Manipulating macrophage polarization with nanoparticles to control metastatic behavior in heterotypic breast cancer micro-tissues via exosome signaling†
Mustafa
Sungu
a,
Melis
Isik
 a,
Ülkü
Güler
b,
Cemil Can
Eylem
c,
Hakan
Eskizengin
d,
Emirhan
Nemutlu
a,
Ülkü
Güler
b,
Cemil Can
Eylem
c,
Hakan
Eskizengin
d,
Emirhan
Nemutlu
 c,
Bekir
Salih
b and
Burak
Derkus
c,
Bekir
Salih
b and
Burak
Derkus
 *ae
*ae
aStem Cell Research Lab, Department of Chemistry, Faculty of Science, Ankara University, 06560 Ankara, Turkey. E-mail: bderkus@ankara.edu.tr
bDepartment of Chemistry, Faculty of Science, Hacettepe University, 06800 Ankara, Turkey
cAnalytical Chemistry Division, Faculty of Pharmacy, Hacettepe University, Ankara 06230, Turkey
dDepartment of Biology, Faculty of Science, Ankara University, 06560 Ankara, Turkey
eNeuroscience and Neurotechnology Excellence Joint Application and Research Center (NEUROM), 06560 Ankara, Turkey
First published on 5th December 2023
Abstract
This study aimed to investigate the effects of nanoparticles on macrophage polarization and their subsequent influence on post-tumorigenic behavior. Initially, seven different nanoparticles were applied to macrophages, and Zn–Ni–FeO (100 nm) and palladium nanoparticles (PdNPs, ∼25 nm) were found to induce M1-polarization in macrophages. A co-culture experiment was then conducted to examine the effects of macrophages on MCF-7 breast cancer micro-tissues. The M2-macrophages promoted tumor proliferation, while M1- and PdNPs-induced macrophages showed anti-tumor effects by suppressing cell proliferation. To reveal the mechanisms of effect, exosomes isolated from M1 (M1-Exo), M0 (M0-Exo), M2 (M2-Exo), and PdNPs-induced (PdNPs-Exo) macrophages were applied to the heterotypic tumor micro-tissues including MCF-7, human umbilical vein endothelial cells (HUVECs), and primary human dermal fibroblasts (phDFs). M2-Exo was seen to promote the migration of cancer cells and induce epithelial–mesenchymal transition (EMT), while M1-Exo suppressed these behaviors. PdNPs-Exo was effective in suppressing the aggressive nature of breast cancer cells similar to M1-Exo, moreover, the efficacy of 5-fluorouracil (5-FU) was increased in combination with PdNPs-Exo in both MCF-7 and heterotypic micro-tissues. In conclusion, PdNPs-Exo has potential anti-tumor effects, can be used as a combination therapy to enhance the efficacy of anti-cancer drugs, as well as innovative implants for breast cancer treatment.
1. Introduction
The prevalence of cancer has reached alarming levels, posing a significant threat to public health worldwide. Recent data from the International Agency for Research on Cancer (IARC) reveals that one in five individuals will be diagnosed with cancer during their lifetime. In 2020 alone, an estimated 19.3 million new cancer cases were reported, leading to approximately 10 million cancer-related deaths.1 Among various types of cancer, breast cancer is the most frequently diagnosed case, affecting 2.3 million individuals. Shockingly, the IARC data indicates that around 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 breast cancer cases resulted in fatalities globally. This distressing trend highlights the urgent need for effective strategies to combat breast cancer and reduce its devastating impact on individuals and societies.
000 breast cancer cases resulted in fatalities globally. This distressing trend highlights the urgent need for effective strategies to combat breast cancer and reduce its devastating impact on individuals and societies.
The tumor microenvironment (TME) plays a crucial role in shaping the challenges associated with cancer. This intricate and dynamic environment, where tumors thrive, encompasses a diverse array of cells and non-cellular components. Immune cells, including T and B lymphocytes, dendritic cells, tumor-associated macrophages, and natural killer cells, coexist with cancer stem cells, cancer-associated fibroblasts, and vascular endothelial cells that drive angiogenesis. Moreover, non-cellular elements such as extracellular matrix (ECM), growth factors, cytokines, chemokines, and extracellular vesicles contribute to the growth-promoting milieu within the TME.2 Conventional two-dimensional (2D) culture systems, commonly employed in in vitro studies, fall short in capturing the intricacies of the TME as they fail to replicate critical aspects such as cell–matrix interactions, cell–cell interactions, and phagocytosis, which are vital for cellular function. In contrast, three-dimensional (3D) culture systems, offer a more representative model that closely mimics the complex in vivo conditions by fostering cell–cell interactions and simulating the microenvironment.3
Macrophages, abundant in TME, are highly effective at killing tumor cells in cases where tumor antigens are scarce. These versatile cells, known for their plasticity, excel in various roles, including defending against pathogens, regulating tissue homeostasis, and aiding in wound healing.4 Macrophages can adopt two distinct phenotypes: the classically activated pro-inflammatory (M1) and the alternatively activated anti-inflammatory (M2), depending on the cues provided by their microenvironment. The M1 macrophages release signal transducer and activator of transcription-1 (STAT-1), STAT3, hypoxia-inducible factor 1-alpha (HIF-1α), activation protein-1 (AP-1), and NF-kappa B (NF-κB). Additionally, predictive cytokines for the M1 macrophage phenotype include interleukin-1beta (IL-1β), IL-6, nitric oxide synthase 2 (NOS2), and tumor necrosis factor (TNF-α). On the other hand, the M2 macrophages express high levels of surface adhesion proteins (CD163, CD206, MMPs, Arg-1, IL-10) and synthesize growth factors like transforming growth factor-beta 1 (TGF-β1), platelet-derived growth factor (PDGF), and vascular endothelial growth factor (VEGF). Furthermore, the M2 macrophages secrete chemokines (CXCL1, CXCL2, and CXCL5) that promote angiogenesis.5–7
Certain biomaterials currently in development have the potential to transform M2 macrophages, which promote tumor growth and spread, into M1 macrophages that can effectively defend against cancer cells. These remarkable biomaterials are known as immunomodulatory biomaterials. In a study by Movva et al., they introduced a specific immunomodulatory biomaterial called the backpack, which securely adheres to the surface of macrophages. The backpack continuously releases IFN-γ, effectively polarizing macrophages towards the M1 phenotype.8 Nanoparticles also show great promise in regulating macrophage phenotypes. When nanoparticles enter the body or cell culture, macrophages recognize them as foreign bodies and internalize them through endocytosis or phagocytosis, facilitated by surface opsonisation.9 The response of macrophages to nanoparticles can be influenced by factors such as dosage, size, composition, and surface properties of the nanoparticles. For example, metallic nanoparticles like titanium nanoparticles (TiNPs), zinc oxide nanoparticles (ZnONPs), super-paramagnetic iron oxide nanoparticles (SPIONs), and palladium nanoparticles (PdNPs) can polarize macrophages towards the M1 phenotype through the mitogen-activated protein kinases (MAPK) or toll-like receptors (TLR) signaling pathways.10–15 On the other hand, liposomal nanoparticles induce macrophages to adopt the M2 phenotype.16 Thus, in this work, we focused on mostly metallic nanoparticles to assess their M1-polarizing potentials, that will enable to develop pro-inflammatory and anti-tumorigenic platforms to manage post-tumorigenic process.
Extracellular vesicles (EVs), particularly exosomes, play a crucial role in cell–cell communication within the TME. These exosomes, ranging from 30 to 150 nm in diameter, are released by various cell types, including stem cells, endothelial cells, macrophages, and cancer cells.17 They serve as carriers for several molecules, such as proteins, lipids, DNA, mRNA, and microRNAs.18 These bioactive molecules are transported to macrophages, regulating multiple signaling pathways and ultimately inducing the polarization of macrophages towards the M2 phenotype through the Phosphatidylinositol-3-kinase (PI3K) and AKT pathways.19,20 Similarly, exosomes derived from M2 macrophages have been shown to enhance the invasive, migratory, and angiogenic properties of cancer cells. For instance, Song et al. investigated the effects of exosomes derived from M2 macrophages on breast cancer. Their findings revealed that these exosomes promoted invasion by activating the miR-223/Mef2c/β-catenin pathway.21 Wu et al. reported that exosomes derived from M2 macrophages increased migration in Hepatocellular carcinoma (HCC) cells by upregulating MMP9 expression.22 Moreover, studies have demonstrated that M2 macrophage exosomes contribute to cell growth, angiogenesis, and chemoresistance within the TME.23–25 On the other hand, there are various studies demonstrating the pro-inflammatory and anti-tumorigenic functions of M1 macrophage exosomes. With a smooth approach, it was showed that pro-inflammatory environment created by M1 macrophage-derived exosomes could potentiate the anti-tumour effects of chemotherapeutics.26 Mechanistically, M1 exosomes were found to suppress proliferation, migration and invasion but induce apoptosis of cancer cells by upregulating TLR5/NF-κB signaling pathway via their long non-coding RNA content.27 Ultimatey, exosomes obtained from M1 macrophages are capable of repolarizing M2 macrophages into M1 phenotype, thereby, they inhibit the development of endometriosis.28
However, studies on how to manipulate these effects of macrophages and how to harness the potential of macrophages or their exosomes in therapies are still in the early stages. Developing therapies to overcome the M1-suppressive function of M2 macrophage exosomes by immunomodulatory nanoparticle-based strategies will contribute to the cancer immuno therapies in the future.
This study aimed to investigate the potentials of nanoparticles-induced macrophage exosomes in cancer immunotherapy. To this end, we first assessed the effects of various nanoparticles, such as zinc–nickel–iron (Zn–Ni–FeO), copper(II) oxide (Cu(II)O), palladium (Pd), platinum (Pt), gold (Au), graphene oxide (GO), and CdSeS/Zn quantum dots (QDs), on macrophage polarization. Based on the promising results observed with PdNPs in promoting pro-inflammatory (M1) macrophage polarization, the impact of PdNPs-induced macrophage cells on the tumorigenic properties of breast cancer cells (MCF-7) was studied using a co-culture system. Additionally, the anti-tumorigenic functions of PdNPs-induced macrophages exosomes (PdNPs-Exo) on MCF-7 tumor micro-tissues, as well as multicellular micro-tissues consisting of MCF-7, human umbilical vein endothelial cells (HUVECs), and primary human dermal fibroblasts (phDFs) were examined in comparison with exosomes derived from M0 macrophages (M0-Exo), M1 macrophages (M1-Exo), and M2 macrophages (M2-Exo). Lastly, the effect of M0-Exo, M1-Exo, and M2-Exo on the resistance of MCF-7 micro-tissues and heterotypic micro-tissues to 5-fluorouracyl (5-FU) was also investigated.
2. Experimental section
2.1. Cell culture
Raw 267.4 and HUVEC cells (kindly provided by Prof. Orhan Adalı, Middle East Technical University, Turkey) were cultured in Dulbecco's modified Eagle's medium-High Glucose (DMEM-HG, Thermo Fisher, USA) supplemented with fetal bovine serum (FBS, 10% v/v, Biological Industries, Germany) and penicillin–streptomycin (P/S, 1% v/v, Thermo Fisher, USA). MCF-7 cells (kindly provided by Dr Ahmet Acar, Middle East Technical University, Turkey) were cultured in DMEM-HG supplemented with FBS (20% v/v) and P/S (1% v/v). Primary human dermal fibroblasts (kindly provided by Prof. Orhan Adalı, Middle East Technical University, Turkey) were cultured in DMEM-F12 (Thermo Fisher, USA) supplemented with FBS (10% v/v) and P/S (1% v/v) in a CO2 incubator (Panasonic, Japan) at 37 °C and 5% CO2 conditions. The media were refreshed every 3 days until the cells reached the confluence.2.2. Polarization of macrophages
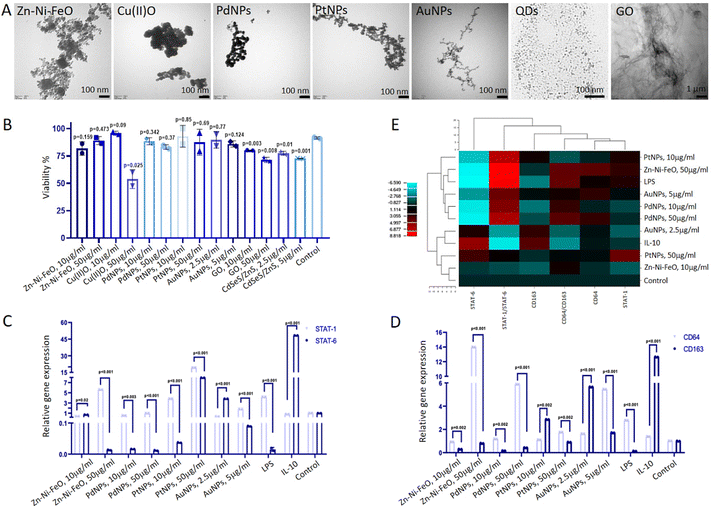
Raw 264.7 cells (3.5 × 104 cells per well) were seeded in a 24-well plate. Following 1-day incubation to ensure cell adhesion, nanoparticles namely Zn–Ni–FeO (100 nm, Sigma-Aldrich, USA), Cu(II)O (50 nm, Sigma-Aldrich, USA), Pd (25 nm, Sigma-Aldrich, USA), Pt (10 nm, Sigma-Aldrich, USA), Au (10 nm, Sigma-Aldrich, USA), GO (Sigma-Aldrich, USA), CdSeS/ZnS Quantum Dots (6 nm, Sigma-Aldrich, USA) introduced to the cells in two different concentrations. The nanoparticles, except for AuNPs and CdSeS/ZnS, were used in 10 μg ml−1 (low) and 50 μg ml−1 (high) concentrations. AuNPs and CdSeS/ZnS were used in 2,5 μg ml−1 (low) and 5 μg ml−1 (high). As positive controls, lipopolysaccharide (LPS form E. coli, Sigma Aldrich L3129, 200 ng ml−1) and IFN-γ (20 ng ml−1) were used to induce M1 polarization, while IL-10 (20 ng ml−1) was used to induce M2 polarization.2.3. Cell proliferation and viability
XTT test was applied to examine the cytotoxic effects of nanoparticles at two different concentrations. For this purpose, Raw 264.7 cells were seeded in a 96-well plate (1 × 104 cells per well) and nanoparticles were loaded to the cells at low and high doses. After 2-days of induction, media were changed to fresh medium (100 μl). Then, cells treated with XTT reagent (50 μL) and incubated for 2 hours. Absorbance values were recorded at 490 nm with a microplate spectrophotometer (Multiskan Sky, Thermo Fisher, USA). For a live/dead assay, Calcein-AM (4 μM, Molecular Probes, Thermo Fisher, UK)/ethidium homodimer-1 (EthD-1, 2 μM, Molecular Probes, Thermo Fisher, UK) were added to wells and the cells were observed under a fluorescent microscope (Leica DMIL, Germany) at 488 nm (green) and 527 nm (red) wavelengths.2.4. Monitoring of nanoparticle uptake by macrophages
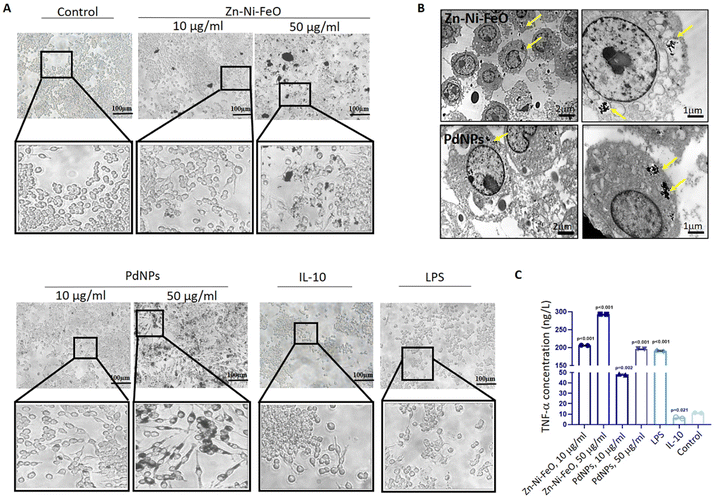
To validate the uptake of nanoparticles by macrophages, sections of cells loaded with nanoparticles were examined using a TEM (JEOL JEM 100CX II, Tokyo, Japan). Raw 264.7 cells were cultured in a T75 flask until they reached confluence. Subsequently, the cells were transferred to a centrifuge tube and centrifuged at 1200 rpm for 5 minutes. Following centrifugation, the supernatant was carefully removed. The cell pellet was then fixed in glutaraldehyde (4%) for 24 hours. After fixation, the pellet was washed with PBS and subsequently stained with uranyl acetate, prepared in 70% ethanol, for 1 hour. Following the staining process, the samples were sequentially washed with 80% ethanol for 10 minutes, 90% ethanol for 10 minutes, 96% ethanol for 10 minutes, and 100% ethanol for 10 minutes (repeated twice). Subsequently, the samples were treated with propylene oxide for another 30 minutes. To prepare the samples for embedding, they were immersed in a mixture of 25% Araldite and 75% propylene oxide for 3 hours, followed by a mixture of 50% Araldite and 50% propylene oxide for 3 hours. The samples were then transferred to a solution containing 75% Araldite and 25% propylene oxide, where they were kept overnight. Finally, the samples were left in pure Araldite for 6 hours. After the embedding process, the samples were placed in an oven and incubated at 45 °C for 24 hours, followed by an additional incubation at 60 °C for 24 hours. Ultrathin sections, ranging from 30 to 60 nm, were meticulously cut using a Reichert OM-U3 ultramicrotome (Austria). These sections were subsequently stained with 1% uranyl acetate for 5 minutes. Finally, the samples were examined using TEM.2.5. Gene expression analysis of nanoparticle-induced macrophage polarization
The impact of nanoparticles on the polarization of macrophages was investigated using RT-qPCR. Specifically, the expressions of M1 marker genes (CD64, STAT-1) and M2 marker genes (CD163, STAT-6) were examined in cells subjected to induction with various nanoparticles. After a 48-hour induction period, total RNA was isolated using a kit (GeneDireX, USA) and subsequently converted into cDNA using a kit from Bio-Rad (USA). RT-qPCR analysis was performed on a thermal cycler (Bio-Rad CFX96, USA). The specific primer sequences utilized for the amplification of target genes are provided in the ESI† and were obtained from Oligomer Biotechnology Jsc. (Turkey).2.6. Characterization of M1 macrophages at protein level
In light of the gene expression data obtained from the screening of different nanoparticles, the impact of Zn–Ni–FeO and Pd nanoparticles at two different doses (10 μg ml−1 and 50 μg ml−1) on TNF-α cytokine levels in macrophages – an indicator of M1 polarization – was investigated using the ELISA method. After a 48-hour induction period, the supernatant was collected for cytokine level measurement, which was performed utilizing the TNF-α ELISA Kit from Bioassay Technology Laboratory (China) adhering to the user manual.2.7. Generation of tumor micro-tissues and establishment of co-culture system
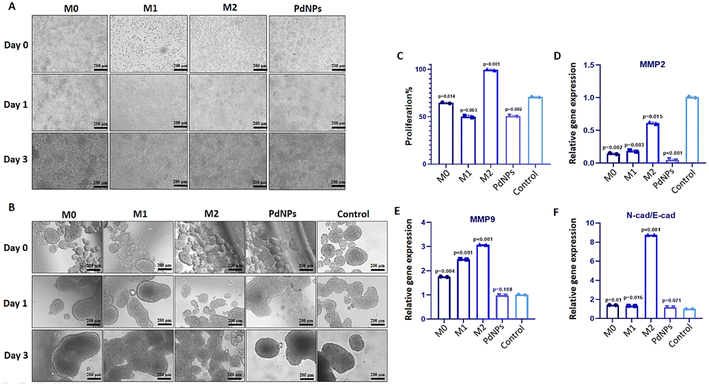
To investigate the potential influence of polarized macrophages on the post-tumorigenic conditions of breast tumor micro-tissues through paracrine mechanisms, a transwell system with two compartments separated by a 0.4 μm membrane was employed. This system facilitated molecular diffusion and vesicle trafficking. MCF-7 cells (3 × 105 cells per well) were seeded in a 6-well ultra-low attachment plate (ULAP) and allowed to incubate for 48 hours to form micro-tissues ranging from 400 to 600 μm in size. Simultaneously, raw 264.7 cells were seeded into inserts (3 × 104 cells per insert) and incubated for one day to promote cell adhesion. Subsequently, the macrophages were treated with LPS/IFN- γ for M1 induction, IL-10 for M2 induction, and PdNPs for M1-like induction for a period of 48 hours. MCF-7 micro-tissues (n = 100) cultivated in 6-ULAPs were then transferred to the bottom chamber of the transwell system, which was coated with 1% (w/v) agar to prevent micro-tissue adhesion, while the macrophages were retained in the top chamber. The co-culture was maintained for up to 72 hours at 37 °C, 5% CO2, and 95% humidity. Tumor micro-tissues were subsequently retrieved from the bottom chamber and prepared for downstream analyses. Each group consisted of 100 micro-tissues, and three biological replicates were used for each group. The number and size of micro-tissues were periodically measured using Image J software (version 1.54c).Following a 3-day co-culture of polarized macrophages (M0, M1, M2, and PdNPs-induced) with tumor micro-tissues, the post-tumorigenic response of the micro-tissues to the polarized macrophages was investigated by examining gene expression levels using RT-qPCR. Specifically, genes associated with metastasis (MMP2 and MMP9) and epithelial–mesenchymal transition (E-Cad and N-Cad) were analyzed. RNA isolation, cDNA synthesis, and RT-qPCR were conducted as previously described. The primary sequences used are provided in the ESI.† Each quantitative experiment was conducted in triplicate.
2.8. Exosome isolation and characterization
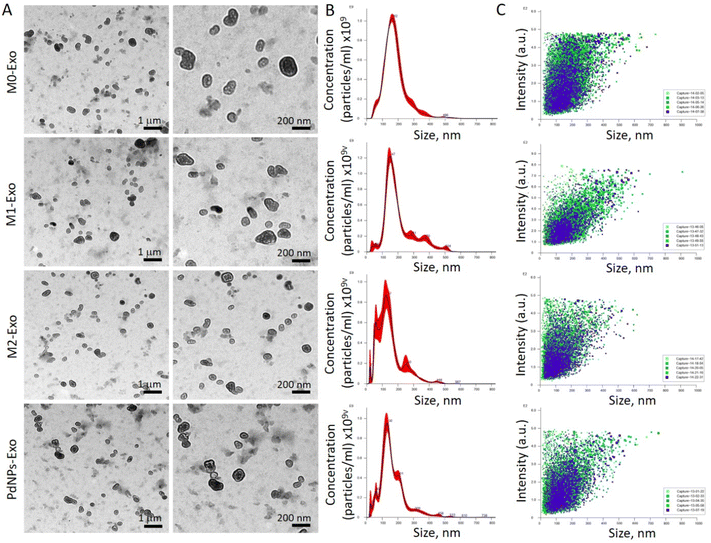
Exosomes were isolated from polarized (M1, M2, PdNPs-induced) and non-polarized macrophages (M0), using a previously described protocol, which has been successfully applied in various studies.29–31 Initially, serum-free conditioned media (CM) were collected from different macrophage groups: (i) unpolarized (M0), (ii) M1 induced by LPS (200 ng ml−1) and IFN-γ (20 ng ml−1), (iii) M2 induced by IL-10 (20 ng ml−1), and (iv) M1-like induced by PdNPs (50 μg ml−1). The CM was then subjected to a series of centrifugation steps at 300g (5 min), 3000g (15 min), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g (15 min) at 4 °C. Subsequently, the CM was filtered through a 0.22 μm syringe filter and further ultracentrifuged (Himac CP100WX, Hitachi, USA) at 120
000g (15 min) at 4 °C. Subsequently, the CM was filtered through a 0.22 μm syringe filter and further ultracentrifuged (Himac CP100WX, Hitachi, USA) at 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 minutes at 4 °C to isolate the exosome pellets. The isolated exosome pellets were dissolved in PBS and characterized using various methods. The protein content was measured using a bicinchoninic acid (BCA) test kit (Thermo Fisher Scientific, USA), the morphology and size were examined using TEM, and the size distribution was confirmed using NTA.
000g for 70 minutes at 4 °C to isolate the exosome pellets. The isolated exosome pellets were dissolved in PBS and characterized using various methods. The protein content was measured using a bicinchoninic acid (BCA) test kit (Thermo Fisher Scientific, USA), the morphology and size were examined using TEM, and the size distribution was confirmed using NTA.
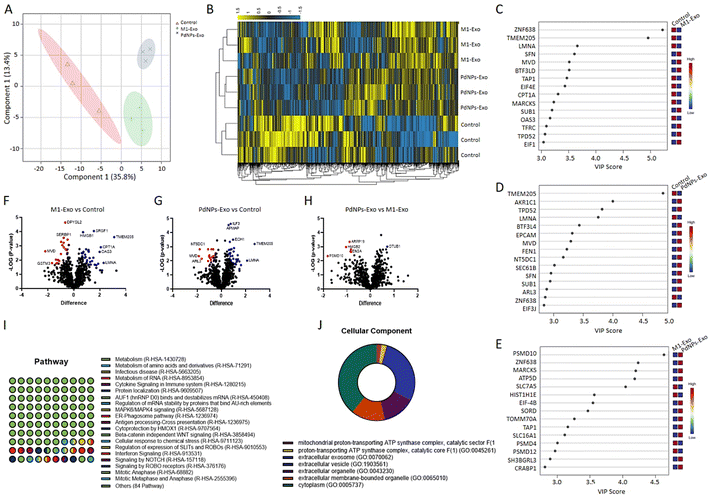
2.9. Proteomic analysis of exosomes using nano-LC-MS
Proteome analysis was conducted as we previously reported.32 The output data from the MS analysis were processed using MaxQuant (ver. 2.1.4.0). Proteins were identified using the UniProt Human Database in the presence of the potential contaminants (Taxonomy ID: 9606).33 Volcano plots were generated in Perseus software and visualized in GraphPad Prism (ver. 9.0) to identify the significant proteins. PLS-DA, heatmap, and VIP graphs were plotted for data interpretation. GO analysis was performed to reveal the significant cellular components, and pathways in the Panther platform using a statistical overrepresentation test.2.10. In vitro wound healing assay
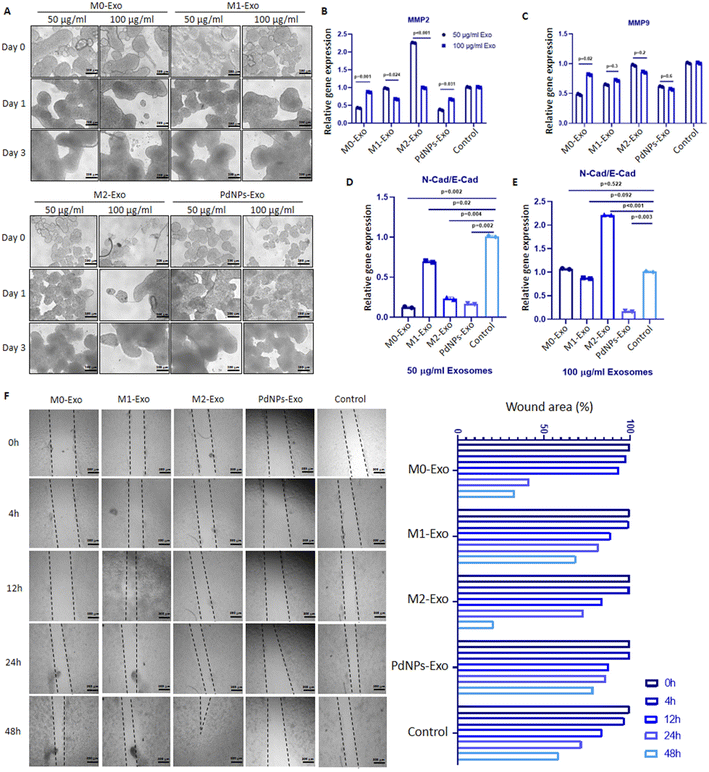
In order to investigate the impact of exosomes derived from M0, M1, M2, and PdNPs-induced macrophages on the invasion of MCF-7 cells, a wound-healing assay was conducted. MCF-7 cells (3 × 105) were seeded in 24-well plates (n = 2) and incubated under optimal conditions of 37 °C and 5% CO2 until they reached complete confluence. Subsequently, the media were removed, gentle scratches were made in the center of each well using a pipette tip, and fresh media with M0-Exo, M1-Exo, M2-Exo, or PdNPs-Exo at 100 μg ml−1 were added to the wells. The changes in wound sizes over time were calculated from the acquired inverted phase/contrast images using the Image J software (version 1.54c).2.11. Testing exosome-mediated post-tumorigenic activity of macrophages
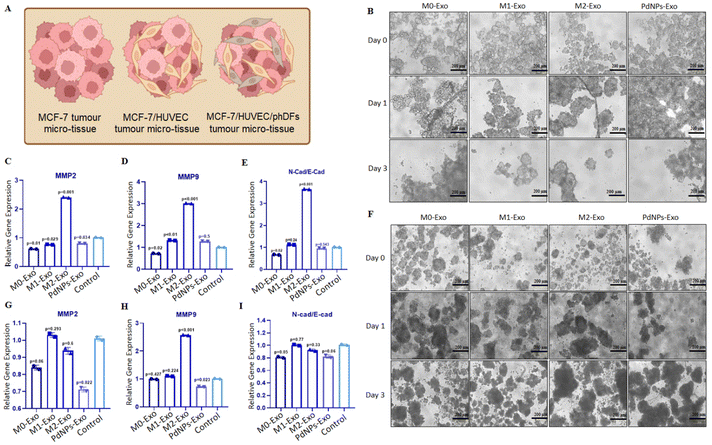
The impact of exosome treatment on gene expression changes in MCF-7 micro-tissues and heterotypic micro-tissues was examined by analyzing markers associated with metastasis (MMP2 and MMP9) and EMT (E-Cad and N-Cad). For this purpose, micro-tissues composed of MCF-7 cells(1.5 × 105 cells)/HUVECs(1.5 × 105 cells) and MCF-7 cells(1.5 × 105 cells)/HUVECs(1.5 × 105 cells)/phDFs(5 × 104 cells) were constructed in 6-ULAP. Subsequently, the micro-tissues were transferred to 24-well plates coated with 1% agar (100 micro-tissues per well), and incubated with medium supplemented with M0-Exo, M1-Exo, M2-Exo, or PdNPs-Exo (100 μg ml−1). Following a 72-hour induction period, changes in the expression levels of metastatic and EMT markers were investigated using RT-qPCR.2.12. Drug testing study
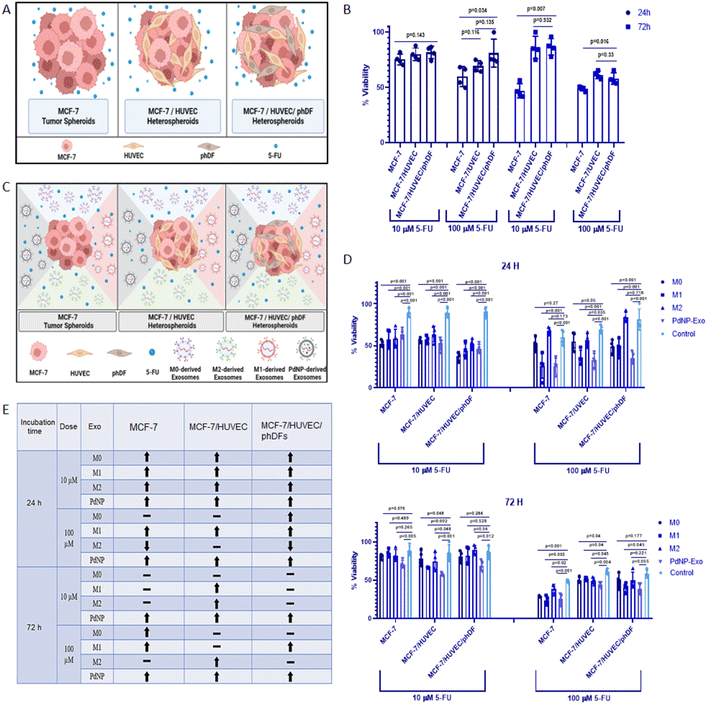
The effect of exosomes on drug resistance was assessed by co-loading exosomes with 5-FU to the micro-tissues. MCF-7 micro-tissues and heterotypic micro-tissues were generated by seeding the cells into 96-ULAP. After a 24-hour culture period, exosomes (100 μg ml−1) were applied to each well. Subsequently, the micro-tissues were treated with either a low dose (10 μg ml−1) or a high dose (100 μg ml−1) of 5-FU. Following a 24-hour or 72-hour exposure to 5-FU, the cell proliferation assay was performed on the micro-tissues.3. Results
3.1. Rationale of study
The aim of this study is to investigate the immunostimulatory properties of nanoparticles and develop an approach that allows for the induction of M1 polarization process with an external induction, such as LPS. Nanoparticles have the ability to polarize macrophages into M1 and M2 phenotypes.34 In this study, the M1-inducing abilities of a range of inorganic (Zn–Ni–FeO, Cu(II)O, Pd, Pt, Au, and CdSeS/ZnS QDs) and organic (graphene oxide, GO) nanoparticles were tested. The hypothesis of the study is based on the possibility that the anti-tumorigenic properties of M1 macrophages may be mediated by exosomes. Therefore, it is expected that exosomes collected from macrophages induced by M1-inductive nanoparticles will also exhibit this property. For this purpose, we selected two different doses of nanoparticles, low (10 μg ml−1) and high (50 μg ml−1), falling in the range of doses most commonly used in the literature.35,36 CdSeS/ZnS and AuNPs, on the other hand, were used at lower concentrations (low = 2.5 μg ml−1, high = 5 μg ml−1) due to their very small sizes and potential genotoxicity.35 The ultimate goal of the study is to stimulate the secretion of M1 exosomes through the manipulation of macrophages and utilize them as anti-cancer agents. This approach can be used as an alternative to LPS for in vitro production of exosomes with anti-cancer properties and can be evaluated as a support for cancer treatments within the scope of cancer immunotherapy after its biocompatibility has been tested.3.2. Screening of nanoparticles as immunomodulators in macrophage polarization
To assess the M1 polarization of macrophages induced by Zn–Ni–FeO and Pd nanoparticles, an ELISA test was employed to determine the secretion of TNF-α, a pro-inflammatory cytokine. The levels of TNF-α cytokine produced were found to increase approximately 20-fold in Zn–Ni–FeO (10 μg ml−1) induced macrophages, 30-fold in Zn–Ni–FeO (50 μg ml−1) induced macrophages, 5-fold in PdNPs (10 μg ml−1) induced macrophages, and 20-fold in PdNPs (50 μg ml−1) induced macrophages, in comparison to non-induced macrophages (Fig. 2C). Notably, the level of TNF-α produced in PdNPs (50 μg ml−1) induced macrophages was found to be similar to that induced by LPS. These results collectively support the successful polarization of macrophages towards the M1 phenotype by both nanoparticles. However, due to the observed precipitation of Zn–Ni–FeO nanoparticles during cell culture (Fig. 2A), which may potentially co-precipitate during subsequent exosome isolation steps, and the slightly more efficient uptake of PdNPs, further experiments were conducted using PdNPs. The potential negative effects of PdNPs on cell viability and proliferation were assessed before further investigations, which showed no toxic effects (Fig. S2†).
3.3. Post-tumorigenic effects of polarized macrophages on MCF-7 breast cancer micro-tissues
Tumor micro-tissues and polarized macrophages (M0, M1, M2, and PdNPs-induced) (Fig. 3A) were co-cultured in transwell systems to investigate the possible anti-tumorigenic or pro-metastatic effects of macrophages on MCF-7 breast cancer micro-tissues. As shown in the Fig. 3A, all types of macrophages exhibited a good proliferative ability. The XTT assay results following the co-culture experiment revealed that M0, M1, and PdNP-induced macrophages exhibited a notable reduction in the viability of MCF-7 tumor micro-tissues. Conversely, macrophages in the M2 polarization state demonstrated a significant increase in the viability of tumor micro-tissues (Fig. 3B and C).The expression of MMP2 was found to be downregulated in tumor micro-tissues co-cultured with M0 macrophages (p = 0.002), M1 macrophages (p = 0.003), PdNPs-induced macrophages (p < 0.001), and even M2 macrophages (p = 0.015) (Fig. 3D). On the other hand, the expression of MMP9, another metastatic gene of clinical importance, was observed to increase in tumor micro-tissues co-cultured with M0 macrophages (by 1.8-fold, p = 0.004), M1 macrophages (by 2.5-fold, p = 0.002), and M2 macrophages (by 3.1-fold, p = 0.001) compared to the control group. However, the activation of MMP9 was not affected by PdNPs-induced macrophages (p = 0.108) (Fig. 3E). To further examine the effect of co-culture on metastasis, the expressions of E-cad and N-cad, which are associated with epithelial–mesenchymal transition (EMT), were investigated. It was observed that M0 macrophages, M1 macrophages, and PdNPs-induced macrophages had no effect on the N-Cad/E-Cad ratio. However, tumor micro-tissues co-cultured with M2 macrophages showed a significant increase in the N-Cad/E-Cad ratio, indicating that M2-macrophages promote metastasis in breast tumor micro-tissues (Fig. 3F).
3.4. Pro-tumorigenic effects of macrophage-derived exosomes
3.5. Anti-tumorigenic effects of PdNPs-Exo on MCF-7/HUVEC and MCF-7/HUVEC/phDFs heterotypic tumor micro-tissues
To evaluate the anti-tumorigenic potential of PdNPs-Exo within a realistic TME, heterotypic micro-tissues composed of MCF-7 cells and HUVECs were generated (Fig. 7A). These micro-tissues were then co-cultured with PdNPs-Exo, M0-Exo, M1-Exo, and M2-Exo at a concentration of 100 μg ml−1 for a duration of 3 days (Fig. 7B). Following the 3-day induction period, the expression levels of MMP2 and MMP9 in the bicellular micro-tissues were examined. Notably, PdNPs-Exo (p = 0.034), M0-Exo (p = 0.01), and M1-Exo (p = 0.029) were found to downregulate MMP2 expression, while M2-Exo demonstrated an increase in MMP2 expression (p = 0.001) (Fig. 7C). In terms of MMP9 expression, there was no significant change observed in PdNPs-Exo treated micro-tissues (p = 0.5). However, M0-Exo treatment led to a downregulation in MMP9 expression (p = 0.02). On the other hand, M1-Exo showed a slight upregulation (1.2-fold, p < 0.01), and M2-Exo induced a significant upregulation (∼3-fold, p = 0.001) in MMP9 expression (Fig. 7D). Furthermore, the ratio of N-cad/E-cad expression was assessed as an indicator of metastatic behavior. There was no significant change observed in the N-Cad/E-Cad ratio with PdNPs-Exo (p = 0.54) and M1-Exo (p = 0.24) treatment. However, M0-Exo (p = 0.02) resulted in a decrease in the N-cad/E-cad ratio, while M2-Exo led to a substantial increase (3.7-fold, p = 0.001). These findings provide further evidence that PdNPs-Exo effectively suppresses the metastatic behavior of cancer cells even in a heterotypic TME (Fig. 7E).To introduce additional complexity to the tumor model, three-cellular heterotypic micro-tissues comprising MCF-7 cells, HUVECs, and phDFs were generated and treated with exosomes for a duration of 3 days (Fig. 7F). Notably, PdNPs-Exo (p = 0.022) and M0-Exo (p = 0.06) exhibited significant downregulation of MMP2 expression, while M1-Exo (p = 0.29) and M2-Exo (p = 0.6) did not show any significant effects on MMP2 expression (Fig. 7G). Conversely, the expression of MMP9 was downregulated by PdNPs-Exo (p = 0.023), upregulated by M2-Exo (p = 0.001), but remained unaffected by treatment with M0-Exo (p = 0.427) and M1-Exo (p = 0.224) (Fig. 7H). To further investigate the potential of exosomes in modulating metastasis, we assessed the N-Cad/E-Cad ratio in the three-cellular micro-tissues. PdNPs-Exo (p = 0.06) and M1-Exo (p = 0.05) were found to decrease the N-Cad/E-Cad ratio, indicating a potential suppression of metastatic behavior. However, the N-Cad/E-Cad ratio was not significantly affected by treatment with M0-Exo (p = 0.77) and M2-Exo (p = 0.33). These results provide further evidence of the modulation of metastasis by PdNPs-Exo in the context of a complex tumor model.
3.6. Effects of exosomes on anti-cancer drug response
Prior to investigating the role of exosomes in the response to anti-cancer drugs, we conducted an assessment of the distinct drug response in unicellular, bicellular, or tricellular tumor micro-tissues (Fig. 8A). The proliferation of cells within these micro-tissues, treated with 5-fluorouracil (5-FU) for 24 and 72 hours, was evaluated using the XTT assay. In the case of 24-hour treatment with 10 μM of 5-FU, we observed weak efficacy on MCF-7 micro-tissues (cell proliferation = 76%), MCF-7/HUVECs micro-tissues (cell proliferation = 80%), and MCF-7/HUVECs/phDFs micro-tissues (cell proliferation = 82%) (Fig. 8B). Increasing the drug dose to 100 μM resulted in a decrease in the proliferation rate of MCF-7 and MCF-7/HUVECs micro-tissues to 58% and 69%, respectively. However, interestingly, the proliferation rate of MCF-7/HUVECs/phDFs heterotypic micro-tissues remained relatively high at 81%. When the drug exposure time was extended to 72 hours, low-dose 5-FU (10 μM) exhibited an impact on MCF-7 micro-tissues (cell proliferation = 47%). However, the drug activity was diminished in the heterotypic micro-tissues of MCF-7/HUVECs (cell proliferation = 86%) and MCF-7/HUVECs/phDFs (cell proliferation = 87%). Moreover, a decrease in proliferation rate was observed in MCF-7 micro-tissues (cell proliferation = 49%), MCF-7/HUVECs micro-tissues (cell proliferation = 61%), and MCF-7/HUVECs/phDFs micro-tissues (cell proliferation = 58%) after treatment with 5-FU (100 μM) for 72 hours (Fig. 8B).In order to investigate the potential of PdNPs-Exo in reducing drug resistance, we co-administered 5-FU at low (10 μM) or high (100 μM) doses, along with exosomes (PdNPs-Exo, M0-Exo, M1-Exo, and M2-Exo) at a concentration of 100 μg ml−1, in unicellular, bicellular, or tricellular tumor micro-tissues (Fig. 8C). Subsequently, the proliferation rates of these micro-tissues were determined by the XTT assay after 24 and 72 hours of treatment (Fig. 8D). The XTT results revealed that low-dose 5-FU treatment significantly inhibited cell proliferation at 24 hours in MCF-7, MCF-7/HUVECs, and MCF-7/HUVECs/phDFs micro-tissues (Fig. 8D). However, at the high dose (100 μM) of 5-FU, it was observed that PdNPs-Exo, M0-Exo, and M1-Exo enhanced the drug efficacy in all types of micro-tissues, thereby confirming their anti-tumorigenic functions. Conversely, M2-Exo demonstrated resistance to the inhibitory effect of 5-FU, as there was no inhibition in cell proliferation in any of the micro-tissues, thus reaffirming the pro-metastatic function of M2-Exo. After 72 hours of 5-FU treatment, the effectiveness of exosomes on the activity of low-dose 5-FU diminished (Fig. 8D). This may be attributed to the suboptimal dose of 5-FU, which allows cells to resume proliferation. When comparing MCF-7 and MCF-7/HUVECs micro-tissues, it was evident that these cells were more sensitive to 5-FU, resulting in a slight inhibition of proliferation. Importantly, PdNPs-Exo exhibited positive effects on drug sensitivity in all three types of micro-tissues, further confirming its anti-tumorigenic function. Lastly, when all micro-tissues were treated with a high dose of 5-FU for 72 hours, cell proliferation in each of them fell below 50%. Once again, micro-tissues treated with PdNPs-Exo demonstrated the most effective behavior in increasing drug sensitivity across all three types of micro-tissues. The findings obtained in the drug testing studies were tabulated in the Fig. 8E. As clearly seen, regardless of the duration and dose, PdNPs-Exo reduces drug resistance in all three types of tumor micro-tissues.
4. Discussion
In the initial phase of the study, we screened a spectrum of nanoparticles to investigate their immunomodulatory potentials. The selection of nanoparticle concentrations and incubation times was based on previous studies in the literature.37–39 Upon analyzing gene expression results and M1/M2 ratios, it was revealed that Zn–Ni–FeO and PdNPs induced M1 polarization in macrophages (Fig. 1C–E). Furthermore, PdNPs-induced macrophages exhibited an M1 macrophage phenotype, consistent with previous literature studies,40,41 supporting the efficient M1 polarization induced by PdNPs (Fig. 2A). Evaluation of TEM images and ELISA results, employed to evaluate the effects of Zn–Ni–FeO and PdNPs-induced macrophages on the secretion of TNF-α, confirmed the successful M1 polarization induced by both types of nanoparticles (Fig. 2B and C). It is worth noting that macrophages that received a higher quantity of nanoparticles (PtNPs) did not exhibit a higher level of M1-polarization compared to the macrophages received a lower quantity of nanoparticles (Zn–Ni–FeO). This indicates that there is no correlation between the quantity of nanoparticle uptake and M1 polarization. On the other hand, Zn–Ni–FeO nanoparticles were observed to form precipitates in the culture medium due to their relatively larger size, rendering them unsuitable for the subsequent co-culture system (Fig. 2A). Consequently, PdNPs at 50 μg ml−1 concentration was chosen for further experimentation.It was observed that PdNPs-induced macrophages suppressed MMP2 gene expression and tumor proliferation16,42 (Fig. 3D). Furthermore, while the expression of MMP9 gene increased in tumor micro-tissues co-cultured with macrophages, except PdNPs-induced macrophages (Fig. 3E). Based on these findings, it can be concluded that PdNPs-induced macrophages effectively suppress metastatic activity in tumor micro-tissues. The function of PdNPs on constraining EMT via inhibition of the TGF-β signaling has been previously reported,43,44 which highly overlaps with our findings.
The reciprocal communication between immune cells and cancer cells is well-established,45 significantly influencing post-tumorigenic mechanisms. This interaction can lead to either anti-tumorigenic or post-metastatic effects. Among the various mediators involved, exosomes have emerged as key players in these processes.46 M1 macrophages, known for their anti-tumorigenic effects and metastasis suppression through diverse mechanisms, offer a promising avenue for cancer intervention. In light of this, our study aimed to elucidate the functional role of exosomes derived from PdNPs-induced macrophages in the pro-tumorigenic phase. To achieve this, we successfully isolated exosomes from different macrophage subsets, including M0, M1, M2, and PdNPs-induced macrophages. These exosomes were thoroughly characterized using advanced techniques such as TEM and NTA (Fig. 4), ensuring their integrity and proper identification. To gain a comprehensive understanding of the biomolecular content of the exosomes, which would shed light on their potential anti-tumorigenic or pro-metastatic mechanisms, a nano-LC-MS-based proteomic analysis was performed. Remarkably, a significant overlap was observed in the proteomic profiles of PdNP-Exo and M1-Exo (Fig. 5). This finding supports our hypothesis that PdNPs-induce M1 polarization, thereby endowing PdNP-Exo with similar functional properties to M1-Exo. Moreover, our analysis revealed meaningful connections between the proteins enriched in M1-Exo and PdNP-Exo and post-tumorigenic processes. For instance, HMGB147 and PDXK,48 found to be enriched in M1-Exo, have been reported to trigger the inflammatory M1 switch in macrophages. Notably, PDXK has also been associated with reduced resistance to chemotherapy,49 a critical aspect addressed in our study. Additionally, TMEM205, a transmembrane protein, has been implicated in drug resistance by modulating the levels of immunosuppressive cells (M2 macrophages and Tregs) and facilitating the infiltration of cytotoxic T cells into the tumor microenvironment.50 Another protein, CPTIA, enriched in M1-Exo, exerts its anti-apoptotic function through interaction with Bcl-2.51 Furthermore, APMAM, enriched in PdNP-Exo, has been linked to phagocytosis and drug uptake.52 These findings provide valuable insights into the potential mechanisms underlying the functions of PdNP-Exo and M1-Exo, shedding light on their involvement in post-tumorigenic processes and potentials as anti-cancer agents. Gene ontology analysis additionally supported these findings and highlighted many pathways; a big part of these pathways were related to immune system, infection, and caner. For example, RUNX3 signaling,53 AXIN1 signaling,54 noncanonical NF-kB signaling,55 and TCR signaling56 have been linked to immune-mediated pro-tumorigenic processes.
To evaluate the anti-tumorigenic potential of PdNPs-Exo compared to M0-Exo, M1-Exo, and M2-Exo, we conducted gene expression analysis. Remarkably, PdNPs-Exo exhibited suppressive effects on key metastatic genes MMP2 and MMP9, as well as on the EMT indicator wN-Cad/E-Cad ratio (Fig. 6B–D). Similarly, M1-Exo led to a downregulation of metastatic gene expression. These findings align with existing literature reports indicating that exosomes derived from M1 macrophages possess the ability to restrain cancer cell migration and invasion.57–59 The inhibition of metastasis by M1 macrophage exosomes can potentially occur through various mechanisms, including the reciprocal transfer of miRNAs between macrophages and cancer cells,57 potentiation of immune checkpoint receptors,58 or the involvement of long non-coding RNAs (lncRNAs) that upregulate the NF-kB pathway.59 Conversely, our observations revealed that M2-Exo promoted EMT in cancer cells. Consistent with these findings, previous studies have reported that M2 macrophage-derived exosomes facilitate the invasion and migration of breast cancer cells.60–63 The pro-metastatic activities of M2-macrophage exosomes can be attributed to their ability to deliver tumour-promoting miRNAs60–62 or proteins such as integrin αVβ3, which enhance cell motility and metastasis.63 Furthermore, it is noteworthy that M2-derived exosomes also provide VEGF, facilitating the establishment of a sufficient blood supply and the provision of energy molecules for the accelerated proliferation of cancer cells. However, it is important to acknowledge that there are still unknown mechanisms that may trigger or promote metastasis, including the activation of proto-oncogenes. Considering these findings collectively, the efficacy of exosomes derived from PdNPs-induced macrophages in suppressing the aggressive behavior of breast cancer represents a promising development. This finding holds potential for their utilization in combination drug therapies aimed at limiting cancer progression and its associated behaviors.
To further validate the anti-tumorigenic effects of PdNPs-Exo in comparison to M0-Exo, M1-Exo, and M2-Exo within the context of more complex multicellular micro-tissues (MCF-7/HUVECs and MCF-7/HUVECs/phDFs), we conducted additional gene expression analyses targeting metastasis and EMT-related genes. In the MCF-7/HUVECs model, PdNPs-Exo exhibited a suppressive effect on metastatic genes MMP2 and MMP9, as well as on the N-Cad/E-Cad ratio, indicating a reduction in EMT (Fig. 7). Conversely, M2-Exo upregulated the expression of these genes, suggesting a pro-metastatic effect. When examining the MCF-7/HUVECs/phDFs micro-tissue model, we observed no significant difference in MMP2 expression among the groups. The level of MMP9 was slightly decreased compared to the MCF-7/HUVECs model, and there was no significant increase in the N-Cad/E-Cad ratio. These findings suggest that the inclusion of fibroblasts in the TME leads to a more aggressive phenotype. These observations are consistent with previous literature demonstrating that the presence of HUVECs in the TME enhances cell invasion,64 while fibroblasts can remodel the microenvironment of breast cancer, promoting a more aggressive phenotype.65 Also, collagens produced by fibroblast can be closely associated with cancer cell invasion, proliferation, and metastasis.66 Overall, these findings indicate that M1 or M1-like exosomes function as anti-tumorigenic agents. However, they may not be sufficient to completely halt metastasis, as evidenced by the persistence of some metastatic gene expression.
To test the responses of the constructed heterotypic micro-tissues to anti-cancer drug 5-FU, we performed a drug testing study on MCF-7, MCF-7/HUVECs, and MCF-7/HUVECs/phDFs. When comparing the viability of monocellular, bicellular, and tricellular micro-tissues, no significant effect on the efficacy of the 10 μM 5-FU drug over a 24-hour period was observed (Fig. 8B). However, it was noted that the viability of MCF-7 micro-tissues significantly decreased (p = 0.034), while it gradually increased in MCF-7/HUVECs and MCF-7/HUVECs/phDFs micro-tissues as the dose of 5-FU increased to 100 μM. These findings are consistent with previous studies that have investigated the proliferative abilities of multicellular tumor micro-tissues in response to anti-cancer drugs.67 As previously mentioned, the presence of endothelial and connective tissue cells can promote cell invasion and proliferation, which may explain the limited effectiveness of drug treatment in complex tumor micro-tissues. Additionally, collagens produced by connective cells within tumor tissues may also influence the response to anti-cancer drugs.68 The observation that viability remained above 50% after 24 hours of drug treatment even at high doses, suggests that the 24 hours of treatment period may be insufficient. However, the higher viability of multicellular micro-tissues compared to monocellular micro-tissues following drug administration can be attributed to the selectivity of the 5-FU anti-cancer drug, specifically its ability to target and eliminate cancer cells.69 When examining the 72 hours of drug testing, the efficacy of the 10 μM 5-FU dose on tumor micro-tissues was found to be much higher than on multicellular micro-tissues. This difference cannot be solely explained by the aforementioned drug selectivity, suggesting that another factor may be at play, such as healthy cells within multicellular heterotypic micro-tissues potentially promoting resistance of cancer cells to the drug. However, a decrease in the viability of multicellular micro-tissues was also observed after 72 hours of treatment with 100 μM 5-FU. This demonstrates that the selective 5-FU anti-cancer drug, which exhibits an effect on cancer micro-tissues at low doses or short exposure times, loses its selectivity with prolonged exposure times and high doses.
Furthermore, an experimental setup was devised to investigate the impact of PdNPs-Exo on the efficacy of 5-FU anti-cancer drugs at two different doses (10 μM and 100 μM) at 24 hours and 72 hours. Remarkably, all tested exosomes, including PdNPs-Exo, M0-Exo, M1-Exo, and M2-Exo, enhanced the effectiveness of the drug on all three types of tumor micro-tissues after 24 hours of treatment with 10 μM 5-FU (Fig. 8D). Specifically, the proliferation rate significantly decreased in MCF-7 tumor micro-tissues following 24 hours of treatment at high drug doses, indicating that PdNPs-Exo and M1-Exo sensitize the tumor cells to the anti-cancer drug. Notably, PdNPs-Exo exhibited a consistent ability to decrease chemoresistance after 24 hours of drug treatment, irrespective of the dose administered. After 72 hours of treatment with 100 μM 5-FU, a substantial decrease in the viability of tumor micro-tissues was observed. PdNPs-Exo, M0-Exo, and M1-Exo significantly increased the efficacy of the drug compared to the control group. Additionally, the proliferation rates of micro-tissues decreased when treated with lower drug doses, suggesting a toxic effect of the 5-FU anti-cancer drug on healthy cells at higher doses and longer treatment periods. Another factor influencing the chemoresistant or sensitizing properties of exosomes is their cargo of biomolecules. For instance, PDXK and APMAP, two proteins enriched in M1-Exo and PdNPs-Exo, have been associated with chemoresistance. The downregulation of PDXK, leading to reduced synthesis of active vitamin B6, confers resistance to chemotherapy with cisplatin, highlighting the significance of PDXK in chemoresistance.70 The upregulation of PDXK observed in our study may provide a potential explanation for the enhanced drug activity mediated by M1-Exo and PdNPs-Exo.
In summary, this study investigated the impact of exosomes derived from macrophages with different phenotypes on the pre- or pro-tumorigenic effects of breast cancer micro-tissues, as well as efficacy of 5-FU anti-cancer drug on the breast cancer micro-tissues. The results demonstrate that the effects vary depending on the complexity of the micro-tissues, as well as the dose and duration of drug treatment. However, our findings consistently indicate that exosomes derived from PdNPs-induced macrophages enhance the efficacy of the anti-cancer drug under all tested conditions. Additionally, it is crucial to consider the dosage of the drug to ensure cancer-specific treatment while minimizing the impact on healthy cells. This aspect was effectively addressed through the utilization of the developed heterotypic micro-tissues in this study. In conclusion, the use of heterotypic tumor micro-tissues provides a more physiologically relevant and predictive model for evaluating the response to chemotherapeutic treatments compared to monocellular micro-tissues.
5. Conclusion
In conclusion, this study elucidated the effects of different nanoparticles on macrophage polarization and their subsequent impact on breast cancer micro-tissues. Through a series of screening steps, it was determined that Cu(II)O, GO, and QDs nanoparticles exhibited cytotoxicity towards macrophages and were therefore excluded from further analysis. On the other hand, Zn–Ni–FeO and Pd nanoparticles were found to induce M1 polarization in macrophages with the highest efficiency, as demonstrated by gene expression analysis and cytokine secretion. Across these nanoparticles, PdNPs were deemed a useful tool to induce M1 polarization of macrophages and create an anti-cancer environment. Exosomes obtained from PdNPs-induced macrophages reflected the anti-tumorigenic potential of M1 macrophages, and downregulated the metastasis-related genes even in heterotypic tumor micro-tissues. Moreover, PdNPs-Exo was shown to down-regulate MMP2 and MMP9, as well as the ratio of N-Cad/E-Cad, while PtNPs-induced macrophages downregulated MMP2, confirming that exosomes have superior functions over their corresponding cells due to their enriched biochemical content. Overall, this study provides valuable insights into the intricate interplay between nanoparticles, macrophages, and cancer cells. The findings highlight the potential of PdNP-induced macrophages and their exosomes as therapeutic agents to modulate tumor behavior and enhance the efficacy of anti-cancer drugs. Further investigations are warranted to fully understand the mechanisms underlying these effects and explore their clinical applications in cancer treatment.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This paper has been produced with the data presented in M. Sungu's Master's Thesis. B. Derkus and B. Salih acknowledge the Turkish Academy of Science (TUBA) for their support. We thank the Fatih Inci and Nedim Haciosmanoglu (Bilkent University, Turkey) for their kind help in NTA analysis. We also thank the Prof. Orhan Adalı and Dr Merve Akkulak (Middle East Technical University, Turkey) for their helps in ultracentrifugation.References
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros and A. Znaor,
et al.
, Int. J. Cancer, 2020, 149(4), 778–789 CrossRef
.
- L. Bejarano, M. J. Jordāo and J. A. Joyce, Cancer Discovery, 2021, 11(4), 933–959 CrossRef CAS PubMed
.
- K. H. Lee and T. H. Kim, Biosensors, 2021, 11(11), 445 CrossRef CAS
.
- T. A. Wynn, A. Chawla and J. W. Pollard, Nature, 2013, 496(7446), 445–455 CrossRef CAS
.
- F. B. Stentz, G. E. Umpierrez, R. Cuervo and A. E. Kitabchi, Diabetes, 2004, 53(8), 2079–2086 CrossRef CAS
.
- Y. Liu and T. Segura, Frontiers. Bioeng. Biotechnol., 2020, 1428 Search PubMed
.
- A. Vishwakarma, N. S. Bhise, M. Evangelista, J. Rouwkema and M. R. Dokmeci,
et al.
, Trends Biotechnol., 2016, 34(6), 470–482 CrossRef CAS PubMed
.
- R. H. Movva, S. R. Yarraguntla and V. K. K. Paravastu, GSC Biol. Pharm. Sci., 2022, 20(1), 126–133 CrossRef CAS
.
- D. Reichel, M. Tripathi and J. M. Perez, Nanotheranostics, 2019, 3(1), 66 CrossRef PubMed
.
- A. M. Scherbart, J. Langer, A. Bushmelev, D. van Berlo, P. Haberzettl and F. J. van Schooten,
et al.
, Part. Fibre Toxicol., 2011, 8(1), 1–19 CrossRef
.
- J. Wang, J. S. Lee, D. Kim and L. Zhu, ACS Appl. Mater. Interfaces, 2017, 9(46), 39971–39984 CrossRef CAS
.
- V. Kodali, M. H. Littke, S. C. Tilton, J. G. Teeguarden, L. Shi and C. W. Frevert,
et al.
, ACS Nano, 2013, 7(8), 6997–7010 CrossRef CAS
.
- S. Gurunathan, M. H. Kang, M. Jeyaraj and J. H. Kim, Int. J. Nanomed., 2021, 16, 2849 CrossRef
.
- X. Ma, Y. Wang, X. L. Liu, H. Ma, G. Li and Y. Li, Nanoscale Horiz., 2019, 4(6), 1450–1459 RSC
.
- R. Rajan, M. K. Sabnani, V. Mavinkurve, H. Shmeeda, H. Mansouri and S. Bonkoungou,
et al.
, J. Controlled Release, 2018, 271, 139–148 CrossRef CAS
.
- X. Chen, Z. Jia, Y. Wen, Y. Huang, X. Yuan and Y. Chen,
et al.
, Acta Biomater., 2022, 151, 537–548 CrossRef CAS
.
- B. Bondhopadhyay, S. Sisodiya, F. A. Alzahrani, M. A. Bakhrebah, A. Chikara and V. Kasherwal, Cancers, 2021, 13(18), 4672 CrossRef CAS PubMed
.
- N. Javeed and D. Mukhopadhyay, J. Biomed. Res., 2017, 31(5), 386 CrossRef CAS
.
- C. Han, C. Zhang, H. Wang and L. Zhao, OncoImmunology, 2021, 10(1), 1887552 CrossRef
.
- E. Vergadi, E. Ieronymaki, K. Lyroni, K. Vaporidi and C. Tsatsanis, J. Immunol., 2017, 198(3), 1006–1014 CrossRef CAS
.
- M. Yang, J. Chen, F. Su, B. Yu, F. Su and L. Lin,
et al.
, Mol. Cancer, 2011, 10(1), 1–13 Search PubMed
.
- J. Wu, W. Gao, Q. Tang, Y. Yu, W. You and Z. Wu,
et al.
, Hepatology, 2021, 73(4), 1365–1380 CrossRef CAS
.
- A. A. El-Arabey, M. Denizli, P. Kanlikilicer, R. Bayraktar, C. Ivan and M. Rashed,
et al.
, Cell. Signalling, 2020, 68, 109539 CrossRef CAS
.
- Z. Yin, T. Ma, B. Huang, L. Lin, Y. Zhou and J. Yan,
et al.
, J. Exp. Clin. Cancer Res., 2019, 38(1), 1–20 CrossRef CAS
.
- Y. Yang, Z. Guo, W. Chen, X. Wang, M. Cao, X. Han and Z. Qiu, Mol. Ther., 2021, 29(3), 1226–1238 CrossRef CAS PubMed
.
- P. Wang, H. Wang, Q. Huang, C. Peng, L. Yao, H. Chen, Z. Qiu, Y. Wu, L. Wang and W. Chen, Theranostics, 2019, 9(6), 1714–1727 CrossRef CAS PubMed
.
- H. Jiang, L. Zhou, N. Shen, X. Ning, D. Wu, K. Jiang and X. Huang, Cell Death Dis., 2022, 183 CrossRef CAS
.
- Q. Li, M. Yuan, X. Jiao, Y. Huang, J. Li, D. Li, M. Ji and G. Wang, Front. Immunol., 2021, 12, 707784 CrossRef CAS
.
- B. Derkus, J. Mater. Sci.: Mater. Med., 2021, 32, 2 CrossRef CAS PubMed
.
- M. Isik, I. Vargel, E. Ozgur, S. B. Cam, P. Korkusuz, E. Emregul, S. Odabas and B. Derkus, Mater. Today Commun., 2023, 36, 106869 CrossRef CAS
.
- B. Derkus, M. Isik and C. C. Eylem,
et al.
, Adv. Biol., 2022, 6(6), 2101317 CrossRef CAS PubMed
.
- D. Barut, E. Manga and B. Derkus,
et al.
, Mol. Omics, 2020, 19, 174–181 RSC
.
- E. Boutet, D. Lieberherr, M. Tognolli, M. Schneider and A. Bairoch, UniPortKB/Swiss-Prot, Methods Mol. Biol., 2007, 406, 89–112 CAS
.
- R. P. Nishanth, R. G. Jyotsna, J. J. Schlager, S. M. Hussain and P. Reddanna, Nanotoxycology, 2011, 5, 4 Search PubMed
.
- J. Ma, R. Liu, X. Wang, Q. Liu and Y. Chen,
et al.
, ACS Nano, 2015, 9(10), 10498–10515 CrossRef CAS PubMed
.
- J. Lovric, H. S. Bazzi and Y. Cuie,
et al.
, J. Mol. Med., 2005, 83, 377–385 CrossRef
.
- J. R. Muvva, V. R. Parasa, M. Lerm, M. Svensson and S. Brighenti, Front. Immunol., 2020, 10, 3157 CrossRef
.
- A. R. B. Ribeiro, E. C. Oliveira Silva and P. M. C. Araujo,
et al.
, Spectrochim. Acta, Part A, 2022, 265, 120328 CrossRef CAS PubMed
.
- F. Gatto, R. Cagliani, T. Catelani, D. Guarnieri, M. Moglianetti, P. P. Pompa and G. Bardi, Nanomaterials, 2017, 7(10), 332 CrossRef PubMed
.
- M. J. Feito, R. Diez-Orejas, M. Cicuéndez, L. Casarrubios, J. M. Rojo and M. T. Portolés, Colloids Surf., B, 2019, 176, 96–105 CrossRef CAS
.
- N. G. Bastús, E. Sánchez-Tilló, S. Pujals, C. Farrera, C. López and E. Giralt,
et al.
, ACS Nano, 2009, 3(6), 1335–1344 CrossRef
.
- C. B. Rodell, S. P. Arlauckas, M. F. Cuccarese, C. S. Garris, R. Li and M. S. Ahmed,
et al.
, Nat. Biomed. Eng., 2018, 2(8), 578–588 CrossRef CAS PubMed
.
- T. Schmitz, M. Jannasch, T. Weigel, C. Moseke, U. Gbureck and J. Groll,
et al.
, Materials, 2020, 13(5), 1142 CrossRef CAS
.
- C. Yunna, H. Mengru, W. Lei and C. Weidong, Eur. J. Pharmacol., 2020, 877, 173090 CrossRef PubMed
.
- S. Wang, J. Li and M. Chen, Natl. Sci. Rev., 2021, 8(7), nwaa226 CrossRef CAS
.
- J. Elsey, J. A. Bubley and El Zhu,
et al.
, Sci. Rep., 2019, 9, 3255 CrossRef PubMed
.
- H. Garner and K. E. deVisser, Nat. Rev. Immunol., 2020, 20, 483–497 CrossRef CAS
.
- W. Yan and S. Jiang, Trends Cancer, 2020, 6(6), 506–517 CrossRef CAS
.
- T. Liu, A. Xiang, T. Peng, A. C. Doran, K. J. Tracey, B. J. Barnes, I. Tabas and M. Son, Proc. Natl. Acad. Sci. U. S. A., 2022, 116(46)), 23254–23263 Search PubMed
.
- Y. Ye, H.-W. Zhang and H.-X. Mei,
et al.
, J. Cell. Mol. Med., 2020, 24(18), 10604–10614 CrossRef CAS PubMed
.
- A. Joseph, J. Pan and J. Michels,
et al.
, OncoImmunology, 2021, 10(1), 1950954 CrossRef
.
- J. Rao, X. Wu and X. Zhou,
et al.
, Front. Genet., 2020, 11, 575776 CrossRef CAS PubMed
.
- L. Gu, R. Surolia and J. L. Larson-Casey,
et al.
, Cell Death Differ., 2022, 29, 118–132 CrossRef CAS
.
- J. Lotem, D. Levanon, V. Negreanu and O. Bauer,
et al.
, Adv. Exp. Med. Biol., 2017, 962, 369–393 CrossRef CAS PubMed
.
- R. Sanson, S. L. Lazzara and D. Cune,
et al.
, Cell. Mol. Gastroenterol. Hepatol., 2023, 15(3), 689–715 CrossRef CAS
.
- S. C. Sun, Nat. Rev. Immunol., 2017, 17, 545–558 CrossRef CAS PubMed
.
- K. Shah, A. Al-Haidari, J. Sun and J. U. Kazi, Signal Transduction Targeted Ther., 2021, 6, 412 CrossRef
.
- M. Moradi-Chaleshtori, S. Shojaei, S. Mohammadi-Yeganeh and S. M. Hashemi, Life Sci., 2021, 282, 119800 CrossRef CAS
.
- Y. W. Choo, M. Kang and H. Y. Kim,
et al.
, ACS Nano, 2018, 12(9), 8977–8993 CrossRef CAS
.
- H. Jiang, L. Zhou and N. Shen,
et al.
, Cell Death Dis., 2022, 13, 183 CrossRef CAS
.
- K. Wei, Z. Ma and F. Yang,
et al.
, Cancer Lett., 2022, 526, 205–216 CrossRef CAS PubMed
.
- J. Lan, L. Sun, F. Xu, L. Liu, F. Hu and D. Song,
et al.
, Cancer Res., 2019, 79(1), 146–158 CrossRef CAS PubMed
.
- X. Mi, R. Xu and S. Hong,
et al.
, Mol. Ther. – Nucleic Acids, 2020, 22, 779–790 CrossRef CAS
.
- L. Huang, F. Wang and X. Wang,
et al.
, MedComm, 2022, 4(1), e191 CrossRef
.
- G. Benton, G. DeGray, H. K. Kleinman, J. George and I. Arnaoutova, PLoS One, 2015, 10(4)), e0123312 CrossRef
.
- H. Dhandapani, A. Siddiqui, S. Karadkar and P. Tayalia,
et al.
, Adv. Healthcare Mater., 2023, 12(21), 2300164 CrossRef CAS
.
- S. Xu, H. Xu and W. Wang,
et al.
, J. Transl. Med., 2019, 17, 309 CrossRef
.
- K. S. Hsu, J. M. Dunleavey and C. Szot,
et al.
, Nat. Commun., 2022, 13, 7078 CrossRef CAS PubMed
.
- D. B. Longley, D. P. Harkin and P. G. Johnston, Nat. Rev. Cancer, 2003, 3(5), 330–338 CrossRef CAS PubMed
.
- L. Galluzzi, I. Vitale, L. Senovilla, K. A. Olaussen, G. Pinna, T. Eisenberg, A. Goubar, I. Martins, J. Michels and G. Kratassiouk, Cell Rep., 2012, 2(2), 257–269 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04980a |
| This journal is © The Royal Society of Chemistry 2024 |