Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
New sorbent-based hydrophobic alginic acid derivatives for fat removal in multi-pesticide residues: analysis of a fatty food sample
Omar A. Thabet ab,
Fahad K. Alenzib,
Maha A. Alshubramya,
Khalid A. Alamry
ab,
Fahad K. Alenzib,
Maha A. Alshubramya,
Khalid A. Alamry *a,
Mahmoud A. Hussein
*a,
Mahmoud A. Hussein *ac and
Richard Hoogenboom
*ac and
Richard Hoogenboom d
d
aDepartment of Chemistry, Faculty of Science, King Abdulaziz University, Jeddah 21589, Saudi Arabia. E-mail: kaalamri@kau.edu.sa; mahussein74@yahoo.com; maabdo@kau.edu.sa
bSaudi Food and Drug Authority, Jeddah 22311, Saudi Arabia
cChemistry Department, Faculty of Science, Assiut University, Assiut 71516, Egypt
dSupramolecular Chemistry Group, Department of Organic and Macromolecular Chemistry, Centre of Macromolecular Chemistry (CMaC)Ghent University, Krijgslaan 281 S4, 9000 Ghent, Belgium
First published on 12th January 2024
Abstract
Hydrophobic alginic acid derivatives were synthesized with various aliphatic hydrocarbon chains for fat removal in an analysis of multi-pesticide residues in a fatty food sample. First, alginic acid was chemically modified using eco-friendly ultrasound-assisted esterification with different alcohols, namely, hydrophobic alginic acid–methanol (HAA-C1), hydrophobic alginic acid–butanol (HAA-C4), and hydrophobic alginic acid–octadecanol (HAA-C18). The degree of esterification (DE) was determined by titration, and the results ranged from 57.3% to 63.7%. The physicochemical properties of the synthesized hydrophobic alginic acids (HAAs) were studied using FT-IR, XRD, TGA, and FE-SEM. Subsequently, the performance of the HAAs was checked and evaluated for the removal of fat from a fatty food sample by calculating the fat removal percentage and the determination of 214 pesticide residues in the fatty food sample. For the fat removal percentage application, the HAAs were able to efficiently remove between 77% and 83% of the fat; HAA-C18 had the highest percentage. Regarding the pesticide residue application, HAAs were also able to remove the fat content from the fatty food sample without a significant effect on the pesticide substances. The recoveries of the detected pesticide compounds were between 80% and 120% for all HAAs. However, there were various missing pesticide compounds for HAAs. The number of missing pesticide compounds was 19, 6, and 33 for HAA-C1, HAA-C4, and HAA-C18, respectively. HAA-C4 had medium hydrophobicity and it lost fewer pesticides than the other HAAs. This was because the multi-pesticide mixture had various classes of chemical structure; hence, it had different polarity powers. We concluded that HAAs are developable and applicable to be safely used as a green material in diverse fatty food sample analysis applications.
1. Introduction
Alginic acid is a heteropolysaccharide linear copolymer consisting of β-D-mannuronic acid (M) and α-L-guluronic acid (G) residues, which are connected through (1,4) glycosidic bonds and are arranged in either repeating GG (MM) blocks or alternating MG structures. Alginic acid is mainly derived from brown seaweed; it has been widely studied and used in different biomedical fields due to its remarkable characteristics, including its low cost, non-toxicity, superior biodegradability, and biocompatibility1,2. Despite the proven interest, alginic acid use remains limited due to the abundance of carboxyl and hydroxyl groups on the molecular chains. These dramatically increase the formation of intramolecular hydrogen bonds in alginate chains, making the structure rigid and stretched. The resulting difficult-to-load hydrophobic materials possess poor compatibility toward hydrophobic compounds. An abundance of hydrophilic groups decreases the stability of alginic acid in biological buffers, exhibiting unexpected and uncontrollable kinetic degradation and extensive water uptake properties3,4.Recently, researchers have focused on the development of hydrophobic materials from alginates through covalent modifications; this field of research is rapidly growing. One approach involves the esterification of carboxyl groups by incorporating long-chain alkyl groups or aromatic rings. Alginates are then esterified using carbodiimide derivatives as a coupling reagent.1 Benzoyl chloride was used to substitute the hydroxy group with the benzoyl group.2 It is also possible to obtain alginate esters by treating tetrabutylammonium salts of alginic acid with alkyl halides. Alginates have been modified by direct esterification through Fischer esterification with selected alcohols in the presence of a suitable catalyst.5 However, the limited use of the Fischer esterification reaction is related to the degradation of the polysaccharide chain in the presence of strongly acidic conditions and high temperatures within the long-time reaction necessary to obtain a high degree of conversion. Research has been developed to overcome this limitation by decreasing the reaction temperature. Broderick et al. investigated the possibility of obtaining alginate butyl esters by applying sodium alginate, butanol, and H2SO4 as a catalyst at room temperature for 18 h.6 Murdzheva exploited ultrasonic irradiation to obtain methylated, ethylated, and isopropylated derivatives of alginic acid. Their finding revealed that an ultrasound-assisted synthesis performed at 45 kHz reduced the reaction time to 2 hours in contrast to esterification performed under conventional conditions of heating.7
There are many studies focused on the synthesis of lipophilic esters using polysaccharide biopolymers to enhance their properties and increase their potential for use in various applications.8 Chitin, cellulose, and agarose are other types of polysaccharide biopolymers that have been developed to obtain lipophilic esters. In chitin, chitin acyl ester compounds were synthesized with short and long chains of fatty acids to distinguish emulsifying,9 crystallinity, and thermal properties.10 Regarding the cellulose and cellulose derivatives, the modification by esterification process has taken place to produce lipophilic cellulose ester.11 In a previous study, carboxymethyl cellulose (CMC) was modified using various hydrocarbon chains by esterification via an eco-friendly ultrasound-assisted procedure to remove fat content in food analysis applications.12 In addition, cellulose ester was modified with extracted fatty acids from sunflower oil to be used as an additive in resins for vat photopolymerization.13 Agarose fatty acid esters are another modified biopolymer produced by the esterification process and are used as hydrocolloid surfactants to investigate their emulsifying ability.14 Also, agarose was modified using gallic acid to be used as coated films for seafood preservation in food packaging.15
Pesticides are multiple; various chemical groups have the ability to kill, repel, or mitigate animals, insects, or cultivated plant pests.16 Although pesticides save and protect crops and agricultural production worldwide, they pose a critical threat to human health when they are misused and can be transferred to the human body by consuming foodstuff.17 Therefore, there have been global collaborative efforts to limit the spread and misuse of pesticides; many regulations have been set for this purpose and monitoring methods have been increased/developed, whether via professional laboratories or farm inspections. Pesticides including insecticides, herbicides, and fungicides are classified into four main groups, based on their chemical composition, namely, organophosphate, organochlorine, pyrethroid, and carbamate.18 Their diversity in structure consequently affects the polarity strength of each group based on the attached function groups.
One of the most challenging issues in the detection of pesticides in foodstuff is the fat content in fatty food samples. Fat has several negative effects on the extraction process of pesticides as well as on the analytical instruments used, including contamination, carryover, and the blockage of columns or pipes.19 Many well-known techniques have been used to remove the fat from fatty food samples, for instance, liquid–liquid extraction with a strong non-polar solvent, Soxhlet extraction, microwave-assisted extraction, and freezing precipitation.20 Despite these attempts, there remains a requirement for a simple, low-cost, and efficient method to effectively perform the task. The QuEChERS (quick, easy, cheap, effective, rugged, and safe) method is a recent and widely used method used to extract pesticide residue from foodstuff,21 but it does not measure the fat content and is not able to clean the sample extractor of the fat content.22
Therefore, we aimed to synthesize a hydrophobic alginic acid to be used as a sorbent clean-up material to be then used in the pesticide residue detection of a fatty food sample. Initially, the chemical modification of alginic acid was conducted using eco-friendly ultrasound-assisted esterification with different alcoholic hydrocarbon chains. Subsequently, the efficiency of the fabricated material was evaluated by calculating the fat removal percent and by analyzing a real animal product sample, which was spiked with pesticide substances.
2. Experimental procedure
2.1 Materials and equipment
All solvents and chemicals used were of an analytical grade. Alginic acid powder (Sigma-Aldrich), methanol (Fisher; 99.8%), 1-butanol (Fisher; 99.8%), 1-octadecanol (Alfa Aesar; 97%), sulfuric acid (PanReac; 96%), ethanol (VWR; 99.7%), acetone (BDH; 99.5%), an LC multi-residue pesticide standard (RESTEK; P/N 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 971), a QuEChERS extraction kit (Agilent; P/N 5982-5650), DI water (Millipore; 18.2 MΩ cm), acetonitrile (ROMIL; 99.9%), formic acid (Sigma-Aldrich; 99%), and an ammonium format (VWR) were used in the experiment. Other equipment was used for the polymer synthesis and application procedure, including an ultrasonic water bath (Elmasnoic; S 60H), ESI-LC-MS/MS Triple Quad (Sciex 6500; Germany), an analytical balance (Mettler Toledo; 6 digit), a vortex (Fisher), a centrifuge (HERMLE; 13
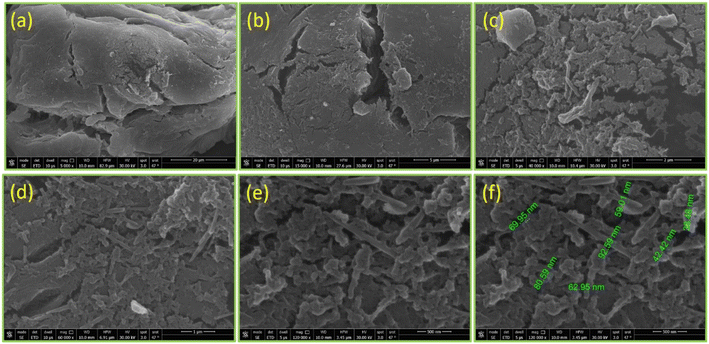
971), a QuEChERS extraction kit (Agilent; P/N 5982-5650), DI water (Millipore; 18.2 MΩ cm), acetonitrile (ROMIL; 99.9%), formic acid (Sigma-Aldrich; 99%), and an ammonium format (VWR) were used in the experiment. Other equipment was used for the polymer synthesis and application procedure, including an ultrasonic water bath (Elmasnoic; S 60H), ESI-LC-MS/MS Triple Quad (Sciex 6500; Germany), an analytical balance (Mettler Toledo; 6 digit), a vortex (Fisher), a centrifuge (HERMLE; 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm), and HPLC vials (Agilent; 2 mL). Regarding the characterization techniques, Fourier-transform infrared spectroscopy (FT-IR) (Thermo Scientific; Nicolet iS50 FT-IR) was used to identify the function groups. X-ray diffraction (XRD) (Bruker Co; D8 Discover; Cu target 40 kV; 40 mA; wavelength 1.54 A) was used to study the crystallographic structure. A thermogravimetric analysis (TGA) (Setaram; Themys one+) was used to study the response of the synthesized biopolymers to thermal stability. The samples were measured at a heating rate of 10° per minute under an argon atmosphere. Field-emission scanning electron microscopy (FE-SEM) (QUANTA FEG 205) was used to observe the microstructure image of the biopolymers.
000 rpm), and HPLC vials (Agilent; 2 mL). Regarding the characterization techniques, Fourier-transform infrared spectroscopy (FT-IR) (Thermo Scientific; Nicolet iS50 FT-IR) was used to identify the function groups. X-ray diffraction (XRD) (Bruker Co; D8 Discover; Cu target 40 kV; 40 mA; wavelength 1.54 A) was used to study the crystallographic structure. A thermogravimetric analysis (TGA) (Setaram; Themys one+) was used to study the response of the synthesized biopolymers to thermal stability. The samples were measured at a heating rate of 10° per minute under an argon atmosphere. Field-emission scanning electron microscopy (FE-SEM) (QUANTA FEG 205) was used to observe the microstructure image of the biopolymers.
2.2 Synthesis of hydrophobic alginic acid (HAA)
Alginic acid was modified using environmentally friendly ultrasound-assisted esterification with three different alcohols (methanol, butanol, and octadecanol). The synthesis of HAAs was adopted from previous work7,23. The reaction took place under heterogeneous conditions. Briefly, three conical flasks with caps were prepared, then 1 g alginic acid (AA) was weighed into each one. Next, 200 mL methanol was added to first sample (HAA-C1), 450 mL butanol was added to the second sample (HAA-C4), and 4.5 g octadecanol and 200 mL methanol as medium (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole) were added to the third sample (HAA-C18). The molar ratio of AA and all alcohols was 1
1 mole) were added to the third sample (HAA-C18). The molar ratio of AA and all alcohols was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Subsequently, 1 mL sulfuric acid at a concentration of 96% was added to each sample, then all samples were placed in a constant ultrasonic water bath for 2 h at 40 °C. Finally, each new synthesized HAA was filtered then washed twice using 70% ethanol, followed by acetone. The new products were then dried in an oven at 40 °C for 1 h. The degree of esterification (DE) was determined by titration, as illustrated in the Food Chemical Codex.24 The physical and chemical properties of the synthesized biopolymers were checked using characterization tools (FT-IR, XRD, TGA, and SEM).
1. Subsequently, 1 mL sulfuric acid at a concentration of 96% was added to each sample, then all samples were placed in a constant ultrasonic water bath for 2 h at 40 °C. Finally, each new synthesized HAA was filtered then washed twice using 70% ethanol, followed by acetone. The new products were then dried in an oven at 40 °C for 1 h. The degree of esterification (DE) was determined by titration, as illustrated in the Food Chemical Codex.24 The physical and chemical properties of the synthesized biopolymers were checked using characterization tools (FT-IR, XRD, TGA, and SEM).
2.3 Determination of fat% removal
To investigate the performance of the synthesized hydrophobic AAs from a fat removal aspect, 2 g of chicken (a fat sample) was weighed in a 50 mL centrifuge tube, and then 10 mL of acetonitrile was added. After shaking for 2 minutes and centrifuging at 4000 rpm for 5 minutes at room temperature, the liquid layer (acetonitrile) was transferred to another tube and evaporated under the N2 evaporation system until dryness. The remaining fat matrix was weighed and recorded, then dissolved again using 10 mL of acetonitrile. At this stage, the HAAs were added, shaken for 10 minutes, and centrifuged again in the same condition as mentioned above to precipitate the HAAs with fat matrix content. After that, the acetonitrile layer was moved to a new tube and then evaporated until dryness, and the remaining fat was weighed and recorded again. The fat% removal was calculated by dividing the wt fat after using HAAs over the wt. fat before, then multiplying by 100. This process was repeated three times, and the average percentage was reported for each HAA.2.4 Determination of pesticide residues in a fat sample using HAAs
The sample preparation and the extraction of pesticide residues from a fat sample were adopted from an international method to minimize the error sources.25 Briefly, 2 g of a homogenized chicken sample was weighed in a 50 mL centrifuge tube and spiked with 30 μg kg−1 of a mixed pesticide standard. Subsequently, 10 mL acetonitrile was added and shaken for 2 min. The extraction salt (QuEChERS) was then added to the mixture and it was manually shaken for 2 min, then centrifuged at 4000 rpm for 5 min at room temperature. Next, 1 mL of the extractor (upper layer) was transferred to a new 2 mL Eppendorf tube. HAA biopolymers were added to remove the fat content. The mixture was shaken for 5 min, then centrifuged again for 5 min at 4000 rpm at 4 °C. Finally, the upper layer was moved to an HPLC vial and injected into LC-MS/MS.The analysis was performed using a liquid chromatography-mass spectrometer 6500 triple quad instrument (±ESI-LC-MS/MS) and a 1290 Infinity UPLC system with ideal conditions. The analytical column used was reverse phase (C18; 2.1 × 150 mm; 1.8 μm). The mobile phase consisted of water; each had a 5 mM ammonium format with 1% formic acid. The MS parameters were set for optimized conditions, with an ion source temperature of 550 °C, ion spray potential of 5500 V, and input potential of 10 V. The quantification of the measured pesticides was calculated using a solvent calibration curve (external calibration strategy).
3. Results and discussion
3.1 Chemistry
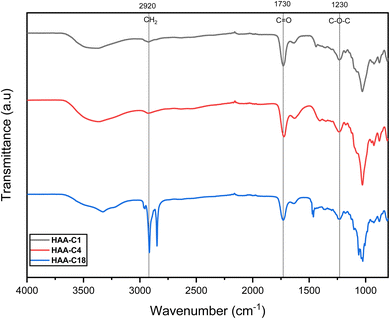
The modified ester of alginic acid HAAs was successfully synthesized using a simple, fast, cost-effective, and eco-friendly method by combining acid catalyzed with ultrasound-assisted irradiation. Methyl, butyl, and octadecyl alginates were esterified, as shown in Scheme 1. The heterogeneous synthesis of HAAs in the (methanol/H+) depended on the surface interaction between the solid phase (alginic acid) and the liquid medium (methanol/H+). The use of ultrasound in the irradiation process significantly reduced the reaction time from 72 h to just 2 h.7 The ultrasound also generated gas microbubbles that implosively disrupted, forming cavitation bubbles at incredibly high temperatures and pressures. These actions resulted in a significant acceleration of the reaction. In HAA-C18, methanol was used as a medium because the octadecanol is in powder form as well as alginic acid, so there is a possibility of esterification with methanol instead of octadecanol. In spite of that, octadecanol competed with methanol in this interaction due to its higher molecular weight. FT-IR spectroscopy supported this interpretation. Thereafter, the degree of esterification of AA was determined and provided at 63.7 ± 4.6%, 57.3 ± 2.7%, and 59.4 ± 3.1% for HAA-C1, HAA-C4, and HAA-C18, respectively.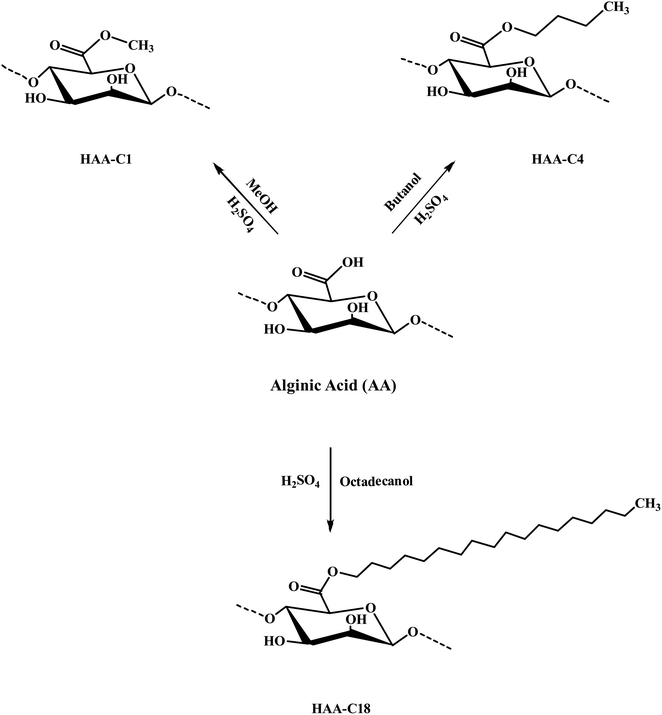 | ||
| Scheme 1 Preparation mechanism of the modified hydrophobic alginic acids HAA-C1, HAA-C4, and HAA-C18. | ||
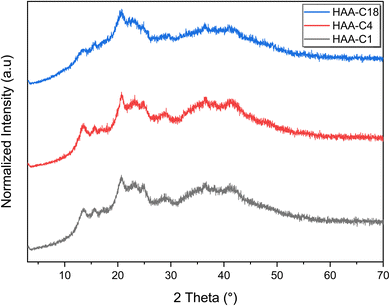
3.2 Characterization confirmation
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O on the ester groups. The other new signals that appeared at 1245, 1243, and 1244 cm−1 were attributed to the stretching vibration of C–O on the ester group. These results indicated that the methyl, butyl, and octadecyl groups had successfully grafted onto alginate via the esterification reaction to generate the HAAs.
O on the ester groups. The other new signals that appeared at 1245, 1243, and 1244 cm−1 were attributed to the stretching vibration of C–O on the ester group. These results indicated that the methyl, butyl, and octadecyl groups had successfully grafted onto alginate via the esterification reaction to generate the HAAs.
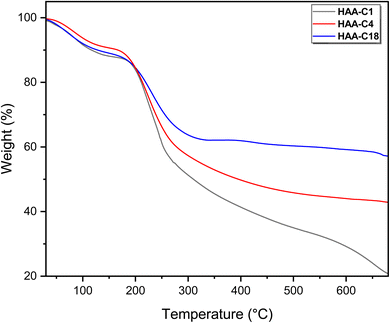
The second weight loss occurred at an inflection point of 200 °C. This degradation step was the main step (fastest step) for all modified derivatives (HAA-C1, HAA-C4, and HAA-C18). This behavior designated the decomposition of the alginate by dehydrating the saccharide rings, breaking the C–H and C–O–C glycoside bonds in the main polysaccharide chain. The data in Table 1 demonstrate the thermal behavior of the modified structure, where 10%, 20%, 30%, and 40% represent the weight loss. No significant change between AA and HAA-C1 was observed at 10%; at 20% to 40%, the degradation decreased with an increase in temperature as the side chain of ester rose. HAA-C4 and HAA-C18 modified with a flexible side chain showed steady state thermal stability started at 450 and 300 °C, respectively, compared with AA and HAA-C1, which had fewer thermal stability characteristics.
Finally, the thermograms showed a highly thermal stable structure in the third degradation step of HAA-C18, caused by the decrease in the carboxyl groups of the polymer and the formation of ester bonds that destroyed the polymeric intramolecular hydrogen bond. All the outcomes confirmed that the esterification of alginic acid was successfully introduced. The final degradation temperature (FDT) refers to the temperature at which the decomposition is completed or becomes final. The literature confirms that pure AA is completely decomposed at 700 °C.31 Whereas, the other materials show significantly higher FDT values in comparison to pure AA. The FDT values are in the following order: HAA-C1 = HAA-C4 < HAA-C18. Such observations are attributed to the measurement conditions under an argon atmosphere, which reduce the thermal degradation process.
3.3 Fat% removal application
The physical appearance of the final extractor was an initial indicator of HAAs working; it became clearer and finer after applying HAAs in the extraction procedure. HAAs were able to remove approximately 77–83% of the fat in the clean-up step. HAA-C18 had the highest removal percent of approximately 83%; the lowest was achieved by HAA-C4. Table 2 shows the fat% removal of each HAA. In general, all HAAs were able to efficiently remove fat and the results were close to each other. Therefore, the results were accepted for the intended purpose, and are promising for future developments.| Hydrophobic alginic acids | Wt fat (mg) | Removal (%) | |
|---|---|---|---|
| Before | After | ||
| HAA-C1 | 5.00 | 0.93 | 79.4 |
| HAA-C4 | 4.51 | 1.01 | 77.6 |
| HAA-C18 | 4.51 | 0.89 | 82.2 |
3.4 Pesticide residue application
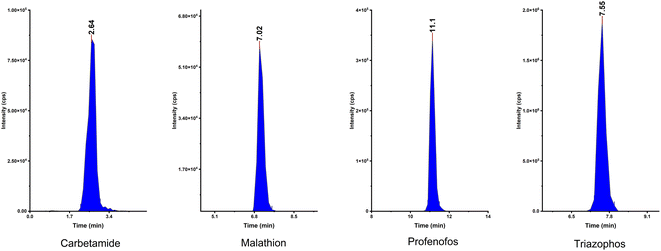
The ability of the fabricated HAAs to remove fat from the sample was checked using HAAs as a clean-up in a pesticide residue application of a fatty sample. Pesticide compounds were chosen due to their diversity of chemical structures; they have wide groups of semi-polar and non-polar substances. Therefore, this method was appropriate to investigate the performance of HAAs to rend off the fat without affecting the extraction and recovery of analytes.An LC mixture standard containing 214 compounds from various chemical pesticide classes such as organochlorine, organophosphorous, carbamate, and pyrethroid was used and spiked in the fatty animal product sample. First, pesticide compounds were analyzed without the HAAs to check their stability and activity. Subsequently, HAAs were applied as a clean-up step and the results were evaluated. Fig. 5 shows the integrated peak shapes for a few of the representative pesticide compounds.
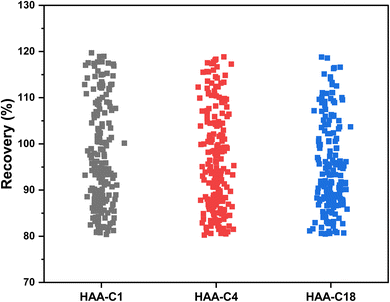
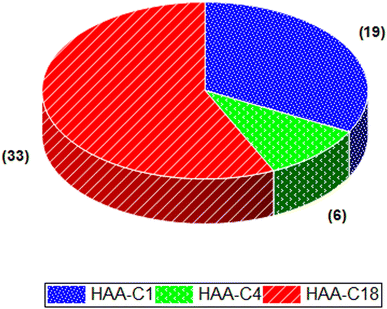
The effect of HAA-C1, HAA-C4, and HAA-C18 on the pesticide compounds was individually evaluated; the analyte recoveries are reported in Table 3. All HAAs provided satisfactory recoveries for the detected pesticide compounds. The recovery ranged from 80% to 120%, which met the requirement specifications according to SANTE/2017.23 Fig. 6 illustrates the recovery ranges for the pesticide compounds after applying the three HAAs. There were various missing pesticides after using HAAs as a fat sorbent. The number of missing pesticide compounds were 19, 6, and 33 for HAA-C1, HAA-C4, and HAA-C18, respectively, as displayed in the pie chart in Fig. 7. The un-unified missing pesticide compounds among HAA sorbents could be interpreted as being due to the difference in the chemical interactions between pesticides and HAAs. The highest number of missing compounds appeared after using HAA-C18, where 33 pesticides were lost (including 4,4′-dichlorobenzophenon, cycloxydim, and carbaryl). After checking the structure of the missed pesticide compounds, it was clearly observed that they had high hydrophobicity properties that could chemically interact with the most hydrophobic HAA-C18; hence, they were probably removed with the fat in the clean-up stage. HAA-C1 and HAA-C4 which had short hydrocarbon chains and, hence, fewer hydrophobicity properties—lost fewer pesticide compounds. It was observed that HAA-C1 lost a greater number of compounds than HAA-C4 (e.g., flufenoxuron), possibly due to the high polarity. In conclusion, HAAs could be used as an efficient removal sorbent for fat in pesticide residue applications, but we highly recommend that the various structures of the wide groups of pesticide compounds and the consequential effects are considered.
| Pesticides name | Spiking level (μg kg−1) | Average recovery (%) (±CV% for n = 3) | ||
|---|---|---|---|---|
| HAA-C1 | HAA-C4 | HAA-C18 | ||
| (Monceren) pencycuron | 30 | ND | 82.3 ± 4 | 80.8 ± 4 |
| 3,4-Dichloroaniline | 30 | 106.5 ± 7 | 106.3 ± 7 | 103.3 ± 7 |
| 4,4′-Dichlorobenzophenon (4.4-DBP) | 30 | ND | 83.4 ± 6 | ND |
| Acibenzolar-S-methyl | 30 | 82.9 ± 6 | 82.2 ± 6 | ND |
| Aclonifen | 30 | 88.6 ± 12 | 86.8 ± 12 | 88.3 ± 12 |
| Ametryn | 30 | 82.1 ± 9 | 85.1 ± 10 | ND |
| Amitraz | 30 | 92.6 ± 9 | 93.0 ± 10 | 85.9 ± 9 |
| Atrazine | 30 | 82.5 ± 9 | 82.0 ± 9 | ND |
| Azaconazole | 30 | 104.3 ± 18 | 104.8 ± 17 | 99.0 ± 18 |
| Azinphos-methyl | 30 | 85.1 ± 13 | 84.6 ± 13 | 83.2 ± 10 |
| Azoxystrobin | 30 | 99.0 ± 11 | 97.5 ± 12 | 95.0 ± 12 |
| Beflubutamid | 30 | 112.9 ± 9 | 102.2 ± 6 | 93.2 ± 4 |
| Benalaxyl | 30 | 100.2 ± 2 | 101.8 ± 6 | 86.7 ± 5 |
| Bendiocarb | 30 | 91.3 ± 12 | 88.4 ± 11 | 99.7 ± 13 |
| Bentazone | 30 | 89.6 ± 17 | 98.3 ± 19 | 91.1 ± 18 |
| Benzovindiflupyr | 30 | 113.3 ± 8 | 107.7 ± 8 | 95.5 ± 8 |
| Benzoximate | 30 | 98.8 ± 18 | 89.4 ± 16 | 96.6 ± 18 |
| Boscalid | 30 | 115.1 ± 15 | 115.6 ± 15 | 112.9 ± 15 |
| Bromacil | 30 | 93.4 ± 14 | 92.6 ± 18 | 89.3 ± 17 |
| Bromfenvinfos | 30 | 82.7 ± 13 | 85.8 ± 13 | 91.5 ± 14 |
| Bromucanozole isomer | 30 | 103.1 ± 13 | 96.3 ± 12 | 96.3 ± 12 |
| Bupirimate | 30 | 88.9 ± 21 | 81.0 ± 20 | 80.6 ± 19 |
| Butafenacil | 30 | 117.5 ± 13 | 114.3 ± 13 | 104.2 ± 12 |
| Butylate | 30 | 85.5 ± 12 | 83.7 ± 12 | ND |
| Carbaryl | 30 | 81.3 ± 14 | 81.5 ± 14 | ND |
| Carbetamide | 30 | 95.8 ± 22 | 95.1 ± 22 | 90.4 ± 21 |
| Carbofuran | 30 | 89.9 ± 18 | 88.6 ± 18 | 89.3 ± 18 |
| Carboxin | 30 | 84.1 ± 20 | 86.5 ± 20 | 80.7 ± 19 |
| Carfentrazone-ethyl | 30 | 107.3 ± 18 | 111.5 ± 19 | 105.1 ± 18 |
| Chlorantraniliprole | 30 | 100.4 ± 23 | 99.9 ± 23 | 95.9 ± 22 |
| Chlorbromuron | 30 | 94.0 ± 21 | 92.0 ± 20 | 90.8 ± 20 |
| Chlorfenvinphos | 30 | 81.9 ± 10 | 87.7 ± 11 | 88.5 ± 11 |
| Chlorfluazuron | 30 | 104.5 ± 12 | 97.9 ± 11 | 90.5 ± 10 |
| Chlorotoluron | 30 | 90.4 ± 14 | 91.6 ± 14 | 86.8 ± 14 |
| Chloroxuron | 30 | 93.9 ± 17 | 95.6 ± 17 | 91.8 ± 16 |
| Chlorpropham | 30 | 89.9 ± 3 | 88.1 ± 3 | 88.2 ± 3 |
| Chlorpyrifos-methyl | 30 | 89.0 ± 15 | 90.6 ± 15 | 81.2 ± 13 |
| Chlortoluron | 30 | 91.7 ± 14 | 92.0 ± 14 | 86.8 ± 14 |
| Clethodim isomer | 30 | ND | 80.6 ± 11 | ND |
| Clodinafop-propargyl | 30 | 117.8 ± 13 | 110.5 ± 12 | 104.1 ± 12 |
| Clofentezine | 30 | 87.6 ± 2 | 86.5 ± 2 | ND |
| Clomazone | 30 | 94.9 ± 12 | 93.7 ± 12 | 90.6 ± 12 |
| Cloquintocet-mexyl | 30 | 89.0 ± 11 | 88.1 ± 11 | ND |
| Coumaphos | 30 | 86.8 ± 17 | 94.8 ± 18 | 96.0 ± 18 |
| Cyantraniliprole | 30 | 113.0 ± 11 | 109.8 ± 11 | 110.0 ± 11 |
| Cyazofamid | 30 | 84.0 ± 9 | 91.6 ± 9 | 81.6 ± 8 |
| Cycloate | 30 | ND | ND | 80.6 ± 18 |
| Cycloxydim | 30 | 86.1 ± 13 | 80.6 ± 12 | ND |
| Cycluron | 30 | 95.9 ± 19 | 93.1 ± 18 | 90.0 ± 18 |
| Cyflumetofen | 30 | 110.7 ± 3 | 107.2 ± 3 | 114.5 ± 3 |
| Cymoxanil | 30 | 97.7 ± 10 | 83.7 ± 9 | 87.0 ± 9 |
| Cyproconazole isomer | 30 | 100.2 ± 18 | 93.9 ± 22 | 89.0 ± 21 |
| Cyprodinil | 30 | 82.9 ± 19 | 90.6 ± 21 | 82.8 ± 19 |
| Desmedipham | 30 | 97.7 ± 19 | 98.4 ± 20 | 91.2 ± 18 |
| Diazinon | 30 | 93.2 ± 13 | 86.9 ± 12 | ND |
| Dichlorvos | 30 | 93.2 ± 9 | 94.2 ± 12 | 89.1 ± 10 |
| Diclobutrazol | 30 | 81.2 ± 20 | 83.4 ± 20 | 87.4 ± 21 |
| Diethofencarb | 30 | 109.7 ± 8 | 110.9 ± 5 | 105.2 ± 7 |
| Difenoconazole isomer | 30 | 100.2 ± 17 | 102.0 ± 17 | 95.6 ± 16 |
| Difenzoquat metilsulfate | 30 | 91.1 ± 13 | 91.3 ± 13 | 89.7 ± 13 |
| Diflubenzuron | 30 | 114.3 ± 22 | 112.4 ± 22 | 102.4 ± 20 |
| Dimethachlor | 30 | 94.6 ± 13 | 95.3 ± 13 | 91.1 ± 13 |
| Dimethomorph isomer | 30 | ND | 117.4 ± 27 | 116.5 ± 27 |
| Dimoxystrobin | 30 | 96.1 ± 12 | 91.0 ± 15 | 88.6 ± 11 |
| Diniconazole | 30 | 85.7 ± 18 | 93.4 ± 20 | 88.2 ± 19 |
| Disulfoton sulfoxid | 30 | 93.4 ± 13 | 94.0 ± 13 | 91.4 ± 13 |
| Edifenphos | 30 | 80.9 ± 12 | 80.5 ± 12 | ND |
| EPN | 30 | 112.8 ± 19 | 103.9 ± 15 | 90.1 ± 20 |
| Epoxiconazole | 30 | 107.0 ± 13 | 99.3 ± 14 | 96.5 ± 14 |
| Etaconazole isomer | 30 | 93.6 ± 4 | 104.6 ± 8 | 95.9 ± 5 |
| Ethion | 30 | 85.0 ± 4 | 81.7 ± 4 | 80.5 ± 3 |
| Ethofumesate | 30 | 108.9 ± 7 | 103.9 ± 6 | 106.3 ± 6 |
| Ethoprophos | 30 | 88.0 ± 14 | 87.5 ± 14 | 87.0 ± 13 |
| Fenamidone | 30 | 100.3 ± 20 | 100.6 ± 20 | ND |
| Fenamiphos | 30 | 106.1 ± 11 | 101.5 ± 14 | 101.1 ± 12 |
| Fenamiphos-sulfoxide | 30 | 101.4 ± 9 | 101.0 ± 9 | 95.3 ± 8 |
| Fenarimol | 30 | 112.0 ± 14 | 109.7 ± 14 | 111.1 ± 14 |
| Fenbuconazole | 30 | ND | 114.6 ± 13 | 103.7 ± 11 |
| Fenhexamid | 30 | 106.7 ± 23 | ND | 112.6 ± 24 |
| Fenitrothion | 30 | 108.6 ± 19 | 114.9 ± 20 | 110.9 ± 19 |
| Fenobucarb | 30 | 89.8 ± 19 | 88.0 ± 19 | 84.6 ± 18 |
| Fenoxycarb | 30 | 86.1 ± 15 | 100.0 ± 17 | 94.9 ± 16 |
| Fenpropimorph | 30 | 88.0 ± 20 | 89.1 ± 21 | 84.8 ± 20 |
| Fensulfothion | 30 | 112.9 ± 16 | 115.5 ± 16 | 111.1 ± 15 |
| Fenthion | 30 | 114.8 ± 20 | 104.8 ± 18 | 97.3 ± 17 |
| Fenthion-sulfoxid | 30 | 112.3 ± 19 | 113.2 ± 20 | 109.2 ± 19 |
| Fipronil | 30 | 117.8 ± 6 | 113.4 ± 6 | 106.0 ± 5 |
| Flamprop-M-isopropyl | 30 | 115.3 ± 22 | 104.3 ± 20 | 102.9 ± 20 |
| Flamprop-M-methyl | 30 | 103.8 ± 17 | 99.3 ± 16 | 96.3 ± 16 |
| Flufenacet | 30 | 93.6 ± 24 | 85.9 ± 22 | 91.6 ± 24 |
| Flufenoxuron | 30 | ND | 112.5 ± 11 | 100.5 ± 10 |
| Flumioxazin | 30 | 116.9 ± 12 | ND | 107.1 ± 11 |
| Fluometuron isomer | 30 | 97.0 ± 21 | 96.0 ± 21 | 87.8 ± 19 |
| Fluopicolide | 30 | 102.4 ± 19 | 99.1 ± 19 | 99.1 ± 19 |
| Fluopyram | 30 | 97.6 ± 20 | 99.1 ± 20 | 98.5 ± 20 |
| Fluoxastrobin | 30 | 88.8 ± 21 | 98.3 ± 23 | 93.2 ± 22 |
| Fluquinconazole | 30 | 93.4 ± 12 | 101.8 ± 13 | 91.8 ± 12 |
| Fluridone | 30 | 94.2 ± 11 | 94.2 ± 11 | 88.5 ± 11 |
| Flutolanil | 30 | 100.8 ± 14 | 97.5 ± 14 | 95.2 ± 14 |
| Flutriafol | 30 | 114.6 ± 24 | 111.1 ± 24 | 111.1 ± 24 |
| Fluxapyroxad | 30 | 112.1 ± 23 | 110.7 ± 23 | 107.5 ± 22 |
| Fonofos | 30 | 90.2 ± 18 | 82.3 ± 17 | 82.8 ± 17 |
| Forchlorfenuron | 30 | 94.4 ± 16 | 93.2 ± 16 | 89.6 ± 16 |
| Fosthiazate | 30 | 88.1 ± 20 | 89.7 ± 21 | 86.6 ± 20 |
| Furalaxyl | 30 | 92.9 ± 9 | 93.6 ± 9 | 90.8 ± 9 |
| Furathiocarb | 30 | 80.9 ± 14 | 87.8 ± 15 | ND |
| Haloxyfop | 30 | 117.3 ± 8 | 109.9 ± 5 | 114.1 ± 7 |
| Hexaconazole | 30 | 98.5 ± 9 | 104.5 ± 10 | 92.0 ± 9 |
| Hexaflumuron | 30 | 103.3 ± 10 | 99.0 ± 9 | 109.6 ± 10 |
| Hydramethylnon | 30 | ND | 83.5 ± 6 | 94.7 ± 7 |
| Imazalil | 30 | 86.8 ± 8 | 87.2 ± 8 | 84.2 ± 8 |
| Indoxacarb | 30 | 88.9 ± 16 | 106.6 ± 19 | 103.6 ± 19 |
| Ipconazole | 30 | 107.7 ± 7 | 105.9 ± 7 | 85.4 ± 8 |
| Iprodione | 30 | 86.2 ± 9 | 96.8 ± 8 | 93.4 ± 7 |
| Iprovalicarb isomer | 30 | 92.6 ± 9 | 106.4 ± 10 | 96.2 ± 9 |
| Isazofos | 30 | 88.1 ± 2 | 85.6 ± 2 | 82.7 ± 2 |
| Isoprocarb | 30 | 94.9 ± 5 | 94.6 ± 5 | 91.6 ± 5 |
| Isoprothiolane | 30 | ND | 82.9 ± 14 | ND |
| Isoproturon | 30 | 90.2 ± 20 | 91.4 ± 21 | 87.0 ± 20 |
| Isopyrazam | 30 | 83.8 ± 21 | 85.7 ± 22 | 91.4 ± 23 |
| Kresoxim-methyl | 30 | 85.6 ± 15 | 90.1 ± 16 | 100.6 ± 18 |
| Lenacil | 30 | 93.3 ± 14 | 92.6 ± 14 | 90.2 ± 14 |
| Linuron | 30 | 98.9 ± 10 | 99.8 ± 10 | 95.4 ± 9 |
| Lufenuron | 30 | 116.9 ± 10 | 113.2 ± 10 | 96.2 ± 8 |
| Malaoxon | 30 | 80.8 ± 8 | 81.0 ± 8 | ND |
| Malathion | 30 | 102.7 ± 33 | 98.9 ± 32 | 97.6 ± 32 |
| Mandipropamid | 30 | 107.6 ± 16 | 101.0 ± 15 | 101.2 ± 16 |
| Mefenacet | 30 | 87.8 ± 16 | 82.9 ± 15 | ND |
| Mepronil | 30 | 95.9 ± 13 | 99.0 ± 13 | 93.5 ± 13 |
| Metaflumizone | 30 | ND | 81.5 ± 8 | ND |
| Metalaxyl | 30 | 96.3 ± 11 | 97.5 ± 11 | 93.4 ± 11 |
| Metazachlor | 30 | 87.2 ± 21 | 84.5 ± 20 | 84.1 ± 20 |
| Metconazole | 30 | 103.4 ± 4 | 100.4 ± 3 | 88.7 ± 9 |
| Methabenzthiazuron | 30 | 88.0 ± 23 | 88.4 ± 24 | 83.9 ± 22 |
| Methiocarb | 30 | 87.0 ± 13 | 87.6 ± 13 | 87.7 ± 13 |
| Methoprene | 30 | 101.5 ± 13 | 104.1 ± 13 | ND |
| Methoprotryne | 30 | 88.3 ± 5 | 91.6 ± 6 | 86.7 ± 4 |
| Metobromuron | 30 | 98.0 ± 10 | 98.5 ± 10 | 94.3 ± 9 |
| Metolachlor | 30 | 84.0 ± 9 | 88.0 ± 9 | 86.7 ± 9 |
| Metrafenone | 30 | 113.3 ± 16 | 101.9 ± 14 | 91.0 ± 13 |
| Monolinuron | 30 | 94.0 ± 7 | 93.3 ± 7 | 89.4 ± 6 |
| Myclobutanil | 30 | 111.9 ± 13 | 118.2 ± 13 | 108.4 ± 12 |
| Neburon | 30 | 99.6 ± 19 | 103.0 ± 20 | 110.1 ± 21 |
| Novaluron | 30 | 94.9 ± 3 | 101.7 ± 5 | 101.1 ± 5 |
| Nuarimol | 30 | 117.4 ± 5 | ND | 113.3 ± 5 |
| Oxycarboxin | 30 | ND | 80.3 ± 11 | ND |
| Paclobutrazol | 30 | 109.1 ± 19 | 110.4 ± 19 | 107.2 ± 19 |
| Paraoxon-methyl | 30 | 93.0 ± 17 | 93.9 ± 17 | 86.6 ± 16 |
| Parathion | 30 | 108.9 ± 22 | 104.5 ± 21 | 99.9 ± 21 |
| Penconazole | 30 | 91.1 ± 23 | 86.2 ± 22 | 82.2 ± 21 |
| Phenmedipham | 30 | 95.8 ± 13 | 91.7 ± 13 | 94.8 ± 13 |
| Phenthoate | 30 | 82.8 ± 21 | 86.4 ± 22 | 87.9 ± 22 |
| Phosalone | 30 | 112.6 ± 33 | 94.8 ± 28 | 93.8 ± 28 |
| Phoxim | 30 | 84.0 ± 7 | 90.7 ± 7 | 84.9 ± 7 |
| Picoxystrobin | 30 | 88.5 ± 23 | 116.7 ± 30 | 106.2 ± 27 |
| Piperonyl butoxide | 30 | 87.8 ± 24 | 84.9 ± 24 | ND |
| Pirimiphos-ethyl | 30 | 89.7 ± 13 | 84.9 ± 12 | ND |
| Pirimiphos-methyl | 30 | 81.3 ± 17 | 83.0 ± 17 | ND |
| Prochloraz | 30 | 90.7 ± 12 | 98.6 ± 13 | 85.3 ± 11 |
| Profenofos | 30 | 101.8 ± 12 | 92.2 ± 11 | 115.1 ± 14 |
| Promecarb | 30 | 86.7 ± 23 | 87.9 ± 23 | 85.3 ± 23 |
| Prometon | 30 | 84.1 ± 15 | 86.0 ± 15 | 81.1 ± 14 |
| Prometryne | 30 | 81.9 ± 20 | m ± 20 | ND |
| Propachlor | 30 | 92.1 ± 7 | 92.8 ± 7 | 89.5 ± 6 |
| Propanil | 30 | 108.1 ± 3 | 109.2 ± 3 | 100.6 ± 3 |
| Propaquizafop | 30 | 115.8 ± 18 | 107.1 ± 16 | 87.5 ± 13 |
| Propetamphos | 30 | 103.9 ± 4 | 98.7 ± 3 | 88.5 ± 3 |
| Propiconazole | 30 | 101.5 ± 7 | 96.5 ± 7 | 91.4 ± 6 |
| Propyzamide | 30 | 102.4 ± 13 | 100.5 ± 13 | 99.1 ± 12 |
| Prothioconazole-desthio | 30 | 81.0 ± 4 | 83.7 ± 4 | 81.1 ± 4 |
| Pyraclofos | 30 | 114.9 ± 15 | 108.2 ± 14 | 95.6 ± 12 |
| Pyraclostrobin | 30 | 91.8 ± 20 | 91.3 ± 20 | 83.1 ± 18 |
| Pyraflufen-ethyl | 30 | 110.9 ± 18 | 112.0 ± 18 | 109.7 ± 18 |
| Pyrazophos | 30 | 108.4 ± 8 | 105.9 ± 7 | 109.3 ± 8 |
| Pyrethrins | 30 | ND | ND | 103.7 ± 27 |
| Pyridaphenthion | 30 | 113.5 ± 16 | 112.3 ± 16 | 105.2 ± 15 |
| Pyrimethanil | 30 | 82.0 ± 10 | 85.6 ± 11 | ND |
| Quinalphos | 30 | 100.3 ± 17 | 93.9 ± 16 | 88.5 ± 15 |
| Rotenone | 30 | 104.6 ± 15 | 105.6 ± 15 | 102.2 ± 15 |
| Secbumeton | 30 | 83.2 ± 8 | 85.1 ± 8 | 80.6 ± 8 |
| Siduron | 30 | 97.3 ± 17 | 97.9 ± 17 | 95.7 ± 17 |
| Spinetoram | 30 | 118.9 ± 10 | 117.6 ± 10 | 102.2 ± 9 |
| Spinosad (spinosyn A) | 30 | 89.0 ± 2 | 95.1 ± 3 | 97.1 ± 3 |
| Spirotetramat | 30 | ND | 118.9 ± 3 | 116.3 ± 3 |
| Spiroxamine isomer | 30 | 87.3 ± 18 | 88.4 ± 18 | ND |
| Sulfosulfuron | 30 | 106.4 ± 20 | 108.0 ± 20 | 106.0 ± 19 |
| Sulfotep | 30 | ND | 84.8 ± 15 | 88.0 ± 16 |
| Sulprofos | 30 | 98.9 ± 9 | 88.9 ± 8 | 83.2 ± 8 |
| Tebuconazole | 30 | 93.7 ± 15 | 87.6 ± 14 | 88.0 ± 15 |
| Tebufenozide | 30 | 93.9 ± 6 | 89.8 ± 5 | 100.7 ± 8 |
| Tebufenpyrad | 30 | 100.6 ± 18 | 91.2 ± 16 | 83.0 ± 15 |
| Tebuthiuron | 30 | 80.4 ± 10 | 80.9 ± 10 | ND |
| Teflubenzuron | 30 | ND | 86.9 ± 20 | 85.1 ± 19 |
| Temephos | 30 | 119.7 ± 21 | ND | 108.9 ± 19 |
| Terbufos sulfoxid | 30 | ND | 88.4 ± 10 | ND |
| Terbumeton | 30 | 85.5 ± 10 | 87.3 ± 10 | 81.9 ± 9 |
| Terbutryn | 30 | ND | 82.0 ± 23 | ND |
| Tetraconazole | 30 | 112.9 ± 17 | 110.1 ± 17 | 104.2 ± 16 |
| Tetramethrin | 30 | 94.7 ± 20 | 98.2 ± 21 | 93.3 ± 20 |
| Thidiazuron | 30 | 119.0 ± 20 | 117.3 ± 20 | 118.8 ± 20 |
| Thiobencarb | 30 | 89.3 ± 9 | M ± 8 | ND |
| Thiodicarb | 30 | ND | 82.2 ± 12 | ND |
| Thiophanate-methyl | 30 | 109.3 ± 2 | 115.7 ± 2 | 118.6 ± 3 |
| Tolclofos-methyl | 30 | 84.3 ± 3 | 92.6 ± 3 | 86.3 ± 3 |
| Tolfenpyrad | 30 | 105.7 ± 9 | 99.6 ± 7 | 84.1 ± 9 |
| Triadimefon | 30 | 117.6 ± 14 | 117.8 ± 14 | 116.6 ± 13 |
| Triadimenol | 30 | 111.8 ± 12 | 116.0 ± 14 | 109.0 ± 14 |
| Triazophos | 30 | 100.9 ± 12 | 93.1 ± 11 | 88.7 ± 11 |
| Trifloxystrobin | 30 | 110.1 ± 33 | 99.1 ± 30 | 91.1 ± 27 |
| Triflumizole | 30 | 85.7 ± 16 | 109.0 ± 20 | ND |
| Triflumuron | 30 | 83.9 ± 19 | 92.2 ± 21 | 90.1 ± 20 |
| Trinexapac-ethyl | 30 | 117.0 ± 13 | 113.6 ± 13 | 109.8 ± 12 |
| Triticonazole | 30 | ND | 118.4 ± 18 | ND |
| Zoxamide | 30 | 85.4 ± 12 | 93.2 ± 13 | 91.9 ± 13 |
It is crucial to mention that HAAs should not be renewed or reused since they react with the food sample's fat matrix and will precipitate and be discarded at the conclusion of the analysis. Rewashing or eliminating the fat content with an organic solvent may cause structural damage to HAAs or eliminate it along with fat molecules.
4. Conclusion
In this project, alginic acid was chemically modified using an eco-friendly esterification procedure with three alcohols (methanol, butanol, and octadecanol) to obtain hydrophobic alginic acids (HAAs). The synthesis of HAAs occurred and was confirmed by FT-IR, TGA, XRD, and SEM. Subsequently, the performance of these biopolymers was evaluated by measuring their ability to remove the fat content of a fatty food sample. The fat removal percent was calculated for each HAA. The highest percent was recorded for HAA-C18, but all materials were successful, with approximate values between 77% and 83%. The fabricated HAAs were used in a real fatty food sample analysis to determine multi-pesticide residues. All HAAs were satisfactorily able to remove fat from a final extractor, and pesticide compounds were successfully recovered. HAA-C4 provided fewer missing multi-class pesticide compounds compared with HAA-C1 and HAA-C18 due to its medium hydrophobicity. Therefore, HAAs are developable and a promising and efficient method to be used in food analysis applications.Disclaimer
“The views expressed in this paper are those of the author(s) and do not necessarily reflect those of the SFDA or its stakeholders. Guaranteeing the accuracy and the validity of the data is a solely the responsibility of the research team”.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, Saudi Arabia, funded this project under grant no. KEP-Phd-46-130-1443. The authors, therefore acknowledge with thanks DSR technical and financial support. Furthermore, the authors are grateful to the Jeddah lab, Saudi Food and Drug Authority (SFDA), Saudi Arabia, for providing the study equipment and obtaining permission to publish this work.References
- L. Yang, B. Zhang, L. Wen, Q. Liang and L.-M. Zhang, Amphiphilic cholesteryl grafted sodium alginate derivative: Synthesis and self-assembly in aqueous solution, Carbohydr. Polym., 2007, 68(2), 218–225, DOI:10.1016/j.carbpol.2006.12.020.
- S. Mosad, H. Wang, T. Tamer and Y. You, Enhancement of Antioxidant and Hydrophobic Properties of Alginate via Aromatic Derivatization: Preparation, Characterization, and Evaluation, Polymers, 2021, 13, 2575, DOI:10.3390/polym13152575.
- X. Chen, Q. Zhu, C. Liu, D. Li, H. Yan and Q. Lin, Esterification of Alginate with Alkyl Bromides of Different Carbon Chain Lengths via the Bimolecular Nucleophilic Substitution Reaction: Synthesis, Characterization, and Controlled Release Performance, Polymers, 2021, 13, 3351, DOI:10.3390/polym13193351.
