Open Access Article
Open Access ArticleUse of Raman spectroscopy to study rat lung tissues for distinguishing asphyxia from sudden cardiac death†
Kai Zhang‡
ab,
Ruina Liu‡c,
Xin Weib,
Zhenyuan Wang *b and
Ping Huang*ad
*b and
Ping Huang*ad
aShanghai Key Lab of Forensic Medicine, Key Lab of Forensic Science, Ministry of Justice, China, Academy of Forensic Science, Shanghai, People's Republic of China. E-mail: huangp@ssfjd.cn
bDepartment of Forensic Pathology, College of Forensic Medicine, NHC Key Laboratory of Forensic Science, Xi'an Jiaotong University, Xi'an, People's Republic of China. E-mail: wzy218@xjtu.edu.cn
cCenter for Translational Medicine, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, People's Republic of China
dInstitute of Forensic Science, Fudan University, Shanghai, People's Republic of China
First published on 14th February 2024
Abstract
Determining asphyxia as the cause of death is crucial but is based on an exclusive strategy because it lacks sensitive and specific morphological characteristics in forensic practice. In some cases where the deceased has underlying heart disease, differentiation between asphyxia and sudden cardiac death (SCD) as the primary cause of death can be challenging. Herein, Raman spectroscopy was employed to detect pulmonary biochemical differences to discriminate asphyxia from SCD in rat models. Thirty-two rats were used to build asphyxia and SCD models, with lung samples collected immediately or 24 h after death. Twenty Raman spectra were collected for each lung sample, and 640 spectra were obtained for further data preprocessing and analysis. The results showed that different biochemical alterations existed in the lung tissues of the rats that died from asphyxia and SCD and could be used to distinguish between the two causes of death. Moreover, we screened and used 8 of the 11 main differential spectral features that maintained their significant differences at 24 h after death to successfully determine the cause of death, even with decomposition and autolysis. Eventually, seven prevalent machine learning classification algorithms were employed to establish classification models, among which the support vector machine exhibited the best performance, with an area under the curve value of 0.9851 in external validation. This study shows the promise of Raman spectroscopy combined with machine learning algorithms to investigate differential biochemical alterations originating from different deaths to aid determining the cause of death in forensic practice.
Introduction
The determination of the cause of death holds significant importance in forensic practice, predominantly relying on conventional morphological methods. However, in certain cases, distinctive morphological indicators may be insufficient to unquestionably determine the cause of death. For example, in cases where an individual has died of asphyxia due to a pillow covering the mouth and nose, hands choking the neck, or inadequate oxygen in the environment or in cases where the body is examined after several days of autolysis and decomposition, morphological signs such as petechial hemorrhage, ligature grooves, throttling marks, and neck muscle hemorrhage, which help to determine the cause of death as asphyxia, may be absent or difficult to identify.1 Nevertheless, asphyxia can be ascertained as the cause of death through systematic autopsy, toxicology examination, and scene investigation by ruling out alternative causes. However, it becomes harder when the deceased has an underlying heart disease that can trigger death because sudden cardiac death (SCD) is also a cause of death that should be concluded by excluding other causes. Unfortunately, this is not rare because asphyxia is a common form of violence and SCD is a significant public health issue worldwide.2–4 Therefore, it is imperative to research means to differentiate between asphyxia and SCD considering the major forensic importance and social value of doing so.Current studies have mainly focused on finding differentially expressed mRNAs, miRNAs, proteins, or metabolites during asphyxia5–8 or SCD.9,10 Although these biomarkers have shown promising sensitivity, their specificity remains insufficiently studied for their practical applications. In particular, further research is needed on their altered content during after-death autolysis and decomposition as well as their determination efficiency. We previously reported that mass spectrometry-based non-targeted metabolomics can be used to identify differential metabolites between asphyxia and SCD.11–13 However, the sample pretreatment and data-collection procedures related to molecular biology techniques and mass spectrometry are laborious, time-consuming, and expensive.
Raman spectroscopy has emerged as a highly promising analytical technique for rapid, non-invasive, non-destructive, and non-label detection, especially in biomedical research, due to its minimal sample requirements, sample preparation, and high resistance to water interference.14,15 Moreover, Raman spectroscopy has gained increasing attention within the forensic field, such as for identifying illegal drugs,16 examining questioned documents,17,18 detecting gunshot residue,19 analyzing body fluid trace evidence acquired from crime scenes,19–21 and estimating the postmortem interval.22,23 Raman spectroscopy can examine subtle biochemical alterations in certain pathophysiological mechanisms to diagnose associated diseases.24,25 Infrared spectroscopy, another vibrational spectroscopy technique complementary to Raman spectroscopy, is also widely used in forensic research to identify injuries, histopathological changes,26,27 and determine the cause of death.28,29 In addition, the technical advantages of Raman spectroscopy make it more suitable for forensic practice than other molecular biological techniques. Therefore, although no relevant research has been reported yet to the best of our knowledge, we believe that Raman spectroscopy can be utilized to determine complex causes of death.
Herein, we established rat death models of asphyxia and SCD. Raman spectra of the lung tissues collected at 0 h and 24 h after death were acquired. The differences in biochemical changes in the lung tissues during death between asphyxia and SCD were analyzed by chemometrics, and multiple classification models were built and compared to determine the cause of death accurately.
Materials and methods
Animal experiments and sample preparation
Thirty-two male Sprague Dawley rats (weighing 200–250 g) were obtained from the Experimental Animal Center of Xi'an Jiaotong University. The rats were housed in a controlled environment at 25 °C with a fixed light–dark cycle of 12 h, provided with unrestricted access to food and water before the modeling, and randomly and equally allocated into asphyxia and SCD groups (n = 16) based on the death-induction method. Subsequently, each group was divided into immediate and 24 h after-death subgroups (n = 8) based on the sample collection time. Before being euthanized, all the rats were anesthetized by the intraperitoneal injection of urethane (20%, 1.0–1.2 g kg−1) to minimize potential suffering. Our animal operations were approved by the Committee of the Ethics of Animal Experiments of Xi'an Jiaotong University and followed the Institutional Animal Care and Use Committee of Xi'an Jiaotong University Guideline.In the asphyxia group, the model was constructed by strangulating by ligature; whereby a cotton thread noose was placed around the neck of each rat, and a small stick was inserted into the noose at the back of the neck. Subsequently, the noose was tightened by rotating the stick to induce asphyxiation in each rat, maintaining constant pressure until death. In the SCD group, acute myocardial ischemia was induced through coronary ligation of the left anterior descending coronary artery to simulate cardiac death.
In the immediate-after-death subgroup, the rat lung was collected and stored at −80 °C until use, while in the 24 h after-death subgroup, the rat cadavers were placed inside an incubator (25 ± 3 °C; 50% ± 5% humidity) for 24 h. Then, the lung of each rat, which had decomposed, was collected and stored at −80 °C until use. Subsequently, all the lung samples were taken from the freezer and sectioned into 10 μm-thick sections with a cryo-microtome. The sections were placed on calcium fluoride (CaF2) slides (Raman grade) before the Raman spectra acquisition. In addition, optical images of the hematoxylin-eosin (HE)-stained lung tissues are shown in Fig. S1 in the ESI.†
Raman spectra acquisition and preprocessing
Raman spectra of the lung samples were acquired using an inVia confocal micro-Raman spectrometer (Renishaw, Gloucestershire, UK) equipped with a 532 nm excitation laser and a cooled charged-couple device (CCD) detector coupled to a Leica microscope. Renishaw Wire (Windows-based Raman Environment) 4.2 software was used to operate the instrument. Twenty points were randomly chosen in the lung tissue area to acquire 20 spectra for each sample using a 100× objective over a 10 s exposure time at 50 mW (50% of the total power). Three scans were accumulated for each spectral acquisition for a suitable signal-to-noise enhancement. The spectra were obtained in the range of 400–4000 cm−1 with a resolution of 1 cm−1. The equipment was calibrated to ensure Raman signal accuracy, setting the 520.5 cm−1 silicon line before the detection.Sharp spikes with narrow bandwidths resulting from cosmic rays frequently occur in Raman spectra, significantly distorting the Raman spectra and impeding the accurate acquisition of attribute data from the measured samples. Additionally, minor alterations in the optical path of the instrument and the measurement environment during detection can introduce data noise. Furthermore, Raman spectra are susceptible to tissue autofluorescence, leading to the emergence of an additive featureless background in the raw Raman spectra. Consequently, the preprocessing procedures for the Raman raw spectra typically include spectral cosmic ray removal, smoothing, and fluorescence background subtraction,30 performed using built-in functions in the WiRE 4.2 software. Moreover, the Raman spectra were normalized (1-Norm, area = 1) to remove the influence from the excitation intensity fluctuation or changes in the focusing. Finally, the Raman spectra were truncated to the bio-fingerprint region (600–1800 cm−1) and the high-wavenumber region (2800–3100 cm−1) before the data analysis.
Per the experimental grouping, the Raman spectral data set included fresh (immediate after death) and decomposed sample subsets (24 h after death). Before establishing and verifying the classification models, of each eight-rat group, five rats were randomly selected with their spectral data for the training set, while the three other rats were assigned to the test set.
Data analysis
Principal component analysis (PCA), a widely used dimension reduction method,31 was applied to the fresh subset to project the samples onto the two-dimensional PC space to visualize a spectral dataset for exploring the spectral variations. Moreover, some spectra outliers were identified and removed based on the leverage values and Q-residuals in the PCA before subsequent analysis.32Partial least-squares discriminant analysis (PLS-DA), a classical supervised pattern recognition algorithm, was performed to further analyze the spectral differences after the preliminary exploration by PCA. A PLS-DA classification model was established using a 10-fold cross-validation to discriminate the spectra of the fresh lung tissues of rats that had died from asphyxia and SCD. The differential spectral variables contributing to the distinction were shown according to the regression coefficients. Subsequently, the relative peak intensities of the main differential spectral variables were compared at different postmortem intervals (PMIs) after overall normalization. Statistical significance differences were assessed by the Mann–Whitney U test (a nonparametric method) since the data did not pass the normality test and the homogeneity test of variance. Statistical analysis was performed using IBM SPSS Statistics Version 20 (IBM corporation, NY, USA). P < 0.05 indicated a significant difference. Consequently, the differential spectral variables that remained significantly different throughout the 24 h after-death period were selected to build classification models to determine the cause of death preliminarily.
Seven conventional machine learning algorithms, including support vector machine (SVM), K-nearest neighbor (KNN), artificial neural network (ANN), logistic regression (LR), random forest (RF), extreme gradient boosting (XGB), and PLS-DA, were adopted to construct the models based on the training set. The proper classification approach was determined by accessing five evaluation metrics based on the confusion matrix (TP: true positive; TN: true negative; FP: false positive; FN: false negative) during the prediction/test process, including the accuracy ((TP + TN)/(TP + TN + FP + FN)), precision (TP (TP + FP)), recall/sensitivity (TP/(TP + FN)), specificity (TN/(FP + TN)), and the area under the curve (AUC) in the receiver operating characteristics (ROC) curve analysis. Data preprocessing and chemometric methods were performed using MATLAB R2017b (MathWorks, MA, USA) equipped with the MIA Toolbox 1.0 (Eigenvector Research, WA, USA) and Python 3.10.11 (Python Software Foundation), with the third-party packages including scikit-learn,33 numpy,34 pandas,35 and matplotlib.36
Results
Preliminary Raman spectral analysis
Fig. 1a shows the Raman spectra of the fresh sample subset in the asphyxia and SCD groups after preprocessing. The solid lines indicate the average spectra, and the shaded lines represent one standard deviation. The spectra showed significant Raman peaks in the regions of 600–1800 cm−1 and 2800–3100 cm−1. The peaks spanning from 2800 to 3100 cm−1 originated from CH, CH2, and CH3 symmetric and antisymmetric stretching of lipids and/or proteins.37 The region of 1630–1700 cm−1 corresponded to the amide I peak, which was due to carbonyl group stretching.38 The peak centered at 1585 cm−1 was attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration in proteins, while the peak at 1558 cm−1 was due to the N–H bending and C–N stretching modes of amide II in proteins.38 The peaks ranging from 1050 to 1500 cm−1 originated from various vibration modes of biological macromolecules, such as the amide III in proteins, the C–H bending and C–C stretching vibrations in lipids and proteins, the ring breathing modes of the DNA/RNA bases, and phosphates vibrations in nucleic acids.38 The peaks in the region of 600–900 cm−1 were mainly attributed to nucleic acids.38
C stretching vibration in proteins, while the peak at 1558 cm−1 was due to the N–H bending and C–N stretching modes of amide II in proteins.38 The peaks ranging from 1050 to 1500 cm−1 originated from various vibration modes of biological macromolecules, such as the amide III in proteins, the C–H bending and C–C stretching vibrations in lipids and proteins, the ring breathing modes of the DNA/RNA bases, and phosphates vibrations in nucleic acids.38 The peaks in the region of 600–900 cm−1 were mainly attributed to nucleic acids.38
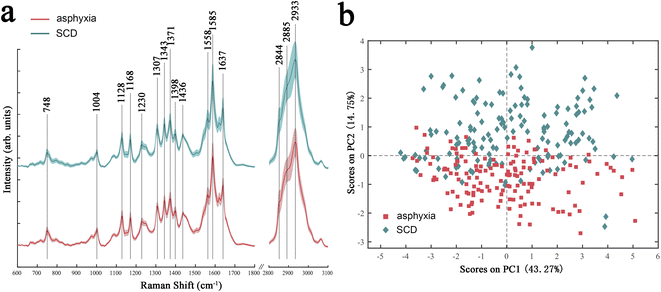 | ||
| Fig. 1 Average spectra (a) and PCA score plot (b) of the lung tissues of rats died from asphyxia (red) or SCD (green). SCD: sudden cardiac death. | ||
The overall variances in the spectra between asphyxia and SCD were revealed by PCA. Fig. 1b presents the PCA score results: each point in the two-dimensional space represents a single spectrum containing rich biochemical information. Although there was a slight overlap, a significant separation tendency of the spectra between the two groups could be seen in the PC-1 direction, which accounted for 14.75% of the total variances.
