Open Access Article
Open Access ArticleTryptophan intercalation in siRNA drives the formation of polymeric micelles with enhanced delivery efficiency†
Yuki
Nakashima‡
a,
Wenqian
Yang‡
a,
Pengwen
Chen
a,
Keita
Masuda
a,
Teahun
Hong
b and
Horacio
Cabral
 *a
*a
aDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan. E-mail: horacio@bmw.t.u-tokyo.ac.jp
bDepartment of Molecular Pathology, Tokyo Medical University, Tokyo, Japan
First published on 2nd July 2024
Abstract
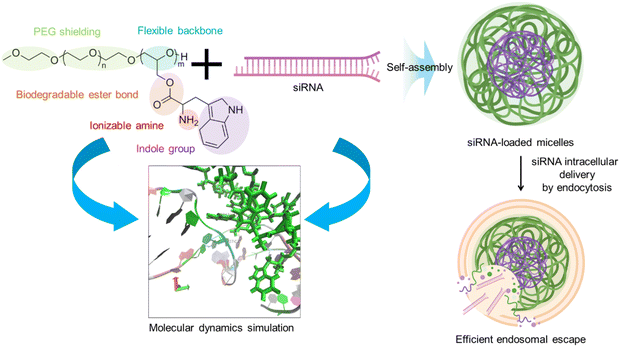
The construction of effective carrier systems is essential for delivering therapeutic small interfering RNA (siRNA). In this study, we present an innovative approach using tryptophan intercalation with siRNA to drive the formation of polymeric micelles. Through a facile yet robust method, the siRNA molecules are encapsulated within polymeric micelles formed by flexible poly(ethylene glycol)-poly(glycerol) (PEG-PG) block copolymers bearing biocompatible tryptophane units (PEG-PGTrp). Molecular dynamics (MD) simulations indicated the significance of the indole group, demonstrating its crucial role in fostering favorable interactions through π–π stacking. Moreover, the tryptophan moieties not only aid in the formation of stable micelles, but also contributed to intracellular trafficking and endosomal escape, thereby augmenting siRNA delivery. In vitro studies showed that the PEG-PGTrp-based micelles promoted intracellular delivery of siRNA, leading to enhanced gene knockdown.
1 Introduction
Gene silencing has emerged as a powerful approach to treat a wide range of diseases.1–4 Small interfering RNA (siRNA) consisting of short double-stranded RNA molecules has been effectively used for this purpose. siRNA can exert RNA interference (RNAi) in the cytosol by precisely binding to the target mRNA in a sequence specific manner to induce its degradation after being loaded in the RNA-induced silencing complex (RISC).5,6 The therapeutic ability has been demonstrated in the clinic, as two types of gene silencing medicines using siRNA have been approved by the U.S. Food and Drug Administration (FDA).7 While these clinical observations are promising, they have also highlighted the need for sophisticated carrier systems capable of improving the stability of nucleic acids, and the specificity and efficiency of the delivery to address the evolving challenges encountered in clinical applications.Various delivery systems, including virus-, lipid- and polymer-based approaches, are being explored for improving siRNA delivery.8,9 Among these systems, polyion complex (PIC) micelles formed in aqueous conditions after mixing nucleic acids and block catiomers having a neutral hydrophilic segment and a polycationic block can provide protection of the cargo loaded in the core, promote targeted delivery by controlling the pharmacokinetics and tissue distribution through size control and ligand installation.10–13 Moreover, the possibility to precisely engineer the micelle-forming polymers allows increasing the stability of the formulations in extracellular spaces and promoting intracellular delivery.8,11,14–17 Particularly, the conjugation of DNA intercalating molecules to polymers has shown high potential for stabilizing formulations through π–π stacking and hydrophobic interactions between carriers and double stranded siRNA.18–21 Nevertheless, the use of intercalation agents raises concerns regarding potential adverse effects in host cells, underscoring a pressing need for non-cytotoxic strategies.
Tryptophan (Trp) is an essential amino acid that can intercalate with double stranded DNA.22–24 Thus, the introduction of Trp moieties in block catiomers could insert in siRNA to enhance stability by π–π stacking and hydrophobic interactions, while maintaining low cytotoxicity levels. In this regard, we have recently shown that the conjugation of tyrosine and phenylalanine to block copolymers enhanced the interaction with single stranded messenger RNA (mRNA) to promote the assembly of micelles with enhanced delivery efficiency.25,26 Encouraged by these findings, herein, we synthesized poly(ethylene glycol)-poly(glycerol) (PEG-PG) block copolymers modified with glycine (PEG-PGGly) or Trp (PEG-PGTrp) for understanding the effects of the indole moieties in the side chain (Scheme 1). PEG-PGGly and PEG-PGTrp were mixed with siRNA to formulate micelles (PEG-PGGly-based structures and PEG-PGTrp/m, respectively). To gain insights into the interactions, we assessed the binding between these polymers and siRNA using molecular dynamics (MD) simulations toward rationalizing our design. The results showed that the PGTrp induced more van der Waals energies, less electrostatic energies with siRNA than PGGly, and π–π stacking between PGTrp and the nucleobases on siRNA. Thus, PEG-PGTrp enhanced the delivery performance, promoting intracellular delivery of siRNA. Our results indicate a viable strategy for enhancing siRNA using polymers bearing aromatic amino acid residues.
