Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interaction of full-length Tau with negatively charged lipid membranes leads to polymorphic aggregates†
Vicky
Ury-Thiery
,
Yann
Fichou
,
Isabel
Alves
,
Michael
Molinari
 ,
Sophie
Lecomte
and
Cécile
Feuillie
,
Sophie
Lecomte
and
Cécile
Feuillie
 *
*
Univ. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, F-33600 Pessac, France. E-mail: cecile.feuillie@u-bordeaux.fr
First published on 22nd August 2024
Abstract
The Tau protein is implicated in various diseases collectively known as tauopathies, including Alzheimer's disease and frontotemporal dementia. The precise mechanism underlying Tau pathogenicity remains elusive. Recently, the role of lipids has garnered interest due to their implications in Tau aggregation, secretion, uptake, and pathogenic dysregulation. Previous investigations have highlighted critical aspects: (i) Tau's tendency to aggregate into fibers when interacting with negatively charged lipids, (ii) its ability to form structured species upon contact with anionic membranes, and (iii) the potential disruption of the membrane upon Tau binding. In this study, we examine the disease-associated P301L mutation of the 2N4R isoform of Tau and its effects on membranes composed on phosphatidylserine (PS) lipids. Aggregation studies and liposome leakage assays demonstrate Tau's ability to bind to anionic lipid vesicles, leading to membrane disruption. Attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR) reveals the accumulation of Tau on the membrane surface without protein insertion, structuration, or lipid removal. Plasmon waveguide resonance (PWR) demonstrates a strong binding of Tau on PS bilayers with an apparent Kd in the micromolar range, indicating the deposition of a thick protein layer. Atomic force microscopy (AFM) real-time imaging allows the observation of partial lipid solubilization and the deposition of polymorphic aggregates in the form of thick patches and fibrillary structures resembling amyloid fibers, which could grow from a combination of extracted anionic phospholipids from the membrane and Tau protein. This study deepens our understanding of full-length Tau's multifaceted interactions with lipids, shedding light on potential mechanisms leading to the formation of pathogenic Tau assemblies.
1. Introduction
The tubulin associated unit (Tau) protein is an intrinsically disordered protein that binds to microtubules.1 Tau has other physiological functions maintaining cell integrity. In healthy neurons, besides regulating microtubules dynamics, Tau may regulate axonal transport through various mechanisms (ref. 2 and references therein). For instance, some Tau protein locates in the nucleus, where it might play a role in maintaining the integrity of genomic DNA. The longest Tau isoform, referred to as 2N4R, contains 441 amino acids and is composed of an N-terminal region rich in acidic residues, a proline-rich region, and 4 repeat domains in the microtubule binding region (Fig. 1A). Both the repeat regions and the proline rich region are positively charged, leading to a positive net charge for 2N4R at physiological pH (+2).Tau plays a pivotal role in a class of diseases called tauopathies, which includes Alzheimer's disease and frontotemporal dementia.2 In pathological conditions, Tau can misfold, aggregate and accumulate in the form of amyloid aggregates forming neurofibrillary tangles.3 A hallmark of these aggregates is the characteristic cross-β structure found in amyloids, where β-sheets are ordered in an amyloid core. In the case of Tau, the amyloid core encompasses the third and fourth repeat regions as well as part of the C terminus, although the exact location of the amyloid core is disease specific.4 The remainder of Tau forms a fuzzy coat structure, in majority unfolded, with a few short and transient elements of secondary structure.5 Various single mutations present in the microtubule binding domain have been shown to promote Tau pathology in frontotemporal dementia linked with chromosome 17.6 Among these mutations, P301L (proline replaced by leucine) is one of the most aggressive and widely studied. At a molecular level, this substitution was shown to promote aggregation by modulating the conformation of the protein and exposing hydrophobic regions.7,8
In vitro, polyanionic co-factors such as heparin are widely used to induce the aggregation of Tau.9 If heparin is the most used co-factor in vitro, others, like RNA10 or negatively charged lipids,3,11–13 also have the propensity to induce the aggregation of Tau. The lipid membrane and its interactions with Tau are increasingly studied, as Tau-lipid interactions could be involved not only in Tau aggregation but also in Tau-induced cell toxicity and Tau spreading.14 Tau has been shown to interact with the plasma membrane of neurons.15,16 Indeed, intracellular Tau can be translocated directly through the membrane in a free form and taken up by nearby cells via different pathways.12,15–17 Tau can also directly interact with lipids such as Phosphatidylinositol 4,5-bisphosphate (PIP2),16 cholesterol, and sphingomyelin15 in cells. Interestingly, lipids were found to be enriched in purified brain homogenates containing paired helical filaments (PHF) from Alzheimer's disease patients.18 With Tau neurofibrillary tangles being intracellular, the effect of anionic lipids, predominant on the inner leaflet of neuronal membranes – especially phosphatidylserine (POPS) – is of particularly interest. Studies have demonstrated that patients with Alzheimer's disease display modifications in both the proportion and content of cellular lipids.19 This includes an increase in POPS content, particularly in the hippocampus and temporal lobe,20 areas correlated with the progression of Alzheimer's disease,21 making it a lipid target of choice.
The mechanism of interactions between Tau protein and negative lipids is far from being fully understood. Existing literature on Tau's interaction with negatively charged lipids has allowed to bring partial understanding of the processes at play. Lipid-induced protein aggregation has been reported in in vitro studies of Tau interactions with negatively charged lipid vesicles,22 with formation of protein/lipid complexes.12,22,23 Full-length Tau 2N4R was shown to form pore-like structures when in interaction with phosphatidylethanolamine (PE)/phosphatidylglycerol (PG) planar bilayers.24 Insertion into the membranes is however not always observed, as in the case of K19 interacting with PS-containing membranes.23 Structural reorganization was observed upon membrane binding: K19, a truncated version of Tau composed of the R1, R3 and R4 domains, for instance has been shown to adopt β-structures upon binding to membrane vesicles composed of dimyristoylphosphatidylcholine (DMPC)/dimyristoylphosphoserine (DMPS) at a molar ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1,25 but has been shown to fold into α-helices upon binding to palmitoyl-oleoyl phosphatidic acid acid (POPA)/palmitoyl-oleoyl phosphatidylcholine (POPC) membranes26 or POPS-containing SUVs.27 Compaction of the full-length Tau protein has also been reported upon selective insertion into DMPG monolayers.28 Tau-induced membrane disruption has been observed in some cases, for instance for K18, a fragment of Tau composed of the four repeat regions, on fluid supported membranes,29 or for full-length Tau on DMPG monolayers.28 Some contradiction remains regarding the disruptive capacities of Tau's interaction with membranes, which seem to depend on lipid composition.25,29 Membrane curvature could also be involved, as K18 was reported to not disrupt the bilayer integrity of PS/PC vesicles,22 but to partly solubilize PS-containing supported membranes locally.29 Overall, many studies have focused on the interactions between membranes and short fragments of Tau, like the microtubule binding domain K18,22,29 the K19 construct,25–27 the microtubule binding domain with the proline-rich P2 region P2R,23 or individual repeat domains.30 The N-terminal region of the protein could nonetheless play a crucial role in Tau – membrane interactions, as indicated by cellular studies,31 and must be considered in Tau assembly and membrane interaction studies.32 As the overall charges of K18 or K19 (respectively +10 and +7 at physiological pH) are higher compared to full-length Tau 2N4R (+2), one can also raise the question of the driving force of the interaction between full-length Tau and membranes compared to shorter fragments.
1,25 but has been shown to fold into α-helices upon binding to palmitoyl-oleoyl phosphatidic acid acid (POPA)/palmitoyl-oleoyl phosphatidylcholine (POPC) membranes26 or POPS-containing SUVs.27 Compaction of the full-length Tau protein has also been reported upon selective insertion into DMPG monolayers.28 Tau-induced membrane disruption has been observed in some cases, for instance for K18, a fragment of Tau composed of the four repeat regions, on fluid supported membranes,29 or for full-length Tau on DMPG monolayers.28 Some contradiction remains regarding the disruptive capacities of Tau's interaction with membranes, which seem to depend on lipid composition.25,29 Membrane curvature could also be involved, as K18 was reported to not disrupt the bilayer integrity of PS/PC vesicles,22 but to partly solubilize PS-containing supported membranes locally.29 Overall, many studies have focused on the interactions between membranes and short fragments of Tau, like the microtubule binding domain K18,22,29 the K19 construct,25–27 the microtubule binding domain with the proline-rich P2 region P2R,23 or individual repeat domains.30 The N-terminal region of the protein could nonetheless play a crucial role in Tau – membrane interactions, as indicated by cellular studies,31 and must be considered in Tau assembly and membrane interaction studies.32 As the overall charges of K18 or K19 (respectively +10 and +7 at physiological pH) are higher compared to full-length Tau 2N4R (+2), one can also raise the question of the driving force of the interaction between full-length Tau and membranes compared to shorter fragments.
Investigating the interaction of full-length Tau with POPS, predominant anionic lipid of the cytosolic inner leaflet of the plasma membrane, offers a promising lead to better understand the interaction of Tau with negatively charged lipids from the cell interior, while deepening the understanding of the mechanisms of aggregation and toxicity of Tau on neuronal membranes. In this study, the interactions between the Tau protein (2N4R) harboring the mutation P301L (Tau-P301L) were investigated on pure POPS or DOPS bilayers. Various biophysical techniques were used to probe the interaction between PS bilayers and Tau-P301L protein at the micro and nanoscale to better decipher the mechanisms by which Tau might induce damages on lipid membranes and aggregate in close contact of the membrane. Thioflavin T fluorescence assays and transmission electron microscopy confirmed the formation of amyloid fibers in presence of POPS liposomes. Calcein leakage assays showed a disruptive effect of Tau-P301L on POPS liposomes. Polarized attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR) spectroscopy and Plasmon waveguide resonance (PWR) revealed the accumulation of Tau-P301L on supported POPS membranes. Finally, Atomic Force Microscopy (AFM) revealed different morphologies of aggregated Tau P301L in contact with supported DOPS membranes.
2. Materials and methods
2.1. Protein production and purification
Tau protein was purified following a previously established protocol.33 In summary, protein expression was conducted in Escherichia coli BL21 (DE3) transformed with a pet28 plasmid containing the 2N4R P301L gene N-terminally tagged with polyhistidine.Cells were initially inoculated into 1 L of LB medium with kanamycin supplementation at 50 μg mL−1. Cultures were maintained at 37 °C with shaking at 200 rpm until they reached an OD600 of 0.6–0.8. Protein expression was induced by adding 1 mM isopropylß-D-488 thiogalactoside (IPTG, Sigma Aldrich), followed by further incubation at 37 °C and 200 rpm for 3 hours. Cells were harvested by centrifugation at 5000g for 20 minutes at 4 °C. The resulting cell pellets were resuspended in a lysis buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 10 mM Imidazole, 0.5 mM DTT, 0.1 mM EDTA), supplemented with 1 mM PMSF, 20 μg ml−1 DNase, 10 mM MgCl2, and 1 Pierce protease inhibitor tablet (Thermo Fisher EDTA-Free, Sigma 88266), and subsequently stored at −20 °C for later use. Cell lysis was initiated with the addition of lysozyme (2 mg ml−1), and the cell suspension was agitated on an orbital shaker at room temperature for 30 minutes. To disrupt the cells, a freeze–thaw cycle was repeated three times using liquid nitrogen. After cell debris removal by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes at 4 °C, the supernatant was heated at 90 °C for 12 minutes, followed by cooling on ice for 20 minutes before another centrifugation at 10
000 rpm for 10 minutes at 4 °C, the supernatant was heated at 90 °C for 12 minutes, followed by cooling on ice for 20 minutes before another centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes at 4 °C to eliminate precipitated proteins. The supernatant, containing Tau protein, was loaded onto a 5 ml Ni-NTA agarose column pre-equilibrated with buffer A (20 mM sodium phosphate pH 7.0, 500 mM NaCl, 10 mM imidazole, 100 μM EDTA). The column was washed successively with 10 column volumes (CV) of buffer A, 8 CV of buffer B (20 mM sodium phosphate pH 7.0, 1 M NaCl, 20 mM imidazole, 100 μM EDTA), and 2 CV of buffer A before elution with 8 CV of buffer C (20 mM sodium phosphate pH 7.0, 500 mM imidazole, 0.5 mM DTT, 100 mM NaCl). Elution progress was monitored using absorbance at 280 nm and SDS-PAGE. Peak fractions were combined and concentrated through centrifugal filters (MWCO 10 kDa, Sigma) for subsequent gel filtration chromatography on a Superdex 200 pg HiLoad 16–600 column equilibrated with a final buffer (20 mM HEPES pH 7.0, 100 mM NaCl). Fractions containing purified protein were further concentrated, and the final protein concentration was determined via UV-vis absorption at 274 nm, employing an extinction coefficient of 7.5 cm−1 mM−1. The proteins were aliquoted into 100 μL tubes and stored at −20 °C for future use.
000 rpm for 10 minutes at 4 °C to eliminate precipitated proteins. The supernatant, containing Tau protein, was loaded onto a 5 ml Ni-NTA agarose column pre-equilibrated with buffer A (20 mM sodium phosphate pH 7.0, 500 mM NaCl, 10 mM imidazole, 100 μM EDTA). The column was washed successively with 10 column volumes (CV) of buffer A, 8 CV of buffer B (20 mM sodium phosphate pH 7.0, 1 M NaCl, 20 mM imidazole, 100 μM EDTA), and 2 CV of buffer A before elution with 8 CV of buffer C (20 mM sodium phosphate pH 7.0, 500 mM imidazole, 0.5 mM DTT, 100 mM NaCl). Elution progress was monitored using absorbance at 280 nm and SDS-PAGE. Peak fractions were combined and concentrated through centrifugal filters (MWCO 10 kDa, Sigma) for subsequent gel filtration chromatography on a Superdex 200 pg HiLoad 16–600 column equilibrated with a final buffer (20 mM HEPES pH 7.0, 100 mM NaCl). Fractions containing purified protein were further concentrated, and the final protein concentration was determined via UV-vis absorption at 274 nm, employing an extinction coefficient of 7.5 cm−1 mM−1. The proteins were aliquoted into 100 μL tubes and stored at −20 °C for future use.
2.2. Liposome preparations
1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (16:0, 18:1 POPS, Avanti polar lipids 840034) or 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (18:1 DOPS, Avanti polar lipids 840035, used for AFM experiment) previously dissolved in chloroform, were dried using a nitrogen stream to create a thin film on the inner wall of an Eppendorf tube and placed in a desiccator under vacuum overnight to remove any remaining chloroform traces. The lipid films were then re-solubilized at 2 mg ml−1 in 20 mM HEPES, pH7, 100 mM NaCl. This buffer was used for all AFM and ATR-FTIR experiments.Small unilamellar vesicles (SUVs), used for aggregation and supported lipid bilayers (SLB) formation, were obtained by sonication of the lipid suspensions until clarity was achieved.
For calcein leakage experiments, large unilamellar vesicles (LUVs) containing calcein were obtained using lipid films of pure POPS, re-suspended in 20 mM HEPES pH7, with 70 mM calcein, and subjected to five freeze/thaw cycles. Subsequently, lipid suspensions were extruded 21 times using an Avanti Polar Lipids extruder set, through a membrane with a pore size of 200 nm. To separate LUVs containing calcein from the free dye, samples were passed through a Sephadex G50 size exclusion column that had been pre-equilibrated with 20 mM HEPES pH7 150 mM NaCl. The concentration of phospholipids was then determined using phosphorus quantification according to a previous protocol.34 The LUVs containing calcein were then stored at room temperature and protected from light for a maximum of 2 weeks.
2.3. Thioflavin T fluorescence aggregation assay
Thioflavin T (ThT) fluorescence assays were conducted using 20 μM ThT and 20 μM protein, incubated either alone or with cofactors in a buffer comprising 20 mM HEPES at pH 7, 100 mM NaCl, and 5 mM DTT for 3 days at 37 °C with vigorous shaking. Various protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid ratios were tested from 1
lipid ratios were tested from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to 1
5 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. To induce or examine protein aggregation, either 200 μM of small unilamellar vesicle (SUV) suspension of POPS or 5 μM of heparin was introduced. Fluorescence emission at 485 nm was measured using a TECAN Infinite M1000 pro plate reader with an excitation wavelength at 450 nm.
30. To induce or examine protein aggregation, either 200 μM of small unilamellar vesicle (SUV) suspension of POPS or 5 μM of heparin was introduced. Fluorescence emission at 485 nm was measured using a TECAN Infinite M1000 pro plate reader with an excitation wavelength at 450 nm.
2.4. Transmission electron microscopy
Aggregation products were imaged by transmission electron microscopy on a Philips CM120 electron microscope at 80 kV. Images were collected by 2 K × 2 K Gatan ss CCD Camera. The sample (4 μl) was deposited onto glow-discharged carbon-coated copper 300 mesh grids (Sigma) for 1 min and stained using uranyl acetate 1%.2.5. Calcein leakage assay
Fluorescence measurements were conducted at 25 °C with a TECAN Infinite M1000 pro plate reader, utilizing an excitation wavelength of 485 nm, and detecting the fluorescence emission at 515 nm. Liposomes were added at a concentration of 10 μM in triplicate wells of a 96-well plate (Microfluor1), and the initial fluorescence (F0) for each well was recorded. Subsequently, increasing concentrations of Tau-P301L (0.01 μM, 0.1 μM, 0.2 μM, 0.5 μM, 1 μM, and 5 μM) were added with fixed volumes (total volume of 100 μl), and the fluorescence was monitored over time (FT). After 14 hours, 10 μl of Triton X-100 (Sigma) was added to achieve complete liposome leakage (Fmax). The percentage of calcein released (Lt %) was calculated using the following equation: Lt % = [(FT − F0)/(Fmax − F0)] × 100. The mean and standard deviation were calculated from 2 independent experiments.2.6. Plasmon-waveguide resonance spectroscopy
PWR spectra are produced by resonant excitation of conduction electron oscillation (plasmons) by light from a polarized CW laser (He–Ne) incident on the back surface of a thin metal film (Ag) deposited on a glass prism and coated with a layer of SiO2. The experimental configuration employed a custom-built instrument equipped with a laser at 632 nm, featuring linear polarization at a 45° angle, all the details are reported in a previous study.35 This polarization setup allows for simultaneous acquisition of p-polarization (light parallel to the incident beam) and s-polarization (light perpendicular to the incident beam) data within a single angular scan. In addition, the total internal reflection angle (TIR) is also monitored, it allows the discrimination of events occurring in close proximity to the sensor surface from those occurring in the bulk.36,37 Experiments were conducted in a controlled room temperature environment.The sensor consisted of a prism coated with a 50 nm-thick layer of silver and overcoated with a 460 nm-thick silica layer. The sensor, in direct contact with the sample cell, was positioned on a rotating table supported by a corresponding motion controller (Newport, Motion controller XPS; ≤1 mdeg resolution).
To form a supported lipid bilayer (SLB) on the sensor's surface, we introduced 250 μl of a POPS SUV suspension at a concentration of 2 mg ml−1, supplemented with 1 mM CaCl2 (freshly added), into a Teflon compartment in contact with the silica-coated prism (cell sample compartment). The spontaneous formation of a lipid bilayer occurred after 30-minutes liposome incubation period, after which the cell was rinsed to remove excess of lipids. The absence of angle shift in the TIR angle after rinsing when comparing with the angle observed with buffer, confirms that all excess liposomes has been removed from the bulk.
Subsequently, Tau-P301L was introduced into the cell incrementally from 0.1 to 1 μM. As the sensor is mounted parallel to the cell sample, there is no problem regarding potential protein settling on the sensor surface. The modifications in the plasmon resonance characteristics reflecting protein/lipid interactions were monitored over time. Here we mainly focus on the shift in the resonance angle position that is correlated with mass deposition, reflecting the quantity of bound protein.
2.7. Polarized ATR-FTIR spectroscopy
Polarized ATR-FTIR experiments were conducted in a temperature-controlled room set at 25 °C using a Nicolet iS50 FTIR spectrometer (Thermo Fisher Scientific) equipped with a liquid nitrogen-cooled mercury-cadmium-telluride (MCT) detector. The ATR element (Ge, single reflection) comes from Specac. All spectra were acquired with a spectral resolution of 4 cm−1 and 200 interferograms were added.Prior to sample measurements, background spectra for both s and p polarizations were recorded. Subsequently, 20 μl of buffer (20 mM Hepes at pH 7 with 100 mM NaCl) was added to the liquid cell created using an o-ring, and the buffer spectra were recorded for both s and p polarizations for later subtraction from the sample spectra.
20 μl of a POPS SUV suspension at a concentration of 1 mg ml−1, in buffer supplemented with 1 mM CaCl2, were deposited and incubated for 5 minutes to facilitate SLB formation. The SLB was then rinsed three times by the buffer to remove any remaining SUV. Polarized ATR-FTIR spectra of the SLB were recorded before Tau-P301L addition, as well as after a 3 h incubation, either alone for control conditions, or with 1 μM of Tau-P301L directly added inside the liquid cell. Final s and p polarized ATR-FTIR spectra after incubation were collected following three rinses with buffer. Each experiment was conducted three times.
Polarized ATR-FTIR spectra of SLB alone and Tau protein interacting with the SLB were processed using Omnic Software (Thermo Fisher Scientific), by subtracting the buffer spectrum at each polarization. Additionally, the baseline was corrected.
2.8. Atomic force microscopy imaging
Supported lipid bilayers (SLB) were prepared for AFM experiments by depositing 100 μl of a SUV suspension of DOPS containing 1 mM of CaCl2 (added just before deposition) at 1 mg ml−1 on freshly cleaved mica and incubated at 60 °C in a humid chamber. Special care was taken to keep the SLB hydrated, by adding buffer every 15 min. After incubation, the sample was allowed to cool down to room temperature for 30 min, and rinsed multiple times (∼10 times) with buffer (20 mM Hepes, pH 7, 100 mM NaCl) prior to observation in 100 μL of buffer.AFM imaging was performed using the Dimension FastScan setup (Bruker) operating in PeakForce Quantitative Nano-Mechanics (PF-QNM) mode in liquid environment at room temperature (∼20 °C). SNL-C probes (Bruker, Silicon tips on silicon nitride cantilevers) were individually calibrated for each experiment, with a nominal spring constant of 0.24 N m−1, a resonance frequency of 56 kHz, and a tip radius ranging from 2 to 12 nm. SLB were imaged with a scan rate of 0.8–1 Hz, a resolution of 256 × 256 pixels, and a constant setpoint force of approximately 0.5 nN. To ensure the stability of the SLB, an area was imaged for 1 to 3 hours before introducing the protein at the desired concentration directly inside the liquid droplet. Images were recorded immediately after the injection of Tau every 3–5 minutes until the system stabilized, typically within 1–2 hours. Different concentrations were tested to maintain consistency with other experiments in terms of protein/lipid ratio, and 400 or 500 nM were chosen to enable the observation of defects without introducing a mechanism that is too rapid. Image analysis was carried out using Gwyddion, and the sizes were determined using the cross-section tool. Each experiment was repeated 3 times.
3. Results
3.1. POPS induces aggregation of Tau-P301L similarly to Tau 2N4R
Fig. 1B displays the Thioflavine T (ThT) fluorescence emission intensity of Tau-P301L, alone or in presence of heparin or small unilamellar vesicles (SUV) of pure POPS. When incubated alone, Tau-P301L did not exhibit an increase of ThT fluorescence (Fig. 1B), as expected in the absence of cofactors, indicating the absence of fiber formation. In contrast, heparin induced a rapid increase in fluorescence, yielding a sigmoidal kinetic profile without a discernible lag time. The observed fluorescence emission of ThT is characteristic of dye binding to cross β structures of amyloid fibers. Subsequent exponential aggregation exhibited a half-time of 4.5 hours for Tau-P301L, before reaching a maximum fluorescence plateau. With POPS, a prolonged lag time of approximately 10 hours was observed for Tau-P301L aggregation, with the maximum fluorescence intensity obtained being fivefold lower than that induced by heparin.The presence of fibers was confirmed by transmission electron microscopy (TEM, Fig. 1C and D). Characteristic amyloid fibers were observed in the presence of both heparin and POPS, displaying long filaments, straight or twisted. No discernible differences in size or morphology were noted based on the cofactors. AFM imaging was used in buffer to characterize the morphologies of POPS-induced Tau-P301L filaments in more details (Fig. 1E). Fibers present lengths of several micrometers, with straight and twisted fibers (Fig. 1E for cross-sections of low and high points of a fiber), as observed for heparin-induced Tau 2N4R fibers.38
As a reference, the aggregation behavior of Tau-WT was also studied (Fig. S1†). In accordance with the results obtained for Tau-P301L, the same increase in fluorescence without lag-time was observed for Tau-WT incubated with heparin, with a half-time of 3 hours but an intensity 3 times lower compared to Tau-P301L. A very low fluorescence intensity was observed with POPS with a lag time of 7 hours, and the maximum fluorescence was threefold lower than that of Tau-P301L, and 4.5 times lower compared to the heparin condition. Furthermore, a greater abundance of fibers was observed in both conditions for Tau-P301L compared to Tau-WT, consistent with the mutant's heightened propensity for aggregation.39 These experiments confirm that POPS is capable of inducing the aggregation of Tau-P301L protein, leading to the formation of fibers. To delve deeper into POPS-induced Tau aggregation and POPS-Tau interactions, we explored the effect of Tau-P301L monomer on POPS lipid membranes.
3.2. Tau-P301L alters POPS liposomes integrity
To explore the impact of Tau-P301L on lipid membranes composed of POPS, calcein leakage assays were performed using a constant concentration of POPS LUVs containing a calcein dye. The increase in fluorescence intensity, indicative of dye release from the liposomes, was followed after addition of different concentrations of Tau-P301L. In absence of Tau-P301L, no leakage was detected after 3 h (Fig. 2). This confirms the stability of POPS liposomes in the absence of Tau-P301L. Upon protein addition, fluorescence increased rapidly, indicating high calcein leakage, with a dose-dependent effect depending on the protein concentration. Over 50% of leakage was observed above 1 μM of Tau-P301L, which may be due to membrane perturbation or disruption. | ||
| Fig. 2 Calcein leakage of POPS liposomes (10 μM) after 3 h of incubation with increasing concentrations of Tau-P301L. Error bars represent standard deviation over 2 repeats. | ||
3.3. Tau-P301L accumulates on POPS supported lipid bilayers
Polarized ATR-FTIR spectroscopy was employed to go further on the effect of Tau-P301L protein on POPS membranes. Fig. 3 shows ATR-FTIR spectra obtained for POPS bilayers alone (grey spectra) and after a 3-hour incubation of 1 μM of Tau-P301L and rinses (green spectra), with focus on 2 spectral ranges: (i) the spectral range between 2800 and 3000 cm−1 that allows monitoring lipid quantity and organization thanks to the vibrational modes of CH2 or CH3 groups, and (ii) the spectral range 1500–1760 cm−1 that also allows monitoring protein quantity and structure thanks to the amide I and II bands (Fig. 3).A POPS SLB alone shows the antisymmetric and symmetric stretching modes of CH2 groups νas(CH2) and νs(CH2) at 2923 cm−1 and 2850 cm−1 respectively, and the νas(CH3) and νs(CH3) modes less intense appear as shoulders at 2950 cm−1 and 2870 cm−1, respectively. The position of the νas(CH2) at 2923 cm−1 indicates disordered chains as expected for fluid membranes, and a value of absorbance at 1.6 × 10−3 (for p-polarization) is consistent with the formation of a single bilayer on the surface of the ATR crystal.40 The band at 1740 cm−1 assigned to the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ester group of the phospholipid (Fig. 3, grey spectra) also confirms the formation of a lipid bilayer. As a control, the stability of the supported POPS bilayers was evaluated by following the absorbance of the band at 2923 cm−1 over time (Fig. 3 inset, grey column). After 3 hours, only a small decrease can be observed, revealing bilayer stability over this timescale.
O) ester group of the phospholipid (Fig. 3, grey spectra) also confirms the formation of a lipid bilayer. As a control, the stability of the supported POPS bilayers was evaluated by following the absorbance of the band at 2923 cm−1 over time (Fig. 3 inset, grey column). After 3 hours, only a small decrease can be observed, revealing bilayer stability over this timescale.
After 3 hours of incubation with Tau-P301L (Fig. 3), both the absorbance of νas(CH2) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ester bands decreased but in the same way as without protein (Fig. 3), with 80 ± 8% of lipids remaining after Tau incubation, compared to 87 ± 16% in the control condition. This suggest that the integrity of the membrane is not significantly affected by its interaction with the Tau protein. The accumulation of Tau-P301L on the POPS membrane is revealed by the observation of the Amide I and Amide II bands centered at 1640 and 1550 cm−1 respectively (Fig. 3). The broad band centered at 1640 cm−1 indicates the presence of various elements of secondary structure, with a high percentage of random coil, as expected for the intrinsically disordered protein (IDP) Tau protein in buffer.41 Upon increase of the protein concentration to 5 μM (see Fig. S2†), similar effect was observed with no diminution of lipid content, suggesting the absence of membrane damage. An increase in the amount of adsorbed protein was however revealed by the increase in intensity of the amide II band (×4). Moreover, the dichroic ratio (Ab νsCH2 p-pol/Ab νsCH2 s-pol) did not vary with or without the addition of the protein, indicating that the protein did not deeply insert into the lipid membrane.
O) ester bands decreased but in the same way as without protein (Fig. 3), with 80 ± 8% of lipids remaining after Tau incubation, compared to 87 ± 16% in the control condition. This suggest that the integrity of the membrane is not significantly affected by its interaction with the Tau protein. The accumulation of Tau-P301L on the POPS membrane is revealed by the observation of the Amide I and Amide II bands centered at 1640 and 1550 cm−1 respectively (Fig. 3). The broad band centered at 1640 cm−1 indicates the presence of various elements of secondary structure, with a high percentage of random coil, as expected for the intrinsically disordered protein (IDP) Tau protein in buffer.41 Upon increase of the protein concentration to 5 μM (see Fig. S2†), similar effect was observed with no diminution of lipid content, suggesting the absence of membrane damage. An increase in the amount of adsorbed protein was however revealed by the increase in intensity of the amide II band (×4). Moreover, the dichroic ratio (Ab νsCH2 p-pol/Ab νsCH2 s-pol) did not vary with or without the addition of the protein, indicating that the protein did not deeply insert into the lipid membrane.
Overall, ATR-FTIR results suggest that Tau-P301L protein accumulates on the POPS without strong damage on the membrane and without strong modifications of the Tau-P301L structure in a 3 hours timescale.
3.4. Tau-P301L binds to POPS with a micromolar affinity
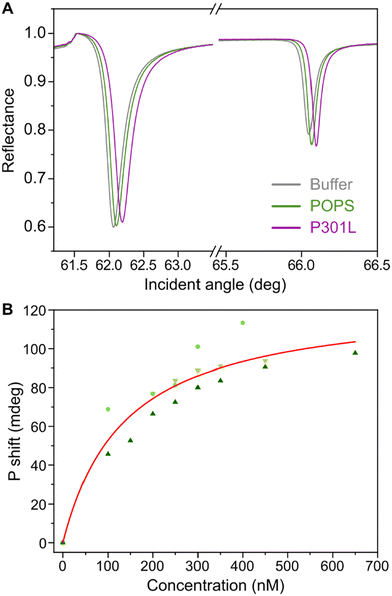
Fig. 4A displays the PWR spectra of buffer, POPS bilayer alone and after addition of Tau-P301L (350 nM). First the formation of the SLB is followed. The positive shifts of the resonance minimum position observed in p- (+66 ± 25 mdeg) and s- (+25 ± 16 mdeg) polarization after deposition of the lipids compared to the buffer were characteristic of a well-organized lipid bilayer.35 Addition of Tau-P301L induced a positive shift (88 mdeg for p- and 35 mdeg for s-, after addition of 350 nM of Tau as presented in Fig. 4B), indicating an increase in mass, resulting from the mass contribution of the protein that is potentially accompanied by a structural reorganization of the SLB. The shifts in p-polarization observed at equilibrium (spectral shifts stabilized) upon incremental protein addition (from 50 to 650 nM, with steps of 50–100 nM) are shown in Fig. 4B. The absence of an increase in the shift of the TIR (total internal reflection) peak confirms that the shifts observed for p- and s- polarization were occurring near the membrane, close to the surface, and not in the bulk solution. Graphical analysis of the p- and s-polarizations shifts ratio upon protein addition42 reveals that the observed positive shift stems mainly from an increase in mass rather than structural reorganization. The preservation of the spectral shape without the presence of asymmetry or shoulders excludes lateral heterogeneities larger than a few hundred nanometers (instrument and sensor lateral resolution). The results are compatible with a homogeneous distribution of Tau-P301L on the membrane surface. A hyperbolic fit of the p-polarization shifts of three independent experiments enabled the determination of an apparent dissociation constant (Kd) of 0.14 ± 0.04 μM. After the end of the titration experiment, a rinsing step of the PWR cell was performed to eliminate any weakly or non-specifically bound protein, retaining only strong interactions between the protein and the bilayer. The presence of protein after rinsing suggests that the interaction is robust and non-reversible.Taken together, ATR-FTIR and PWR data suggest that Tau-P301L accumulates on the POPS surface with a strong binding to the phospholipid surface, without inserting or damaging the lipids present in the supported bilayer.
3.5. Tau-P301L forms aggregated species on phosphatidylserine membranes
Atomic force microscopy imaging was used to probe the interaction of Tau on a PS membrane in real time and at the nanoscale. Achieving the formation of a homogeneous supported lipid bilayer with minimal defects proved challenging. The deposition of pure POPS (16:0, 18:1) SUVs with calcium resulted in the formation of heterogeneous multilayers onto the mica substrate, impeding proper investigation of the impact of Tau. We therefore used DOPS (18:1), which formed homogeneous lipid bilayers on the mica substrate. Though differing in one of their fatty acid chains, with 1 additional unsaturation and 2 additional carbons for DOPS compared to POPS, both lipids share the same anionic headgroup and are fluid at room temperature, with melting temperatures of −11 and 14 °C for DOPS and POPS, respectively.43 Previous studies focusing on the impact of different phosphatidylserine (PS)-based phospholipids, such as DMPS (14:0), DSPS (18:0), DOPS (18:1), and POPS (16:0, 18:1) on protein aggregation have demonstrated that the most significant difference observed was between the saturated phospholipids (DMPS and particularly DSPS) and the unsaturated lipids (DOPS and POPS), with both DOPS and POPS exhibiting similar effects.44,45 Additional polarized ATR-FTIR measurements of DOPS bilayers alone (Fig. S3†) and after 3 h of incubation with P301L tau (Fig. S4†) confirmed that both POPS and DOPS bilayers are fluid and behave similarly in interaction with the protein.The deposition of a DOPS SLB resulted in a homogeneous bilayer with an approximate thickness of 3.0 ± 1.1 nm, as illustrated by the cross-section of a defect in Fig. 5A and D. To ensure the stability of the SLB, areas of the DOPS bilayer were imaged over time to monitor any time-dependent disruption and ensure that no AFM tip-induced disruption occurred (Fig. S6†). DOPS bilayers remained stable over the experimental timescale of 1 to 3 hours, showing no additional holes or significant modifications.
Fig. 5 presents a sequence of images obtained for the interaction of a DOPS bilayer with 400 nM of Tau-P301L as a function of time. Following the addition of Tau-P301L in the experimental volume, significant changes were immediately observed. After 6 min (Fig. 5B and E), pre-existing defects in the bilayer have widened, suggesting partial solubilization of the membrane at the edges of the initial defects. Interestingly, after incubation with Tau-P301L, the height difference between the mica and the remaining lipid layer is only about 1.5 nm, rather than the expected 3–3.5 nm for the SLB thickness, as shown in Fig. 5B and E at the location of the green arrowhead. This height would be consistent with a monolayer of lipids, hinting at membrane thinning and lipid removal. In addition to an apparent membrane disruption and loss of lipids, the rapid apparition of both intermediate height and high large flat patches of approximately 7.8 ± 0.5 nm and 16.5 ± 0.8 nm thickness was observed, depositing onto the remaining lipid layer, and reminding of multiple lipid bilayers.46 This suggests a possible re-deposition of solubilized phospholipids, associated or not with the protein on the mica and remaining lipid layer. Observed heights for the flat patches would be consistent with the deposition of 2 to 5 bilayers. Over time, the deposited patches appear to break and reorganize, as smaller patches were visualized after 2 hours (Fig. 5C). The major effect of Tau-P301L on the membrane occurred within the first 30 minutes of interaction and subsequently stabilized rapidly without further changes (Fig. 5C).
The observed effect on the DOPS surface appears to be correlated with the concentration of Tau-P301L added to the bilayer surface. Higher concentrations of Tau-P301L led to comparable effects regarding membrane disruption as well as deposition of flat thick patches, but different morphologies in the deposited patches were observed. Fig. 6 presents a sequence of images happening before and after addition of 500 nM of Tau-P301L to a DOPS bilayer. Initially, the SLB appeared homogeneous (Fig. 6A). After 15 min of incubation with Tau-P301L at 500 nM, thick flat patches of 15–20 nm were observed, but also elongated structures that seem to extend from the patches (see green arrowheads on Fig. 6B and C). Moreover, these filamentous structures exhibited growth over short timescales, with significant growth in 8 minutes evidenced by arrowheads on Fig. 6C. This suggests that these filaments could be Tau aggregates in the process of fiber formation.
This concentration-dependent effect was also noted with the incremental addition of proteins, with defects widening at 300 nM with holes consistent with the removal of only a monolayer of lipids (Fig. S7†), patch redeposition at 400 nM (Fig. 5 and Fig. S8B†), and the growth of fibrillar structures at 500 nM (Fig. 6 and Fig. S8C†). Two types of filaments were observed, presenting thicknesses of ∼5 nm (Fig. 6B, C, E and F) or ∼13 nm (Fig. S8D and E†). In comparison to POPS-induced Tau filaments obtained in solution, that showed a thickness of ∼17 nm (Fig. 1E), the filaments observed in interaction with DOPS supported bilayers appear to be much thinner. However, these lower thicknesses could be consistent with protofibrils, that could assemble into thicker filaments later in the fibrillization process, as was observed previously for heparin-induced filaments.38
Overall, AFM data suggest that the incubation of Tau on the anionic SLB leads to partial solubilization of the membrane, with patchy aggregates deposition on the remaining bilayer, and eventual growth of fibrillary structures extruding from the patches.
4. Discussion
The interaction of Tau with lipids, lipid bilayers, and membranes is gaining interest in the last decades, due to its implications in Tau aggregation, secretion, uptake, and pathogenic dysregulation.14,17,32 Tau has been observed to interact with certain lipids of the membrane in vivo15,16,31,47,48 and to form amyloid fibers when incubated with anionic vesicles in vitro.11–13,22 However, the mechanisms of interaction between Tau and anionic lipids are not completely understood. In addition, studies often focus on truncated forms of the Tau protein (K18, K19, PHF6) with a higher density of positive charges than the full-length protein. The nature and organization of the anionic lipids also differ between studies, often focusing on PG or PS in micelles, vesicles, monolayers at the air–water interface or supported bilayer forms. Many aspects of the Tau – lipid membrane interactions are documented, with lipid-induced fiber formation,23 structuration of the protein in contact with the lipids27 or the effect of the protein on membrane integrity,29 yet all effects are mostly considered separately.Our study aims at providing a comprehensive answer to all these questions for the interaction of P301L-Tau, a more aggressive form of full-length Tau, with negatively charged lipids. As the inner neuronal membrane contains a high content of POPS of about 10–20% of its total lipid content,20,49,50 we chose to focus on this anionic lipid. An array of biophysical methods was used to study the interaction at the global and local scale, to investigate whether Tau-P301L is capable of interacting with the POPS with focus on the strength of the interaction and the mechanisms involved, and to assess if this interaction can impact the lipid organization of the membrane.
4.1 Tau interacts strongly with phosphatidylserine lipids and forms fibers
Our results demonstrate Tau-P301L capacity to form amyloid fibers when incubated with SUVs composed of pure POPS. This is consistent with previous in vitro studies that used negative lipids to trigger Tau full length aggregation.11,13 The aggregation of Tau truncated form (K18) was also observed in presence of POPS mixed with POPC.22 A decrease in aggregation was reported at higher pH, leading to charge neutralization and consequently decreasing the interaction of K18 with the anionic lipids.22 In the same way the aggregation of the full-length protein with anionic lipids appears to be governed by electrostatic interactions even though its charge is only +2 at the experimental pH 7.4. We can assume that the protein is sufficiently unfolded for the highly positive (+10) charged parts (4R replication domain) to be in contact with the negatively charged PS heads, without the fuzzy coat inhibiting the interaction. The apparent binding affinity (Kd) determined in our study supports this hypothesis. The obtained Kd value of 0.14 ± 0.04 μM indicates a strong binding between POPS and Tau-P301L and falls within a consistent range when compared to prior studies.13 Yao et al. determined a Kd of 0.32 ± 0.12 μM for the interaction of pure DMPS and the full-length Tau protein. Künze et al. measured a Kd value at 0.22 ± 0.04 μM for the binding of Tau truncated form (K19), highly positively charged, with pure DMPS liposomes. The K19 interaction with purely zwitterionic POPC vesicles led to an even higher Kd of 53.3 ± 18.3 μM.25 This underscores the significant role of electrostatic interactions in mediating the binding of Tau with anionic vesicles. Thus, the presence of the negatively charged N-terminal region of 2N4R does not seem to have a significant impact on the Tau-lipid interaction strength. This is in agreement with the comparison of interaction abilities of full length Tau and K18 with Porcine Brain PhosphatidylSerine (BPS) vesicles, that suggested the N-terminal region is not essential but may modulate the capacity of Tau to form Tau-lipid complexes.124.2 Tau-P301L is not totally folded in contact with POPS membranes
Our ATR-FTIR results indicate an extended form of the Tau-P301L protein on PS membranes. Indeed, the amide I band is centered at 1640 cm−1, which corresponds to a majority of random coil form, without any hydrogen bonds, as expected for an IDP. The width of the amide I band suggests that there are some secondary structure elements (turn, αhelix, β sheet) but in very small proportions. At the short timescale studied (3 h), we did not observe any strong β-sheet structuring of Tau-P301L that might indicate fiber formation. The adsorption of Tau-P301L and its accumulation on the POPS membrane do not drastically alter the organization of the membrane, which remains fluid. Our results are in agreement with other studies, comparing similar forms of Tau (full-length) and lipid systems (phosphatidylserine-containing membranes). Yao et al. also detected an extended form of Tau in contact with DMPS vesicles using FRET allowing to detect intramolecular distances between parts of the protein.13 They also established that the contact between the protein and the DMPS was located in the microtubule binding domain.13 The study of interactions between truncated forms (K19) and anionic lipids showed the formation of α helices, observed by CD26 and electron spin resonance spectroscopy.27 Georgieva et al.27 presented an interaction model for K19 with 3 short alpha helices belonging to the replication domains (253–251, 315–323 and 346–355) lying on the surface of PC-PS lipid membranes. These are the same domains involved in binding to microtubules.12,13,25–28,38 Ait-Bouziad et al. incubated K18 for 48 hours with BPS vesicles to be able to detect a structuration of the peptide by CD, with an increase in secondary structure content.12Finally, in previous studies, structuration was observed only for shorter constructs of Tau (K18, K19, P2R…).12,23,25–27 However, the full-length Tau protein, particularly 2N4R, possesses a significant portion of disordered regions.2,51 Our results show thatTau-P301L in interaction with POPS is mainly in random coil (IDP) and accumulates on the surface of the POPS membrane. Adsorption could take place via 3 small α helices, as shown for K19 interacting with PS-containing membranes, but it is impossible to detect this in the midst of a large majority of unstructured amide groups.
4.3 Tau-P301L makes the PS membrane permeable
Our calcein leakage results show that Tau-P301L interacts with POPS LUVs, leading to Tau-P301L aggregation and liposome perturbation. Tau-induced membrane damage was observed for other forms of Tau as for example the 0N4R isoform on PIP2 LUVs.16 Individual R domains were also shown to possess lytic activity on DOPC:DOPG liposomes, linked to their ability to interact as demonstrated by ITC experiments.30 Overall, these results seem to confirm that the Tau protein has the ability to permeabilize the membrane leading to calcein leakage. Disruption of small DMPS vesicles, less than 40 nm in diameter, was induced by K19.13 In that case the small size of SUV supposes a high curvature of the vesicles and protein adsorption can lead to disruption. Interestingly the authors observe intact membrane fragments suggesting that the membranes are not fully destroyed.Indeed, our ATR-FTIR and PWR results support the idea that P301L induced perturbation rather than destruction of the POPS membrane. ATR and PWR data point also to a strong binding and accumulation of the protein on the bilayer surface, but without significant membrane disruption or lipid loss. How can we explain the discrepancy between what happens on a flat lipid membrane vs. what happens with lipid vesicles? One could invoke membrane curvature. For α-synuclein, the effect of membrane curvature was studied with varied lipid vesicle compositions,52 revealing a much stronger affinity of the protein for small vesicles of POPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPC (1
POPC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) compared to large ones. If SUV present more packing defects that LUVs,53 we could attribute this difference in interaction to the accessibility of the hydrophobic tails of the lipids, made more easily available in a vesicular context. In the case of positively charged P301L-Tau, it would be consistent with a shift of the interaction mechanism from an electrostatic to a hydrophobic interaction. However, some local membrane disruption was also observed by AFM imaging. The fact that we do not observe pore-like structure tends to suggest that the membrane leakage observed in liposomes is linked to accumulation of peptides on their surface.
1) compared to large ones. If SUV present more packing defects that LUVs,53 we could attribute this difference in interaction to the accessibility of the hydrophobic tails of the lipids, made more easily available in a vesicular context. In the case of positively charged P301L-Tau, it would be consistent with a shift of the interaction mechanism from an electrostatic to a hydrophobic interaction. However, some local membrane disruption was also observed by AFM imaging. The fact that we do not observe pore-like structure tends to suggest that the membrane leakage observed in liposomes is linked to accumulation of peptides on their surface.
4.4 Tau-P301L forms polymorphic aggregates on PS supported lipid bilayers
To fully understand the interaction mechanism between Tau-P301L and PS-containing membranes, a question remains as to the form adopted by Tau-P301L bound to the membrane. No insertion in the membrane was discernible by ATR-FTIR, as evidenced by the maintained dichroic ratio. PWR also allows us to probe the microscale organization, as the appearance of additional resonances could be attributed to the formation of heterogeneities with different mass and/or organization,35i.e. microdomains. We do not observe such additional resonances, which supports the absence of protein insertion in the bilayer, and indicates a lack of microdomains of more than a few hundred nanometers, as it could be observed in the case of Aβ1–40 peptide in a DOPC/SM/cholesterol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) bilayer54 or M8 mutant of HET-s peptide in a DOPG bilayer.35 These observations suggest the creation of a uniform adsorption on the surface, supported by the significant shift values observed in PWR after Tau addition, indicative of a substantial protein layer. The protein accumulation both observed in ATR-FTIR and PWR may induce a small perturbation of the membrane explaining the observed calcein leakage, without totally breaking down the membrane.
1) bilayer54 or M8 mutant of HET-s peptide in a DOPG bilayer.35 These observations suggest the creation of a uniform adsorption on the surface, supported by the significant shift values observed in PWR after Tau addition, indicative of a substantial protein layer. The protein accumulation both observed in ATR-FTIR and PWR may induce a small perturbation of the membrane explaining the observed calcein leakage, without totally breaking down the membrane.
AFM allowed us to go further in the understanding of the processes at play, by observing the local effects of Tau-P301L interaction with PS membranes at the nanoscale, using DOPS bilayers. Though differing in one of their fatty acid chains, with 1 additional unsaturation and 2 additional carbons for DOPS compared to POPS, both lipids share the same anionic headgroup and are fluid at room temperature, with melting temperatures of −11 and 14 °C for DOPS and POPS, respectively.43 Polarized ATR-FTIR data (Fig. S3†) confirms that both DOPS and POPS bilayers are fluid in our experimental conditions. We also showed (Fig. S4†) that both bilayers behave similarly in interaction with Tau-P301L, thus validating the use of DOPS for AFM experiments. Our AFM results suggest that Tau-P301L has two effects on DOPS membranes, the disruption of the supported membrane, and a simultaneous self-assembly on the DOPS surface.
The bilayer solubilization effect observed in our experiments is consistent with previous reports for the interaction of shorter Tau fragment K18 with mixed POPC:POPS or POPC:PIP2 membranes.29 In the case of membranes containing PIP2 especially, large portions of the membrane showed similar thinning as in Tau-P301L/DOPS interaction after incubation with the protein.29 The lower thickness observed locally after incubation with the protein could be consistent with a remaining monolayer of lipids in both cases. When defects are present in the bilayer, solubilization seems to be starting at the defects, supporting a hydrophobic interaction with the hydrophobic tails of the lipids. However, we note that in some cases, holes are forming upon protein addition in areas of the bilayer that did not contain defects (Fig. S7†).
Regarding Tau self-assembly on the DOPS surface, two types of aggregates are observed, patches and fibrillary structures originating from the patches. Our findings suggest that the morphology of the deposit is concentration-dependent, and that varying concentrations of Tau can elicit different effects on the membrane surface, with fibers only observed at concentrations of 500 nM and higher. Flat patches of amorphous aggregates of 2N4R Tau have been previously reported on membranes composed of brain total lipid extracts by Mari et al., for concentrations ranging from 100 to 300 nM.55 The surface area of the reported “flat islands” increases over time and is dependent on the concentration of Tau. However, the reported patches were much thinner than the ones we report, with thicknesses of ∼2 nm, and showed the presence of protrusions. This could be partly explained by the lipid composition of the supported membranes considered. In the present study, we use 100% DOPS, whereas Mari et al. used brain total lipid extract, which although overall negatively charged is heterogeneous in composition. Therefore, a charge-mediated interaction of Tau with membranes would be much stronger in fully anionic membranes, and could lead to bigger aggregates. Our observations of fibers on PS membranes is consistent with previous observations of K18 interactions on POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PIP2 (4
PIP2 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) SLB,29 where fibers appeared after 6 hours of incubation and also seemed to emerge from patchy aggregates on the membranes. However, fibers appeared much faster in the case of Tau-P301L interacting with DOPS, i.e. within 15 minutes of incubation, with subsequent observable fiber growth in a 1-hour timescale, although the concentration of protein was 2 times lower (500 nM vs. 1 μM in a previous study29). The faster growth we observed in the case of Tau-P301L could be attributed to the use of pure anionic membranes in our case, which could accelerate the impact of Tau on the membrane, as opposed to mixed SLB with zwitterionic lipids.29
1) SLB,29 where fibers appeared after 6 hours of incubation and also seemed to emerge from patchy aggregates on the membranes. However, fibers appeared much faster in the case of Tau-P301L interacting with DOPS, i.e. within 15 minutes of incubation, with subsequent observable fiber growth in a 1-hour timescale, although the concentration of protein was 2 times lower (500 nM vs. 1 μM in a previous study29). The faster growth we observed in the case of Tau-P301L could be attributed to the use of pure anionic membranes in our case, which could accelerate the impact of Tau on the membrane, as opposed to mixed SLB with zwitterionic lipids.29
Our observations that fibers assemble on negatively charged lipid surfaces that show membrane disruption and potential lipid loss suggests that lipid removal and fiber growth are two linked processes, which would agree with previous reports indicating recruitment of PIP2 inside a K18 fiber.56
4.5 Proposed interaction mechanisms for Tau-P301L interacting with PS-containing membranes
Based on our multi-technique and multi-scale approach, we are able to propose interactions mechanisms for Tau-P301L and PS membranes.The incubation of Tau-P301L with small liposomes of POPS resulted in the aggregation of Tau, leading to the formation of amyloid fibers. This aggregation was closely associated with liposome leakage, that could be attributed to a different interaction mechanism due to membrane curvature, potentially linked to easier access to hydrophobic tails of lipids in a vesicular context. Through the utilization of ATR-FTIR and PWR techniques, which provide a global scale assessment of the sample, we observed a robust and irreversible binding of Tau to the POPS surface without the emergence of microdomains, insertion in the bilayer, or lipid removal. This binding appears to create a homogeneous and thick layer of protein on the surface, resembling the carpet mechanism (Fig. 7A) previously identified in other amyloids.57,58 Carpeting is consistent with the morphologies observed by AFM imaging on DOPS bilayers, where the formation of large flat patches was observed. Additionally, we found no evidence of a predominant protein structure, indicating the potential coexistence of different species. This potential polymorphism was further confirmed at the nanoscale by AFM, where both thick patches and fibrillary structures were observed. The fibrillary forms resembled amyloid fibers, and appeared to extend and grow from the flat patches at higher concentrations of Tau-P301L (Fig. 7B). Considering the role of the anionic membrane in interacting with Tau, it has the ability to concentrate Tau at its surface, possibly triggering various disruptive mechanisms that result in the extraction of anionic lipids and the accumulation of polymorphic aggregates possibly composed of both proteins and lipids.
We propose that Tau-P301L interacts with PS-containing membranes by first establishing electrostatic interactions, that leads to accumulation of the protein in a thick layer without insertion. Carpeting leads to partial membrane disruption, a process that depends on membrane curvature, possibly accelerated by hydrophobic interactions with the lipid tails. Because ATR-FTIR did not show lipid depletion, we propose that the deposited flat patches could be a mixed composition of solubilized phospholipids and proteins, which could reconcile ATR-FTIR and AFM results, considering the timescales of both experiments. Above a threshold concentration of Tau-P301L, the aggregate structure shifts towards fibrillary structures that would be consistent with amyloid fibers, and would be consistent with the observed amyloid fibers obtained after incubation of Tau-P301L with small liposomes of POPS. Such objects are extremely interesting, both in secondary structure and chemistry, and a nanoscale investigation using AFM coupled to infrared spectroscopy could provide complementary information to confirm these fibers hold a beta-sheet content consistent with amyloids, and assess whether they contain lipids.
5. Conclusions
In this study, we employed a comprehensive approach, integrating aggregation studies, vesicle leakage assays, ATR-FTIR, plasmon waveguide resonance, and atomic force microscopy, to investigate the interaction of the disease-associated mutant P301L of Tau (2N4R) with phosphatidylserine, a negatively charged lipid of the inner neuronal membrane. Our findings reveal that Tau-P301L has the capacity to assemble into amyloid fibers in the presence of POPS SUVs, associated with vesicle damage. However, within a supported lipid bilayer context, ATR-FTIR did not show a significant disruption of the membrane but rather an accumulation of the Tau-P301L protein on its surface without insertion, thus hinting at a carpeting mechanism. The interaction of Tau with POPS demonstrated robust binding, with an apparent Kd of 0.14 ± 0.04 μM, also unveiling the deposition of a thick and homogeneous carpet on the bilayer's surface. Finally, AFM real time imaging showed a nanoscale impact of Tau-P301L on DOPS bilayers, with partial membrane disruption associated to the deposition of patches. Higher Tau concentration led to the growth of fibrillar structures from the patches, consistent with amyloid fibers. Because ATR-FTIR did not show lipid depletion, we propose that the deposited patches could be of a mixed composition of solubilized phospholipids and proteins, which could reconcile ATR-FTIR and AFM results, considering the timescales of both experiments. Our results bring additional input into Tau's potential ability to extract anionic lipids from membranes to form amyloid species, thus contributing to its deleterious effects. In alignment with previous studies highlighting the role of the lipid bilayer in amyloid pathogenicity,14,32,58,59 this study contributes to a better understanding of the potential role of the cellular membrane in Tau's toxicity.Author contributions
V. U.-T. performed all the experiments. Y. F. supervised production of Tau protein. I. A. supervised the PWR experiments.M. M., C. F. and S. L. managed the project. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
All the authors are grateful to the Agence Nationale de la Recherche for financial support under contract ANR-20-CE29-0004. The authors thank the VIbrAFM platform on which the AFM and ATR-FTIR experiments were performed. We also thank Clara Piersson for assistance with protein production.References
- M. D. Weingarten, A. H. Lockwood, S. Y. Hwo and M. W. Kirschner, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 1858–1862 CrossRef CAS PubMed.
- Y. Wang and E. Mandelkow, Nat. Rev. Neurosci., 2016, 17, 22–35 CrossRef CAS.
- K. S. Kosik, C. L. Joachim and D. J. Selkoe, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 4044–4048 CrossRef CAS.
- Y. Shi, W. Zhang, Y. Yang, A. G. Murzin, B. Falcon, A. Kotecha, M. Van Beers, A. Tarutani, F. Kametani, H. J. Garringer, R. Vidal, G. I. Hallinan, T. Lashley, Y. Saito, S. Murayama, M. Yoshida, H. Tanaka, A. Kakita, T. Ikeuchi, A. C. Robinson, D. M. A. Mann, G. G. Kovacs, T. Revesz, B. Ghetti, M. Hasegawa, M. Goedert and S. H. W. Scheres, Nature, 2021, 598, 359–363 CrossRef CAS PubMed.
- C. M. Wischik, M. Novak, H. C. Thøgersen, P. C. Edwards, M. J. Runswick, R. Jakes, J. E. Walker, C. Milstein, M. Roth and A. Klug, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 4506–4510 CrossRef CAS.
- L. N. Clark, P. Poorkaj, Z. Wszolek, D. H. Geschwind, Z. S. Nasreddine, B. Miller, D. Li, H. Payami, F. Awert, K. Markopoulou, A. Andreadis, I. D'Souza, V. M. Lee, L. Reed, J. Q. Trojanowski, V. Zhukareva, T. Bird, G. Schellenberg and K. C. Wilhelmsen, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 13103–13107 CrossRef CAS.
- D. Chen, K. W. Drombosky, Z. Hou, L. Sari, O. M. Kashmer, B. D. Ryder, V. A. Perez, D. R. Woodard, M. M. Lin, M. I. Diamond and L. A. Joachimiak, Nat. Commun., 2019, 10, 2493 CrossRef PubMed.
- K. Pounot, C. Piersson, A. K. Goring, F. Rosu, V. Gabelica, M. Weik, S. Han and Y. Fichou, JACS Au, 2024, 4, 92–100 CrossRef CAS.
- G. Limorenko and H. A. Lashuel, Neurobiol. Dis., 2021, 161, 105536 CrossRef CAS.
- T. Kampers, P. Friedhoff, J. Biernat, E.-M. Mandelkow and E. Mandelkow, FEBS Lett., 1996, 399, 344–349 CrossRef CAS.
- C. N. Chirita, M. Necula and J. Kuret, J. Biol. Chem., 2003, 278, 25644–25650 CrossRef CAS PubMed.
- N. Ait-Bouziad, G. Lv, A.-L. Mahul-Mellier, S. Xiao, G. Zorludemir, D. Eliezer, T. Walz and H. A. Lashuel, Nat. Commun., 2017, 8, 1678 CrossRef.
- Q.-Q. Yao, J. Wen, S. Perrett and S. Wu, Nanoscale, 2022, 14, 4604–4613 RSC.
- E. Bok, E. Leem, B.-R. Lee, J. M. Lee, C. J. Yoo, E. M. Lee and J. Kim, Front. Cell Dev. Biol., 2021, 9, 653815 CrossRef.
- M. Merezhko, C. A. Brunello, X. Yan, H. Vihinen, E. Jokitalo, R.-L. Uronen and H. J. Huttunen, Cell Rep., 2018, 25, 2027–2035 CrossRef CAS.
- T. Katsinelos, M. Zeitler, E. Dimou, A. Karakatsani, H.-M. Müller, E. Nachman, J. P. Steringer, C. Ruiz de Almodovar, W. Nickel and T. R. Jahn, Cell Rep., 2018, 23, 2039–2055 CrossRef CAS.
- C. A. Brunello, M. Merezhko, R.-L. Uronen and H. J. Huttunen, Cell. Mol. Life Sci., 2020, 77, 1721–1744 CrossRef CAS PubMed.
- G. P. Gellermann, T. R. Appel, P. Davies and S. Diekmann, Biol. Chem., 2006, 387, 1267–1274 CAS.
- G. Di Paolo and T.-W. Kim, Nat. Rev. Neurosci., 2011, 12, 284–296 CrossRef CAS PubMed.
- K. Wells, A. A. Farooqui, L. Liss and L. A. Horrocks, Neurochem. Res., 1995, 20, 1329–1333 CrossRef CAS.
- D. S. Knopman, H. Amieva, R. C. Petersen, G. Chételat, D. M. Holtzman, B. T. Hyman, R. A. Nixon and D. T. Jones, Nat. Rev. Dis. Primers, 2021, 7, 33 CrossRef PubMed.
- S. Elbaum-Garfinkle, T. Ramlall and E. Rhoades, Biophys. J., 2010, 98, 2722–2730 CrossRef CAS PubMed.
- N. El Mammeri, O. Gampp, P. Duan and M. Hong, Commun. Biol., 2023, 6, 467 CrossRef CAS PubMed.
- N. Patel, S. Ramachandran, R. Azimov, B. L. Kagan and R. Lal, Biochemistry, 2015, 54, 7320–7325 CrossRef CAS PubMed.
- G. Künze, P. Barré, H. A. Scheidt, L. Thomas, D. Eliezer and D. Huster, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 2302–2313 CrossRef.
- P. Barré and D. Eliezer, J. Mol. Biol., 2006, 362, 312–326 CrossRef.
- E. R. Georgieva, S. Xiao, P. P. Borbat, J. H. Freed and D. Eliezer, Biophys. J., 2014, 107, 1441–1452 CrossRef CAS PubMed.
- E. M. Jones, M. Dubey, P. J. Camp, B. C. Vernon, J. Biernat, E. Mandelkow, J. Majewski and E. Y. Chi, Biochemistry, 2012, 51, 2539–2550 CrossRef CAS PubMed.
- M. Azouz, C. Feuillie, M. Lafleur, M. Molinari and S. Lecomte, Nanoscale Adv., 2021, 3, 4244–4253 RSC.
- S. S. Dicke, L. Tatge, P. E. Engen, M. Culp and L. R. Masterson, Biochem. Biophys. Res. Commun., 2017, 493, 1504–1509 CrossRef CAS.
- R. Brandt, J. Léger and G. Lee, J. Cell Biol., 1995, 131, 1327–1340 CrossRef CAS PubMed.
- C. A. Sallaberry, B. J. Voss, J. Majewski, J. Biernat, E. Mandelkow, E. Y. Chi and C. M. Vander Zanden, Front. Cell Dev. Biol., 2021, 9, 725241 CrossRef.
- Y. Fichou, Z. R. Oberholtzer, H. Ngo, C.-Y. Cheng, T. J. Keller, N. A. Eschmann and S. Han, Front. Neurosci., 2019, 13, 1339 CrossRef PubMed.
- K. Itaya and M. Ui, Clin. Chim. Acta, 1966, 14, 361–366 CrossRef CAS.
- E. Harté, N. Maalouli, A. Shalabney, E. Texier, K. Berthelot, S. Lecomte and I. D. Alves, Chem. Commun., 2014, 50, 4168–4171 RSC.
- P. Calmet, M. De Maria, E. Harté, D. Lamb, M. Serrano-Vega, A. Jazayeri, N. Tschammer and I. D. Alves, Sci. Rep., 2016, 6, 36181 CrossRef CAS PubMed.
- I. D. Alves and S. Lecomte, Acc. Chem. Res., 2019, 52, 1059–1067 CrossRef CAS.
- S. Wegmann, Y. J. Jung, S. Chinnathambi, E.-M. Mandelkow, E. Mandelkow and D. J. Muller, J. Biol. Chem., 2010, 285, 27302–27313 CrossRef CAS.
- P. Nacharaju, J. Lewis, C. Easson, S. Yen, J. Hackett, M. Hutton and S.-H. Yen, FEBS Lett., 1999, 447, 195–199 CrossRef CAS PubMed.
- S. Castano and B. Desbat, Biochim. Biophys. Acta, Biomembr., 2005, 1715, 81–95 CrossRef CAS.
- J. Berriman, L. C. Serpell, K. A. Oberg, A. L. Fink, M. Goedert and R. A. Crowther, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9034–9038 CrossRef CAS PubMed.
- Z. Salamon and G. Tollin, Biophys. J., 2004, 86, 2508–2516 CrossRef CAS PubMed.
- R. A. Demel, F. Paltauf and H. Hauser, Biochemistry, 1987, 26, 8659–8665 CrossRef CAS PubMed.
- A. Ali, K. Zhaliazka, T. Dou, A. P. Holman and D. Kurouski, J. Biol. Chem., 2023, 299, 10583 Search PubMed.
- A. Frese, C. Goode, K. Zhaliazka, A. P. Holman, T. Dou and D. Kurouski, Protein Sci., 2023, 32, e4717 CrossRef CAS.
- S. D. Connell and D. A. Smith, Mol. Membr. Biol., 2005, 23, 17–28 CrossRef.
- A. M. Pooler, A. Usardi, C. J. Evans, K. L. Philpott, W. Noble and D. P. Hanger, Neurobiol. Aging, 2012, 33, 431.e27–431.e38 CrossRef CAS PubMed.
- M. Arrasate, M. Pérez and J. Avila, Neurochem. Res., 2000, 25, 43–50 CrossRef CAS.
- H.-Y. Kim, B. X. Huang and A. A. Spector, Prog. Lipid Res., 2014, 1–18 CrossRef PubMed.
- H. I. Ingólfsson, M. N. Melo, F. J. van Eerden, C. Arnarez, C. A. Lopez, T. A. Wassenaar, X. Periole, A. H. de Vries, D. P. Tieleman and S. J. Marrink, J. Am. Chem. Soc., 2014, 136, 14554–14559 CrossRef.
- G. Limorenko and H. A. Lashuel, Chem. Soc. Rev., 2022, 51, 513–565 RSC.
- E. R. Middleton and E. Rhoades, Biophys. J., 2010, 99, 2279–2288 CrossRef CAS.
- M. S. Terakawa, Y. Lin, M. Kinoshita, S. Kanemura, D. Itoh, T. Sugiki, M. Okumura, A. Ramamoorthy and Y.-H. Lee, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1741–1764 CrossRef CAS PubMed.
- S. Devanathan, Z. Salamon, G. Lindblom, G. Gröbner and G. Tollin, FEBS J., 2006, 273, 1389–1402 CrossRef CAS PubMed.
- S. A. Mari, S. Wegmann, K. Tepper, B. T. Hyman, E.-M. Mandelkow, E. Mandelkow and D. J. Müller, Nano Lett., 2018, 18, 3271–3281 CrossRef CAS.
- D. Talaga, W. Smeralda, L. Lescos, J. Hunel, N. Lepejova-Caudy, C. Cullin, S. Bonhommeau and S. Lecomte, Angew. Chem., Int. Ed., 2018, 57, 15738–15742 CrossRef CAS.
- K. Berthelot, C. Cullin and S. Lecomte, Biochimie, 2013, 95, 12–19 CrossRef CAS PubMed.
- O. Press-Sandler and Y. Miller, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1889–1905 CrossRef CAS.
- F. Collin, O. Cerlati, F. Couderc, B. Lonetti, J.-D. Marty and A.-F. Mingotaud, TrAC, Trends Anal. Chem., 2020, 132, 116059 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr01343c |
| This journal is © The Royal Society of Chemistry 2024 |