Screening of the spectral features for determining the cause of death
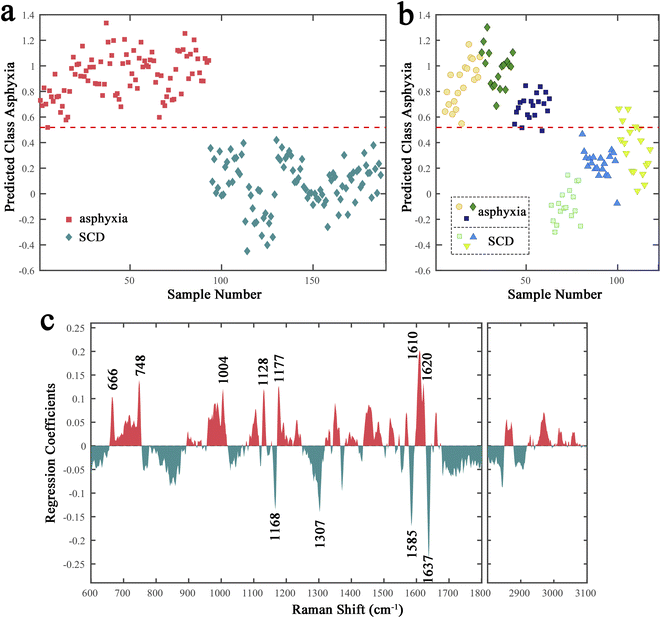
A binary PLS-DA model was further established to distinguish the spectra in the fresh sample subset between asphyxia and SCD. A venetian blind cross-validation procedure was applied to select an optimal number of latent variables (LVs), which was necessary for the model establishment and is a key step in determining the discrimination performance. Fig. S2a† illustrates the classification error rate plot versus the LV number. Four LVs were ultimately used to build the PLS-DA model; Fig. 2a presents the calibration score plot: the red dotted line represents the model threshold that decides each spectrum class. All the asphyxia group spectra were split entirely from that of the SCD group. Additionally, a distinction close to exactly correct was achieved in the external validation (Fig. 2b). Fig. S2b and c in the ESI† depict the corresponding ROC curve analysis results (AUC: 0.9903) and confusion matrix (accuracy: 0.9469).However, the results were unsatisfactory when we tried to put the decomposed samples spectra into the PLS-DA model as the external validation (Fig. S3†). The classification model failed to distinguish the decomposed samples spectra collected at 24 h after death between the asphyxia and SCD groups. Therefore, we tried to screen the potential spectral features whose significant differences could be maintained during after-death autolysis and decomposition to achieve the postmortem determination of asphyxia from SCD. The regression coefficient plot demonstrates the degree to which the spectral features contribute to the discrimination (Fig. 2c). The higher the Raman peak coefficient absolute value, the more important their contribution to the classification model. The Raman peaks with positive and negative coefficients, marked red and green, mean that their relative intensities were higher in the asphyxia and SCD groups, respectively. The negatively correlated peaks for the SCD group were mainly at 1637, 1585, 1307, and 1168 cm−1, while the positively correlated peaks for the asphyxia group were mainly at 1620, 1610, 1177, 1128, 1004, 748, and 666 cm−1. The chemical signatures of these main Raman peaks were assigned, and are listed in Table 1.38–47
| Peak (cm−1) | Assignment |
|---|---|
| a ν = stretching vibration; δ = bending vibration. | |
| 1637 | ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of amide I O) of amide I |
| 1620 | Tryptophan, ν(C–C) of porphyrin or hemoglobin |
| 1610 | Cytosine |
| 1585 | Hydroxyproline, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) of aromatic amino acids (including phenylalanine, tyrosine, and tryptophan), porphyrin or hemoglobin, cytochrome c C) of aromatic amino acids (including phenylalanine, tyrosine, and tryptophan), porphyrin or hemoglobin, cytochrome c |
| 1307 | δ(CH2/CH3) of collagens |
| 1177 | Cytosine, guanine |
| 1168 | Amino acids, carbohydrates |
| 1128 | ν(C–N) of proteins, ν(C–O) of carbohydrates, cytochrome c |
| 1004 | Phenylalanine |
| 748 | Red blood cell or hemoglobin, cytochrome c, DNA |
| 666 | Guanine, thymine |
Next, the relative peak intensities of these 11 potential spectral features in fresh and decomposed samples (training set) were further compared (Fig. 3). The relative peak intensities of these spectral features varied with after-death autolysis and decomposition. Among them, three spectral features, although still significantly different 24 h after death, showed an opposite quantitative relationship between the asphyxia and SCD groups. Eight spectral features maintained the significance and direction of their differences 24 h after death. Therefore, excluding the peaks at 1177, 748, and 666 cm−1, the other eight spectral features could be used to distinguish the spectra between the asphyxia and SCD groups within 24 h after death.
Establishment and evaluation of classification models
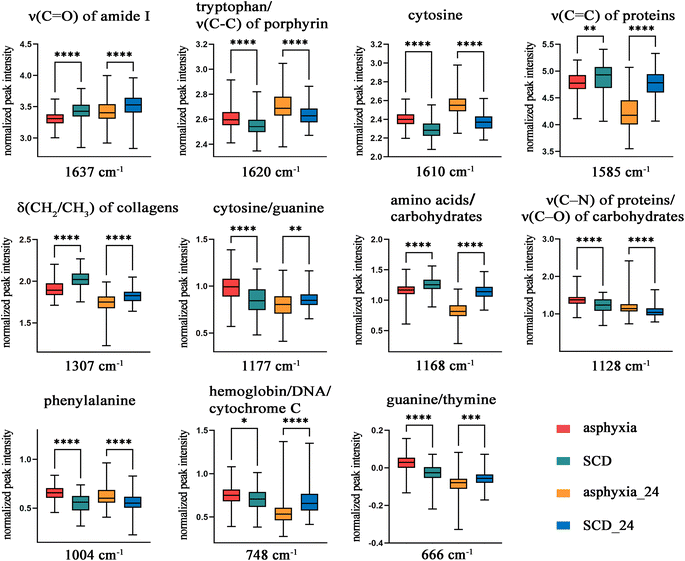
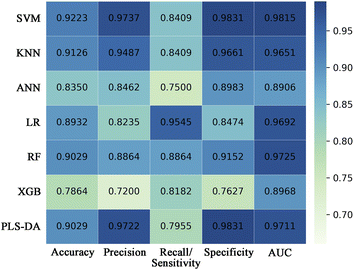
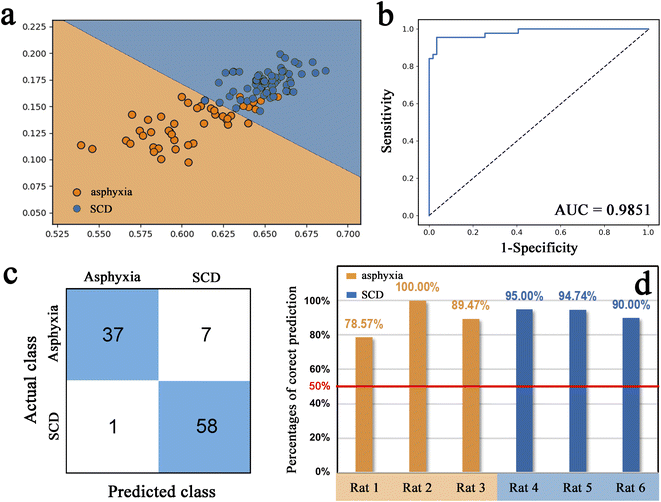
Seven conventional machine learning classification algorithms, namely SVM, KNN, ANN, LR, RF, XGB, and PLS-DA, were used to build classification models with the selected eight spectral features on the training set of the decomposed sample subset. Each model was validated in the test set. Fig. 4 displays a heatmap of the external validation results with five evaluation metrics. The SVM method showed the highest accuracy, precision, specificity, and AUC value but barely satisfactory sensitivity. The LR approach had the highest sensitivity, but its precision and specificity were insufficient. The overall classification performances of the ANN and XGB methods were unsatisfactory. KNN, RF, and PLS-DA performed well but slightly worse than SVM.Fig. 5a presents the visual prediction results of the SVM classifier. Each point represents a single spectrum, and different colors represent different groups. The classification results were represented through different colors of the spaces in which the points were located. Most spectra were correctly classified. Seven spectra in the asphyxia group were misclassified as SCD, and one in the SCD group was misclassified as asphyxia, as the confusion matrix also shows in Fig. 5c. The model accuracy was 0.9223 in the test process. Fig. 5b shows the ROC curve with an AUC of 0.9851. The SVM classifier gave a sensitivity of 0.8409 and a specificity of 0.9831 at the optimal threshold. Fig. 5d illustrates the overall correct prediction percentages for each rat in the test set when we set the classification criterion as “the class/group to which more than 50% of the spectrum belongs”. The cause of death of each rat in the test set was determined correctly.
Discussion
This study detected the Raman spectra of lung tissues, collected at 0 h and 24 h after death, of rats that had died from asphyxia and SCD. The spectral differences in the Raman profiles were analyzed and the spectral features that maintained their significant differences during the 24 h after-death period were screened. Seven machine learning algorithms were employed to build classification models to distinguish between the two complex causes of death.The mean preprocessed Raman spectra showed that the fluorescence background was eliminated from the baseline and the random signal noises and cosmic ray spikes were successfully reduced. The Raman spectra provided comprehensive biochemical information of the lung tissues, including nucleic acids, proteins, and lipids. However, the spectral shape and the characteristic spectral positions between the asphyxia and SCD groups were similar, making the visual discrimination challenging. Nevertheless, a clear discrimination trend emerged between the spectra of the two groups in the PCA score plot. These results demonstrate subtle yet detectable biochemical differences in the lung tissues of rats that had died from asphyxia and SCD, obtained by Raman spectroscopy combined with chemometrics.
The supervised pattern recognition method PLS-DA was further utilized to differentiate the spectra between the asphyxia and SCD groups. The spectra of the two groups in the training set were completely distinguished, and most spectra in the test set were correctly classified, yielding an AUC value of 0.9903. This indicates that the classification model was effectively constructed to determine the cause of death by classifying the Raman spectra.
By examining the regression coefficients, the primary spectral features that contributed to the discrimination could be analyzed and the underlying biochemical changes explored. The Raman peak with the highest absolute regression coefficient was the negatively correlated peak at 1637 cm−1; this peak may be assigned to the amide I band, which was due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of proteins (both α-helix and β-sheet structure).38,39 The sum of intensities of the peaks at 1637, 1558 (C–N stretching and N–H bending vibrations of amide II), and 1230 cm−1 (amide III) were compared in the two groups, representing the protein content level (Fig. S4†). These results may indicate that the lung tissues of SCD rats, compared with those of asphyxiated rat, had a higher total protein content, with a difference in protein composition or conformation.