2 Materials and methods
2.1 Materials
Toluene (super dehydrated, purity >99.5+%) were purchased from Kanto Chemical, Co., Inc. (Tokyo, Japan). α-Methoxy-poly(ethylene glycol) (MeO-PEG-OH; molecular weight (MW): 10 kDa) was purchased from Nanosoft Polymers (Winston-Salem, NC, USA). Ethanol, LDH-Cytotoxic Test, and piperidine (purity >98.0%) were purchased from Fujifilm Wako Pure Chemical, Co., Inc. (Tokyo, Japan). Phosphazene base P4-t-Bu solution (P4-t-Bu), palladium on carbon (Pd/C), N-[(9H-fluoren-9-ylmethoxy)carbonyl]-glycine(FMOC-Gly) (purity >98.0%), and N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-Tryptophan (FMOC-Trp) (purity >98.0%), HEPES buffer, and MES were purchased from Sigma-Aldrich (St Louis, MO, U.S.A.). Triethylsilane (TES), N,N-dimethylformamide (DMF) (purity >99.5%), N-(4-pyridyl)dimethylamine (DMAP) (purity >99.0%), and 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC) (purity >98.0%) were purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). RNA, Cy5-labelled RNA, and 5′-FITC-labelled RNA were purchased from Hokkaido System Science Co., Ltd (Hokkaido, Japan). The sequence of siRNA used is to target firefly GL3 luciferase with sense: 5′-CUU ACG CUG AGU ACU UCG AdTdT-3′ and anti-sense: 5′-UCG AAG UAC UCA GCG UAA GdTdT-3′. Dextran sulfate sodium salt (MW = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was purchased from Biosynth Co., Ltd (Staad, Switzerland). Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium, Penicillin–Streptomycin, LysoTracker Green and Triton X-100 (TX-100) were purchased from Thermo Fisher Scientific (Waltham, MA, U.S.A.). HeLa-luc cell line was purchased from Caliper LifeScience (Hopkinton, Massachusetts, U.S.A.).
000) was purchased from Biosynth Co., Ltd (Staad, Switzerland). Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium, Penicillin–Streptomycin, LysoTracker Green and Triton X-100 (TX-100) were purchased from Thermo Fisher Scientific (Waltham, MA, U.S.A.). HeLa-luc cell line was purchased from Caliper LifeScience (Hopkinton, Massachusetts, U.S.A.).
2.2 Polymer synthesis
α-Methoxy-poly(ethylene glycol)-block-poly(glycerol) copolymer (PEG-PG) was synthesized as follows: Briefly, toluene (500 mL) and 1 g of MeO-PEG-OH (0.1 mmol) were mixed in a two-neck flask, toluene and water were distilled off after reflux at 175 °C for 30 min. Super dehydrated toluene (2 mL) and benzyl glycidyl ether (1.53 mL, 10 mmol, 100 eq.) were then added to the system under argon atmosphere. MeO-PEG-OH was reacted with benzyl glycidyl ether in the presence of phosphazene base P4-t-Bu solution (160 μmol, 200 μL) at 37 °C for 26 h under argon atmosphere. The catalyst was removed by alumina column. The polymer solution was precipitated twice in cold ether and dried under vacuum to obtain MeO-PEG-benzyl glycidyl ether block copolymer. The polymer was characterized by 1H NMR (400 MHz, JEOL ECS-400, JEOL, Tokyo, Japan) in DMSO and GPC (TOSOH HLC-8220, Tokyo, Japan) analysis. TES was dropwise from a pressure-equalizing drooping funnel to a stirred solution of MeO-PEG-benzyl glycidyl ether (200 mg) and palladium 10% on carbon powder (20% by weight) in MeOH–DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (2 mL) under an argon-dilled balloon. After 1 hour, the mixture was filtered through Celite column, and the solvent was removed by vacuum. Finally, the obtained PEG-PG was dissolved in benzene and collected after evaporation under vacuum. The polymer was characterized by 1H NMR (DMSO; 80 °C) and GPC analysis.
1) (2 mL) under an argon-dilled balloon. After 1 hour, the mixture was filtered through Celite column, and the solvent was removed by vacuum. Finally, the obtained PEG-PG was dissolved in benzene and collected after evaporation under vacuum. The polymer was characterized by 1H NMR (DMSO; 80 °C) and GPC analysis.
To obtain α-methoxy-poly(ethylene glycol)-block-poly(glycidyl-amino acids), PEG-PG was reacted with [(9-Fluorenylmethyl)oxy]carbonyl (FMOC) amino acids through coupling reaction. Briefly, 50 mg of PEG-PG was dissolved in DMF at concentration of 10 mg mL−1, then FMOC-amino acid was added (FMOC-Gly: 1.52 mmol, 616 mg; FMOC-Trp: 1.52 mmol, 616 mg) to the system, followed by EDC·HCl (2.8 mmol, 536 mg) and DMAP (0.28 mmol, 34.21 mg). The mixture was stirred in water bath at 37 °C for 26 h with aluminium foil shield, and then dialysis against DMF to remove unreacted FMOC-amino acid and catalyst. The deprotection of FMOC group was conducted by dissolve polymer in DMF at 10 mg ml−1, then piperidine was drop to the system at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of DMF
1 of DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) piperidine (v/v), then react at 37 °C for 1 h and dialysis against DMF. Final polymer was collected by benzene freezing dry under vacuum. The composition and molecular weight distribution were determined by 1H NMR (DMSO; 80 °C) and GPC analysis.
piperidine (v/v), then react at 37 °C for 1 h and dialysis against DMF. Final polymer was collected by benzene freezing dry under vacuum. The composition and molecular weight distribution were determined by 1H NMR (DMSO; 80 °C) and GPC analysis.
2.3 Preparation of siRNA-loaded micelles
siRNA-loaded micelles were prepared by mixing block copolymers (1 mg mL−1) and siRNA (0.0135 mg mL−1) dissolved in 10 mM HEPEs buffer (pH7.4) at different amine/phosphate (N/P) ratio. The siRNA used here is GL3, and sequence is UCGAAGUACUCAGCGUAAGdTdT. Micelle size distribution was determined by dynamic light scattering (DLS) measurement at 25 °C by Zeta Sizer Nano ZS (Malvern Instruments Ltd, UK). In addition, the feature of micelles was directly observed by transmission electron microscopy (TEM) imaging, followed by being stained by uranyl acetate and placed on 400-mech copper grids for TEM observation (JEM-1400, JEOL).2.4 Molecular simulation
siRNA was modelled using a 19-base double strand, the length matches the siRNA referred to as GL3. One sequence is 5′-UCG AAG UAC UCA GCG UAA G-3′ and the other is 3′-AGC UUC AUG AGU CGC AUU C-5′. The starting coordinates were built to be a canonical A-form using the AMBER NAB tool. Poly(glycidol glycine) (PGGly) and poly(glycidol tryptophan) (PGTrp) polymers with 10 repeating monomer units were used in the study. The primary amines were set with positive charges at physiological conditions, the polymer was charged +10. We then created models of the polymer in complex with siRNA in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, which was considered to be a reliable model of binding. All simulations and data analyses were performed using the AMBER 16 suite of programs. The OL3 (chiOL3) force field was chosen to describe siRNA, and the General Amber Force Field (GAFF) and TIP3P were used for polymers and water respectively. The topology file of the polymer was generated by connecting the monomer head by head, and the complex was then run via tleap (terminal LEap) module incorporated in AMBER. Each polymer chain was placed at 10 Å from RNA at the beginning of the simulations. Polymer and siRNA were in the fully extended conformation, solvated in TIP3P water box, Cl− and Na+ ions (∼0.15 M) were added to neutralize the system. Minimization of the system was carried out in two steps. The simulation process followed the method described in our previous publication.16 As briefly explanation, First, energy minimization was carried out on water molecules by 10
1 ratio, which was considered to be a reliable model of binding. All simulations and data analyses were performed using the AMBER 16 suite of programs. The OL3 (chiOL3) force field was chosen to describe siRNA, and the General Amber Force Field (GAFF) and TIP3P were used for polymers and water respectively. The topology file of the polymer was generated by connecting the monomer head by head, and the complex was then run via tleap (terminal LEap) module incorporated in AMBER. Each polymer chain was placed at 10 Å from RNA at the beginning of the simulations. Polymer and siRNA were in the fully extended conformation, solvated in TIP3P water box, Cl− and Na+ ions (∼0.15 M) were added to neutralize the system. Minimization of the system was carried out in two steps. The simulation process followed the method described in our previous publication.16 As briefly explanation, First, energy minimization was carried out on water molecules by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps (including 5000 steps of steepest descent and 5000 steps of conjugate gradient) with the RNA and polymer being restricted. Next, the system was left free to move, and the whole system was minimized by 10
000 steps (including 5000 steps of steepest descent and 5000 steps of conjugate gradient) with the RNA and polymer being restricted. Next, the system was left free to move, and the whole system was minimized by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps (including 5000 steps of steepest descent and 5000 steps of conjugate gradient). The system was then heated from 0 k to 300 k for 50 ps, then followed by a density equilibration run (50 ps) under NPT condition (constant particle number (N), regulated pressure (P) and constant temperature (T)). The production dynamic lasted for 100 ns for each system under NPT condition. Finally, CPPTRAJ model of AMBER 18 was used to analyse the dynamic trajectories (including RMSD, RMSF and Radius of gyration), and molecular visualisation was done using the Visual Molecular Dynamics (VMD) package. The binding free energies of RNA-polymer complex was carried out using the Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method, which was described through the following equation:
000 steps (including 5000 steps of steepest descent and 5000 steps of conjugate gradient). The system was then heated from 0 k to 300 k for 50 ps, then followed by a density equilibration run (50 ps) under NPT condition (constant particle number (N), regulated pressure (P) and constant temperature (T)). The production dynamic lasted for 100 ns for each system under NPT condition. Finally, CPPTRAJ model of AMBER 18 was used to analyse the dynamic trajectories (including RMSD, RMSF and Radius of gyration), and molecular visualisation was done using the Visual Molecular Dynamics (VMD) package. The binding free energies of RNA-polymer complex was carried out using the Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method, which was described through the following equation:| ΔGbind = ΔHbind − TΔSbind | (1) |
| ΔHbind = ΔEMM + ΔGPB + ΔGSA | (2) |
| ΔEMM = ΔEbond − ΔEangle + ΔEtorsion + ΔEvdw + ΔEele | (3) |
The gas–phase molecular mechanics interaction energy between RNA and polymer ΔEMM includes the internal bone energy ΔEbond, angle energy ΔEangle and torsion energy ΔEtorsion, van der Waals energies ΔEvdw and electrostatic ΔEele. ΔGPB/GB, PB/GB models were used for estimating the polar contribution, and ΔGSA is used for calculating energies from non-electrostatic solvation components (non-polar contribution).
The radius of gyration (Rg) was analysed to compute the atomic motion in the RNA–polymer complex from their shared centre of gravity, applying eqn (4):
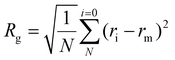 | (4) |
2.5 Fluorescence quenching
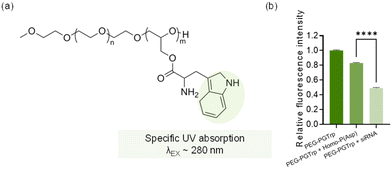
Homo-P(Asp) was prepared following our previous study.16 50 μL of 1 mg mL−1 PEG-PGTrp solution was placed in 96-well plates. Next, siRNA (20 μM) and homo-P(Asp) solution was applied to each well at molar ratios of primary amines in polymer to phosphates in the siRNA/homo-P(Asp) in the homo-P(Asp) of 1.0. The siRNA used here is GL3, and sequence is UCGAAGUACUCAGCGUAAGdTdT. As Trp is known to absorb light at the near ultraviolet region, around 280 nm. The fluorescence intensity of the solution was evaluated at the excitation wavelength (λex) of 280 nm and emission wavelength (λex) of 350 nm by multimode microplate reader (Tecan Group Ltd, Switzerland).2.6 Measurement of counter polyanion exchange
The stability of siRNA assemblies against counter polyanion was estimated by mixing the micelles with a dextran sulfate solution. The mixture was done at different ratios of sulfate moieties in the dextran sulfate to phosphate groups in the siRNA (S/P). The micelles containing Cy5-labelled siRNA (33.3 ng μL−1) were incubated with the dextran sulfate for 30 minutes. Then, fluorescence corelation spectroscopy (FCS) measurements were performed using an LSM-880 confocal laser scanning microscopy (CLSM; Carl Zeiss AG, Oberkochen, Germany) with He–Ne laser (633 nm) scanning, following our previous research.16 The increase of diffusion time (D) of the samples presents to the decrease of diffusion coefficient (DH) according to Stokes–Einstein equation. The decline of diffusion time of the micelles refers to the decomposition of the nanostructures to free polymers and siRNA. In this experiment, the autocorrelation curve for each sample was obtained from 10 measurements at 10 seconds. The measurements were repeated 10 times. The diffusion coefficient was obtained from the Stokes–Einstein equation following our previous research (5),16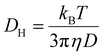 | (5) |
2.7 In vitro evaluation of endosomal escape
HeLa-luc cells (3 × 104) were cultured on 8 Chambered 1.0 Borosilicate Cover glass system (Lab Tek) and incubated for 24 h. Next, naked siRNA, FITC labelled siRNA-loaded micelles were applied to each well (final concentration = 100 nM siRNA per well). Uptake of micelles were evaluated after 1 h, 5 h, 9 h and 24 h. Microscopy was completed using an LSM-780 confocal laser scanning microscopy (CLSM; Carl Zeiss AG, Oberkochen, Germany) with a 40X objective (C-Apochromat, Carl Zeiss, Germany) for LysoTracker Red (green), FITC (green), and Hoechst 33342 (blue), respectively. To evaluate the intracellular distributions of siRNA loaded on PIC micelles, the rate of colocalization of FITC-siRNA with LysoTracker Red was quantified.2.8 LDH leakage assay
To study the disruption of cellular membranes by the polymers, HepG2 cells were seeded in 96-well plates at 1 × 104 viable cells per well and incubated overnight at 37 °C and 5% CO2. The cells were exposed to PEG-PGGly or PEG-PGTrp solutions in HEPES (pH 7.4) or MES (pH 5.5) buffers for 15 minutes at 37 °C. After incubation, the supernatant was collected and lactate dehydrogenase (LDH) content in the solution was measured using as LDH-cytotoxic test following the included protocol. TX-100 was used as a positive control to cause the disruption of cellular membrane in both pH situations. The UV absorbance (A560) was read out with an Infinite M200 microplatereader (Tecan, Männendorf, Switzerland). Relative LDH leakage was calculated as (A560 − A560,n.c.)/(A560,p.c. − A560,n.c.).2.9 In vitro gene silencing
HeLa-luc cells were plated onto 96-well plates (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well), followed by 24 h incubation in DMEM containing 10% FBS. Next, naked siRNA, siRNA-loaded micelles based on PEG-PGGly or PEG-PGTrp were applied to each well (20 nM, 100 nM, 200 nM siRNA per well). Also, a scrambled sequence (siScramble) was used to verify the sequence specificity of siRNA. After 24 h, the cells were washed with 0.1 mL of PBS twice and lysed with 0.02 mL of cell culture lysis buffer (Promega). The luciferase activity of the lysates was determined from the photo luminescence intensity using the Luciferase Assay System (Promega).
000 cells per well), followed by 24 h incubation in DMEM containing 10% FBS. Next, naked siRNA, siRNA-loaded micelles based on PEG-PGGly or PEG-PGTrp were applied to each well (20 nM, 100 nM, 200 nM siRNA per well). Also, a scrambled sequence (siScramble) was used to verify the sequence specificity of siRNA. After 24 h, the cells were washed with 0.1 mL of PBS twice and lysed with 0.02 mL of cell culture lysis buffer (Promega). The luciferase activity of the lysates was determined from the photo luminescence intensity using the Luciferase Assay System (Promega).
3 Results and discussion
3.1 Block copolymer synthesis
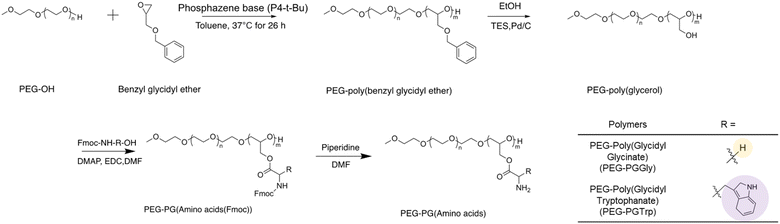
PEG-PGGly and PEG-PGTrp were synthesized according to Fig. 1. MeO-PEG-OH was used as the initiator for the ring opening reaction of benzyl glycidyl ether. The degree of polymerization was determined to be 78 by 1H NMR analysis (ESI Fig. S1†), and unimodal molecular weight distribution was confirmed in GPC analysis (ESI Fig. S2†). The obtained polymer was treated with palladium 10% carbon powder for removing the benzyl group. The complete deprotection and the quality of the polymer were confirmed by 1H NMR and GPC analysis (ESI Fig. S3 and S4†). The hydroxyl groups of the PEG-PG were then modified with Fmoc-protected glycine or tryptophan via condensation reaction to form hydrolysable ester bonds. The protecting groups were removed by 20% piperidine treatment, resulting in PEG-PGGly and PEG-PGTrp. The conjugation rates of glycine and tryptophan were calculated to be 84.3% and 89.7% by 1H NMR analysis, respectively (Table 1 and ESI Fig. S5, S6†). Besides, these block copolymers prepared with narrow molecular weight distributions (Mw/Mn) (Table 1).3.2 Characterization of nanostructures
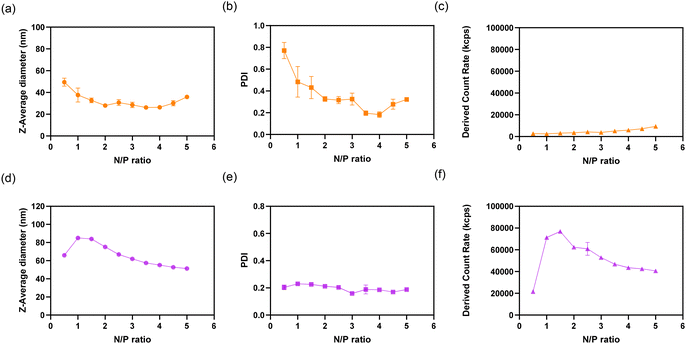
PEG-PGGly and PEG-PGTrp were mixed with siRNA in 10 mM HEPES buffer (pH 7.4) at different N/P ratios, and the formation of self-assembled nanostructures was evaluated by assessing the diameter, the polydispersity index (PDI) and the light scattering intensity using DLS. While PEG-PGGly showed the formation of relatively small nanostructures with large PDI and low scattered light (Fig. 2a, b and c), PEG-PGTrp formed nanostructures of approximately 60 nm even at a low N/P ratio of 0.5 (Fig. 2d) with narrow PDI (Fig. 2e). The nanostructures from PEG-PGTrp presented a maximum size of 80 nm at N/P = 1. Moreover, the diameter of the PEG-PGTrp/siRNA self-assemblies decreased as higher N/P and reached a plateau at 60 nm value at N/P ≥ 3. The normalized derived count rate, which corresponds to the size and concentration of the self-assemblies, showed a maximum at N/P = 1, and gradually decreased as the diameter diminished at higher N/P ratios (Fig. 2f). At N/P = 3, PEG-PGTrp micelles showed bigger size and higher normalized derived count rate compared to PEG-PGGly (Table 2). The shape of the assemblies at N/P = 3 was evaluated by TEM, following uranyl acetate staining. The results showed that PEG-PGGly formed small assemblies with siRNA, whereas spherical nanostructures observed for the PEG-PGTrp system (Fig. 3). As the PEG block is not visible in the TEM observation with uranyl acetate staining, we calculated the size of the core of the nanostructures (Table 2). The results showed that PEG-PGGly showed small cores of around 10 nm, whereas PEG-PGTrp made structures with a core of more than 42 nm. By subtracting the size of the core to the size obtained by DLS, the size of the PEG shell can be calculated nanostructures (Table 2). The results indicate a dense PEG shielding for both structures, with PEG-PGTrp showing a larger PEG shell. These findings indicate that PEG-PGGly and PEG-PGTrp are forming disparate nanostructures with siRNA. The system based on PEG-PGGly appear to be forming stable unit polyion complexes at the studied N/P ratios without being able to form multimolecular micellar assemblies.27 On the other hand, the indole hydrophobic moieties could reinforce the interaction with siRNA and between the siRNA/polymer complexes, leading to the assembly of core–shell micelles.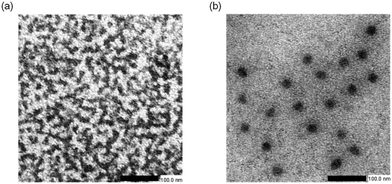 | ||
| Fig. 3 TEM observation of siRNA-assemblies prepared with PEG-PGGly (a) and PEG-PGTrp (b) at N/P = 3. | ||
3.3 Molecular dynamic simulation of polymer siRNA interaction
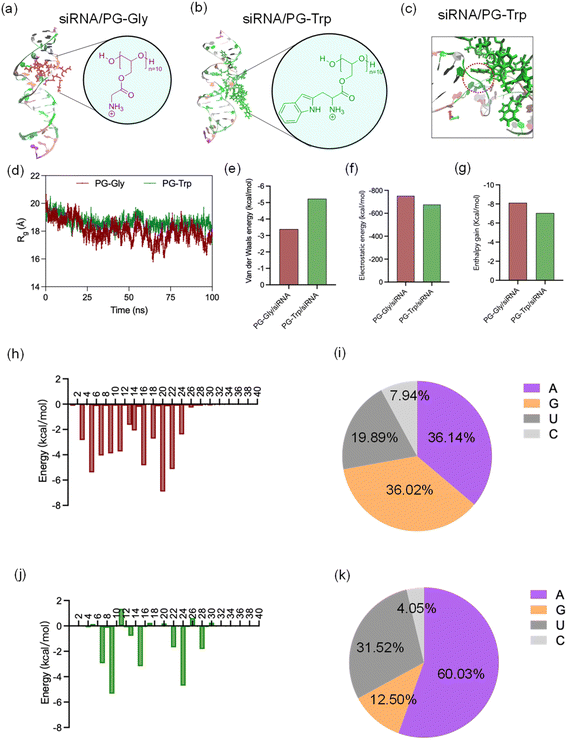
MD simulation was used to explore siRNA-polymer interactions at the atomic level. PGTrp/siRNA and PGGly/siRNA systems were simulated to determine the effects of the indole groups in the complexation. The model contains 1 siRNA and 1 polycationic segment. For each system, appropriate amount of Na+ and Cl− ions were added to simulate the salt concentration of 150 mM during assembly. The front view snapshots taken from the dynamic simulation of PGGly/siRNA and PGTrp/siRNA are shown in Fig. 4a and b, respectively. The residue uracil 27 base form π-stacking interactions with Trp 43 (Fig. 4c). These interactions are present for almost the entire simulation indicating their high stability. To better understand the complex, we monitored the radius of gyration (Rg) over the entire trajectories. Rg is the root mean square distance of each atom from the center of mass of the structure considered (equation was in above section 2.2), which was used as an indicator of the structure compactness.28 PGTrp/siRNA showed a higher Rg compared with PGGly/siRNA during the simulation trajectories (Fig. 4d), suggesting a loose complexation. Higher Rg value in the PGTrp/siRNA complex may also result from a more significant steric hindrance of PGTrp compared to PGGly. To comparatively evaluate the effect of π-stacking interaction on the binding ability of polymer to RNA, the binding free energies are decomposed into van der Waals interactions (ΔEvdw), electrostatics interactions (ΔEele), enthalpy contributions. The results indicate the electrostatic interaction of PGTrp to siRNA is weakened compared to that of Gly to the siRNA (Fig. 4f). However, the ΔEvdw of PGTrp to siRNA are enhanced compared to PGGly (Fig. 4e). Overall, the PG-PGTrp/siRNA complex resulted in less enthalpy gain than that of PGGly (Fig. 4g). To further complement the results derived from the energetic study, the polymer in the complex with siRNA (bound at major groove site) was further subjected to MM/GBSA free energy decomposition analysis to determine the contribution of each residue to the total energy of the complex (Fig. 4h–k). In the PGGly/siRNA complex, the contribution ratio to the total binding free energy of the complex were 36.14%, 36.02%, 19.89%, and 7.94% from adenine, guanine, uracil, and cytosine respectively (Fig. 4i). The siRNA binding modes are different in the PGTrp/siRNA system, reflected by the considerably higher occupancy of adenine (60.03%) (Fig. 4k). The π–π interaction between the indole group and uracil group was also observed during the dynamic run as mentioned above, implying its strong moiety ability. Thus, the MD simulation provides a clear view of the insertion of the Trp units in siRNA and allows us to differentiate the enhanced stability of PEG-PGTrp/siRNA complex. Considering the differences of the sidechain moieties between PGGly and PGTrp, the synergic effect of electrostatic interactions and π–π interactions contributed to the more stabilized structures of PEG-PGTrp/siRNA complex.3.4 π–π stacking assessment by Trp fluorescence quenching
The π–π stacking effect between PEG-PGTrp and siRNA was investigated by measuring the fluorescence quenching of Trp moieties in the polymer. The indole group in Trp has fluorescence at λex/λem = ∼280 nm/∼350 nm, and π–π stacking could quench the fluorescence by electron transfer.29,30 Then, the fluorescence of Trp groups in PEG-PGTrp was measured to determine the difference between PEG-PGTrp polymer and micelles formed with siRNA. Additionally, negatively-charged homo-poly(aspartic acid) was used for assembling micelles with PEG-PGTrp by electrostatic interactions. According to the measurements of the emission intensity of indole groups, a significant decrease of the fluorescence was detected when PEG-PGTrp was complexed with siRNA (Fig. 5b). Moreover, as homo-poly(aspartic acid) cannot make π–π stacking, the fluorescence was less quenched that with siRNA. The decrease in the fluorescence observed of PEG-PGTrp plus homo-poly(aspartic acid) may be related to self-quenching by the interaction between Trp units. These results indicate that Trp moieties in PEG-PGTrp contributed to π–π stacking interactions between the polymer and siRNA.3.5 Stability against polyanions
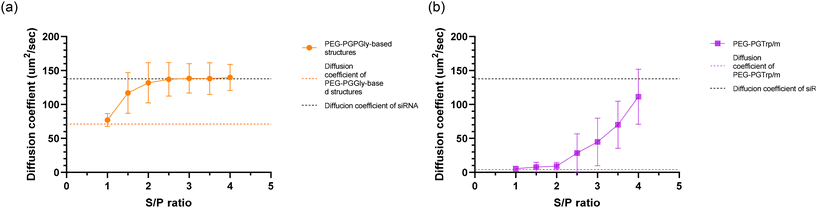
Carrier systems based on PICs are vulnerable to dissociation via polyion exchange upon exposure to anionic macromolecules.31,32 Consequently, the stabilities of PEG-PGGly-based and PEG-PGTrp/m-based structures were assessed under polyanion-rich conditions. Samples were incubated with negatively charged dextran sulfate at varying S/P ratios for 30 minutes. The dissociation of these nanostructures was evaluated by monitoring changes in the diffusion coefficient of Cy5-labelled siRNA using FCS measurements. When siRNA is encapsulated in the PEG-PGGly-based structures and PEG-PGTrp/m, the diffusion coefficient is lower than that of free siRNA. Additionally, due to the smaller size of PEG-PGGly-based structures compared to PEG-PGTrp/m, these nanostructures exhibit different diffusion coefficients (Fig. 6).16 For PEG-PGGly-based structures, the diffusion coefficient began to increase at S/P = 1.5, with significant siRNA release observed at S/P = 2 (Fig. 6a), indicating the structures are completely dissociated at S/P = 2. In contrast, PEG-PGTrp/m remained stable at S/P = 2, with siRNA displacement occurring above S/P = 4 (Fig. 6b). These findings indicate that PEG-PGTrp/m retains siRNA more effectively than the PEG-PGGly-based structures, likely due to enhanced stability facilitated by π–π stacking interactions between PEG-PGTrp and siRNA.3.6 In vitro activity
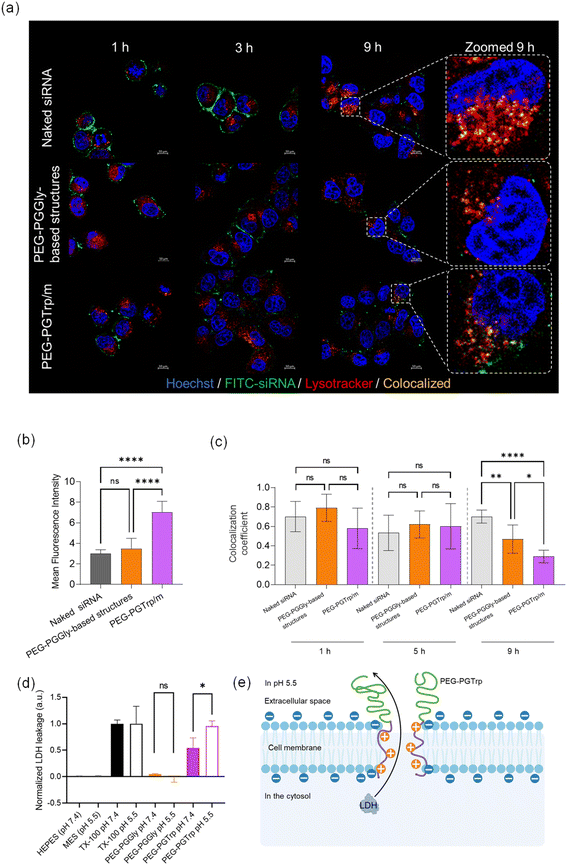
The performance of the siRNA-loaded micelles was explored in vitro. HeLa-luc cells were used for investigating the ability of the micelles to deliver siRNA and enhance gene silencing. The uptake of siRNA was visualized by CLSM using micelles loading FITC-labelled siRNA at different time points (Fig. 7a). After 1 hour incubation of naked siRNA, a green signal from labelled siRNA was observed on the cell membrane surface, and the same behaviour was confirmed even after 3 hours. Additionally, the similar trend was observed in PEG-PGGly-based structures. It could reveal that much of naked siRNA and PEG-PGGLy-based structures were trapped by the cellular membrane, and few of them reached the cytosol. It means that PEG-PGGLy-based structures might be collapsed, then loaded-siRNA could behave like naked siRNA. On the other hand, PEG-PGTrp/m showed relatively weak green signals on the cell membrane in both one hour and 3 hours. This effect might be assisted by the stable formulation of siRNA-loaded micelles with PEG-PGTrp by π–π stacking interactions. Furthermore, 9 hours after these treating, the highest increase in cellular uptake of siRNA was observed in the micelles made by PEG-PGTrp (Fig. 7b). It suggests that the stable formulation of micelles with PEG-PGTrp could allow the loaded-siRNA to be taken up by cells through overcoming the cellular membrane.Escaping from endosomes after cellular uptake is also an important factor in siRNA delivery. Thus, the ability of the delivery systems to achieve endosomal escape was studied by endosomal colocalization method. The endo/lysosomes were labelled with LysoTracker Red, and the siRNA in endosomes was visualized as yellow pixels in the CLSM images (Fig. 7c). The colocalization rate of siRNA and endo/lysosomes was quantified by the colocalization coefficient, and a low colocalization coefficient indicates a high endosomal escape ability. After 9 hours incubation of the micelles with the HeLa cells, a decrease of colocalization coefficient was observed in PEG-PGTrp/m compared to naked siRNA and PEG-PGGly-based structures, indicating critical endosomal escape of siRNA was facilitated by PEG-PGTrp. In this regard, hydrophobic moieties and aromatic groups have been showed destabilize endosomal membrane to enhanced endosomal escape of various payloads.25,26,33–35 Moreover, we have recently reported that the benzyl group of the amino acids conjugated to PEG-PG has a pKa of 5.9, which indicates that most of the amines are not protonated when the micelles are at extracellular physiological pH (pH 7.4), but will protonate within acidic endosomal compartments (pH 4.5–6.5).26 Thus, inside the endosomes, the number of polymers interacting with siRNA in the micelles may decrease to keep the charge balance. Then, the protonated polymers detached from the micelles could destabilize the membrane of the endosomes promoting endosomal escape.26 Thus, it is likely that the Trp groups in PEG-PGTrp are ionized in the endosomes to effectively disrupt the endosomal membrane for promoting the cytosolic access of siRNA.
The ability of PEG-PGTrp to disrupt the endosomal membranes was assessed by studying the pH-dependent leakage of cytosolic LDH protein. The PEG-PGTrp showed significantly higher LDH leakage at pH 5.5 than at pH 7.4 (Fig. 7d), suggesting the capability of the polymer for disrupting endosomal membranes. In contrast, PEG-PGGly did not induce LDH leakage in both pH 7.4 and pH 5.5 (Fig. 7d). These results suggest that PEG-PGTrp may create pores in the cell membrane large enough to allow the passage of LDH (Fig. 7e), which ranges from 35 to 140 kDa. Thus, this feature would be beneficial for the cytosolic delivery of siRNA.
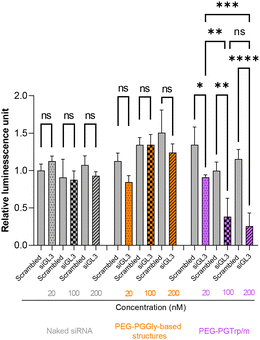
The gene silencing ability of the micelles was also studied in HeLa-luc cells by delivering GL3 siRNA. After 24 h incubation, the reporter protein expression in the treated cells was quantified by measuring their luminescence signal. According to the results, PEG-PGTrp/m achieved dose dependent gene silencing and significant luciferase knockdown effects (Fig. 8). The control groups, i.e., naked siRNA and PEG-PGGly-based system, showed negligible gene silencing at any dose. The trends in gene knockdown efficiency were consistent with the cellular uptake and endosomal escape ability of the micelles made from PEG-PGTrp.
4 Conclusion
Our research showed that critical siRNA delivery was achieved by promoting π–π stacking interactions between the PEG-PGTrp and siRNA, resulting in effective micelle formation. The incorporation of indole moieties played a pivotal role in augmenting cellular uptake, facilitating endosomal escape and promoting gene silencing of siRNA in vitro. Notably, the observed impact of side chain groups within the polymer aligns consistently with simulated estimations. Consequently, this study underscores the significance of employing simulations to predict the characteristics of polymers intended for siRNA delivery. Moreover, our findings emphasize the potential of a rational design approach for polymeric materials based on simulated data, offering a pathway to achieve enhanced siRNA delivery with heightened efficiency.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.† Raw data that support the findings of this study are available from the corresponding auhtor, upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Project for Cancer Research and Therapeutic Evolution (P-CREATE) (H.C.) from the Japan Agency for Medical Research and Development (AMED). The work was also supported by the AMED Seeds A grant (23ym0126805j0002; 8H.C.). The study was partially supported by Grants-in-Aid for Scientific Research A (23H00546; H.C.), Grants-in-Aid for Scientific Research B (22H03967; Y.M) Grants-in-Aid for Exploratory Research (22K19541; H.C.), and the Fund for the Promotion of Joint International Research (Fostering Joint International Research (B), 21KK0197; H.C.), from the Japan Society for the Promotion of Science (JSPS).References
- B. L. Davidson and P. B. McCray Jr, Nat. Rev. Genet., 2011, 12, 329–340 CrossRef CAS PubMed.
- D. H. Kim and J. J. Rossi, Nat. Rev. Genet., 2007, 8, 173–184 CrossRef CAS PubMed.
- G. R. Devi, Cancer Gene Ther., 2006, 13, 819–829 CrossRef CAS PubMed.
- C. Chakraborty, A. R. Sharma, G. Sharma, C. G. Doss and S. S. Lee, Mol. Ther. – Nucleic Acids, 2017, 8, 132–143 CrossRef CAS PubMed.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- H. O. Iwakawa and Y. Tomari, Mol. Cell, 2022, 82, 30–43 CrossRef CAS PubMed.
- B. Hu, L. Zhong, Y. Weng, L. Peng, Y. Huang, Y. Zhao and X. J. Liang, Signal Transduction Targeted Ther., 2020, 5, 101 CrossRef CAS PubMed.
- Y. Dong, D. J. Siegwart and D. G. Anderson, Adv. Drug Delivery Rev., 2019, 144, 133–147 CrossRef CAS PubMed.
- K. Gao and L. Huang, Mol. Pharm., 2009, 6, 651–658 CrossRef CAS PubMed.
- S. Uchida, F. Perche, C. Pichon and H. Cabral, Mol. Pharm., 2020, 17, 3654–3684 CrossRef CAS PubMed.
- H. J. Kim, A. Kim, K. Miyata and K. Kataoka, Adv. Drug Delivery Rev., 2016, 104, 61–77 CrossRef CAS PubMed.
- W. Yang, L. Mixich, E. Boonstra and H. Cabral, Adv. Healthcare Mater., 2023, 12, 2202688 CrossRef CAS PubMed.
- H. Cabral, K. Miyata, K. Osada and K. Kataoka, Chem. Rev., 2018, 118, 6844–6892 CrossRef CAS PubMed.
- H. Wang, S. Zhang, J. Lv and Y. Cheng, View, 2021, 2, 20200026 CrossRef CAS.
- T. Miyazaki, S. Uchida, S. Nagatoishi, K. Koji, T. Hong, S. Fukushima, K. Tsumoto, K. Ishihara, K. Kataoka and H. Cabral, Adv. Healthcare Mater., 2020, 9, 2000538 CrossRef CAS PubMed.
- W. Yang, T. Miyazaki, P. Chen, T. Hong, M. Naito, Y. Miyahara, A. Matsumoto, K. Kataoka, K. Miyata and H. Cabral, Sci. Technol. Adv. Mater., 2021, 22, 850–863 CrossRef CAS PubMed.
- J. Liu, H. Cabral and P. Mi, Adv. Drug Delivery Rev., 2024, 115239 CrossRef CAS PubMed.
- K. Zhou, P. Kos, Y. Yan, H. Xiong, Y. L. Min, K. A. Kinghorn, J. T. Minnig, J. B. Miller and D. J. Siegwart, Chem. Commun., 2016, 52, 12155–12158 RSC.
- X. Huang, G. Wu, C. Liu, X. Hua, Z. Tang, Y. Xiao, W. Chen, J. Zhou, N. Kong, P. Huang, J. Shi and W. Tao, Nano Lett., 2021, 22, 9706–9714 CrossRef PubMed.
- W. Chen, Y. Yuan, D. Cheng, J. Chen, L. Wang and X. Shuai, Small, 2014, 10, 2678–2687 CrossRef CAS PubMed.
- D. Cheng, N. Cao, J. Chen, X. Yu and X. Shuai, Biomaterials, 2012, 33, 1170–1179 CrossRef CAS PubMed.
- M. R. Rajeswari, T. Montenay-Garestier and C. Helene, Biochemistry, 1987, 26, 6825–6831 CrossRef CAS PubMed.
- M. R. Rajeswari, H. S. Bose, S. Kukreti, A. Gupta, V. S. Chauhan and K. B. Roy, Biochemistry, 1992, 31, 6237–6241 CrossRef CAS PubMed.
- M. R. Rajeswari, J. Biomol. Struct. Dyn., 1996, 14, 25–30 CrossRef CAS PubMed.
- W. Yang, T. Miyazaki, Y. Nakagawa, E. Boonstra, K. Masuda, Y. Nakashima, P. Chen, L. Mixich, K. Barthelmes, A. Matsumoto, P. Mi, S. Uchida and H. Cabral, Sci. Technol. Adv. Mater., 2023, 24, 2170164 CrossRef PubMed.
- L. Mixich, E. Boonstra, K. Masuda, S. W. Li, Y. Nakashima, F. Meng, M. Sakata, T. Goda, S. Uchida and H. Cabral, Biomacromolecules, 2024, 25, 1058–1067 CrossRef CAS PubMed.
- S. Watanabe, K. Hayashi, K. Toh, H. J. Kim, X. Liu, H. Chaya, S. Fukushima, K. Katsushima, Y. Kondo, S. Uchida, S. Ogura, T. Nomoto, H. Takemoto, H. Cabral, H. Kinoh, H. Y. Tanaka, M. T. Kano, Y. Matsumoto, H. Fukushima, S. Uchida, M. Nangaku, K. Osada, N. Nishiyama, K. Miyata and K. Kataoka, Nat. Commun., 2019, 10, 1894 CrossRef PubMed.
- T. Yanao, W. S. Koon, J. E. Marsden and I. G. Kevrekidis, J. Chem. Phys., 2007, 126, 124102 CrossRef PubMed.
- X. J. Gan, S. P. Liu, Z. F. Liu, X. L. Hu, J. Tian and J. X. Xue, Luminescence, 2013, 28, 265–269 CrossRef CAS PubMed.
- X. Gan, S. Liu, Z. Liu and X. Hu, J. Fluoresc., 2012, 22, 129–135 CrossRef CAS PubMed.
- J. E. Zuckerman, C. H. J. Choi, H. Han and M. E. Davis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 3137–3142 CrossRef CAS PubMed.
- M. Ruponen, S. Ylä-Herttuala and A. Urtti, Biochim. Biophys. Acta, Biomembr., 1999, 1415, 331–341 CrossRef CAS PubMed.
- W. B. Kauffman, S. Guha and W. C. Wimley, Nat. Commun., 2018, 9, 2568 CrossRef PubMed.
- F. Wang, K. Hu and Y. Cheng, Acta Biomater., 2016, 29, 94–102 CrossRef CAS PubMed.
- G. Creusat and G. Zuber, ChemBioChem, 2008, 9, 2787–2789 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4pm00142g |
| ‡ There authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |