Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tailoring lipid nanoparticle dimensions through manufacturing processes†
Caitlin
McMillan
 a,
Amy
Druschitz
b,
Stephen
Rumbelow
b,
Ankita
Borah
a,
Burcu
Binici
a,
Zahra
Rattray
a,
Amy
Druschitz
b,
Stephen
Rumbelow
b,
Ankita
Borah
a,
Burcu
Binici
a,
Zahra
Rattray
 a and
Yvonne
Perrie
*a
a and
Yvonne
Perrie
*a
aUniversity of Strathclyde, Glasgow, UK. E-mail: yvonne.perrie@strath.ac.uk
bCroda International Plc and Avanti Polar Lipids, Alabaster, AL, USA
First published on 23rd September 2024
Abstract
Lipid nanoparticles (LNPs), most commonly recognised for their role in COVID-19 mRNA vaccines, are important delivery vehicles for nucleic acid (mRNA, siRNA) therapies. The physicochemical attributes, such as size, nucleic acid encapsulation and electric charge, may have a significant impact on the efficacy of these medicines. In this study, adjustments to aqueous to lipid phase ratios were assessed for their impact on LNP size and other critical quality attributes (CQAs). It was observed that minor adjustments of aqueous-to-organic lipid phase ratios can be used to precisely control the size of ALC-0315-formulated LNPs. This was then used to evaluate the impact of phase ratio and corresponding size ranges on the in vitro and in vivo expression of these LNPs. In HEK293 cells, larger LNPs led to higher expression of the mRNA cargo within the LNPs, with a linear correlation between size and expression. In THP-1 cells this preference for larger LNPs was observed up to 120 d.nm after which there was a fall in expression. In BALB/c mice, however, LNPs at the lowest phase ratio tested, >120 d.nm, showed reduced expression compared to those of range 60–120 d.nm, within which there was no significant difference between sizes. These results suggest a robustness of LNP expression up to 120 d.nm, larger than those <100 d.nm conventionally used in medicine.
1. Introduction
Lipid nanoparticles (LNPs) lie within the family of lipid-based nanoparticles, which also includes liposomes, and other nanoscale lipid complexes. What differentiates LNPs from other lipid-based nanoparticle formats is the presence of a solid core created from nucleic acid complexed to a permanent or ionisable cationic lipid. Thus, LNPs are the vector of choice for the delivery of RNA-based therapeutics and vaccines.1–3 The COVID-19 pandemic placed LNPs in the spotlight, hugely increasing interest and recognition of their potential in drug and vaccine delivery, which is evidenced by a doubling of published literature on LNPs in the last 5 years.4Optimising LNP manufacture has emerged as valuable research, especially concerning production speeds, scalability, stability and efficacy of the end product.5,6 The growth of this field is driven by challenges faced during the rapid mass production of LNP-based mRNA vaccines during the pandemic. One of the biggest challenges was producing formulations rapidly to meet demand. Furthermore, manufacturing formulation parameters have been widely explored in the field of liposomes,7,8 so translating this research to LNPs is valuable. In addition to optimisation of the manufacturing process, testing the robustness of parameters used is vital, as this aids in understanding the formulation, as well as defining the limits within formulations, and expands the knowledge base in nanomedicine formulation strategy. For example, there is evidence that size impacts LNP immunogenicity and mRNA expression, impacting therapeutic efficacy.3,9,10 However, the impact varies based on the indication and pharmacokinetics; while larger LNPs (>100 d.nm) may have higher mRNA expression,10 there is evidence that smaller LNPs (<100 d nm) are more readily absorbed from the injection site.9
Microfluidics is a precise, reproducible, and scalable method for manufacturing LNPs. It has easily adjustable parameters, which can be precisely controlled with relative ease in comparison with other methods of LNP manufacturing.11 Parameters, such as mixing speed and flow-rate ratio, can significantly impact the critical quality attributes of the produced LNPs. The flow-rate ratio is equivalent to the ratio of nucleic acid-containing aqueous phase to organic phase dissolved in ethanol. Previous work has shown that higher proportions of organic phase and ethanol increase particle size in liposomes and some formulations of LNPs.3,7,11,12 In this study we assess different formulations of LNPs and correlate their in vitro and in vivo responses, with a specific focus on the LNP mRNA expression.
The aim of this study was to assess whether the phase ratio to size relationship translates to two clinically used LNP formulations, i.e. those LNPs formulated with ALC-0315 and SM-102 as the ionisable lipid, and evaluate the impact of phase ratio and LNP particle size on in vitro and in vivo expression.
2. Materials and methods
2.1. Materials
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) was obtained from Lipoid (Ludwigshafen, Germany). 1,2-Dioleoyl-3-trimethylammoniumpropane (chloride salt) (DOTAP), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG 2000) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Cholesterol, citric acid, sodium citrate tribasic dehydrate, polyadenylic acid (PolyA), 6-(p-toluidino)-2-naphthalenesulfonic acid sodium salt, sodium phosphate monobasic (NaH2PO4·H2O), and sodium phosphate dibasic (anhydrous) (Na2HPO4) were acquired from Sigma-Aldrich (St Louis, MO, USA). Phosphate-buffered saline tablets (PBS pH 7.4) were acquired from Oxoid Ltd (Basingstoke, UK). Tris base and ethanol (EtOH) were obtained from Fisher Scientific (Loughborough, UK). Methanol and propan-2-ol (isopropanol, IPA) were purchased from VWR Chemicals (Lutterworth, UK). The ionisable lipids [(4-hydroxybutyl)azanediyl]di(hexane-6,1-diyl) bis(2-hexyldecanoate) (ALC-0315) and 9-heptadecanyl 8-((2-hydroxyethyl)[6-oxo-6-(undecyloxy)-hexyl]amino)octanoate (SM-102), and the PEG-lipid 2-[(polyethylene glycol)-2000]-N,N-ditetradecylacetamide, 1,2-distearoyl-sn-glycero-3-phosphocholine (ALC-0159), were purchased from BroadPharm (San Diego, CA, USA). Minimum Essential Medium, Trypsin-EDTA (0.05%), PBS, pH 7.4 solution, Quant-it™ RiboGreen RNA Assay Kit, RiboGreen RNA Reagent, and RediPlate™ 96 RiboGreen™ RNA Quantitation Kit were purchased from Fisher Scientific UK Ltd (Renfrew, UK). EZ Cap™ Firefly Luciferase mRNA was purchased from ApexBio Technology (Houston, TX, USA). mGreenLantern was provided by CPI via the Intracellular Drug Delivery Centre (IDDC) research programme. ONE-Glo™ Luciferase Assay System was acquired from Promega UK (Southampton, UK). All solvents and other chemicals were of analytical grade, and Milli-Q-water was provided by an in-house system.2.2. Aqueous and organic phase preparation
The aqueous phase containing mRNA and the organic phase were prepared separately before microfluidics. The aqueous phase was prepared using 50 mM citrate buffer (pH 4), dissolving PolyA, or Firefly Luciferase (FLuc) mRNA for in vitro and in vivo experiments, at mRNA concentrations required to maintain a nitrogen-to-phosphate (N/P) ratio of 6, the molar ratio of ionisable lipid containing positively charged amines (N) to the mRNA containing negatively charged phosphates (P). Therefore, the concentrations were adjusted for different aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase ratios and when different lipids or mRNA were used, to maintain this N/P ratio.
organic phase ratios and when different lipids or mRNA were used, to maintain this N/P ratio.
The lipid molar ratio of the LNPs was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5% (structural lipid
1.5% (structural lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ionizable lipid
ionizable lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEGylated lipid). This led to the following combinations utilised in the experiments: (1) DSPC
PEGylated lipid). This led to the following combinations utilised in the experiments: (1) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-0315
ALC-0315![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000, (2) DSPC
DMG-PEG2000, (2) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-0315
ALC-0315![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-PEG and (3) DSPC
ALC-PEG and (3) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SM-102
SM-102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000. The lipid stocks were prepared and stored at concentrations of 5–20 mg mL−1, dissolving each lipid in ethanol. Unless otherwise stated, the organic phase injected into the microfluidic device has a concentration of 5 mg mL−1.
DMG-PEG2000. The lipid stocks were prepared and stored at concentrations of 5–20 mg mL−1, dissolving each lipid in ethanol. Unless otherwise stated, the organic phase injected into the microfluidic device has a concentration of 5 mg mL−1.
2.3. Manufacture of the LNPs using microfluidics
The NanoAssemblr benchtop (Precision Nanosystems Inc., Vancouver, Canada) was used in all experiments in this paper. The ethanol-based organic phase and the citrate buffer-based aqueous phase containing mRNA in the form of PolyA or FLuc mRNA were fed through syringes into a microfluidics chip with a herringbone mixer design. The phase ratio (aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase) was adjusted using the computer interface of the machine, varying between the limits of 1
organic phase) was adjusted using the computer interface of the machine, varying between the limits of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–3
1–3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 aqueous
1 aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase, respectively.
organic phase, respectively.
2.4. Purification methods
For purification by dialysis buffer exchange, the LNP sample in the original ethanol/citrate buffer was exchanged into 200 mL of PBS (pH 7.4) for every mL of sample. Samples manufactured with PolyA as the internal nucleic acid and mRNA samples for in vitro experiments were dialysed for 1 hour at ambient temperature, and for 24 hours at 4 °C for in vivo studies. Spin columns of 100 kDa MWCO were used for centrifugation and purification of LNPs alongside dialysis in optimisation experiments. LNPs were diluted 40× in PBS. The samples were spun at 2000g at 4 °C for 20–40 minutes (time varying with formulation) until the sample reached the original pre-purification volume.2.5. Measuring critical quality attributes (CQAs)
Nanoparticle tracking analysis was performed on samples post-purification using the Malvern Panalytical NanoSight Pro (Malvern Panalytical, Malvern, Worcestershire, UK) and the NS Xplorer software was used for data analysis. Nanoparticle size distribution and estimated particle concentration were measured using a 488 nm laser block and videos were captured using a high sensitivity sCMOS camera with a light-scatter filter. Samples were diluted to 0.05–0.1 μg mL−1 and analysed under constant flow (1–1.5 μL min−1) using a syringe driver, with the temperature set to 20 °C. Five 11.5 s videos were captured for each sample and optimised using the automated optimal settings employed by the software. Between 2700 and 5100 particle trajectories (valid tracks) were analysed depending on the sample to gather the NTA data. These values were 2774, 3623, 4976 and 5056 for the aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase ratios of 1.3
organic phase ratios of 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1.5
1, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively, which equate to the total number of particles measured per sample (Table S1†).
1, respectively, which equate to the total number of particles measured per sample (Table S1†).
When analysing the results, the total RNA for each sample from the Triton wells and free (unentrapped) RNA from the TE-only wells were obtained. The following equations were used to calculate encapsulation efficiency (EE%) and mRNA recovery as follows:
| EE% = 100 × (CT − CF)/CT | (1) |
| mRNA recovery (%) = 100 × CT/CI | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in media and cultured for 72 hours, after which LNPs were added. LNPs were formulated as above, with EZ Cap™ Firefly Luciferase mRNA as the internal RNA. LNP formulations were added to the cells, replacing the growth media, and diluted in media at total mRNA concentrations of 2 μg mL−1, 1 μg mL−1, 0.5 μg mL−1 and 0.25 μg mL−1, using three wells per concentration and leaving three wells free from LNPs (control wells). Cells were incubated with LNPs for 24 hours prior to the addition of the luciferase reagent. The luciferase reagent, prepared as per the manufacturer's instructions, was added to each well of 100 μL of media and cells containing LNPs from the previous day, creating a total volume of 200 μL, mixed thoroughly and left to incubate for 3 minutes. The luminescence of the plates was read using the GloMax plate reader (Promega, UK).
000 cells per well in media and cultured for 72 hours, after which LNPs were added. LNPs were formulated as above, with EZ Cap™ Firefly Luciferase mRNA as the internal RNA. LNP formulations were added to the cells, replacing the growth media, and diluted in media at total mRNA concentrations of 2 μg mL−1, 1 μg mL−1, 0.5 μg mL−1 and 0.25 μg mL−1, using three wells per concentration and leaving three wells free from LNPs (control wells). Cells were incubated with LNPs for 24 hours prior to the addition of the luciferase reagent. The luciferase reagent, prepared as per the manufacturer's instructions, was added to each well of 100 μL of media and cells containing LNPs from the previous day, creating a total volume of 200 μL, mixed thoroughly and left to incubate for 3 minutes. The luminescence of the plates was read using the GloMax plate reader (Promega, UK).
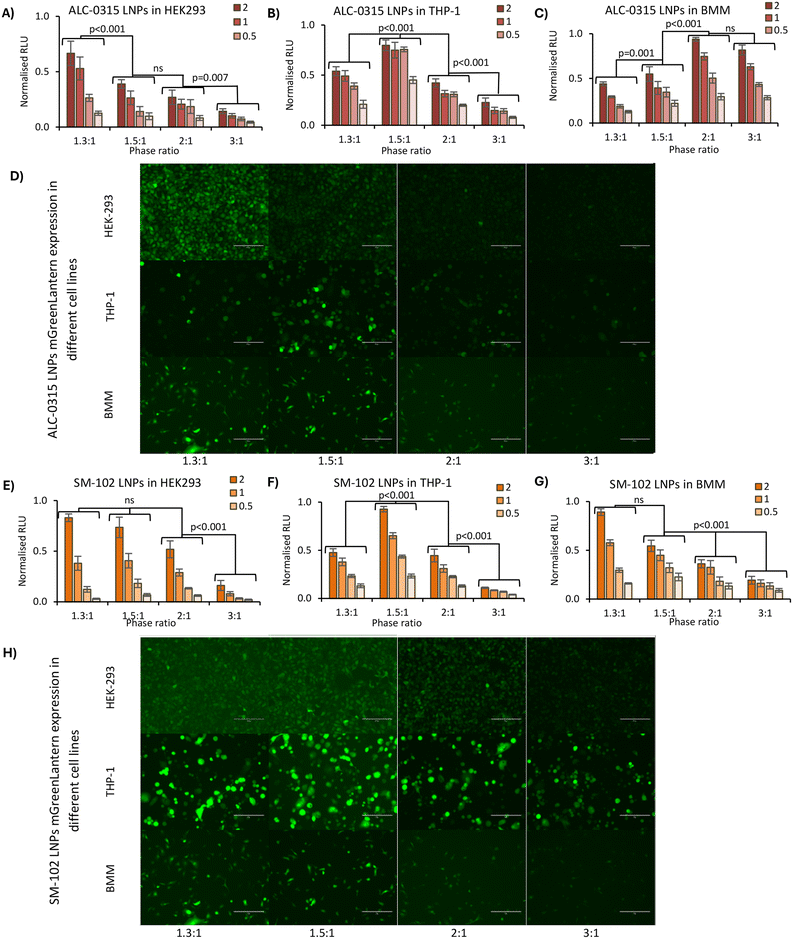
LNP FLuc mGreenLantern expression was analysed in HEK293, THP-1 and BMM (bone marrow-derived macrophages) cells using Green Lantern mRNA for taking fluorescence images of LNP expression. Cells were cultured as above in 6-well plates and transfected with LNPs at a total mRNA concentration of 2 μg mL−1. Cells were incubated with LNPs for 24 hours prior to imaging using the GFP filter on the EVOS M5000 microscope (Thermo Fisher Scientific, UK). The total fluorescence of these images was quantified using ImageJ, using 5 images per sample.
2.6. Statistical analysis
The mean and standard deviation were calculated for all experiments, performed in independent triplicate batches unless otherwise stated, with each batch manufactured on a different day to account for inter-day variation. For hydrodynamic size, polydispersity, and zeta potential, three measurements were taken for each batch and purification status; therefore these groups for statistical analysis were n = 9. For statistical comparison of groups, one way (for one independent variable) or two/three-way (for two and three independent variables, respectively) analysis of variance (ANOVA) was performed with Tukey's HSD post-hoc for pairwise comparisons. Unpaired t-tests were used when comparing just two groups, and paired t-tests were used for comparing the same group/batch over time or treatment. In the graphs, statistical significance is indicated by individual p values for p < 0.05, whereas (ns) denotes no statistical significance.3. Results and discussion
3.1. The effect of altering manufacturing parameters on the physicochemical attributes of lipid nanoparticles (LNPs)
To confirm that one hour of dialysis was sufficient for LNP sample purification, samples manufactured at an aqueous-to-organic phase ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were purified by both dialysis and spin columns (Table 1). This showed that although spin columns slightly reduced the size of these LNPs, the other CQAs were unaffected by the differing purification methods. It was necessary to use dialysis for the purification of LNPs in the upper size range. Based on these results, dialysis was used as the purification method for all subsequent samples and extended to 24 hours for samples in vivo to optimise buffer exchange.
1 were purified by both dialysis and spin columns (Table 1). This showed that although spin columns slightly reduced the size of these LNPs, the other CQAs were unaffected by the differing purification methods. It was necessary to use dialysis for the purification of LNPs in the upper size range. Based on these results, dialysis was used as the purification method for all subsequent samples and extended to 24 hours for samples in vivo to optimise buffer exchange.
| Purification status | Size (nm) | PDI | Zeta potential (mV) | EE (%) | mRNA recovery (%) |
|---|---|---|---|---|---|
| Before purification | 89 ± 13 | 0.15 ± 0.04 | n/a | n/a | n/a |
| Dialysis | 88 ± 10 | 0.18 ± 0.03 | −1.4 ± 0.3 | 91 ± 2 | 72 ± 7 |
| Spin column | 72 ± 5* | 0.12 ± 0.04 | −0.4 ± 1.3 | 95 ± 1 | 86 ± 6 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) organic phase ratio across different LNP formulations.
The impact of the proportion of aqueous phase to organic phase on the LNP physicochemical attributes, i.e. size, polydispersity, zeta potential and nucleic acid recovery, was evaluated across three different LNP formulations. These were (1) DSPC
organic phase ratio across different LNP formulations.
The impact of the proportion of aqueous phase to organic phase on the LNP physicochemical attributes, i.e. size, polydispersity, zeta potential and nucleic acid recovery, was evaluated across three different LNP formulations. These were (1) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-0315
ALC-0315![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000, (2) DSPC
DMG-PEG2000, (2) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-0315
ALC-0315![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ALC-0159 and (3) DSPC
ALC-0159 and (3) DSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SM-102
SM-102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMG-PEG2000, each at a molar ratio of 10
DMG-PEG2000, each at a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.5
38.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 for each of the lipids, respectively. LNPs composed of ALC-0315 as the ionisable lipid and DMG-PEG2000 as the PEGylated lipid, were initially used. Seven different aqueous-to-organic phase ratios were tested, and the results showed a trend of decreasing LNP size as the proportion of aqueous phase to organic phase increased (Fig. 1B). For samples manufactured at ratios of 1
1.5 for each of the lipids, respectively. LNPs composed of ALC-0315 as the ionisable lipid and DMG-PEG2000 as the PEGylated lipid, were initially used. Seven different aqueous-to-organic phase ratios were tested, and the results showed a trend of decreasing LNP size as the proportion of aqueous phase to organic phase increased (Fig. 1B). For samples manufactured at ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.2
1 and 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the polydispersity was high, with values of 0.58 and 0.76, suggesting a highly heterogeneous population. These more heterogeneous LNP batches also tended to show more variability in terms of zeta potential, encapsulation efficiency (EE%) and mRNA recovery (%) (Table 2). Due to this, the mixing ratios of 1
1, the polydispersity was high, with values of 0.58 and 0.76, suggesting a highly heterogeneous population. These more heterogeneous LNP batches also tended to show more variability in terms of zeta potential, encapsulation efficiency (EE%) and mRNA recovery (%) (Table 2). Due to this, the mixing ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.2
1 and 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were not further investigated. The ratio of 1.8
1 were not further investigated. The ratio of 1.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was also not included in future experiments for simplification purposes.
1 was also not included in future experiments for simplification purposes.
| DSPC:Chol: | Aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase ratio organic phase ratio |
Zeta potential (mV) | EE% | mRNA recovery (%) |
|---|---|---|---|---|
| ALC-0315:DMG-PEG2000, PolyA | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
8.3 ± 8.3 | 70 ± 14 | 37 ± 17 |
1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6.0 ± 6.8 | 61 ± 19 | 68 ± 4 | |
1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
4.6 ± 5.0 | 75 ± 23 | 65 ± 10 | |
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1.8 ± 3.8 | 96 ± 1 | 68 ± 10 | |
1.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.8 ± 3.0 | 96 ± 1 | 79 ± 10 | |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−1.7 ± 1.3 | 94 ± 1 | 93 ± 6 | |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−2.6 ± 1.2 | 98 ± 1 | 91 ± 13 | |
| ALC-0315:ALC-0159, PolyA | 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.4 ± 6.4 | 94 ± 2 | 97 ± 4 |
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
4.0 ± 3.9 | 90 ± 7 | 81 ± 2 | |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1.4 ± 1.7 | 89 ± 4 | 75 ± 8 | |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−1.4 ± 0.3 | 91 ± 2 | 72 ± 7 | |
| ALC-0315:ALC-0159, mRNA | 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−1.3 ± 2.2 | 61 ± 18 | 82 ± 4 |
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−0.2 ± 2.1 | 79 ± 7 | 86 ± 7 | |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−1.5 ± 1.0 | 89 ± 2 | 83 ± 12 | |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−0.9 ± 1.4 | 91 ± 2 | 94 ± 13 | |
| SM-102:DMG-PEG2000, PolyA | 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
5.6 ± 0.8 | 97 ± 2 | 78 ± 5 |
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
3.7 ± 3.0 | 98 ± 1 | 89 ± 5 | |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
4.6 ± 1.5 | 99 ± 1 | 95 ± 8 | |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
3.4 ± 1.2 | 99 ± 1 | 87 ± 0 | |
| SM-102:DMG-PEG2000, mRNA | 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
−0.9 ± 12 | 95 ± 4 | 93 ± 11 |
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
5.2 ± 1.2 | 94 ± 5 | 82 ± 11 | |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
4.8 ± 1.4 | 96 ± 3 | 84 ± 11 | |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
3.4 ± 1.5 | 96 ± 3 | 100 ± 20 |
The second LNP formulation tested used ALC-0315 in combination with ALC-0159 as the PEGylated lipid, similar to the Pfizer/BioNTech Comirnaty® mRNA vaccine lipid composition. The aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase ratios tested were 1.3
organic phase ratios tested were 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1.5
1, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The same trend of size decreasing as the aqueous phase increased was observed (Fig. 1A). The PDI remained low through all phase ratios of approximately <0.2, and the zeta potential remained neutral (−5 to 5 mV) (Table 2).
1. The same trend of size decreasing as the aqueous phase increased was observed (Fig. 1A). The PDI remained low through all phase ratios of approximately <0.2, and the zeta potential remained neutral (−5 to 5 mV) (Table 2).
This pattern was repeated with LNPs prepared using SM-102 and DMG-PEG2000 (a mimic of the Moderna Spikevax® lipid composition). These formulations were measured using both PolyA and mRNA as the nucleic acid cargo, both of which gave the same phase ratio–size relationship. A two-way ANOVA with Tukey's post hoc analysis found significant differences in size output for all phase ratios, with average sizes being approximately 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = 70 d.nm, 2
1 = 70 d.nm, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = 80 d.nm, 1.5
1 = 80 d.nm, 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = 110 d.nm and 1.3
1 = 110 d.nm and 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = 145 d.nm (aqueous
1 = 145 d.nm (aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase) (Fig. 1A). Within the phase ratios tested, the PDI remained low (<0.2), the zeta potential was near neutral and EE% and mRNA recovery% were >60% (Table 2). This correlation between phase ratio and LNP particle size results from the rate of polarity change induced by ethanol dilution with the aqueous phase. Because lipids are amphiphilic, as ethanol is diluted (and polarity increases), the lipids self-assemble to form LNPs. In higher proportions of aqueous phase to ethanol concentrations, the ethanol is diluted faster, the lipids have less opportunity to aggregate, and hence, this drives the formation of smaller LNPs.14
organic phase) (Fig. 1A). Within the phase ratios tested, the PDI remained low (<0.2), the zeta potential was near neutral and EE% and mRNA recovery% were >60% (Table 2). This correlation between phase ratio and LNP particle size results from the rate of polarity change induced by ethanol dilution with the aqueous phase. Because lipids are amphiphilic, as ethanol is diluted (and polarity increases), the lipids self-assemble to form LNPs. In higher proportions of aqueous phase to ethanol concentrations, the ethanol is diluted faster, the lipids have less opportunity to aggregate, and hence, this drives the formation of smaller LNPs.14
The size of different phase ratios was also measured using nanoparticle tracking analysis (Fig. 2). This technique allowed for the number of particles to be estimated in each sample (Fig. 2A), showing per 1 mg mL−1 lipid concentration there were more particles for smaller LNPs and fewer particles of the larger LNPs, which makes sense, due to the same amount of material being present in each sample. All of the samples tested had a similar span of between 0.9 and 1.1, indicating the same particle size distribution in the formulations tested. Interestingly, the same number of particles was found in the 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.3
1 and 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 samples, despite a 20 nm difference in their z-average size, suggesting that this difference in particle size is not solely dependent on the number of lipid molecules within each LNP. Comparing the intensity-to-frequency plots (Fig. 2B–E) showed a similar size distribution given by both techniques. The smaller particles (2
1 samples, despite a 20 nm difference in their z-average size, suggesting that this difference in particle size is not solely dependent on the number of lipid molecules within each LNP. Comparing the intensity-to-frequency plots (Fig. 2B–E) showed a similar size distribution given by both techniques. The smaller particles (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 90 d.nm and 70 d.nm) map (Fig. 2D and E), whereas the larger particles (1.3
1, 90 d.nm and 70 d.nm) map (Fig. 2D and E), whereas the larger particles (1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.5
1 and 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 140 d.nm and 120 d.nm) show a shift in the DLS intensity readings towards higher particle size. This is likely due to larger particles contributing more to the overall intensity in scattered light for DLS measurements,15,16 resulting in a greater intensity value in the intensity distributions shown. Overall, these data confirm the ability to precisely control LNP size through adjustments in the aqueous-to-organic phase ratio for both ALC-0315 and SM-102 ionisable lipid-containing LNPs, as previously seen with liposomes and LNPs.3,7,9,10,17
1, 140 d.nm and 120 d.nm) show a shift in the DLS intensity readings towards higher particle size. This is likely due to larger particles contributing more to the overall intensity in scattered light for DLS measurements,15,16 resulting in a greater intensity value in the intensity distributions shown. Overall, these data confirm the ability to precisely control LNP size through adjustments in the aqueous-to-organic phase ratio for both ALC-0315 and SM-102 ionisable lipid-containing LNPs, as previously seen with liposomes and LNPs.3,7,9,10,17
To further investigate LNPs made at different sizes, the corresponding pKa values for these LNPs were measured using the TNS assay (Fig. 3) in a method adapted from previous studies.11,13 The pKa values of LNPs generally fall within the range of 6–7.18,19 Using the Henderson–Hasselbalch equation, this signifies that under pH conditions equal to those of the pKa, the ionisable lipid is 50% protonated, while at one pH unit below the pKa it is 90% protonated, and at two units it is 99% protonated. This indicates that at a manufacturing pH of 4, the ionisable lipid is almost entirely cationic, and therefore, this has been used as a reliable manufacturing pH for LNPs in research and commercially.20,21 The TNS fluorescence is a direct indicator of the % protonation of the ionizable lipid. Upon fixing the pH at which the TNS emits 50% of the maximum fluorescence in the presence of the ALC-0315 LNPs, the pKa value can be estimated. For both small (phase ratio = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and large (phase ratio = 1.3
1) and large (phase ratio = 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LNPs, the pKa was approximately 6.1, in agreement with prior literature18,19 and confirms that the lipid is sufficiently protonated under manufacturing conditions (pH 4) for full mRNA encapsulation. We also observe that the manufacturing phase ratio and size of the LNPs have no impact on the pKa and so this variable should not impact the mRNA encapsulation and expressing potential of the LNPs. To examine any potential physical differences between the different-sized LNPs, cryo-TEM was performed to image each phase ratio of the LNPs (Fig. 4), and this presented similar filled-circular structures for the first three smaller ratios. However, at the 1.3
1) LNPs, the pKa was approximately 6.1, in agreement with prior literature18,19 and confirms that the lipid is sufficiently protonated under manufacturing conditions (pH 4) for full mRNA encapsulation. We also observe that the manufacturing phase ratio and size of the LNPs have no impact on the pKa and so this variable should not impact the mRNA encapsulation and expressing potential of the LNPs. To examine any potential physical differences between the different-sized LNPs, cryo-TEM was performed to image each phase ratio of the LNPs (Fig. 4), and this presented similar filled-circular structures for the first three smaller ratios. However, at the 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 phase ratio, a double-lamellar LNP was found to be the most-commonly presented structure. This shows that there are structural differences between this size of LNP and that of the smaller LNPs, and that these are perhaps necessary to form a stable lipid nanoparticle of this size. Furthermore, as stated, during the formation of these LNPs where the ethanol concentration is higher, there is more opportunity for lipids to aggregate and form larger LNPs. This may result in different morphological structures with the potential for phase separation of the dense mRNA and ionisable lipid core, leaving the other lipids to form extra layers, similar to what has been observed in the formation of bleb-rich LNPs.22 Furthermore, this double-layer morphology has previously been observed in larger LNPs and liposomes.3,23
1 phase ratio, a double-lamellar LNP was found to be the most-commonly presented structure. This shows that there are structural differences between this size of LNP and that of the smaller LNPs, and that these are perhaps necessary to form a stable lipid nanoparticle of this size. Furthermore, as stated, during the formation of these LNPs where the ethanol concentration is higher, there is more opportunity for lipids to aggregate and form larger LNPs. This may result in different morphological structures with the potential for phase separation of the dense mRNA and ionisable lipid core, leaving the other lipids to form extra layers, similar to what has been observed in the formation of bleb-rich LNPs.22 Furthermore, this double-layer morphology has previously been observed in larger LNPs and liposomes.3,23
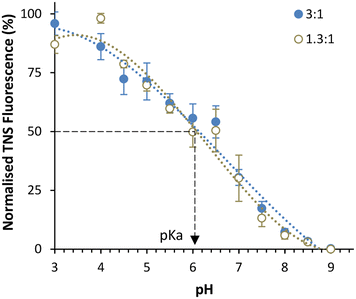 | ||
Fig. 3 Measuring the pKa of ALC-0315 ionisable lipid LNPs. The normalised fluorescence values of TNS when incubated with ALC-0315 LNPs titrated in buffers over a pH range of 3–9. Citrate buffer was used for pH 3–6, sodium phosphate buffer was used for pH 6.5–8, and Tris buffer was used for pH 8.5–9. The readings were normalised per each maximum value for each LNP batch. Each data set is representative of 3 batches of LNPs manufactured at an aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid phase ratio of 3 lipid phase ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (blue solid circles) or 1.3 1 (blue solid circles) or 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (grey open circles). The pKa value was found by taking the pH value at 50% of the maximum fluorescence. To fit a sigmoidal curve to the data, polynomial lines of best fit to the order of 3 were used. The LNPs were manufactured at an FRR of 3 1 (grey open circles). The pKa value was found by taking the pH value at 50% of the maximum fluorescence. To fit a sigmoidal curve to the data, polynomial lines of best fit to the order of 3 were used. The LNPs were manufactured at an FRR of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with an N/P ratio of 6 from 5 mg mL−1 lipid stock, and pH 7.4 PBS was used as the exchange buffer.8 1 with an N/P ratio of 6 from 5 mg mL−1 lipid stock, and pH 7.4 PBS was used as the exchange buffer.8 | ||
3.2. The impact of aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) organic phase ratio and LNP size on in vitro and in vivo expression
organic phase ratio and LNP size on in vitro and in vivo expression
As the aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic phase ratio reliably and reproducibly enabled the control of LNP size, this was then used to infer the impact of LNP size on expression, both in vitro and in vivo. LNPs encapsulating firefly luciferase mRNA were prepared, which when expressed catalyses luciferin into a bioluminescent product. The LNPs were added to HEK293, THP-1 and BMM cells, as three distinct cell lines at four standard concentrations in the 0.25–2 mg mL−1 test concentration range.24–26We observed that in HEK-293 cells the largest LNPs resulted in a 4 times higher expression than the smallest LNPs (Fig. 5A and E), with a pattern of gradually decreasing comparative expression as the phase ratio increases (and size decreases). This was true for both ALC-0315 and SM-102 LNPs adjusted to four different concentrations of mRNA. This same trend of higher expression for larger LNPs versus standard-size LNPs has previously been observed in LNPs manufactured with Dlin-MC3-DMA as the ionizable lipid in NCI-H358 cells.10 Larger LNPs were more likely to be internalised by cells via macropinocytosis instead of receptor-dependent endocytosis and it was discussed how larger particles encapsulate higher mRNA content, which is supported by our findings (Fig. 2A), showing fewer particles with larger-sized LNPs, whilst containing the same amount of mRNA within the sample. A reason why fewer particles with higher mRNA content may be more efficient for mRNA delivery and expression than more small particles containing less mRNA may be due to limited cell uptake. Though the uptake of labelled mRNA was found to be the same for each LNP size, expression differences were found to be independent of internalization, perhaps owing to the differing effects of LNP composition and internalisation mechanisms on downstream intracellular processes. In THP-1 cells, a similar pattern was observed, except for slightly lower expression in the 1.3
organic phase ratio reliably and reproducibly enabled the control of LNP size, this was then used to infer the impact of LNP size on expression, both in vitro and in vivo. LNPs encapsulating firefly luciferase mRNA were prepared, which when expressed catalyses luciferin into a bioluminescent product. The LNPs were added to HEK293, THP-1 and BMM cells, as three distinct cell lines at four standard concentrations in the 0.25–2 mg mL−1 test concentration range.24–26We observed that in HEK-293 cells the largest LNPs resulted in a 4 times higher expression than the smallest LNPs (Fig. 5A and E), with a pattern of gradually decreasing comparative expression as the phase ratio increases (and size decreases). This was true for both ALC-0315 and SM-102 LNPs adjusted to four different concentrations of mRNA. This same trend of higher expression for larger LNPs versus standard-size LNPs has previously been observed in LNPs manufactured with Dlin-MC3-DMA as the ionizable lipid in NCI-H358 cells.10 Larger LNPs were more likely to be internalised by cells via macropinocytosis instead of receptor-dependent endocytosis and it was discussed how larger particles encapsulate higher mRNA content, which is supported by our findings (Fig. 2A), showing fewer particles with larger-sized LNPs, whilst containing the same amount of mRNA within the sample. A reason why fewer particles with higher mRNA content may be more efficient for mRNA delivery and expression than more small particles containing less mRNA may be due to limited cell uptake. Though the uptake of labelled mRNA was found to be the same for each LNP size, expression differences were found to be independent of internalization, perhaps owing to the differing effects of LNP composition and internalisation mechanisms on downstream intracellular processes. In THP-1 cells, a similar pattern was observed, except for slightly lower expression in the 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
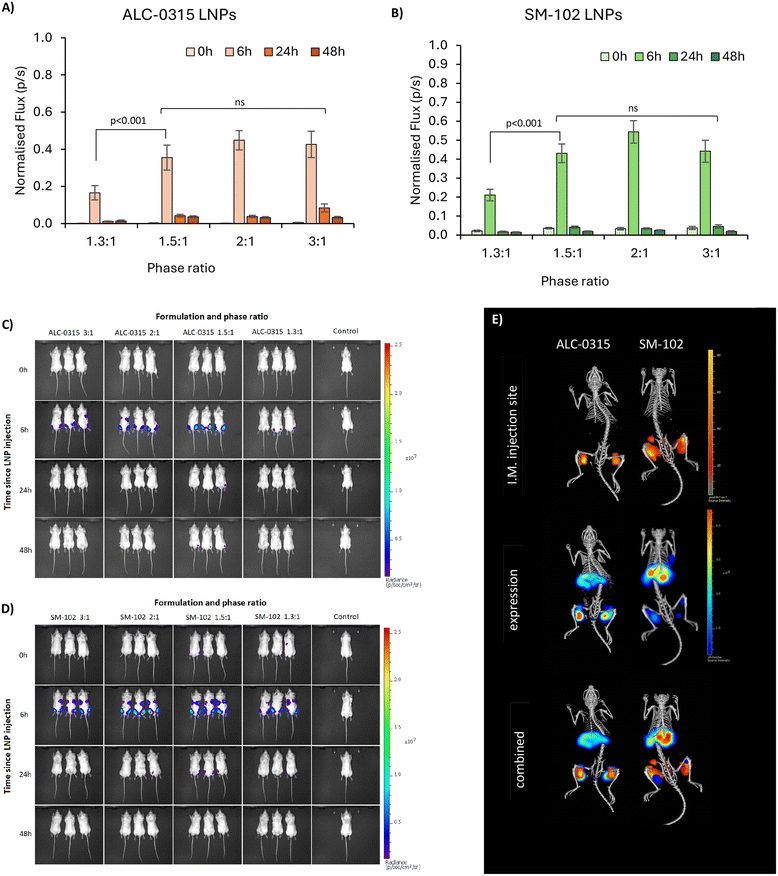
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 phase ratio samples (Fig. 5B and F), which may be due to less uptake of these larger LNPs into this cell type. For BMM cells, a differing trend was seen between ALC-0315 and SM-102 manufactured LNPs (Fig. 5C and G); the SM-102 samples exhibited the same preference for larger particles as shown in the other cell lines, but the ALC-0315 data showed an opposite trend. It is unknown as to why the expression pattern is different in this cell line, specifically for one ionisable lipid, where the LNPs manufactured with different ionisable lipids have previously shown the same pattern. This shows that the trend of larger LNPs expressing more is true for most, but not all formulations of LNPs in the tested cell lines. To visually observe differences in mRNA expression in these three cell lines, LNPs were manufactured with mGreenLantern mRNA for capturing images of this green fluorescent protein when expressed in cells (Fig. 5D and H); these showed the same expression patterns as the FLuc mRNA LNPs, with the exception being the ALC-0315 LNPs in BMM again (quantified in Fig. S1†). This is observed in Fig. 6E, showing a CT scan and 3D biodistribution and expression of ALC-0315 and SM-102 LNPs. Furthermore, contrary to most of the in vitro results, the largest LNPs had the lowest expression in vivo, while the other sizes showed no significant difference in expression, which was found to be variable (Fig. 6). These largest LNPs manufactured at a phase ratio of 1.3
1 phase ratio samples (Fig. 5B and F), which may be due to less uptake of these larger LNPs into this cell type. For BMM cells, a differing trend was seen between ALC-0315 and SM-102 manufactured LNPs (Fig. 5C and G); the SM-102 samples exhibited the same preference for larger particles as shown in the other cell lines, but the ALC-0315 data showed an opposite trend. It is unknown as to why the expression pattern is different in this cell line, specifically for one ionisable lipid, where the LNPs manufactured with different ionisable lipids have previously shown the same pattern. This shows that the trend of larger LNPs expressing more is true for most, but not all formulations of LNPs in the tested cell lines. To visually observe differences in mRNA expression in these three cell lines, LNPs were manufactured with mGreenLantern mRNA for capturing images of this green fluorescent protein when expressed in cells (Fig. 5D and H); these showed the same expression patterns as the FLuc mRNA LNPs, with the exception being the ALC-0315 LNPs in BMM again (quantified in Fig. S1†). This is observed in Fig. 6E, showing a CT scan and 3D biodistribution and expression of ALC-0315 and SM-102 LNPs. Furthermore, contrary to most of the in vitro results, the largest LNPs had the lowest expression in vivo, while the other sizes showed no significant difference in expression, which was found to be variable (Fig. 6). These largest LNPs manufactured at a phase ratio of 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 have notably different morphology with potential phase separation of the lipids and this, combined with their larger size, could be impacting on in vivo performance as a result of different biological interactions and/or stability. This lack of correlation between in vitro and in vivo results with respect to LNPs and other nanomedicines is not uncommon,27–30 as there are many impacting factors in vivo, such as biological milieu interactions,31 immune system interactions,32 circulation and movement to target site amongst others. This presents major challenges in screening LNPs and the down-selection from in vitro to in vivo studies. Furthermore, homogeneous cell cultures do not represent the heterogeneous complexity of a living organism, and the different pharmacokinetic barriers, which could impact the stability and potency of different formulations. This indicates major limitations in relying on these in vitro models to predict in vivo outcomes and highlights the importance of in vivo testing. However, there is also potential for discrepancies between preclinical trials in murine models and clinical outcomes, and therefore additional models are required to better predict clinical outcomes. Typically the optimal size for LNPs is in the 50–100 nm size range depending on the formulation,3,33 and previous studies have suggested that smaller LNPs may be better indicated for siRNA-based therapies, and larger LNPs for those that are mRNA-based, in conjunction with the molecular sizes of the nucleic acid cargo,10 so it may be that larger sizes can more efficiently pack mRNA, but smaller LNPs are more stable, creating an optimal range for LNP expression. Our data suggest that LNPs between approximately 60 and 120 d.nm have equivalent mRNA expression after intramuscular injection. This suggests that particle size is a key critical quality attribute and is sensitive to small changes in the microfluidic mixing processes; however, it is less influential on potency in small-animal studies. Indeed, this has also been shown in mRNA vaccine studies as Hassett et al.3 looked at immune responses in mice and non-human primates, where all tested sizes of SM-102 ionisable lipid LNPs from 60–150 d.nm gave consistent results. This study supports and further investigates this work by specifically looking at mRNA expression in in vitro and in vivo across the same size range in both SM-102 and ALC-0315 LNPs.
1 have notably different morphology with potential phase separation of the lipids and this, combined with their larger size, could be impacting on in vivo performance as a result of different biological interactions and/or stability. This lack of correlation between in vitro and in vivo results with respect to LNPs and other nanomedicines is not uncommon,27–30 as there are many impacting factors in vivo, such as biological milieu interactions,31 immune system interactions,32 circulation and movement to target site amongst others. This presents major challenges in screening LNPs and the down-selection from in vitro to in vivo studies. Furthermore, homogeneous cell cultures do not represent the heterogeneous complexity of a living organism, and the different pharmacokinetic barriers, which could impact the stability and potency of different formulations. This indicates major limitations in relying on these in vitro models to predict in vivo outcomes and highlights the importance of in vivo testing. However, there is also potential for discrepancies between preclinical trials in murine models and clinical outcomes, and therefore additional models are required to better predict clinical outcomes. Typically the optimal size for LNPs is in the 50–100 nm size range depending on the formulation,3,33 and previous studies have suggested that smaller LNPs may be better indicated for siRNA-based therapies, and larger LNPs for those that are mRNA-based, in conjunction with the molecular sizes of the nucleic acid cargo,10 so it may be that larger sizes can more efficiently pack mRNA, but smaller LNPs are more stable, creating an optimal range for LNP expression. Our data suggest that LNPs between approximately 60 and 120 d.nm have equivalent mRNA expression after intramuscular injection. This suggests that particle size is a key critical quality attribute and is sensitive to small changes in the microfluidic mixing processes; however, it is less influential on potency in small-animal studies. Indeed, this has also been shown in mRNA vaccine studies as Hassett et al.3 looked at immune responses in mice and non-human primates, where all tested sizes of SM-102 ionisable lipid LNPs from 60–150 d.nm gave consistent results. This study supports and further investigates this work by specifically looking at mRNA expression in in vitro and in vivo across the same size range in both SM-102 and ALC-0315 LNPs.
4. Conclusions
In this study, the impact of the aqueous![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) organic buffer phase ratio was evaluated for its ability to alter ALC-0315- and SM-102-formulated LNP critical quality attributes. We observed that the phase ratio correlated with LNP size leading to the manufacture of LNPs with differing sizes ranging from 60 to 140 nm in diameter for further evaluation. LNPs over the size of 120 d.nm showed a different morphology to smaller LNPs observed using CryoTEM, with a double-lamellar structure being the most common. Particle size is a well-recognised critical quality attribute for LNPs and critical to the quality and efficacy of an LNP product. Generally, LNPs used in the clinic are in the 50–100 nm size range and manufactured at a phase ratio of 3
organic buffer phase ratio was evaluated for its ability to alter ALC-0315- and SM-102-formulated LNP critical quality attributes. We observed that the phase ratio correlated with LNP size leading to the manufacture of LNPs with differing sizes ranging from 60 to 140 nm in diameter for further evaluation. LNPs over the size of 120 d.nm showed a different morphology to smaller LNPs observed using CryoTEM, with a double-lamellar structure being the most common. Particle size is a well-recognised critical quality attribute for LNPs and critical to the quality and efficacy of an LNP product. Generally, LNPs used in the clinic are in the 50–100 nm size range and manufactured at a phase ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The manufacture of smaller size ranges and larger LNPs can be used to infer a more detailed view of the impact LNP particle size has on expression. Our in vitro experiments in three distinct cell lines revealed that larger LNPs usually resulted in higher mRNA expression than smaller sized LNPs. This has been attributed to different uptake mechanisms and the impact on downstream intracellular processes. However, when tested in vivo, this was no longer the case. In vivo, we saw the largest LNPs give the lowest expression, and the other sizes were not significantly different from each other.
1. The manufacture of smaller size ranges and larger LNPs can be used to infer a more detailed view of the impact LNP particle size has on expression. Our in vitro experiments in three distinct cell lines revealed that larger LNPs usually resulted in higher mRNA expression than smaller sized LNPs. This has been attributed to different uptake mechanisms and the impact on downstream intracellular processes. However, when tested in vivo, this was no longer the case. In vivo, we saw the largest LNPs give the lowest expression, and the other sizes were not significantly different from each other.
Here we show that small changes in manufacturing will impact LNP particle size, but this does not necessarily translate to changes in their efficacy. Furthermore, the issue regarding the lack of correlation between in vitro and in vivo results elucidates that although in vitro studies are fundamental for assessing toxicity and baseline expression of formulations, in vivo studies are crucial when evaluating the efficacy of formulations.
Author contributions
Conceptualization, methodology, formal analysis, investigation, data curation, writing – original draft preparation, writing – reviewing and editing, visualisation, CMcM, AD, SR, AB, BB, ZR, YP; supervision, AD, SR, ZR, YP; project administration, funding acquisition, AD, SR, YP. All authors have read and agreed to the published version of the manuscript.Data availability
The supporting data set is available online at https://doi.org/10.15129/fb395621-acf1-4edc-aab0-fc5275d48bdf.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Malvern Panalytical for providing access to the Malvern Panalytical NanoSight Pro, as well as Simon Eggleston and Yu-Su Chen for their input and technical support. We also acknowledge the Scottish Centre for Macromolecular Imaging (SCMI) and James Streetley for assistance with cryo-EM experiments and access to instrumentation, funded by the MRC (MC_PC_17135, MC_UU_00034/7) and SFC (H17007). This research is funded by the BBSRC & CRODA (BB/X511006/1). Institutional Review Board Statement: all animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of the UK Home Office Animals Scientific Procedures Act of 1986 in accordance with an internal ethics board and UK government-approved project licence (Project licence PP1650440; Granted: 29 May 2020).References
- S. H. Kiaie, N. Majidi Zolbanin, A. Ahmadi, R. Bagherifar, H. Valizadeh and F. Kashanchi, et al., Recent advances in mRNA-LNP therapeutics: immunological and pharmacological aspects, J. Nanobiotechnol., 2022, 20(1), 276 CrossRef CAS.
- C. Zhang, Y. Ma, J. Zhang, J. C. Kuo, Z. Zhang and H. Xie, et al., Modification of Lipid-Based Nanoparticles: An Efficient Delivery System for Nucleic Acid-Based Immunotherapy, Molecules, 2022, 27(6), 1943 CrossRef CAS PubMed.
- K. J. Hassett, J. Higgins, A. Woods, B. Levy, Y. Xia and C. J. Hsiao, et al., Impact of lipid nanoparticle size on mRNA vaccine immunogenicity, J. Controlled Release, 2021, 335, 237–246 CrossRef CAS PubMed.
- NCBI, Lipid+nanoparticle – search results – PubMed: United States National Library of Medicine, 2023 available from: https://pubmed.ncbi.nlm.nih.gov/?term=lipid%2Bnanoparticle.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Lipid nanoparticles for mRNA delivery, Nat. Rev. Mater., 2021, 6(12), 1078–1094 CrossRef CAS PubMed.
- S. J. Shepherd, X. Han, A. J. Mukalel, R. El-Mayta, A. S. Thatte and J. Wu, et al., Throughput-scalable manufacturing of SARS-CoV-2 mRNA lipid nanoparticle vaccines, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(33), e2303567120 CrossRef CAS.
- C. B. Roces, E. C. Port, N. N. Daskalakis, J. A. Watts, J. W. Aylott and G. W. Halbert, et al., Rapid scale-up and production of active-loaded PEGylated liposomes, Int. J. Pharm., 2020, 586, 119566 CrossRef CAS.
- S. Shah, V. Dhawan, R. Holm, M. S. Nagarsenker and Y. Perrie, Liposomes: Advancements and innovation in the manufacturing process, Adv. Drug Delivery Rev., 2020, 154–155, 102–122 CrossRef CAS PubMed.
- J. Di, Z. Du, K. Wu, S. Jin, X. Wang and T. Li, et al., Biodistribution and Non-linear Gene Expression of mRNA LNPs Affected by Delivery Route and Particle Size, Pharm. Res., 2022, 39(1), 105–114 CrossRef CAS PubMed.
- M. R. Hunter, L. Cui, B. T. Porebski, S. Pereira, S. Sonzini and U. Odunze, et al., Understanding Intracellular Biology to Improve mRNA Delivery by Lipid Nanoparticles, Small Methods, 2023, e2201695 CrossRef PubMed.
- C. B. Roces, G. Lou, N. Jain, S. Abraham, A. Thomas and G. W. Halbert, et al., Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics, Pharmaceutics, 2020, 12(11), 1095 CrossRef CAS PubMed.
- G. Lou, G. Anderluzzi, S. Woods, C. W. Roberts and Y. Perrie, A novel microfluidic-based approach to formulate size-tuneable large unilamellar cationic liposomes: Formulation, cellular uptake and biodistribution investigations, Eur. J. Pharm. Biopharm., 2019, 143, 51–60 CrossRef CAS.
- H. Tanaka, N. Takata, Y. Sakurai, T. Yoshida, T. Inoue and S. Tamagawa, et al., Delivery of Oligonucleotides Using a Self-Degradable Lipid-Like Material, Pharmaceutics, 2021, 13(4), 544 CrossRef CAS PubMed.
- M. Maeki, Y. Fujishima, Y. Sato, T. Yasui, N. Kaji and A. Ishida, et al., Understanding the formation mechanism of lipid nanoparticles in microfluidic devices with chaotic micromixers, PLoS One, 2017, 12(11), e0187962 CrossRef PubMed.
- V. Filipe, A. Hawe and W. Jiskoot, Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates, Pharm. Res., 2010, 27(5), 796–810 CrossRef CAS.
- A. Kim, W. B. Ng, W. Bernt and N. J. Cho, Validation of Size Estimation of Nanoparticle Tracking Analysis on Polydisperse Macromolecule Assembly, Sci. Rep., 2019, 9(1), 2639 CrossRef PubMed.
- I. V. Zhigaltsev, N. Belliveau, I. Hafez, A. K. Leung, J. Huft and C. Hansen, et al., Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing, Langmuir, 2012, 28(7), 3633–3640 CrossRef CAS PubMed.
- Y. Suzuki and H. Ishihara, Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs, Drug Metab. Pharmacokinet., 2021, 41, 100424 CrossRef CAS PubMed.
- P. Patel, N. M. Ibrahim and K. Cheng, The Importance of Apparent pKa in the Development of Nanoparticles Encapsulating siRNA and mRNA, Trends Pharmacol. Sci., 2021, 42(6), 448–460 CrossRef CAS.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement, ACS Nano, 2021, 15(11), 16982–17015 CrossRef PubMed.
- J. A. Kulkarni, M. M. Darjuan, J. E. Mercer, S. Chen, R. van der Meel and J. L. Thewalt, et al., On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA, ACS Nano, 2018, 12(5), 4787–4795 CrossRef.
- M. H. Y. Cheng, J. Leung, Y. Zhang, C. Strong, G. Basha and A. Momeni, et al., Induction of Bleb Structures in Lipid Nanoparticle Formulations of mRNA Leads to Improved Transfection Potency, Adv. Mater., 2023, 35(31), e2303370 CrossRef.
- C. Webb, S. Khadke, S. T. Schmidt, C. B. Roces, N. Forbes and G. Berrie, et al., The Impact of Solvent Selection: Strategies to Guide the Manufacturing of Liposomes Using Microfluidics, Pharmaceutics, 2019, 11(12), 653 CrossRef PubMed.
- N. Sciolino, S. Reverdatto, A. Premo, L. Breindel, J. Yu and G. Theophall, et al., Messenger RNA in lipid nanoparticles rescues HEK 293 cells from lipid-induced mitochondrial dysfunction as studied by real time pulse chase NMR, RTPC-NMR, spectroscopy, Sci. Rep., 2022, 12(1), 22293 CrossRef PubMed.
- L. Cui, S. Renzi, E. Quagliarini, L. Digiacomo, H. Amenitsch and L. Masuelli, et al., Efficient Delivery of DNA Using Lipid Nanoparticles, Pharmaceutics, 2022, 14(8), 1698 CrossRef.
- J. Long, C. Yu, H. Zhang, Y. Cao, Y. Sang and H. Lu, et al., Novel Ionizable Lipid Nanoparticles for SARS–CoV–2 Omicron mRNA Delivery, Adv. Healthcare Mater., 2023, 12(13), 2202590 CrossRef.
- X. Cheng and R. J. Lee, The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery, Adv. Drug Delivery Rev., 2016, 99(Pt A), 129–137 CrossRef PubMed.
- R. A. Meyer, S. Y. Neshat, J. J. Green, J. L. Santos and A. D. Tuesca, Targeting strategies for mRNA delivery, Mater. Today Adv., 2022, 14(2), 100240 CrossRef.
- K. Paunovska, C. D. Sago, C. M. Monaco, W. H. Hudson, M. G. Castro and T. G. Rudoltz, et al., A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation, Nano Lett., 2018, 18(3), 2148–2157 CrossRef.
- P. Jain, R. S. Pawar, R. S. Pandey, J. Madan, S. Pawar and P. K. Lakshmi, et al., In vitro in vivo correlation (IVIVC) in nanomedicine: Is protein corona the missing link?, Biotechnol. Adv., 2017, 35(7), 889–904 CrossRef.
- V. Francia, R. M. Schiffelers, P. R. Cullis and D. Witzigmann, The Biomolecular Corona of Lipid Nanoparticles for Gene Therapy, Bioconjugate Chem., 2020, 31(9), 2046–2059 CrossRef PubMed.
- Y. Lee, M. Jeong, J. Park, H. Jung and H. Lee, Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics, Exp. Mol. Med., 2023, 55(10), 2085–2096 CrossRef PubMed.
- L. Zhang, K. R. More, A. Ojha, C. B. Jackson, B. D. Quinlan and H. Li, et al., Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability, npj Vaccines, 2023, 8(1), 156 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4pm00128a |
| This journal is © The Royal Society of Chemistry 2024 |