Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Physicochemical surface properties of Chlorella vulgaris: a multiscale assessment, from electrokinetic and proton uptake descriptors to intermolecular adhesion forces†
Nicolas
Lesniewska
*a,
Jérôme F. L.
Duval
 *a,
Céline
Caillet
*a,
Céline
Caillet
 a,
Angelina
Razafitianamaharavo
a,
José P.
Pinheiro
a,
Angelina
Razafitianamaharavo
a,
José P.
Pinheiro
 a,
Isabelle
Bihannic
a,
Isabelle
Bihannic
 a,
Renaud
Gley
a,
Renaud
Gley
 a,
Hélène
Le Cordier
a,
Varun
Vyas‡
a,
Hélène
Le Cordier
a,
Varun
Vyas‡
 a,
Christophe
Pagnout
b,
Bénédicte
Sohm
a,
Christophe
Pagnout
b,
Bénédicte
Sohm
 b and
Audrey
Beaussart§
b and
Audrey
Beaussart§
 *a
*a
aUniversité de Lorraine, CNRS, LIEC, F-54000 Nancy, France. E-mail: nicolas.lesniewska@univ-lorraine.fr; jerome.duval@univ-lorraine.fr; audrey.beaussart@cnrs.fr
bUniversité de Lorraine, CNRS, LIEC, F-57000, Metz, France
First published on 18th January 2024
Abstract
Given the growing scientific and industrial interests in green microalgae, a comprehensive understanding of the forces controlling the colloidal stability of these bioparticles and their interactions with surrounding aqueous microenvironment is required. Accordingly, we addressed here the electrostatic and hydrophobic surface properties of Chlorella vulgaris from the population down to the individual cell levels. We first investigated the organisation of the electrical double layer at microalgae surfaces on the basis of electrophoresis measurements. Interpretation of the results beyond zeta-potential framework underlined the need to account for both the hydrodynamic softness of the algae cells and the heterogeneity of their interface formed with the outer electrolyte solution. We further explored the nature of the structural charge carriers at microalgae interfaces through potentiometric proton titrations. Extraction of the electrostatic descriptors of interest from such data was obscured by cell physiology processes and dependence thereof on prevailing measurement conditions, which includes light, temperature and medium salinity. As an alternative, cell electrostatics was successfully evaluated at the cellular level upon mapping the molecular interactions at stake between (positively and negatively) charged atomic force microscopy tips and algal surface via chemical force microscopy. A thorough comparison between charge-dependent tip-to-algae surface adhesion and hydrophobicity level of microalgae surface evidenced that the contribution of electrostatics to the overall interaction pattern is largest, and that the electrostatic/hydrophobic balance can be largely modulated by pH. Overall, the combination of multiscale physicochemical approaches allowed a drawing of some of the key biosurface properties that govern microalgae cell–cell and cell–surface interactions.
Introduction
Over the past decades, microalgae have been the subject of growing interest both from fundamental and industrial points of view.1 As a representative of oil-accumulating cells, microalgae are considered as a promising sustainable resource for a biofuel production capable of replacing fossil fuel.2,3 Given their high yield in proteins, carbohydrates, vitamins and pigments, microalgae could also serve as a basis for e.g. food supplements and feeds, nutraceuticals, cosmetics or fertilizers.4,5 Nevertheless, the bottleneck in commercial exploitation of microalgae is related to the high energy and operational costs currently associated with their harvesting and the extraction of their high-value by-products.6–9 A challenge relates to the control of the algal cell-wall properties because this charged and rigid structure prevents natural flocculation of the cells and limits the possibility of intracellular content extraction.From an environmental perspective, microalgae are basic elements of the food chain in aquatic media. Due to their short life cycle and ease of cultivation, they are commonly employed as a bioindicator for the evaluation of toxicants impacts (e.g. nanoparticles10 or metals11) and quality of aqueous environments. They have also emerged as a potential substrate in bioremediation processes of wastewater and polluted ecosystems through their capacity to adsorb and accumulate toxic compounds.12–14 In order to enhance their contaminant removal efficiency, various strategies have been proposed, such as cells immobilization or development of algal consortia in biofilm-based cultures (cf. e.g. review ref. 15 and references therein). Here again, the physicochemical surface properties of microalgae come into the picture as they drive the magnitude of algae homo-interactions and that of their hetero-interactions with other cells, abiotic supports or macromolecular pollutants.
In view of the above elements, a mechanistic assessment of the properties of the microalgae cell-wall is a prerequisite for proper biotechnological exploitation of algal resources. However, to date, studies dealing with the physicochemical characterization of microalgae surfaces remain relatively scarce16,17 and, most often, retrieved descriptors of cell surface properties cannot be considered as intrinsic attributes but, instead, adjustable variables that strongly depend on cell growth conditions and environmental factors.18–20 Among them, pH is one of the key parameters that influences microalgae reactivity. As an illustration, strong variations of pH, as met in acid mine drainage, can dramatically affect the bioremediation capacity of microalgae due to unfavourable change in their electrostatic interactions with heavy metals.12 Besides, proper modification of pH condition in cell culture media is one of the possible microalgae harvesting method employed to generate auto-flocculation21–23 or enhance the effects of flocculants.24,25
Motivated by the need to control colloidal stability of microalgae suspensions or microalgae interactions with various ions or (macro)molecules of interest in aqueous media, several research teams have attempted to evaluate microalgae surface charge properties as a function of pH and/or ionic strength of the surrounding solution.6,16,26 To that end, and along the lines detailed in most of the literature work quoted above, authors relied notably on electrophoresis measurements interpreted according to classical Smoluchowski representation of charged surfaces with electrostatics expressed in terms of zeta-potential value. These electrokinetic results are further considered to establish predictions of microalgae interactions on the basis of standard DLVO theory.7,27,28
However, many studies have underlined the strict applicability of zeta-potential concept to so-called hard particles (cf. e.g. reviews ref. 29 and 30 and references therein), i.e. particles that are impermeable to electrolyte ions and to the electroosmotic flow developed under electrophoresis measuring conditions. This concept becomes meaningless for soft (i.e. ions- and flow-permeable) (bio)surfaces that are generally covered by polyelectrolyte-like material carrying 3D-distributed charges.29,30 For such interfacial systems, the a priori location of a well-defined slip plane is impossible, and the conversion of measured electrophoretic mobility values into zeta-potential irrelevant.31,32 As an alternative, theory for electrokinetics of soft surfaces and particles has been reported29–32 and its merits largely documented with e.g. the successful interpretation of the peculiar electrokinetic and electric double layer properties of bacteria,33 yeasts34 and, very recently, microalgae.35 In turn, ignoring the soft nature of algae interface in the analysis of electrophoresis data may generate incorrect biosurface electrostatic descriptors and, therewith, lead to misevaluation of the electrostatic component of e.g. cell–cell or cell–surface interactions.36
In addition, to get a comprehensive picture of the physicochemical interactions involving microalgae, electrostatics of the algal cell surface should be considered along with other contributions that can balance cell–cell or cell–surface electrostatic repulsion/attraction, in particular hydrophobic effects and/or specific key-lock biomolecular interactions. Interestingly, variation of algae growth conditions37 or environmental factors like pH38 can change the nature of the dominant interactions in cell adhesion process. In that sense, recent studies have highlighted the crucial role played by algal surface hydrophobicity in cell/cell or cell/substrate interactions,39 and in the adhesion of microalgae to air bubbles during harvesting flotation process.28,40 Although both electrostatic and hydrophobic cell-wall properties can impact the stability of microalgae in aqueous media, very few techniques allow a proper quantitative assessment of their respective contributions depending on environmental conditions.
Among the eukaryotic green microalgae with high potential for biotechnological applications, Chlorella vulgaris is one of the most studied species. Due to its fast replication in freshwaters, C. vulgaris, an easy-to-grow cell model, is an excellent candidate for industrial lipid extraction. On an academic level, C. vulgaris has also been largely used as a convenient microorganism model to address fundamental issues on aquatic contaminants toxicity. This species is further commonly employed in standardized ecotoxicological bioassays and considered as a suitable system for water bioremediation.26,41
In the current study, we addressed the physicochemical surface properties, including electrostatics, of C. vulgaris at various scales and for different environmental conditions. Electrophoresis measurements on suspensions of microalgae cells, interpreted by electrokinetic theory for diffuse soft particles,42 provided some surface- and cell-averaged indications on the overall density and spatial organisation of the structural charges carried by the algae as a function of electrolyte concentration and solution pH. To further assess the quantity of structural charges carried by functional groups operative at the microalgae interface, we performed potentiometric proton titration experiments. We evidenced that interpretation of these results is obscured by ongoing physiological processes and associated transmembrane proton-exchange equilibria other than those governing the surface concentration and dissociation characteristics of charge-determining functional groups. Finally, at the molecular scale, atomic force microscopy (AFM)-based force spectroscopy measurements were monitored in liquid according to so-called chemical force microscopy (CFM) mode, between the surface of individual algal cells and nanometric tips featuring controlled electrostatic or hydrophobic coatings.43 The obtained tip-to-cell adhesion maps revealed the spatial distribution of the electrostatic and hydrophobic reactive sites/domains of the cell wall, they qualified the heterogeneity of sites distribution at the single cell and molecular scales, and force measurements further shed light on the typical range of hydrophobic interactions depending on pH.
Results
In the following developments, the electrostatic properties of microalgae were evaluated in aqueous medium versus electrolyte concentration and solution pH. We adopted C. vulgaris (C211-11B), a microalgae strain from the branch of the Chlorophyta. C. vulgaris are unicellular eukaryotic and photosynthetic microorganisms possessing a cell membrane formed by a double lipidic layer surrounded by a cell wall (without appendages) whose dimension and density increase during growth.44,45 The cell wall of C. vulgaris is mostly composed of (poly)saccharides, with the additional presence of proteins and lipids,46 and there are few indications in literature about the nature of the charge-carrying components, about their surface concentration and distribution at the algal interface formed with the outer aqueous medium.16,46 In addition, as evidenced by recent work,35 microalgae can be viewed as soft particles, i.e. particles permeable to electrolyte ions and/or electroosmotic flow32 (Fig. 1a). The electrohydrodynamic properties of these particles can be retrieved upon exploitation of (i) electrophoresis data measured as a function of salt concentration in solution and (ii) force spectroscopy measurements (cf. e.g. ref. 47 and 48).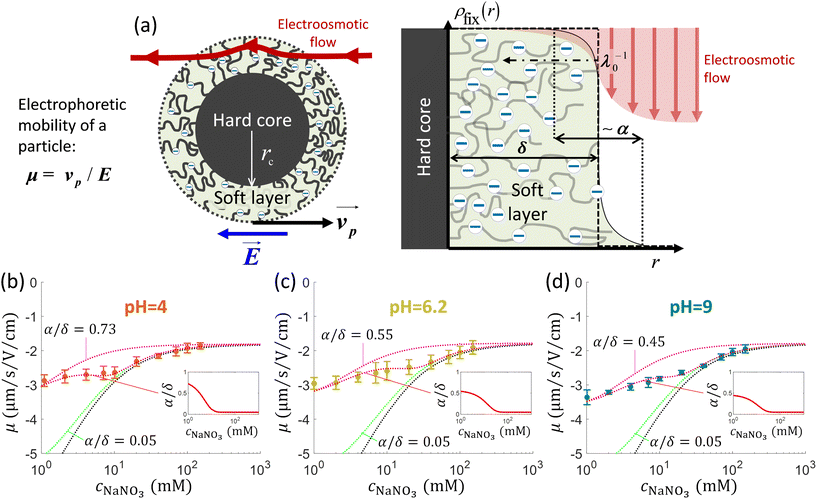 | ||
Fig. 1 (a) Schematics of electrophoresis of a core/soft shell particle,31 composed of a hard core of radius rc (m), and a surrounding soft layer with thickness δ (m) permeable to ions and to the electroosmotic flow generated by the interaction between the applied electric field 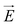 (in V m−1) and the interfacial electric double layer. The soft layer features a 3D distribution of fixed (immobile) structural charges (charge-carrying groups represented by the symbol ⊖) with a resulting position-dependent charge density, ρfix(r) (C m−3), and a spatial diffuseness (or heterogeneity) subsumed in the dimensionless ratio α/δ, where r (m) is the radial coordinate (origin set at the particle centre) and α (m) is the interfacial heterogeneity length scale. The radial dependence of ρfix typically corresponds to a sigmoid-like function decreasing with distance, and the case of homogeneous charge distribution within the shell is captured by the limit α/δ → 0 (black dotted curve). λ0−1 (m) is the reciprocal of the hydrodynamic softness of the cell interface, and it defines the extent of flow penetration within the particle shell. The electrophoretic velocity of the soft particle, denoted as (in V m−1) and the interfacial electric double layer. The soft layer features a 3D distribution of fixed (immobile) structural charges (charge-carrying groups represented by the symbol ⊖) with a resulting position-dependent charge density, ρfix(r) (C m−3), and a spatial diffuseness (or heterogeneity) subsumed in the dimensionless ratio α/δ, where r (m) is the radial coordinate (origin set at the particle centre) and α (m) is the interfacial heterogeneity length scale. The radial dependence of ρfix typically corresponds to a sigmoid-like function decreasing with distance, and the case of homogeneous charge distribution within the shell is captured by the limit α/δ → 0 (black dotted curve). λ0−1 (m) is the reciprocal of the hydrodynamic softness of the cell interface, and it defines the extent of flow penetration within the particle shell. The electrophoretic velocity of the soft particle, denoted as 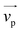 (m s−1), is indicated as well as the applied electric field (m s−1), is indicated as well as the applied electric field 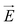 . The electrophoretic mobility of the particles is defined by . The electrophoretic mobility of the particles is defined by 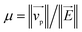 (m2 s−1 V−1). Measured electrophoretic mobility μ (symbols) of C. vulgaris as a function of NaNO3 concentration denoted as cNaNO3 at (b) pH = 4, (c) pH = 6.2 and (d) pH = 9 (indicated). In panels (b–d): black dotted curves are fits of electrophoretic mobility data using well-known analytical Ohshima expression,31,42 valid here at sufficiently high NaNO3 concentrations (above ca. 30 mM). This expression assumes homogeneous charge distribution throughout the shell (α/δ = 0). Green and pink dotted curves correspond to predictions from Duval–Ohshima model42 with α/δ = 5 × 10−2 (green) and with fixing the value α/δ to that at 1 mM NaNO3 (pink) where interfacial heterogeneity is most pronounced. The red dashed lines in (b–d) are fits of data according to Duval–Ohshima theory42 by adjustment of the dependence of α/δ on electrolyte concentration (specified in the insets), with adopting here a cell radius of 2 μm and a shell thickness δ of 20 nm which is of the order of the cell wall thickness (estimation from TEM imaging on C. vulgaris44). Each reported electrophoretic mobility data point for a given NaNO3 concentration is the average of 6 electrophoretic mobility acquisitions on 3 different batches of microalgae per tested pH condition, with one replicate per batch (cf. details in Methodology section). The error bars for each data point represent the standard deviations over the 6 acquired μ values at a given salinity. (m2 s−1 V−1). Measured electrophoretic mobility μ (symbols) of C. vulgaris as a function of NaNO3 concentration denoted as cNaNO3 at (b) pH = 4, (c) pH = 6.2 and (d) pH = 9 (indicated). In panels (b–d): black dotted curves are fits of electrophoretic mobility data using well-known analytical Ohshima expression,31,42 valid here at sufficiently high NaNO3 concentrations (above ca. 30 mM). This expression assumes homogeneous charge distribution throughout the shell (α/δ = 0). Green and pink dotted curves correspond to predictions from Duval–Ohshima model42 with α/δ = 5 × 10−2 (green) and with fixing the value α/δ to that at 1 mM NaNO3 (pink) where interfacial heterogeneity is most pronounced. The red dashed lines in (b–d) are fits of data according to Duval–Ohshima theory42 by adjustment of the dependence of α/δ on electrolyte concentration (specified in the insets), with adopting here a cell radius of 2 μm and a shell thickness δ of 20 nm which is of the order of the cell wall thickness (estimation from TEM imaging on C. vulgaris44). Each reported electrophoretic mobility data point for a given NaNO3 concentration is the average of 6 electrophoretic mobility acquisitions on 3 different batches of microalgae per tested pH condition, with one replicate per batch (cf. details in Methodology section). The error bars for each data point represent the standard deviations over the 6 acquired μ values at a given salinity. | ||
Electrophoresis on C. vulgaris
Fig. 1b–d display the variation of the electrophoretic mobility μ of C. vulgaris with changing NaNO3 concentration (denoted hereafter as cNaNO3) and solution pH. For all three pH conditions tested (pH = 4, 6.2 and 9) μ is negative, which indicates that the net density of surface charges of the microalgae probed by electrokinetics is negative. Different functional groups have been identified at C. vulgaris surfaces such as carboxyl, phosphoryl, amine and hydroxyl groups,16 and anionic components are seemingly predominant. This result agrees with the reported composition of C. vulgaris cell-wall which hosts many polysaccharidic compounds (and therewith carbo/hydroxyl groups).45,46Fig. 1b–d further show that |μ| decreases with increasing cNaNO3 as a result of screening of cell charges by electrolyte ions. More remarkably, μ levels off to reach a non-zero plateau value for cNaNO3 exceeding ca. 100 mM. For each pH condition, this non-zero plateau value reached asymptotically by μ is the most obvious electrokinetic signature of soft particles: it is explained by the finite flow penetration within the charged particle shell component (draining process), as extensively discussed by Ohshima and co-workers.31,42 In the developments below, following the classical representation of soft particles31 and previous modelling of cell electrophoresis data whose lines are adopted here,49 we understand hereafter by shell the peripheral part of the microalgae and assume that this soft structure includes – at least partially – the cell wall.In a first approach, experimental data were fitted using the classical Ohshima model,31,35,50 therefore assuming that the structural charges are homogeneously distributed within the shell and that Donnan electrostatics representation holds at the shell/solution interface (the reader is referred to e.g. ref. 35 and 42 for details on the limits of Ohshima model). In turn, data fitting led to the evaluation of two quantities: the density of cell charges, ρ0, here expressed as an equivalent concentration of anionic charges (mM), and the hydrodynamic softness of the soft algal interface (Fig. 1a), λ0 (m−1), which corresponds to the reciprocal of the characteristic flow penetration length scale within the shell31 (Table 1). As expected, Fig. 1b–d evidence that fitting of electrophoresis data to Ohshima model is possible only for sufficiently large cNaNO3 (typically above 30 mM), which is in agreement with some of the approximations underlying the applicability of Ohshima's expression for the electrophoretic mobility of soft particles, i.e. electric double layer polarization is ignored, the shell layer (thickness δ) is thick as compared to the Debye layer thickness (denoted hereafter as 1/κ) and to λ0−1, and the distribution of the structural charges is homogeneous in the shell. Related to the latter point, we recall that charge distribution heterogeneity in the radial dimension impacts all the more particle electrophoretic mobility as salt concentration decreases.42
| pH | ρ 0 (mM) | λ 0 −1 (nm) |
|---|---|---|
| 4 | −17 ± 1.7 | 3.3 ± 0.3 |
| 6.2 | −41 ± 4.1 | 2.1 ± 0.2 |
| 9 | −41 ± 4.2 | 2.1 ± 0.2 |
To refine interpretation of the electrokinetic properties of microalgae, we confronted data to predictions from Duval–Ohshima formalism (cf. details in ref. 42 and 51) where interface diffuseness (radial heterogeneity) and electric double layer polarisation are accounted and, unlike Ohshima model, the theory does not suffer from any approximation on the relative magnitudes of κ−1, δ and λ0−1 while providing a rigorous solution to the key coupled electrostatic and hydrodynamic equations driving the migration of soft particles under applied DC field condition.31 In detail, interface diffuseness is modelled here by a sigmoid-like distribution for the concentration of charge-carrying groups across the shell, with the characteristic lengths ratio α/δ where α (m) corresponds to the distance over which the density of structural charges decreases from bulk shell value to 0 (Fig. 1a). Within Duval–Ohshima theory, data fitting then requires the only adjustment of α/δ as a function of cNaNO3 with adopting the limit α/δ → 0 at high salt concentrations where data are properly reconstructed by Ohshima model. Accordingly, the relevant ρ0 and λ0−1 parameters involved in the refined data modelling exercise are those retrieved from data analysis done on the basis of the approximate analytical expression by Ohshima. The reader is referred to ref. 49 (Fig. S2† therein) and ref. 42 for further modelling details.
As expected, at large cNaNO3, predictions derived from full numerical evaluation of the relevant electrohydrodynamic equations governing the electrophoresis of soft particles42 converge to Ohshima's results, with a reduced impact of the interface diffuseness α/δ on cell mobility μ as cNaNO3 increases. The fitting of the electrophoretic mobility data for all pH conditions requires an adjusted increase of α/δ with decreasing cNaNO3 (Fig. 1b–d and insets thereof) because the corresponding heterogeneous extension of the shell (cf.Fig. 1a) leads to the required decrease of |μ| as compared to the outcomes of Ohshima model that overestimates experimental |μ| values.42 The reduction of |μ| with increasing α/δ at given pH stems from the associated dominant increase of the hydrodynamic drag exerted by the particle on the electroosmotic flow.42 Whereas this heterogeneity probed by electrokinetics increases with decreasing cNaNO3 (due to possible swelling of the interfacial region following increased repulsion between neighbouring charged groups52), data modelling suggests that it further slightly increases with decreasing pH, as shown in the insets of Fig. 1b–d with α/δ values ranging from 0.45 at pH 9 to 0.73 at pH 4 at cNaNO3 = 1 mM. The values of ρ0 and λ0−1 fitted by Duval–Ohshima formalism are collected in Table 1 and hereafter discussed. For the sake of comparison, Gomes et al.35 reported – from the analysis (using Ohshima's model) of electrophoretic mobility measurements performed on C. vulgaris – the following electrohydrodynamic parameter values ρ0 = −33 mM and λ0−1 = 1.6 nm, under neutral pH condition (presumably, as the pH value is not specified in the article). Pagnout et al.49 reported for different Escherichia coli strains values of ρ0 and λ0−1 ranging from −110 mM to −185 mM and from 0.76 nm to 0.79 nm, respectively, at pH = 6.7.
Comparison between predictions of μ at the three pH conditions (Fig. S1, ESI†) at fixed salt concentration shows that |μ| basically decreases with pH. This finding is in agreement with results previously published (cf. e.g. ref. 16 where μ is converted into zeta-potential, a physically meaningless parameter for soft interfaces), and with the found decrease of |ρ0| upon decreasing pH (Table 1) due to weaker dissociation of hydroxyl and carboxyl end groups. In line with the latter argument, |ρ0| is (within experimental error) identical at pH = 6.2 and 9 as functional shell groups are then fully dissociated in this pH range. The decrease of |μ| with decreasing pH at fixed electrolyte concentration is further associated with an increase in the shell heterogeneity α/δ at cNaNO3 < 10 mM. Last, the characteristic flow penetration length scale within the shell material, λ0−1, increases with decreasing pH (Table 1). This finding may suggest an increase in cell surface roughness when switching from basic to acidic pH conditions, which qualitatively supports the companion increase of interface heterogeneity invoked above. However, the relatively large uncertainty in the experimental data prevents from drawing firm conclusions on the pH-dependence of λ0−1.
Sequential proton titrations on C. vulgaris
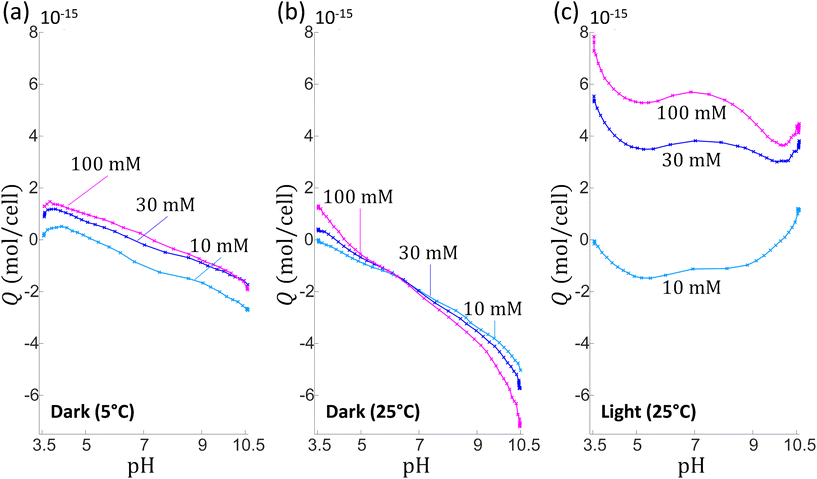
Whereas the electrophoretic mobility reflects the electrohydrodynamic properties of an outer (electrokinetically active) particle shell region,30,51,52 potentiometric proton titrations allow, in principle, the evaluation of all structural charges at the particle surface. Providing that these charges display well differentiated dissociation properties, hypotheses on their nature may be further advanced from proper analysis of proton affinity spectra obtained from differentiation of titration data with respect to pH.53 We performed potentiometric titrations series on C. vulgaris in electrolyte solution to determine the mean amount of charges per microalgae, Q (in mol per cell), as a function of pH for a stepwise increase in NaNO3 concentration (10, 30 and 100 mM, see Material and methods for details). To ensure that variation in titrated charge at different cNaNO3 is not caused by differences among microalgae batches, the sequential titrations at cNaNO3 = 10, 30 and 100 mM were carried out on a unique microalgae batch, and the titration process was then replicated on several batches for repeatability purpose. As a part of the titration measurements, charge titrations by addition of 10 mM NaOH at cNaNO3 = 30 and 100 mM were each preceded by a ‘backward titration’ via the addition of acid solution (10 mM HNO3) at the desired cNaNO3. Doing so, the extent of hysteresis in the forward and backward titration data could be addressed and, therewith, possible ongoing degradation of titrated material detected.53 Additionally, we varied light and temperature conditions in order to assess how cell physiology impacted (or not) the amount of interfacial cell charges.Fig. 2 shows representative results (Q versus pH and salinity) of three series of titrations on C. vulgaris. For given light and temperature conditions, the overall pattern describing qualitatively the change in Q with pH and cNaNO3 were found to be well consistent from one cell batch to the other, but high variability in Q (ca. 1 to 2 units in Q) was found due to cell physiology (detailed later) that apparently differs significantly among tested batches. Consequently, no marked quantitative trends in the dependence of Q on pH were measured, which renders impossible any attempt to identify the nature of the groups at the origin of the cell surface charge. Under dark and cold (5 °C) conditions (Fig. 2a), the positioning of the titration curves versus cNaNO3 is not according to expectation as |Q| does not increase significantly with cNaNO3 over the whole pH range. Remarkably, when increasing temperature from 5 °C to 25 °C (Fig. 2b), the aspects of the pH-dependent titration curves completely changed in terms of magnitude (increase in |Q|) with the apparition of a common intersection point between curves pertaining to the three cNaNO3-conditions tested. Titration data suggested a possible reversal of the sign of the charge with varying pH at fixed cNaNO3 and with varying cNaNO3 at fixed pH. In addition, there was a marked hysteresis between backward and forward titrations at cNaNO3 = 30 mM and 100 mM (Fig. S2 in ESI†), which indicates that chemical equilibria other than protonation/deprotonation of shell functional groups take place during titration. The apparent ‘loss’ of charges titrated between sequential addition of acid (pH 10.5 to 3.5) and that of base solution (pH 3.5 to 10.5) is the possible signature of a release of dissolved CO2 by C. vulgaris,54 leading to a carbonatation of the medium (at basic pH values). At 25 °C and in presence of light (Fig. 2c), Q is positive over the entire pH range at cNaNO3 = 30 mM and 100 mM, and it increases strongly with cNaNO3.
Reversal of the sign of the titrated charge evidenced in Fig. 2b (and, to some extent, in Fig. 2a and c depending on pH and salt concentration conditions) is unexpected in view of the electrophoresis results that pinpoint a negative (electrokinetic) charge for pH between 4 and 9. It may be argued that this apparent ‘inconsistency’ originates from the different time scales of the experiments (up to 8 hours for proton titrations compared to few minutes for electrophoresis), which possibly defines different algae response to pH stress. Reports evidence indeed that Chlorella microalgae in contact with an ‘unusual’ pH-environment can regulate their internal pH as well as the pH in their phycosphere55 around neutral value. To cope with such a pH stress, cells can deploy various metabolic strategies, e.g. inter-organelle proton exchanges, protons release via dedicated efflux pumps,56 and for chlorophyte microorganisms, the efficiency of these adaptative mechanisms depends intrinsically on light conditions.57 In particular, internal pH regulation for C. vulgaris in media whose pH is comprised between 4 and 9 is a few hours-long process that gains efficiency under light-exposure conditions.56,58 Under harsh pH conditions (typically for pH below 3 and above 10) cells viability drops dramatically under both dark and light-exposure conditions56,58 as a result of important intracellular pH fluctuation and/or unregulated ion exchanges between inner and outer cell components.56,58 In view of the above elements, we hypothesise that the increasing quantity of positive charges measured under light conditions (Fig. 2c) is related to the response of C. vulgaris to imposed variations of pH and electrolyte concentration, a response that necessarily differs when titration is operated in dark (Fig. 2a and b). Among physiological changes reported for microalgae subjected to pH variation, the modification of pigment production appears as an important factor.59–62 During proton-titration experiments, no color alteration of the Chlorella suspension could be observed by eye. However, UV-visible absorbance spectra of C. vulgaris cells measured under the pH conditions adopted in AFM (cf. below) and electrokinetic experiments (i.e. pH 4.5 and 6.2) (Fig. S3†) reveal that spectra profiles were severally modified at pH 4.5 after 8 hours, and that spectra modifications were even more pronounced after 24 hours with a quasi-complete extinction of the chlorophyll signal (at ca. 700 nm). This finding confirms that important physiological cell regulations are operational during proton-titration experiments measured as a function of solution pH.
Force spectroscopy measurements on microalgae
To further explore the electrostatics of microalgae soft interface, we detail below molecular interactions measured at the surface of single cells by chemical force spectroscopy (CFM technique63–65), between C. vulgaris and different AFM tips (Fig. 3). Using controlled charged AFM probes, we evaluate and map the electrostatic properties of algal cell surface with a molecular resolution at pH close to physiological condition (Fig. 4 and 5). Following a similar strategy, we address the hydrophobicity level of microalgae surface and compare the corresponding tip-to-cell surface adhesion features to those measured with electrostatic AFM probes so as to unravel their respective contributions to interactions involving microalgae (Fig. 6). Finally, based on the outcomes from the above measurements, we shed light on the effect of acidic pH on microalgae surface properties (Fig. 4–6). In the following, we first describe the experimental methodology and data analysis approach that we used independently of the tips functionalization, and we then successively discuss the results obtained with the NH2-, COOH- and CH3-modified AFM tips.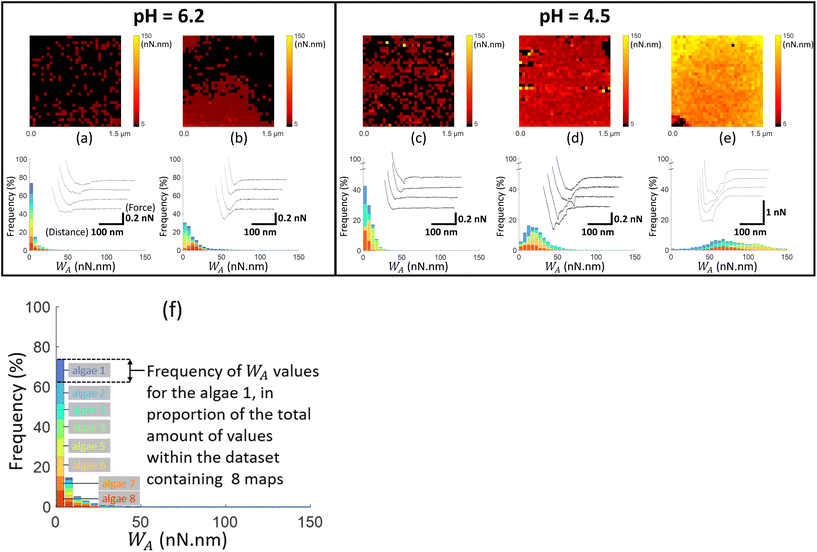 | ||
| Fig. 4 Work of adhesion WA of AFM tips coated by thiol-NH2 on C. vulgaris surface, in 10 mM NaNO3 solution, for (a and b) pH = 6.2 and (c–e) pH = 4.5. The histograms (a–e) represent the cumulative distributions of WA values for different sets of WA-maps sorted according to similarity in WA values distributions. The histograms in (a–e) represent the cumulative statistical distribution of WA values from sets of 8, 8, 5, 5 and 5 WA-maps (each C. vulgaris cell has been mapped only once by a given functionalized tip: 31 microalgae were probed by tips-NH2), respectively. Each color in the histograms corresponds to the contribution of one WA-map measured on a given cell to the overall WA-histogram. For each histogram, a representative WA-map is provided (1.5 μm × 1.5 μm, 32 × 32 pixels) and a collection of four illustrative force–distance curves is given in the inset of each histogram, with specified scales for the distance and force axes. The schematics in (f) illustrates the composition of the histograms presented in Fig. 4–6, with considering the histogram (a) as an illustrative example. For a given dataset, each color in the graphic (8 in total) corresponds to the statistical distribution of WA values for a single WA-map of a microalgae surface which was probed only once. For a given WA value, each colored bar represents the number of occurrences of that value in the associated WA-map in proportion to the total number of values (pixels) in the dataset (here 8 × 1024 pixels total). The envelope of the histogram corresponds to the cumulative statistical distribution of WA values for the given dataset. | ||
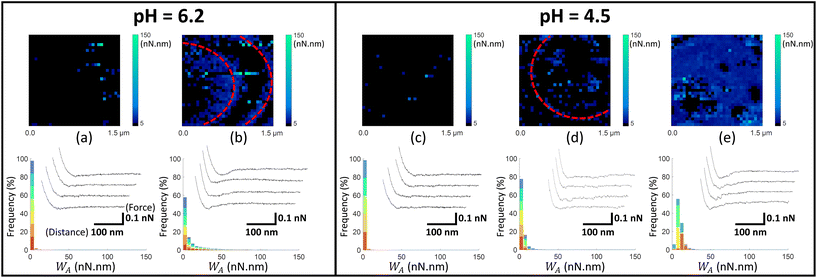 | ||
| Fig. 5 Work of adhesion WA of AFM tips coated by thiol-COOH on C. vulgaris surface, in 10 mM NaNO3 solution, for (a and b) pH = 6.2 and (c–e) pH = 4.5. The histograms (a–e) represent the cumulative distributions of WA values for different sets of WA-maps sorted according to similarity in WA values distributions. The histograms in (a–e) represent the cumulative statistical distribution of WA values from sets of 7, 7, 5, 6 and 4 WA-maps (each selected C. vulgaris cell has been mapped only once by a given functionalized tip: 29 microalgae were probed by tips-COOH), respectively. Each color in the histograms corresponds to the contribution of one WA-map measured on a given cell to the overall WA-histogram (cf.Fig. 4f). In (b and d), the red dotted ellipses highlight circular adhesion patterns on microalgae surfaces. For each histogram, a representative WA-map is provided (1.5 μm × 1.5 μm, 32 × 32 pixels) and a collection of four illustrative force–distance curves is given in the inset of each histogram, with specified scales for the distance and force axes. | ||
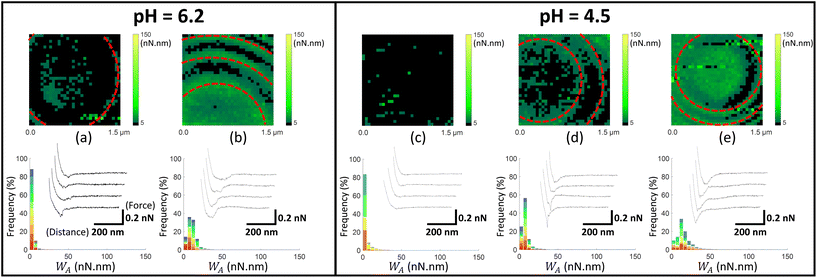 | ||
| Fig. 6 Work of adhesion WA of AFM tips coated by thiol-CH3 on C. vulgaris surface, in 10 mM NaNO3 solution, for (a and b) pH = 6.2 and (c–e) pH = 4.5. The histograms (a–e) represent the cumulative distributions of WA values for different sets of WA-maps sorted according to similarity in WA values distributions. The histograms in (a–e) represent the cumulative statistical distribution of WA values from sets of 10, 10, 4, 10 and 6 WA-maps (each selected C. vulgaris cell has been mapped only once by a given functionalized tip: 40 microalgae were probed by tips-CH3), respectively. Each color in the histograms corresponds to the contribution of one WA-map measured on a given cell to the overall WA-histogram (cf.Fig. 4f). In (a, b, d and e), the red dotted ellipses highlight circular adhesion patterns on microalgae surfaces. For each histogram, a representative WA-map is provided (1.5 μm × 1.5 μm, 32 × 32 pixels) and a collection of four illustrative force–distance curves is given in the inset of each histogram, with specified scales for the distance and force axes. | ||
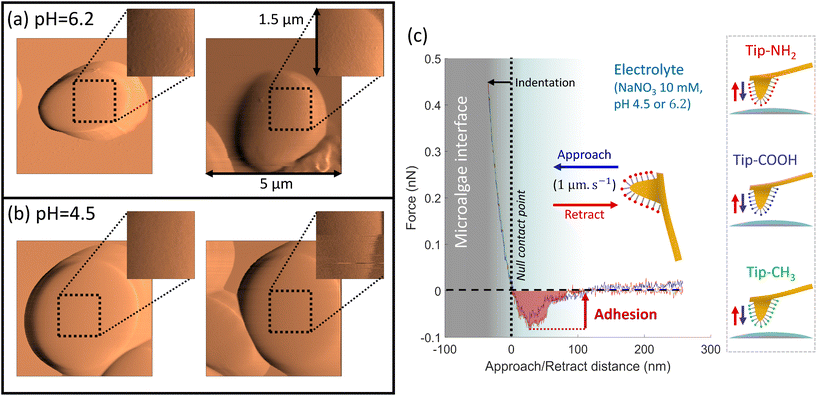
To assess the spatial distribution of the cell surface properties by CFM, we work in so-called force-volume mode where a virtual mesh of 32 × 32 pixels (which corresponds to 1.5 μm × 1.5 μm surface area) was generated at the cell surface. Approach and retract curves were then recorded at each pixel (Fig. 3c). This process then provides a 3D map of the interaction force operational between functionalized tip and algal surface. After contact between tip and cell surface, inspection of the force-retraction regime (Fig. 3c, red curve) allows to state whether or not the functionalized tip adhere to the algal cell-wall, to evaluate the adhesion force required to detach the tip from the biosurface and to monitor the (possible) unfolding of the biomolecules involved in the interaction when withdrawing the tip from the cell surface. To prevent contamination, the functionalized AFM tips were replaced every 4 to 6 maps. To ensure that the cellular surface was not getting damaged by pH effects during AFM experiments, microalgae attached to PEI–substrate were not exposed to a given pH condition for more than 2 hours. During this period, viability and membrane integrity of Chlorella cells were not significantly affected by pH stress, as confirmed by independent flow cytometry measurements with propidium iodine cell staining (Fig. S4†).
Fig. 4–6 report maps of the adhesion of functionalised AFM probes on C. vulgaris surfaces, at pH = 4.5 and 6.2, and fixed electrolyte concentration cNaNO3 = 10 mM. Looking at the profiles of the force–distance curves forming these maps, we found that, independently of the tip chemistry, most of the curves displayed ‘blunt’ peaks (cf. insets in Fig. 4–6) whose exact positioning and magnitude were difficult to interpret using conventional analysis methods.66 These peaks stem from unspecific interaction forces that induce unfolding of several biomolecules at C. vulgaris surface (cf. e.g. CFM on cellular membranes67). Consequently, as illustrated in Fig. 3, we chose to evaluate the work of adhesion, denoted as WA (nN nm), at every probed pixel of the cell surface, thereby converting the force vs. distance-maps into WA-maps. Further considering the characteristic signal-to-noise ratio of the measurements, we determined a cut-off value of WA = 5 nN nm below which we consider that there is no tip-to-cell adhesion.
For each tip functionalisation and pH condition adopted, we acquired several WA-maps (at least 15 cells were considered per examined condition, each cell being probed only once). Given the heterogeneity of the obtained maps, we decided to classify them in different ‘sets’, according to their similarities in terms of statistical distribution of WA values and/or spatial distribution of these values over the cell surface. In addition, we computed the cumulative statistical distribution of WA values for each identified set of similar maps (histograms in Fig. 4–6), which gives an overall indication of the adhesion capacity of microalgae surfaces. For the sake of illustration, in Fig. 4–6 each set of similar WA-maps is represented by one illustrative WA map with a sample of 4 force–distance curves recorded upon tip retraction (see insets in Fig. 4–6). Additional examples of maps included in each set can be found in Fig. S5.†
To detect the negative interfacial charges of C. vulgaris, we mapped microalgae surfaces with AFM tips coated by thiols terminated by amine groups (tip-NH2), which act as positively charged probes.68 At pH = 6.2 (Fig. 4a and b), the force distance curves generally display a peak of around 100 to 200 pN over a distance of few tens nanometres from which WA can be evaluated. The obtained WA-maps can be divided into 2 sets of profiles: maps (shown in Fig. 4a, representative of 8 maps) displaying randomly distributed adhesion sites over the microalgae surfaces, with WA value of ca. 10 nN nm (corresponding to 10 aJ), and maps (shown in Fig. 4b, representative of 8 maps) that feature adhesion domains where WA values are slightly higher than in Fig. 4a.
At lower pH value (pH = 4.5; Fig. 4c–e), the force–distance curves display multi-peaks profiles where both interaction force and interaction distance increased as compared to those corresponding to higher pH. The corresponding WA-maps can be categorized according to 3 types of profiles (Fig. 4c–e) and they highlight that WA is higher at pH 4.5 as compared to 6.2, with some microalgae even displaying remarkably strong adhesive surface events (Fig. 4e, representative of 5 maps). A higher intra- and inter-cellular heterogeneity is also noticed at pH 4.5, in the sense that the distributions in WA values for a given cell and among cells are broader than at pH 6.2. Still, the adhesion sites are rather homogenously distributed on the maps, and the adhesive patches observed at pH 6.2 are no longer distinguishable at pH 4.5.
At neutral pH, the thiol terminal groups are (weakly) protonated into –NH3+ – the acidity pK constant of the terminal groups of cysteamine thiols is ca. 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68 – which promotes electrostatic attraction between coated AFM tips and the negatively charged microalgae surfaces (Fig. 1). At lower pH, the surface charge of the tips-NH2 increases due to the protonation of the terminal groups of cysteamine thiols,68 in agreement with the increase in electrostatic attraction suggested by Fig. 4 (panels (a) and (b) vs. (d) and (e)). Interestingly, few force curves feature the unfolding of some cell wall components upon tip retraction, which is identified from the succession of multiple adhesion peaks at relatively large distance (>100 nm) prior to final rupture (cf. insets Fig. 4d and e).
68 – which promotes electrostatic attraction between coated AFM tips and the negatively charged microalgae surfaces (Fig. 1). At lower pH, the surface charge of the tips-NH2 increases due to the protonation of the terminal groups of cysteamine thiols,68 in agreement with the increase in electrostatic attraction suggested by Fig. 4 (panels (a) and (b) vs. (d) and (e)). Interestingly, few force curves feature the unfolding of some cell wall components upon tip retraction, which is identified from the succession of multiple adhesion peaks at relatively large distance (>100 nm) prior to final rupture (cf. insets Fig. 4d and e).
In Fig. 5, we report the equivalent of Fig. 4 with use here of electrostatic AFM tips coated by thiols terminated by carboxyl groups (tip-COOH), which are commonly employed as negatively charged probes.69 At pH 6.2, two sets of WA-maps profiles can be distinguished (Fig. 5a and b): a first set (shown in Fig. 5a, representative of 7 maps), for which the adhesion of tips-COOH to cell surface is insignificant as there is no detection of adhesion peaks (cf. representative curves in Fig. 5a), and a second set (shown in Fig. 5b, representative of 7 maps) corresponding to cells that feature slightly adhesive patches (with WA ∼ 10 nN nm). At pH 4.5 (Fig. 5c–e), the microalgae surface adhesion to tips-COOH remains weak as judged by the corresponding low WA values. The spatial distribution of the adhesion events can be categorized in 3 sets: maps without or with few adhesive events detected over the cell surface (Fig. 5c, representative of 5 maps), maps featuring few adhesive patches (Fig. 5d, representative of 6 maps) and others displaying a homogeneously adhesive surface (Fig. 5e, representative of 4 maps). Overall, the weak adhesion measured at both pH 6.2 and 4.5 (Fig. 5) is consistent with a dominant electrostatic repulsion between the negatively charged microalgae surface and the tips coating where thiol terminal groups COOH are deprotonated.70 With decreasing pH from 6.2 to 4.5, both the algal shell and the tip coating get increasingly protonated, thereby decreasing the contribution of electrostatics to the overall measured interaction. Accordingly, the adhesion events featured in Fig. 5b, d and e likely originate from interaction processes other than electrostatic in nature.
Considering the tip coating properties under acidic pH conditions, protonated –COOH terminal groups are indeed prone to form hydrogen bonds with molecular partners of the cell wall.71 This is confirmed by additional control measurements between tips and gold surfaces, both coated with thiols-COOH, which shows an increase of adhesion from pH 6.2 to 4.5 (Fig. S6a and b†). Hence, with lowering pH, the decrease of the tip-to-cell electrostatic repulsion and the possible formation of hydrogen bonds between tip-COOH and cell wall components like carboxyl end groups16 could explain the observed increase in the occurrence of adhesion events. For some situations where a significant adhesion was measured between tip-COOH and cell surface (Fig. 5b and d), adhesion patches appear in the form of circular patterns centred on the top of C. vulgaris with respect to the sample support. Inspection of the topographic image associated with each FV confirmed that these patterns were neither due to topographic features that could change the contact area between tip and biosurface, nor to an experimental drift of the tip toward the PEI-coated glass substrate (Fig. S7†). The corresponding WA-maps may thus suggest a difference in the nature of the cell wall compounds that interact with tips-COOH and tips-NH2 (Fig. 4 and 5).
Following the investigation of the electrostatic (and H-bonds) contributions of tip-to-cell adhesion, we proceed with the determination of the surface hydrophobicity of microalgae, described in literature as an important component of their interactions with their surrounding environment.16,39,40 Accordingly, Fig. 6 reports CFM measurements performed on C. vulgaris surface using methyl-terminated thiol coated tips (tip-CH3) – serving as hydrophobic probes39,63,69 – under similar pH and salt conditions as those prevailing in Fig. 4 and 5.
Overall, at pH = 6.2 the centred value of the WA-distributions (Fig. 6a and b) is slightly higher than that determined with tips-NH2 (Fig. 4). Decreasing solution pH from 6.2 to 4.5 (Fig. 6c–e) hardly impacts the overall adhesion of the cells surface. However (and similarly to Fig. 4 and 5), this decrease leads to a larger heterogeneity in the statistical distribution of WA values among the probed microalgae surfaces (cf. histograms in Fig. 6). We further controlled how pH affected the adhesion of tip-CH3 on planar gold surfaces coated with the very same thiols (Fig. S6c and d†) as those used for tip functionalisation. We observed that WA decreases when decreasing pH, a trend we assign to proton binding by-/absorption on-the thiols.43,72 This pH-dependence of WA as revealed by controlled experiments is however not reflected by the data in Fig. 6 as the decrease of pH from 6.2 to 4.5 does not clearly induce a decrease in the hydrophobic tip-to-cell adhesion. Comparison between Fig. 6 and 4 further indicates that the hydrophobic contribution to the interactions involving C. vulgaris surface at pH 4.5 is lower (both in terms of adhesion force and frequency of adhesive events) than the electrostatic contribution.
Interestingly, regardless of pH, the WA-maps displayed in Fig. 6a, b, d and e show that the distributions of adhesion sites at the microalgae surface take the form of circular and concentric patterns, similar to those identified with the tips-COOH (Fig. 5b and d). These patterns reflect a peculiar spatial distribution of hydrophobic compounds at the cell wall of C. vulgaris under the measuring conditions adopted in this work. The corresponding spatial heterogeneities over the cell surface are not distinguishable on the topographic images of C. vulgaris presented in Fig. 3a and b and on those reported elsewhere.19 We further note that similar circular and concentric patterns are discernible in some of the CFM maps reported in literature for other types of algae.37 Remarkably, these concentric patterns are systematically centred on the top of the microalgae surface (dome) with respect to the sample support on which C. vulgaris is attached. This property might be a surface phenotype of C. vulgaris cell wall, which constrains the orientation of the microalgae on the supporting PEI-coated glass surface. Conversely, we could hypothesize that this pattern is a result of microalgae immobilization onto PEI and associated modification of cell surface tension.73 It cannot be excluded that such a distribution pattern of hydrophobic compounds is also related to a specific repartition of lipids within the C. vulgaris cell wall/membrane.46,74,75 At this stage, the above assumptions are obviously largely speculative, and their validation requires additional analysis that goes beyond the scope of this work.
Discussion
The aim of this study is to determine the electrostatic properties of C. vulgaris and to evaluate how they are impacted by the pH of the surrounding solution. The work thus covers both fundamental and applicative dimensions, given the paramount importance of electrostatics in defining the homo- and hetero-interactions cells experience in various industrial and environmental processes. Our conclusions are based on results obtained by means of three types of experiments performed at various spatial and time scales: electrophoresis measurements on suspensions of microalgae cells, interpreted by electrokinetic theory for diffuse soft particles, potentiometric proton titration experiments, and AFM-based force spectroscopy measurements at the individual cell level. This original combination of methodologies allows us to infer some correlations between the information extracted from the data obtained with each of these techniques. It also brings to light important limitations (often overlooked in literature) in applying these techniques to biological samples, while highlighting some guidelines required to achieve a proper interpretation of the data.Electrophoresis measurements provide useful insights into the electrostatics of C. vulgaris soft interface with the estimation of the densities of structural charges it carries, and they further evidence a marked radial heterogeneity of the interface at low pH and/or under low salt concentration conditions (Fig. 1 and Table 1). Obviously, these results do not inform on the 3D heterogeneity properties of the interface nor on its composition, having further in mind that cell electrophoretic mobility is necessarily a surface-averaged indicator of the electrohydrodynamic properties of the ensemble of cells that experience the applied electric field.
Accordingly, we report potentiometric proton titrations to further address the dissociation features of the structural charges of C. vulgaris cells. However, the poor (quantitative) repeatability of the titration data and their strong dependence on illumination and temperature conditions suggest that during titration experiments (i.e. up to 8 hours) complex biological processes are involved in the regulation of the interfacial charge of C. vulgaris, which adds a difficulty to a proper definition of the electrostatic cell surface properties.
This urges us to consider spatially resolved CFM measurements at the cell surface and shorter measurement timescale so as to minimize the influence of physiological cell regulations. Accordingly, we perform CFM measurements to further address the dissociation features of the structural charges of C. vulgaris cells and their repartition at the cellular scale. Via the use of chemically modified AFM tips, we estimate the contributions of different force components (electrostatic, hydrogen-bonds and hydrophobic) to the overall algal adhesion. As electrokinetic analysis reveals that C. vulgaris are negatively charged (Fig. 1), we first determine the electrostatic forces operative between the biosurface and positively charged AFM probes (i.e. amine-functionalized tips) and we show that the corresponding tip-to-cell adhesion is higher at acidic pH as compared to that prevailing at ca. neutral pH (Fig. 4).
However, we can question the extent to which this effect is dominated by the variations of cell surface charges and/or the charges carried by the functionalized tip itself. The electrokinetic analysis of the biosurface evidences a decrease in the average density (in absolute value) of the structural cell charges with decreasing pH (Table 1). The underlying pH dependence of |ρ0|, if solely considered, would thus lead to a decreasing adhesion in CFM measurements between the amine-tip and the biosurface from pH 6.2 to 4.5. Accordingly, the variation of the charge of the tip with pH dominates apparently the one pertaining to the cell-wall and it governs, at least qualitatively, the way in which WA changes with decreasing pH (Fig. 4).
There is another cell surface property to be considered for a more complete overview of the processes that determine the electrostatic interactions between the cell surface and the amine tips as addressed by CFM as a function of pH: it relates to the way the constitutive charged components of the cell wall in interaction with the tips are distributed over space. Such an information is qualitatively retrieved from analysis of electrophoretic data, with the conclusion that the diffuseness (or heterogeneity in the radial dimension) of an individual algal interface increases with decreasing salt concentration at fixed pH and increases at 1 mM cNaNO3 with decreasing pH (Fig. 1 and Fig. S1†). Unfortunately, the assessment of the interface diffuseness operational during tip retraction cannot be straightforwardly compared to that obtained from electrokinetics as the very indentation of the charged tip into the cell prior to retraction has modified the distribution of cell structural charges in CFM experiments. In contrast, connections between electrostatics of diffuse interfaces as evaluated from analysis of electrokinetic data and from AFM can be drawn for the case of tip-to-cell force curves measured when approaching the tip towards the cell before contact.76 In the current work, such approach-force curves are not considered because the corresponding measured attractive force is found to be of the same order of magnitude than that of the background noise.
Further CFM measurements using carboxyl- and methyl-coated tips allow to estimate the importance of hydrogen-bonds and hydrophobic effect as compared to electrostatic interaction component under different pH conditions. This CFM-methodology with molecular scale resolution reveals remarkable circular chemical patterns at the cell surface (Fig. 5 and 6). However, such patterning cannot be directly interpreted through the interfacial diffuseness parameters (α/δ) involved in the Duval–Ohshima formalism42 (adopted to fit electrophoretic data of Fig. 1) as this parameter refers to the radial distribution of functional groups (and density of cell material that carries them) at the cell/solution interface and not to their lateral arrangement.
Potentiometric titration data (Fig. 2) turn to be decisive as they highlight the difficulty to decipher the physicochemical surface properties of the cells and the impacts of their response to pH- and/or salinity-induced stress on these properties. In that respect, we cannot a priori exclude that the strong electrostatic adhesion measured by AFM under acidic pH condition (Fig. 4) stems, at least partly, from physiological processes that could lead to the release of e.g. metabolites or polysaccharides as the latter biomolecules could then contribute to the cell surface–AFM tips interaction. However, current literature reports that such cell response occurs only at extreme basic pH values.22
Finally, transmembrane proton exchange/release in the phycosphere – i.e. in the close vicinity of the algal envelope – may modify the local pH and ionic strength conditions prevailing near the cell surface, with possible significant differences between such surface conditions and those holding in the bulk solution. Obviously, such intricate interfacial processes may considerably complicate data interpretation, as evidenced by the here-reported proton-titration data which underline an obvious alteration of the phycosphere. The typical delay adopted here for the incubation of cells in solution prior to electrokinetic and AFM data acquisition (1 to 2 h at most) is significantly shorter than that required to complete the proton titration experiments (up to 8 h). In turn, this minimises possible severe biology-mediated effects (discussed in Fig. 2via proton titration data) on cell electrophoretic mobility data and on measured AFM force–separation distance curves.
Conclusions and perspectives
In this work, we address the interfacial properties of microalgae at various relevant scales of biological organization, from the population level via electrophoresis and proton titration experiments, down to the cellular and molecular scale by CFM techniques, as a function of environmental conditions including pH. Analysis of the electrophoretic features of C. vulgaris cells evidences a marked heterogeneity of the microalgae interface as electrolyte concentration and/or pH get lower, due to possible diffuse swelling of cell peripheral region and/or increase in cell surface heterogeneity (roughness) under acidic conditions. We further evidence that potentiometric proton titrations cannot provide quantitative information on cell double layer charging process as interfering biological processes largely contribute to proton charge balance at the cell/solution interface. Using functionalized AFM tips, the electrostatic, hydrophobic and H-bonds contributions to tip-to-cell adhesion features are evaluated, and connections (if relevant) between electrostatic descriptors of the algae interface derived from electrokinetics (population scale) and CFM (single cell and molecular scales) are discussed. CFM results further suggest that, depending on solution pH, electrostatics can dominate over hydrophobic and hydrogen-bonds contributions to the overall tip/cell interaction. Interestingly, CFM measurements collected with use of –CH3 and –COOH coated tips reveal the existence of spatialized cell wall (hydrophobic) patterns.While evidencing the multiscale heterogeneity of C. vulgaris interfaces (from the population to the single cell level, and over the surface of a given individual) and underlining the possible role(s) played by cell physiology in regulation of interfacial charges, our work provides insights into electrostatics and hydrophobicity features of C. vulgaris. The results may serve as a new basis for the interpretation of microalgae interactions with their ionic and/or particulate environment beyond approximate zeta-potential concept and DLVO theory in the framework of which particles are incorrectly viewed as hard and homogeneous systems. We believe that such fundamental understanding of the interfacial properties governing cell behaviour would contribute to the improvement of industrial or environmental exploitation of microalgal resources.
Material and methods
Culture of the microalgae
Chlorella vulgaris (C211-11B) were cultivated in 250 mL beakers (corked with air-filter cap) containing 100 mL of Lefebre-Czarda (LC) medium, inside an incubator Innova 42 (Eppendorf) thermostated at 23 °C, under day/night cycle of 16 h/8 h under permanent agitation at 94 rpm. The cell density was controlled via measurement of the optical density (OD) using spectrometer UV-2501PC (Shimadzu). From cell counting experiments, we determined that an OD value of unity at an absorption wavelength of 686.5 nm corresponds to 2.47 × 107 cells per millilitre. The microalgae used for all measurements in this work were harvested at 6 days of growth, during the mid-log growth phase.Electrophoresis
The electrophoretic mobility of Chlorella vulgaris (C211-11B) microalgae was measured as a function of pH (4, 6.2 and 9) and concentration of NaNO3 (Sigma-Aldrich, purity >99%) in the range 1 mM to 250 mM at room temperature using a Zetaphoremeter IV device (CAD Instruments). Prior to measurements, cells were washed twice by centrifugation–resuspension (720g for 6 min) in 10 mM NaNO3. Further dilution by ultrapure water or salt addition were made to obtain NaNO3 solution at the desired concentrations, with a final OD686.5nm value of 0.07. The cell density was chosen in order to optimize the measurement statistics of the electrophoretic mobility distribution of the cells in the different conditions adopted in this work. pH values were adjusted by proper addition of HNO3 (0.1 M, Titrapur, Sigma-Aldrich) and NaOH (0.1 M, Carl Roth) solutions. Each reported data point for a given NaNO3 concentration is the average of 6 mobility acquisitions on 3 different batches of microalgae per tested pH condition, with one replicate per batch.Potentiometric proton titrations
Chlorella vulgaris (C211-11B) microalgae were titrated at different NaNO3 concentrations in a closed container using a TITRANDO 809 (Metrohm) controlled by tiamo 2.4 software. 20 mL of microalgae culture suspension were harvested after 6 days of growth, centrifugated during 6 min at 720g using Centrifuge 5804 R (Eppendorf) and rinsed with 30 mL NaNO3 (10 mM) solution. Centrifugation and rinsing were repeated to get rid of LC growth medium. From the rinsed cell suspension, we prepared a 40 mL dilution in NaNO3 10 mM at pH 3.5 defined by a cell density of 0.7. The sequential titration process consisted into 5 successive titrations performed on a given microalgae sample, under light or dark conditions, in thermostated environment at 5 °C or 25 °C, and under a permanent flux of argon to avoid external sample contamination by carbon dioxide. The first titration in NaNO3 10 mM was made by addition of NaOH 10 mM (Carl Roth) until pH value stabilised to 10.5. The electrolyte concentration was then increased to 30 mM by addition of 1 M NaNO3 (Sigma-Aldrich) while maintaining pH to 10.5. The second and third titrations corresponded to a backward titration from pH 10.5 to 3.5 upon addition of 10 mM HNO3 (Titrapur, Sigma-Aldrich), and to the forward titration from pH 3.5 to 10.5, before a new adjustment of electrolyte concentration to 100 mM. The fourth and fifth titration then followed, from pH 10.5 to 3.5 and pH 3.5 to 10.5, respectively. The pH range over which samples were titrated was chosen so as to lead to a complete (de)protonation of the –OH and –COOH chemical groups carried by chlorophyte microalgae surface.16,45 ‘Blank’ sequential titrations (i.e. in the absence of cells) were also performed following the above protocol in order to subtract the contribution from the electrolyte dispersing medium. The results displayed in Fig. 2 are the titrated charges collected on a single C. vulgaris batch sample by addition of NaOH at 10 mM, 30 mM and 100 mM NaNO3 concentration after subtracting the background electrolyte contribution measured from corresponding ‘blank’ experiments.Preparation of microalgae for AFM measurement
C. vulgaris were harvested after 6 days of cultivation in LC medium. After 6 min at 720g centrifugation using centrifuge 5804 R (Eppendorf), the microalgae samples were rinsed in NaNO3 (10 mM) solution buffered by MES (1 mM), at pH 4.5 or 6.2 (depending on the pH condition tested). 2 cm × 2 cm rectangular glass slides were incubated for 45 min in RBS-25 detergent (0.1%) at 60 °C, rinsed abundantly with ultrapure-water, dry with N2, and finally incubating in PEI (0.1%, Sigma Mw = 750![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) solution during 20 min. After rinsing the PEI-coated substrate, a drop of 1 mL NaNO3-microalgae suspension was deposited during approximately 15 min, allowing time for the microalgae to adhere on the surface of the substrate. Finally, the glass slides covered by microalgae were rinsed with NaNO3 solution at the ionic strength and pH value tested.
000 g mol−1) solution during 20 min. After rinsing the PEI-coated substrate, a drop of 1 mL NaNO3-microalgae suspension was deposited during approximately 15 min, allowing time for the microalgae to adhere on the surface of the substrate. Finally, the glass slides covered by microalgae were rinsed with NaNO3 solution at the ionic strength and pH value tested.
Preparation of thiol coated AFM tips
Oxide-sharpened microfabricated silicon-nitride cantilevers with gold coating (NPG-10, Bruker Corporation) were used and their spring constants (of nominal values 0.06 N m−1) were accurately determined on the basis of the thermal noise method.77 Prior to functionalisation, AFM tips were cleaned for 5 minutes by UV–ozone treatment, rinsed in ethanol and dried with N2. To perform amine tip functionalisation, tips were immersed for 2 hours in a 20 mM cysteamine thiol solution in 0.1 M MES buffer and rinsed twice in NaNO3 solution. To perform carboxyl tip functionalisation, tips were immersed overnight in a 1 mM 16-mercaptohexadenoic acid (16-MHDA) solution in ethanol absolute anhydrous and rinsed with ethanol. To perform methyl tip functionalisation, tips were immersed overnight in a 1 mM dodecanethiol solution in absolute anhydrous ethanol and rinsed with ethanol.Atomic force microscopy measurements
AFM force-volume measurements and contact imaging were performed at room temperature using a dimension ICON set up (Bruker Corporation) with Nanoscope operation software (Bruker Corporation). In Fig. 3a and b, peak-force measurements were performed to provide topographic maps (5 μm × 5 μm and 1.5 μm × 1.5 μm) of C. vulgaris surfaces, using silicon-nitride cantilevers without coating. Acquiring larger images after the 1.5 μm-image confirmed that the set-up was not drifting.Concerning force spectroscopy measurements, prior to all force-maps acquisitions, images were taken with bare tips to check the state of the cells. Then, the bare tip was replaced by a functionalized tip, and only very low-resolution images with a minimum amount of scan lines were collected with the functionalized tip to locate the cell before rapidly switching to force spectroscopy measurements. Force–separation distance curves for interacting thiol-coated tips/microalgae were obtained in NaNO3 solution (10 mM), buffered with MES (1 mM) at pH 4.5 and pH 6.2. For statistical analysis purpose, at least two tips were used per microalgae sample, and cells from several C. vulgaris batches were probed per pH- and tip coating-condition. For each pH condition and tip coating tested, adhesion maps were obtained by recording multiple (32 × 32 pixels) force–distance curves on 1.5 μm × 1.5 μm areas of microalgae. No gradual decrease/increase of adhesion appeared during the acquisition of a given map (especially following the scan direction), which could have indicated tip contamination. Additionally, no particular evolution (neither decrease nor increase) in the frequency of adhesion events was observed between successive maps, and the different sets of profiles were randomly obtained with different functionalized tips independently of the scanning order.
Unless otherwise stated, all force curves were obtained using an applied force of 500 pN and approach and retraction speeds of 1 μm s−1 with a ramp size between 300 and 500 nm. Control measurements performed with tip-COOH or tip-CH3, and –COOH/–CH3 gold coated silicon wafers were performed at pH 6.2 and 4.5 (Fig. S6†).
Author contributions
Nicolas Lesniewska: methodology, software, formal analysis, validation, investigation, writing – original draft. Jérôme F. L. Duval: conceptualization, software, investigation, formal analysis, writing – review & editing, supervision. Céline Caillet: methodology, writing – review & editing. Angelina Razafitianamaharavo: methodology, software, writing – review & editing. José P. Pinheiro: methodology, investigation, software, writing – review & editing. Isabelle Bihannic: investigation, writing – review & editing. Renaud Gley: methodology, technical support, writing – review. Hélène Le Cordier: methodology, technical support, writing – review & editing. Varun Vyas: methodology, technical support, writing – review & editing. Christophe Pagnout: writing – review. Bénédicte Sohm: methodology, investigation, writing – review. Audrey Beaussart: conceptualization, methodology, software, formal analysis, investigation, writing – review & editing, supervision.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by ANR grant ANR-20-CE34-0005-01 to AB. This work was partly done with resources from the Pôle de Compétences en Physico-Chimie de l'Environnement as well as the Pôle de Compétences en Biologie Environnementale, ANATELo, LIEC Laboratory, UMR 7360 CNRS – Université de Lorraine.References
- J. A. Garrido-Cardenas, F. Manzano-Agugliaro, F. G. Acien-Fernandez and E. Molina-Grima, Algal Res., 2018, 35, 50–60 CrossRef.
- J. Milano, H. C. Ong, H. H. Masjuki, W. T. Chong, M. K. Lam, P. K. Loh and V. Vellayan, Renewable Sustainable Energy Rev., 2016, 58, 180–197 CrossRef.
- A. Raheem, P. Prinsen, A. K. Vuppaladadiyam, M. Zhao and R. Luque, J. Cleaner Prod., 2018, 181, 42–59 CrossRef CAS.
- M. F. De Jesus Raposo, R. M. S. C. De Morais and A. M. M. B. De Morais, Mar. Drugs, 2013, 11, 233–252 CrossRef PubMed.
- J. Benemann, Energies, 2013, 6, 5869–5886 CrossRef.
- T. D. P. Nguyen, M. Frappart, P. Jaouen, J. Pruvost and P. Bourseau, Environ. Technol., 2014, 35, 1378–1388 CrossRef CAS PubMed.
- T. Ndikubwimana, X. Zeng, N. He, Z. Xiao, Y. Xie, J. S. Chang, L. Lin and Y. Lu, Biochem. Eng. J., 2015, 101, 160–167 CrossRef CAS.
- N. Fayad, T. Yehya, F. Audonnet and C. Vial, Algal Res., 2017, 25, 1–11 CrossRef.
- S. Li, T. Hu, Y. Xu, J. Wang, R. Chu, Z. Yin, F. Mo and L. Zhu, Renewable Sustainable Energy Rev., 2020, 131, 110005 CrossRef CAS.
- F. Wang, W. Guan, L. Xu, Z. Ding, H. Ma, A. Ma and N. Terry, Appl. Sci., 2019, 9, 1534 CrossRef CAS.
- C. M. Monteiro, S. C. Fonseca, P. M. L. Castro and F. X. Malcata, J. Appl. Phycol., 2011, 23, 97–103 CrossRef CAS.
- Â. Almeida, J. Cotas, L. Pereira and P. Carvalho, Phycology, 2023, 3, 186–201 CrossRef.
- A. Abdelfattah, S. S. Ali, H. Ramadan, E. I. El-Aswar, R. Eltawab, S. H. Ho, T. Elsamahy, S. Li, M. M. El-Sheekh, M. Schagerl, M. Kornaros and J. Sun, Environ. Sci. Ecotechnology, 2023, 13, 100205 CrossRef CAS PubMed.
- Y. K. Leong and J. S. Chang, Bioresour. Technol., 2020, 303, 122886 CrossRef CAS PubMed.
- R. K. Goswami, K. Agrawal, M. P. Shah and P. Verma, Lett. Appl. Microbiol., 2022, 75, 701–717 CrossRef CAS PubMed.
- S. Hadjoudja, V. Deluchat and M. Baudu, J. Colloid Interface Sci., 2010, 342, 293–299 CrossRef CAS PubMed.
- A. Ozkan and H. Berberoglu, Colloids Surf., B, 2013, 112, 287–293 CrossRef CAS PubMed.
- R. Soto-Ramírez, M. G. Lobos, O. Córdova, P. Poirrier and R. Chamy, J. Hazard. Mater., 2021, 411, 125059 CrossRef PubMed.
- J. F. L. Duval, A. Razafitianamaharavo, I. Bihannic, M. Offroy, N. Lesniewska, B. Sohm, H. Le Cordier, C. Mustin, C. Pagnout and A. Beaussart, Algal Res., 2023, 69, 102955 CrossRef.
- L. Xia, R. Huang, Y. Li, S. Song and M. D. Lambreva, PLoS One, 2017, 12, e0186434 CrossRef PubMed.
- F. Yang, W. Xiang, J. Fan, H. Wu, T. Li and L. Long, J. Appl. Physiol., 2016, 28, 747–756 CAS.
- M. Castrillo, L. M. Lucas-Salas, C. Rodríguez-Gil and D. Martínez, Bioresour. Technol., 2013, 128, 324–329 CrossRef CAS PubMed.
- L. Pérez, J. L. Salgueiro, R. Maceiras, Á. Cancela and Á. Sánchez, Biomass Bioenergy, 2017, 97, 20–26 CrossRef.
- J. A. Gerde, L. Yao, J. Y. Lio, Z. Wen and T. Wang, Algal Res., 2014, 3, 30–35 CrossRef.
- I. Demir, J. Blockx, E. Dague, P. Guiraud, W. Thielemans, K. Muylaert and C. Formosa-Dague, ACS Appl. Bio Mater., 2021, 3, 8446–8459 CrossRef PubMed.
- Y. Li, L. Xia, R. Huang, C. Xia and S. Song, RSC Adv., 2017, 7, 34600–34608 RSC.
- C. Wei, Y. Huang, Q. Liao, A. Xia, X. Zhu and X. Zhu, Bioresour. Technol., 2020, 304, 123012 CrossRef CAS PubMed.
- K. Xu, Y. Li, X. Zou, H. Wen, Z. Shen and X. Ren, Biochem. Eng. J., 2018, 137, 294–304 CrossRef CAS.
- J. F. L. Duval and F. Gaboriaud, Curr. Opin. Colloid Interface Sci., 2010, 15, 184–195 CrossRef CAS.
- P. P. Gopmandal and J. F. L. Duval, Curr. Opin. Colloid Interface Sci., 2022, 60, 101605 CrossRef CAS.
- H. Ohshima, Adv. Colloid Interface Sci., 1995, 62, 189–235 CrossRef CAS.
- K. Makino and H. Ohshima, Sci. Technol. Adv. Mater., 2011, 12, 023001 CrossRef PubMed.
- R. Bos, H. C. Van Der Mei and H. J. Busscher, Biophys. Chem., 1998, 74, 251–255 CrossRef CAS PubMed.
- R. J. Karreman, E. Dague, F. Gaboriaud, F. Quilès, J. F. L. Duval and G. G. Lindsey, Biochim. Biophys. Acta, Proteins Proteomics, 2007, 1774, 131–137 CrossRef CAS PubMed.
- P. A. Gomes, J.-B. d'Espinose de Lacaillerie, B. Lartiges, M. Maliet, V. Molinier, N. Passade-Boupat and N. Sanson, Langmuir, 2022, 38, 14044–14052 CrossRef CAS PubMed.
- J. F. L. Duval, J. Merlin and P. A. L. Narayana, Phys. Chem. Chem. Phys., 2011, 13, 1037–1053 RSC.
- F. Pillet, E. Dague, J. Pečar Ilić, I. Ružić, M. P. Rols and N. Ivošević DeNardis, Bioelectrochemistry, 2019, 127, 154–162 CrossRef CAS PubMed.
- H. Yuan, X. Zhang, Z. Jiang, X. Wang, X. Chen, L. Cao and X. Zhang, Colloids Surf., B, 2019, 177, 479–486 CrossRef CAS PubMed.
- M. Laviale, A. Beaussart, J. Allen, F. Quilès and S. El-Kirat-Chatel, ACS Appl. Mater. Interfaces, 2019, 11, 48574–48582 CrossRef CAS PubMed.
- I. Demir-Yilmaz, P. Guiraud and C. Formosa-Dague, Algal Res., 2021, 60, 102506 CrossRef.
- K. Suresh Kumar, H. U. Dahms, E. J. Won, J. S. Lee and K. H. Shin, Ecotoxicol. Environ. Saf., 2015, 113, 329–352 CrossRef CAS PubMed.
- J. F. L. Duval and H. Ohshima, Langmuir, 2006, 22, 3533–3546 CrossRef CAS PubMed.
- D. V. Vezenov, A. Noy and P. Ashby, J. Adhes. Sci. Technol., 2005, 19, 313–364 CrossRef CAS.
- M. Yamamoto, M. Fujishita, A. Hirata and K. Shigeyuki, J. Plant Res., 2004, 117, 257–264 Search PubMed.
- P. H. Baudelet, G. Ricochon, M. Linder and L. Muniglia, Algal Res., 2017, 25, 333–371 CrossRef.
- I. Demir-Yilmaz, M. Schiavone, J. Esvan, P. Guiraud and C. Formosa-Dague, Algal Res., 2023, 72, 103102 CrossRef.
- A. Beaussart, C. Beloin, J. M. Ghigo, M. P. Chapot-Chartier, S. Kulakauskas and J. F. L. Duval, Nanoscale, 2018, 10, 12743–12753 RSC.
- A. Beaussart, C. Caillet, I. Bihannic, R. Zimmermann and J. F. L. Duval, Nanoscale, 2018, 10, 3181–3190 RSC.
- C. Pagnout, R. M. Présent, P. Billard, E. Rotureau and J. F. L. Duval, Sens. Actuators, B, 2018, 270, 482–491 CrossRef CAS.
- H. Ohshima, Electrophoresis, 2006, 27, 526–533 CrossRef CAS PubMed.
- J. R. S. Martin, I. Bihannic, C. Santos, J. P. S. Farinha, B. Demé, F. A. M. Leermakers, J. P. Pinheiro, E. Rotureau and J. F. L. Duval, Langmuir, 2015, 31, 4779–4790 CrossRef CAS PubMed.
- M. Moussa, C. Caillet, R. M. Town and J. F. L. Duval, Langmuir, 2015, 31, 5656–5666 CrossRef CAS PubMed.
- J. P. Pinheiro, E. Rotureau and J. F. L. Duval, J. Colloid Interface Sci., 2021, 583, 642–651 CrossRef CAS PubMed.
- J. A. Raven, Ann. Bot., 1976, 40, 587–602 CrossRef CAS.
- M. Lavoie, J. F. L. Duval, J. A. Raven, F. Maps, B. Béjaoui, D. J. Kieber and W. F. Vincent, Environ. Sci. Technol., 2018, 52, 9403–9411 CrossRef CAS PubMed.
- K. A. Gehl and B. Colman, Plant Physiol., 1985, 77, 917–921 CrossRef CAS PubMed.
- S. Ihnken, J. Beardall, J. C. Kromkamp, C. G. Serrano, M. A. Torres, J. Masojídek, I. Malpartida, R. Abdala, C. G. Jerez, J. R. Malapascua, E. Navarro, R. M. Rico, E. Peralta, J. P. F. Ezequil and F. L. Figueroa, Aquat. Biol., 2014, 22, 95–110 CrossRef.
- A. E. Lane and J. E. Burris, Plant Physiol., 1981, 68, 439–442 CrossRef CAS PubMed.
- S. Boussiba, W. Bing, J.-P. Yuan, A. Zarka and F. Chen, Biotechnol. Lett., 1999, 21, 601–604 CrossRef CAS.
- J. Masojídek, G. Torzillo, J. K. Kopecký, M. Koblížek, L. Nidiaci, J. Komenda, A. Lukavská and A. Sacchi, J. Appl. Phycol., 2000, 12, 417–426 CrossRef.
- Z. Pavlinska, D. Chorvat, A. Mateasik, M. Jerigova, D. Velic, N. Ivošević DeNardis and A. Marcek Chorvatova, J. Biotechnol., 2020, 324S, 100018 CrossRef PubMed.
- A. Marcek Chorvatova, M. Uherek, A. Mateasik and D. Chorvat, Methods Appl. Fluoresc., 2020, 8, 024007 CrossRef CAS PubMed.
- D. Alsteens, E. Dague, P. G. Rouxhet, A. R. Baulard and Y. F. Dufrêne, Langmuir, 2007, 23, 11977–11979 CrossRef CAS PubMed.
- E. Dague, D. Alsteens, J. P. Latgé, C. Verbelen, D. Raze, A. R. Baulard and Y. F. Dufrêne, Nano Lett., 2007, 7, 3026–3030 CrossRef CAS PubMed.
- M. E. McConney, S. Singamaneni and V. V. Tsukruk, Polym. Rev., 2010, 50, 235–286 CrossRef CAS.
- M. I. Giannotti and G. J. Vancso, ChemPhysChem, 2007, 8, 2290–2307 CrossRef CAS PubMed.
- P. R. Laskowski, M. Pfreundschuh, M. Stauffer, Z. Ucurum, D. Fotiadis and D. J. Müller, ACS Nano, 2017, 11, 8292–8301 CrossRef CAS PubMed.
- V. Molinero and E. J. Calvo, J. Electroanal. Chem., 1998, 445, 17–25 CrossRef CAS.
- A. Beaussart, T. C. Ngo, S. Derclaye, R. Kalinova, R. Mincheva, P. Dubois, P. Leclère and Y. F. Dufrêne, Nanoscale, 2014, 6, 565–571 RSC.
- R. Schweiss, C. Werner and W. Knoll, J. Electroanal. Chem., 2003, 540, 145–151 CrossRef CAS.
- F. Ahimou, F. A. Denis, A. Touhami and Y. F. Dufrêne, Langmuir, 2002, 18, 9937–9941 CrossRef CAS.
- C. Dicket and G. Hähner, J. Am. Chem. Soc., 2002, 124, 12619–12625 CrossRef PubMed.
- S. Hamla, P. Y. Sacré, A. Derenne, B. Cowper, E. Goormaghtigh, P. Hubert and E. Ziemons, Spectrochim. Acta, Part A, 2021, 262, 120109 CrossRef PubMed.
- T. Fujimoto and R. G. Parton, Cold Spring Harbor Perspect. Biol., 2011, 3, 1–17 Search PubMed.
- I. Levental and S. L. Veatch, J. Mol. Biol., 2016, 428, 4749–4764 CrossRef CAS PubMed.
- F. Gaboriaud, M. L. Gee, R. Strugnell and J. F. L. Duval, Langmuir, 2008, 24, 10988–10995 CrossRef CAS PubMed.
- J. te Riet, A. J. Katan, C. Rankl, S. W. Stahl, A. M. van Buul, I. Y. Phang, A. Gomez-Casado, P. Schön, J. W. Gerritsen, A. Cambi, A. E. Rowan, G. J. Vancso, P. Jonkheijm, J. Huskens, T. H. Oosterkamp, H. Gaub, P. Hinterdorfer, C. G. Figdor and S. Speller, Ultramicroscopy, 2011, 111, 1659–1669 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04740g |
| ‡ Current address: Department of Biotechnology, School of Engineering and Applied Sciences, Bennett University, Greater Noida, India. |
| § Current address: Univ. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, F-33600 Pessac, France. |
| This journal is © The Royal Society of Chemistry 2024 |