Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Characterization of a unique attachment organelle: Single-cell force spectroscopy of Giardia duodenalis trophozoites†
Gubesh
Gunaratnam
 *a,
Ricarda
Leisering
b,
Ben
Wieland
*a,
Ricarda
Leisering
b,
Ben
Wieland
 a,
Johanna
Dudek
c,
Nicolai
Miosge
c,
Sören L.
Becker
a,
Johanna
Dudek
c,
Nicolai
Miosge
c,
Sören L.
Becker
 a,
Markus
Bischoff
a,
Markus
Bischoff
 a,
Scott C.
Dawson
a,
Scott C.
Dawson
 d,
Matthias
Hannig
d,
Matthias
Hannig
 c,
Karin
Jacobs
c,
Karin
Jacobs
 ef,
Christian
Klotz
ef,
Christian
Klotz
 b,
Toni
Aebischer
b,
Toni
Aebischer
 b and
Philipp
Jung
b and
Philipp
Jung
 a
a
aInstitute of Medical Microbiology and Hygiene, Saarland University, Homburg, Germany. E-mail: gubesh.gunaratnam@uni-saarland.de
bDepartment of Infectious Diseases, Unit 16 Mycotic and Parasitic Agents and Mycobacteria, Robert Koch-Institute, Berlin, Germany
cClinic of Operative Dentistry and Periodontology, Saarland University, Homburg, Germany
dDepartment of Microbiology and Molecular Genetics, University of California Davis, Davis, USA
eExperimental Physics, Saarland University, Saarbrücken, Germany
fMax Planck School, Matter to Life, Heidelberg, Germany
First published on 19th March 2024
Abstract
The unicellular parasite Giardia duodenalis is the causative agent of giardiasis, a gastrointestinal disease with global spread. In its trophozoite form, G. duodenalis can adhere to the human intestinal epithelium and a variety of other, artificial surfaces. Its attachment is facilitated by a unique microtubule-based attachment organelle, the so-called ventral disc. The mechanical function of the ventral disc, however, is still debated. Earlier studies postulated that a dynamic negative pressure under the ventral disc, generated by persistently beating flagella, mediates the attachment. Later studies suggested a suction model based on structural changes of the ventral discs, substrate clutching or grasping, or unspecific contact forces. In this study, we aim to contribute to the understanding of G. duodenalis attachment by investigating detachment characteristics and determining adhesion forces of single trophozoites on a smooth glass surface (RMS = 1.1 ± 0.2 nm) by fluidic force microscopy (FluidFM)-based single-cell force spectroscopy (SCFS). Briefly, viable adherent trophozoites were approached with a FluidFM micropipette, immobilized to the micropipette aperture by negative pressure, and detached from the surface by micropipette retraction while retract force curves were recorded. These force curves displayed novel and so far undescribed characteristics for a microorganism, namely, gradual force increase on the pulled trophozoite, with localization of adhesion force shortly before cell detachment length. Respective adhesion forces reached 7.7 ± 4.2 nN at 1 μm s−1 pulling speed. Importantly, this unique force pattern was different from that of other eukaryotic cells such as Candida albicans or oral keratinocytes, considered for comparison in this study. The latter both displayed a force pattern with force peaks of different values or force plateaus (for keratinocytes) indicative of breakage of molecular bonds of cell-anchored classes of adhesion molecules or membrane components. Furthermore, the attachment mode of G. duodenalis trophozoites was mechanically resilient to tensile forces, when the pulling speeds were raised up to 10 μm s−1 and adhesion forces increased to 28.7 ± 10.5 nN. Taken together, comparative SCSF revealed novel and unique retract force curve characteristics for attached G. duodenalis, suggesting a ligand-independent suction mechanism, that differ from those of other well described eukaryotes.
1 Introduction
The unicellular parasite Giardia duodenalis (syn.: G. lamblia, G. intestinalis) and causal agent of giardiasis exists as a surface-colonizing trophozoite in the small intestine of humans. It adopts a unique teardrop-like morphology that involves a microtubule cytoskeleton. Among the extended cytoskeleton components are eight flagella, which are responsible for the free- and planar swimming behaviour of the trophozoites and for the positioning of the adhering cell on the substratum.1,2 Furthermore, a unique microtubule organelle, the cup shaped ventral disc, consisting of a three-dimensional spiral array of microtubules with a single overlap zone and an inner bare area, is crucial for reversible attachment to human host structures.3,4 In the early phase of attachment, the periphery of the ventral disc is surrounded by an electron dense structure, namely the lateral crest. Additionally, in the later stage of attachment, ventral cell body regions, the lateral shield and the plasma membrane of the bare area, contact the surface.1,2 Depending on ventral disc function, G. duodenalis is capable of binding to different kinds of artificial surfaces including plain, positively charged, or hydrophobic glass substrates.5,6 Importantly, the trophozoite is able to establish tight attachment under shear stress conditions, believed to resist peristalsis in the intestine.1,7Different theoretical approaches have been suggested to describe the mode of attachment for Giardia species. Particular interest were on two simultaneously beating ventral flagella and the ventral disc. In a first model, it was suggested that the beating action of the ventral flagella draws water molecules out of the ventral disc cup, generating a dynamic negative pressure below the ventral disc, which results in a suction-based attachment.8,9 This is in line with analysis from light micrographs of trophozoites in early studies, and from sophisticated light microscopy set-ups utilized later on, where attached trophozoites show regular fast-beating ventral flagella while being attached to the surface.4,8 Furthermore, a directed liquid flow below or adjacent to the ventral disc was identified by particle image velocimetry that was thought to be caused by a possible displacement-pump function of the regular beating of the ventral flagella.2,10 However, the theoretical framework for this active hydrodynamic mode of attachment was challenged when beating flagella-defective trophozoites were observed to resist detachment forces during a centrifugation experiment,1 suggesting that the hydrodynamic suction-based attachment is probably not the only mode employed by this parasite to adhere to a surface.
Recent studies demonstrated that the ventral disc is a non-rigid, highly dynamic organelle and capable of mechanical adaptions. High-resolution live cell imaging revealed domed and flat disc conformations of the ventral disc, and disc dynamics, such as disc compression, overlap zone sliding, or rotational constriction of the ventral disc's lateral crest, which is an electron-dense peripheral rim of the ventral disc.11 Furthermore, the ventral disc is associated with more than 80 disc-associated proteins (DAPs), which are likely important for the disc architecture.12 Thus, in a second model, it was proposed that the suction force is generated by dynamic changes of the ventral disc, in an interplay with associated DAP proteins and microtubule cross bridges, hence, allowing for an attachment force that is independent from flagellar motility.9,11
Other work indicates that G. duodenalis attachment to a biological or abiotic surface might be mediated by a clutching or grasping mechanism that depends on the ventral disc. This had been indicated by circular imprints on intestinal brush borders after trophozoite detachment by SEM micrographs. Notably, morphological changes of the ventral disc had been proposed13,14 even before DAPs were described as molecular contributors to the contracting, tightening and the changing of the disc diameter. Moreover, a more specific lectin-type ligand–receptor binding mechanism to glycosylated cell-surface components of enterocytes was proposed15 but has been not identified to date.
To experimentally investigate the physical mode of attachment of G. duodenalis or any other candidate microbial species, analysis of adhesive properties at a single-cell level is required.16 During the last two decades, atomic force microscopy (AFM) has emerged as a tremendously valuable tool to study the adhesive properties of single bacterial, fungal or mammalian cells.16–27 In this approach, a biofunctionalized AFM probe (i.e. a cantilever harbouring a cell on its one end) allows to analyse specific adhesion and detachment characteristics between microbes and a substratum and to read out adhesion parameters from force–distance curves. One important advance in AFM-based adhesion studies was the combination of AFM with microfluidics into fluidic force microscopy (FluidFM).28 In FluidFM, the AFM cantilever is equipped with an inside microchannel and is connected to a pressure control system to exert positive or negative pressure. Via an opening at the probing region of the cantilever, negative pressure can be applied to grab and detach an individual cell from the surface. This technique named AFM-based single-cell force spectroscopy (SCFS) allows quantifying crucial detachment parameters. Such parameters are the maximum detachment force (which is commonly known as “adhesion force”, a terminology continued to be used in this study), the cell detachment length (i.e. the movement of the probe in z-length to detach the cell from the surface), and the respective localization of the adhesion force towards cell detachment length.
Here, we investigated the mode of G. duodenalis attachment by studying detachment characteristics of trophozoites adhering to a flat glass surface, which is to our knowledge a novelty that SCFS was used to address a suction-based microbial attachment mechanism. Additionally, we aimed to compare the G. duodenalis way of attachment to previously described adhesion mechanisms of the yeast species Candida albicans and human keratinocytes as other classes of eukaryotic cells. Those mechanisms are based on molecular bonds of individual cell wall- or cell membrane-anchored adhesion molecules, and their biophysical properties while the cell is pulled from the surface. We emphasize that the investigation of an organelle-based attachment mechanism in G. duodenalis represents a new biophysical approach to study microbial adhesion.
2 Results
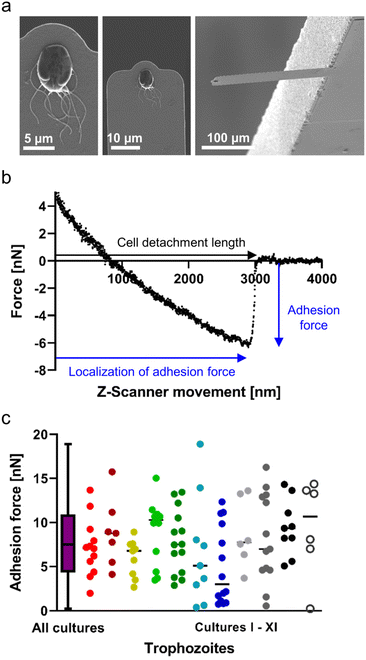
Since FluidFM-based SCFS with G. duodenalis trophozoites is a completely new approach, we decided to visualise the uptake and orientation of the trophozoite at the micropipette, after successfully pulling and detaching it from the glass substratum, using SEM (Fig. 1a). Here, the ventral side of the trophozoite and the species-unique ventral disc were clearly visible. The visual appearance of the ventral disc further suggests that this microtubule organelle was involved in the attachment to the glass surface and did not break away from the cell body during detachment. Releasing the trophozoites again into the liquid environment resulted in their active swimming or in their reattachment to the glass surface, both, confirming cell viability during force–distance measurements (ESI Fig. S1†). During the retraction of the micropipette (caused by the Z-scanner movement), we observed a distinct major bond rupture of the trophozoite (Fig. 1b). This major peak was the adhesion force whose localization closely precedes the cell detachment length, which marks the last contact of the pulled trophozoite on the surface. The occurrence of that major force peak indicated the presence of a substantial proportion of cell area, i.e. the ventral disc, involved in attachment to the glass substratum, and was found for all trophozoites investigated in this study.To learn whether there is any impact of the trophozoite “head-to-tail” polarization on the adhesion force as determined by FluidFM technology, we investigated a representative number of trophozoites in different orientations. We found that the rotational orientation of the attached trophozoites (of similar sized cells), relative to the fixed movement of the micropipette, neither influenced the adhesion forces nor produced technical bias in force curve characteristics (ESI Fig. S2†). However, one orientation (see “up”, ESI Fig. S2†) occasionally produced a lateral vertical movement of the trophozoites (due to the inbuilt angle of the micropipette) and was excluded from further measurements. Hence, for the total number of trophozoites of all biological cultures, tested by SCFS with a pulling speed of 1 μm s−1, we found normally distributed adhesion forces with a mean of 7.7 ± 4.2 nN (n = 111, Fig. 1c). Here, the high deviation of adhesion forces from the mean was apparent, and we wondered whether this occurred due to the “adhesive strength” of trophozoites between different experiments (i.e. biological cultures), or whether there was a heterogeneous distribution of adhesive strength in trophozoites in each tested population. Highlighting the data of each single experiment in a particular colour (Fig. 1c, right), we found that each experiment contained trophozoites with variable adhesive strength, covering a large range of adhesion forces, displaying 1–3 nN for some trophozoites, to almost 20 nN for others. These tested individual trophozoites strongly indicated the existence of different trophozoite culture subpopulations (i.e. members with a distinct adhesive strength of unknown origin) within each biological culture. This observation was true for all investigated Giardia cultures (Fig. 1c, individual colours for each experiment).
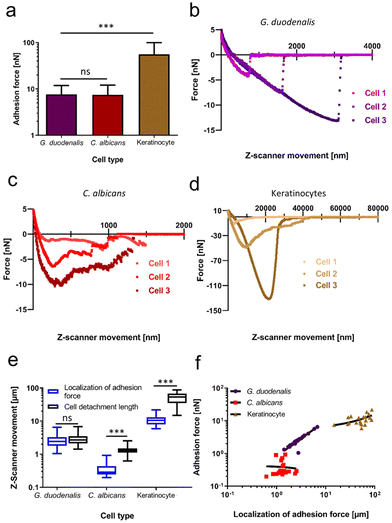
To examine potential cell-geometrical reasons as the cause for this distribution of force values, we investigated whether the total cell length and the maximum width of the teardrop-shaped trophozoites correlated with the quantified adhesion forces (ESI Fig. 3†). We found that the trophozoites displayed a typical length between 17.5–21 μm which had a weak linear correlation with the adhesion forces (R2 = 0.16; ESI Fig. 3a†). Interestingly, a higher degree of correlation was identified between the maximum cell width (left to right diameter of the cell body) of 10 μm–14 μm and the adhesion forces (R2 = 0.37; ESI Fig. 3b†). Compared to other eukaryotic cell types or pathogens, which mediate adhesion/attachment to the substratum by the tethering of cell surface-exposed macromolecules, the prevailing biophysical attachment mode of G. duodenalis trophozoites is unknown. Thus, to compare with well-known adhesion mechanisms, we evaluated the adhesion forces and the retract force-curve characteristics of C. albicans yeast cells and human (oral) keratinocytes. Both cell types are well characterized in terms of adhesion and had been subject of various studies investigating their attachment mechanisms.18–20,22,23,29,30 To allow for comparisons, both cell types were allowed to adhere to the same glass surface, which was used to study the attachment of Giardia trophozoites. For these two types of eukaryotic species, we selected shorter attachment times (15–60 min), if compared to the rather extended attachment time for G. duodenalis of 48 hours. In this way, we reduced an uncontrolled increase of the adhesive strength over time, viz. bond strengthening of cell-wall anchored adhesins in C. albicans,31 or increase in contact area in human cells.23 Adhesion forces recorded for C. albicans were 7.5 ± 4.6 nN (n = 20), while oral keratinocytes displayed adhesion forces of 55.8 ± 43.9 nN (n = 20, Fig. 2a). These quantitative data showed that the adhesion forces obtained with C. albicans after detachment from glass were in a similar range as the adhesion forces of G. duodenalis (7.7 ± 4.2 nN), and that the early spreading of human keratinocytes on glass already surpassed the values of both pathogens. Superior adhesive strength of keratinocytes was also valid, when we compared the work of adhesion, which was not significantly different between G. duodenalis and C. albicans but was the largest for the keratinocytes (ESI Fig. 4†). Analysing the cell detachment lengths identified further differences. Typical cell detachment lengths in the retraction curves are depicted in Fig. 2b (G. duodenalis), Fig. 2c (C. albicans) and Fig. 2d (Keratinocytes). While for the human keratinocytes a long cell detachment length of 54.2 ± 20.4 μm was found, which can be explained by tethering and unbinding of cell membrane components, and known high mammalian cell elasticities,23,32G. duodenalis exhibited intermediate values of 3.0 ± 1.4 μm, and C. albicans cells produced the lowest values of 1.4 ± 0.5 μm (Fig. 2e, black boxes). Although cell detachment lengths were in a similar range for G. duodenalis and C. albicans, representative force curves of C. albicans showed a characteristic pattern with multiple peaks of different force (Fig. 2c), indicative of multiple molecular bond breakages at the interface of cell and substratum, which was clearly not the case for G. duodenalis trophozoites (Fig. 2b). Instead, as mentioned above, all G. duodenalis trophozoites uniformly had a single-major force peak in their retraction curves, which indicated the breakage of only one major attachment event between the trophozoite and the glass surface. Furthermore, force curve characteristics of G. duodenalis were in stark contrast to those of keratinocytes (Fig. 2d). Force curves of keratinocytes had shape variations among individual cells and had inherent characteristics where the adhesion forces was followed by smaller detachment forces and force plateaus (Fig. 2d, cell 2). These phenomena were absent in G. duodenalis trophozoites.
A further closer look at each cell type's individual retraction curves revealed insights into the localization of the adhesion force (Fig. 2e, blue boxes) in relation to the cell detachment length. For G. duodenalis trophozoites the mean localization of the adhesion forces was at 2.81 ± 1.32 μm and corresponds to 93% of the cell detachment length (3.01 ± 1.36 μm), from which it was not significantly different (P = 0.49). This clearly demonstrates that the strongest resistance to detachment of the trophozoites coincided with the detachment of the whole cell from the glass surface. Importantly, such phenomenon was observed neither for C. albicans cells nor for the oral keratinocytes. The localization of adhesion force for C. albicans arose at a relative short distance of 0.38 ± 0.22 μm, and this major peak position was followed by numerous, smaller detachment events until the cell detachment length was reached at a significantly higher distance at 1.36 ± 0.46 μm. For oral keratinocytes, the localization of adhesion force was at 11.31 ± 0.41 μm, and, in a similar manner as for the yeast, significantly shorter than their cell detachment length (54.19 ± 20.43 μm). Additionally, for high localization of adhesion force, we observed a strong positive correlation (R2 = 0.95) with adhesion forces for G. duodenalis trophozoites (Fig. 2f), in contrast to the stark discrepancy seen with C. albicans (R2 = −0.004) and oral keratinocytes (R2 = 0.26), respectively, suggesting a more dynamic attachment behaviour for trophozoites than for the other cell types.
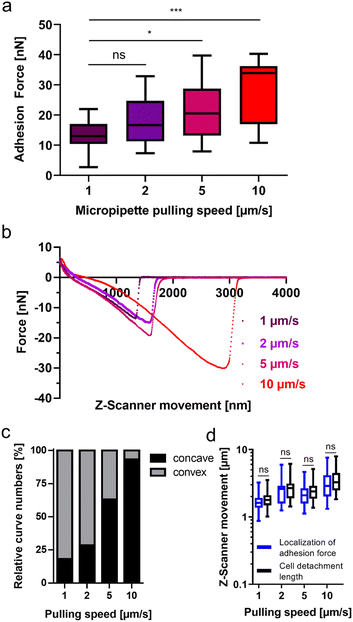
Like other non-invasive intestinal microorganisms, G. duodenalis must withstand mechanical stress at its infection site in the human body. This mechanical stress on G. duodenalis trophozoites is believed to be caused by peristalsis in the small intestine. Therefore, after having identified attachment characteristics of this microbial species on glass, we tried to learn more about the resilience of the attachment to mechanical stress. To achieve this, we varied the micropipette pulling speed, which is known to increase the tensile stress on the attached cell.33 In this set of experiments, with the same pulling speed (1 μm s−1) as for the experiment depicted in Fig. 1c, we determined adhesion forces of 13.3 ± 5.2 nN (n = 22, Fig. 3a, left box) for the trophozoites. When the pulling speed was increased to 2 μm s−1, 5 μm s−1, and 10 μm s−1, adhesion forces increased to 18.5 ± 7.7 nN (n = 15), 21.5 ± 9.1 nN (n = 19), and 28.7 ± 10.5 nN (n = 15), respectively (Fig. 3a). Retraction force curves of G. duodenalis trophozoites demonstrated that the characteristic major force peak of the retract curve was retained for all four different pulling speeds of the micropipette (Fig. 3b). However, during initial pulling phases of the trophozoites from the surface, the shape of the retraction curves changed from a mainly convex (1 μm s−1) to a concave form (10 μm s−1) at increasing pulling speeds (Fig. 3c), and thus a slower force transmission of the restoring force of the micropipette on the majority of the trophozoites at 5 and 10 μm s−1. Unaffected by this, at the end of their course, retract curves resulted in the characteristic major force peak for all tested pulling speeds, demonstrating the resilience of this attachment mode to mechanical stress. This observation is supported by the maintained close correlation of the localization of adhesion force and cell detachment length for all pulling speeds (Fig. 3d).
3 Discussion
In this study, we introduced FluidFM-based SCFS as a tool to study parasite attachment and investigated the detachment forces and characteristics of G. duodenalis trophozoites in comparison with the well-known adhesion mechanisms of other eukaryotic single cell types, which were the yeast species C. albicans, and human oral keratinocytes. Interestingly, compared to the force pattern with force peaks of different values (indicative of multiple bond breakages) during the detachment of C. albicans, G. duodenalis trophozoites showed a major bond breakage on the glass surface. A force pattern with force peaks of different values can be attributed to the rupture of multiple cell-wall anchored adhesins, such as the Als1 protein in C. albicans, or cell-wall anchored proteins in bacterial species, like Staphylococcus aureus, which play an important role in irreversible adhesion to biological and artificial surfaces and subsequent biofilm formation.34–36 For G. dudodenalis, an attachment model solely relying on a putative tethering of macromolecules to the glass surface is doubted by us since the occurrence of a single major bond breakage would require that all involved tetherings would simultaneously detach from the surface. This is very unlikely, because it would require same-length proteinaceous, or polymeric surface macromolecules with identical mechanical stretching properties. Such properties were not described, so far, for rare surface proteins in G. duodenalis, such as antigenic variant-specific surface proteins (VSPs) or potential glycoproteins. More likely, an adhesion model that depends on the mechanical properties of the cup-like ventral disc and not on surface membrane properties best describe our observations on attachment by G. duodenalis to flat glass surfaces. The function of this cup might rely on the sealing of the ventral disc by the electron-dense (fibrillar and globular) lateral crest,1,37 followed by a conformational change of the ventral disc, from a flat towards a domed conformation.38 The breakage of this seal, mediated by the unique attachment organelle, and thereby overcoming the force it transmitted to the glass surface could explain why all tested trophozoites showed the uniform retract force curve with the single-major bond rupture. Furthermore, the release of the whole cup at once from the glass surface could be reason of the close correlation of the localization of the adhesion force and the cell detachment length.Compared to the observations with keratinocytes, we observed no force plateaus in the force curves during the detachment of G. duodenalis from the glass substrate. During detachment of mammal or human cells, lipid membrane tethers of the phospholipid bilayer are created by the adhesion between single points of the cell membrane and the surface. Membrane tethers are long (tens of microns), nanometer-thick membrane tubes, which are released from the underlying cell cytoskeleton (e.g. microtubules or actin cortex) through interaction with the surface, and are detected in force spectroscopy.39 The absence of such tether formation in retraction curves of G. duodenalis indicate that the ventral cell membrane is strongly cross-linked with the peripheral ventral disc cytoskeleton. This observation is in accordance with earlier micrographs showing a close proximity of the microtubule at the periphery of the ventral disc with the ventral cell membrane in G. muris.8 One further apparent difference of the detachment characteristics between the keratinocytes and the trophozoite, was the significant distance between the localization of adhesion force and cell detachment length. This clearly indicates for the keratinocytes that a number of different interactions were still involved after the adhesion force was reached, i.e. specific receptor–ligand interactions between cell receptors and sedimented proteins (of the cell culture medium and/or secreted by the keratinocytes) on the glass substratum; or weak, unspecific interactions.40 Similar specific interactions, e.g. lectin-type binding, of G. duodenalis on the glass surface were not directly visible in our force retraction curves, indicating their absence or only minor contribution to the adhesion force in the attachment of trophozoites to glass.
We observed that the adhesion force of G. duodenalis were comparable to those values of C. albicans and were much smaller than those values of the keratinocytes. This strongly suggest that the lower nN-force adhesive strength of G. duodenalis is comparable to the initial attachment strength of C. albicans yeast cells and other microbes,41,42 and is substantially different to the adhesion forces observed with adherent and spreading keratinocytes. We assume that the high adhesion forces observed with keratinocytes are due to an overall larger contact area of the keratinocyte with multiple focal adhesion spots, which in sum exceeded the contact areas produced by G. duodenalis and C. albicans on this type of surface, respectively. Interestingly, our mean adhesion forces for trophozoites were significantly higher than those values (2–3 nN range) derived from centrifugation assays,5 or from microfluidic devices (0.9 nN on glass, based on suction force and van der Waals forces, or 1.2 nN, considering clutching forces as well).5,43 Importantly, those experiments are not necessarily in contrast with our experiments, because in those experimental approaches, EC50 values were calculated, which are the forces that could detach 50% of the trophozoites from the surface. Consequently, the determined values represent the minimum requirements for the remaining trophozoites to stay attached to the substratum and could, thus, be significantly higher (several nN as determined by us) than calculated.
The attachment characteristics of G. duodenalis are mediated and controlled by the dynamics of the ventral disc. One of our groups showed that 93% of wildtype trophozoites formed a lateral crest seal and a bare area contact, and the rest only formed the seal but not the bare area contact.44 Findings based on high resolution TIRF microscopy indicated that trophozoite attachment is initialized by serial attachment events of different body components surrounding the ventral disc45 could last for seconds to hours, and ends with detachment of the ventral disc, surrounding body components and thus leading to detachment of the complete cell, (attachment – detachment cycle). Individual stages within this cycle, likely, reflect a regulated process that depends on biochemical signals and stages could explain differences in adhesion force for individual trophozoites as observed in this study. For example, most of the trophozoites might form a bare area contact and produce high adhesion forces, and a smaller fraction of trophozoites, which demonstrated lower nanonewton values, might be in an earlier stage of attachment. This indicates that we might have probed trophozoites at different stages of their natural attachment cycle during our SCFS experiments. Moreover, other factors, such as the cell cycle and division, or trophozoite fitness might contribute to individual adhesion-force differences as well.4 Additionally, we found a better correlation between cell width and adhesion force than for the cell length. Recent works11,44 with immunostained Giardia trophozoites showed that the ventral disc diameter (7.5–9.5 μm) varied among trophozoites, to a similar degree as did the cell width in this study. Hence, it is possible that the cell width is a good approximation for the disc diameter, and the disc diameter is correlated with adhesion forces. However, as the force of attachment is also dependent on the stage of attachment cycle this is likely not a mutually exclusive conclusion and the exact contribution of disc size (body diameter sizes) and stage of cycle need to be explored in future studies.
Our observations indicate that the attachment mode of G. duodenalis is resilient to mechanical stress, since adhesion forces increased to several tens of nanonewton as a function of the pulling speed. It is of major interest to unravel, which cell mechanical factors allow such great resistance to tensile forces without a significant breakage of the single-major attachment bond observed in our force curves. It is possible that DAPs are playing a key role at all the tested pulling speeds by giving a hyperstability to the ventral disc.12 In addition, lateral shield and bare area contacts might contribute to attachment forces as well. Our force data contribute to the well-known fact that mechanical stress, i.e. shear forces, can effectively be resisted by the attached trophozoites.7 Interestingly, a positive correlation between the pulling speed on the trophozoite and the adhesion force was detected in our SCFS studies. The detachment of the trophozoite at high pulling speeds might be in stark contrast to the rather slow ‘natural’ separation (up to 15 s) of lateral shield and bare zone area contacts with the surface.45 Hence, one could interpret this high adhesion forces because of a limitation of biochemical signals that regulate trophozoite adhesive strength and detachment under physiological conditions and time intervals. Reduction of adhesive force likely is regulated by signalling which may be limited by diffusion of signalling molecules, e.g. by cytoplasmic protein activity and their diffusion coefficients.46 Considering a distance of >3 μm between the quickly pulled dorsal cell membrane (by FluidFM) and the ventral disc, the reaction time of the trophozoite might be too long to modulate the signal transmission from the mechanically stressed dorsal cell end towards the release of adherent bodily structures at the other, ventral end. Interestingly, we observed an increasing number of concave force curves at high pulling speeds, indicating a non-linear and weaker force response to the pulling of the FluidFM micropipette. Further experiments will be needed in the future to investigate, whether this phenomenon correlates with adaptations in cell elasticity adaptions of the trophozoites at higher pulling speeds, or with the viscosity of the trophozoites that modulate the diffusion coefficient of macromolecules in the cytoplasm.46
4 Conclusions
Our SCFS-based study is a new approach to describe the microbial detachment characteristics and attachment forces of G duodenalis, a protozoan that has evolved a unique attachment organelle, the ventral disc. Our results are compatible with both, a suction based or a clutching/grasping based attachment mechanism but are incompatible with a significant contribution of cell-surface-ligand receptor interactions that underlay adhesion processes of other eukaryotic cells tested. Surface attachment by a specific microtubule-based organelle, represents a completely new attachment strategy at the single cell level, adding to the multifunctional nature of microtubule structures in eukaryotic cells.5 Material and methods
5.1 Giardia duodenalis, Candida albicans, and oral keratinocytes cultivation and sample preparation
G. duodenalis assemblage AI isolate WBC6 (ATCC 50803) was grown as trophozoites in TYI-S-33 medium, containing adult bovine serum, adapted from a protocol previously described.47 Trophozoites were cultivated in T25 cell culture dishes at 37 °C under anaerobic conditions in a container system containing a rectancular jar (Mitsubishi Gas Chemical Company, Japan) and an indicator sachet (BD GasPak™ EZ, Beckton Dickinson, USA). Trophozoite monolayers showing 80–90% confluency were passaged every 2–3 days by detaching them at 4 °C for 20 minutes, and transferring them in a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 or 1
100 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 into fresh TYI-S-33 medium. Prior to SCFS or SEM experiments, detached cells were seeded 1
1000 into fresh TYI-S-33 medium. Prior to SCFS or SEM experiments, detached cells were seeded 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2500 in fresh TYI-S-33 medium in glass-bottomed cell culture dishes (FD5040, World Precision Instruments, Friedberg, Germany), and were cultivated for 48 hours at 37 °C under anaerobic conditions (as described above).
2500 in fresh TYI-S-33 medium in glass-bottomed cell culture dishes (FD5040, World Precision Instruments, Friedberg, Germany), and were cultivated for 48 hours at 37 °C under anaerobic conditions (as described above).
The oral gum keratinocyte cell line K2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 was cultivated in KGM2 medium (Promocell, Germany) at 5% CO2 and 37 °C, and was regularly passaged at 80–90% cell confluency. After trypsination of cells and resuspension in fresh KGM2 medium, the now round-shaped cells were left to sediment and adhere on FD5040 dishes for 15 (minimum)–60 min (maximum) prior to SCFS experiments.
48 was cultivated in KGM2 medium (Promocell, Germany) at 5% CO2 and 37 °C, and was regularly passaged at 80–90% cell confluency. After trypsination of cells and resuspension in fresh KGM2 medium, the now round-shaped cells were left to sediment and adhere on FD5040 dishes for 15 (minimum)–60 min (maximum) prior to SCFS experiments.
The yeast C. albicans (Robin) Berkhout strain DSM 1386 (syn. ATCC 10231) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). The yeast cultivation and preparation protocol was described earlier.20 DSM 1386 was cultivated on solid trypticase soy agar (TSA) plates with 5% sheep blood. For SCFS experiments, yeast-phase cells were grown in yeast extract peptone dextrose (YPD) liquid medium (Becton Dickinson GmbH, Germany) for 19 hours at 150 RPM, 37 °C, and aerobic conditions with a flask to medium ration of 10 to 1. For SCFS experiments, 1 μl of a diluted (OD600 of ∼0.3) yeast liquid culture was pipetted on the surface of PBS-covered FD5040 glass-bottom dishes. Yeast cells were allowed to sediment and adhere for 15–60 min prior to SCFS experiments.
5.2 Single-cell force spectroscopy
SCFS experiments were performed on a Flex-Bio atomic force microscope (Nanosurf GmbH, Liestal, Switzerland) with a FluidFM® module (Cytosurge, Glattbrugg, Switzerland) mounted on a Zeiss Observer Z1 microscope (Carl Zeiss, Jena, Germany). Single adherent G. duodenalis trophozoites were approached with the FluidFM micropipette. After contact establishment (for details see below), micropipette retraction was used to detach the individual trophozoite from the borosilicate glass surface. All experiments were conducted in pre-conditioned anaerobic TYI-S-33 medium for a maximum experiment time of 60 min under microaerophilic conditions at room temperature. In detail, tipless cantilever-type FluidFM micropipettes (Cytosurge AG, Glattbrugg, Switzerland) with a nominal spring constant of 0.3 N m−1 and an aperture of 2 μm were used. Adherent trophozoites were brought into optical focus (500-fold magnification), and the micropipette was positioned in a close z-distance (20 μm) above the cell body of the trophozoite. Negative pressure was set to −5 mbar and the micropipette was moved towards the adherent cell with a speed of 1 μm s−1. A loading force of 5 nN and a surface contact time of 100 ms were applied. After establishing the initial contact, negative pressure was automatically increased to −75 mbar (−150 mbar in pulling experiments to test mechanical properties of the attachment) and was kept until the complete detachment of the trophozoite. The detachment was initialized by cantilever retraction at 1 μm s−1 (additionally 2, 5 and 10 μm s−1 in pulling experiments).Single adherent C. alibcans yeast cells and adherent oral keratinocytes were approached with the FluidFM micropipette with the negative pressure turned off. An approach and pulling speed of 1 μm s−1, a loading force of 5 nN and 10 nN, respectively, and a surface contact time of 100 ms were applied. After establishing the initial contact, negative pressure was increased to −350 mbar and −700 mbar, respectively, before the pulling and detachment of the cell from the surface were initiated.
The retraction part of the resulting force–distance curves were quantified for maximum detachment force (adhesion force), cell detachment length, and localization of adhesion force with SPIP software version 6.6.2 (Image Metrology, Hørsholm, Denmark) and OriginPro 2019b (OriginLab, Northampton, MA, USA).
5.3 Scanning electron microscopy of G. duodenalis trophozoites
G. duodenalis trophozoites were approached with a micropipette and detached (as described above), and treated with a fixative while being immobilized on the micropipette. For the fixation of the trophozoite at the cantilever, the TYI-S-33 medium covering the cantilever was supplemented with a glutaraldehyde/0.1 M – Cacodylat solution to reach a final concentration of 2% glutaraldehyde. Fixation was done for 1 h, while the negative pressure of the FluidFM was maintained at −150 mbar. Subsequently, the negative pressure was turned off, and the micropipette with the immobilized trophozoite was transferred into the dry manufacturer's original micropipette packaging. For scanning electron microscopy (SEM) preparation, following steps were conducted in the liquid reservoir of the original packing: the immobilized trophozoite was incubated in a second fixation step in 4% glutaraldehyde/0.1 M Cacodylat for 1 h, and was then washed three times for 10 min with 1 ml 0.1 M Cacodylat buffer. Dehydration steps for SEM were done with 2 × 5 min in 50% ethanol (EtOH), 1 × 5 min in 70% EtOH, 1 × 5 min in 80% EtOH, 1 × 5 min in 90% EtOH, and 2 × 10 min in 100% EtOH. Sample drying was done by adding 2 × 10 min 1 ml hexamethyldisilazane (HMDS), then removing HMDS, and air-drying overnight in a fume hood. The micropipette containing the fixated trophozoite was glued to a SEM specimen stub, using adhesive tape. Sputtering was carried out with carbon (Bal-tec SCD 030 sputter coater, Leica Microsystems, Vienna, Austria) and gold (Bal-tec SCD 005 sputter coater, Leica Microsystems, Vienna, Austria). Imaging was performed with a Philips/FEI XL30 ESEM FEG microscope (FEI, Eindhoven, Netherlands).5.4 Statistical analysis
Statistical evaluation of data sets was assessed by Mann–Whitney U test (comparison of two groups) or a Kruskal–Wallis test followed by multiple comparison (comparison of three and more groups) in GraphPad Prism 9 (Boston, MA, USA). Identified p-values <0.05 were considered statistically significant. Normal distribution of data was analysed by D'Agostino & Pearson test in GraphPad Prism 9.Author contributions
S. C. D., K. J., C. K., T. A. and P. J. conceptualised the study. S. L. B., M. B., M. H., K. J., C. K., T. A. and P. J. obtained the fundings. N. M., M. B., M. H. and T. A. provided resources. G. G., B. W., J. D., R. L. and P. J. conducted the experiments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the German Research Foundation (DFG) for funding within the framework of Priority Programme (SPP) 2332 (project 1) and within the Collaborative Research Center (SFB) 1027 (project B2 and B3). We further thank Dr Anne Holtsch for valuable support in graphical design, and Karin Hilgert and Norbert Pütz for excellent technical assistance.References
- S. A. House, D. J. Richter, J. K. Pham and S. C. Dawson, PLoS Pathog., 2011, 7, e1002167 CrossRef CAS PubMed.
- S. C. Lenaghan, C. A. Davis, W. R. Henson, Z. Zhang and M. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, E550–E558 CrossRef CAS.
- M. C. Sousa, C. A. Gonçalves, V. A. Bairos and J. Poiares-Da-Silva, Clin. Diagn. Lab. Immunol., 2001, 8, 258–265 CrossRef CAS.
- J. M. van Rijn, J. Eriksson, J. Grüttner, M. Sundbom, D.-L. Webb, P. M. Hellström, S. G. Svärd and M. E. Sellin, mBio, 2021, 13, e0002222 CrossRef PubMed.
- W. R. Hansen, O. Tulyathan, S. C. Dawson, W. Z. Cande and D. A. Fletcher, Eukaryotic Cell, 2006, 5, 781–783 CrossRef CAS PubMed.
- K. D. Hagen, S. G. McInally, N. D. Hilton and S. C. Dawson, Adv. Parasitol., 2020, 107, 25–96 Search PubMed.
- D. J. Woessner and S. C. Dawson, Eukaryotic Cell, 2012, 11, 292–301 CrossRef CAS.
- D. V. Holberton, J. Cell Sci., 1973, 13, 11–41 CrossRef CAS.
- D. V. Holberton, J. Exp. Biol., 1974, 60, 207–221 CrossRef CAS.
- T. J. Picou III, The Hydrodynamic Model of Giardia lamblia Attachment, Georgetown University, 2017 Search PubMed.
- C. Nosala, K. D. Hagen, S. L. Guest, N. A. Hilton, A. Müller, M. Laue, C. Klotz, A. Aebischer and S. C. Dawson, bioRxiv, 2023, 2023.07.04.547600.
- C. Nosala, K. D. Hagen, N. Hilton, T. M. Chase, K. Jones, R. Loudermilk, K. Nguyen and S. C. Dawson, J. Cell Sci., 2020, 133, jcs227355 CrossRef CAS.
- H. G. Elmendorf, S. C. Dawson and J. M. McCaffery, Int. J. Parasitol., 2003, 33, 3–28 CrossRef.
- D. E. Feely, J. V. Schollmeyer and S. L. Erlandsen, Exp. Parasitol., 1982, 53, 145–154 CrossRef CAS.
- D. Magne, L. Favennec, C. Chochillon, A. Gorenflot, D. Meillet, N. Kapel, D. Raichvarg, J. Savel and J. G. Gobert, Parasitol. Res., 1991, 77, 659–662 CrossRef CAS.
- E. Maikranz, C. Spengler, N. Thewes, A. Thewes, F. Nolle, P. Jung, M. Bischoff, L. Santen and K. Jacobs, Nanoscale, 2020, 12, 19267–19275 RSC.
- A. Beaussart, S. El-Kirat-Chatel, R. M. A. Sullan, D. Alsteens, P. Herman, S. Derclaye and Y. F. Dufrêne, Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS PubMed.
- T. J. Dauben, C. Dewald, I. Firkowska-Boden, C. Helbing, H. Peisker, M. Roth, J. Bossert and K. D. Jandt, Colloids Surf., B, 2020, 194, 111177 CrossRef CAS.
- S. El-Kirat-Chatel and Y. F. Dufrêne, Nanoscale Horiz., 2016, 1, 69–74 RSC.
- G. Gunaratnam, J. Dudek, P. Jung, S. L. Becker, K. Jacobs, M. Bischoff and M. Hannig, Microorganisms, 2021, 9, 2213 CrossRef PubMed.
- M. Mathelié-Guinlet, F. Viela, J. Dehullu, S. Filimonava, J. M. Rauceo, P. N. Lipke and Y. F. Dufrêne, Commun. Biol., 2021, 4, 33 CrossRef.
- P. Jung, C. E. Mischo, G. Gunaratnam, C. Spengler, S. L. Becker, B. Hube, K. Jacobs and M. Bischoff, Virulence, 2020, 11, 1453–1465 CrossRef CAS.
- E. Potthoff, O. Guillaume-Gentil, D. Ossola, J. Polesel-Maris, S. LeibundGut-Landmann, T. Zambelli and J. A. Vorholt, PLoS One, 2012, 7, e52712 CrossRef CAS.
- C. Spengler, F. Nolle, J. Mischo, T. Faidt, S. Grandthyll, N. Thewes, M. Koch, F. Müller, M. Bischoff, M. A. Klatt and K. Jacobs, Nanoscale, 2019, 11, 19713–19722 RSC.
- N. Thewes, P. Loskill, C. Spengler, S. Hümbert, M. Bischoff and K. Jacobs, Eur. Phys. J. E, 2015, 38, 140 CrossRef.
- M. Benoit, D. Gabriel, G. Gerisch and H. E. Gaub, Nat. Cell Biol., 2000, 2, 313–317 CrossRef CAS PubMed.
- P.-H. Puech, K. Poole, D. Knebel and D. J. Muller, Ultramicroscopy, 2006, 106, 637–644 CrossRef CAS.
- A. Meister, M. Gabi, P. Behr, P. Studer, J. Vörös, P. Niedermann, J. Bitterli, J. Polesel-Maris, M. Liley, H. Heinzelmann and T. Zambelli, Nano Lett., 2009, 9, 2501–2507 CrossRef CAS.
- J. Dehullu, C. Valotteau, P. Herman-Bausier, M. Garcia-Sherman, M. Mittelviefhaus, J. A. Vorholt, P. N. Lipke and Y. F. Dufrêne, Nano Lett., 2019, 19, 3846–3853 CrossRef CAS PubMed.
- A. M. Dranginis, J. M. Rauceo, J. E. Coronado and P. N. Lipke, Microbiol. Mol. Biol. Rev., 2007, 71, 282–294 CrossRef CAS.
- A. L. J. Olsson, H. C. van der Mei, H. J. Busscher and P. K. Sharma, Langmuir, 2010, 26, 11113–11117 CrossRef CAS PubMed.
- M. Sun, J. S. Graham, B. Hegedüs, F. Marga, Y. Zhang, G. Forgacs and M. Grandbois, Biophys. J., 2005, 89, 4320–4329 CrossRef CAS PubMed.
- Y. F. Dufrêne, A. Viljoen, J. Mignolet and M. Mathelié-Guinlet, Cell. Microbiol., 2021, 23, e13324 CrossRef PubMed.
- P. W. Doll, K. Doll, A. Winkel, R. Thelen, R. Ahrens, M. Stiesch and A. E. Guber, ACS Omega, 2022, 7, 17620–17631 CrossRef CAS PubMed.
- V. Ho, P. Herman-Bausier, C. Shaw, K. A. Conrad, M. C. Garcia-Sherman, J. Draghi, Y. F. Dufrene, P. N. Lipke and J. M. Rauceo, mBio, 2019, 10, e01766–e01719, DOI:10.1128/mbio.01766-19.
- N. Thewes, A. Thewes, P. Loskill, H. Peisker, M. Bischoff, M. Herrmann, L. Santen and K. Jacobs, Soft Matter, 2015, 11, 8913–8919 RSC.
- A. P. R. Gadelha, M. Benchimol and W. de Souza, Histochem. Cell Biol., 2022, 157, 251–265 CrossRef CAS PubMed.
- C. Nosala and S. C. Dawson, BioRxiv Prepr. Serv. Biol., 2017, 213421.
- J. Helenius, C.-P. Heisenberg, H. E. Gaub and D. J. Muller, J. Cell Sci., 2008, 121, 1785–1791 CrossRef CAS PubMed.
- R. Harjumäki, X. Zhang, R. W. N. Nugroho, M. Farooq, Y.-R. Lou, M. Yliperttula, J. J. Valle-Delgado and M. Österberg, ACS Appl. Bio Mater., 2020, 3, 1406–1417 CrossRef PubMed.
- M. Cavalheiro, D. Pereira, C. Formosa-Dague, C. Leitão, P. Pais, E. Ndlovu, R. Viana, A. I. Pimenta, R. Santos, A. Takahashi-Nakaguchi, M. Okamoto, M. Ola, H. Chibana, A. M. Fialho, G. Butler, E. Dague and M. C. Teixeira, Commun. Biol., 2021, 4, 886 CrossRef CAS PubMed.
- G. Gunaratnam, C. Spengler, S. Trautmann, P. Jung, J. Mischo, B. Wieland, C. Metz, S. L. Becker, M. Hannig, K. Jacobs and M. Bischoff, Sci. Rep., 2020, 10, 20992 CrossRef CAS PubMed.
- Y. Wang, L. Lu, G. Zheng and X. Zhang, ACS Nano, 2020, 14, 9861–9872 CrossRef CAS PubMed.
- K. D. Hagen, C. Nosala, N. A. Hilton, A. Müller, D. Holthaus, M. Laue, C. Klotz, A. Aebisher and S. C. Dawson, bioRxiv, 2023, 2023.07.02.547441.
- C. Nosala, K. D. Hagen and S. C. Dawson, Trends Cell Biol., 2018, 28, 99–112 CrossRef CAS.
- M. Kumar, M. S. Mommer and V. Sourjik, Biophys. J., 2010, 98, 552–559 CrossRef CAS.
- D. B. Keister, Trans. R. Soc. Trop. Med. Hyg., 1983, 77, 487–488 CrossRef CAS.
- A. Aljohmani, H. Heinze, F. G. Gharzia, B. Reda, A. M. M. Abdrabou, S. Becker, M. Bischoff, M. Hannig and D. Yildiz, BioRxiv Prepr. Serv. Biol., 2023, 2023.07.21.550016 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Viability and reattachment of G. duodenalis trophozoites following FluidFM measurements, analysis of different pulling orientations of trophozoites and adhesion force, correlation of cell geometry and adhesion force, analysis of de-adhesion work. See DOI: https://doi.org/10.1039/d4nr00122b |
| This journal is © The Royal Society of Chemistry 2024 |