Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Doxorubicin-loaded DNA origami nanostructures: stability in vitreous and their uptake and toxicity in ocular cells†
Anna
Klose
 *a,
Zahra
Gounani
*a,
Zahra
Gounani
 a,
Heini
Ijäs
a,
Heini
Ijäs
 b,
Tatu
Lajunen
b,
Tatu
Lajunen
 ac,
Veikko
Linko
ac,
Veikko
Linko
 *bd and
Timo
Laaksonen
*bd and
Timo
Laaksonen
 *ae
*ae
aDrug Research Program, Division of Pharmaceutical Biosciences, Faculty of Pharmacy, University of Helsinki, Viikinkaari 5, 00790 Helsinki, Finland. E-mail: anna.klose@helsinki.fi; timo.laaksonen@helsinki.fi
bBiohybrid Materials, Department of Bioproducts and Biosystems, Aalto University, P.O. Box 16100, 00076 Aalto, Finland
cSchool of Pharmacy, University of Eastern Finland, Yliopistonrinne 3, 70210 Kuopio, Finland
dInstitute of Technology, University of Tartu, Nooruse 1, 50411, Tartu, Estonia. E-mail: veikko.pentti.linko@ut.ee
eChemistry and Advanced Materials, Faculty of Engineering and Natural Sciences, Tampere University, Korkeakoulunkatu 8, 33720 Tampere, Finland
First published on 27th August 2024
Abstract
Biocompatibility and precise control over their size and shape make DNA origami nanostructures (DONs) promising for drug delivery applications. Whilst many investigations have focused on cancer treatment, this might not be the best fit for DONs that get degraded by nucleases in blood. In comparison, an eye is a uniquely isolated target organ, which could benefit from DONs to achieve and maintain therapeutic concentrations in diseases that threaten the eyesight of millions of patients every year. We investigated the loading of doxorubicin (DOX) as a model drug into three distinct DONs and tested their stability upon storage. Further, we chose one structure (24HB) to probe its stability under physiological conditions in cell media and porcine vitreous, before examining the uptake and effect of DOX-loaded 24HB (24HB-DOX) on the cell viability in a retinal cell line (ARPE-19). Similar to previous reports, the tested low μM loading concentrations of DOX resulted in high drug loadings of up to 34% (m/m), and remained mostly intact in water for at least 2 months at 4 °C. In cell media and porcine vitreous at 37 °C, however, 24HB required additional Mg2+ supplementation to avoid degradation and the loss of the attached fluorophores. With added Mg2+, 24HB remained stable in vitreous for 7 days at 37 °C. The treatment with 24HB-DOX was well tolerated by ARPE-19 cells, compared to the observed higher toxicity of free DOX. Uptake studies revealed, however, that in contrast to free DOX, very little 24HB-DOX was taken up by the cells. Instead, the particles were observed to attach around the cells. Hence, our results suggest that since the uptake seems to be the bottleneck for therapies using DONs, further strategies such as adding ocular targeting moieties are necessary to increase the uptake and efficacy of doxorubicin-loaded DONs.
Introduction
DNA origami nanostructures (DONs) are attractive nanocarriers in drug delivery applications because their dimensions and functionalities can be precisely designed, controlled and modified according to need.1–5 A single-stranded DNA scaffold strand of known sequence is folded into a desired DON shape by annealing with dozens of short, specifically designed, base-complementary staple strands under simple and reproducible conditions.6–8 Hence, DONs can be easily tailored to therapeutic applications, for instance, by fine-tuning their shape, size, surface charge and functionality to the preferences of the target cells.3,9 Furthermore, on their surface, DONs can display in a spatially-controlled manner targeting moieties, such as peptides,10 ligands,11,12 and aptamers,13–18 to enhance selective uptake and boost their drug delivery performance in vitro11,12,14,15,19,20 and in vivo.17,18 Yet, most importantly for biomedical applications, DONs are biodegradable and usually well tolerated in in vitro and in vivo systems.21–23However, the stability of DONs under physiological conditions, namely in low-magnesium environments (<1 mM) and their susceptibility against nucleases in the blood, still remains a concern and needs to be investigated.1 Because DONs are most commonly made up of densely packed DNA helices, the negative charge of their phosphate backbone requires sufficient magnesium concentrations to screen the electrostatic repulsion and maintain their structural integrity.24,25 DONs’ stability against magnesium depletion and digestion by enzymes is, however, superstructure-dependent, and hence, can necessitate addition of magnesium to the media,24 structural redesign26 or coating strategies.27–30
Furthermore, while DONs can carry various therapeutic molecules, such as antibody fragments31 or nucleic acids,10,32,33 the anti-cancer drug doxorubicin (DOX) is one of the most widely investigated.34 As DOX is a DNA-intercalating drug,35 it allows straightforward formation of complexes with DONs (DOX-DONs), achieving relatively high drug loadings. DOX-DONs also have been effective in circumventing drug resistance,36 whilst exhibiting little toxicity in vivo.37 This is especially promising as to avoid the severe, dose-limiting side effects of DOX, which include cardiotoxicity.38 Yet, it remains important to always ensure proper characterization DOX-DONs as drug nanocarriers, especially considering that reported loading and purification protocols for DOX-DONs vary largely and often lack comparability, with a tendency to overestimate drug loading if no suitable purification method is employed.39
Thus far, investigations of DONs for drug delivery have mainly centered around systemic administration in cancer applications,40 where DOX-DONs can accumulate non-specifically through enhanced permeability and retention effects,37 but are continuously exposed to nucleases and immune cells in the entire body. We propose that DONs could be more compatible with local administration to an isolated, immune-privileged target organ like the eye.41,42 Because overcoming the blood-retinal barrier in systemic applications is challenging, and penetration upon topical instillation is poor,41,43 local intravitreal injections into the eye ball remain the most used approach to deliver therapeutics to the back of the eye. Upon such intravitreal injection, not only are DONs shielded from nucleases, but specifically their intrinsic negative charge and controlled size could be advantageous to their mobility in the vitreous body,44 a gel-like matrix (>98% water) from collagen and glycosaminoglycans that fills the inner eye.45 Hence, DONs could be promising for treating ocular diseases, but studies to assess their compatibility and applicability are necessary.
Currently, 4.3 million people worldwide are affected by blindness, but the total number could increase to 61 million by 2050.46 Amongst the leading causes of blindness, age-related macular degeneration (AMD) alone is affecting 67 million patients in Europe, and due to an aging population, is expected to impair 77 million by 2050, displaying similar trends globally.47–49 Intravitreal injections of anti-VEGF (vascular endothelial growth factor) treatments are effective in reducing neovascularization in the disease progress of AMD, or diabetic macular edema (DME). As a downside, however, they still need to be administered indefinitely every other month to uphold therapeutic concentrations.50,51 Besides the risk for side effects and burden on the healthcare system, these regular injections restrict and impact patients physically and psychologically, albeit necessary to preserve their eyesight.43,52,53 Hence, there is a need to find suitable, long-lasting, and effective therapeutic strategies and nanocarriers,41 and to this aim, also continue to explore and characterize novel nanocarriers like DONs with unique characteristics that have not been evaluated before for ocular implementations.
Thus, the control over their specific size and shape, and charge makes DNA-based nanocarriers interesting for investigations into ocular drug delivery and its different barriers. For instance, tetrahedral framework nucleic acids of defined size successfully reached the retina after periocular application.54 Here, we were interested to see whether larger and more dense DONs could also serve as suitable nanocarriers for ocular drug delivery. Currently, DOX is not very common for treating ocular diseases, but reports have shown its beneficial effects against choroidal neovascularization55,56 as a HIF1α-inhibitor in reducing pro-angiogenic factors, which is promising against AMD,56,57 or in its application against retinoblastoma.58–62 Also in these applications, encapsulating DOX into drug carriers can increase its efficiency and reduce local toxicity.56–58,63 Taking advantage of their multiple, densely packed DNA helices, DONs are expected to show high drug loading capacities of DNA-intercalating drugs such as DOX, and hence, here we investigated and characterized the DOX-loading into three differently shaped DONs (24-helix bundle (24HB), 60-helix bundle (60HB) and a rectangular plate structure) and their storage stability. Further, to elucidate their structural stability under physiological conditions of low Mg2+ concentrations, fluorophore-labeled 24HB was exposed and tested in cell medium and porcine vitreous. Thirdly, we evaluated the impact of DOX-loaded 24HB and free DOX on cell viability and studied their uptake into an ocular cell line (ARPE-19), to better understand the opportunities and challenges of using drug-loaded DONs for ocular delivery.
Materials and methods
DNA origami (DONs) preparation
To study the effect of DON superstructure (shape and size) on DOX-loading, three DONs (24HB, 60HB and the plate) were folded and purified from excess staples via polyethylene glycol (PEG) precipitation (ESI section SI1†).30,39,64,65 Details of the DONs are listed in Fig. 1a. For confocal imaging and stability testing, Atto488-labeled 24HBs were prepared. First, 24 staple strands were replaced in the 24HB folding reaction with staple strands that were extended at the 3′ end.66 Then in a second step, complementary strands carrying Atto488 at the 5′ end were annealed to these extensions by cooling from 40 °C to 20 °C in steps of – 0.1 °C/40 s, followed by the removal of excess Atto488-strands via PEG-precipitation (ESI section SI1†).65,66 Two versions of Atto488-labeled 24HB were tested, using either 17 nt long (24HB-Atto488 short),66 or 23 nt long Atto488-modified-strands (24HB-Atto488) with 16 nt or 22 nt long staple strand extensions, respectively (for details, ESI section SI2 and Table SI2b†). Their melting temperatures were estimated using the IDT OligoAnalyzer™ Tool (ESI Fig. SI2a†). After purification, all DONs were resuspended overnight on a shaker (at 30 °C, 600 rpm) in their respective folding buffer (1× FOB comprising of 1× Tris-acetate-EDTA buffer (1× TAE: 40 mM Tris, 20 mM acetic acid, 1 mM EDTA), and 17.5 mM MgCl2 for 24HB, or 20 mM MgCl2 and 5 mM NaCl for 60HB and the plate).Characterization of DONs
Agarose gel electrophoresis (AGE) and transmission electron microscopy (TEM) verified proper folding and purification, Atto488 attachment and stability of DONs (ESI section SI1 and Fig. SI3, 4†). Mixed with 6× loading dye solution (Sigma Aldrich), samples were loaded and run in 2% agarose gels with 0.46 μg mL−1 ethidium bromide (EtBr) or 1× SYBR Safe (Invitrogen) at 90 V for 50 min (running buffer: 1× TAE, 11 mM MgCl2). Gels were visualized under UV light (EtBr) or blue light (Atto488 and SYBR Safe) using BioRad ChemiDoc MP imaging System or Universal Hood II.For TEM imaging, samples, fixated on plasma-cleaned Formvar carbon-coated copper grids (FCF-400-CU, Electron Microscopy Sciences), were negatively stained with 2% (w/v) uranyl formate solution (adjusted with 25 mM NaOH, ESI section SI1†),8 and imaged on FEI Tecnai 12 Bio-Twin electron microscope (120 kV acceleration voltage) or on Hitachi HT7800 Transmission Electron Microscope (100 kV acceleration voltage). Fiji-Image J was used to adjust the contrast or crop TEM images.
The hydrodynamic volume (intensity-weighed Z-average) and polydispersity index (PDI) of DONs was determined via dynamic light scattering (DLS) using an automated plate sampler (Zetasizer APS, Malvern Instruments).
DON concentration c (in mol L−1) was determined from its absorbance at 260 nm (A260), according to Lambert–Beer law (A = εcl, with a pathlength of l = 0.05 cm (for Take3™ plate reader) or nominally l = 10 mm (for NanoDrop One)). Based on the number of hybridized and non-hybridized nucleotides, the molar extinction coefficient ε was estimated for 24HB (1.05 × 108 M cm−1), 24HB-Atto488 short (1.08 × 108 M cm−1), 24HB-Atto488 (1.10 × 108 M cm−1), 60HB (0.91 × 108 M cm−1), and the plate (1.04 × 108 M cm−1).67
Buffer exchange and DOX loading into DONs
To reduce agglomeration, DONs were kept overnight on a shaker (30 °C, 600 rpm) before exchanging FOB to deionized water. Adapted from Kielar et al.,25 DONs (175 μL) were spun with water (175 μL) in pre-rinsed Amicon Ultra 0.5 mL centrifugal filters with 100 kDa molecular weight cut-off (MWCO) (Merck Millipore, 6000g, 10 min). After a second washing with water (314 μL, 6000g, 10 min), DONs were recovered (1000g, 2 min), diluted, and their concentration analyzed.Doxorubicin hydrochloride (DOX, 579.99 g mol−1, CRS, European Pharmacopoeia Reference Standard) stocks in water (10 mM) were thawed and diluted to 100 μM working solutions. DOX loading reactions in water, consisting of 2 nM of DONs with 2.5 μM, 5 μM, or 15 μM DOX, were vortexed before their incubation at room temperature for at least an hour. DONs without DOX, and free DOX at corresponding concentrations served as references of similar composition. Before purifying DOX-DONs from excess DOX, their absorbance spectra (240–650 nm) were recorded in a quartz cuvette (l = 1 cm, Varian Cary 50 UV-Vis spectrophotometer). DOX-DONs (480 μL) were centrifuged in pre-rinsed Amicon Ultra 0.5 mL centrifugal filter with 100 kDa MWCO (6000g, 10 min).39 After two washes with water (480 μL, 6000g, 10 min), purified DOX-DONs were recovered (1000g, 2 min), and rediluted with water (460 μL). After purification, their absorbance spectra were measured again. Taking into account losses in spin-filtration and the absorption of DOX at 543 nm and of DONs at 260 nm, DOX-DONs drug loading (m/m%), drug loading efficiency, and DOX packing density were calculated (for details see ESI Fig. SI5 and Table SI6†).
DON stability in water at 4 °C
DONs with or without loaded DOX were checked after a minimum of 6 weeks for signs of instability via AGE and TEM. DLS was used to monitor major aggregation and instability events over the course of 7 weeks (n = 3 with 2–3 measurements per sample).Porcine vitreous extraction
Porcine eyes on ice were procured from an abattoir (HKScan Finland Oyj, Finland) and their vitreous was isolated as previously described.44 In short, after removal of excess tissue and washing in 70% ethanol, the eyes were rinsed and stored in 1× Dulbecco's phosphate-buffered saline (DPBS, Gibco™) at 4 °C. The following day, the anterior part including the lens was excised with a scalpel to collect the vitreous. The vitreous was then homogenized on ice with a glass tissue homogenizer and centrifuged to sediment and remove melanin (3200g, 4 °C, 1 h). To the same aim, the decanted vitreous was also filtered through a 0.45 μM and 0.22 μM sterile filter (Sartorius, 16537 and K16532 K) before freezing aliquots at −80 °C. Before use, the vitreous was thawed overnight in the fridge.24HB stability in cell medium and vitreous
Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12, HEPES, Gibco™, suitable for ARPE-19 cells, in the following abbreviated as cell medium (CM)), was adjusted with MgCl2 to CM-x with varying final Mg2+ concentrations (n. = no additional Mg2+, x = final Mg2+-concentration in mM, composition: 2% (v/v) Mg2+/H2O, 98% (v/v) CM). Then, buffer-exchanged 24HB in water (10% (v/v)) were incubated in CM-x (90% (v/v)) for 4 h or 24 h, at 37 °C or room temperature, respectively. 24HB-DOX, 24HB-Atto488 short and 24HB-Atto488 were also tested. Additionally, shorter incubations such as 2 h, varying dilution ratios (25% or 50%), and corresponding references in 1× FOB or water were prepared and investigated.Further, homogenized porcine vitreous (V, 70 or 77% (v/v)) was incubated for up to 7 days with 24HB(-Atto488) in water (30 or 23% (v/v)) at 4 °C and 37 °C. The incubated samples were adjusted with MgCl2 to different to V-x (n. = no supplementation, x = final Mg2+-concentration in mM, without considering the preexisting Mg2+ content in vitreous). To check the integrity of 24HB after incubation, all samples were adjusted to similar final Mg2+ concentrations before running AGE. The vitreous samples were diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to reduce aggregation in the gel pocket before loading onto the gel. Selected samples were also fixated and visualized under TEM.
1) to reduce aggregation in the gel pocket before loading onto the gel. Selected samples were also fixated and visualized under TEM.
Cell culturing
The ARPE-19 cells (human retinal pigment epithelial cell line) were a kind gift from Prof. Arto Urtti at the University of Eastern Finland, and originally purchased from the American Type Culture Collection (ATCC). The ARPE-19 cells were cultured at 37 °C with 5% or 7% CO2 in TC-treated T75 flasks (Sarstedt) with CM (DMEM/F12, HEPES, Gibco™), supplemented with 10% fetal bovine serum (FBS, Gibco™) and 1% penicillin–streptomycin (PS, Gibco™).Cell viability assay
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded overnight into TC-treated 96-well plates (Sarstedt). The next day, the cells were washed with 1× DPBS, exposed to treatments for 4 h, and after washing with 1× DPBS, kept again overnight in CM (10% FBS, 1% PS). After 20 h, the CM was replaced with 1× alamarBlue™ Cell Viability Reagent (Invitrogen™) in CM (1% PS), and incubated for 3 h at 37 °C. The alamarBlue mix was transferred to a black-walled wellplate (Thermo Scientific™, 265301) for fluorescence read-out (Varioskan LUX, Thermo Scientific™, top optics, λexc = 560 nm, λdet = 590 nm). The assay was repeated three times on separate days with each three technical replicates, and the cell viability was expressed in % of viability in relation to the CM ref treatment (CM, 1% PS). The controls and treatments contained 24HB, free DOX or 24HB-DOX in CM with 1% PS, adjusted to a Mg2+-concentration of 3 mM after final dilution. CM 25% and CM 50% served as CM controls of similar composition but without any DONs or DOX. For 24HB-DOX, 24HB was loaded with 5 μM DOX, purified, upconcentrated and pooled after spin-filtration. For CM 25%, 24HB (5.3 nM), DOX (1–10 μM), and 24HB-DOX (1–10 μM), the treatments consisted of 25% (v/v) 24HB/DOX/H2O in 75% (v/v) CM-4 (4 mM MgCl2), while for CM 50%, 24HB (10.6 nM), DOX (20 μM), and 24HB-DOX (20 μM), they consisted of 50% (v/v) treatment and 50% (v/v) CM-6 (6 mM MgCl2). Triton X-100 1% in 1× DPBS served as a positive control.
000 cells per well were seeded overnight into TC-treated 96-well plates (Sarstedt). The next day, the cells were washed with 1× DPBS, exposed to treatments for 4 h, and after washing with 1× DPBS, kept again overnight in CM (10% FBS, 1% PS). After 20 h, the CM was replaced with 1× alamarBlue™ Cell Viability Reagent (Invitrogen™) in CM (1% PS), and incubated for 3 h at 37 °C. The alamarBlue mix was transferred to a black-walled wellplate (Thermo Scientific™, 265301) for fluorescence read-out (Varioskan LUX, Thermo Scientific™, top optics, λexc = 560 nm, λdet = 590 nm). The assay was repeated three times on separate days with each three technical replicates, and the cell viability was expressed in % of viability in relation to the CM ref treatment (CM, 1% PS). The controls and treatments contained 24HB, free DOX or 24HB-DOX in CM with 1% PS, adjusted to a Mg2+-concentration of 3 mM after final dilution. CM 25% and CM 50% served as CM controls of similar composition but without any DONs or DOX. For 24HB-DOX, 24HB was loaded with 5 μM DOX, purified, upconcentrated and pooled after spin-filtration. For CM 25%, 24HB (5.3 nM), DOX (1–10 μM), and 24HB-DOX (1–10 μM), the treatments consisted of 25% (v/v) 24HB/DOX/H2O in 75% (v/v) CM-4 (4 mM MgCl2), while for CM 50%, 24HB (10.6 nM), DOX (20 μM), and 24HB-DOX (20 μM), they consisted of 50% (v/v) treatment and 50% (v/v) CM-6 (6 mM MgCl2). Triton X-100 1% in 1× DPBS served as a positive control.
Confocal imaging
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded overnight to the Cellview cell culture slide (TC, glass bottom, 10 wells, Greiner, ref: 543078). The following day, the cells were washed with 1× DPBS and treated for 4 h or 24 h. The treatments contained 24HB-Atto488 (5.3 nM or 10.6 nM), Atto488-strands (254 nM, corresponding to the concentration for 24HB-Atto488 at 10.6 nM), free DOX (20 μM) or 24HB-Atto488-DOX (20 μM) in CM with 1% PS, adjusted to a Mg2+-concentration of 3 mM after final dilution. CM with similar dilution and composition without 24HB-Atto488 or DOX served as a reference. For 24HB-Atto488-DOX, 24HB-Atto488 was loaded with 5 μM DOX, purified, upconcentrated and pooled after spin-filtration. The treatments for 24HB-Atto488 at 5.3 nM concentration consisted of 25% (v/v) 24HB-Atto488/DOX/H2O and 75% CM-4 (4 mM MgCl2), or for 10.6 nM (and/or 20 μM DOX) of 50% (v/v) treatment and 50% CM-6 (6 mM MgCl2). The cells were stained at the end of the incubation time for 30 min with Hoechst 33258 pentahydrate (Invitrogen™), for 10 min with CellMask™ Deep Red plasma membrane stain (Invitrogen™) or for 1 h with LysoTracker™ Deep Red (Invitrogen™). After removal of the treatments, the cells were washed with 1× DPBS and replaced with CM (reference).
000 cells per well were seeded overnight to the Cellview cell culture slide (TC, glass bottom, 10 wells, Greiner, ref: 543078). The following day, the cells were washed with 1× DPBS and treated for 4 h or 24 h. The treatments contained 24HB-Atto488 (5.3 nM or 10.6 nM), Atto488-strands (254 nM, corresponding to the concentration for 24HB-Atto488 at 10.6 nM), free DOX (20 μM) or 24HB-Atto488-DOX (20 μM) in CM with 1% PS, adjusted to a Mg2+-concentration of 3 mM after final dilution. CM with similar dilution and composition without 24HB-Atto488 or DOX served as a reference. For 24HB-Atto488-DOX, 24HB-Atto488 was loaded with 5 μM DOX, purified, upconcentrated and pooled after spin-filtration. The treatments for 24HB-Atto488 at 5.3 nM concentration consisted of 25% (v/v) 24HB-Atto488/DOX/H2O and 75% CM-4 (4 mM MgCl2), or for 10.6 nM (and/or 20 μM DOX) of 50% (v/v) treatment and 50% CM-6 (6 mM MgCl2). The cells were stained at the end of the incubation time for 30 min with Hoechst 33258 pentahydrate (Invitrogen™), for 10 min with CellMask™ Deep Red plasma membrane stain (Invitrogen™) or for 1 h with LysoTracker™ Deep Red (Invitrogen™). After removal of the treatments, the cells were washed with 1× DPBS and replaced with CM (reference).
Samples were imaged using an inverted confocal laser scanning microscope (Leica TCS SP8 STED 3X CW 3D STED (Stimulated Emission Depletion), Germany). The 405 nm -diode laser was used for exciting Hoechst 33258, the argon 483 nm laser was used for exciting doxorubicin68 and Atto488, and the HeNe 633 nm laser was used for exciting LysoTracker™ Deep Red or CellMask™ Deep Red Plasma Membrane Stain. Images were obtained using a 63×/1.20 (water, wd 0.3 mm) objective, and analyzed using LAS X software version 3.7.4.23463 for Leica SP8.
Results and discussion
DOX-loading into DNA origami and DON storage stability
To investigate their drug loading capacity, three 3D DNA origami nanostructures (DONs) of different sizes and shapes, 24-helix bundle (24HB), 60-helix bundle (60HB), and plate, were loaded with the DNA-intercalating drug doxorubicin (DOX). After the incubation of 2 nM DONs in water with 2.5 μM, 5 μM or 15 μM DOX, and their subsequent purification from excess DOX via spin-filtration, the amount of DOX in DOX-loaded DONs (DOX-DONs) was determined via absorption measurements (ESI Fig. SI5†). Considering the recovery of DOX-DONs, drug loading was expressed in mass percent (m/m%) and compared for different DOX loading concentrations in different DONs.In agreement with previous reports, similar drug loading was achievable for all DONs structures, regardless of their differences in size, shape, aspect ratio, or DNA sequence (Fig. 1a).39 This was possibly due to alike GC-contents of tested DONs, offering similar amounts of intercalation sites per structure.35 Most importantly, an increase in the initial DOX loading concentration resulted in higher drug loading after purification (Fig. 1b):15,39 A six-fold increase in the initial DOX loading concentration from 2.5 μM to 15 μM at least doubled the final drug loading, for instance for 60HB from 16% (m/m) to 34% (m/m), corresponding to a denser packing of DOX molecules per base pairs (ESI Table SI6†). In the case of 60HB with an initial 15 μM DOX loading concentration (before purification corresponding to one DOX molecule for every 0.7 base pairs), the previously reported maximum of one intercalated DOX molecule every two to three base pairs was exceeded even after purification, with one DOX molecule available every 1.5 base pairs.39 The intercalation capacity for 60HB was nearly maxed out already with one DOX molecule every 3 base pairs after purifying a ∼5 μM DOX loading reaction (before purification corresponding to one DOX molecule every 2.2 base pairs). Therefore, the observed result suggested another mode of DOX binding and aggregation on top of DONs, contributing in part to its loading capacity.35 In line with these findings, loading efficiencies which expressed how much of the initially added DOX was loaded into the DONs, decreased with higher DOX loading concentrations from ∼76–84% for 2.5 μM DOX to ∼35–40% for 15 μM DOX (ESI Table SI6†), congruent with free DOX being removed in the purification step. Our final loaded DOX concentrations were in good agreement with reports that used similar loading conditions.39,69
While high loading efficiencies for DOX-DONs are widely acknowledged, the reported efficiencies vary between 7–71%.13–15,36,37,69–71 For instance, introducing specific twists into DONs can allow fine-tuning of DOX encapsulation and release.72 Yet, the variety in the ways for reporting DOX loadings – such as loading efficiency, DOX concentration, DOX per DON, or DOX per base pair – in combination with different and underreported DOX loading, purification and detection methods, limits and complicates the direct comparability.39 In comparison to our spin-filtering method, removal of free DOX via centrifugation often risks retaining DOX aggregates alongside of DOX-DONs and overestimating the actual DOX loading.39 In such case, DOX loading efficiencies would be skewed, being rather a reflection of the initial DOX loading concentration, and loading and purification conditions, than of the actual DOX-loading capacity of DONs. To ensure that all DOX was bound to DNA in further in vitro studies, we accepted lower total DOX loadings by using DOX loading concentrations within low μM, instead of mM range,13,14,36,37,70 and by opting to report drug loading (m/m%, ESI Fig. SI6†).
Importantly, most DONs and DOX-DONs maintained their structural integrity and electrophoretic mobility in deionized water at 4 °C for at least 6 weeks, as observed via agarose gel electrophoresis (AGE) and under the transmission electron microscope (TEM, Fig. 1c, d and ESI Fig. SI7–9†).25,64 The same discrete bands on the agarose gels for DONs and purified DOX-DONs confirmed that most of our DONs remained intact, and no shift for our DOX-DONs band was observable upon DOX loading (Fig. 1c). Only for the DOX-plate, a widened band hinted at some aggregation and some slight staple strand loss during storage.
While some imperfect and unraveled DONs were visible in TEM (Fig. 1d and ESI Fig. SI7, 8†), possibly also due to mechanical stress during spin-filtration, most retained their shape after DOX-loading,10,70 even though intercalated DOX can exert strain on DONs.12,72 In addition, buffer exchanges to low Mg2+ environments, such as water or cell media, can destabilize DONs,25 because sufficient Mg2+ ions are vital to screen the negative charges of their DNA phosphate backbones, reducing repulsion effects and allowing for compact DONs. In such case, more rigid DON structures like the 24HB, that are structurally supported by many crossovers in their staple and scaffold design, are more dependent on Mg2+ ions to ensure their stability.26,73 While some other ions and polymers can also stabilize DONs,74 low Mg2+ concentrations can cause local degradation in DONs, which, however, can self-repair once favorable ionic conditions are attained again.75
Additionally, no major aggregation and instability events, were detectable via dynamic light scattering (DLS). The polydispersity index (PDI) for all DONs started increasing only after 4 weeks of storage (ESI Fig. SI10†). This increase possibly indicated that during longer storage, DONs started interacting more with each other and forming aggregates,64 which depending on the structure could be harder to redisperse and lead also to multiple bands on AGE. Since 24HB appeared the most stable, it was selected for further in vitro studies.
24HB stability testing in cell media and porcine vitreous
In preparation for in vitro studies, 24HB with Atto488-fluorophores, attached via extended staple strands that hybridized with Atto488 strands (24HB-Atto488, ESI section SI2†), was probed for its structural integrity at 37 °C. To mimic the low-nuclease environment of the eye,76 24HB-Atto488 underwent 24 h-incubation at 37 °C in various dilutions of cell media (CM), revealing that additional Mg2+ supplementation to a minimum of 3 mM concentration (CM-3) was necessary to effectively stabilize the 24HB structure (Fig. 2a, EtBr-channel).24 23 nt-long staple extensions were required for a stabile attachment of the Atto488 strands under incubation at 37 °C in CM-3 (Fig. 2a, Atto-channel, ESI section SI2†). Shorter extensions (17 nt) led to significant losses of Atto488 strands already after 4 h at 37 °C in CM-3 (SI11†). Apart from 24HB itself requiring Mg2+ supplementation for stabilization, Mg2+ addition was still relevant to maintain the fluorophore attachment at 37 °C in CM. Increasing the DNA melting temperature in complex, high ionic environments by simply elongating fluorophore attachment strands is therefore a useful but not complete solution, although attachments of similar or even shorter length have been reported.3,12,18,24,77,78 Another possible solution to reduce the loss of fluorophores could be incorporating the fluorophore directly into the DONs by attaching it to the staple strands, though no systematic investigations in that regard are available.79Hahn et al. described similarly that DONs’ stability in CM (with <1 mM Mg2+) is time- and structure-dependent, but structural integrity and fluorophore attachment could be maintained with Mg2+ addition at 6 mM concentration, without any impact on in vitro studies.24 Since no Mg2+ addition was necessary if the 24HB-Atto488 in CM was incubated at room temperature, the various ions in cell media together with the higher temperature led to destabilizing interactions, if not sufficient Mg2+ was available to counterbalance them, like in FOB for instance. Remarkably though, 24HB-Atto488 was stable in water at 37 °C without any additional Mg2+. As previously discussed, low Mg2+ buffers can contribute to the destabilization of DONs if certain components in buffers like phosphate ions or EDTA can chelate Mg2+ ions.25 In the case of CM, however, the phosphate concentration is only ∼0.9 mM. Instead, abundant ion species in CM, such as sodium ions (∼136 mM) can be exchanged with Mg2+ ions on the phosphate backbone, thus, destabilizing the DON in a similar manner,25,26,74 but not in water. We similarly carried out stability testing for DOX-loaded 24HB-Atto488 structures: 24HB-Atto488-DOX appeared also stable in CM with 3 mM Mg2+, but not in water at 37 °C, illustrating that at higher temperatures, there was a more pronounced, destabilizing effect of DOX on the DONs structure (ESI Fig. SI12†). Further stabilization strategies to address this issue could involve increasing the crossover spacing in DONs to enhance their flexibility,26 or providing alternative stabilizing ions or polymeric coatings that can interact with the DNA phosphate backbone.3,27,74,80 Even though the stability of DONs is, in general, highly structure-dependent,25 stability testing is often alas superficially performed and reported, and sometimes does not reflect the subsequent in vitro conditions. This is especially relevant for uptake studies with fluorophore-labeled DONs, in which loss and possible uptake of degraded and free fluorophores can lead to misinterpretation.81
For more biorelevant stability testing, porcine vitreous at 37 °C was used. Here, the 24HB-Atto488 similarly required a minimum of 3 mM Mg2+ addition for stabilization (Fig. 2b and ESI Fig. SI13†). This is without considering any contribution of pre-existing Mg2+ in the vitreous (in humans around 0.84–0.96 mM Mg2+)82 that could amount to an additional ∼0.6–0.7 mM Mg2+ per sample. Upon 3 mM Mg2+ supplementation in vitro, however, 24HB-Atto488 even remained stable in vitreous for up to seven days (ESI Fig. SI14†), which is a reasonable residence time in the eye. While the reported residence times (hours to months) vary for nanoparticles depending on their properties and the animal model, 50 nm liposomes in rabbit eyes exhibited for instance an elimination half-life of 8 days.83 However, it seems difficult to suggest Mg2+ supplementation as a feasible stabilization strategy for DONs in in vivo settings, as small Mg2+ ions will be cleared out quicker than DONs. This would mean that for intravitreal application, selecting DONs that can better withstand low Mg2+ conditions, or further protective coatings could be needed to ensure structural integrity.27,74
TEM further revealed that at 37 °C, 24HB in CM without additional Mg2+ (CM n.) degraded into smaller, yet distinct fragments within 4 h (Fig. 3, red frame). Possibly, 24HB has some particularly rigid segments, where, under low Mg2+ conditions, the electrostatic repulsion between the DNA helices strained specific staple strands. This could have resulted in their dissociation, and the characteristic fragment pattern.73 In comparison, supplementing CM and vitreous with 3 mM Mg2+ mostly preserved the 24HB structure for 24 h. Interestingly, regardless of the widened band on the gel (ESI Fig. SI12†), 24HB-DOX in CM n. appeared less fractured under TEM, compared to the 24HB without DOX. This was unexpected since DOX as an intercalator, often does introduce strain into DONs,72 and on our agarose gels, appeared to be rather destabilizing like in the case of 24HB-DOX in CM n. and water (ESI Fig. SI12†), indicating that additional Mg2+ is necessary for stabilization. After investigating its structural integrity under in vitro conditions, 24HB-DOX was next screened in ARPE-19 cells for any toxic effect and their uptake behavior.
Treatment of an ocular cell line with DOX-loaded 24HB
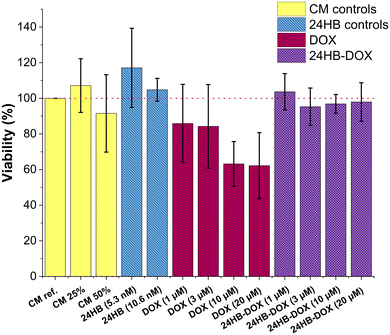
To assess the effect on their cell viability, ARPE-19 cells, a human retinal pigment epithelial cell line, were treated for 4 h with pooled 24HB-DOX, achieving final DONs concentration of up to 10.6 nM and DOX concentrations between 1–20 μM. The metabolic activity of the cells was then analyzed the following day via alamarBlue assay (Fig. 4). Further, ARPE-19 treated with undiluted CM with 1% PS (CM ref., yellow) was set to 100% viability, while CM treatments accounting for dilution and Mg2+ supplementation (CM 25%, 50%, also in yellow), and plain 24HB in two concentrations (24HB (5.3 nM, 10.6 nM), blue) served as controls, revealing that the chosen conditions did not impact the viability of the cells. This concurred with previous reports that plain DONs are well tolerable for cells.12,14 In contrast, ARPE-19 cells treated with free DOX (red) displayed at concentrations above 10 μM reduced cell viability by approximately 40%. At corresponding DOX concentrations, however, none of the 24HB-DOX treatments (purple) led to a decrease in cell viability, and instead, reduced the toxicity of DOX. Despite DOX not being a very commonly used drug in ocular diseases, when it is applied, a major concern is reducing its toxicity by avoiding high local concentrations and precipitation on the retinal surface.56 This is attainable by encapsulating DOX into particles, which also could positively improve the residence time in the eye.56,57,63,84Generally, reported toxicities for DOX-DONs vary because of the differences in experimental conditions, concerning the type of assay, treatment duration (hours to days), or drug-susceptibility of the cell line. Here, the short-time exposure of the ocular cell line ARPE-19 to DOX in FBS-free media did not significantly affect its metabolic activity. Focusing first on the toxic effect of free DOX in ARPE-19, some reports showed viabilities in a similar range.57,85,86 On the other hand, some report IC50 values between 0.13–1 μM DOX after 48–72 h DOX exposure, which is considerably more toxic, but understandable, as longer exposure can lead to higher uptake and therefore also higher toxicity.87–89 However, there are differences between cell lines, and in some cases DOX-DONs have performed similarly14,37,70 or worse15 than free DOX, while in others, DOX-DONs even increased toxicity by promoting intracellular DOX accumulation.36,72,79
To elucidate its drug-delivering capacities, we investigated the uptake 24HB-Atto488 into the ARPE-19 cells via live confocal imaging. After 4 h, no colocalization between 24HB-Atto488 and the lysosomes was observable, but instead, 24HB-Atto488 was clearly visible outside of the cells (ESI Fig. SI15†). Similarly, even after 24 h, the uptake of 24HB-Atto488-DOX (20 μM) was limited (Fig. 5 and ESI Fig. SI16†). Only few 24HB-Atto488 (green) could breach the cell membrane (red) within 24 h, while it posed no barrier to free DOX (20 μM, yellow) that was localized in the nucleus as expected. In the case of 24HB-Atto488-DOX (20 μM), some DOX was also evident in the nucleus (faint yellow signal), but visibly less than in the case of free DOX treatment. While this could stem from some premature DOX release, we have observed previously using steady-state and time-resolved fluorescence anisotropy measurements that our DOX-loading and purification protocols ensure that all DOX present in the sample remained bound to DONs.90 24HB-Atto488 were mostly seen at the bottom surface of the wells around the cells (ESI Fig. SI17–19†). In contrast to our observations, many publications have reported that DONs end up in the cytosol after escaping the lysosomes, while DOX then becomes apparent in the nucleus.14,36,70,72,77,91 Since close to none of the Atto488-strands by itself were taken up by cells (ESI Fig. SI20†), it confirmed that the green signal inside of the cells came from uptaken 24HB-Atto488, and their adhesion to the cell membrane was not mediated by the fluorophore. This, in combination with the low toxicity observed for 24HB-DOX, underlined that the uptake of 24HB-Atto488 was simply not very efficient, and hence contributed to the reduced DOX toxicity.
This limited uptake is plausible, as the inherent negative charge of DONs can hinder their uptake.11,12,79 While the uptake behavior of certain cells such as macrophages is unaffected by charge,3 often polymeric or protein coatings on DONs are necessary to facilitate cell entry.3,27,28,92 Generally, uptake behaviors and kinetics can depend on the cell line, with for instance specialized immune cells outperforming epithelial or endothelial cells by far.3,9 This complicates inferring from DONs uptake studies performed in other cell lines than ARPE-19, but it would offer an explanation for the low efficiency in cell entry that needs to be improved.
The dimensions of DONs is another factor influencing their cellular uptake: systematic investigations showed that cells seem to prefer compact and big DONs for endocytosis, with a low aspect ratio and directly structure-incorporated fluorophores.9 While 24HB-Atto488 is compact, and DON of similar dimensions have been internalized by cells,78 it is also quite stretched with Atto488 annealed via staple strand extensions, and hence, not exactly matching these criteria. While uptake kinetics are cell line-dependent and uptake increases for some after longer incubation,9,12,78 this was not observable for ARPE-19 within 24 h. While ARPE-19 cells tolerated the prolonged Mg2+-supplemented DONs treatment, the addition of FBS would be advisable to maintain cell health for monitoring the uptake over even longer periods. But, since ARPE-19 can effectively take up folate-targeted nanoparticles of up to 250 nm in size within 4 h,93 this rather suggests a low internalization efficiency of negatively charged 24HB itself, and the missing colocalization with lysosomes possibly hinted at the lack of receptor-mediated uptake.
Unfortunately, our knowledge on how DONs enter cells is still lacking. Some of the uncertainty is due to the variability of experimental conditions in vitro in published works, varying choice of DONs and cell lines, and originate from underreporting. However, in the case of effectively uptaken DONs, receptor-mediated uptake pathways,94via scavenger receptors,79,95 or caveolin-dependent,78,79 or lipid-raft mediated14 endocytosis have been suggested as uptake route. Hence, incorporating targeting moieties, such as aptamers,14–19,69 folate,12,20 and cell penetrating peptides,10 can improve uptake, which could be also a promising approach for our DONs to ARPE-19 cells and possibly other ocular cell lines.55,93 Especially since through receptor binding, spatial distribution of aptamers on DONs can regulate cell uptake and increase target specificity, which can offer unique optimization opportunities.13
In summary, we showed that DONs could effectively be loaded with DOX, independent of their superstructure, and remained stable under physiological conditions in cell media and in vitreous upon Mg2+ supplementation. Further, in ARPE-19 cells, DONs and DOX-DONs were well tolerated compared to free DOX. However, the uptake of DOX-DONs into cells was very low, warranting additional strategies such as coatings and targeting moieties to increase the efficiency of cell uptake for such DONs.
Conclusions
DONs are promising nanocarriers in drug delivery applications, for the versatility and predictability of their structure allows precise tailoring and optimization to improve their drug delivering capacities. This makes DONs also interesting for intraocular drug delivery, where therapeutic drug concentrations are hard to achieve and maintain, but where the eye presents a uniquely isolated target organ. DONs intercalated with DOX allowed for desirably high drug loadings, whilst also demonstrating structural integrity over an extended storage period. Stability testing revealed emerging structural defects under physiological conditions at 37 °C in cell media and porcine vitreous of the eye, that were, however, addressable with additional Mg2+ supplementation. DOX-DONs showed no toxicity in a retinal cell line (ARPE-19), compared to free DOX, which could be favorable in reducing irritation and side-effects during application. Their uptake into ARPE-19 cells was, however, quite limited, warranting additional strategies to improve their uptake. We envision for instance, that incorporating specific targeting moieties onto DONs could help overcome this limited uptake and even contribute to selective uptake by specific target cells, making them more efficient for ocular drug delivery.Data availability
The data supporting this article has been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support by the Emil Aaltonen Foundation, the Sigrid Jusélius Foundation, the Jane and Aatos Erkko Foundation, ERA Chair MATTER from the European Union's Horizon 2020 research and innovation programme under grant agreement No 856705, Mobilitas 3.0 Programme under the framework of the JPIAMR – Joint Programming Initiative on Antimicrobial Resistance, European Research Council Consolidator Grant (PADRE, 101001016), Academy of Finland (project no. 323669, flagship GeneCellNano), and the Finnish Cultural Foundation. We also thank the Light Microscopy Unit (LMU), and Electron Microscopy Unit (EMBI) at the Institute of Biotechnology, supported by HiLIFE and Biocenter Finland, at the University of Helsinki. We acknowledge the provision of facilities and technical support by Aalto University at OtaNano – Nanomicroscopy Center (Aalto-NMC). A. K. would like to thank the Biohybrid Materials Group under Prof. Mauri Kostiainen at Aalto University for their support and access to their facilities. Part of the graphical abstract was created with BioRender.com.References
- A. Keller and V. Linko, Angew. Chem., Int. Ed., 2020, 59, 15818–15833 CrossRef CAS.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Nucleic Acids Res., 2009, 37, 5001–5006 CrossRef CAS PubMed.
- M. M. Koga, A. Comberlato, H. J. Rodríguez-Franco and M. M. C. Bastings, Biomacromolecules, 2022, 23, 2586–2594 CrossRef CAS PubMed.
- S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan and P. Zhan, Nat. Rev. Methods Primers, 2021, 1, 13 CrossRef CAS.
- T. Tørring, N. V. Voigt, J. Nangreave, H. Yan and K. V. Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS.
- S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 Search PubMed.
- C. E. Castro, F. Kilchherr, D.-N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe and H. Dietz, Nat. Methods, 2011, 8, 221–229 CrossRef CAS PubMed.
- M. M. C. Bastings, F. M. Anastassacos, N. Ponnuswamy, F. G. Leifer, G. Cuneo, C. Lin, D. E. Ingber, J. H. Ryu and W. M. Shih, Nano Lett., 2018, 18, 3557–3564 CrossRef CAS.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang, F. Liu, S. Liu, S. Zhao, Z. Han, J. Sun, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2021, 60, 2594–2598 CrossRef CAS.
- Z. Ge, L. Guo, G. Wu, J. Li, Y. Sun, Y. Hou, J. Shi, S. Song, L. Wang, C. Fan, H. Lu and Q. Li, Small, 2020, 16, 1904857 CrossRef CAS PubMed.
- S. Pal and T. Rakshit, Front. Chem., 2021, 9, 721105 CrossRef CAS PubMed.
- K. Liu, C. Xu and J. Liu, J. Mater. Chem. B, 2020, 8, 6802–6809 RSC.
- Q. Pan, C. Nie, Y. Hu, J. Yi, C. Liu, J. Zhang, M. He, M. He, T. Chen and X. Chu, ACS Appl. Mater. Interfaces, 2020, 12, 400–409 CrossRef CAS PubMed.
- S. Chaithongyot, R. Duangrat, C. Wootthichairangsan, R. Hanchaina, A. Udomprasert and T. Kangsamaksin, Mater. Lett., 2020, 260, 126952 CrossRef CAS.
- P. Sun, N. Zhang, Y. Tang, Y. Yang, J. Zhou and Y. Zhao, RSC Adv., 2018, 8, 26300–26308 RSC.
- J. Liu, L. Song, S. Liu, S. Zhao, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2018, 57, 15486–15490 CrossRef CAS PubMed.
- J. Liu, L. Song, S. Liu, Q. Jiang, Q. Liu, N. Li, Z.-G. Wang and B. Ding, Nano Lett., 2018, 18, 3328–3334 CrossRef CAS PubMed.
- R. Chen, P. Sun, X. Chu, X. Pu, Y. Yang, N. Zhang and Y. Zhao, Int. J. Nanomed., 2020, 15, 1309–1320 CrossRef CAS PubMed.
- N. Navarro, A. Aviñó, Ò. Domènech, J. H. Borrell, R. Eritja and C. Fàbrega, Nanomedicine, 2024, 55, 102722 CrossRef CAS PubMed.
- R. R. Du, E. Cedrone, A. Romanov, R. Falkovich, M. A. Dobrovolskaia and M. Bathe, ACS Nano, 2022, 16, 20340–20352 CrossRef CAS PubMed.
- C. R. Lucas, P. D. Halley, A. A. Chowdury, B. K. Harrington, L. Beaver, R. Lapalombella, A. J. Johnson, E. K. Hertlein, M. A. Phelps, J. C. Byrd and C. E. Castro, Small, 2022, 18, 2108063 CrossRef CAS PubMed.
- V. J. Schüller, S. Heidegger, N. Sandholzer, P. C. Nickels, N. A. Suhartha, S. Endres, C. Bourquin and T. Liedl, ACS Nano, 2011, 5, 9696–9702 CrossRef PubMed.
- J. Hahn, S. F. J. Wickham, W. M. Shih and S. D. Perrault, ACS Nano, 2014, 8, 8765–8775 CrossRef CAS PubMed.
- C. Kielar, Y. Xin, B. Shen, M. A. Kostiainen, G. Grundmeier, V. Linko and A. Keller, Angew. Chem., Int. Ed., 2018, 57, 9470–9474 CrossRef CAS PubMed.
- Y. Xin, P. Piskunen, A. Suma, C. Li, H. Ijäs, S. Ojasalo, I. Seitz, M. A. Kostiainen, G. Grundmeier, V. Linko and A. Keller, Small, 2022, 18, 2107393 CrossRef CAS PubMed.
- N. Ponnuswamy, M. M. C. Bastings, B. Nathwani, J. H. Ryu, L. Y. T. Chou, M. Vinther, W. A. Li, F. M. Anastassacos, D. J. Mooney and W. M. Shih, Nat. Commun., 2017, 8, 15654 CrossRef CAS PubMed.
- H. Auvinen, H. Zhang, Nonappa, A. Kopilow, E. H. Niemelä, S. Nummelin, A. Correia, H. A. Santos, V. Linko and M. A. Kostiainen, Adv. Healthcare Mater., 2017, 6, 1700692 CrossRef PubMed.
- S. D. Perrault and W. M. Shih, ACS Nano, 2014, 8, 5132–5140 CrossRef CAS.
- S. Julin, Nonappa, B. Shen, V. Linko and M. A. Kostiainen, Angew. Chem., Int. Ed., 2021, 60, 827–833 Search PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, Nat. Mater., 2021, 20, 421–430 CrossRef CAS PubMed.
- X. Wu, Q. Liu, F. Liu, T. Wu, Y. Shang, J. Liu and B. Ding, Nanoscale, 2021, 13, 12848–12853 RSC.
- Y. Zhang, X. Tian, Z. Wang, H. Wang, F. Liu, Q. Long and S. Jiang, Front. Mol. Biosci., 2023, 10, 1239952 CrossRef CAS PubMed.
- C. Pérez-Arnaiz, N. Busto, J. M. Leal and B. García, J. Phys. Chem. B, 2014, 118, 1288–1295 CrossRef PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z.-G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- Q. Zhang, Q. Jiang, N. Li, L. Dai, Q. Liu, L. Song, J. Wang, Y. Li, J. Tian, B. Ding and Y. Du, ACS Nano, 2014, 8, 6633–6643 CrossRef CAS PubMed.
- V. B. Pai and M. C. Nahata, Drug Saf., 2000, 22, 263–302 CrossRef CAS PubMed.
- H. Ijäs, B. Shen, A. Heuer-Jungemann, A. Keller, M. A. Kostiainen, T. Liedl, J. A. Ihalainen and V. Linko, Nucleic Acids Res., 2021, 49, 3048–3062 CrossRef.
- S. Zhang, X.-Y. Lou, L. Liu and Y.-W. Yang, Adv. Healthcare Mater., 2023, 12, 2301066 CrossRef CAS PubMed.
- E. M. Del Amo, A.-K. Rimpelä, E. Heikkinen, O. K. Kari, E. Ramsay, T. Lajunen, M. Schmitt, L. Pelkonen, M. Bhattacharya, D. Richardson, A. Subrizi, T. Turunen, M. Reinisalo, J. Itkonen, E. Toropainen, M. Casteleijn, H. Kidron, M. Antopolsky, K.-S. Vellonen, M. Ruponen and A. Urtti, Prog. Retinal Eye Res., 2017, 57, 134–185 CrossRef CAS PubMed.
- J. V. Forrester and H. Xu, Front. Immunol., 2012, 3, 338 CAS.
- D. F. Martin, Am. J. Ophthalmol., 2018, 191, xli–xlviii CrossRef CAS PubMed.
- S. Tavakoli, O. K. Kari, T. Turunen, T. Lajunen, M. Schmitt, J. Lehtinen, F. Tasaka, P. Parkkila, J. Ndika, T. Viitala, H. Alenius, A. Urtti and A. Subrizi, Mol. Pharm., 2021, 18, 699–713 CrossRef CAS PubMed.
- B. T. Käsdorf, F. Arends and O. Lieleg, Biophys. J., 2015, 109, 2171–2181 CrossRef PubMed.
- R. Bourne, J. D. Steinmetz, S. Flaxman, P. S. Briant, H. R. Taylor, S. Resnikoff, R. J. Casson, A. Abdoli, E. Abu-Gharbieh, A. Afshin, H. Ahmadieh, Y. Akalu, A. A. Alamneh, W. Alemayehu, A. S. Alfaar, V. Alipour, E. W. Anbesu, S. Androudi, J. Arabloo, A. Arditi, M. Asaad, E. Bagli, A. A. Baig, T. W. Bärnighausen, M. Battaglia Parodi, A. S. Bhagavathula, N. Bhardwaj, P. Bhardwaj, K. Bhattacharyya, A. Bijani, M. Bikbov, M. Bottone, T. Braithwaite, A. M. Bron, Z. A. Butt, C.-Y. Cheng, D.-T. Chu, M. V. Cicinelli, J. M. Coelho, B. Dagnew, X. Dai, R. Dana, L. Dandona, R. Dandona, M. A. Del Monte, J. P. Deva, D. Diaz, S. Djalalinia, L. E. Dreer, J. R. Ehrlich, L. B. Ellwein, M. H. Emamian, A. G. Fernandes, F. Fischer, D. S. Friedman, J. M. Furtado, A. M. Gaidhane, S. Gaidhane, G. Gazzard, B. Gebremichael, R. George, A. Ghashghaee, M. Golechha, S. Hamidi, B. R. Hammond, M. E. R. Hartnett, R. K. Hartono, S. I. Hay, G. Heidari, H. C. Ho, C. L. Hoang, M. Househ, S. E. Ibitoye, I. M. Ilic, M. D. Ilic, A. D. Ingram, S. S. N. Irvani, R. P. Jha, R. Kahloun, H. Kandel, A. S. Kasa, J. H. Kempen, M. Keramati, M. Khairallah, E. A. Khan, R. C. Khanna, M. N. Khatib, J. E. Kim, Y. J. Kim, S. Kisa, A. Kisa, A. Koyanagi, O. P. Kurmi, V. C. Lansingh, J. L. Leasher, N. Leveziel, H. Limburg, M. Majdan, N. Manafi, K. Mansouri, C. McAlinden, S. F. Mohammadi, A. Mohammadian-Hafshejani, R. Mohammadpourhodki, A. H. Mokdad, D. Moosavi, A. R. Morse, M. Naderi, K. S. Naidoo, V. Nangia, C. T. Nguyen, H. L. T. Nguyen, K. Ogundimu, A. T. Olagunju, S. M. Ostroff, S. Panda-Jonas, K. Pesudovs, T. Peto, Z. Quazi Syed, M. H. U. Rahman, P. Y. Ramulu, S. Rawaf, D. L. Rawaf, N. Reinig, A. L. Robin, L. Rossetti, S. Safi, A. Sahebkar, A. M. Samy, D. Saxena, J. B. Serle, M. A. Shaikh, T. T. Shen, K. Shibuya, J. I. Shin, J. C. Silva, A. Silvester, J. A. Singh, D. Singhal, R. S. Sitorus, E. Skiadaresi, V. Skirbekk, A. Soheili, R. A. R. C. Sousa, E. E. Spurlock, D. Stambolian, B. W. Taddele, E. G. Tadesse, N. Tahhan, M. I. Tareque, F. Topouzis, B. X. Tran, R. S. Travillian, M. K. Tsilimbaris, R. Varma, G. Virgili, Y. X. Wang, N. Wang, S. K. West, T. Y. Wong, Z. Zaidi, K. A. Zewdie, J. B. Jonas and T. Vos, Lancet Global Health, 2021, 9, e130–e143 CrossRef PubMed.
- W. L. Wong, X. Su, X. Li, C. M. G. Cheung, R. Klein, C.-Y. Cheng and T. Y. Wong, Lancet Global Health, 2014, 2, e106–e116 CrossRef PubMed.
- J. D. Steinmetz, R. R. A. Bourne, P. S. Briant, S. R. Flaxman, H. R. B. Taylor, J. B. Jonas, A. A. Abdoli, W. A. Abrha, A. Abualhasan, E. G. Abu-Gharbieh, T. G. Adal, A. Afshin, H. Ahmadieh, W. Alemayehu, S. A. S. Alemzadeh, A. S. Alfaar, V. Alipour, S. Androudi, J. Arabloo, A. B. Arditi, B. B. Aregawi, A. Arrigo, C. Ashbaugh, E. D. Ashrafi, D. D. Atnafu, E. A. Bagli, A. A. W. Baig, T. W. Bärnighausen, M. Battaglia Parodi, M. S. Beheshti, A. S. Bhagavathula, N. Bhardwaj, P. Bhardwaj, K. Bhattacharyya, A. Bijani, M. Bikbov, M. Bottone, T. M. Braithwaite, A. M. Bron, S. A. Burugina Nagaraja, Z. A. Butt, F. L. L. Caetano Dos Santos, V. L. J. Carneiro, R. J. Casson, C.-Y. J. Cheng, J.-Y. J. Choi, D.-T. Chu, M. V. M. Cicinelli, J. M. G. Coelho, N. G. A. Congdon, R. A. A. Couto, E. A. M. Cromwell, S. M. Dahlawi, X. Dai, R. Dana, L. Dandona, R. A. Dandona, M. A. Del Monte, M. Derbew Molla, N. A. Dervenis, A. A. P. Desta, J. P. Deva, D. Diaz, S. E. Djalalinia, J. R. Ehrlich, R. R. Elayedath, H. R. B. Elhabashy, L. B. Ellwein, M. H. Emamian, S. Eskandarieh, F. G. Farzadfar, A. G. Fernandes, F. S. Fischer, D. S. M. Friedman, J. M. Furtado, S. Gaidhane, G. Gazzard, B. Gebremichael, R. George, A. Ghashghaee, S. A. Gilani, M. Golechha, S. R. Hamidi, B. R. R. Hammond, M. E. R. K. Hartnett, R. K. Hartono, A. I. Hashi, S. I. Hay, K. Hayat, G. Heidari, H. C. Ho, R. Holla, M. J. Househ, J. J. E. Huang, S. E. M. Ibitoye, I. M. D. Ilic, M. D. D. Ilic, A. D. N. Ingram, S. S. N. Irvani, S. M. S. Islam, R. Itumalla, S. P. Jayaram, R. P. Jha, R. Kahloun, R. Kalhor, H. Kandel, A. S. Kasa, T. A. Kavetskyy, G. A. H. Kayode, J. H. Kempen, M. Khairallah, R. A. Khalilov, E. A. C. Khan, R. C. Khanna, M. N. A. Khatib, T. A. E. Khoja, J. E. Kim, Y. J. Kim, G. R. Kim, S. Kisa, A. Kisa, S. Kosen, A. Koyanagi, B. Kucuk Bicer, V. P. Kulkarni, O. P. Kurmi, I. C. Landires, V. C. L. Lansingh, J. L. E. Leasher, K. E. LeGrand, N. Leveziel, H. Limburg, X. Liu, S. Madhava Kunjathur, S. Maleki, N. Manafi, K. Mansouri, C. G. McAlinden, G. G. M. Meles, A. M. Mersha, I. M. R. Michalek, T. R. Miller, S. Misra, Y. Mohammad, S. F. A. Mohammadi, J. A. H. Mohammed, A. H. Mokdad, M. A. A. Moni, A. A. R. Montasir, A. R. F. Morse, G. F. C. Mulaw, M. Naderi, H. S. Naderifar, K. S. Naidoo, M. D. Naimzada, V. Nangia, S. M. Narasimha Swamy, D. M. Naveed, H. L. Negash, H. L. Nguyen, V. A. Nunez-Samudio, F. A. Ogbo, K. T. Ogundimu, A. T. E. Olagunju, O. E. Onwujekwe, N. O. Otstavnov, M. O. Owolabi, K. Pakshir, S. Panda-Jonas, U. Parekh, E.-C. Park, M. Pasovic, S. Pawar, K. Pesudovs, T. Q. Peto, H. Q. Pham, M. Pinheiro, V. Podder, V. Rahimi-Movaghar, M. H. U. Y. Rahman, P. Y. Ramulu, P. Rathi, S. L. Rawaf, D. L. Rawaf, L. Rawal, N. M. Reinig, A. M. Renzaho, A. L. Rezapour, A. L. Robin, L. Rossetti, S. Sabour, S. Safi, A. Sahebkar, M. A. M. Sahraian, A. M. Samy, B. Sathian, G. K. Saya, M. A. Saylan, A. A. A. Shaheen, M. A. T. Shaikh, T. T. Shen, K. S. Shibuya, W. S. Shiferaw, M. Shigematsu, J. I. Shin, J. C. Silva, A. A. Silvester, J. A. Singh, D. S. Singhal, R. S. Sitorus, E. Y. Skiadaresi, V. Y. A. Skryabin, A. A. Skryabina, A. B. Soheili, M. B. A. R. C. Sorrie, R. A. R. C. Sousa, C. T. Sreeramareddy, D. G. Stambolian, E. G. Tadesse, N. I. Tahhan, M. I. Tareque, F. X. Topouzis, B. X. Tran, G. K. Tsegaye, M. K. Tsilimbaris, R. Varma, G. Virgili, A. T. Vongpradith, G. T. Vu, Y. X. Wang, N. H. Wang, A. H. K. Weldemariam, S. K. G. West, T. G. Y. Wondmeneh, T. Y. Wong, M. Yaseri, N. Yonemoto, C. S. Yu, M. S. Zastrozhin, Z.-J. R. Zhang, S. R. Zimsen, S. Resnikoff and T. Vos, Lancet Global Health, 2021, 9, e144–e160 CrossRef PubMed.
- J. Q. Li, T. Welchowski, M. Schmid, M. M. Mauschitz, F. G. Holz and R. P. Finger, Br. J. Ophthalmol., 2020, 104, 1077–1084 CrossRef PubMed.
- U. Schmidt-Erfurth, V. Chong, A. Loewenstein, M. Larsen, E. Souied, R. Schlingemann, B. Eldem, J. Monés, G. Richard and F. Bandello, Br. J. Ophthalmol., 2014, 98, 1144–1167 CrossRef.
- U. Schmidt-Erfurth, J. Garcia-Arumi, F. Bandello, K. Berg, U. Chakravarthy, B. S. Gerendas, J. Jonas, M. Larsen, R. Tadayoni and A. Loewenstein, Ophthalmologica, 2017, 237, 185–222 CrossRef PubMed.
- R. Wang, C. K. McClard, S. Laswell, R. Mahmoudzadeh, M. Salabati, M. Ammar, J. Vannavong, A. A. Aziz, A. Ewald, A. V. Calvanese, E. B. Lehman, S. Fried, V. Windham, A. Strutt, N. Saroj, A. M. Khanani, D. A. Eichenbaum, C. Regillo and C. C. Wykoff, BMJ Open Ophthalmol., 2022, 7, e001188 CrossRef PubMed.
- C. K. McClard, R. Wang, V. Windham, J. Munoz, S. Gomez, S. Fried, N. Saroj, C. Regillo, C. C. Wykoff and A. M. Strutt, BMJ Open Ophthalmol., 2021, 6, e000669 CrossRef PubMed.
- R. Wang, Y. Liu, W. Xiao, Q. Yi, M. Jiang, R. Guo, L. Song, M. Li, F. Li, D. Shi, L. Zhao, W. Huang, X. Zuo and X. Mao, ACS Appl. Mater. Interfaces, 2023, 15, 541–551 CrossRef CAS PubMed.
- J. Wang, Y. Liu, Y. Li, W. Dai, Z. Guo, Z. Wang and Q. Zhang, Invest. Ophthalmol. Visual Sci., 2012, 53, 7348–7357 CrossRef CAS PubMed.
- T. Iwase, J. Fu, T. Yoshida, D. Muramatsu, A. Miki, N. Hashida, L. Lu, B. Oveson, R. Lima e Silva, C. Seidel, M. Yang, S. Connelly, J. Shen, B. Han, M. Wu, G. L. Semenza, J. Hanes and P. A. Campochiaro, J. Controlled Release, 2013, 172, 625–633 CrossRef CAS PubMed.
- S. J. Kelly, K. Halasz, R. Smalling and V. Sutariya, Drug Dev. Ind. Pharm., 2019, 45, 715–723 CrossRef CAS.
- W. Farhat, V. Yeung, F. Kahale, M. Parekh, J. Cortinas, L. Chen, A. E. Ross and J. B. Ciolino, Bioengineering, 2022, 9, 671 CrossRef CAS PubMed.
- S. H. S. Boddu, J. Jwala, M. R. Chowdhury and A. K. Mitra, J. Ocul. Pharmacol. Ther., 2010, 26, 459–468 CrossRef CAS PubMed.
- K. Long, Y. Yang, W. Lv, K. Jiang, Y. Li, A. C. Y. Lo, W. C. Lam, C. Zhan and W. Wang, Adv. Sci., 2021, 8, 2101754 CrossRef CAS PubMed.
- R. Gao, R. N. Mitra, M. Zheng, K. Wang, J. C. Dahringer and Z. Han, Adv. Funct. Mater., 2018, 28, 1806248 CrossRef PubMed.
- M. Li, X. Bian, X. Chen, N. Fan, H. Zou, Y. Bao and Y. Zhou, Drug Deliv., 2022, 29, 519–533 CrossRef CAS PubMed.
- T. Hu, Q. Le, Z. Wu and W. Wu, J. Pharm. Biomed. Anal., 2007, 43, 263–269 CrossRef CAS PubMed.
- V. Linko, B. Shen, K. Tapio, J. J. Toppari, M. A. Kostiainen and S. Tuukkanen, Sci. Rep., 2015, 5, 15634 CrossRef CAS PubMed.
- E. Stahl, T. G. Martin, F. Praetorius and H. Dietz, Angew. Chem., Int. Ed., 2014, 53, 12735–12740 CrossRef CAS PubMed.
- I. Seitz, H. Ijäs, V. Linko and M. A. Kostiainen, ACS Appl. Mater. Interfaces, 2022, 14, 38515–38524 CrossRef CAS PubMed.
- A. M. Hung, C. M. Micheel, L. D. Bozano, L. W. Osterbur, G. M. Wallraff and J. N. Cha, Nat. Nanotechnol., 2010, 5, 121–126 CrossRef CAS PubMed.
- J. Liang, Z. Zhang, H. Zhao, S. Wan, X. Zhai, J. Zhou, R. Liang, Q. Deng, Y. Wu and G. Lin, RSC Adv., 2018, 8, 15621–15631 RSC.
- A. Udomprasert, C. Wootthichairangsan, R. Duangrat, S. Chaithongyot, Y. Zhang, R. Nixon, W. Liu, R. Wang, M. Ponglikitmongkol and T. Kangsamaksin, ACS Appl. Bio Mater., 2022, 5, 2262–2272 CrossRef CAS PubMed.
- Y. Zeng, J. Liu, S. Yang, W. Liu, L. Xu and R. Wang, J. Mater. Chem. B, 2018, 6, 1605–1612 RSC.
- M. Li, G. Yang, Y. Zheng, J. Lv, W. Zhou, H. Zhang, F. You, C. Wu, H. Yang and Y. Liu, J. Nanobiotechnol., 2023, 21, 186 CrossRef CAS PubMed.
- Y.-X. Zhao, A. Shaw, X. Zeng, E. Benson, A. M. Nyström and B. Högberg, ACS Nano, 2012, 6, 8684–8691 CrossRef CAS.
- M. Hanke, E. Tomm, G. Grundmeier and A. Keller, ChemBioChem, 2023, 24, e202300338 CrossRef CAS PubMed.
- J. A. L. Roodhuizen, P. J. T. M. Hendrikx, P. A. J. Hilbers, T. F. A. de Greef and A. J. Markvoort, ACS Nano, 2019, 13, 10798–10809 CrossRef CAS PubMed.
- A. Bednarz, S. M. Sønderskov, M. Dong and V. Birkedal, Nanoscale, 2023, 15, 1317–1326 RSC.
- L. Peeters, N. N. Sanders, K. Braeckmans, K. Boussery, J. Van de Voorde, S. C. De Smedt and J. Demeester, Invest. Ophthalmol. Visual Sci., 2005, 46, 3553–3561 CrossRef PubMed.
- T. Xu, S. Yu, Y. Sun, S. Wu, D. Gao, M. Wang, Z. Wang, Y. Tian, Q. Min and J.-J. Zhu, Small, 2021, 17, 2101780 CrossRef CAS PubMed.
- P. Wang, M. A. Rahman, Z. Zhao, K. Weiss, C. Zhang, Z. Chen, S. J. Hurwitz, Z. G. Chen, D. M. Shin and Y. Ke, J. Am. Chem. Soc., 2018, 140, 2478–2484 CrossRef CAS PubMed.
- Y. Wang, E. Benson, F. Fördős, M. Lolaico, I. Baars, T. Fang, A. I. Teixeira and B. Högberg, Adv. Mater., 2021, 33, 2008457 CrossRef CAS PubMed.
- V. Linko and A. Keller, Small, 2023, 19, 2301935 CrossRef CAS PubMed.
- A. Lacroix, E. Vengut-Climent, D. De Rochambeau and H. F. Sleiman, ACS Cent. Sci., 2019, 5, 882–891 CrossRef CAS PubMed.
- J. Kokavec, S. H. Min, M. H. Tan, J. S. Gilhotra, H. S. Newland, S. R. Durkin, J. Grigg and R. J. Casson, Clin. Exp. Ophthalmol., 2016, 44, 597–609 CrossRef PubMed.
- A. Sadeghi, M. Ruponen, J. Puranen, S. Cao, R. Ridolfo, S. Tavakoli, E. Toropainen, T. Lajunen, V.-P. Ranta, J. van Hest and A. Urtti, Int. J. Pharm., 2022, 621, 121800 CrossRef CAS PubMed.
- H.-K. Kuo, Y.-H. Chen, P.-C. Wu, Y.-C. Wu, F. Huang, C.-W. Kuo, L.-H. Lo and J. Shiea, Invest. Ophthalmol. Visual Sci., 2012, 53, 3167–3174 CrossRef PubMed.
- Z. Pan, H. Zhu, Y. Zhang, Q. Liao, Y. Sun, E. Wu, Y. Wang, K. Shi, Y. Zhang, L. Chen, M. Ye and W. Wu, Anal. Chem., 2023, 95, 5095–5108 CrossRef CAS PubMed.
- S. O. Mohammadi, M. C. LaRocca, C. D. Yang, J. Jessen, M. C. Kenney and K. Y. Lin, J. Clin. Transl. Ophthalmol., 2023, 1, 107–116 Search PubMed.
- G. A. Zelada-Guillén, J. A. Ríos-Arce, M. A. Leyva-Peralta, M. Flores-Álamo, J. C. Gálvez-Ruíz, K. Calderón and M. V. Escárcega-Bobadilla, Chem. Biodivers., 2023, 20, e202200972 CrossRef PubMed.
- Z. Shen, J. Shao, J. Dai, Y. Lin, X. Yang, J. Ma, Q. He, B. Yang, K. Yao and P. Luo, Toxicol. Rep., 2016, 3, 78–86 CrossRef CAS PubMed.
- G. D. García-Olaiz, E. Alcántar-Zavala, A. Ochoa-Terán, A. Cabrera, R. Muñiz-Salazar, J. Montes-Ávila, A. J. Salazar-Medina, E. Alday, C. Velazquez, J. L. Medina-Franco and R. Laniado-Laborín, Bioorg. Chem., 2020, 95, 103483 CrossRef PubMed.
- E. S. Lisitsyna, A. Klose, E. Vuorimaa-Laukkanen, H. Ijäs, T. Lajunen, K. Suhling, V. Linko and T. Laaksonen, bioRxiv, 2024, preprint, DOI:10.1101/2024.06.20.599777.
- Z. Guo, H. Song, Y. Tian, J. Xu, G. Zhang, Y. Guo, R. Shen and D. Wang, Int. J. Nanomed., 2024, 19, 171–188 CrossRef CAS PubMed.
- J. Mikkilä, A.-P. Eskelinen, E. H. Niemelä, V. Linko, M. J. Frilander, P. Törmä and M. A. Kostiainen, Nano Lett., 2014, 14, 2196–2200 CrossRef PubMed.
- W.-L. L. Suen and Y. Chau, J. Pharm. Pharmacol., 2014, 66, 564–573 CrossRef CAS PubMed.
- C. M. Green, D. Mathur and I. L. Medintz, J. Mater. Chem. B, 2020, 8, 6170–6178 RSC.
- P. Wang, M. A. Rahman, Z. Zhao, K. Weiss, C. Zhang, Z. Chen, S. J. Hurwitz, Z. G. Chen, D. M. Shin and Y. Ke, J. Am. Chem. Soc., 2018, 140, 2478–2484 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr01995d |
| This journal is © The Royal Society of Chemistry 2024 |