Open Access Article
Open Access ArticleAntimicrobial sesquiterpenes from the cultured mycobiont Diorygma pruinosum against methicillin-resistant Staphylococcus aureus isolated from Vietnamese street foods†
Thi-Kim-Dung Le ab,
Thuc-Huy Duong*c,
Huy Truong Nguyenb,
Nguyen-Kim-Tuyen Phamd,
Thi-Phi-Giao Voe,
Ngoc-Hong Nguyenf,
Nakorn Niamnontg,
Jirapast Sichaemh and
Thi-Minh-Dinh Tran*i
ab,
Thuc-Huy Duong*c,
Huy Truong Nguyenb,
Nguyen-Kim-Tuyen Phamd,
Thi-Phi-Giao Voe,
Ngoc-Hong Nguyenf,
Nakorn Niamnontg,
Jirapast Sichaemh and
Thi-Minh-Dinh Tran*i
aLaboratory of Biophysics, Institute for Advanced Study in Technology, Ton Duc Thang University, Ho Chi Minh City 700000, Vietnam
bFaculty of Pharmacy, Ton Duc Thang University, Ho Chi Minh City 700000, Vietnam
cDepartment of Chemistry, Ho Chi Minh City University of Education, 280 An Duong Vuong Street, District 5, Ho Chi Minh City 700000, Vietnam. E-mail: huydt@hcmue.edu.vn
dFaculty of Environment, Sai Gon University, 273 An Duong Vuong, Ward 3, District 5, Ho Chi Minh City 700000, Vietnam
eFaculty of Biology and Biotechnology, University of Science, Vietnam National University Ho Chi Minh City, Ho Chi Minh City 700000, Vietnam
fCirTech Institute, HUTECH University, 475 A Dien Bien Phu Street, Binh Thanh District, Ho Chi Minh City 700000, Vietnam
gOrganic Synthesis, Electrochemistry & Natural Product Research Unit, Department of Chemistry, Faculty of Science, King Mongkut's University of Technology Thonburi, Bangkok 10140, Thailand
hResearch Unit in Natural Products Chemistry and Bioactivities, Faculty of Science and Technology, Thammasat University Lampang Campus, Lampang 52190, Thailand
iDepartment of Biology, Ho Chi Minh City University of Education, 280 An Duong Vuong Street, District 5, Ho Chi Minh City 700000, Vietnam. E-mail: dinhttm@hcmue.edu.vn
First published on 6th February 2024
Abstract
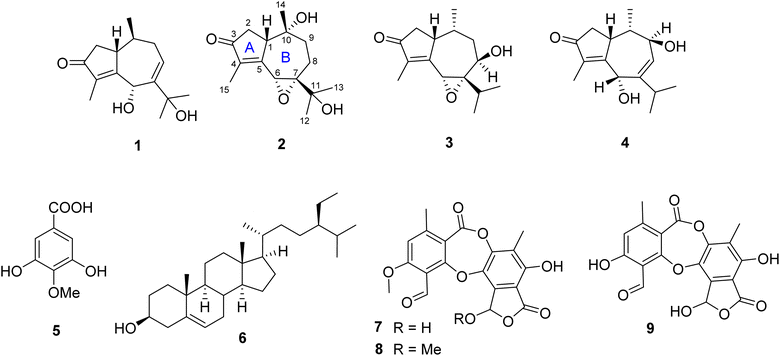
Traditionally, lichen has been used for many purposes, but there remains a lack of understanding regarding the chemical composition and antimicrobial characteristics of Diorygma pruinosum, a lichen native to Vietnam. In this study, four sesquiterpenes, diorygmones B–E (1–4), one phenolic compound, 3,5-dihydroxy-4-methoxybenzoic acid (5), and one sterol, β-sitosterol (6), were isolated and structurally elucidated from the cultured mycobiont of the lichen Diorygma pruinosum. Additionally, two compounds, stictic acid (7) and norstictic acid (8), were also isolated from the lichen D. pruinosum. Compounds 2–4 were new compounds. Their chemical structures were established using comprehensive spectroscopic data, and the absolute configurations were confirmed through the analysis of NOESY and electronic circular dichroism (ECD). Moreover, Staphylococcus aureus, a Gram-positive bacterium, has been responsible for various infections, including food poisoning. Herein, we identified and isolated 13 strains of S. aureus from street food sources. Among these strains, one was identified as a multidrug-resistant variant, designated as SAX15, and was subsequently used for further antimicrobial testing. Compounds 1–3 produced zones of inhibition against S. aureus SAX15 (each 5 mm) in comparison to commercial drugs such as penicillin, ciprofloxacin, gentamicin, cefoxitin, and clarithromycin, which displayed inhibitory zones of 7, 5, 10, 9.7, and 7 mm, respectively.
1. Introduction
Staphylococcus aureus is a Gram-positive bacterium that produces staphylococcal enterotoxins, which can cause gastroenteritis worldwide.1 Staphylococcal enterotoxins are found in a diverse range of food items, including meat and meat products, poultry, eggs, dairy products, salads, baked goods like cream-filled pastries and cakes, as well as sandwich fillings.2 In addition, S. aureus is also a leading cause of sepsis, infective endocarditis, bone, skin, and soft tissue infections, pleura, and medical device-related infections.3 The treatment of diseases caused by S. aureus is facing many difficulties due to the increasing rate of antibiotic resistance in S. aureus bacteria.4 Even more concerning, multidrug-resistant strains of S. aureus have been detected in a variety of foods, including meat, poultry, seafood, and dairy products.5 These strains are considered a potential route for the transmission of antibiotic-resistant bacteria to humans.6Lichens represent a symbiotic partnership between fungi and cyanobacteria and/or green algae.7 Lichens are employed for various purposes by traditional peoples, including their utilization as medicines, cosmetics, aphrodisiacs, and food.8 Lichens are well known to produce a wide range of characteristic secondary metabolites, some of which exhibit antibiotic, antifungal, antiviral, antitumor, and anticancer properties.7,9 Vietnam has a tropical climate and is home to various common crustose lichens, yet only a few Vietnamese lichens have undergone chemical examination.10 Researchers believe that lichen-derived mycobionts hold promise as valuable sources for discovering new compounds.11–13 Nevertheless, the understanding of the chemical composition and antimicrobial properties of Diorygma pruinosum, a lichen native to Vietnam, remains quite limited. To date, only three phytochemical investigations have been carried out on the Diorygma genus, uncovering the presence of five new compounds, funiculosone,14 pruinosone, hydroxypruinosone,13 and diorygmones A–B.15
2. Results and discussion
In this investigation, we described the isolation of methicillin-resistant S. aureus strains derived from street food collected in Ho Chi Minh City, Vietnam. Additionally, we reported the isolation of three new diorygmones C–E (2–4) together with three known compounds, diorygmone B (1), 3,5-dihydroxy-4-methoxybenzoic acid (5), and β-sitosterol (6) from the cultured mycobiont of the lichen D. pruinosum. The structures and absolute configurations of new compounds were established via extensive analyses of spectroscopic data and high-resolution electrospray ionization mass spectrometry data as well as by comparison with literature values. Compounds 1–4, 7, and 8 were evaluated for their antimicrobial activity against the selected methicillin-resistant S. aureus strain, namely SAX15.The isolation and identification of these compounds hold significant value in advancing our comprehension of the chemical composition of D. pruinosum, a lichen species that has previously received limited attention in the realm of scientific investigation. The discovery of new compounds 2–4, carries particular importance, as they may possess distinctive properties and potential applications. These findings contribute to the broader field of natural product discovery and could have implications for the development of novel bioactive compounds with potential uses in various areas, including medicine and agriculture.
2.1. Staphylococcus aureus isolation
In the process of isolating S. aureus from street food samples collected in Ho Chi Minh City, a specific approach was employed due to the unique characteristics of S. aureus. This bacterium has the ability to ferment mannitol, leading to a distinctive change in the color of the pH indicator phenol red when grown on mannitol salt phenol red agar (MSA) medium. As a result, colonies that exhibited a yellow color with surrounding yellow zones on MSA were carefully selected. These selected colonies were then transferred onto Baird-Parker agar with tellurite egg yolk, a medium conducive to the growth of S. aureus. Notably, 18 colonies displaying typical characteristics of S. aureus were identified during this process. These characteristics included the colonies being convex, shiny, black, and encompassed by a halo of lightening egg yolk (Fig. S1†).This isolation and selection methodology underscores the precision and importance of identifying S. aureus in street food samples, as it capitalizes on the bacterium's distinctive fermentation properties. Subsequent experiments, such as Gram staining and biochemical tests, were conducted on these colonies to confirm their identity as S. aureus.
2.2. Biochemical tests for identification of S. aureus strains
In the biochemical tests conducted for the identification of S. aureus strains, all 18 presumptive S. aureus strains exhibited Gram-positive staphylococci characteristics when observed under the microscope, displaying distinct purple, round clusters. However, a subset of 5 out of the 18 strains tested negative for methyl red (MR) and Voges–Proskauer (VP) reactions, exhibited oxidase positivity, and were coagulase-negative. These atypical biochemical reactions did not align with the typical profile of S. aureus.Conversely, 13 out of the 18 strains displayed consistent biochemical reactions, which included positive results for catalase, methyl red (MR), Voges–Proskauer (VP), carbohydrate fermentation, and coagulase tests. Additionally, these strains tested negative for indole and oxidase tests. Based on these consistent reactions, these 13 strains were conclusively identified as genuine S. aureus strains (Table 1 and Fig. S2–S9†).
| Strain | Gram | Catalase | Indole | MR | VP | Oxidase | Carbohydrate fermentation | Coagulase |
|---|---|---|---|---|---|---|---|---|
| SAG1 | + | + | − | + | + | − | + | + |
| SAG2 | + | + | − | + | + | − | + | + |
| SAG3 | + | + | − | + | + | − | + | + |
| SAG4 | + | + | − | + | + | − | + | + |
| SAG5 | + | + | − | + | + | − | + | + |
| SAG6 | + | + | − | + | + | − | + | + |
| SAG7 | + | + | − | + | + | − | + | + |
| SAG8 | + | + | − | − | − | − | + | − |
| SAG9 | + | + | − | + | + | − | + | + |
| SAG10 | + | + | − | − | − | − | + | − |
| SAG11 | + | + | − | + | + | − | + | + |
| SAG12 | + | + | − | + | + | − | + | + |
| SAX13 | + | + | − | + | + | + | + | − |
| SAX14 | + | + | − | + | + | − | + | + |
| SAX15 | + | + | − | + | + | − | + | + |
| SAX16 | + | + | − | + | + | − | + | + |
| SAX17 | + | + | − | − | + | + | + | − |
| SAX18 | + | + | − | + | + | + | + | − |
2.3. Antibiotic susceptibility
The results of the antibiotic susceptibility tests revealed that the majority of the strains demonstrated sensitivity to the tested antibiotics. Specifically, 9 strains displayed susceptibility to penicillin, 12 to cefoxitin, 5 to clarithromycin, 12 to tetracycline, 10 to ciprofloxacin, and 11 to gentamicin (Table 2).| Strain | Inhibition zone diameter (mm) | |||||
|---|---|---|---|---|---|---|
| Penicillina | Cefoxitinb | Clarithromycinc | Tetracyclined | Ciprofloxacine | Gentamicinf | |
| a Penicillin: ≥29: susceptible, <28: resistant.b Cefoxitin: ≥22: susceptible, ≤21: resistant.c Clarithromycin: ≥18: susceptible, 14–17: intermediate, ≤13: resistant.d Tetracycline: ≥19: susceptible, 15–18: intermediate, ≤14: resistant.e Ciprofloxacin: ≥21: susceptible, 16–20: intermediate, ≤15: resistant.f Gentamicin ≥15: susceptible, 13–14: intermediate, ≤12: resistant. | ||||||
| SAG1 | 30.7 ± 0.6 | 30.3 ± 0.6 | 21.3 ± 0.6 | 21.7 ± 0.6 | 19.7 ± 0.6 | 20.3 ± 0.6 |
| SAG2 | 6.7 ± 1.2 | 23.7 ± 1.5 | 7.7 ± 0.6 | 21.7 ± 0.6 | 19.7 ± 0.6 | 6.3 ± 0.6 |
| SAG3 | 30.3 ± 0.6 | 30.3 ± 0.6 | 20.7 ± 0.6 | 24.7 ± 0.6 | 21.3 ± 0.6 | 16.0 ± 0.0 |
| SAG4 | 39.3 ± 1.2 | 34.0 ± 1.7 | 14.7 ± 0.6 | 12.7 ± 0.6 | 27.3 ± 2.1 | 19.0 ± 1.0 |
| SAG5 | 42.3 ± 0.6 | 35.0 ± 0.0 | 22.3 ± 0.6 | 26.0 ± 1.0 | 29.3 ± 1.2 | 27.3 ± 0.6 |
| SAG6 | 15.0 ± 1.0 | 31.0 ± 2.6 | 11.3 ± 0.6 | 23.0 ± 3.5 | 30.0 ± 0.0 | 29.0 ± 1.0 |
| SAG7 | 41.0 ± 1.0 | 34.0 ± 1.0 | 16.0 ± 1.0 | 25.7 ± 1.2 | 29.0 ± 1.7 | 32.3 ± 2.9 |
| SAG9 | 42.3 ± 2.1 | 34.7 ± 0.6 | 15.7 ± 1.2 | 23.7 ± 1.2 | 26.7 ± 1.5 | 32.3 ± 2.9 |
| SAG11 | 36.7 ± 1.5 | 26.3 ± 1.5 | 20.7 ± 0.6 | 25.7 ± 0.6 | 29.0 ± 1.0 | 34.0 ± 1.0 |
| SAG12 | 42.0 ± 2.6 | 26.7 ± 0.6 | 16.7 ± 0.6 | 24.3 ± 0.6 | 27.3 ± 1.2 | 33.3 ± 0.6 |
| SAX14 | 26.7 ± 0.6 | 37.3 ± 0.6 | 15.0 ± 1.0 | 29.3 ± 0.6 | 25.0 ± 0.0 | 32.7 ± 1.2 |
| SAX15 | 7.0 ± 0.0 | 9.7 ± 2.1 | 7.0 ± 0.0 | 22.3 ± 0.6 | 5.0 ± 0.0 | 10.0 ± 0.0 |
| SAX16 | 57.3 ± 1.2 | 39.7 ± 0.6 | 22.7 ± 0.6 | 21.7 ± 0.6 | 32.7 ± 2.1 | 25.3 ± 0.6 |
However, 5 out of the 13 strains exhibited resistance to at least one of the tested antibiotics, and 2 of these strains were classified as multidrug-resistant S. aureus (MDR-SA) due to their resistance to antibiotics from three different classes. Particularly noteworthy was the strain SAX15, which displayed resistance to 5 out of 6 tested antibiotics, including penicillin, cefoxitin, gentamicin, ciprofloxacin, and clarithromycin. The resistance to cefoxitin strongly suggested that it was a putative methicillin-resistant S. aureus (MRSA).
Further confirmation of its identity as an MRSA strain was obtained through the positive detection of mecA (Fig. S10†). MRSA is associated with significant clinical challenges, marked by persistently high morbidity and mortality rates.16 Consequently, this MRSA strain was selected for subsequent experiments focused on screening for antibacterial compounds.
2.4. Phytochemical identification of 2–4
The extract of the cultured mycobiont of the lichen D. pruinosum was evaluated for its antimicrobial activity against S. aureus SAX15 and exhibited an inhibition zone of 7 mm against this strain. This extract was further selected for phytochemical investigation. Compounds 1–5 were obtained from the most bioactive fraction, EA5. β-Sitosterol (6) was also detected as a major component of the studied mycobiont.| No. | 2 | 3 | 4 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | 3.04 (d, 9.0) | 50.3 | 3.05 (m) | 43.9 | 3.93 (m) | 37.0 |
| 2 | 2.51 (d, 18.0) | 45.3 | 2.42 (dd, 18.5, 7.0) | 40.8 | 2.46 (dd, 18.5, 7.5) | 39.3 |
| 2.29 (dd, 18.5, 7.0) | 2.03 (m) | 2.08 (m) | ||||
| 3 | 206.4 | 206.4 | 208.3 | |||
| 4 | 141.3 | 138.7 | 135.7 | |||
| 5 | 165.7 | 165.6 | 169.3 | |||
| 6 | 4.20 (s) | 57.0 | 3.76 (s) | 54.4 | 4.95 (d, 8.5) | 68.9 |
| 7 | 70.7 | 73.2 | 151.6 | |||
| 8 | 1.93 (m) | 23.8 | 4.23 (ddd, 11.5, 7.0, 3.5) | 66.4 | 5.79 (d, 7.5) | 128.4 |
| 1.63 (m) | ||||||
| 9 | 2.26 (m) | 38.6 | 2.03 (m) | 40.4 | 4.17 (td, 7.5, 6.0) | 70.8 |
| 1.96 (m) | 1.88 (m) | |||||
| 10 | 72.1 | 2.08 (m) | 32.5 | 2.36 (hex, 7.5) | 41.3 | |
| 11 | 70.5 | 2.80 (m) | 29.1 | 2.40 (m) | 37.3 | |
| 12 | 1.29 (s) | 26.1 | 0.86 (d, 7.0) | 16.2 | 1.09 (d, 7.0) | 21.5 |
| 13 | 1.23 (s) | 26.0 | 1.10 (d, 7.0) | 20.0 | 1.08 (d, 6.5) | 22.3 |
| 14 | 0.78 (s) | 19.2 | 0.57 (d, 7.0) | 11.3 | 0.56 (d, 7.0) | 10.5 |
| 15 | 1.76 (d, 1.5) | 8.0 | 1.78 (d, 1.5) | 7.7 | 1.67 (d, 1.5) | 8.2 |
| 6-OH | 5.38, (d, 8.5) | |||||
| 8-OH | 3.94 (d, 7.0) | |||||
| 9-OH | 5.19, (d. 5.5) | |||||
| 10-OH | 3.78 (s) | |||||
| 11-OH | 3.60 (s) | |||||
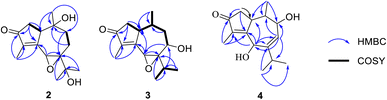
Compound 2 was isolated as a colorless oil. A molecular formula of C15H22O4 was determined by the HRESIMS ion observed at m/z 267.1594 ([M + H]+, calcd for C15H23O4, 267.1518), indicating five degrees of unsaturation. The 1H NMR showed a signal of an oxymethine [δH 4.20 (1H, s)], a methine [δH 3.04 (1H, d, J = 9.0 Hz)], four methyl groups [δH 0.78 (3H, s), 1.23 (3H, s), 1.29 (3H, s), and 1.76 (3H, d, J = 1.5 Hz)], and three methylenes in the range of 1.60–2.51 ppm. The 13C NMR data with the aid of the HSQC spectrum revealed the presence of a carbonyl ketone (δC 206.4), two methines (δC 57.0 and 50.3), three methylenes (δC 45.3, 38.6, and 23.8), four methyl groups (δC 26.1, 26.0, 19.2, and 8.0), and five quaternary carbons (δC 165.7, 141.3, 72.1, 70.7, and 70.5). The HMBC correlations of two methyls H3-12 (δH 1.29) and H3-13 (δH 1.23) to C-7 (δC 70.7) and C-11 (δC 70.5) confirmed an isopropyl moiety linked to C-7 (Fig. 2). Moreover, HMBC correlations seen from H3-15 (δH 1.76) to C-3 (δC 206.4), C-4 (δC 141.3), and C-5 (δC 165.7), and from H3-14 (δH 0.78) to C-1 (δC 50.3), C-9 (δC 38.6), and C-10 (δC 72.1) indicated that 2 possessed a guaiane-type sesquiterpenoid core.
The HMBC spectrum gave the correlations of H3-15 to C-3, C-4, and C-5, of H-2a (δH 2.51) to C-3 and C-5, and of both H-2b (δH 2.29) and H3-14 (δH 0.78) to C-1, helping to define the existence of a 2-methylcyclopent-2-en-1-one moiety (ring A). In addition, HMBC correlations of proton H-6 (δH 4.20) to C-5 and C-7, and of proton H-9a (δH 2.26) to C-7 indicated the location of a quaternary carbon C-7. A hydroxyl group forming at C-10 was concluded from key HMBC correlations of H3-14 to C-1, C-9, and C-10 (δC 72.1). Furthermore, the HMBC cross-peaks seen from H2-8 (δH 1.63 and 1.93) to C-10 established the connection in ring B. Lastly, the direct linkage between rings A and B of 2 was determined at C-1 and C-5 due to HMBC correlations of H3-14 to C-1 and H-6 to C-5. Comparison of the 1H and 13C NMR data of 2 and hydroxypruinosone (recorded in the same deuterated solvent, acetone-d6, Table S1†) revealed a close similarity between them, suggesting they share the same skeleton.13 However, the chemical shifts of H-6 and C-6 of 2 were shifted upfield when compared to those of hydroxypruinosone. Therefore, it suggests that epoxidation could have occurred at positions C-6 and C-7, and this was backed up by the molecular formula of 2.
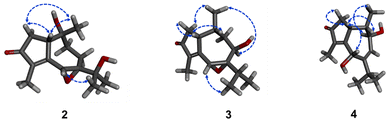
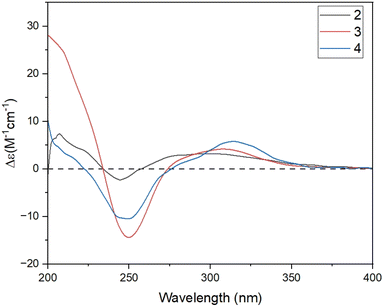
The relative configuration of 2 was characterized via NOESY data. In particular, the NOESY correlations of H-6/H3-12/H3-13 and of H-2a/H3-14/H-1 suggested these protons were cis-configured (Fig. 3). The ECD spectrum of 2 (Fig. 4) showed a positive Cotton Effect (CE) at 297 nm and negative CE at 244 nm, which were similar to diorygmone B (1) in previous report,15 supporting a 1S, 6R, 7S, 10R configuration of 2. Accordingly, the structure of 2 was elucidated as shown in Fig. 1. This is a new compound, namely diorygmone C.
Compound 3 exhibited the molecular formula C15H22O3, as established by an analysis of HRESIMS data (m/z 251.1647 [M + H]+, calcd for C15H23O3, 251.1649). The 1H NMR spectrum of 3 showed the presence of four methyl groups at δH 1.78, 1.10, 0.86, and 0.57, two oxygenated methines at δH 4.23 and 3.76, along with other signals corresponding to three methines and two methylenes (Table 3). The 13C NMR and HSQC spectra revealed 15 carbon resonances assigned to four methyls, two methylenes, five methines, and four quaternary carbons, including one keto carbonyl, two olefinic, and one saturated carbon. These data suggest that 3 has a guaiane-sequiterpene skeleton.
A comparison of the 1D NMR spectra of 2 and 3 indicated that their structures were nearly identical, except for the disappearance of a hydroxyl group at C-10 (δC 32.5) and C-11 (δC 29.1) and the presence of a hydroxyl group at C-8 (δC 66.4) in 3. This finding was confirmed by the 1H NMR, which showed the signals of a proton at C-10 (δH 2.08), a proton at C-11 (δH 2.80), and the absence of one proton at H-8. Moreover, the 13C NMR spectrum of 3 revealed the upfield shift of C-10 (Δδ − 39.6), C-11 (Δδ − 41.4), and the downfield shift of C-8 (Δδ + 37.6) compared to those of 2. The HMBC correlations of H3-14 (δH 0.57) to C-10, C-1 (δC 43.9), of H3-12 (δH 0.86) and H3-13 (δH 1.10) to C-11 and C-7 (δC 73.2), and of both H-6 (δH 3.76) and H-11 to C-8 confirmed the structure change in 3. Besides, the 1H–1H COSY correlations of H-8/H-9a, H-1/H-2a, H3-14/H-10, and H-11/H3-12/H3-13 supported the proposed structure. Moreover, the planar structure of 3 was also proved by comparing its 1D NMR with pruinosone (Table S2†).13
The stereochemistry of 3 was defined using NOESY correlations, ECD data, and comparison with the data in the literature. At first, the upfield 1H NMR chemical shift of H3-14 (δH/δC 0.57/11.3 in 3 vs. 0.78/19.2 in 2 and 0.81/19.2 in 1) indicated anti-configuration of H-1 and H3-14, further supported by a lacked NOESY correlation between H-1 and H3-14. These data were identical with those of stelleranoids K and M reported by Pan et al. (2021).18 The NOESY correlations of H-8/H3-14, of H3-14/H-2b, of H-6/H3-12, of H-1/H-2a, and of H-1/H-10, helped to assign the relative configuration of 3. These results suggested the cis-orientation of H-1, H-6, and the isopropyl moiety, and of H-8 and H3-14. Moreover, the high consistency in the ECD data of 2 and 3 (Fig. 4) allowed the determination of the absolute configuration of 3. These ECD data were identical with those of the enantiomer of stelleranoids K and M,18 suggesting the absolute (1R,10R) configuration. As shown in Fig. 1, the structure of 3 was confirmed.
Compound 4 was isolated as a colorless oil. The molecular formula of 4 was established by its HRESIMS to be C15H22O3 based on the sodium adduct ion at m/z 273.1463 ([M + Na]+, calcd for C15H22O3Na, 273.1467). This molecular formula was consisted with 15 carbon resonances present in the J-modulated 13C NMR spectrum. In the 1H NMR spectrum, four methyl groups [δH 1.67 (3H, d, J = 1.5 Hz), 1.09 (3H, d, J = 7.0 Hz), 1.08 (3H, d, J = 6.5 Hz), and 0.56 (3H, d, J = 7.0 Hz)] were observed. The 13C NMR revealed the presence of a ketone carbon (δC 208.3), four olefinic carbons (δC 169.3, 151.6, 135.7, and 128.4), two oxygenated carbons (δC 70.8 and 68.9), and four methyls (δC 22.3, 21.5, 10.5, and 8.2), which are characteristic signals of a guaiane-type sesquiterpene. Comparison of the 1H and 13C NMR data of 4 with those of hydroxypruinosone13 (Table S1†) indicated the absence of one hydroxyl group at C-9 or C-11.
The HMBC spectrum of 4 showed correlations from both H-1 (δH 3.93) and H-8 (δH 5.79) to C-6 (δC 68.9) and C-9 (δC 70.8), confirming that OH groups were located at C-6 and C-9.
Furthermore, detailed analysis of 2D NMR spectra of 4 allowed to confirm its structure. The existance of a 2-methylcyclopent-2-en-1-one unit (ring A) was confirmed by the HMBC spectrum, which showed correlations from both H3-15 (δH 1.69) and H2-2 (δH 2.46 and 2.08) to C-3 (208.3), C-4 (δC 135.7), and C-5 (δC 169.3). In addition, HMBCs from H3-14 (δH 0.56) to C-1 (δC 37.0), C-9, and C-10 (δC 37.3); from H-6 (δH 4.94) to C-1, C-5, C-7 (δC 151.6), and C-8 (δC 128.4); from H-8 to C-6, C-9, and C-10; and from H-9 (δH 4.15) to C-1, C-7, and C-8, revealed that ring B consists of seven carbons including C-1 and C-5–C-10. The HMBC correlations of the two methyl signals H3-12 (δH 1.09), H3-13 (δH 1.08), and of the olefin proton H-6 with C-7 and C-11 (δC 41.3), indicated that the isopropyl group was located at C-7. The linkage between rings A and B at C-1–C-5 was determined based on the key HMBC correlations from both H-6 and H-2 to C-1 and C-5. Therefore, the planar structure of 1 was deduced as shown in Fig. 1.
The correlations of H-1/H-2a (δH 2.46)/H-6/H-10, and the opposite correlations of H-2b (δH 2.08)/H-9/H3-14 in the NOESY spectrum (Fig. 3) suggested that 4 has a relative configuration as 1R*,6S*,9R*,10S*. The ECD data of 4 was highly similar to that of 3, indicating that they shared the same stereochemistry at C-1 and C-10. Accordingly, the complete structure of 4 was defined, as shown in Fig. 1. Notably, comparison of the ECD data of 2–4 gave the same negative Cotton effect at 250 nm. This negative CE might be an indicator for determining the stereocenter at C-1 of guaiane-type sesquiterpenes.18,19 Thus, the absolute configuration of 4 was 1R,6S,9R,10S.
2.5. Phytochemical data of 7–9 from the lichen D. pruinosum
The crude acetone extract of the lichen D. pruinosum was prepared and subsequently analyzed using the thin-layer chromatography (TLC) method, revealing the presence of three components, stictic acid (7), norstictic acid (8), and 9′-O-methylstictic acid (9), with 7 as the major component. The isolation of this extract led to the purification of 7 and 8 (Fig. 1).2.6. Biological activity of isolated compounds
Compounds 1–3 exhibited a moderate inhibitory effect against multidrug-resistant S. aureus SAX15, with each displaying an inhibition zone of 5 mm, while compounds 4, 7, and 8 were inactive. Penicillin, ciprofloxacin, gentamicin, cefoxitin, and clarithromycin exhibited inhibition zones of 7, 5, 10, 9.7, and 7 mm, respectively. The activity of 5 was previously reported.16 β-Sitosterol (6) is considered a potent antimicrobial component.21 Thus, compound 6 might be the most active compound in the studied mycobiont. These findings suggest that compounds derived from D. pruinosum could serve as promising candidates for alternative or complementary antimicrobial agents, particularly considering their effectiveness against a multidrug-resistant bacterial strain. Further research and exploration of these compounds may unveil their therapeutic potential and pave the way for the development of novel antimicrobial agents.3. Material and methods
3.1. S. aureus isolation
In September 2022, street food samples, including stir-fried rice noodles, sticky rice with assorted toppings, and spring rolls, were randomly purchased from street vendors in Ward 4, District 5, Ho Chi Minh City.Isolation of S. aureus from street food samples was carried out as the method reported previously.22 First, 10 g of each sample was transferred separately to a sterile Stomacher bag and 90 mL of NaCl (Merck, Germany) 0.9% solution were added. The sample underwent homogenization in a stomacher (Seward, England) operating at 200 rpm for 5 min. Then, a 10-fold dilution of the sample was prepared using sterile NaCl (Merck, Germany) 0.9%, and 0.1 mL of various dilutions was spread onto the surface of mannitol salt phenol red agar (Merck, Germany) plates. The plates were then incubated at 35 ± 2 °C for 24 h. Yellow colonies with yellow zones were transferred to Baird-Parker agar (Merck, Germany) medium with tellurite egg yolk (Merck, Germany). Subsequently, the plates were incubated at 35 ± 2 °C for 24 h. Black, shiny, convex colonies surrounded by a lightening halo of the egg yolk were used for biochemical tests, including Gram stain, catalase test, indole test, Methyl red-Voges Proskauer test, oxidase test, test for fermentation of glucose, lactose, and sucrose, and coagulase test.
3.2. Biochemical test for identification of S. aureus strains
Biochemical tests for the identification of S. aureus were carried out as previously mentioned.23–29 For Gram stain, a thin smear of every isolate was prepared on every sterilized glass slide, followed by heat fixation. Next, the slides were stained with crystal violet (Merck, Germany) for 1 min, followed by the addition of Gram's iodine for 1 min. After that, they were decolored with alcohol for 10–20 s. Lastly, they were counterstained with fuchsin (Merck, Germany). Gram-positive bacteria appeared purple, while Gram-negative bacteria appeared red to pink under the microscope.23The catalase test was conducted using the test tube method. A sterile bamboo stick was used to collect a small number of bacterial colonies, which were then placed in the test tube containing 2 mL of 3% hydrogen peroxide (OPC Pharma, Vietnam). The tube was next placed against a black background, and the immediate formation of bubbles at the end of the bamboo stick was observed. The bacterial strain that caused immediate bubble formation was considered a catalase-positive strain. No bubbles formed, indicating that the strain was negative for catalase.26
The Indole test was detected by the test tube method. Bacteria were cultured in test tubes containing 4.5 mL of tryptone broth (Merck, Germany) at 35 ± 2 °C for 24–48 h. Next, add 5 drops of Kovács reagent (amyl alcohol 150 mL, p-dimethylaminobenzaldehyde 10 g, and HCl 50 mL) directly to the tube. The formation of a pink to red color in the reagent layer on top of the medium within sec of adding the reagent indicated a positive indole test. The reagent layer remaining yellow or becoming slightly cloudy indicated a negative indole test.24
For the MR and VP tests, bacteria were cultured in test tubes containing 5 mL of MR-VP broth (Merck, Germany) for 18–24 h. Next, every culture was divided into aliquots of 2.5 mL in new sterile test tubes. Then, 5 drops of the methyl red (Merck, Germany) reagent were added to the test tube. MR-positive strains showed a red coloration as a result of high acid production and a decrease in the pH of the culture medium to 4.4. The MR-negative culture showed a yellow color, indicating a less acidic medium. The remaining 2.5 mL of every culture grown in MR-VP broth was used for the VP test. An amount of 0.6 mL of Barritt's reagent A was added to each test tube, followed by 0.2 mL of Barritt's reagent B. Then, the tubes were carefully shaken for 30 s to 1 minute to expose the medium to atmospheric oxygen (necessary for oxidation of acetoin to obtain a color reaction). After that, the tubes were allowed to stand for at least 30 min. VP-positive cultures showed red coloration on top of the culture, whereas VP-negative isolates had a yellowish color.25
The oxidase test was detected by the test tube method. Bacteria were grown in 4.5 mL of nutrient broth for 18–24 h. Next, 0.2 mL of 1% α-naphthol (Merck, Germany) was added to every test tube, followed by 0.3 mL of 1% p-aminodimethylaniline oxalate (Merck, Germany) (Gaby and Hadley reagents). Microorganisms are oxidase positive when the color changes to blue within 15 to 30 s. Microorganisms are delayed oxidase positive when the color changes to purple within 2 to 3 min. Microorganisms are oxidase negative if the color does not change.27
Bacteria were tested for the fermentation of carbohydrates, including glucose, lactose, and sucrose. They were cultured in phenol red carbohydrate broth (proteose peptone 10 g, sodium chloride 5 g, beef extract 1 g, phenol red 0.018 g, distilled water 1000 mL, carbohydrate 10 g) at 35 to 37 °C for 18 to 24 hours. A yellow color indicates that enough acid products have been produced by fermentation of the sugar to lower the pH to 6.8 or less. A reddish or pink color indicates a negative reaction.28
The coagulase test was detected by the test tube method. Bacteria were cultured in test tubes containing 5 mL of brain heart infusion (BHI) broth (Merck, Germany) for 18–24 h. Lyophilized rabbit plasma (BD BBL™, United States) was transferred into these test tubes. Next, they were incubated at 35 ± 2 °C. Clot formation was observed every 4 h until 24 h. The test was recorded as positive if a clot was observed. No clots indicated a negative reaction.29
3.3. Antimicrobial susceptibility testing
The resistance of isolates to penicillin, gentamicin, and ciprofloxacin was tested via disk diffusion assay using Mueller–Hinton agar as instructed by the guidelines of the Clinical and Laboratory Standards Institute (CLSI 2021). Suspensions equivalent to 0.5 McFarlands were streaked on MHA (Merck, Germany), followed by placing antibiotic discs on the surface of the MHA medium. Next, the plates were incubated upside down at 35 °C for 18–24 h. After that, inhibition zones were measured and compared with the CLSI guidelines to report the result as susceptible (S), intermediate (I), or resistant (R). The antibiotic discs tested include penicillin (10 U), gentamicin (10 μg), ciprofloxacin (5 μg), cefoxitin (30 μg), clarithromycin (15 μg), and tetracycline (30 μg). The antibiotic discs were supplied by Nam Khoa Biotek, Vietnam.Cefoxitin-resistant isolates were tested for the mecA gene using a specific primer pair (MECA_F: AGCGACTTCACATCTATTAGG, MECA_R: TGTTATTTAACCCAATCATTGCTGTT) in a conventional PCR assay. The primers were synthesized by PhuSa Genomics JSC (Can Tho province, Viet Nam). To prepare the DNA template, a single colony from each strain was collected using a sterile toothpick and placed in a sterile Eppendorf tube containing 50 μL of buffer (0.025 N NaOH, 0.123% SDS). The tubes were then heated at 95 °C for 5 min, followed by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. The supernatant was used as the DNA template for the PCR reaction. The PCR reaction components consisted of 1.5 μL of 10X Taq buffer, 1.5 μL of 2 mM dNTP, forward and reverse primers at a final concentration of 5 pM each, 0.5 U of Taq DNA polymerase (Thermo Fisher Scientific, USA), 1.5 μL of DNA template, and sterilized Milli-Q water to reach a total volume of 15 μL. PCR reactions were carried out in a PCR Thermal Cycler (AGILENT SureCycler 8800, USA) with the following thermocycle: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 30 s, and a final extension step at 72 °C for 5 min. PCR products were mixed with loading dye (containing 0.25% bromophenol blue, 0.25% xylene cyanol, and 40% glycerol in TAE buffer) and transferred into a well on a 2% agarose gel. DNA electrophoresis was performed using the Cleaver Scientific DNA electrophoresis apparatus at a voltage of 120 V and an amperage of 300 mA for 15 min.
000 rpm for 5 min. The supernatant was used as the DNA template for the PCR reaction. The PCR reaction components consisted of 1.5 μL of 10X Taq buffer, 1.5 μL of 2 mM dNTP, forward and reverse primers at a final concentration of 5 pM each, 0.5 U of Taq DNA polymerase (Thermo Fisher Scientific, USA), 1.5 μL of DNA template, and sterilized Milli-Q water to reach a total volume of 15 μL. PCR reactions were carried out in a PCR Thermal Cycler (AGILENT SureCycler 8800, USA) with the following thermocycle: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 30 s, and a final extension step at 72 °C for 5 min. PCR products were mixed with loading dye (containing 0.25% bromophenol blue, 0.25% xylene cyanol, and 40% glycerol in TAE buffer) and transferred into a well on a 2% agarose gel. DNA electrophoresis was performed using the Cleaver Scientific DNA electrophoresis apparatus at a voltage of 120 V and an amperage of 300 mA for 15 min.
3.4. Source of the lichen material
The lichen Diorygma pruinosum was collected from tree bark in Quang Ngai City (15.104 N; 108.806 E), Vietnam (approx. 9 m alt.), in March 2022. The voucher specimens were identified by one of the authors (T.-P. G. Vo) and deposited at University of Sciences, Ho Chi Minh City, Vietnam (registration no. UE-L007).3.5. Mycobiont culture
The methods for mycobiont culture followed those described in previous papers related to the cultivation of mycobionts derived from Diorygma lichens.11,133.6. Extraction and isolation of 1–6 from the mycobiont
The harvested colonies, with a dry weight of 18 g, were ground and then extracted with EtOAc (10 × 100 mL each) at room temperature. The combined extracts were concentrated under reduced pressure to yield 198 mg of residue. The EtOAc extract was dissolved in acetone to obtain both a solid (22 mg) and a solution. The solid was identified as 6. The solution was subjected to silica gel column chromatography (CC) and eluted with n-hexane–chloroform–acetone–methanol–water (400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, v/v/v/v/v) to yield ten fractions (EA1–EA10). Fraction EA5 (32 mg) was further purified using silica gel CC with the same solvent system mentioned earlier. Subsequently, it was applied to C18-reverse phase silica gel CC, eluted with acetone–water (10
0.1, v/v/v/v/v) to yield ten fractions (EA1–EA10). Fraction EA5 (32 mg) was further purified using silica gel CC with the same solvent system mentioned earlier. Subsequently, it was applied to C18-reverse phase silica gel CC, eluted with acetone–water (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to afford 1 (4.3 mg), 2 (1.3 mg, 0.66%), 3 (1.7 mg, 0.86%), and 5 (2.5 mg, 1.26%). Fraction EA6 (17 mg) was purified using silica gel CC and eluted with n-hexane–EtOAc (2
1, v/v) to afford 1 (4.3 mg), 2 (1.3 mg, 0.66%), 3 (1.7 mg, 0.86%), and 5 (2.5 mg, 1.26%). Fraction EA6 (17 mg) was purified using silica gel CC and eluted with n-hexane–EtOAc (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to afford 4 (3.5 mg, 1.77%).
1, v/v) to afford 4 (3.5 mg, 1.77%).
3.7. Structural elucidation of the compounds
ECD spectra were obtained on a JASCO J-815 circular dichroism spectrometer (JASCO, Easton, MD, USA). 1D and 2D NMR spectra were acquired on a Bruker AVANCE III 500 MHz spectrometer in acetone-d6. Chemical shifts in ppm are referenced to the residual solvent signal (acetone-d6: δH = 2.05, δC = 29.8 ppm). The HRESIMS spectra were recorded using a MicrOTOF-Q mass spectrometer on an LC-Agilent 1100 LC-MSD Trap spectrometer. Silica gel 60 (0.040–0.063 mm, Himedia) were used for column chromatography. Analytical TLC was carried out on aluminum plates precoated with silica gel 60 F254 or silica gel 60 RP-18 F254S (Merck), and eluted zones were visualized by spraying with 10% H2SO4 solution, followed by heating.3.8. Phytochemical analysis of the lichen D. pruinosum
The lichen material (95 mg) was ground and then extracted with acetone (10 × 10 mL each) at room temperature. The extract was analyzed by the TLC method, using the specific solvent systems for identification of lichen metabolites reported by Orange and co-workers (2001).30 The combined extracts were concentrated under reduced pressure to yield 18 mg of residue. This extract was subjected to silica gel CC and eluted with n-hexane–chloroform–acetone–EtOAc–water (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01, v/v/v/v/v) to yield 7 (3.3 mg, 18.33%) and 8 (1.7 mg, 9.44%).
0.01, v/v/v/v/v) to yield 7 (3.3 mg, 18.33%) and 8 (1.7 mg, 9.44%).
3.9. Antimicrobial activity assay
The antibacterial activity of the isolated compounds against multidrug-resistant S. aureus was investigated using the disk diffusion method. Compounds 1–4, 7, and 8 were dissolved in DMSO, and 6 mm paper discs were saturated with 100 μg of each compound. Subsequently, the paper discs were allowed to dry in a clean bench. Bacteria were cultured in Brain Heart Infusion broth at 35 °C overnight. The culture was diluted with sterile 0.9% NaCl to obtain a bacterial suspension with an OD600 ranging from 0.08 to 0.1. Then, 100 mL of this suspension was spread on a Mueller–Hinton agar plate, and the paper discs were placed on the agar surface, which had been seeded with bacteria. The plates were incubated at 35 °C for 16–18 h, and the antibacterial activity of each compound was assessed by measuring the diameters of the inhibition zones surrounding the discs.4. Conclusions
The present study focuses on the isolation of methicillin-resistant S. aureus obtained from street food samples collected in Ho Chi Minh City, Vietnam. The strain, known as SAX15, was subsequently utilized in antimicrobial assay. Three new sesquiterpenes, diorygmones C–E (2–4), along with three known compounds, diorygmone B (1), 3,5-dihydroxyl-4-methylbenzoic acid (5), and β-sitosterol (6), were isolated from the cultured mycobiont of D. pruinosum. In addition, two known compounds, stictic acid (7) and norstictic acid (8), were also isolated from the lichen D. pruinosum. Compounds 1–3 exhibited 5 mm zones of inhibition against S. aureus SAX15, while compound 6, known for its antimicrobial effectiveness, was also identified. These findings contribute to the understanding of the chemical constituents of D. pruinosum and their antimicrobial property.Conflicts of interest
No potential conflict of interest was reported by the authors.Acknowledgements
This study was supported by the Thammasat University Research Unit in Natural Products Chemistry and Bioactivities (chemicals and NMR recording). This research is funded by Ho Chi Minh city University of Education Foundation for Science and Technology under grant number CS2021.19.40.References
- P. Zimmet, K. Alberti and J. Shaw, Global and societal implications of the diabetes epidemic, Nature, 2001, 414(6865), 782–787, DOI:10.1038/414782a.
- J. Kadariya, T. C. Smith and D. Thapaliya, Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health, BioMed Res. Int., 2014 DOI:10.1155/2014/827965.
- S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler Jr, Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management, Clin. Microbiol. Rev., 2015, 28(3), 603–661 CrossRef CAS PubMed.
- Y. Guo, G. Song, M. Sun, J. Wang and Y. Wang, Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus, Front. Cell. Infect. Microbiol., 2020, 10, 107 CrossRef PubMed.
- D. Gutiérrez, S. Delgado, D. Vázquez-Sánchez, B. Martínez, M. L. Cabo, A. Rodríguez, J. J. Herrera and P. García, Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces, Appl. Environ. Microbiol., 2012, 78(24), 8547–8554 CrossRef PubMed.
- I. Ahmad, H. A. Malak and H. H. Abulreesh, Environmental antimicrobial resistance and its drivers: a potential threat to public health, J. Global Antimicrob. Resist., 2021, 27, 101–111 CrossRef PubMed.
- G. Shrestha and L. L. St. Clair, Lichens: a promising source of antibiotic and anticancer drugs, Phytochem. Rev., 2013, 12, 229–244 CrossRef CAS.
- M. Abdel-Hameed, R. L. Bertrand, M. D. Piercey-Normore and J. L. Sorensen, Identification of 6-hydroxymellein synthase and accessory genes in the lichen Cladonia uncialis, J. Nat. Prod., 2016, 79(6), 1645–1650 CrossRef CAS PubMed.
- M. S. Maier, M. L. Rosso, A. T. Fazio, M. T. Adler and M. D. Bertoni, Fernene triterpenoids from the lichen Pyxine berteriana, J. Nat. Prod., 2009, 72(10), 1902–1904 CrossRef CAS PubMed.
- A. Aptroot and L. B. Sparrius, Additions to the lichen flora of Vietnam, with an annotated checklist and bibliography, Bryologist, 2006, 358–371 CrossRef.
- S. Huneck, I. Yoshimura, S. Huneck and I. Yoshimura, Identification of Lichen Substances, Springer, 1996 Search PubMed.
- T.-H. Duong, H.-H. Nguyen, T.-T. Le, T.-N. Tran, J. Sichaem, T.-T. Nguyen, T.-P. Nguyen, D.-T. Mai, H.-H. Nguyen and H.-D. Le, Subnudatones A and B, new trans-decalin polyketides from the cultured lichen mycobionts of Pseudopyrenula subnudata, Fitoterapia, 2020, 142, 104512 CrossRef CAS PubMed.
- T.-H. Do, T. Aree, H.-H. Nguyen, H.-C. Nguyen, T.-P. G. Vo, N.-H. Nguyen and T.-H. Duong, Two new guaiane-sesquiterpenes from the cultured lichen mycobiont of Diorygma pruinosum, Phytochem. Lett., 2022, 52, 59–62 CrossRef CAS.
- S. Padhi, M. Masi, A. Cimmino, A. Tuzi, S. Jena, K. Tayung and A. Evidente, Funiculosone, a substituted dihydroxanthene-1, 9-dione with two of its analogues produced by an endolichenic fungus Talaromyces funiculosus and their antimicrobial activity, Phytochemistry, 2019, 157, 175–183 CrossRef CAS PubMed.
- H.-H. Nguyen, T. Aree, H. T. Nguyen, T.-M.-D. Tran, T.-P. Nguyen, T.-P. G. Vo, N.-H. Nguyen and T.-H. Duong, Diorygmones AB, two new guaiane-sesquiterpenes from the cultured lichen mycobiont of Diorygma sp, Nat. Prod. Res., 2023, 1–6 Search PubMed.
- T.-H. Do, T.-H. Duong, H. T. Nguyen, T.-H. Nguyen, J. Sichaem, C. H. Nguyen, H.-H. Nguyen and N. P. Long, Biological activities of lichen-derived monoaromatic compounds, Molecules, 2022, 27(9), 2871 CrossRef CAS PubMed.
- N. Kamal, C. Clements, A. I. Gray and R. Edrada-Ebel, Anti-infective activities of secondary metabolites from Vitex pinnata, J. Appl. Pharm. Sci., 2016, 6(1), 102–106 CrossRef CAS.
- J. Pan, J.-C. Su, Y.-H. Liu, B. Deng, Z.-F. Hu, J.-L. Wu, R.-F. Xia, C. Chen, Q. He and J.-C. Chen, Stelleranoids A–M., guaiane-type sesquiterpenoids based on [5, 7] bicyclic system from Stellera chamaejasme and their cytotoxic activity, Bioorg. Chem., 2021, 115, 105251 CrossRef CAS PubMed.
- F. Liu, C. Liu, W. Liu, Z. Ding, H. Ma, N. P. Seeram, L. Xu, Y. Mu, X. Huang and L. Li, New sesquiterpenoids from Eugenia jambolana seeds and their anti-microbial activities, J. Agric. Food Chem., 2017, 65(47), 10214–10222 CrossRef CAS PubMed.
- F. Lohézic-Le Dévéhat, S. Tomasi, J. A. Elix, A. Bernard, I. Rouaud, P. Uriac and J. Boustie, Stictic acid derivatives from the lichen Usnea articulata and their antioxidant activities, J. Nat. Prod., 2007, 70(7), 1218–1220 CrossRef PubMed.
- L. P. Luhata and T. Usuki, Antibacterial activity of β-sitosterol isolated from the leaves of Odontonema strictum (Acanthaceae), Bioorg. Med. Chem. Lett., 2021, 48, 128248 CrossRef PubMed.
- T. A. Taylor and C. G. Unakal, Staphylococcus aureus, StatPearls Publishing, 2022 Search PubMed.
- A. C. Smith and M. A. Hussey, “Gram Stain Protocols”, ASM.org, 2005 Search PubMed.
- M. P. MacWilliams, “Indole Test”, ASM.org, 2009 Search PubMed.
- S. McDevitt, “Methyl Red and Voges-Proskauer Test Protocols”, ASM.org, 2009 Search PubMed.
- Catalase Test Protocol Search PubMed.
- P. Shields and L. Cathcart, “Oxidase Test”, ASM.org, 2010 Search PubMed.
- K. Reiner, “Carbohydrate Fermentation Protocol”, ASM.org, 2012 Search PubMed.
- D. Sue Katz, “Coagulase Test Protocol,” Coagulase Test Protocol, 2010 Search PubMed.
- A. Orange, P. W. James and F. White, Microchemical Methods for the Identification of Lichens, British Lichen Society, 2001 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07112j |
| This journal is © The Royal Society of Chemistry 2024 |