Mater. Sci., 2021, 56, 17254–17267 CrossRef CAS.
- K. Buvanesvaragurunathan, J. Ganesh, S. N. Kumar, V. Porchezhiyan, A. Radha, P. Azhahianambi, P. Pandikumar and S. Ignacimuthu, Exp. Parasitol., 2022, 236, 108258 CrossRef PubMed.
- N. Dewake, X. Ma, K. Sato, S. Nakatsu, K. Yoshimura, Y. Eshita, H. Fujinaka, Y. Yano, N. Yoshinari and A. Yoshida, Med. Microbiol. Immunol., 2021, 65, 343–351 CrossRef CAS PubMed.
- R. Paduch and M. Kandefer-Szerszen, Mini-Rev. Org. Chem., 2014, 11, 262–268 CrossRef CAS.
- D. Elebeedy, W. F. Elkhatib, A. Kandeil, A. Ghanem, O. Kutkat, R. Alnajjar, M. A. Saleh, A. I. Abd El Maksoud, I. Badawy and A. A. Al-Karmalawy, RSC Adv., 2021, 11, 29267–29286 RSC.
- J.-Y. Pu, L. He, S.-Y. Wu, P. Zhang and X. Huang, J. Virol., 2013, 29, 673–679 CAS.
- C. Huan, Y. Xu, W. Zhang, T. Guo, H. Pan and S. Gao, Front. Pharmacol, 2021, 12, 680674 CrossRef CAS PubMed.
- H. Sato, W. Goto, J.-i. Yamamura, M. Kurokawa, S. Kageyama, T. Takahara, A. Watanabe and K. Shiraki, Antiviral Res., 1996, 30, 171–177 CrossRef CAS PubMed.
- M. E. Hardy, J. M. Hendricks, J. M. Paulson and N. R. Faunce, Virol. J., 2012, 9, 96 CrossRef CAS PubMed.
- X. Wang, F. Xie, X. Zhou, T. Chen, Y. Xue and W. Wang, Pharmaceut. Biol., 2021, 59, 1096–1103 CrossRef CAS PubMed.
- J.-C. Lin, J.-M. Cherng, M.-S. Hung, L. A. Baltina, L. Baltina and R. Kondratenko, Antiviral Res., 2008, 79, 6–11 CrossRef CAS PubMed.
- K. Fukuchi, N. Okudaira, K. Adachi, R. Odai-Ide, S. Watanabe, H. Ohno, M. Yamamoto, T. Kanamoto, S. Terakubo and H. Nakashima, In Vivo, 2016, 30, 777–785 CrossRef CAS PubMed.
- C. F. Yeh, K. C. Wang, L. C. Chiang, D. E. Shieh, M. H. Yen and J. S. Chang, J. Ethnopharmacol., 2013, 148, 466–473 CrossRef.
- L. A. Baltina, H.-C. Lai, Y.-C. Liu, S.-H. Huang, M.-J. Hour, L. A. Baltina, T. R. Nugumanov, S. S. Borisevich, L. M. Khalilov and S. F. Petrova, Bioorg. Med. Chem., 2021, 41, 116204 CrossRef CAS.
- M. A. Zígolo, M. Salinas, L. Alché, A. Baldessari and G. G. Liñares, Bioorg. Chem., 2018, 78, 210–219 CrossRef.
- S. Liang, M. Li, X. Yu, H. Jin, Y. Zhang, L. Zhang, D. Zhou and S. Xiao, Eur. J. Med. Chem., 2019, 166, 328–338 CrossRef CAS PubMed.
- S. Liang, X. Ma, M. Li, Y. Yi, Q. Gao, Y. Zhang, L. Zhang, D. Zhou and S. Xiao, Front. Chem., 2022, 10, 836955 CrossRef CAS PubMed.
- H. Ding, W. Deng, L. Ding, X. Ye, S. Yin and W. Huang, J. Med. Virol., 2020, 92, 2200–2204 CrossRef CAS PubMed.
- I. S. Alanazi, M. Emam, M. Elsabagh, S. Alkahtani and M. M. Abdel-Daim, Environ. Sci. Pollut. Res., 2021, 28, 58322–58330 CrossRef CAS PubMed.
- R. Melekoglu, O. Ciftci, S. Eraslan, S. Alan and N. Basak, BioMed Res. Int., 2018, 2018, 5421308 Search PubMed.
- M. F. u. Rehman, S. Akhter, A. I. Batool, Z. Selamoglu, M. Sevindik, R. Eman, M. Mustaqeem, M. S. Akram, F. Kanwal and C. Lu, Antibiotics, 2021, 10, 1011 CrossRef CAS PubMed.
- S. K. Hasan, R. Khan, N. Ali, A. Q. Khan, M. U. Rehman, M. Tahir, A. Lateef, S. Nafees, S. J. Mehdi and S. Rashid, Hum. Exp. Toxicol., 2015, 15, 104187 Search PubMed.
- S.-y. Wu, S.-c. Cui, L. Wang, Y.-t. Zhang, X.-x. Yan, H.-l. Lu, G.-z. Xing, J. Ren and L.-k. Gong, Acta Pharmacol. Sin., 2018, 39, 1865–1873 CrossRef CAS PubMed.
- Z. Wang, J. Ma, Y. He, K. K. Miu, S. Yao, C. Tang, Y. Ye and G. Lin, Phytomedicine, 2022, 102, 154162 CrossRef CAS PubMed.
- Q. Wang, G.-C. Song, F.-Y. Weng, B. Zou, J.-Y. Jin, D.-M. Yan, B. Tan, J. Zhao, Y. Li and F.-R. Qiu, Front. Pharmacol, 2022, 13, 881231 CrossRef CAS PubMed.
- Z. Wang, J. Ma, S. Yao, Y. He, K.-K. Miu, Q. Xia, P. P. Fu, Y. Ye and G. Lin, Front. Pharmacol, 2022, 13, 850859 CrossRef CAS PubMed.
- S. Wu, H. Lu, W. Wang, L. Song, M. Liu, Y. Cao, X. Qi, J. Sun and L. Gong, Cell Death Dis., 2021, 12, 480 CrossRef CAS PubMed.
- T. Moro, Y. Shimoyama, M. Kushida, Y. Y. Hong, S. Nakao, R. Higashiyama, Y. Sugioka, H. Inoue, I. Okazaki and Y. Inagaki, Life Sci., 2008, 83, 531–539 CrossRef CAS PubMed.
- M. Ablise, B. Leininger-Muller, C. D. Wong, G. Siest, V. Loppinet and S. Visvikis, Chem. Pharm. Bull., 2004, 52, 1436–1439 CrossRef CAS PubMed.
- D. Maitraie, C.-F. Hung, H.-Y. Tu, Y.-T. Liou, B.-L. Wei, S.-C. Yang, J.-P. Wang and C.-N. Lin, Bioorg. Med. Chem., 2009, 17, 2785–2792 CrossRef CAS PubMed.
- Q. Zhang, E. A. H. Mohammed, Y. Wang, Z. Bai, Q. Zhao, D. He and Z. Wang, Bioorg. Chem., 2020, 99, 103804 CrossRef CAS PubMed.
- L. Zou, Q. Li, Y. Hou, M. Chen, X. Xu, H. Wu, Z. Sun and G. Ma, Food Funct., 2022, 13, 12487–12509 RSC.
| This journal is © The Royal Society of Chemistry 2024 |