Polyurethane foam acidolysis with carboxylic acids: acid structure dictates N-containing product distribution and kinetics†
Zach
Westman
a,
Manasa
Perikala
b,
Xinyi
Wu
a,
Kelsey
Richardson
 a,
Christopher S.
Letko
c,
Vojtech
Vlcek
bd,
Phillip
Christopher
a,
Christopher S.
Letko
c,
Vojtech
Vlcek
bd,
Phillip
Christopher
 *a and
Mahdi M.
Abu-Omar
*a and
Mahdi M.
Abu-Omar
 *ab
*ab
aDepartment of Chemical Engineering, University of California, Santa Barbara, Santa Barbara, CA 93106-5080, USA. E-mail: pchristopher@ucsb.edu; mabuomar@ucsb.edu
bDepartment of Chemistry and Biochemistry, University of California, Santa Barbara, Santa Barbara, CA 93106-9510, USA
cThe Dow Chemical Company, Midland, Michigan 48642, USA
dDepartment of Materials, University of California, Santa Barbara, Santa Barbara, CA 93016-5050, USA
First published on 9th November 2024
Abstract
Obtaining circularity will be essential in managing plastic waste and moving towards sustainable materials. Chemical recycling offers a pathway to obtain valuable molecules from plastic waste, closing the loop on what is currently a linear economy. Here, we report on the chemical recycling of polyurethane foam (PUF) via acidolysis with dicarboxylic acids (DCAs) to release value-added molecules. While previous work has explored recovery of the recycled polyol (repolyol), we focus in this report on elucidating the product distribution and kinetics of the nitrogen-containing products from the acidolysis reaction. Using Nuclear Magnetic Resonance (NMR) spectroscopy and Ultra-High Pressure Liquid Chromatography – Mass Spectroscopy (UPLC-MS), we demonstrate how acid loading and structure influence product distribution of acidolysis. The use of excess acid can eliminate oligomeric content and aromatic amines from the product mixture. With DCAs composed of 2 or 3 carbons between the carboxylic acid groups, we observe the formation of imide products during acidolysis, which has only very recently been reported in the literature. Furthermore, the kinetics of imide formation were investigated and modeled for glutaric acid (GA) and succinic acid (SA), which form 6- and 5-membered cyclic imides, respectively. Di-imide formation with SA proceeds without accumulation of intermediates and is an order of magnitude faster than imide formation with GA, for which an amide-imide intermediate is detected and the rate of reaction is sensitive to steric hindrance. This report offers fundamental insights into the N-containing products formed during acidolysis, which will aid scale-up of closed-loop chemical recycling processes.
Introduction
Polyurethanes (PUs) are versatile condensation polymers produced from the reaction of isocyanates and polyols. PUs are the 6th most produced polymer worldwide and the basis of many important consumer products, including mattresses, automotive components, and insulation, among others.1,2 Global annual PU production reached 25.8 million metric tons (Mt) in 2022 and is expected to increase to 31.2 Mt by 2030.3 The rapid increase in plastic production and consumption is outpacing waste management strategies; only 10% of plastic waste is recycled, and an estimated 4900 Mt – a staggering 60% of all plastic ever produced – has been discarded and is accumulating in landfills and the environment.4–6 50% of PU waste is currently landfilled.7As the world transitions to more sustainable practices, it is critical to develop circularity in plastic waste management. Landfilling and incineration result in the loss of the material and any value associated with it. Recycling offers a route to recover value from PU waste but is not yet sufficiently developed. Products of mechanical PU recycling possess inferior physical properties and reduced recyclability compared to the virgin plastic.7–10 Thus, mechanical recycling alone does not achieve full circularity, as resins cannot be used past a certain number of cycles.4,11 Reprocessing of PU thermoset materials using carbamate exchange to partially decompose and re-crosslink the polymer network has received recent interest in the literature as an alternative to mechanical recycling.12–17 The addition of carbamate exchange catalysts such as dibutyl tin dilaurate (DBTDL) during melt-state processing such as compression molding and twin-screw extrusion can enable the reconstruction of covalent adaptable networks (CANs) in PU thermosets to form new materials with robust mechanical properties, circumventing some of the limitations of mechanical recycling. However, carbamate exchange reprocessing is still underdeveloped and has limitations at present, including the necessity of a catalyst and difficulty processing PUFs; notably, foam-to-foam recycling has only very recently been demonstrated on a laboratory scale.13
Chemical recycling is an emerging strategy to manage plastic waste. In contrast to mechanical recycling, chemical recycling uses catalysts and/or reagents (i.e. chemolysis) to cleave polymer bonds, resulting in valuable monomers that can subsequently be used to make new plastic materials. Unlike polyolefins, PU materials contain reactive C–O and C–N bonds that allow for facile polymer deconstruction with reagents such as glycols, acids, amines, and water without an additional depolymerization catalyst.18 Chemical recycling via depolymerization with glycols (glycolysis) and carboxylic acids (acidolysis) have both been explored to a limited extent on an industrial scale as a method to recover polyol from polyurethane foams (PUFs) for use in new PU formulations.19–22 However, the design space for both reactions remains underdeveloped, particularly related to the influence of chemical reagent structure on acidolysis rates and product scope, and more work must be done to advance towards a circular PU economy.
Acidolysis is the reaction of PUF with an acid, typically a dicarboxylic acid (DCA), to produce polyol, CO2, H2O, and nitrogen-containing byproducts that result from reaction of the isocyanate segment of PUF with the depolymerization reagent. Because the polyol is the major mass component of most PUF formulations recovery of polyol has been the focus of most acidolysis studies. For example, we recently found that the proximity of a DCA's two carboxyl groups influences the rate of polyol release during acidolysis of flexible PUF, providing insights into DCA selection for optimizing polyol release kinetics.23 However, isocyanates represent approximately 30–40% of the mass of a typical flexible PUF formulation, and thus it is important to consider utilizing fractions originally coming from isocyanate species. Few studies have focused on recovering and valorizing isocyanates and isocyanate derivatives from PU chemical recycling schemes.
Previous literature analyzing N-containing products from PUF acidolysis has predominantly reported amide products,24–26 although some studies have reported aromatic amine formation as well.27 We have previously demonstrated that amides obtained from PUF acidolysis with maleic acid (MA) can be separated from the reaction mixture via liquid–liquid extraction and subsequently hydrolyzed to generate toluene diamine (TDA), a precursor to toluene diisocyanate (TDI).26 A very recent report described the recovery of anilines from a combined acidolysis-hydrolysis process.28 Isocyanate recovery from non-acidolysis chemical recycling schemes is gaining traction as well. For example, a β-chlorocatecholborane-mediated depolymerization was recently used to directly regenerate isocyanates (mainly methylene diphenyl diisocyanate (MDI)) from PU thermoset and thermoplastic materials.29 In addition, tert-amyl alcohol-mediated solvolysis was shown to produce polyol and aromatic diamines from a variety of PU materials, including flexible and rigid foams.30 Nevertheless, the N-containing products from acidolysis have generally received little attention in literature. As a result, the influence of acid chemical structure and reaction conditions on the formation kinetics and product distribution of N-containing products is not well understood. Effective separations and utilization of N-containing byproducts from recycled polyol generated during acidolysis is one of the main limitations preventing commercialization of acidolysis processes.18 Controlling the product distribution from PUF acidolysis may aid the separation and eventual utilization of N-containing products.
In this study, we examine the influence of dicarboxylic acid structure on the product distribution and kinetics of N-containing PUF acidolysis products. Prior literature on PUF acidolysis used acid loadings either sub-stoichiometric or near stoichiometric with urethane bond content. By increasing acid loading, we demonstrate that the N-containing product distribution can be simplified; with at least 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DCA
1 DCA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PUF molar loadings, aromatic amines and oligomeric content can be eliminated from the product stream. Notably, in addition to the amine and amide products previously reported in literature, we observe imide products from PUF acidolysis resulting from ring-closing reactions of amides. DCA carboxyl proximity is shown to be relevant to imide formation; performing acidolysis with diacids that have carboxyl groups separated by fewer carbons result in imide products, while diacids with more carbons separating the acid carboxyl groups, as well as monoacids (benzoic acid), only form amide products. Finally, we probe the kinetics of imide formation during PUF acidolysis with glutaric acid (GA) and succinic acid (SA) using Nuclear Magnetic Resonance (NMR) spectroscopy and provide mechanistic insights into the formation of imide products with 5- and 6-membered rings. Thus, this work along with our previous study demonstrates that DCA carboxyl-to-carboxyl distance influences polyol and N-containing product release kinetics and N-containing product distribution during PUF acidolysis.23 This work provides a fundamental framework for understanding the mechanism and kinetics of N-containing product formation from PUF acidolysis, which may facilitate the design of PU chemical recycling schemes for a circular plastic economy.
PUF molar loadings, aromatic amines and oligomeric content can be eliminated from the product stream. Notably, in addition to the amine and amide products previously reported in literature, we observe imide products from PUF acidolysis resulting from ring-closing reactions of amides. DCA carboxyl proximity is shown to be relevant to imide formation; performing acidolysis with diacids that have carboxyl groups separated by fewer carbons result in imide products, while diacids with more carbons separating the acid carboxyl groups, as well as monoacids (benzoic acid), only form amide products. Finally, we probe the kinetics of imide formation during PUF acidolysis with glutaric acid (GA) and succinic acid (SA) using Nuclear Magnetic Resonance (NMR) spectroscopy and provide mechanistic insights into the formation of imide products with 5- and 6-membered rings. Thus, this work along with our previous study demonstrates that DCA carboxyl-to-carboxyl distance influences polyol and N-containing product release kinetics and N-containing product distribution during PUF acidolysis.23 This work provides a fundamental framework for understanding the mechanism and kinetics of N-containing product formation from PUF acidolysis, which may facilitate the design of PU chemical recycling schemes for a circular plastic economy.
Experimental section
Materials
The model PUF (M-PUF) sample was synthesized at The Dow Chemical Company (Dow) following the formulation listed in Table S1† and provided as a white-colored slab. More detailed preparation and characterization of the PUF materials used is provided in our previous publication.26 Succinic acid (>99%), phthalic acid (PA) (>99.5%), glutaric acid (99%), adipic acid (AA) (99%), pimelic acid (PiA) (98%), and benzoic acid (BA) (>99%) were purchased from Sigma-Aldrich. Ethyl acetate (EtOAc, ACS reagent >99.5%) was purchased from Sigma-Aldrich. Hexadeuterated dimethyl sulfoxide (DMSO-d6, 99.9%) was purchased from Cambridge Isotope Laboratories Inc. Industrial grade liquid nitrogen (N2) was purchased from Airgas. Silicone oil for oil baths was purchased from Sigma-Aldrich. All purchased chemicals were used as received.Grinding pre-treatment
Before the acidolysis reaction, both model PUF and EOL-PUF chunks were ground into smaller particles. The chunks of flexible foam were first flash frozen in liquid nitrogen to increase their brittleness and transferred subsequently into a grinder equipped with cross blades. After grinding, the particle size of model PUF was determined by SEM to be between 500–2000 µm, while the EOL-PUF particles were around 155–750 µm.Characterization of M-PUF
We have previously performed characterization of the starting M-PUF material via TGA, SEM, and ATR-FTIR.26 It is worth noting that based on the foam formulation, urethane bonds make up a relatively small fraction of the total composition of the foam (33% molar basis). Most of the bonds (∼67% molar basis) are assumed to be urea bonds formed from the reaction of isocyanate and water. The TGA of the foam was consistent with these ratios, with approximately 71% of the mass of the M-PUF being attributed to release of polyol from decomposition of urethane bonds, followed by decomposition of the polyether ether linkages and volatilization of the resulting decomposition products.Acidolysis experiments
PUF acidolysis was carried out using round-bottom flasks in an oil bath. Briefly, 0.5 g shredded M-PUF and a specified amount of acid (1.5 g unless otherwise stated) were added to a 25 mL round-bottom flask with a stir bar. The flask was shaken using a vortex mixer to ensure reactants were initially well-mixed, then attached to a short-path distillation head to prevent most of the water, acid, and other volatile species from escaping the system during reaction. The gas outlet was connected to Tygon® tubing; the outlet of the tubing was placed in a beaker of water such that atmospheric pressure inside the system could be maintained without introducing air to the system. Before reaction, the entire system was purged with 100 cm3 min−1 N2 for at least 5 minutes to prevent oxidative side reactions. After purging, the flask was lowered into the oil bath and heated at the reaction temperature for the specified reaction time. The magnetic stir bar was set to 350 rpm. After reaction, the flask was removed from the oil bath and quenched in ice water for ca. 5 minutes. Unless otherwise specified, the product mixture (which in most cases was a solid or semisolid due to the large excess of acid) was extracted directly from the round-bottom flask using a spatula and used for further analysis.Isolation of GA N-containing acidolysis products
For detailed characterization of peaks assigned to amides in 1H NMR, N-containing products from a GA acidolysis reaction at 185 °C were isolated for 2D NMR analysis. A reaction time of 4 hours was chosen, as 1H NMR of GA acidolysis products showed the highest concentration of amide-imide intermediates at this time (Fig. S23†). A reaction was run as described above, with the following changes to the procedure: 3 g M-PUF, 3 g GA, 50 mL round-bottom flask. After thermally quenching in an ice water bath, 40 mL ethyl acetate (EtOAc) was added to the reaction flask. The flask was vigorously mixed via vortex mixer and manual shaking for ca. 5 minutes to solubilize excess acid and polyol. The contents of the flask were transferred to a 50 mL centrifuge tube and centrifuged for 10 minutes at 5000 rpm. After centrifugation, the top (EtOAc) phase was extracted via pipette and discarded; the remaining dark brown solid was dried overnight under vacuum. The dry solid was then characterized via NMR (see below).Nuclear magnetic resonance (NMR) spectroscopy analysis
Samples for NMR were prepared as follows: 150 mg of sample was extracted from the round-bottom flask after the reaction was quenched and placed in a 3-dram vial. The sample was then dissolved in 600 µL DMSO-d6; in kinetic experiments, 25.0 mg of maleic acid (MA) was added as an internal standard. All NMR samples were run on a Bruker Avance NEO 500 MHz equipped with a 5 mm CryoProbe Prodigy BBO probe with Z-axis PFG.Ultra-high-pressure liquid chromatography – mass spectrometry (UPLC-MS) analysis
UPLC-MS was carried out on a Waters Acquity H-class Ultra High-Pressure Liquid Chromatography (UPLC) system coupled with a Waters Xevo G2-XS Time-of-Flight Mass Spectrometer. The system employs a Waters Acquity H-class Quaternary Separations Module. Briefly, 1 mg sample was dissolved in 1 mL methanol (MeOH) and placed in the sampler. The injection volume was 0.5 µL, with a solvent (MeOH) flowrate of 0.5 mL min−1. All samples were run on bypass.Kinetic modeling
Kinetic modeling of amide & imide formation was performed using NMR integrals obtained from analysis in MestReNova and fit using MATLAB R2024a. Each 1H NMR spectrum was baselined using a polynomial fit and phase corrected. NMR integrals were found with respect to the internal standard (MA) using MestReNova's line fitting tool. For PUF acidolysis kinetics using GA, the amide region (δ1H = 10.1–9.1 ppm) was considered; for PUF acidolysis kinetics using SA, it was more straightforward to analyze the methyl region (δ1H = 2.5–1.5 ppm). Integrals were converted to moles using the internal standard. Four species were identified for kinetic analysis for PUF acidolysis with GA (2,4- and 2,6-diamide, 2-amide-4-imide, and 2-amide-6-imide) and two were identified for PUF acidolysis with SA (2,4- and 2,6-diimide). Kinetic models were fit using the lsqnonlin function in MATLAB (see ESI† for details on kinetic models). Arrhenius plot linear regressions were calculated using Origin 2024.Results & discussion
To explore the influence of acid loading, acid structure, and temperature on the N-containing product distribution, PU acidolysis studies were carried out using a model flexible polyurethane foam (M-PUF). The M-PUF was synthesized using TDI, which contains an isomeric mixture of 2,4-TDI and 2,6-TDI in a molar ratio of approximately 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively, and VORANOL™ 8136, a three-arm, glycerine-initiated polyether polyol with hydroxyl-equivalent molecular weight of 1030 g mol−1 (see Experimental section, M-PUF synthesis & characterization for details). Experiments were conducted in a 25 mL round-bottom flask at temperatures between 165–195 °C using pre-shredded M-PUF to remove the influence of mass and heat transfer limitations (Experimental section, acidolysis experiments). A similar setup has been demonstrated to be effective for measuring the kinetics of polyol release from M-PUF acidolysis and details of the apparatus are provided in the previous report.23
1, respectively, and VORANOL™ 8136, a three-arm, glycerine-initiated polyether polyol with hydroxyl-equivalent molecular weight of 1030 g mol−1 (see Experimental section, M-PUF synthesis & characterization for details). Experiments were conducted in a 25 mL round-bottom flask at temperatures between 165–195 °C using pre-shredded M-PUF to remove the influence of mass and heat transfer limitations (Experimental section, acidolysis experiments). A similar setup has been demonstrated to be effective for measuring the kinetics of polyol release from M-PUF acidolysis and details of the apparatus are provided in the previous report.23
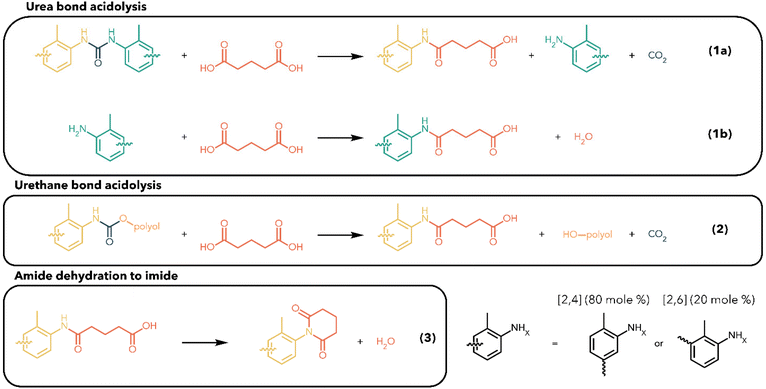
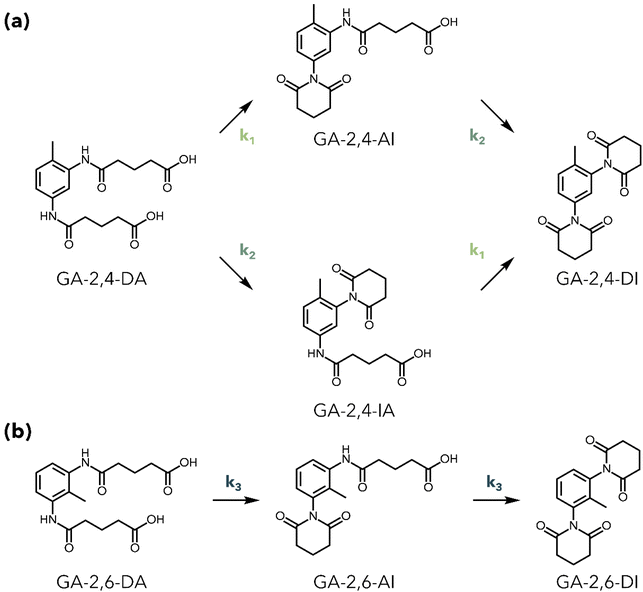
The stoichiometry of M-PUF acidolysis is summarized in Scheme 1, using GA as an example. Both urethane and urea bonds undergo acidolysis. Reaction of the urea bond with GA initially forms an amide, an amine, and CO2 (1a). The amine can subsequently react with GA to produce a second (identical) amide, as well as H2O (1b). Similarly, a urethane bond can react with GA to produce an amide product and a polyol (2). Macroscopic kinetics demonstrate similar rates of acidolysis of urethane and urea bonds by DCAs, although the microkinetics of the reaction may be influenced by the complex physical structure of PUFs. Amides from the decomposition of either bond may undergo a subsequent ring-closing reaction to form an imide and H2O, a step which will be discussed in detail below (3). The polyol –OH groups have been shown to undergo esterification with excess acid to produce a polyol ester (Fig. S1†).25,27,31 Assuming esterification and reaction of amines with DCAs occur on a similar timescale to urethane/urea bond cleavage, this stoichiometry suggests that a minimum 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of DCA to isocyanate –NCO groups in the foam formulation (i.e., to both urethane and urea bonds) is required to guarantee complete decomposition of PUF materials.
1 molar ratio of DCA to isocyanate –NCO groups in the foam formulation (i.e., to both urethane and urea bonds) is required to guarantee complete decomposition of PUF materials.
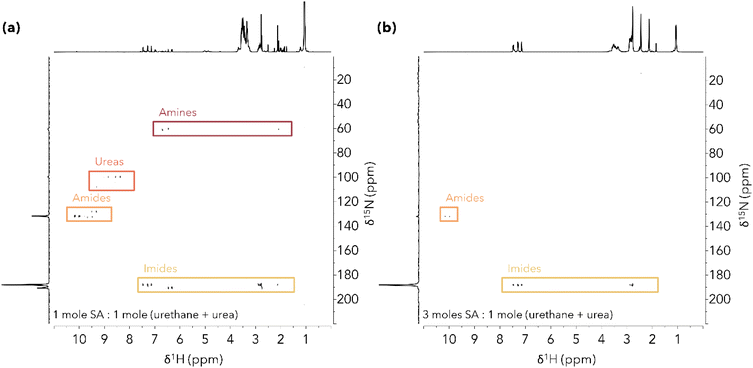
Concentration dependence of N-containing product distribution
To investigate the effect of acid loading on nitrogen-containing product formation, M-PUF acidolysis with SA was run at low acid loadings. Fig. 1 shows 1H–15N Heteronuclear Multiple Bond Correlation (HMBC) NMR spectra of the products from acidolysis of 500 mg M-PUF with 125 mg SA (appx. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 moles SA
1 moles SA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) moles –NCO) versus 375 mg SA (appx. 3
moles –NCO) versus 375 mg SA (appx. 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 moles SA
1 moles SA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) moles –NCO) at 175 °C for 1 hour. At 1
moles –NCO) at 175 °C for 1 hour. At 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar SA loading, four distinct nitrogen environments were observed, Fig. 1(a) (see ESI† for detailed assignments). Signals at δ15N ≈ 60 ppm are attributed to free amines in the reaction mixture.32 The cross signals at δ1H = 6.70, 6.47 & 2.08 ppm are consistent with the aromatic –CH and methyl –CH3 protons of 2,4-TDA, respectively. The direct –NH2 signal of 2,4-TDA was not observed in the HMBC NMR spectrum, possibly due to rapid proton exchange with water generated during the reaction. Signals at δ15N ≈ 100–110 ppm and δ1H ≈ 8.0–10.0 ppm are attributed to –NH groups of oligomeric ureas, indicating that the reaction was incomplete at stoichiometric acid loadings. Signals at δ15N ≈ 125–135 ppm and δ1H ≈ 9.0–10.5 ppm were assigned to –NH amide groups. Finally, signals at δ15N ≈ 187–191 ppm were assigned to imide groups; this assignment will be discussed in detail below. Because imides contain no direct N–H bonds, only the protons of CHx groups 2–3 bonds away from imide N atoms are visible in the HMBC NMR. Aromatic –CH imide groups were observed at δ1H ≈ 6.0–8.0 ppm, aliphatic –CH2 groups were visible at δ1H ≈ 2.6–3.0 ppm, and the peak at δ1H = 2.13 ppm was assigned to the methyl –CH3 of the 2,4-diimide (SA-2,4-DI).
1 molar SA loading, four distinct nitrogen environments were observed, Fig. 1(a) (see ESI† for detailed assignments). Signals at δ15N ≈ 60 ppm are attributed to free amines in the reaction mixture.32 The cross signals at δ1H = 6.70, 6.47 & 2.08 ppm are consistent with the aromatic –CH and methyl –CH3 protons of 2,4-TDA, respectively. The direct –NH2 signal of 2,4-TDA was not observed in the HMBC NMR spectrum, possibly due to rapid proton exchange with water generated during the reaction. Signals at δ15N ≈ 100–110 ppm and δ1H ≈ 8.0–10.0 ppm are attributed to –NH groups of oligomeric ureas, indicating that the reaction was incomplete at stoichiometric acid loadings. Signals at δ15N ≈ 125–135 ppm and δ1H ≈ 9.0–10.5 ppm were assigned to –NH amide groups. Finally, signals at δ15N ≈ 187–191 ppm were assigned to imide groups; this assignment will be discussed in detail below. Because imides contain no direct N–H bonds, only the protons of CHx groups 2–3 bonds away from imide N atoms are visible in the HMBC NMR. Aromatic –CH imide groups were observed at δ1H ≈ 6.0–8.0 ppm, aliphatic –CH2 groups were visible at δ1H ≈ 2.6–3.0 ppm, and the peak at δ1H = 2.13 ppm was assigned to the methyl –CH3 of the 2,4-diimide (SA-2,4-DI).
In contrast, the 1H–15N HMBC NMR spectrum of the reaction run with excess DCA (∼3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 moles SA
1 moles SA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) moles –NCO) showed aromatic and aliphatic peaks associated with only amide and imide products, Fig. 1(b). No urea content was observed, suggesting that the M-PUF was fully decomposed after 1 hour reaction time. Polyol release from PUF acidolysis with SA at 195 °C has been observed via monitoring of CO2 evolution to be complete in ca. 5 minutes, albeit with a larger excess of SA (∼12
moles –NCO) showed aromatic and aliphatic peaks associated with only amide and imide products, Fig. 1(b). No urea content was observed, suggesting that the M-PUF was fully decomposed after 1 hour reaction time. Polyol release from PUF acidolysis with SA at 195 °C has been observed via monitoring of CO2 evolution to be complete in ca. 5 minutes, albeit with a larger excess of SA (∼12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 moles SA
1 moles SA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) moles –NCO).23 Signals corresponding to aromatic amines were also not detected at this acid loading, consistent with the reaction of all amine intermediates with excess SA.
moles –NCO).23 Signals corresponding to aromatic amines were also not detected at this acid loading, consistent with the reaction of all amine intermediates with excess SA.
Overall, decreasing the complexity of the product distribution from PUF chemical recycling schemes is desirable from a process standpoint, as it can make post-reaction separations and treatments easier. In particular, the formation of aromatic amines during PUF acidolysis is undesirable, as there are limits to the level allowed in the recycled polyol. The results in Fig. 1 demonstrate that the use of excess acid is crucial to ensure complete depolymerization during PU acidolysis reactions. It is important to stress that both urethane and urea bonds must be taken into account when calculating acid loading, as both bonds react at similar apparent rates during PUF acidolysis with DCAs,23 and the secondary reactions of amines and polyols with DCAs occur at similar rates to the initial urethane/urea cleavage. Previous studies have almost exclusively used sub-stoichiometric acid loadings.24,25,27,33,34 Using the M-PUF foam formulation studied here as a basis, the minimum required mass ratios to ensure complete PUF decomposition for various DCAs are: 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.1 (SA
2.1 (SA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M-PUF), 1.0
M-PUF), 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.7 (AA
1.7 (AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M-PUF), 1.0
M-PUF), 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 (PA
1.5 (PA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M-PUF). Note that these calculations assume that only one carboxyl group per acid molecule reacts; the validity of this assumption will be discussed in more detail below. As a result, we hypothesize that the presence of aromatic amines, oligomeric ureas, and TDA/amide-terminated polyols reported in previous studies are a result of sub-stoichiometric acid loadings and can be mitigated by the use of excess acid.
M-PUF). Note that these calculations assume that only one carboxyl group per acid molecule reacts; the validity of this assumption will be discussed in more detail below. As a result, we hypothesize that the presence of aromatic amines, oligomeric ureas, and TDA/amide-terminated polyols reported in previous studies are a result of sub-stoichiometric acid loadings and can be mitigated by the use of excess acid.
Imide formation during acidolysis
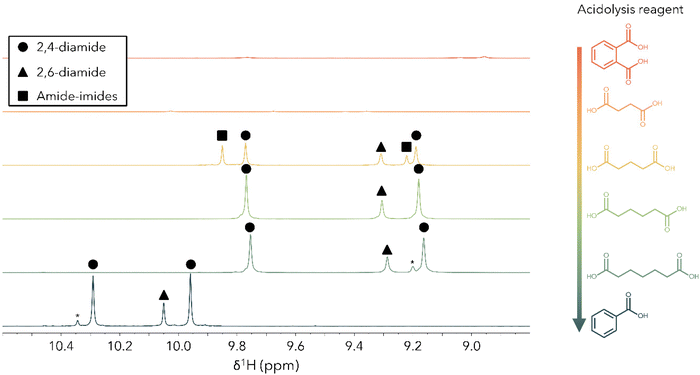
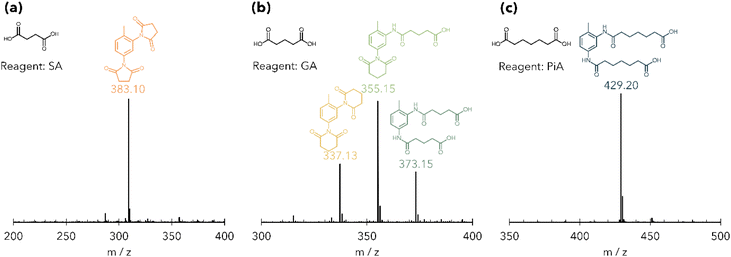
With an understanding of how acid loading influences the product distribution, we turned next to the case where acid is in large excess. To study how the product distribution changes as a function of acid structure, we characterized products of PUF acidolysis reactions using five DCAs and benzoic acid (BA), a monocarboxylic acid (Table S1†) with both NMR and UPLC-MS. Reactions were run for 2 hours at 195 °C using 0.5 g M-PUF and 1.5 g DCA (BA acidolysis was run for 24 hours due to the significantly slower rate of PUF decomposition observed with monocarboxylic acids). Fig. 2 shows the 1H NMR spectra of the resulting M-PUF acidolysis products for all the acids tested. Interestingly, the N-containing product distribution varied with acid structure. When using SA and PA as reagents no signals appear in the amide region (δ1H = 9–11 ppm), indicating transformation of all amide species to imides, while for acidolysis using AA, PiA, and BA peaks corresponding to their respective 2,4- and 2,6-diamides were present. Unlike the other acids, when using GA as an acidolysis reagent five peaks appeared in the amide region. Peaks at δ1H = 9.19 ppm and 9.76 ppm were assigned to GA-2,4-diamide (GA-2,4-DA); the peak at δ1H = 9.30 ppm was assigned to GA-2,6-diamide (GA-2,6-DA). Based on the imide formation observed during SA acidolysis, the other two peaks (δ1H = 9.85 and 9.22 ppm) were assumed to be mixed amide-imide intermediates. To substantiate these assignments, the N-containing products from PUF acidolysis with GA were isolated and characterized via NMR (see Experimental section for details). 1H–13C HMBC NMR was used to assign the peak at δ1H = 9.85 ppm to GA-2-amide-4-imide (GA-2,4-AI) and the peak at δ1H = 9.22 ppm to GA-2-amide-6-imide (GA-2,6-AI) (Fig. S2†). The 2D NMR was also used to distinguish between the –NH signals of GA-2,4-DA. The peak at δ1H = 9.19 ppm was assigned to the para-amide, while the peak at δ1H = 9.76 ppm was assigned to the ortho-amide (see ESI† for explanation of peak assignments). The aromatic and aliphatic regions of the 1H NMR were consistent with the product assignments for all reactions (Fig. S3–S7†).To confirm assignments from 1H NMR, UPLC-MS was also used to characterize acidolysis products. Fig. 3 shows representative UPLC-MS spectra of M-PUF acidolysis reactions that produce only imides (SA reagent), only amides (PiA reagent), and a mixture of amide and imide species (GA reagent). The UPLC-MS spectra of products from the reactions using SA and PA showed species with masses of 309.08 and 383.10 g mol−1, respectively, consistent with the formation of diimide species (Fig. 3(a) and Fig. S8, S9†). The UPLC-MS spectra of products from acidolysis reactions using GA had signals corresponding to not only the diamide species (373.15 g mol−1), but also amide-imide (355.15 g mol−1) and diimide (337.13 g mol−1) species (Fig. 3(b) and Fig. S10†). Finally, the UPLC-MS and NMR spectra of the products of M-PUF acidolysis with PiA, AA, and BA showed only the diamide product at 429.21 g mol−1, 401.17 g mol−1, and 331.14 g mol−1, respectively (Fig. 3(c) and Fig. S11–S13†). It is worth noting that ‘acid-bridged’ amide/imide dimers were also observed in the UPLC-MS of reactions with GA, AA, and PiA (Fig. S10–S12†). These species result from the reaction of both carboxyl groups on a single DCA molecule with urethane/urea/amine moieties. These species are low in concentration relative to monomeric amides & imides, which is expected given the large excess of acid used in these reactions.
Although 1H NMR and UPLC-MS suggested that amides and imides were the predominant products, neither method can directly interrogate the chemical environment of the imide N. To validate assignment of imide species, 1H–15N HMBC NMR was performed on the products of M-PUF acidolysis reactions (Fig. S14–S19†). The 1H–15N HMBC NMR was consistent with the assignments discussed above for products of acidolysis with all acids; for acidolysis reactions using PA and SA, cross signals were observed at δ15N > 160 ppm in the aromatic (δ1H = 6.5–8 ppm) and aliphatic (δ1H = 2–3 ppm) regions of the 2D NMR consistent with imide species, while spectra of acidolysis reaction products using AA, PiA, and BA reagents had cross signals at δ15N = 120–140 ppm in the amide (δ1H = 9–11 ppm), aromatic, and aliphatic regions of the 2D NMR consistent with amide species. The 1H–15N HMBC NMR of the acidolysis reaction products using GA reagents had signals corresponding to both amide and imide species.
Taken together, the UPLC-MS, 1H NMR, and 1H–15N HMBC NMR all suggest a trend between DCA carboxyl proximity and their proclivity to form imide species during M-PUF acidolysis. Performing acidolysis using DCAs with 2 or 3 carbons between their carboxyl groups (SA, PA, & GA) yielded imide products with 5- or 6-membered rings, respectively. When using SA & PA as acidolysis reagents, imide products rapidly formed; by 2 hours at 195 °C, no amide species were detected. On the other hand, M-PUF acidolysis reactions with GA, which form imides with 6-membered rings, had a significant quantity of diamides and amide-imides in the reaction mixture after 2 hours at 195 °C, suggesting that GA imide formation is much slower than SA or PA imide formation. Acidolysis using AA and PiA reagents, which would form imides with 7- and 8-membered rings, respectively, yields only amide products. It is well-established that 7+ membered ring formation reactions can be limited kinetically, thermodynamically, or both.35 BA has no second carboxyl group that could participate in a ring-closing reaction and it is thus not surprising that no imides were formed during M-PUF acidolysis with BA.
Imide formation during PUF acidolysis has only very recently been demonstrated in the literature.28 Our findings agree with the recent report of formation of succinimide and phthalimide products during PUF acidolysis with SA and PA reagents. The formation of imides should be carefully considered when designing acidolysis processes, as their presence may complicate – or simplify – the valorization of products from PU acidolysis. As mentioned above, hydrolysis to amines has been the only method explored for recovery of isocyanate-based products from PUF acidolysis. Interestingly, higher yields from both acid and alkaline hydrolysis of imides to TDA as compared to hydrolysis of AA-based amides was recently reported. Furthermore, the facile separation of succinimide products from the repolyol as compared to phthalimides or the AA-based diamides was observed. While we did not extensively study separation of imides and repolyol here, we found that isolation of GA-amides/imides from repolyol was also simple, as the N-containing products were solids with low solubility in ethyl acetate and were therefore easily removed from the reaction mixture (see Experimental section). On the other hand, some imide moieties offer direct potential for reuse without requiring an intermediate hydrolysis treatment. For example, both PU and non-PU self-healing thermoset materials have been directly synthesized via the reaction of bismaleimides with amine and furan groups.36–38 Imides formed from the reaction of the M-PUF used in this study with MA would form 2,4- and 2,6-bismaleimidotoluene, which could be directly reused in a similar fashion to these studies. Thus, the choice of acid reagent(s) could facilitate further novel uses of imide products from PUF acidolysis processes.
It is also worth noting that acids that form imide products also have faster kinetics for polyol release and therefore constitute more efficient acidolysis reagents.23 Taken together, these results suggest that reaction pathways that facilitate imide formation are desirable over those that form amides. However, the observation of diamide and amide-imide intermediates after 2 hours at 195 °C during M-PUF acidolysis with GA suggests that imide formation occurs on longer timescales than polyol release, which occurs in minutes rather than hours. If elucidated, the difference in timescales between the completion of polyol release and the completion of imide formation offers the potential to tune the N-containing product distribution without affecting recovery of the polyol product. More detailed kinetic studies of PUF acidolysis with DCAs that result in imide formation to better understand the complex evolution of various products are therefore merited.
Kinetics of imide formation
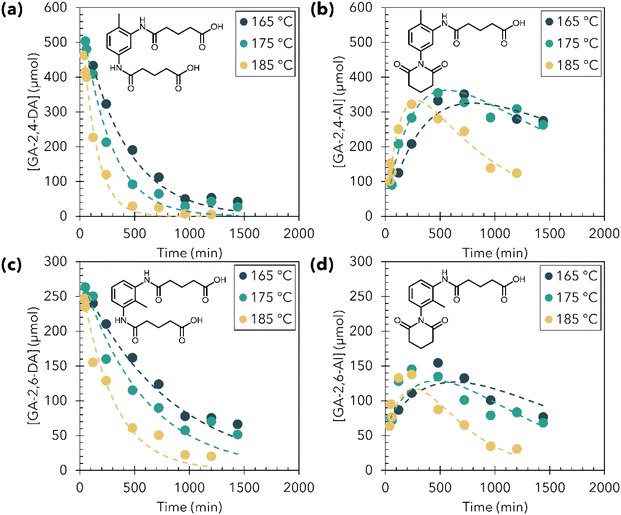
The formation of amide-imides mixed intermediates during M-PUF acidolysis with GA reagents was particularly interesting, as the long timescale of reaction revealed mechanistic details that were not observable during acidolysis using SA and PA reagents. Notably, the GA-2-imide-4-amide (GA-2,4-IA) intermediate was not observed in the 1H NMR of the product mixture at any reaction time tested. This suggested that the reaction of amide groups ortho to the methyl group of TDI-based molecules is sterically hindered, and thus the reaction of amide groups in the 4-position (para to the methyl) proceeds significantly faster. This is further supported by long M-PUF acidolysis experiments using GA; 1H NMR of a reaction run for 120 hours at 195 °C showed peaks corresponding to GA-2,6-DA, GA-2,4-AI, and GA-2,6-AI, while the peaks associated with GA-2,4-DA completely disappeared (Fig. S20†).To evaluate this further and to understand the timescales of imide formation relative to CO2/polyol release, we investigated the kinetics of imide formation during M-PUF acidolysis with GA between 165–195 °C. For GA reagents, M-PUF acidolysis reactions were run at time intervals between 30 minutes and 24 hours. After each reaction, the products were quenched over ice, then extracted with no other post-reaction treatment and analyzed by 1H NMR (see Experimental section for details). The aromatic and aliphatic regions of the NMR were difficult to extract kinetic information from, due to the overlapping signals of many products, although methyl signals from GA-2,4-DI and GA-2,6-DI (δ1H = 1.99 and 1.65 ppm, respectively) were qualitatively observed to increase throughout the reaction, indicating the formation of GA diimide species. However, the amide region proved reasonably simple to deconvolute and was used to perform kinetic studies (Fig. S21–S23†). The concentrations of the four species observable in the amide region of the 1H NMR (GA-2,4-DA, GA-2,6-DA, GA-2,4-AI, GA-2,6-AI) were calculated for each timepoint at each temperature.
Scheme 2 depicts the proposed kinetic model used to fit the time dependent product concentrations from M-PUF acidolysis by GA. It is clear from the accumulation of GA-2,4-AI during the reaction that k1 is faster than k2. Because GA-2,4-IA is not observed during the reaction, a full kinetic model (with six rate constants) will not yield meaningful parameters for reactions involving this species. We overcame this shortcoming by assuming that the reaction rate of an amide on the ring is independent of whether the other N-containing substituent on the ring is an amide or an imide, simplifying the kinetic model to three rate constants. In this model, the predominant rate-controlling factor is assumed to be the steric environment rather than electronic effects from the other (non-reacting) N-containing group on the ring, resulting in rate constants describing sterically similar imide formation reactions. The validity of this assumption is vetted next. An alternative model in which GA-2,4-IA is excluded entirely was also tested (Scheme S1, Fig. S24 and Table S2†).
Fig. 4 shows the concentration of the four measurable amide species (dots), as well as the kinetic model fit (dashed lines) for each species from M-PUF acidolysis reactions by GA run at 165, 175 and 185 °C. We chose to stay well below 200 °C in these reactions as thermal degradation of M-PUF is kinetically accessible above these temperatures on the reaction time scales used here and we wanted to focus specifically on acid mediated reactions. The kinetic model generally provided an excellent fit to the data, in support of the proposed mechanism. The concentration of GA-2,6-AI was slightly underpredicted by the model, but using an additional rate constant for the reaction of GA-2,6-AI to GA-2,6-DI did not provide a significantly better fit. The reaction rate constant k1, representative of the reaction of the amide in the 4-position, was ca. 5× larger than k2 and ca. 2× larger than k3, which represent reactions of amides in the 2-position, consistent with this reaction being less sterically hindered by the methyl group. The kinetic model excluding reactions involving GA-2,4-IA also provided a reasonable fit to the data, but the extracted rate constants had larger uncertainty than the model including GA-2,4-IA (Fig. S24 and Table S2†). This model also slightly overpredicted the concentration of GA-2,4-AI, suggesting that the formation of GA-2,4-IA was a minor but prevalent reaction during imide formation.
The rate constants extracted from the model fits displayed Arrhenius dependence in this temperature range (Fig. S25†). The activation energies (Eas) for k1, k2, and k3 were 91 ± 14 kJ mol−1, 91 ± 28 kJ mol−1, and 82 ± 18 kJ mol−1, respectively (Table S3†). The similar Ea for all three rate constants suggests, as proposed above, that the other substituents on the ring (electronics) do not influence the barrier for the ring closure reaction. The differences in rate constant magnitudes, but similar Ea for imide formation suggest that steric effects primarily influence the apparent activation entropies of these reactions.
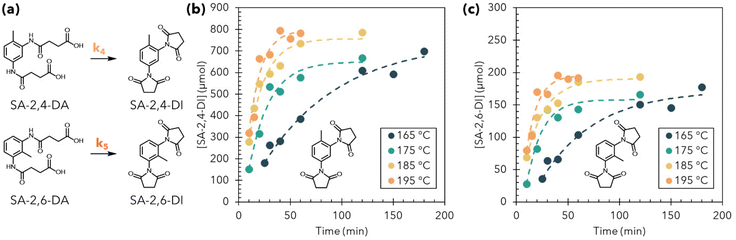
Finally, we investigated the kinetics of imide formation from M-PUF acidolysis with SA, which occurred on a faster timescale than imide formation during M-PUF acidolysis with GA. Even at low temperatures, imide formation during M-PUF acidolysis with SA was complete in 1–3 hours, in contrast to GA, which took >24 hours. As a result, the amide region of the 1H NMR from SA acidolysis had low signal compared to the internal standard, making extraction of quantitative kinetics in this region challenging. On the other hand, the aliphatic region (δ1H = 2.5–1.5 ppm) was uncluttered compared to reactions run with GA reagents due to the lack of amide-imide intermediates and single –CH2 signal from SA moieties, and thus signals corresponding to the methyl –CH3 groups of the 2,4- and 2,6-diimides (SA-2,4-DI & SA-2,6-DI, respectively) could be extracted (Fig. S26–S28†).
Because of the rapid disappearance of amide and amide-imide N-containing products during M-PUF acidolysis by SA, we proposed a pseudo-first order model for imide formation, which is summarized in Fig. 5(a). Fig. 5(b) and (c) show the concentration of SA-2,4-DI and SA-2,6-DI with their respective fits from 165–185 °C. Rate constants k4 and k5 were similar at all temperatures, suggesting that all SA amide bonds react at approximately the same rate (Table S4†). Arrhenius fits of the data resulted in Eas of 112 ± 57 kJ mol−1 and 74 ± 76 kJ mol−1 for k1 and k2, respectively (Table S4 and Fig. S29†), although the kinetic behavior was somewhat non-Arrhenius, which is reflected in the high standard errors for the reported Eas.
Here, in contrast to reactions with GA, there was no observed steric effect from the methyl group, as SA-2,4-DA and SA-2,6-DA reacted at approximately the same rate, and there was no accumulation of amide-imide intermediates. While the uncertainties on the Arrhenius fits for imide formation during SA acidolysis are high, the fits appeared to appropriately model the rate of imide formation at all temperatures. While we will not attempt to draw conclusions from the measured Ea and pre-exponential factors from these reactions, it is clear from the timescale of the reaction and the low concentrations of diamides and amide-imides that the formation of 5-membered-ring imides is significantly faster than the analogous reaction with GA to form a 6-membered-ring imide. It is worth noting that SA has a melting point of 186 °C and therefore transports as a vapor when used as an acidolysis reagent at lower temperatures.31 The phase of the acid might be expected to influence the solvation of amides and imides during the reaction (i.e., acids that are liquids at reaction temperatures can serve as solvents for amides/imides), although the fast rate of imide formation during M-PUF acidolysis with SA as opposed to GA suggests that this plays a minor role in dictating reaction kinetics. Nevertheless, it seems plausible that vapor-phase transport of SA at these reaction temperatures imposes diffusion limitations on the rate of amide formation at the start of the reaction, which could in turn influence the rate of imide formation and result in apparent rate constants for imide formation which do not follow Arrhenius-like behavior.
Notably, imide formation during acidolysis is much slower than deconstruction of the polymer; based on CO2 evolution, decomposition of urethane & urea bonds is complete in ca. 20–60 minutes for M-PUF acidolysis with GA and ca. 10–25 minutes for M-PUF acidolysis with SA at the reaction temperatures tested.23 In contrast, diamides took appx. 8–24 hours to disappear from the reaction mixture during acidolysis with GA, and amide-imide intermediates were still present in high concentrations after >24 hours. Imide formation during M-PUF acidolysis with SA took 2–3 hours. Previous work has demonstrated that polyol/amide release is the rate-limiting step during urethane/urea bond decomposition, and therefore CO2 evolution is a good proxy for the release of amides during the reaction.23 As a result, upon cessation of CO2 evolution it is assumed that amide species are no longer being generated. While imide formation likely begins while depolymerization is still occurring, the rate of these ring-closing reactions is 1–2 orders of magnitude lower than the rate of the polymer decomposition itself. These results suggest that both polyol release and imide formation are correlated with acid intramolecular carboxyl proximity.
Conclusions
In this study, we provide a fundamental framework for understanding the product distribution and kinetics of N-containing products from PUF acidolysis as a function of acid loading, structure, and reaction conditions. We show the use of excess acid eliminates aromatic amines, TDI-terminated polyols, and oligomeric ureas from the product mixture, and estimate that 2 moles DCA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole –NCO is a minimum acid loading to reach full deconstruction of the polyurethane network in the lab. Furthermore, we demonstrate the formation of imide products during acidolysis and provide a relationship between DCA carboxylic acid proximity and the likelihood and timescales of forming imide products. Kinetics of imide formation were measured for M-PUF acidolysis with GA and SA, showing the relative timescales and intermediates during the reaction. Imide formation during acidolysis reactions occurred 1–2 orders of magnitude slower than CO2/polyol release, depending on the acidolysis reagent used. Contrary to prior reports, both polyol release and imide formation occurred at temperatures at least as low as 165 °C, well below temperatures where thermal degradation pathways are relevant, suggesting a mechanism in which urethane and urea bonds are directly cleaved by the DCA. In conjunction with previous work showing the kinetics of polyol release, this manuscript provides a full picture of the reactions occurring during the acidolysis-mediated depolymerization of PUF, providing a complete kinetic framework that could enable modelling and reactor design for scaled-up PUF chemical recycling via acidolysis. Ultimately, we hope that this work will guide process development to create a circular global plastics economy.
1 mole –NCO is a minimum acid loading to reach full deconstruction of the polyurethane network in the lab. Furthermore, we demonstrate the formation of imide products during acidolysis and provide a relationship between DCA carboxylic acid proximity and the likelihood and timescales of forming imide products. Kinetics of imide formation were measured for M-PUF acidolysis with GA and SA, showing the relative timescales and intermediates during the reaction. Imide formation during acidolysis reactions occurred 1–2 orders of magnitude slower than CO2/polyol release, depending on the acidolysis reagent used. Contrary to prior reports, both polyol release and imide formation occurred at temperatures at least as low as 165 °C, well below temperatures where thermal degradation pathways are relevant, suggesting a mechanism in which urethane and urea bonds are directly cleaved by the DCA. In conjunction with previous work showing the kinetics of polyol release, this manuscript provides a full picture of the reactions occurring during the acidolysis-mediated depolymerization of PUF, providing a complete kinetic framework that could enable modelling and reactor design for scaled-up PUF chemical recycling via acidolysis. Ultimately, we hope that this work will guide process development to create a circular global plastics economy.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare the following competing financial interest(s): C.S.L. is an employee of The Dow Chemical Company.Acknowledgements
This work was supported by The Dow Chemical Company. Dr Alan L. Stottlemyer and Dr Nasim Hooshyar from The Dow Chemical Company are gratefully acknowledged for providing guidance and input on the study. Dr Nathan Rau from The Dow Chemical Company is also gratefully acknowledged for providing supporting NMR analysis and expertise. Some experiments were performed using UC Santa Barbara's MRL Shared Experimental Facilities, supported by the MRSEC Program from NSF (award no. DMR 1720256). P.C. and M.M.A.-O. both acknowledge the Mellichamp Academic Initiative in Sustainability at UCSB for support. VORANOL™ is a trademark of The Dow Chemical Company (“Dow”) or an affiliated company of Dow.References
- A. Das and P. Mahanwar, A brief discussion on advances in polyurethane applications, Adv. Ind. Eng. Polym. Res., 2020, 3(3), 93–101 Search PubMed.
- J. O. Akindoyo, et al., Polyurethane types, synthesis and applications–a review, RSC Adv., 2016, 6(115), 114453–114482 RSC.
- Statistica, Market volume of polyurethane worldwide from 2015 to 2022, with a forecast for 2023 to 2030, 2023 5 June 2024]; Available from: https://www.statista.com/statistics/720341/global-polyurethane-market-size-forecast/#:~:text=The%20global%20market%20volume%20of,tons%20in%20the%20year%202030.
- R. Geyer, J. R. Jambeck and K. L. Law, Production, use, and fate of all plastics ever made, Sci. Adv., 2017, 3(7), e1700782 CrossRef PubMed.
- J. R. Jambeck, et al., Plastic waste inputs from land into the ocean, Science, 2015, 347(6223), 768–771 CrossRef CAS PubMed.
- H. Li, et al., Expanding plastics recycling technologies: chemical aspects, technology status and challenges, Green Chem., 2022, 24(23), 8899–9002 RSC.
- A. Kemona and M. Piotrowska, Polyurethane recycling and disposal: Methods and prospects, Polymers, 2020, 12(8), 1752 CrossRef CAS PubMed.
- Z. O. Schyns and M. P. Shaver, Mechanical recycling of packaging plastics: A review, Macromol. Rapid Commun., 2021, 42(3), 2000415 CrossRef CAS PubMed.
- K. M. Zia, H. N. Bhatti and I. A. Bhatti, Methods for polyurethane and polyurethane composites, recycling and recovery: A review, React. Funct. Polym., 2007, 67(8), 675–692 CrossRef CAS.
- G. Rossignolo and G. Malucelli, Recycling of Polyurethanes: where we are and where we are going to, Green Chem., 2024, 26, 1132–1152 RSC.
- Y.-H. Lee, et al., Effect of hot pressing/melt mixing on the properties of thermoplastic polyurethane, Macromol. Res., 2009, 17, 616–622 CrossRef CAS.
- C. Bakkali-Hassani, et al., Transcarbamoylation in polyurethanes: underestimated exchange reactions?, Macromolecules, 2022, 55(18), 7974–7991 CrossRef CAS.
- S. Kim, et al., Circular reprocessing of thermoset polyurethane foams, Adv. Mater., 2023, 35(41), 2305387 CrossRef CAS PubMed.
- S. A. Madbouly, Novel recycling processes for thermoset polyurethane foams, Curr. Opin. Green Sustain. Chem., 2023, 42, 100835 CrossRef CAS.
- J. A. Nettles, et al., Functional Upcycling of Polyurethane Thermosets into Value-Added Thermoplastics via Small-Molecule Carbamate-Assisted Decross-Linking Extrusion, JACS Au, 2024, 4(8), 3058–3069 CrossRef CAS PubMed.
- D. T. Sheppard, et al., Reprocessing postconsumer polyurethane foam using carbamate exchange catalysis and twin-screw extrusion, ACS Cent. Sci., 2020, 6(6), 921–927 CrossRef CAS PubMed.
- L. M. Felsenthal, S. Kim and W. R. Dichtel, Robust Self-Healing Adhesives Based on Dynamic Urethane Exchange Reactions, ACS Appl. Mater. Interfaces, 2024, 16(42), 57687–57694 CrossRef CAS PubMed.
- B. Liu, et al., Opportunities in Closed-Loop Molecular Recycling of End-of-Life Polyurethane, ACS Sustainable Chem. Eng., 2023, 11(16), 6114–6128 CrossRef CAS.
- GMBH, H.S.A., Innovative approach to conversion of flexible PU foam residues into polyol on an industrial scale.
- P. Kanchanapiya, N. Intaranon and T. Tantisattayakul, Assessment of the economic recycling potential of a glycolysis treatment of rigid polyurethane foam waste: A case study from Thailand, J. Environ. Manage., 2021, 280, 111638 CrossRef CAS PubMed.
- M. Sołtysiński, et al., Conversion of polyurethane technological foam waste and post-consumer polyurethane mattresses into polyols–industrial applications, Polimery, 2018, 63(3), 234–238 CrossRef.
- D. Simón, et al., Recycling of polyurethanes from laboratory to industry, a journey towards the sustainability, Waste Manage., 2018, 76, 147–171 CrossRef PubMed.
- Z. Westman, et al., Influence of Carboxylic Acid Structure on the Kinetics of Polyurethane Foam Acidolysis to Recycled Polyol, JACS Au, 2024, 4(8), 3194–3204 CrossRef CAS PubMed.
- N. Gama, et al., Recycling of polyurethane scraps via acidolysis, J. Chem. Eng., 2020, 395, 125102 CrossRef CAS.
- B. Godinho, et al., Recycling of polyurethane wastes using different carboxylic acids via acidolysis to produce wood adhesives, J. Polym. Sci., 2021, 59(8), 697–705 CrossRef CAS.
- B. Liu, et al., Polyurethane Foam Chemical Recycling: Fast Acidolysis with Maleic Acid and Full Recovery of Polyol, ACS Sustainable Chem. Eng., 2024, 12(11), 4435–4443 CrossRef CAS PubMed.
- M. Grdadolnik, et al., Insight into chemical recycling of flexible polyurethane foams by acidolysis, ACS Sustainable Chem. Eng., 2022, 10(3), 1323–1332 CrossRef CAS PubMed.
- T. B. Bech, et al., Chemical separation of polyurethane via acidolysis–combining acidolysis with hydrolysis for valorisation of aromatic amines, Green Chem., 2024, 26, 8395–8404 RSC.
- R. M. O'Dea, et al., Toward Circular Recycling of Polyurethanes: Depolymerization and Recovery of Isocyanates, JACS Au, 2024, 4(4), 1471–1479 CrossRef PubMed.
- M. B. Johansen, et al., tert-Amyl Alcohol-Mediated Deconstruction of Polyurethane for Polyol and Aniline Recovery, ACS Sustainable Chem. Eng., 2022, 10(34), 11191–11202 CrossRef CAS.
- B. Liu, et al., Vapor-Phase Dicarboxylic Acids and Anhydrides Drive Depolymerization of Polyurethanes, ACS Macro Lett., 2024, 13, 435–439 CrossRef CAS PubMed.
- K. A. Thorn, et al., N-15 NMR study of the immobilization of 2, 4-and 2, 6-dinitrotoluene in aerobic compost, Environ. Sci. Technol., 2008, 42(7), 2542–2550 CrossRef CAS PubMed.
- N. Gama, et al., Recycling of polyurethane by acidolysis: The effect of reaction conditions on the properties of the recovered polyol, Polymer, 2021, 219, 123561 CrossRef CAS.
- H. He, et al., Chemical Recycling of Waste Polyurethane Foams: Efficient Acidolysis under the Catalysis of Zinc Acetate, ACS Sustainable Chem. Eng., 2023, 11(14), 5515–5523 CrossRef CAS.
- M. A. Casadei, C. Galli and L. Mandolini, Ring-closure reactions. 22. Kinetics of cyclization of diethyl (. omega.-bromoalkyl) malonates in the range of 4-to 21-membered rings. Role of ring strain, J. Am. Chem. Soc., 1984, 106(4), 1051–1056 CrossRef CAS.
- P. Wu, et al., A self-healing and recyclable polyurethane-urea Diels–Alder adduct synthesized from carbon dioxide and furfuryl amine, Green Chem., 2021, 23(1), 552–560 RSC.
- K. Yasuda, K. Sugane and M. Shibata, Self-healing high-performance thermosets utilizing the furan/maleimide Diels-Alder and amine/maleimide Michael reactions, J. Polym. Res., 2020, 27, 1–11 CrossRef.
- H. Chang, et al., Design of closed-loop recycling production of a Diels–Alder polymer from a biomass-derived difuran as a functional additive for polyurethanes, Green Chem., 2021, 23(23), 9479–9488 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4py01116c |
| This journal is © The Royal Society of Chemistry 2024 |