Open Access Article
Open Access ArticleDe novo designed aliphatic and aromatic peptides assemble into amyloid-like cytotoxic supramolecular nanofibrils†
Satyabrata Samuia,
Soumi Biswasa,
Shubhanwita Basaka,
Shreya Ghosha,
K. Muniyappab and
Jishu Naskar *a
*a
aDepartment of Biochemistry and Biophysics, University of Kalyani, Nadia, WB 741235, India
bDepartment of Biochemistry, Indian Institute of Science, Bangalore, Karnataka 560 012, India. E-mail: jishunaskar@gmail.com
First published on 1st February 2024
Abstract
Peptides are very interesting biomolecules that upon self-association form a variety of thermodynamically stable supramolecular structures of nanometric dimension e.g. nanotubes, nanorods, nanovesicles, nanofibrils, nanowires and many others. Herein, we report six peptide molecules having a general chemical structure, H-Gaba-X-X-OH (Gaba: γ-aminobutyric acid, X: amino acid). Out of these six peptides, three are aromatic and the others are aliphatic. Atomic force microscopic (AFM) studies reveal that except peptide 6 (H-Gaba-Trp-Trp-OH), all the reported peptides adopt nanofibrillar morphology upon aggregation in aqueous medium. These supramolecular assemblies can recognize amyloid-specific molecular probe congo red (CR) and thioflavine t (ThT) and exhibit all the characteristic properties of amyloids. The MTT cell viability assay reveals that the toxicity of both aliphatic and aromatic peptides increases with increasing concentration of the peptides to both cancer (HeLa) and non-cancer (HEK 293) cells. Of note, the aromatic peptides show a slightly higher cytotoxic effect compared to the aliphatic peptides. Overall, the studies highlight the self-assembling nature of the de novo designed aliphatic and aromatic peptides and pave the way towards elucidating the intricacies of pathogenic amyloid assemblies.
Introduction
Self-assembly, a typical phenomenon exists in matters of all scales from molecules to galaxies.1a,b It is an intriguing ‘bottom-up’ strategy to fabricate various supramolecular structures.2a In the self-assembly process the building blocks are held together reversibly by various non-covalent intermolecular forces like H-bonding, hydrophobic and hydrophilic interactions, electrostatic interactions, van der Waals forces and stacking interactions.2b–d The molecular self-assembly is an intermolecular phenomenon leading to various supramolecular structures having distinctive properties compared to the building blocks (molecules).2e,fPeptides are very smart and versatile molecules having both H-bond accepting and donating sites. Self-assembling peptides could independently build up a variety of supramolecular structures e.g. nanotubes, nanorods, nanofibrils, nanovesicles, and many others that usually play a decisive role in the function of the materials.3a–f Among the supramolecular structures adopted by peptide assemblies, nanofibrils in recent years have drawn considerable attention due to their resemblance with amyloid fibrils which are associated with a great variety of diseases.4a–c Several peptides/proteins from unrelated origins are found to form amyloid fibrils in vitro and in vivo.5a–d Although the sequence homology among the different amyloid forming protein/peptide is not at all found, yet the amyloid aggregates show similar morphological and biophysical characteristics.4b,6a There are various factors such as hydrophobicity, charge, secondary structure propensity, sequence patterning etc. which are believed to influence the aggregation of peptides and proteins.6b,7a–d But, the relative contribution of these non-covalent forces in fibrillogenesis process is yet to be understood. In natural amyloid, aromatic residues are quite abundant which leads to the concept that aromatic amino acids play an important role in amyloid formation.8a–c Several studies have shown that π–π interaction, a kind of hydrophobic interaction, originating from aromatic amino acid residues is the key to the genesis and stabilization of amyloid fibrils.8b,c,9a–j A short peptide, NFGAILSS (hIAPP22–29), is found to aggregate into β-sheet containing amyloid fibrils which are very much similar to full-length hIAPP. But, when Phe-23 is substituted with alanine, the peptide fails to show amyloidogenic properties.10a,b On the other hand, if Phe-23 is replaced with tryptophan, the resulting peptide forms amyloid fibrils at much higher rate.11 In another study, it is shown that NFGAIL (hIAPP22–27) has the ability to form amyloid fibrils very much similar to the full-length polypeptide.10a Importantly, the amyloidogenicity of this peptide was completely lost upon the substitution of phenylalanine with alanine, but the substitution of any other amino acid with alanine did not affect the amyloid formation.8a Likewise, the role of aromatic amino acid residues in amyloid β-protein (Aβ) (which has a connection to the pathogenesis of Alzheimer's disease) aggregation has also been studied. From this study, it was found that the substitution of phenylalanine at position 19 of Aβ protein with leucine resulted in a peptide that aggregates at a significantly slower rate than the native one.12 The influence of aromatic rings, the side-chain hydrophobicity, and steric effects on the self-assembly of Aβ16–22 (Aβ derived peptide) have also been studied.13a–c The role of phenylalanine residue in the fibrillation of a model peptide KFFEAAAKKFFE is reported by Marshall et al.14 There are also various peptides reported in the literature which does not contain any aromatic amino acid residue, still can form amyloid like fibrils.15a–g Lakshmanan et al. reported two short aliphatic peptides, LD6 and ID3 which can form amyloid fibrils despite lacking any aromatic amino acid residue.16 Another peptide, GGVVIA, from the transmembrane domain of amyloid-β (Aβ) shows a very high propensity for amyloid aggregation.17
We are continuously investigating the supramoleecular assemblies of various peptide-based molecules and studying the amyloidogenic nature of the nanofibrillar peptide aggregates.18a–e Here we have reported a set of aliphatic and aromatic peptides having general chemical structure (Fig. 1) H-Gaba-X-X-OH (Gaba: γ-aminobutyric acid, X: amino acid) and address its supramolecular structures, amyloidogenic nature and cytotoxicity. The chemical structure of the peptide (H-Gaba-X-X-OH) was designed to investigate the aggregation propensity of different amino acids (X). We have purposefully incorporated γ-aminobutyric acid (Gaba) as the N-terminal residue to induce flexibility in the peptide backbone.
Results and discussion
Morphological analysis
Primarily to assess the aggregation of peptides in aqueous medium dynamic light scattering (DLS) experiment was performed. DLS is a rapid screening method to speculate the size of the supramolecule.19 From DLS study it was observed that for each peptide there was a discrete peak of different intensities at different position of the spectrum. The hydrodynamic diameter of the supramolecular aggregates were 280, 732, 278, 386, 690 and 3433 nm respectively for peptide 1, 2, 3, 4, 5 and 6 (Fig. S35 and Table S1†). Thus DLS study encouraged us to further investigate the morphology of the aggregated peptides by high-end, sophisticated microscope. Now, to study the morphological characteristics of the supramolecular assemblies, atomic force microscopic (AFM) experiment has been performed. AFM study revealed that except peptide 6, all peptides formed nanofibrillar aggregates upon self association in aqueous medium. Interestingly, the length and diameter of the fibrils were unique for each peptide. The diameter of the fibrils ranges from 9 to 50 nm (Fig. 2).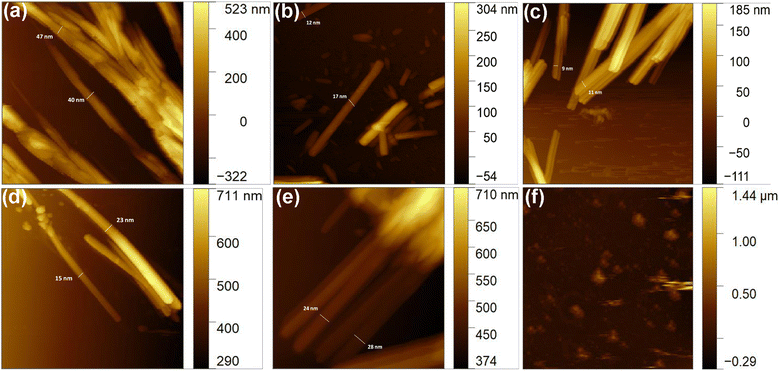 | ||
| Fig. 2 Morphology of the aggregated peptides. (a)–(f) are the AFM images of aggregated peptide 1, 2, 3, 4, 5, and 6 respectively. | ||
Congo red (CR) binding study
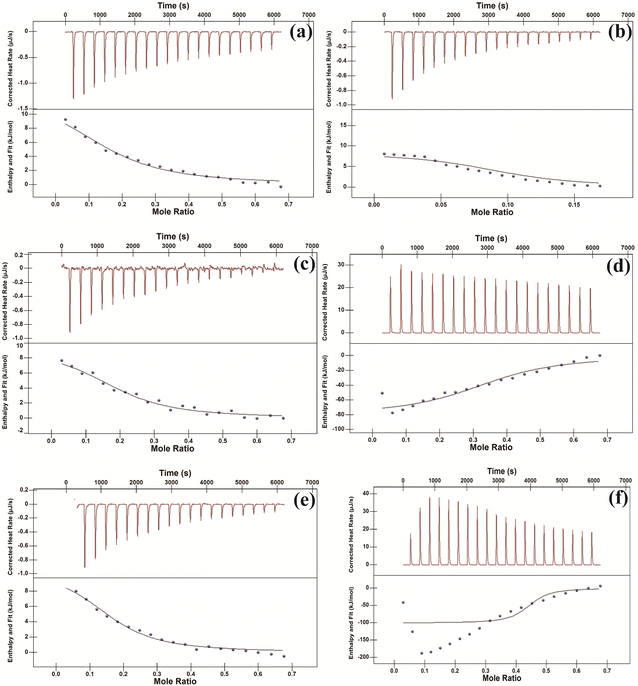
CR is a very well-known dye which is usually used to check the amyloidogenic nature of the peptide/protein aggregate.20a,b Of note, it cannot bind with the monomer of protein/peptide but can do with aggregate.21a,b Firstly, isothermal titration calorimetric (ITC) study has been performed in order to check whether CR can bind with the aggregated peptides. From the study it was observed that CR formed thermodynamically stable conjugate with all peptides (Fig. 3). The binding constant (Ka) values and other thermodynamic parameters for the interactions have been provided in Table S2.† Further, both the CR absorbance and CR birefringence studies have been performed to presume amyloidogenic nature of the aggregate.20a,22 From UV study, an increased molar absorbance (hyperchromicity) was observed when CR binds with the peptide aggregates (Fig. S36†). Interestingly, aggregates of peptide 1, 2, 3, 4 and 5 upon conjugation with CR exhibit characteristic birefringence under polarized light (Fig. 4 and S37†). But the aggregate of peptide 6 was unable to show birefringence property under similar experimental condition. Thus CR binding study primarily indicates the amyloidogenic nature of all peptide aggregates except peptide 6.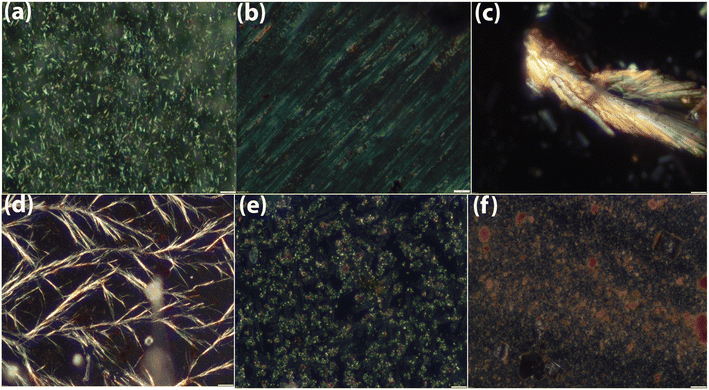 | ||
| Fig. 4 Characteristic birefringence of CR-peptide conjugates under polarized light. (a)–(f) are the representative of peptide 1, 2, 3, 4, 5 and 6. | ||
Thioflavine T (ThT) binding study
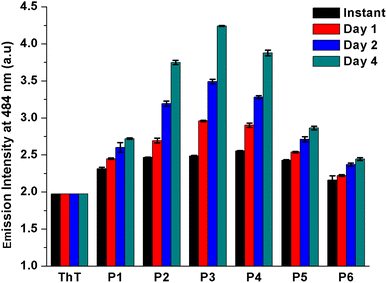
Further, ThT binding study has been performed to validate the amyloidogenic nature of the peptide aggregates. ThT is another amyloid specific dye which particularly binds with the aggregated β-sheet rich structure of amyloid but not with the monomeric protein/peptide.23 The peptides were co-incubated with ThT for 4 days and time-to-time the emission of ThT-peptide complexes were measured upon excitation at 450 nm. From the study, it was observed that for all peptides the emission maxima of peptide-ThT complex was at 484 nm (Fig. S38†) and the emission intensity gradually increased with time, although the rate of increment was not same for all cases (Fig. 5). At instant ThT-peptide 4 conjugate exhibited slightly higher emission intensity than others. But after 4 days of incubation ThT-peptide 3 conjugate exhibited the highest emission intensity and the order of emission intensity after 4 days followed the trend peptide 3 > peptide 4 > peptide 2 > peptide 5 > peptide 1. Form this result it is anticipated that at instant the higher emission intensity of ThT-peptide 4 conjugate is probably due to the higher aggregation propensity of peptide 4 compared to other peptides. But after 4 days, an equilibrium established between monomer and aggregated peptides. ThT binds with the thermodynamically stable aggregates of peptide and shows the characteristic emission. Thus the emission characteristic after 4 days suggests that peptide 3 has the highest amyloidogenic potential. Finally to check the morphology of the aggregated peptides, ThT stained peptide aggregates were viewed under fluorescence microscope (Fig. 6).24a Except peptide 6, all peptides revealed characteristic fibrillar morphology supporting the result obtained from AFM study.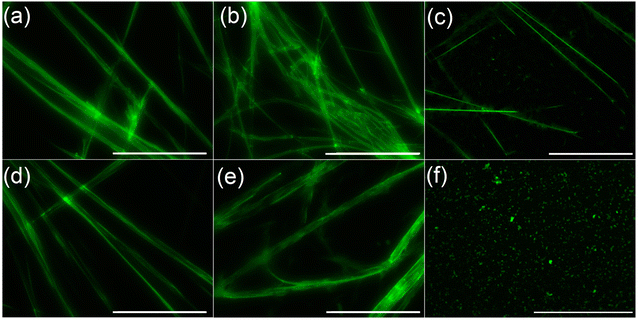 | ||
| Fig. 6 Fluorescence microscopic images of aggregated peptide upon staining with ThT. (a)–(f) stand for peptide 1, 2, 3, 4, 5 and 6 respectively. Scale bar is of 5 micrometer. | ||
Conformational analysis
Fourier-transformed infrared (FT-IR) spectroscopic studies have been carried out to know the secondary structure of the aggregated peptides in solution.24b The seven days aged solutions of all peptides exhibited sharp C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (amide I) band near 1635 cm−1 which indicates the hydrogen bonded supramolecular β-sheet structure of the peptides in the aggregated state (Fig. S34†).
O stretching (amide I) band near 1635 cm−1 which indicates the hydrogen bonded supramolecular β-sheet structure of the peptides in the aggregated state (Fig. S34†).
Cytotoxicity of the aggregated peptides
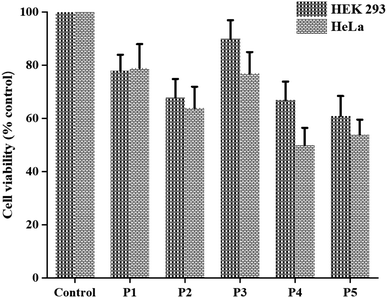
Finally, we evaluated the cytotoxic potential of the aggregated peptides on cancer (HeLa) and non-cancer (HEK 293) cell lines using MTT cell viability assay. To this end, the cell lines were exposed to different concentrations of peptide ranging from 0 to 7.5 mM for 48 h. The results indicated that the reported peptides exert cytotoxic effect in a manner dependent on their concentrations (Fig. S39†). Interestingly, the peptides containing aromatic amino acids displayed slightly higher cytotoxic effect than the peptides containing aliphatic amino acid on both cell lines (Fig. 7 and Table S3†). We are unable to test the cytotoxic effect of peptide 6, as it precipitated under the experimental condition.Conclusion
Here, we report the supramolecular features of the de novo designed aliphatic and aromatic peptides. All the reported peptides (except peptide 6), form nanofibrillar aggregates upon self-association in aqueous medium. CR absorption and birefringence study along with characteristic ThT emission confirm the amyloidogenic nature of the nanofibrillar peptide aggregates. The MTT cell survival assay suggests that the cytotoxicity of the peptides increases with increasing concentration in both cancer and non-cancer cell lines. Interestingly, aromatic peptides are found to have slightly higher cytotoxic property than aliphatic peptides. The studies are in agreement with the generic nature of amyloid and also support the previous report15g that the presence of aromatic residue(s) or aromatic–aromatic interaction is not the sole requirement for the amyloid fibrillogenesis process. Overall the studies pave the way to elucidate the intricacies of pathogenic amyloid assemblies.Materials and methods
The amino acids required for the synthesis of peptides and coupling reagents were purchased from Sisco Research Laboratories Pvt. Ltd (SRL), India. The peptides were synthesized by conventional solution phase methods using racemization free fragment condensation method. The Boc group was used for N-terminal protection and the C-terminus was protected as methylester. Couplings were mediated by dicyclohexylcarbodiimide (DCC) and 1-hydroxybenzotriazole (HOBt). The protecting group from C-terminus was removed by base catalyzed saponification method and the Boc-group from N-terminus was removed upon treatment with trifluoroaceticacid (TFA). All intermediates have been characterized by 1H NMR spectroscopy and the final compounds were fully characterized by NMR, IR spectroscopy and mass spectrometry. Detailed synthetic and experimental procedures and spectral data have been provided in the ESI.†Conflicts of interest
There is no conflict of interest to declare.Acknowledgements
Shubhanwita Basak acknowledges University of Kalyani for URS fellowship. Dr Jishu Naskar is indebted to the Department of Science and Technology (DST) (EEQ/2022/000738) and UGC-DAE (CRS/2021–22/04/634) for funding.References
- (a) J.-M. Lehn, Science, 2002, 295, 2400–2403 CrossRef CAS PubMed; (b) G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef CAS PubMed.
- (a) G. M. Whitesides, J. P. Mathias and C. T. Seto, Science, 1991, 254, 1312–1319 CrossRef CAS PubMed; (b) G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef CAS PubMed; (c) Y. Bai, Q. Luo and J. Liu, Chem. Soc. Rev., 2016, 45, 2756–2767 RSC; (d) C. Gong, S. Sun, Y. Zhang, L. Sun, Z. Su, A. Wu and G. Wei, Nanoscale, 2019, 11, 4147–4182 RSC; (e) T. Sawada, M. Yamagami, K. Ohara, K. Yamaguchi and M. Fujita, Angew. Chem., Int. Ed., 2016, 55, 4519–4522 CrossRef CAS PubMed; (f) M. Yamagami, T. Sawada and M. Fujita, J. Am. Chem. Soc., 2018, 140, 8644–8647 CrossRef CAS PubMed.
- (a) A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Nat. Rev. Chem, 2020, 4, 615–634 CrossRef CAS; (b) P. Janković, I. Šantek, A. S. Pina and D. Kalafatovic, Front. Chem., 2021, 9, 723473, DOI:10.3389/fchem.2021.723473; (c) T. Li, X.-M. Lu, M.-R. Zhang, K. Hu and Z. Li, Bioact. Mater., 2022, 11, 268–282 CAS; (d) E. Gazit, Chem. Soc. Rev., 2007, 36, 1263–1269 RSC; (e) S. L. Manna, C. D. Natale, V. Onesto and D. Marasco, Int. J. Mol. Sci., 2021, 22(23), 12662 CrossRef PubMed; (f) B. Adhikari and A. Banerjee, J. Indian Inst. Sci., 2011, 91, 471–483 CAS.
- (a) N. P. Reynolds, Biointerphases, 2019, 814(4), 040801, DOI:10.1116/1.5098332; (b) S. K. Maji, L. Wang, J. Greenwald and R. Riek, FEBS Lett., 2009, 583, 2610–2617 CrossRef CAS PubMed; (c) S. Mankar, A. Anoop, S. Sen and S. K. Maji, Nano Rev., 2011, 2, 6032, DOI:10.3402/nano.v2i0.6032.
- (a) J. Bieschke, J. Russ, R. P. Friedrich, D. E. Ehrnhoefer, H. Wobst, K. Neugebauer and E. E. Wanker, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7710–7715 CrossRef CAS PubMed; (b) Y. Porat, A. Abramowitz and E. Gazit, Chem. Biol. Drug Des., 2006, 67, 27–37 CrossRef CAS PubMed; (c) H. Shoval, D. Lichtenberg and E. Gazit, Amyloid, 2007, 14, 73–87 CrossRef CAS PubMed; (d) K. Ono, K. Hasegawa, H. Naiki and M. Yamada, J. Neurosci. Res., 2004, 75, 742–750 CrossRef CAS PubMed.
- (a) L. C. Serpell, Biochim. Biophys. Acta, 2000, 1502, 16–30 CrossRef CAS PubMed; (b) I. W. Hamley, Angew. Chem., Int. Ed., 2007, 46, 8128–8147 CrossRef CAS PubMed.
- (a) F. T. Senguen, T. M. Doran, E. A. Anderson and B. L. Nilsson, Mol. BioSyst., 2011, 7, 497–510 RSC; (b) F. Chiti, M. Stefani, N. Taddei, G. Ramponi and C. M. Dobson, Nature, 2003, 424, 805–808 CrossRef CAS PubMed; (c) O. S. Makin, E. Atkins, P. Sikorski, J. Johansson and L. C. Serpell, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 315–320 CrossRef CAS PubMed; (d) F. Bemporad, G. Calloni, S. Campioni, G. Plakoutsi, N. Taddei and F. Chiti, Acc. Chem. Res., 2006, 39, 620–627 CrossRef CAS PubMed.
- (a) R. Azriel and E. Gazit, J. Biol. Chem., 2001, 276, 34156–34161 CrossRef CAS PubMed; (b) E. Gazit, FASEB J., 2002, 16, 77–83 CrossRef CAS PubMed; (c) C. H. Görbitz, Chem. Commun., 2006, 22, 2332–2334 RSC.
- (a) I. M. Stanković, S. Niu, M. B. Hall and S. D. Zarić, Int. J. Biol. Macromol., 2020, 156, 949–959 CrossRef PubMed; (b) S. M. Tracz, A. Abedini, M. Driscoll and D. P. Raleigh, Biochemistry, 2004, 43, 15901–15908 CrossRef CAS PubMed; (c) D. Zanuy, Y. Porat, E. Gazit and R. Nussinov, Structure, 2004, 12, 439–455 CrossRef CAS PubMed; (d) G. W. Platt, K. E. Routledge, S. W. Homans and S. E. Radford, J. Mol. Biol., 2008, 378, 251–263 CrossRef CAS PubMed; (e) A. Frydman-Marom, M. Rechter, I. Shefler, Y. Bram, D. E. Shalev and E. Gazit, Angew. Chem., Int. Ed., 2009, 48, 1981–1986 CrossRef CAS PubMed; (f) D. M. Chung, Y. Dou, P. Baldi and J. S. Nowick, J. Am. Chem. Soc., 2005, 127, 9998–9999 CrossRef CAS PubMed; (g) C. D. Tatko and M. L. Waters, J. Am. Chem. Soc., 2002, 124, 9372–9373 CrossRef CAS PubMed; (h) M. L. Waters, Curr. Opin. Chem. Biol., 2002, 6, 736–741 CrossRef CAS PubMed; (i) G. G. Tartaglia, A. Cavalli, R. Pellarin and A. Caflisch, Protein Sci., 2004, 13, 1939–1941 CrossRef CAS PubMed; (j) T. M. Doran, A. J. Kamens, N. K. Byrnesand and B. L. Nilsson, Proteins, 2012, 80, 1053–1065 CrossRef CAS PubMed.
- (a) K. Tenidis, M. Waldner, J. Bernhagen, W. Fischle, M. Bergmann, M. Weber, M.-L. Merkle, W. Voelter, H. Brunner and A. Kapurniotu, J. Mol. Biol., 2000, 295, 1055–1071 CrossRef CAS PubMed; (b) P. Westermark, U. Engstrӧm, K. H. Johnson, G. T. Westermark and C. Betsholtz, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 5036–5040 CrossRef CAS PubMed.
- Y. Porat, A. Stepensky, F. Naider and E. Gazit, Biopolymers, 2003, 69, 161–164 CrossRef CAS PubMed.
- R. Cukalevski, B. Boland, B. Frohm, E. Thulin, D. Walsh and S. Linse, ACS Chem. Neurosci., 2012, 3, 1008–1016 CrossRef CAS PubMed.
- (a) F. T. Senguen, N. R. Lee, X. Gu, D. M. Ryan, T. M. Doran, E. A. Anderson and B. L. Nilsson, Mol. BioSyst., 2011, 7, 486–496 RSC; (b) N. Chaudhary and R. Nagaraj, J. Pept. Sci., 2011, 17, 115–123 CrossRef CAS PubMed; (c) S. K. Pachahara and R. Nagaraj, Biochem. Biophys. Rep., 2015, 2, 1–13 Search PubMed.
- K. E. Marshall, K. L. Morris, D. Charlton, N. O'Reilly, L. Lewis, H. Walden and L. C. Serpell, Biochemistry, 2011, 50, 2061–2071 CrossRef CAS PubMed.
- (a) S. Ray, A. K. Das, M. G. Drew and A. Banerjee, Chem. Commun., 2006, 4230–4232 RSC; (b) C. A. Hauser, R. Deng, A. Mishra, Y. Loo, U. Khoe, F. Zhuang, D. W. Cheong, A. Accardo, M. B. Sullivan, C. Riekel, J. Y. Ying and U. A. Hauser, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1361–1366 CrossRef CAS PubMed; (c) P. Moscarelli, F. Boraldi, B. Bochicchio, A. Pepe, A. M. Salvi and D. Quaglino, Matrix Biol., 2014, 36, 15–27 CrossRef CAS PubMed; (d) L. L. del Mercato, G. Maruccio, P. P. Pompa, B. Bochicchio, A. M. Tamburro, R. Cingolani and R. Rinaldi, Biomacromolecules, 2008, 9, 796–803 CrossRef CAS PubMed; (e) N. Gour, C. P. Kanth, B. Koshti, V. Kshtriya, D. Shah, S. Patel, R. Agrawal-Rajput and M. K. Pandey, ACS Chem. Neurosci., 2019, 10, 1230–1239 CrossRef CAS PubMed; (f) S. Shaham-Niv, L. Adler-Abramovich, L. Schnaider and E. Gazit, Sci. Adv., 2015, 1, e1500137 CrossRef PubMed; (g) P. Marek, A. Abedini, B. Song, M. Kanungo, M. E. Johnson, R. Gupta, W. Zaman, S. S. Wong and D. P. Raleigh, Biochemistry, 2007, 46, 3255–3261 CrossRef CAS PubMed.
- A. Lakshmanan, D. W. Cheong, A. Accardo, E. Di Fabrizio, C. Riekel and C. A. Hauser, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 519–524 CrossRef CAS PubMed.
- J.-P. Colletier, A. Laganowsky, M. Landau, M. Zhao, A. B. Soriaga, L. Goldschmidt, D. Flot, D. Cascio, M. R. Sawaya and D. Eisenberg, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16938–16943 CrossRef CAS PubMed.
- (a) S. Samui, S. Biswas, K. Roy, I. Deb and J. Naskar, ACS Chem. Neurosci., 2019, 10, 2915–2918 CrossRef CAS PubMed; (b) S. Samui, A. Chakraborty, S. Biswas, G. Singh, S. Mondal, S. Ghosh, A. Bagchi, R. S. Ampapathi and J. Naskar, ChemistrySelect, 2018, 3, 2523–2527 CrossRef CAS; (c) S. Biswas, S. Samui, A. Chakraborty, S. Biswas, D. De, U. Ghosh, A. K. Das and J. Naskar, Biochem. Biophys. Rep., 2017, 11, 112–118 Search PubMed; (d) S. Samui, S. Biswas, K. Muniyappa and J. Naskar, chemistryselect, 2021, 6, 11103–11107 CrossRef CAS; (e) J. Naskar, M. G. B. Drew, I. Deb, S. Das and A. Banerjee, Org. Lett., 2008, 10, 2625–2628 CrossRef CAS PubMed.
- A. Wishard and B. C. Gibb, Supramol. Chem., 2019, 31, 608–615 CrossRef CAS PubMed.
- (a) W. E. Klunk, R. F. Jacob and R. P. Mason, Methods Enzymol., 1999, 309, 285–305 CAS; (b) G. T. Westermark, K. H. Johnson and P. Westermark, Methods Enzymol., 1999, 309, 3–25 CAS.
- (a) S. K. Maji, M. H. Perrin, M. R. Sawaya, S. Jessberger, K. Vadodaria, R. A. Rissman, P. S. Singru, K. P. Nilsson, R. Simon, D. Schubert, D. Eisenberg, J. Rivier, P. Sawchenko, W. Vale and R. Riek, Science, 2009, 325, 328–332 CrossRef CAS PubMed; (b) S. K. Maji, D. Schubert, C. Rivier, S. Lee, J. E. Rivier and R. Riek, PLoS Biol., 2008, 6, e17 CrossRef PubMed.
- R. N. Rambaran and L. C. Serpell, Prion, 2008, 2, 112–117 CrossRef PubMed.
- H. LeVine III, Methods Enzymol., 1999, 309, 274–284 Search PubMed.
- (a) N. N. Jha, A. Anoop, S. Ranganathan, G. M. Mohite, R. Padinhateeri and S. K. Maji, Biochemistry, 2013, 52, 8800–8810 CrossRef CAS PubMed; (b) A. Anoop, S. Ranganathan, B. D. Dhaked, N. N. Jha, S. Pratihar, S. Ghosh, S. Sahay, S. Kumar, S. Das, M. Kombrabail, K. Agarwal, R. S. Jacob, P. Singru, P. Bhaumik, R. Padinhateeri, A. Kumar and S. K. Maji, J. Biol. Chem., 2014, 289, 16884–16903 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07869h |
| This journal is © The Royal Society of Chemistry 2024 |