- E. Broderick, H. Lyons, J. Pembroke, H. Byrne, B. Murray and M. Hall, The characterisation of a novel, covalently modified, amphiphilic alginate derivative, which retains gelling and non-toxic properties, J. Colloid Interface Sci., 2006, 298, 154–161, DOI:10.1016/j.jcis.2005.12.026.
- J.-S. Yang, Y.-J. Xie and W. He, Research progress on chemical modification of alginate: A review, Carbohydr. Polym., 2011, 84(1), 33–39, DOI:10.1016/j.carbpol.2010.11.048.
- E. Broderick, H. Lyons, T. Pembroke, H. Byrne, B. Murray and M. Hall, The characterisation of a novel, covalently modified, amphiphilic alginate derivative, which retains gelling and non-toxic properties, J. Colloid Interface Sci., 2006, 298(1), 154–161, DOI:10.1016/j.jcis.2005.12.026.
- D. Murdzheva and P. Denev, Chemical modification of alginic acid by ultrasonic irradiation, Acta Sci. Nat., 2016, 3, 13–18, DOI:10.1515/asn-2016-0002.
- N. Chopin, X. Guillory, P. Weiss, J. Bideau and S. Colliec-Jouault, Design Polysaccharides of Marine Origin: Chemical Modifications to Reach Advanced Versatile Compounds, Curr. Org. Chem., 2014, 18, 867–895, DOI:10.2174/138527281807140515152334.
- K. Kohori, H. Hirayama, K. Yamamoto and J. Kadokawa, Synthesis of mixed chitin esters with long fatty and bulky acyl substituents in ionic liquid, Int. J. Biol. Macromol., 2021, 190, 763–768, DOI:10.1016/j.ijbiomac.2021.09.044.
- H. Ilyasoglu, S. Anankanbil, M. Nadzieja and Z. Guo, Lipophilization of chitin as novel polymeric stabilizer for improved oil-in-water emulsions, Colloid Polym. Sci., 2018, 296, 1841–1848, DOI:10.1007/s00396-018-4410-z.
- Y. Wang, X. Wang, Y. Xie and K. Zhang, Functional nanomaterials through esterification of cellulose: a review of chemistry and application, Cellulose, 2018, 25, 3703–3731 CrossRef CAS.
- O. Thabet, F. Muzini, A. Atiya, K. Alamry, M. Hussein and R. Hoogenboom, Hydrophobic carboxymethyl cellulose as a clean-up sorbent in the determination of nitrofuran metabolites in animal-fat samples, RSC Adv., 2023, 13, 33221–33230, 10.1039/d3ra07021b.
- M. Maturi, et al., Fatty acid – functionalized cellulose nanocomposites for vat photopolymerization, Addit. Manuf., 2023, 61, 103342, DOI:10.1016/j.addma.2022.103342.
- Q. Xiao, G. Chen and A. Xiao, Preparation, characterization, and emulsification properties of agarose fatty acid derivatives with different hydrophobic chains, Int. J. Biol. Macromol., 2019, 141, 906–918, DOI:10.1016/j.ijbiomac.2019.09.056.
- L. Fu, et al., Bio-based active packaging: Gallic acid modified agarose coatings in grass carp (Ctenopharyngodon idellus) preservation, Int. J. Biol. Macromol., 2024, 255, 128196, DOI:10.1016/j.ijbiomac.2023.128196.
- X. Theurillat, M. Dubois and J. F. Huertas-Pérez, A multi-residue pesticide determination in fatty food commodities by modified QuEChERS approach and gas chromatography-tandem mass spectrometry, Food Chem., 2021, 353, 129039, DOI:10.1016/j.foodchem.2021.129039.
- D. Rojas-Rueda, et al., Environmental Risk Factors and Health: An Umbrella Review of Meta-Analyses, Int. J. Environ. Res. Public Health, 2021, 18, 704, DOI:10.3390/ijerph18020704.
- T. Thorat, B. K. Patle, M. Wakchaure and L. Parihar, Advancements in techniques used for identification of pesticide residue on crops, J. Nat. Pestic. Res., 2023, 4, 100031, DOI:10.1016/j.napere.2023.100031.
- S. W. C. Chung and B. L. S. Chen, Determination of organochlorine pesticide residues in fatty foods: A critical review on the analytical methods and their testing capabilities, J. Chromatogr. A, 2011, 1218(33), 5555–5567, DOI:10.1016/j.chroma.2011.06.066.
- S. Mandal, R. Poi, D. K. Hazra, I. Ansary, S. Bhattacharyya and R. Karmakar, Review of extraction and detection techniques for the analysis of pesticide residues in fruits to evaluate food safety and make legislative decisions: Challenges and anticipations, J. Chromatogr. B, 2023, 1215, 123587, DOI:10.1016/j.jchromb.2022.123587.
- A. Wilkowska and M. Biziuk, Determination of pesticide residues in food matrices using the QuEChERS methodology, Food Chem., 2011, 125(3), 803–812, DOI:10.1016/j.foodchem.2010.09.094.
- P. Brugnerotto, et al., Determination of seven pesticide residues in Mimosa scabrella honeydew honey from Brazil by GC-MS, J. Food Compos. Anal., 2023, 122, 105433, DOI:10.1016/j.jfca.2023.105433.
- T. Taubner, M. Marounek and A. Synytsya, Preparation and characterization of amidated derivatives of alginic acid, Int. J. Biol. Macromol., 2017, 103, 202–207, DOI:10.1016/j.ijbiomac.2017.05.070.
- Institute of Medicine, Food Chemicals Codex, The National Academies Press, Washington, DC, 5th edn, 2003, DOI:10.17226/10731.
- M. Anastassiades, S. J. Lehotay, D. Štajnbaher and F. J. Schenck, Fast and Easy Multiresidue Method Employing Acetonitrile Extraction/Partitioning and ‘Dispersive Solid-Phase Extraction’ for the Determination of Pesticide Residues in Produce, J. AOAC Int., 2003, 86(2), 412–431, DOI:10.1093/jaoac/86.2.412.
- Y. Wang, M. Mortimer, C. Chang and P. Holden, Alginic Acid-Aided Dispersion of Carbon Nanotubes, Graphene, and Boron Nitride Nanomaterials for Microbial Toxicity Testing, Nanomaterials, 2018, 8, 76, DOI:10.3390/nano8020076.
- N. Sanchez-Ballester, B. Bataille, R. Benabbas, B. Alonso and I. Soulairol, Development of alginate esters as novel multifunctional excipients for direct compression, Carbohydr. Polym., 2020, 240, 116280, DOI:10.1016/j.carbpol.2020.116280.
- H. Yan, et al., Synthesis of alginate derivative via the Ugi reaction and its characterization, Carbohydr. Polym., 2015, 136, 757–763, DOI:10.1016/j.carbpol.2015.09.104.
- M. Ionita, A. Pandele and H. Iovu, Sodium alginate/graphene oxide composite films with enhanced thermal and mechanical properties, Carbohydr. Polym., 2013, 94, 339–344, DOI:10.1016/j.carbpol.2013.01.065.
- X. Chen, et al., Synthesis of amphiphilic alginate derivatives and electrospinning blend nanofibers: a novel hydrophobic drug carrier, Polym. Bull., 2015, 72, 3097–3117, DOI:10.1007/s00289-015-1455-8.
- B. Sarker, et al., Fabrication of alginate-gelatin crosslinked hydrogel microcapsules and evaluation of the microstructure and physico-chemical properties, J. Mater. Chem., 2014, 2, 1470–1482, 10.1039/C3TB21509A.
| This journal is © The Royal Society of Chemistry 2024 |