O stretching vibrations of proteins (both α-helix and β-sheet structure).38,39 The sum of intensities of the peaks at 1637, 1558 (C–N stretching and N–H bending vibrations of amide II), and 1230 cm−1 (amide III) were compared in the two groups, representing the protein content level (Fig. S4†). These results may indicate that the lung tissues of SCD rats, compared with those of asphyxiated rat, had a higher total protein content, with a difference in protein composition or conformation.
Another negatively correlated peak at 1585 cm−1 with a high absolute regression coefficient may be attributed to hydroxyproline.38 Hydroxyproline is a main component of collagen, and the connective tissues mainly contained in the lung interstitium have a lot of collagen fibers. This was consistent with the high relative intensity of the 1585 cm−1 peak in Fig. 1a. Senavirathna et al.40 found that hypoxia induces pulmonary fibroblast proliferation, and combined with our results, this may suggest that the lung tissue of asphyxiated rats tended to take up more hydroxyproline to synthesize collagen tissue. Besides, the Raman peak at 1585 cm−1 may also represent the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching of aromatic amino acids, including phenylalanine, tyrosine, and tryptophan.38,41 Another two positively correlated peak at 1004 and 1620 cm−1 could be assigned to phenylalanine38 and tryptophan,42 respectively. It was reported that the amount of these aromatic amino acids in the lung tissue was changed during asphyxia.48 Moreover, phenylalanine and tyrosine are precursors to dopamine, a precursor to epinephrine and norepinephrine. Also, tryptophan is a precursor to 5-hydroxytryptamine (5-HT). The chemical structure of these substances is similar in having a benzene ring structure, which may also show the C
C stretching of aromatic amino acids, including phenylalanine, tyrosine, and tryptophan.38,41 Another two positively correlated peak at 1004 and 1620 cm−1 could be assigned to phenylalanine38 and tryptophan,42 respectively. It was reported that the amount of these aromatic amino acids in the lung tissue was changed during asphyxia.48 Moreover, phenylalanine and tyrosine are precursors to dopamine, a precursor to epinephrine and norepinephrine. Also, tryptophan is a precursor to 5-hydroxytryptamine (5-HT). The chemical structure of these substances is similar in having a benzene ring structure, which may also show the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration peak on a Raman spectrum. Epinephrine has a contractile effect on small pulmonary vessels and a strong bronchial dilation effect. In addition, during hypoxia, the increase in vasoconstricting substances is one of the mechanisms leading to pulmonary vasoconstriction, and 5-HT may be one of these.49 It was also reported that dopamine receptors are expressed in fibroblasts in lung tissue, and even lung tissue itself can produce dopamine.50 Therefore, the difference in these peaks in the Raman spectra between the two groups may be related to the different dopamine, epinephrine, or 5-HT levels with their different physiological effects in lung tissue during the process of asphyxia and myocardial infarction affecting the body and the body response.
C stretching vibration peak on a Raman spectrum. Epinephrine has a contractile effect on small pulmonary vessels and a strong bronchial dilation effect. In addition, during hypoxia, the increase in vasoconstricting substances is one of the mechanisms leading to pulmonary vasoconstriction, and 5-HT may be one of these.49 It was also reported that dopamine receptors are expressed in fibroblasts in lung tissue, and even lung tissue itself can produce dopamine.50 Therefore, the difference in these peaks in the Raman spectra between the two groups may be related to the different dopamine, epinephrine, or 5-HT levels with their different physiological effects in lung tissue during the process of asphyxia and myocardial infarction affecting the body and the body response.
In addition to being attributed to tryptophan, the 1620 cm−1 peak may also be derived from porphyrin42 or haemoglobin.43 Besides, the 1585 cm−1 peak may also be related to porphyrin or hemoglobin due to having a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C structure (Neugebauer et al.43 assigned the peak at 1584 cm−1 to hemoglobin). Furthermore, it was reported that the 750 and 752 cm−1 peaks may be derived from CH2,6 out-of-plane bending of red blood cells (RBCs) and the porphyrin breathing mode in the heme groups of hemoglobin, respectively.38 This suggests that our positively correlated peak at 748 cm−1 may originate from RBC or hemoglobin. Moreover, blood spectral data from some Raman spectroscopy studies based on time since deposition,51 gender,52 and species53 of bloodstains also support that some of the differential peaks (such as 1004, 1128, and 1168 cm−1) may be related to hemoglobin or RBCs. Small vein congestion, enhanced vascular permeability, and elevated blood pressure caused by asphyxia may lead to capillary rupture and bleeding. Therefore, the differences in the Raman spectra may suggest more RBC or hemoglobin in lung tissues of asphyxiated rats, which could be related to pulmonary petechial hemorrhage.
C structure (Neugebauer et al.43 assigned the peak at 1584 cm−1 to hemoglobin). Furthermore, it was reported that the 750 and 752 cm−1 peaks may be derived from CH2,6 out-of-plane bending of red blood cells (RBCs) and the porphyrin breathing mode in the heme groups of hemoglobin, respectively.38 This suggests that our positively correlated peak at 748 cm−1 may originate from RBC or hemoglobin. Moreover, blood spectral data from some Raman spectroscopy studies based on time since deposition,51 gender,52 and species53 of bloodstains also support that some of the differential peaks (such as 1004, 1128, and 1168 cm−1) may be related to hemoglobin or RBCs. Small vein congestion, enhanced vascular permeability, and elevated blood pressure caused by asphyxia may lead to capillary rupture and bleeding. Therefore, the differences in the Raman spectra may suggest more RBC or hemoglobin in lung tissues of asphyxiated rats, which could be related to pulmonary petechial hemorrhage.
Moreover, a 2020 Raman spectroscopy study of lung cancer tissues assigned three Raman peaks at 748, 1129, and 1586 cm−1 to cytochrome c. These three are also Raman peaks with high regression coefficients in our results (the latter two displayed a small deviation in the Raman shift, which was acceptable). Cytochrome c is a heme protein involved in the respiratory chain of mitochondria as an electron carrier.54 Hypoxia causes oxidative respiratory chain dysfunction, and exogenous cytochrome c can play a first aid or auxiliary treatment role in tissue hypoxia. Therefore, the three differential Raman peaks in our results may suggest the differences in the effects of the two causes of death on oxidation–reduction processes in energy metabolism.
The positively correlated peak at 1610 cm−1 was located in the region dominated mainly by the amide I band of proteins. However, this peak was insignificant in Fig. 1a and the 1637 cm−1 peak was assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of amide I, as mentioned above. Therefore, the difference in the 1610 cm−1 peak may be due to other substances. By searching the literature, the 1610 cm−1 peak and another three positively correlated peaks at 1177, 748 (besides RBCs or hemoglobin), and 666 cm−1 may be attributed to nucleic acid.38 Therefore, these results may indicate different nucleic acid metabolisms in the lung tissues of the two groups during the process of asphyxia and myocardial infarction, affecting the body and the body response and causing a higher nucleic acid content in the lung tissues of asphyxiated rats compared with SCD rats.
O stretching vibration of amide I, as mentioned above. Therefore, the difference in the 1610 cm−1 peak may be due to other substances. By searching the literature, the 1610 cm−1 peak and another three positively correlated peaks at 1177, 748 (besides RBCs or hemoglobin), and 666 cm−1 may be attributed to nucleic acid.38 Therefore, these results may indicate different nucleic acid metabolisms in the lung tissues of the two groups during the process of asphyxia and myocardial infarction, affecting the body and the body response and causing a higher nucleic acid content in the lung tissues of asphyxiated rats compared with SCD rats.
The lipid peaks in the range of 3100–2800 cm−1 showed high relative intensities (Fig. 1a) but low regression coefficients (Fig. 2c), indicating that the lipid metabolism difference was not very large and was not an important aspect to distinguish the lung tissues of the two groups. The negatively correlated peak at 1307 cm−1 may be attributed to the CH2/CH3 bending vibration of lipids and/or collagens.38 Due to its higher regression coefficient than lipid peaks, the 1307 cm−1 peak may be related to collagen tissues, as was the 1585 cm−1 peak in this study. Moreover, the 1168 cm−1 peak may be attributed to amino acids or carbohydrates45–47 and the 1128 cm−1 peak may be derived from proteins or carbohydrates.38 The results suggest that in addition to differences in the protein type and content shown also by the 1637 cm−1 peak in the lung tissues of the two groups, there may also be differences in carbohydrates, such as glucose (energy supply or consumption), or substances containing carbohydrates, such as glycolipids or glycoproteins (cell function or structure).
Notably, it is not realistic to make a clear assignment for these Raman peaks due to the simultaneous contribution of various biomolecules to a particular Raman peak and the complex nature of lung tissue as a biological sample containing a wide variety of biomolecules. However, we tried to search previous literature for peak assignments. Moreover, screening the differential spectral features and building classification models to distinguish the causes of death is more important than interpreting the biomolecular differences in this study.
A PLS-DA classification model that can distinguish fresh lung tissues spectra was successfully constructed. However, our attempt to determine the cause of death using this model in the 24 h after-death sample data failed (Fig. S3†). This is because autolysis and putrefaction significantly alter all biomolecular content after death; therefore, some subtle biochemical differences between asphyxia and SCD may be masked. However, we do not intend to develop another model to distinguish the spectra of lung tissue collected 24 h after death because, in practice, postmortem time points are infinite, and it is impossible to build models for every postmortem time point. Therefore, the spectral features that maintained significant differences after death were screened subsequently.
The height of a Raman peak is directly related to the relative content of the corresponding chemical signatures.55 We compared the relative intensity changes of the primary differential spectral features; finding that with after-death autolysis and decomposition, the relative contents of some substances that these spectral features may represent were increased and some were decreased, which might be due to biomacromolecular degradation and microbial behaviors. Importantly, 8 of the 11 spectral features maintained a significant difference up to 24 h after death (the other three, although significant at 24 h after death, had opposite quantitative relationships between the two groups). Therefore, the eight differential spectral features were further used to build classification models.
In preprocessing in some literature, spectral data obtained from various animal individuals were randomly allocated into training and test data sets at a ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. However, it is hypothesized that this may overlook the individual variations, as certain spectra employed for external validation could originate from the same individuals as those utilized for modeling, leading to improved yet inaccurate prediction performance in the classification model. Therefore, a random selection process was employed to assign five rats from each group as the training set, while the remaining three rats were designated as the test set during the data preprocessing stage to enhance the reliability of the findings. The classification model evaluation was solely based on the outcomes of the independent test set, ensuring complete separation from the training set.
3. However, it is hypothesized that this may overlook the individual variations, as certain spectra employed for external validation could originate from the same individuals as those utilized for modeling, leading to improved yet inaccurate prediction performance in the classification model. Therefore, a random selection process was employed to assign five rats from each group as the training set, while the remaining three rats were designated as the test set during the data preprocessing stage to enhance the reliability of the findings. The classification model evaluation was solely based on the outcomes of the independent test set, ensuring complete separation from the training set.
Classification is a fundamental task in machine learning with numerous algorithms; diverse classification algorithms can be explored experimentally to ascertain the most suitable one for a given task or dataset. This study calculated five conventional evaluation metrics for the classification task: accuracy, precision, recall/sensitivity, specificity, and AUC. The first four indicators can only offer a partial assessment of a model's performance, and each has its own limitations, while the AUC indicator exhibits a relatively balanced evaluation. In short, the higher the overall value of these indicators, the better the classification model performance. The SVM classification algorithm has the advantages of good classification performance and high capacity for generalization.56 Meanwhile, its disadvantages are the difficulty handling large-scale samples, solving multi-class problems, and sensitivity to missing data,56 but these problems did not exist in our classification task. These are probably why the SVM classifier performed best here. The classification model, constructed using chosen differential spectral features, displayed exceptional performance, with an AUC value of 0.9851. This result indicates that the most spectra of the decomposed lung tissue from the asphyxia and SCD groups could be accurately differentiated by the model. Furthermore, the biochemical information of each rat's lung tissue was actually represented by more than a dozen spectra rather than a single one. The cause of death in a rat should not be determined by a single spectrum. Therefore, if we set the classification criterion as “the class/group to which more than 50% of the spectrum belongs”, the cause of death of each rat in the test set would be determined correctly, as shown in Fig. 5c.
As previously stated in the Introduction, in most previous studies employing molecular biological techniques to screen biomarkers, asphyxia or SCD were studied separately (the rats sacrificed by cervical dislocation were set as the control group), which lacked investigation of the specificity. Furthermore, the potential impact of after-death autolysis and decomposition on the identified biomarkers was not considered. Herein, we directly took asphyxia and myocardial infarction, which often affect each other's accurate cause of death determination in practical settings, as the experimental group to improve the specificity and practicality of the results. Additionally, our method was designed to determine the causes of death in the presence of after-death autolysis and decomposition. Although we only examined the differences and diagnostic abilities of certain spectral features at 24 h after death, the encouraging positive outcomes inspire and present a novel approach to solving or avoiding the effect after death. Although the Raman spectroscopy technology incomprehensively elucidated the precise biomolecular alterations occurring in various death processes, it offers global biochemical information and could prove highly advantageous for forensic applications as a convenient, rapid, non-destructive, and inexpensive tool. In subsequent investigations, it is imperative to explore a wider PMI range and collect human samples to assess the feasibility and practical significance of this research idea.
Conclusions
Different biochemical alterations in the lung tissues of rats that had died from asphyxia and SCD could be detected and preliminary analyzed using Raman spectroscopy. The differential spectral features representing these subtle biochemical differences were employed to build a classification model to distinguish asphyxia from SCD. Eight out of eleven main differential spectral features maintained their significant differences at 24 h after death and could be utilized to determine the cause of death under decomposition and autolysis. Among the seven frequently used machine learning classification algorithms, SVM performed superior with these given data, achieving an AUC value of 0.9851 in external validation. This study demonstrated the potential of Raman spectroscopy with machine learning algorithms to investigate differential biochemical alterations derived from different death processes to aid determining the cause of death.Author contributions
Kai Zhang and Ping Huang designed the study; Kai Zhang, Ruina Liu, and Xin Wei performed the measurements and analyzed the data; Ping Huang and Zhenyuan Wang provided the instruments; Kai Zhang and Ruina Liu wrote the manuscript; Zhenyuan Wang and Ping Huang supervised the projects. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Shanghai Key Lab of Forensic Medicine (Academy of Forensic Science) Open Projects Fund (No. KF202302), the Fundamental Research Funds for the Central Universities (No. xzy012023107), the NHC Key Laboratory of Forensic Science (Xi'an Jiaotong University) Open Projects Fund (No. 2023FYXH003), and the Shaanxi Province Postdoctoral Research Project Fund.References
- P. Saukko and B. Knight, Knight's Forensic Pathology, CRC press, 2015 Search PubMed.
- M. Hayashi, W. Shimizu and C. M. Albert, Circ. Res., 2015, 116, 1887–1906 CrossRef CAS PubMed.
- J. Ma, H. Jing, Y. Zeng, L. Tao, Y. Yang, K. Ma and L. Chen, J. Forensic Leg. Med., 2016, 42, 19–24 CrossRef PubMed.
- C. X. Wong, A. Brown, D. H. Lau, S. S. Chugh, C. M. Albert, J. M. Kalman and P. Sanders, Heart, Lung Circ., 2019, 28, 6–14 CrossRef PubMed.
- Y. Zeng, L. Tao, J. Ma, L. Han, Y. Lv, P. Hui, H. Zhang, K. Ma, B. Xiao and Q. Shi, J. Leg. Med., 2018, 132, 655–665 Search PubMed.
- E. Gutjahr and B. Madea, Forensic Sci. Int., 2019, 297, 315–325 CrossRef CAS PubMed.
- H. Zhang, Y. Hu, H. Wang, L. Tian, W. Li, L. Han, H. Xu, J. Ma, K. Ma and B. Xiao, Am. J. Transl. Res., 2019, 11, 4568–4583 CAS.
- L. Han, H. Zhang, Y. Zeng, Y. Lv, L. Tao, J. Ma, H. Xu, K. Ma, Q. Shi and B. Xiao, Forensic Sci. Int., 2020, 311, 110293 CrossRef CAS PubMed.
- S. Sabatasso, P. Mangin, T. Fracasso, M. Moretti, M. Docquier and V. Djonov, J. Leg. Med., 2016, 130, 1265–1280 Search PubMed.
- O. Carvajal-Zarrabal, P. M. Hayward-Jones, C. Nolasco-Hipolito, D. M. Barradas-Dermitz, A. L. Calderón-Garcidueñas and N. López-Amador, J. Forensic Sci., 2017, 62, 1332–1335 CrossRef CAS PubMed.
- K. Zhang, H. Yan, R. Liu, P. Xiang, K. Deng, J. Zhang, Y. Tuo, Z. Wang and P. Huang, J. Chromatogr. B, 2021, 1171, 122638 CrossRef CAS PubMed.
- K. Zhang, H. Yan, R. Liu, P. Xiang, J. Zhang, K. Deng, P. Huang and Z. Wang, ACS Omega, 2021, 6, 2100–2109 CrossRef CAS PubMed.
- K. Zhang, Y. Tuo, R. Liu, H. Yan, P. Xiang, Z. Wang and P. Huang, Arabian J. Chem., 2022, 15, 104322 CrossRef CAS.
- N. M. Ralbovsky and I. K. Lednev, Chem. Soc. Rev., 2020, 49, 7428–7453 RSC.
- I. Pence and A. Mahadevan-Jansen, Chem. Soc. Rev., 2016, 45, 1958–1979 RSC.
- C. A. F. de Oliveira Penido, M. T. T. Pacheco, I. K. Lednev and L. Silveira Jr, J. Raman Spectrosc., 2016, 47, 28–38 CrossRef.
- A. Braz, M. López-López and C. García-Ruiz, Forensic Sci. Int., 2013, 232, 206–212 CrossRef CAS PubMed.
- J. Zięba-Palus, A. Wesełucha-Birczyńska, B. Trzcińska, R. Kowalski and P. Moskal, J. Mol. Struct., 2017, 1140, 154–162 CrossRef.
- K. C. Doty and I. K. Lednev, TrAC, Trends Anal. Chem., 2018, 103, 215–222 CrossRef CAS.
- E. Buchan, M. Hardy, P. d. C. Gomes, L. Kelleher, H. O. M. Chu and P. G. Oppenheimer, Appl. Spectrosc. Rev., 2022, 1–38 CrossRef.
- N. George, H. Singh, R. Jotaniya and S. R. Pandya, Forensic Sci. Int., 2022, 340, 111441 CrossRef CAS PubMed.
- G. Falgayrac, R. Vitale, Y. Delannoy, H. Behal, G. Penel, L. Duponchel and T. Colard, Talanta, 2022, 249, 123589 CrossRef CAS PubMed.
- L. Ortiz-Herrero, B. Uribe, L. H. Armas, M. Alonso, A. Sarmiento, J. Irurita, R. Alonso, M. Maguregui, F. Etxeberria and L. Bartolome, Forensic Sci. Int., 2021, 329, 111087 CrossRef CAS PubMed.
- M. Paraskevaidi, B. J. Matthew, B. J. Holly, B. J. Hugh, C. P. Thulya, C. Loren, C. StJohn, G. Peter, G. Callum and K. G. Sergei, Appl. Spectrosc. Rev., 2021, 56, 804–868 CrossRef CAS.
- A. Kozik, M. Pavlova, I. Petrov, V. Bychkov, L. Kim, E. Dorozhko, C. Cheng, R. D. Rodriguez and E. Sheremet, Anal. Chim. Acta, 2021, 1187, 338978 CrossRef CAS PubMed.
- H. Lin, Z. Wang, Y. Luo, Q. Sun, Y. Shen and P. Huang, Spectrochim. Acta, Part A, 2022, 268, 120630 CrossRef CAS PubMed.
- K. Zhang, A. Zhang, R. Liu, H. Zhang, H. Lin, P. Zhang, P. Huang and Z. Wang, Leg. Med., 2020, 47, 101748 CrossRef CAS PubMed.
- H. Lin, Y. Luo, Q. Sun, J. Zhang, Y. Tuo, Z. Zhang, L. Wang, K. Deng, Y. Chen and P. Huang, Anal. Chem., 2018, 90, 2708–2715 CrossRef CAS PubMed.
- K. Zhang, R. Liu, Y. Tuo, K. Ma, D. Zhang, Z. Wang and P. Huang, ACS Omega, 2022, 7, 46859–46869 CrossRef CAS PubMed.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner and P. L. Martin-Hirsch, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- R. Bro and A. K. Smilde, Anal. Methods, 2014, 6, 2812–2831 RSC.
- T. Næs, T. Isaksson, T. Fearn and T. Davies, A User-Friendly Guide to Multivariate Calibration and Classification, NIR Chichester, 2002 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, J Mach Learn Res, 2011, 12, 2825–2830 Search PubMed.
- C. R. Harris, K. J. Millman, S. J. Van Der Walt, R. Gommers, P. Virtanen, D. Cournapeau, E. Wieser, J. Taylor, S. Berg and N. J. Smith, Nature, 2020, 585, 357–362 CrossRef CAS PubMed.
- W. McKinney, Python for high performance and scientific computing, 2011, vol. 14, pp. 1–9 Search PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- S. Mazurek and R. Szostak, J. Pharm. Biomed. Anal., 2006, 40, 1225–1230 CrossRef CAS PubMed.
- A. C. S. Talari, Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2015, 50, 46–111 CrossRef CAS.
- R. J. Lakshmi, V. Kartha, C. Murali Krishna, J. R. Solomon, G. Ullas and P. Uma Devi, Radiat. Res., 2002, 157, 175–182 CrossRef CAS.
- L. K. Senavirathna, C. Huang, X. Yang, M. C. Munteanu, R. Sathiaseelan, D. Xu, C. A. Henke and L. Liu, Sci. Rep., 2018, 8, 2709 CrossRef PubMed.
- F. M. Lyng, D. Traynor, T. N. Q. Nguyen, A. D. Meade, F. Rakib, R. Al-Saady, E. Goormaghtigh, K. Al-Saad and M. H. Ali, PLoS One, 2019, 14, e0212376 CrossRef CAS PubMed.
- Z. Huang, A. McWilliams, H. Lui, D. I. McLean, S. Lam and H. Zeng, Int. J. Cancer, 2003, 107, 1047–1052 CrossRef CAS PubMed.
- U. Neugebauer, T. Bocklitz, J. Clement, C. Krafft and J. Popp, Analyst, 2010, 135, 3178–3182 RSC.
- Y. Oshima, H. Shinzawa, T. Takenaka, C. Furihata and H. Sato, J. Biomed. Opt., 2010, 15, 017009 CrossRef PubMed.
- S. Firdous, M. Nawaz, M. Ahmed, S. Anwar, A. Rehman, R. Rashid and A. Mahmood, Laser Phys., 2012, 22, 1090–1094 CrossRef CAS.
- S. Varghese, A. Reveendran, V. S. Kumar, K. Tm and V. Ranganathan, Asian J. Pharm. Clin. Res., 2018, 11, 176–178 CrossRef.
- E. Kaznowska, K. Łach, J. Depciuch, R. Chaber, A. Koziorowska, S. Slobodian, K. Kiper, A. Chlebus and J. Cebulski, Infrared Phys. Technol., 2018, 89, 282–290 CrossRef CAS.
- N. Philip, H. Pi, M. Gadkari, X. Yun, J. Huetsch, C. Zhang, R. Harlan, A. Roux, D. Graham and L. Shimoda, Pulm. Circ., 2023, 13, e12205 CrossRef CAS PubMed.
- J. T. Sylvester, L. A. Shimoda, P. I. Aaronson and J. P. Ward, Physiol. Rev., 2012, 92, 367–520 CrossRef CAS PubMed.
- A. J. Haak, E. Kostallari, D. Sicard, G. Ligresti, K. M. Choi, N. Caporarello, D. L. Jones, Q. Tan, J. Meridew and A. M. Diaz Espinosa, Sci. Transl. Med., 2019, 11, eaau6296 CrossRef CAS PubMed.
- A. R. Weber and I. K. Lednev, Forensic Chem., 2020, 19, 100248 CrossRef CAS.
- A. Sikirzhytskaya, V. Sikirzhytski and I. K. Lednev, Anal. Chem., 2017, 89, 1486–1492 CrossRef CAS PubMed.
- K. C. Doty and I. K. Lednev, Forensic Sci. Int., 2018, 282, 204–210 CrossRef CAS PubMed.
- W. J. Lennarz and M. D. Lane, Encyclopedia of Biological Chemistry, Academic Press, 2013 Search PubMed.
- A. D. Meade, F. M. Lyng, P. Knief and H. J. Byrne, Anal. Bioanal. Chem., 2007, 387, 1717–1728 CrossRef CAS PubMed.
- J. G. Greener, S. M. Kandathil, L. Moffat and D. T. Jones, Nat. Rev. Mol. Cell Biol., 2022, 23, 40–55 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Plots of the selection of latent variables for the PLS-DA model; ROC curves and confusion matrix for the PLS-DA model; classification results of decomposed lung tissues in the PLS-DA model built with fresh lung tissues; comparison of total protein content between the two groups. See DOI: https://doi.org/10.1039/d3ra07684a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |