Open Access Article
Open Access ArticleGreen synthesis of silver nanoparticles using Phyllanthus emblica extract: investigation of antibacterial activity and biocompatibility in vivo†
Md Monir
Hossain‡
 a,
Amir
Hamza‡
a,
Shakil Ahmed
Polash
ab,
Mehedi Hasan
Tushar
a,
Masato
Takikawa
c,
Anuj Bhowmik
Piash
a,
Chaitali
Dekiwadia
d,
Tanushree
Saha
ef,
Shinji
Takeoka
a,
Amir
Hamza‡
a,
Shakil Ahmed
Polash
ab,
Mehedi Hasan
Tushar
a,
Masato
Takikawa
c,
Anuj Bhowmik
Piash
a,
Chaitali
Dekiwadia
d,
Tanushree
Saha
ef,
Shinji
Takeoka
 g and
Satya Ranjan
Sarker
g and
Satya Ranjan
Sarker
 *a
*a
aDepartment of Biotechnology and Genetic Engineering, Jahangirnagar University, Savar, Dhaka - 1342, Bangladesh. E-mail: satya.sarker@bgeju.edu.bd
bNanobiotechnology Research Laboratory (NBRL), School of Science, RMIT University, Melbourne, Victoria 3001, Australia
cDepartment of Advanced Science and Engineering, Waseda University (TWIns), Shinjuku-ku, Tokyo 162-8480, Japan
dRMIT Microscopy and Microanalysis Facility, RMIT University, Melbourne 3001, Victoria, Australia
eSchool of Engineering, RMIT University, Melbourne 3001, Victoria, Australia
fDepartment of Textile Engineering, Dhaka University of Engineering and Technology, Gazipur 1700, Bangladesh
gDepartment of Life Science and Medical Bioscience, Graduate School of Advanced Science and Engineering, Waseda University (TWIns), Shinjuku-ku, Tokyo 162-8480, Japan
First published on 21st March 2024
Abstract
The application of nanotherapeutics is being considered as one of the most sought-after strategies to combat the threat posed by drug resistant bacteria. One promising type of nanotherapeutic is biogenic silver nanoparticles (bAgNPs) generated through exploiting the reducing potential of plant extracts. Herein, bAgNPs were synthesized at pH 7.4 (bAgNPs) and pH 10 (bAgNPs@pH) through green chemistry approaches using an extract of Phyllanthus emblica fruit as a source of reducing agent. The physicochemical properties, antibacterial potential, and biocompatibility of the as-synthesized bAgNPs were determined. The average size of bAgNPs and bAgNPs@pH was 15.3 and 20.1 nm, respectively, and both types of nanoparticles were negatively charged (i.e., ∼−25 mV). The as-synthesized bAgNPs exhibited excellent antibacterial activity against different bacterial strains such as Bacillus subtilis RBW, Escherichia coli DH5a, Salmonella typhi, Hafnia alvei, enteropathogenic E. coli, Vibrio cholerae, and Staphylococcus aureus. The most effective antibacterial activity of bAgNPs and bAgNPs@pH was observed against Hafnia alvei, a Gram-negative bacterium, with a zone of inhibition (ZOI) of ∼24 and 26 mm in diameter, respectively. The nanoparticles exhibited antibacterial activity through damaging the bacterial cell wall, oxidizing the membrane fatty acids, and interacting with cellular macromolecules to bring about bacterial death. Furthermore, bAgNPs showed excellent hemocompatibility against human red blood cells, and there was no significant toxicity observed in rat serum ALT, AST, γ-GT, and creatinine levels. Thus, bAgNPs synthesized using Phyllanthus emblica fruit extract hold great promise as nanotherapeutics to combat a broad spectrum of pathogenic bacteria. Future directions may involve further exploration of the potential applications of biogenic silver nanoparticles in clinical settings, including studies on long-term efficacy, extensive in vivo toxicity profiles, and scalable production methods for clinical use.
1. Introduction
The development of resistance in pathogenic bacteria to chemically synthesized antibiotics is a threat to the existence of humanity. On the other hand, many therapeutic agents, including antibiotics, can cause systemic toxicity, which is alarming.1 Antibiotics are frequently prescribed to treat severe and potentially fatal infections. However, their abuse has led to the emergence of increased bacterial resistance and side effects such as nausea, diarrhea, and stomach pain, among others.2 Antibiotics also interact with other drugs and make them less effective, which may result in adverse effects, including dehydration. Moreover, antibiotics have been linked to hepatotoxicity and nephrotoxicity such as acute kidney injury, cirrhosis, cholestatic injury, and hepatitis.1,3 Therefore, it is urgent to find alternative but biocompatible antibacterial agents that can efficiently combat a wide range of bacteria.The field of nanoscience has become a highly promising interdisciplinary area of research for developing bioactive nanoparticles for use as therapeutics. The aim is to engineer materials at both the atomic and molecular levels to create nanostructures with superior functionalities.4 Inorganic nanoparticles, such as gold (Au)5–9 and silver (Ag),4,7,10–13 have been widely studied as antibacterial agents due to their excellent physical and chemical properties. Among these, silver nanoparticles (AgNPs) have been extensively utilized as antimicrobial agents in various medical applications, including wound dressings, cardiovascular and orthopedic implants, catheters, and dental composites.14 Various physical, chemical, and biological techniques have been employed to prepare AgNPs.15 However, many of these methods are costly and time-consuming, and involve the use of environmentally and biologically harmful chemicals and solvents, generating toxic chemical byproducts that hinder their clinical applications.15,16 Moreover, chemically synthesized AgNPs are prone to oxidation and aggregation, which leads to instability and toxicity to both human health and the environment.17,18 Therefore, green synthesis is a more economical and eco-friendly approach for synthesizing AgNPs. The size, morphology, and synthesis of AgNPs are influenced by different factors, including the concentration of AgNO3, pH, duration of reaction, and light irradiation.19 pH plays a crucial role in determining the average size and morphology of biologically synthesized nanoparticles by modulating the surface charges of bioactive molecules and capping agents, which affects their capacity to reduce and incorporate metal ions.4,11 The use of plant extracts and various microbes, including bacteria, fungi, microalgae, and cyanobacteria, for the reduction of metals, has gained widespread attention as an alternative to using phytoconstituents for preparing metal nanoparticles.10,20
Biogenic silver nanoparticles (bAgNPs) exhibit potential as antibacterial agents; however, before their use in clinical trials and commercial applications, it is essential to thoroughly assess their biocompatibility. The presence of biocompatible phytoconstituents as capping agents may reduce the cytotoxicity of bAgNPs.4,10 Therefore, the inherent biocompatibility of the conjugated phytoconstituents renders bAgNPs suitable for various clinical applications. Previously, we demonstrated that bAgNPs exhibited excellent hemocompatibility against human and rat red blood cells (RBCs). Furthermore, rats treated with bAgNPs exhibited no significant toxicity in terms of their hematological and biochemical parameters.4,10–12 This is due to the presence of secondary metabolites such as saponins, flavonoids, terpenoids, and quinines in plant extracts, which have strong reducing potential and lead to the production of nanoparticles with better characteristics.4 As a result, silver nanoparticles made from plant extracts have a natural ability to combat various bacterial strains, including those that are resistant to multiple drugs, while also being more biocompatible.11,12
Phyllanthus emblica (syn. Emblica officinalis), also known as emblica or amla (Hindi) or amalaka (Sanskrit) or Indian gooseberry (English), belongs to the family Phyllanthaceae. The fruit of the Phyllanthus plant is used in traditional medicine in Southeast Asia because of its unique properties, which include high levels of antioxidants, and anti-aging, antipyretic, and anti-inflammatory agents.21,22 It exists in two varieties: cultivated (gramya) and wild (vanya). The wild variant is smaller, whereas the cultivated type is larger, smoother, and juicier. Comprising over 80% water, the chemical composition of P. emblica includes large amounts of proteins, carbohydrates, phenolic compounds, glycosides, alkaloids, coumarins, flavonoids, saponins, resins, etc.23 It also contains a variety of isolated compounds with significant medicinal potential. These compounds include ascorbic acid (vitamin C), tannins such as gallotannins and ellagitannins, gallic acid, ellagic acid, and quercetin.24,25 Ascorbic acid serves as a major constituent with potent antioxidant properties, while tannins contribute to the fruit's bitterness and antioxidant effects. Gallic acid exhibits anti-ulcerogenic activity, and ellagic acid offers antioxidant benefits. Quercetin is a flavonoid known for its antioxidant and anti-inflammatory properties.24,25 These compounds have been studied for their diverse pharmacological activities, including anti-ulcerogenic and hepatoprotective activities, highlighting the potential health benefits of P. emblica.24,25 Besides, a comparative analysis revealed that extracts from Phyllanthus emblica demonstrated antibacterial effects against both Gram-positive (Bacillus subtilis) and Gram-negative bacteria (Escherichia coli and Salmonella typhi).23 While there have been numerous investigations into the plant's pharmacognosy and phytochemistry, its potential as a biocompatible material for producing AgNPs is yet to be fully explored.23,26 Previous studies on biogenic silver nanoparticles have overlooked the specific pH effects during synthesis, hindering the optimization of their stability and reactivity.27–29 Additionally, there is a dearth of information regarding the hydrodynamic diameter and zeta potential of nanoparticles synthesized from Phyllanthus emblica extracts, crucial for understanding their surface properties and biological interactions. Investigating the antibacterial activity of these nanoparticles under varied pH conditions could offer insights into enhancing their efficacy. Furthermore, assessing the biocompatibility of these nanoparticles through hemocompatibility and in vivo cytotoxicity studies in animal models is imperative for evaluating their safety and potential clinical applications.
To address this gap, we have used the extract of P. emblica fruit to produce green and biocompatible silver nanoparticles that pose no chemical toxicity related risk. Therefore, P. emblica fruit extract was utilized as a source of reducing agent to synthesize bAgNPs at both physiological (pH 7.4) and alkaline pH (10.0). The as-synthesized bAgNPs were characterized using UV-Vis spectroscopy, Fourier transform infrared (FTIR) spectroscopy, energy dispersive X-ray spectroscopy (EDS), powder X-ray diffraction (XRD), transmission electron microscopy (TEM), and scanning electron microscopy (SEM). Furthermore, the size and zeta potential of bAgNPs were determined using a zeta size analyzer. The antibacterial potential of bAgNPs was investigated against five pathogenic bacterial strains such as Salmonella typhi, Hafnia alvei, enteropathogenic E. coli, Vibrio cholerae, and Staphylococcus aureus. In addition, two nonpathogenic bacterial strains such as Bacillus subtilis RBW and Escherichia coli DH5a were also used in the antibacterial potential assay. The antibacterial potential of bAgNPs was investigated through disk diffusion, broth dilution, and CellTox green assay. The lipid peroxidation (LPO) assay was used to investigate the mechanism of the antibacterial activity of bAgNPs. Moreover, the hemocompatibility of bAgNPs was assessed using human red blood cells (RBCs), while the biocompatibility was tested in vivo using a Wister rat model.
2. Materials and methods
2.1. Materials
Sigma Aldrich (USA) provided absolute ethanol, potassium bromide, EDTA, and silver nitrate (AgNO3). Unichem (China) supplied sodium chloride, yeast extract, and peptone. Titan Biotech Ltd (India) provided agar powder. Promega (USA) provided CellTox. JT Baker (USA) supplied thiobarbituric acid (TBA), while trichloroacetic acid (TCA) was obtained from Merck (Germany). Vitro Scient (Egypt) provided the biochemical analysis kits for determining ALT, AST, and γ GT, while Crescent Diagnostics (The Kingdom of Saudi Arabia) supplied the serum creatinine measurement kits. Both the nonpathogenic bacterial strains (i.e., Bacillus subtilis RBW and Escherichia coli DH5a) and pathogenic bacterial strains (i.e., Salmonella typhi, Hafnia alvei, enteropathogenic E. coli, Vibrio cholerae, Staphylococcus aureus) were obtained from the Department of Biotechnology and Genetic Engineering at Jahangirnagar University, Savar, Dhaka 1342, Bangladesh. Male Wister albino rats weighing between 170 and 180 g were obtained from the Department of Biochemistry and Molecular Biology at Jahangirnagar University, Savar, Dhaka, Bangladesh.2.2. Synthesis of biogenic silver nanoparticles (bAgNPs)
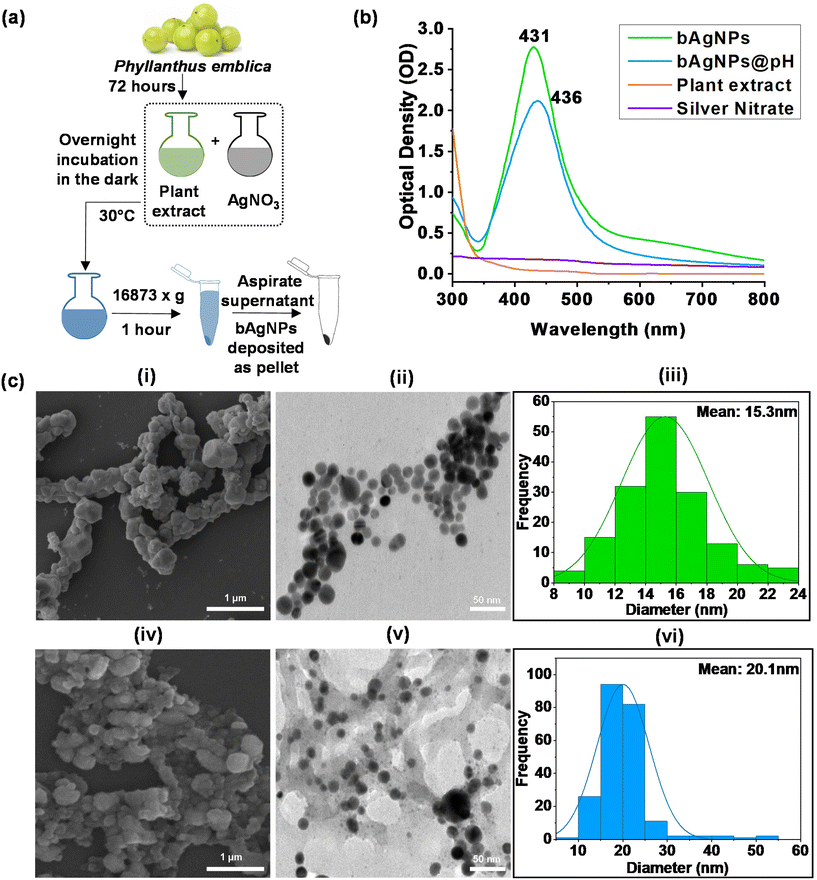
To synthesize biogenic silver nanoparticles (bAgNPs), ethanolic extract of P. emblica flesh was utilized (Fig. 1a). The flesh extract was prepared according to our previously published protocol.12,30–32 Briefly, P. emblica was procured from the local market, Savar, Dhaka-1342, Bangladesh, thoroughly washed with distilled water, sun-dried, and ground to fine powder. Ten grams (10 gm) of the powder was mixed with 100 mL of 70% ethanol and continuously stirred for 72 hours. The resulting mixture was filtered using Whatman No. 1 filter paper, the solvent was evaporated completely, and stored at 4 °C. The extract was resuspended in MQ water to make 1% suspension prior to mixing with 10 mM AgNO3 in two separate Duran glass bottles at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (i.e., 8 mL extract suspension and 72 mL AgNO3). One bottle was left undisturbed while pH of the other bottle was adjusted to 10 using concentrated NaOH solution. Both reaction mixtures were incubated in the dark for 24 h at room temperature with constant stirring to prevent the photoactivation of AgNO3. The phytochemicals present in the extract served as reducing agents and brought about the reduction of Ag+ to Ag0. After incubation, the unconjugated phytoconstituents, and AgNO3 were separated from the reaction mixture by centrifugation at 16
9 (i.e., 8 mL extract suspension and 72 mL AgNO3). One bottle was left undisturbed while pH of the other bottle was adjusted to 10 using concentrated NaOH solution. Both reaction mixtures were incubated in the dark for 24 h at room temperature with constant stirring to prevent the photoactivation of AgNO3. The phytochemicals present in the extract served as reducing agents and brought about the reduction of Ag+ to Ag0. After incubation, the unconjugated phytoconstituents, and AgNO3 were separated from the reaction mixture by centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 873g for an hour. The supernatant was carefully removed to collect the synthesized bAgNPs. Finally, the synthesized bAgNPs were washed twice with distilled water and redispersed using distilled water.
873g for an hour. The supernatant was carefully removed to collect the synthesized bAgNPs. Finally, the synthesized bAgNPs were washed twice with distilled water and redispersed using distilled water.
2.3. Characterization of biogenic silver nanoparticles (bAgNPs)
The generation of bAgNPs was confirmed using various analytical techniques such as UV-Visible (Specord® 205, Analytik Jena, Germany) and FTIR spectroscopy (IRPrestige-21, SHIMADZU, Japan). The hydrodynamic size, and zeta potential of nanoparticles were determined using a zeta size analyzer (Nano-ZS90; Spectris PLC, Egham, England) after sonicating the nanoparticles for 30 minutes in a bath-type sonicator. The shape, morphology, and composition of bAgNPs were analyzed using a scanning electron microscope (SEM) operated at 10 kV with a probe current of 100pA, and a transmission electron microscope (TEM) (HF-2200; Hitachi, Tokyo, Japan) integrated with an energy-dispersive X-ray spectrometer (EDAX Genesis; AMETEK, Pennsylvania, USA) and operated at 200 kV. The nanoparticles were neither stained nor coated with any conductive metal prior to observing under an electron microscope. XRD patterns of bAgNPs were analyzed using a powder X-ray diffractometer (GNR Xray Explorer, Italy) following our previously published protocol.4,10,112.4. Antibacterial activity assay
The antibacterial potential of bAgNPs was assessed against both Gram-positive (i.e., Bacillus subtilis, and Staphylococcus aureus), and Gram-negative bacteria (i.e., Salmonella typhi, Hafnia alvei, Enteropathogenic Escherichia coli (EPEC), Escherichia coli DH5α, and Vibrio cholerae). Among others B. subtilis, and E. coli DH5α are non-pathogenic, while the remaining bacterial strains are pathogenic. The antibacterial potential of bAgNPs was evaluated through minimum inhibitory concentration (MIC), disk diffusion, and CellTox green assays.2.5. Lipid peroxidation (LPO) assay
The lipid peroxidation (LPO) potential of bAgNPs was investigated following our previously published protocol.5 Firstly, 200 μl of bAgNPs was mixed with 1 ml of the respective bacterial culture and incubated for 30 minutes at room temperature. After incubation, 2 ml of 10% trichloroacetic acid (TCA) was added to bAgNP treated bacterial culture followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 416g for 35 minutes at room temperature to remove the insoluble cellular components. The supernatant was then re-centrifuged for 20 minutes at the same relative centrifugal force (g) to remove any protein precipitates and dead cells. The resulting supernatant containing malonaldehyde was mixed with 4 mL of 0.67% TBA solution, heated for 10 minutes in a hot water bath to form the malondialdehyde-TBA adduct, and then cooled down to room temperature. The absorbance of the malondialdehyde-TBA adduct was measured at 532 nm using a UV-Visible spectrophotometer (Specord® 205, Analytik Jena, Germany).
416g for 35 minutes at room temperature to remove the insoluble cellular components. The supernatant was then re-centrifuged for 20 minutes at the same relative centrifugal force (g) to remove any protein precipitates and dead cells. The resulting supernatant containing malonaldehyde was mixed with 4 mL of 0.67% TBA solution, heated for 10 minutes in a hot water bath to form the malondialdehyde-TBA adduct, and then cooled down to room temperature. The absorbance of the malondialdehyde-TBA adduct was measured at 532 nm using a UV-Visible spectrophotometer (Specord® 205, Analytik Jena, Germany).
2.6. In vitro hemocompatibility assay
The compatibility of bAgNPs with human red blood cells (RBCs) was investigated according to our previously published protocol with minor modifications.5,6,12 In brief, 6 mL of human blood was drawn from the donor's left hand following the venipuncture technique and collected in a tube containing 10% EDTA. The blood was then centrifuged at 500g for 10 minutes at room temperature to separate RBCs from serum. The serum was discarded, and the RBCs were resuspended in 5 ml PBS and centrifuged again at 500g for 10 minutes. The supernatant was removed, and the RBCs were washed twice using 150 mM NaCl solution at 3000g for three minutes. Then, 0.1 ml RBC solution was mixed with 0.4 ml of various concentrations (10, 20, 40, and 60 μg) of bAgNPs and incubated at 37 °C, and 150 rpm for 30 minutes. After incubation, the mixture was centrifuged at 1377g for 5 minutes at room temperature, and the supernatant was collected. The absorbance of the supernatant was measured at 570 nm. RBCs treated with PBS, and MQ water were used as the negative and positive controls, respectively.33 The proportion of hemolysis was determined using the following formula:2.7. In vivo cytotoxicity assay
The biocompatibility of bAgNPs was investigated in vivo using rat models. Herein, 30 Wistar rats (male) were used and divided into five groups (six rats per group), as described previously.10 Briefly, the rats were kept in a clean environment with controlled temperature (23 ± 2 °C) and humidity (55 ± 7%), following a 12 h day and 12 h night cycle. Before administering bAgNPs, the rats were allowed to acclimatize to the environment for one week. Based on previous studies, which indicated that smaller AgNPs (approximately 20 nm) have a higher rate of deposition in the tissues like liver, spleen, kidney, and brain.34,35 The highest and the lowest doses used in the study were 6 mg per kg and 3 mg per kg body weight respectively, as the size of the as-synthesized bAgNPs and bAgNPs@pH was 15.3, and 20.1 nm, respectively. The rats were anesthetized using ketamine/xylazine prior to intravenous administration (through the tail vein) of two doses of bAgNPs and bAgNPs@pH. After seven days, blood samples were collected from all the rats, and serum was separated and stored at 4 °C for further analysis. The study was approved by the Biosafety, Biosecurity & Ethical Committee of Jahangirnagar University (BBEC, JU/M 2019 (4)2).To evaluate the potentially toxic effects of bAgNPs on rat tissues, the concentrations of enzymes that usually get elevated in response to any liver and kidney damage were measured. Specifically, the concentrations of serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transferase (γ-GT), and creatinine were analyzed in the experimental rats and compared to those of the control group. The concentrations of biomarkers were measured using commercially available kits following the instructions provided by the manufacturer.
2.8. Statistical analysis
The mean ± standard error of the mean (SEM) was used to present the data. Statistical analysis was performed using one-way ANOVA, and the Dunnett test was used for post-hoc testing. Results were considered statistically significant at P < 0.05.10 The experiments were repeated three times, and the results presented are average measurements with the standard deviation.3. Results and discussion
3.1. Characterization of bAgNPs
The colorless AgNO3 solution turned brown upon the addition of the extract of P. emblica followed by incubation at room temperature for 24 h in the dark (Fig. 1a). The change of color was observed because of the reduction of Ag+ to Ag0 due to the presence of secondary metabolites, including alkaloids, flavonoids, and tannins in the extract.36 The Ag0 then formed nanoclusters and ultimately nanoparticles, known as bAgNPs, which exhibit powerful scattering and absorption capabilities when exposed to light of a certain wavelength.37 To confirm the generation of bAgNPs, analysis of UV-Visible spectra was conducted and the λmax values were 431 and 436 nm for bAgNPs and bAgNPs@pH, respectively, (Fig. 1b) which closely matched with the usual surface plasma resonance (SPR) value of AgNPs (i.e., 420 nm).18 The absorption spectrum of bAgNPs@pH synthesized in alkaline conditions (pH 10) displayed a redshift of the SPR band [i.e., λmax shifted to higher wavelengths (436 nm)] indicating that the nanoparticles had a distinct morphology in terms of size and shape.11 It is generally assumed that any shift in the SPR peak indicates a change in the size of nanoparticles, with a shift towards the shorter wavelength indicating a decrease in the size of the as-prepared bAgNPs (as shown in Fig. 1b and c). The observed spectral changes can be attributed to the reduction of the size of metal nanoparticles. On the other hand, oxidation prevails over the reduction process in a low-pH environment (below 5). In contrast, in a very high-pH environment (≥9), the rate of reduction is very fast, leading to nanoparticle aggregation.11 As a result, the size of bAgNPs synthesized at pH 10 was larger (as depicted in Fig. 1b and c). Our as prepared bAgNPs demonstrated similar SPR bands (ranging from 400 to 450 nm) to that of other AgNPs.11 However, the particle size was different which can be attributed to the presence of various secondary metabolites in P. emblica extract. In addition, we have confirmed the long-term stability of our as-synthesized bAgNPs. For example, our bAgNPs are stable at room temperature for several years (i.e., 3 years) as shown in Fig. S1.†Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) (Fig. 1c) images demonstrate that both the as-synthesized nanoparticles are spherical in shape. The particle size distribution (PSD) graphs of both bAgNPs and bAgNPs@pH were prepared using their respective TEM images. The average size of bAgNPs and bAgNPs@pH were found to be 15.3 nm and 20.1 nm, respectively (Fig. 1c).
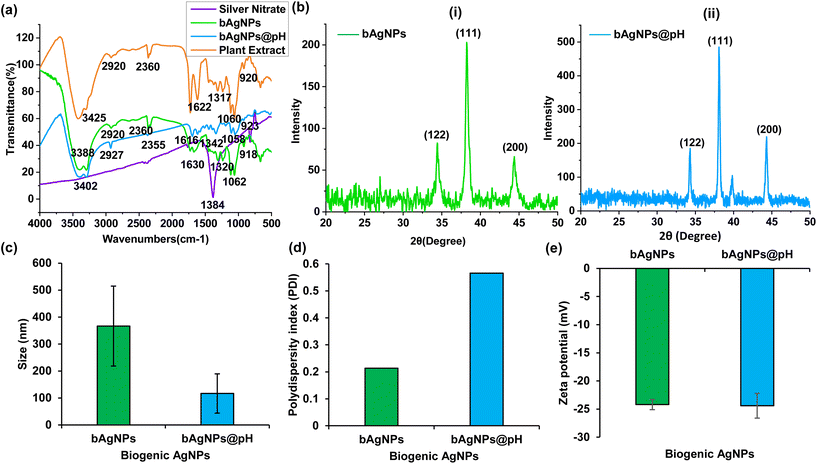
FTIR spectra reveal that bioactive phytoconstituents with diverse functional groups are present in both the plant extract and as-synthesized Ag nanoparticles (Fig. 2a).38,39 The ethanolic extract of P. emblica flesh, and bAgNPs exhibited distinct peaks at 920 and 918, 1060 and 1062, 1317 and 1320, 1622 and 1630, 2360 and 2360, 2920 and 2920, and 3425 and 3388 cm−1, which are attributed to aromatic C–H bending, C–OH stretching, C–N stretching, carbonyl stretching, C–H asymmetric stretching, and N–H stretching, respectively.40 On the other hand, the FTIR spectrum of bAgNPs@pH also demonstrated the presence of prominent peaks at 923, 1058, 1342, 1616, 2355, 2927, and 3402 cm−1 that correspond to aromatic C–H bending, C–OH stretching, C–N stretching, carbonyl stretching, C–H asymmetric stretching, and N–H stretching, respectively.10 The strong signature peak at 1384 cm−1 was observed for silver nitrate.10 The FTIR spectra confirm the presence of hydroxyl (–OH) as well as carboxyl (–COOH) groups in the constituents of P. emblica flesh extracts. The peaks observed in the spectra were mainly due to the presence of flavonoids and other secondary metabolites present in the plant extract.12
The phase, orientation, and crystal size of the bAgNPs in their as-prepared state were determined through powder XRD analysis. Fig. 2b exhibits clear distinction patterns indicative of crystalline silver. These patterns confirmed the presence of face-centered cubic (fcc) crystalline elemental silver, as evidenced by the Braggs peaks at 2θ angles of 34.32, 38.08, and 44.32 degrees which correspond to the Miller indices (122), (111), and (200), respectively, for both the as-synthesized bAgNPs. These results were consistent with the JCPDS database designated file number 04-0783. Furthermore, the crystalline grain size of bAgNPs was also determined using Scherer's equation, where the mean crystalline size of the particle (D) was calculated as D = (Kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ), with a shape factor (K) of 0.9, wavelength of the X-ray radiation (λ) of 0.154 nm, full width at half maximum (FWMH) (β) of (π/180) × FWMH, and Bragg angle (θ). The average size of the particles calculated using Scherer's equation was 15.86 nm and 19.21 nm, respectively. The size of the particles matches closely with the average size of bAgNPs, and bAgNPs@pH obtained from TEM images, that is, 15.3 nm and 20.1 nm, respectively.
θ), with a shape factor (K) of 0.9, wavelength of the X-ray radiation (λ) of 0.154 nm, full width at half maximum (FWMH) (β) of (π/180) × FWMH, and Bragg angle (θ). The average size of the particles calculated using Scherer's equation was 15.86 nm and 19.21 nm, respectively. The size of the particles matches closely with the average size of bAgNPs, and bAgNPs@pH obtained from TEM images, that is, 15.3 nm and 20.1 nm, respectively.
The hydrodynamic diameter of bAgNPs and bAgNPs@pH were 366.4 ± 148.2, and 116.8 ± 72.9 nm, respectively, as measured by the dynamic light scattering (DLS) technique (Fig. 2c and Table S1†). However, the size of bAgNPs obtained from TEM images was smaller than that of the hydrodynamic diameter. This is because TEM images provide information about the nanoparticles’ inorganic core only, whereas the hydrodynamic diameter of nanoparticles is influenced by the interaction between the electric dipole of the solvent and conjugated bioactive molecules of the phytoconstituents.11 The stability of nanoparticles is influenced by the physiochemical properties of the solvents used for their dispersion as well as the type of polymers conjugated/adsorbed on their surface.41 The polydispersity index (PDI) of bAgNPs and bAgNPs@pH was 0.214 and 0.566, respectively (Fig. 2c and Table S1†). A PDI value of less than 0.50 indicates good stability of the particles.42 On the other hand, the zeta potentials of both bAgNPs and bAgNPs@pH were almost the same (i.e., −24.3 mV) (Fig. 2c and Table S1†), which can be attributed to the bioactive molecules adsorbed onto the surface of the nanoparticles.11 Energy dispersive X-ray (EDS) microanalysis confirmed that the nanoparticles were composed of Ag, C, and O with a distinctive Ag peak at about 3 keV (Fig. S2†). Based on the results obtained from electron microscopy (EM) observation and EDS analysis, it was estimated that the peaks corresponding to elements Si and Cu were likely artifacts stemming from the vacuum pump's silicon oil and the copper present in the TEM grid used during the analysis, respectively. However, the key finding was that Ag nanoparticles were coated with bioactive polymers derived from the flesh of P. emblica.
3.2. Antibacterial activity assay
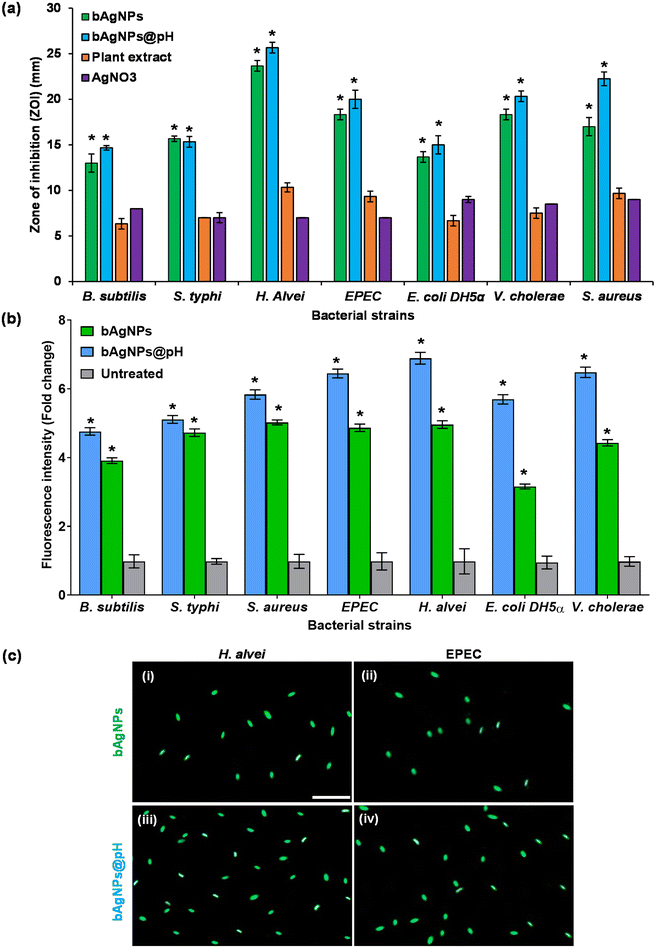
Phyllanthus emblica flesh extracts contain phytoconstituents with long hydrocarbon chains as well as carboxyl and hydroxyl groups which function as hydrophilic moieties. Consequently, interactions between bAgNPs and bacteria are predominantly non-covalent, allowing the negatively charged bAgNPs and bAgNPs@pH to be more readily taken up due to their greater hydrophobicity compared to plant extracts and silver nitrate.10 Molecular crowding may also contribute to the interaction between bAgNPs and bacteria.4 Furthermore, the primary antibacterial mechanism of bAgNPs involves either the release of silver ions or the deposition of bAgNPs inside cells, leading to cell membrane damage, disruption of energy metabolism, oxidative stress from reactive oxygen species (ROS) production, and gene transcription suppression. The released silver ions interact with proteins in the bacterial cell wall and plasma membrane, containing sulfur and phosphorus, through electrostatic interactions.10,12 This interaction results in pores in the bacterial cell membrane, eliminating intracellular bacterial substances and causing permanent cell damage leading to an electrochemical imbalance that causes permanent cell damage.46 The antibacterial efficacy of both bAgNPs and bAgNPs@pH is more pronounced against Gram-negative bacteria like H. alvei compared to Gram-positive bacteria like B. subtilis which is attributed to the distinct cell wall compositions of these bacteria. Gram-negative bacteria have an outer layer of lipopolysaccharides and a thin peptidoglycan layer, which is about 7–8 nanometers thick, while Gram-positive bacteria only have a thick layer of peptidoglycan.47 Although lipopolysaccharides are composed of covalently linked lipids and polysaccharides, there is a lack of strength and rigidity. The negative charges on lipopolysaccharides are attracted toward the weak positive charge available on AgNPs.48 This attraction may lead to enhanced antibacterial activity against Gram-negative bacteria.48–51
Furthermore, antibiotic-resistant bacteria pose a significant threat to public health worldwide. Biogenic nanoparticles offer a promising alternative to conventional antibiotics for combating resistant strains. Their unique mechanisms of action make them less susceptible to microbial resistance mechanisms, such as efflux pumps and enzymatic degradation, which are common among antibiotic-resistant bacteria.52 Consequently, biogenic silver nanoparticles (bAgNPs) have emerged as highly promising contenders owing to their exceptional antibacterial characteristics, crucial for addressing microbial resistance.4,10 The distinctive antimicrobial attributes of bAgNPs highlighted in this study are of significant interest in confronting multidrug-resistant bacteria such as MRSA and VRE.13 These biogenic Ag nanoparticles achieve this by disrupting bacterial cell membranes, inhibiting enzymes, and interfering with DNA replication, and ultimately inducing cell death. Their small size and large surface area enable efficient interaction with bacterial cells, enhancing antimicrobial activity, while their synergy with conventional antibiotics boosts efficacy against resistant strains.12 Moreover, biogenic nanoparticles exhibit comparatively lower toxicity towards animal models, rendering them safer for biomedical applications, and can be customized for targeted delivery to specific bacterial strains or infection sites.4,28 Offering greater stability and durability than their chemically synthesized counterparts, biogenic silver nanoparticles show great promise in addressing multidrug resistance, though further research is imperative to optimize clinical effectiveness.
3.3. Lipid peroxidation (LPO) potential assay
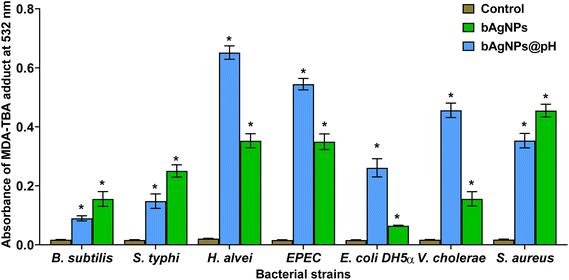
To assess the oxidative potential exerted on bacterial cell membrane fatty acids exposed to bAgNPs, the lipid peroxidation (LPO) assay was carried out. The interaction between nanoparticles and the bacterial cell wall, facilitated by strong hydrophobic interactions upon exposure to bAgNPs and bAgNPs@pH, led to the formation of malondialdehyde-thiobarbituric acid (MDA-TBA) adducts. The highest formation of MDA-TBA adducts was observed when H. alvei, a Gram-negative bacterium, was subjected to treatment with bAgNPs@pH, compared to all other bacterial strains. Conversely, S. aureus, a Gram-positive bacterium, exhibited the highest amount of MDA-TBA adduct formation among all bacterial strains treated with bAgNPs (Fig. 4). This corroborates the similar antibacterial profile of bAgNPs and bAgNPs@pH obtained through both the disk diffusion assay and the CellTox Green assay. The variations in the quantities of MDA-TBA adduct among bacterial strains can be attributed to the diverse interactions of nanoparticles with bacteria, influenced by the distinct compositions of bacterial cell walls across species. When exposed to bAgNPs, bacterial membrane fatty acids undergo oxidation, resulting in the formation of lipid peroxides. This process also triggers the generation of reactive oxygen species (ROS) due to oxidative stress induced by transition metals,10 causing the oxidation of bacterial membrane fatty acids and producing lipid peroxides, with the redox balance favoring oxidation. ROS-induced oxidative stress further disrupts the electron transport chain and interferes with bacterial metabolic reactions,10 ultimately activating apoptotic genes and oxidative proteins, leading to bacterial apoptosis.3.4. In vitro hemocompatibility assay
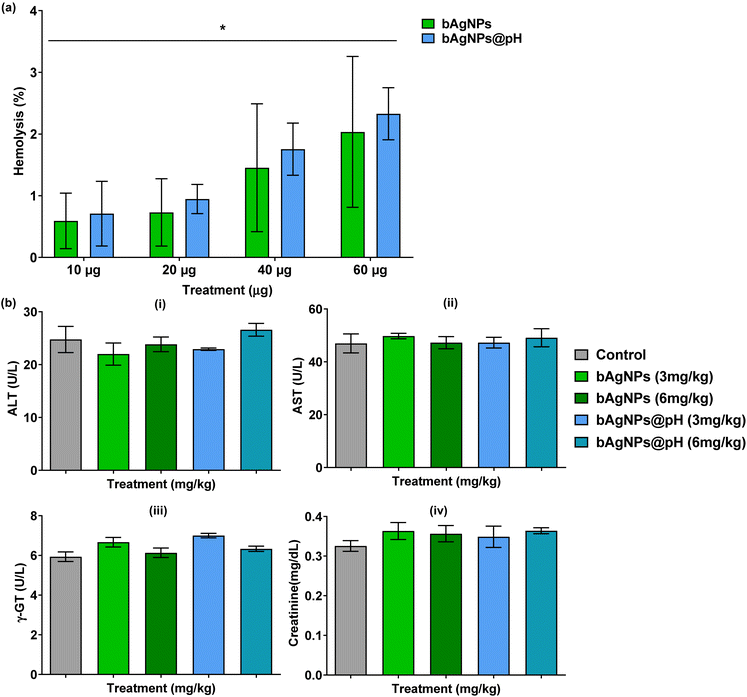
The hemolytic potential of the biologically synthesized silver nanoparticles (bAgNPs) with human red blood cells (RBCs) was evaluated to assess their compatibility. The results presented in Fig. 5a demonstrated that both bAgNPs and bAgNPs@pH exhibited high compatibility with human RBCs. Various concentrations of nanoparticles, ranging from 10–60 μg, were tested. At 60 μg, the percentage of hemolysis for bAgNPs and bAgNPs@pH was 2.23% and 3.07%, respectively. The enhanced hemocompatibility of bAgNPs can be attributed to their more negative zeta potential (i.e., −24.3 mV), resulting in stronger electrostatic repulsion with negatively charged RBCs. On the other hand, the size of bAgNPs is smaller than that of bAgNPs@pH which results in a higher surface-to-volume ratio for bAgNPs. This results in a stronger electrostatic repulsion with the RBCs when compared to bAgNPs@pH. This phenomenon results in a lower percentage of hemolysis for bAgNPs compared to bAgNPs@pH. Importantly, the percentage of hemolytic potential was well below the safe limit of 5% for the therapeutic applications of biomaterials.4 Furthermore, hemolysis was verified using Milli Q water (MQ) as a positive control and Phosphate Buffer Saline (PBS) as a negative control, as illustrated in Fig. S4.†3.5. Evaluation of rat liver and kidney function biomarkers
The potential toxicity of intravenously administered biogenic silver nanoparticles (bAgNPs) and bAgNPs@pH on rat liver and kidneys was investigated. The results, as depicted in Fig. 5b, revealed no significant toxic effects on either organ. Specifically, there was no discernible difference (p > 0.05) in the levels of ALT, AST, γ-GT, and creatinine in the serum of experimental rats compared to control rats. These findings suggest that treatment with bAgNPs (3 and 6 mg kg−1) or bAgNPs@pH (3 and 6 mg kg−1) did not have an impact on these biomarkers. Considering the liver's crucial role in rat physiological functions and the known tendency of colloidal silver nanoparticles to accumulate in the liver, the absence of significant changes in biomarkers indicates the safety of bAgNPs.10To assess liver function, the serum levels of ALT, AST, and γ-GT enzymes were measured following the intravenous administration of bAgNPs. Liver damage is often indicated by a decrease in ATP levels, a high-energy phosphate compound, leading to the release of enzymes (e.g., ALT and AST) into tissues and their subsequent presence in the serum. Additionally, gamma-glutamyltransferase (γ-GT), an enzyme of hepatobiliary origin, brings about cholestasis, a liver disease caused due to impaired bile flow. Our findings suggest that doses of bAgNPs up to 6 mg kg−1 are safe for the liver, as evidenced by the lack of significant changes in these biomarkers.10
The assessment of bAgNPs’ toxicity on the kidneys involved measuring serum creatinine levels. A low level of serum creatinine indicates impaired kidney function in filtering and excreting waste products through urine.10 Our results indicate that administering various doses of bAgNPs up to 6 mg kg−1 for 7 days did not result in a significant difference in serum creatinine levels, suggesting that bAgNPs are not toxic to the kidneys and are compatible with liver function.
Moreover, the serum levels of ALT, AST, and γ-GT fall within the normal reference range (Table S5†). Interestingly, the biologically synthesized AgNPs are compatible to animals as reflected in the data obtained from the in vivo study irrespective of the size, shape and surface charge of the nanoparticles.
4. Conclusion
We have successfully synthesized biogenic silver nanoparticles (bAgNPs) using Phyllanthus emblica flesh extract in a cost-effective and eco-friendly manner. The as-synthesized bAgNPs demonstrated excellent antibacterial activity against both pathogenic and nonpathogenic bacterial strains. The nanoparticles demonstrated antibacterial activity by damaging the bacterial cell wall, oxidizing the cell membrane fatty acids, and interacting with cellular macromolecules (i.e., DNA, proteins etc.) to bring about bacterial death. On an additional note, bAgNPs showed excellent hemocompatibility and biocompatibility with no significant toxicity to the rat liver and kidneys at higher doses. Therefore, our as-synthesized bAgNPs hold great potential to be used as antimicrobials against a wide range of bacteria causing infectious diseases. While the assumption that size, shape, and surface charge do not significantly influence the toxicity of biogenic AgNPs simplifies the understanding, it is essential to acknowledge the nuanced interplay of these factors in determining nanoparticle toxicity. Further research employing sophisticated experimental models and comprehensive toxicity assessments is necessary to elucidate the intricate relationships between nanoparticle characteristics and biological responses extensively.Author contributions
M. M. H. and A. H. have contributed equally to this work. All authors made significant, direct, and intellectual contributions to the research and endorsed it for publication.Data availability
The study's original contributions are accessible in the article/ESI, and additional inquiries can be directed to the corresponding author.Ethics statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Biosafety, Biosecurity & Ethical Committee of Jahangirnagar University.In addition, all experiments were performed in accordance with the Guidelines of Biosafety, Biosecurity & Ethical Committee of Jahangirnagar University, and experiments were approved by the ethics committee of Jahangirnagar university. Informed consent was obtained from the human participants of this study.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Jahangirnagar University Research Grant 2018 and University Grants Commission (UGC)-Jahangirnagar University Joint Research Grant 2019, Government of Bangladesh, provided partial funding for this research project. The authors express their gratitude to Wazed Miah Science Research Center of Jahangirnagar University, Bangladesh, and Waseda University Central Instrument Facility, School of Advanced Science and Engineering, Japan, and RMIT Microscopy and Microanalysis Facility, RMIT University, Australia, for allowing access to their comprehensive facilities and services.References
- G. S. Pazhayattil and A. C. Shirali, Drug-induced impairment of renal function, Int. J. Nephrol. Renovasc. Dis., 2014, 457–468 Search PubMed.
- S. Mohsen, J. A. Dickinson and R. Somayaji, Update on the adverse effects of antimicrobial therapies in community practice, Can. Fam. Physician, 2020, 66, 651–659 Search PubMed.
- M. Robles, E. Toscano, J. Cotta, M. Isabel Lucena and R. J. Andrade, Antibiotic-induced liver toxicity: mechanisms, clinical features and causality assessment, Curr. Drug Saf., 2010, 5, 212–222 CrossRef CAS PubMed.
- M. S. Niloy, M. M. Hossain, M. Takikawa, M. S. Shakil, S. A. Polash, K. M. Mahmud, M. F. Uddin, M. Alam, R. D. Shubhra, M. M. A. K. Shawan, T. Saha, S. Takeoka, M. A. Hasan and S. R. Sarker, Synthesis of Biogenic Silver Nanoparticles Using Caesalpinia digyna and Investigation of Their Antimicrobial Activity and In Vivo Biocompatibility, ACS Appl. Bio Mater., 2020, 3, 7722–7733 CrossRef CAS PubMed.
- S. Ranjan Sarker, S. A. Polash, J. Boath, A. E. Kandjani, A. Poddar, C. Dekiwadia, R. Shukla, Y. Sabri and S. K. Bhargava, Functionalization of elongated tetrahexahedral Au nanoparticles and their antimicrobial activity assay, ACS Appl. Mater. Interfaces, 2019, 11, 13450–13459 CrossRef CAS PubMed.
- S. R. Sarker, S. A. Polash, M. N. Karim, T. Saha, C. Dekiwadia, V. Bansal, Y. Sabri, A. E. Kandjani and S. K. Bhargava, Functionalized concave cube gold nanoparticles as potent antimicrobial agents against pathogenic bacteria, ACS Appl. Bio Mater., 2022, 5, 492–503 CrossRef CAS PubMed.
- M. M. Hossain, S. A. Polash, T. Saha and S. R. Sarker, in Nano-Strategies for Addressing Antimicrobial Resistance: Nano-Diagnostics, Nano-Carriers, and Nano-Antimicrobials, ed. S. I. Publishing, Springer, 2022, pp. 311–351 Search PubMed.
- M. Arifuzzaman, S. A. Polash, M. M. Hossain and S. R. Sarker, in Biogenic Nanomaterial for Health and Environment, CRC Press, 2024, pp. 76–96 Search PubMed.
- S. A. Polash, A. Hamza, M. M. Hossain, C. Dekiwadia, T. Saha, R. Shukla, V. Bansal and S. R. Sarker, Lactoferrin functionalized concave cube Au nanoparticles as biocompatible antibacterial agent, OpenNano, 2023, 100163 CrossRef.
- M. M. Hossain, S. A. Polash, M. Takikawa, R. D. Shubhra, T. Saha, Z. Islam, S. Hossain, M. A. Hasan, S. Takeoka and S. R. Sarker, Investigation of the Antibacterial Activity and in vivo Cytotoxicity of Biogenic Silver Nanoparticles as Potent Therapeutics, Front. Bioeng. Biotechnol., 2019, 7, 239 CrossRef PubMed.
- K. M. Mahmud, M. M. Hossain, S. A. Polash, M. Takikawa, M. S. Shakil, M. F. Uddin, M. Alam, M. M. Ali Khan Shawan, T. Saha, S. Takeoka, M. A. Hasan and S. R. Sarker, Investigation of Antimicrobial Activity and Biocompatibility of Biogenic Silver Nanoparticles Synthesized using Syzigyum cymosum Extract, ACS Omega, 2022, 7, 27216–27229 CrossRef CAS PubMed.
- S. Polash, A. Hamza, M. Hossain, M. Tushar, M. Takikawa, R. Shubhra, N. Saiara, T. Saha, S. Takeoka and S. Sarker, Diospyros malabarica Fruit Extract Derived Silver Nanoparticles: A Biocompatible Antibacterial Agent, Front. Nanotechnol., 2022, 4, 888444 CrossRef.
- S. A. Polash, M. M. Hossain, T. Saha and S. R. Sarker, Biogenic Silver Nanoparticles: A Potent Therapeutic Agent, Emerging Trends in Nanomedicine, Springer Nature, 2021, pp. 81–127 Search PubMed.
- A.-C. Burduşel, O. Gherasim, A. M. Grumezescu, L. Mogoantă, A. Ficai and E. Andronescu, Biomedical applications of silver nanoparticles: an up-to-date overview, Nanomaterials, 2018, 8, 681 CrossRef PubMed.
- S. Poulose and E. Poulose, Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects, Int. Nano Lett., 2012, 2, 1 CrossRef.
- P. Chandra, P. Kumar Maurya, A. P. Kumar, P. Tripathi and A. K. Srivastava, Diagnosis of reumatic infections caused by group A Streptococcus Pyogenes: future investigation by nanotechnology, Digest J. Nanomater. Biostruct., 2009, 4(4), 645–650 Search PubMed.
- A. P. Gondikas, A. Morris, B. C. Reinsch, S. M. Marinakos, G. V. Lowry and H. Hsu-Kim, Cysteine-induced modifications of zero-valent silver nanomaterials: implications for particle surface chemistry, aggregation, dissolution, and silver speciation, Environ. Sci. Technol., 2012, 46, 7037–7045 CrossRef CAS PubMed.
- B. Ramalingam, T. Parandhaman and S. K. Das, Antibacterial effects of biosynthesized silver nanoparticles on surface ultrastructure and nanomechanical properties of Gram-negative bacteria viz. Escherichia coli and Pseudomonas aeruginosa, ACS Appl. Mater. Interfaces, 2016, 8, 4963–4976 CrossRef CAS PubMed.
- P. Traiwatcharanon, K. Timsorn and C. Wongchoosuk, Flexible room-temperature resistive humidity sensor based on silver nanoparticles, Mater. Res. Express, 2017, 4, 085038 CrossRef.
- R. A. Hamouda, M. H. Hussein, R. A. Abo-Elmagd and S. S. Bawazir, Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica, Sci. Rep., 2019, 9, 1–17 CrossRef CAS PubMed.
- G. Dang, R. Parekar, S. Kamat, A. Scindia and N. Rege, Antiinflammatory activity of Phyllanthus emblica, Plumbago zeylanica and Cyperus rotundus in acute models of inflammation, Phytother. Res., 2011, 25, 904–908 CrossRef CAS PubMed.
- S. Pientaweeratch, V. Panapisal and A. Tansirikongkol, Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and silymarin: An in vitro comparative study for anti-aging applications, Pharm. Biol., 2016, 54, 1865–1872 CrossRef CAS PubMed.
- R. Mondal, S. A. Polash, T. Saha, Z. Islam, M. Sikder, N. Alam, M. Hossain and S. R. Sarker, Investigation of the Phytoconstituents and Bioactivity of Various Parts of Wild Type and Cultivated Phyllanthus emblica L., Adv. Biosci. Biotechnol., 2017, 8, 211–227 CrossRef CAS.
- B. Ahmad, N. Hafeez, A. Rauf, S. Bashir, H. Linfang, M.-u. Rehman, M. S. Mubarak, M. S. Uddin, S. Bawazeer and M. A. Shariati, Phyllanthus emblica: A comprehensive review of its therapeutic benefits, S. Afr. J. Bot., 2021, 138, 278–310 CrossRef CAS.
- M. Gul, Z.-W. Liu, R. Rabail, F. Faheem, N. Walayat, A. Nawaz, M. A. Shabbir, P. E. Munekata, J. M. Lorenzo and R. M. Aadil, Functional and Nutraceutical Significance of Amla (Phyllanthus emblica L.): A Review, Antioxidants, 2022, 11, 816 CrossRef CAS PubMed.
- S. A. Polash, T. Saha, M. Hossain and S. R. Sarker, Investigation of the phytochemicals, antioxidant, and antimicrobial activity of the Andrographis paniculata leaf and stem extracts, Adv. Biosci. Biotechnol., 2017, 8, 149–162 CrossRef CAS.
- R. Renuka, K. R. Devi, M. Sivakami, T. Thilagavathi, R. Uthrakumar and K. Kaviyarasu, Biosynthesis of silver nanoparticles using Phyllanthus emblica fruit extract for antimicrobial application, Biocatal. Agric. Biotechnol., 2020, 24, 101567 CrossRef.
- M. M. I. Masum, M. M. Siddiqa, K. A. Ali, Y. Zhang, Y. Abdallah, E. Ibrahim, W. Qiu, C. Yan and B. Li, Biogenic synthesis of silver nanoparticles using Phyllanthus emblica fruit extract and its inhibitory action against the pathogen Acidovorax oryzae strain RS-2 of rice bacterial brown stripe, Front. Microbiol., 2019, 10, 820 CrossRef PubMed.
- V. Nayagam, M. Gabriel and K. Palanisamy, Green synthesis of silver nanoparticles mediated by Coccinia grandis and Phyllanthus emblica: a comparative comprehension, Appl. Nanosci., 2018, 8, 205–219 CrossRef CAS.
- R. D. Shubhra, S. A. Polash, T. Saha, A. Hasan, S. Hossain, Z. Islam and S. R. Sarker, Investigation of the phytoconstituents and antioxidant activity of Diospyros malabarica fruit extracts, Adv. Biosci. Biotechnol., 2019, 10, 431–454 CrossRef CAS.
- R. D. Shubhra, S. A. Polash, M. M. Hossain, A. Hamza, M. M. H. Tushar, T. Saha, M. A. Hasan, M. M. Sikder, N. Alam, Z. Islam, M. S. Hossain and S. R. Sarker, Investigation of the Antimicrobial Activity and in Vivo Cytotoxicity of Diospyros malabarica (Desr.) Kostel. Fruit Extracts, Nat. Sci., 2021, 13, 331–351 CAS.
- R. D. Shubhra, S. A. Polash, M. M. Hossain, A. Hamza, M. M. H. Tushar, T. Saha, M. A. Hasan, M. M. Sikder, N. Alam, Z. Islam, M. S. Hossain and S. R. Sarker, Investigation of the Antimicrobial Activity and in Vivo Cytotoxicity of Diospyros malabarica (Desr.) Kostel. Fruit Extracts, Nat. Sci., 2021, 13, 331–351 CAS.
- X. Li, S. M. Robinson, A. Gupta, K. Saha, Z. Jiang, D. F. Moyano, A. Sahar, M. A. Riley and V. M. Rotello, Functional gold nanoparticles as potent antimicrobial agents against multi-drug-resistant bacteria, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS PubMed.
- K. Dziendzikowska, J. Gromadzka-Ostrowska, A. Lankoff, M. Oczkowski, A. Krawczyńska, J. Chwastowska, M. Sadowska-Bratek, E. Chajduk, M. Wojewódzka, M. Dušinská and M. Kruszewski, Time-dependent biodistribution and excretion of silver nanoparticles in male Wistar rats, J. Appl. Toxicol., 2012, 32, 920–928 CrossRef CAS PubMed.
- L. Yang, H. Kuang, W. Zhang, Z. P. Aguilar, H. Wei and H. Xu, Comparisons of the biodistribution and toxicological examinations after repeated intravenous administration of silver and gold nanoparticles in mice, Sci. Rep., 2017, 7, 3303 CrossRef PubMed.
- P. Singh, A. Garg, S. Pandit, V. Mokkapati and I. J. N. Mijakovic, Antimicrobial effects of biogenic nanoparticles, Nanomaterials, 2018, 8, 1009 CrossRef PubMed.
- W. R. Li, X. B. Xie, Q. S. Shi, H. Y. Zeng, Y. S. Ou-Yang and Y. B. Chen, Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli, Appl. Microbiol. Biotechnol., 2010, 85, 1115–1122 CrossRef CAS PubMed.
- N. Alshaye, M. Elobeid, D. Alkhalifah and A. E. Mohammed, Characterization of biogenic silver nanoparticles by Salvadora persica leaves extract and Its application against some MDR pathogens E. coli and S. Aureus, Res. J. Microbiol., 2017, 12, 74–81 CAS.
- R. M. Elamawi, R. E. Al-Harbi and A. A. Hendi, Biosynthesis and characterization of silver nanoparticles using Trichoderma longibrachiatum and their effect on phytopathogenic fungi, Egypt. J. Biol. Pest Control, 2018, 28, 1–11 CrossRef.
- R. M. Silverstein and G. C. Bassler, Spectrometric identification of organic compounds, J. Chem. Educ., 1962, 39, 546 CrossRef CAS.
- R. F. Domingos, M. A. Baalousha, Y. Ju-Nam, M. M. Reid, N. Tufenkji, J. R. Lead, G. G. Leppard and K. J. Wilkinson, Characterizing manufactured nanoparticles in the environment: multimethod determination of particle sizes. Environmental science & technology, Environ. Sci. Technol., 2009, 43, 7277–7284 CrossRef CAS PubMed.
- M. J. Khan, K. Shameli, A. Q. Sazili, J. Selamat and S. Kumari, Rapid green synthesis and characterization of silver nanoparticles arbitrated by curcumin in an alkaline medium, Molecules, 2019, 24, 719 CrossRef CAS PubMed.
- X. Wang, K. Xu, W. Cui, X. Yang, M. F. Maitz, W. Li, X. Li and J. Chen, Controlled synthesis of mussel-inspired Ag nanoparticle coatings with demonstrated in vitro and in vivo antibacterial properties, Mater. Des., 2021, 208, 109944 CrossRef CAS.
- S. Ahmed, M. Ahmad, B. L. Swami and S. Ikram, Green synthesis of silver nanoparticles using Azadirachta indica aqueous leaf extract, J. Radiat. Res. Appl. Sci., 2016, 9, 1–7 Search PubMed.
- M. M. I. Masum, M. M. Siddiqa, K. A. Ali, Y. Zhang, Y. Abdallah, E. Ibrahim, W. Qiu, C. Yan and B. Li, Biogenic Synthesis of Silver Nanoparticles Using Phyllanthus emblica Fruit Extract and Its Inhibitory Action Against the Pathogen Acidovorax oryzae Strain RS-2 of Rice Bacterial Brown Stripe, Front. Microbiol., 2019, 10, 820 CrossRef PubMed.
- T. Dakal, A. Kumar, R. Majumdar and V. Yadav, Mechanistic basis of antimicrobial actions of silver nanoparticles, Front. Microbiol., 2016, 7, 1831 Search PubMed.
- R. Guerrero, Brock biology of microorganisms, Prentice Hall, Upper Saddle River, NJ, 2000 Search PubMed.
- G. Franci, A. Falanga, S. Galdiero, L. Palomba, M. Rai, G. Morelli and M. Galdiero, Silver nanoparticles as potential antibacterial agents, Molecules, 2015, 20, 8856–8874 CrossRef CAS PubMed.
- Z. Sui, X. Chen, L. Wang, L. Xu, W. Zhuang, Y. Chai and C. J. Yang, Capping effect of CTAB on positively charged Ag nanoparticles, Phys. E, 2006, 33, 308–314 CrossRef CAS.
- A. M. Fayaz, K. Balaji, M. Girilal, R. Yadav, P. T. Kalaichelvan and R. Venketesan, Nanomedicine, 2010, 6, 103–109 CrossRef CAS.
- F. Monedeiro, P. Pomastowski, M. Milanowski, T. Ligor and B. Buszewski, Monitoring of Bactericidal Effects of Silver Nanoparticles Based on Protein Signatures and VOC Emissions from Escherichia coli and Selected Salivary, J. Clin. Med., 2019, 8, 2024 CrossRef CAS PubMed.
- W. C. Reygaert, An overview of the antimicrobial resistance mechanisms of bacteria, AIMS Microbiol., 2018, 4, 482 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3pm00077j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |