Open Access Article
Open Access ArticlePhytochemical and pharmacological properties of the genus Alpinia from 2016 to 2023
Isoo
Youn
 a,
Ah-Reum
Han
b,
Donglan
Piao
a,
Hwaryeong
Lee
a,
Hyunkyung
Kwak
a,
Yeju
Lee
a,
Joo-Won
Nam
a,
Ah-Reum
Han
b,
Donglan
Piao
a,
Hwaryeong
Lee
a,
Hyunkyung
Kwak
a,
Yeju
Lee
a,
Joo-Won
Nam
 c and
Eun Kyoung
Seo
c and
Eun Kyoung
Seo
 *a
*a
aGraduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea. E-mail: yuny@ewha.ac.kr
bAdvanced Radiation Technology Institute, Korea Atomic Energy Research Institute (KAERI), Jeongeup-si, Jeollabuk-do 56212, Republic of Korea
cCollege of Pharmacy, Yeungnam University, Gyeongsan-si, Gyeongsangbuk-do 38541, Republic of Korea
First published on 8th May 2024
Abstract
Covering 2016 up to the end of 2023
Alpinia is the largest genus of flowering plants in the ginger family, Zingiberaceae, and comprises about 500 species. Many Alpinia are commonly cultivated ornamental plants, and some are used as spices or traditional medicine to treat inflammation, hyperlipidemia, and cancers. However, only a few comprehensive reviews have been published on the phytochemistry and pharmacology of this genus, and the latest review was published in 2017. In this review, we provide an extensive coverage of the studies on Alpinia species reported from 2016 through 2023, including newly isolated compounds and potential biological effects. The present review article shows that Alpinia species have a wide spectrum of pharmacological activities, most due to the activities of diarylheptanoids, terpenoids, flavonoids, and phenolics.
1. Introduction
The genus Alpinia in the Zingiberaceae family is large, comprising more than 500 species. According to the work of Ma et al., phytochemical and pharmacological data are available for only 35 Alpinia species:40A. blepharocalyx K.Schum, A. bracteata Roscoe, A. calcarata (Andrews) Roscoe, A. chinensis (Retz.) Roscoe, A. conchigera Griff., A. coriandriodora D. Fang, A. densispicata Hayata, A. densibracteata T. L. Wu and S. J. Chen, A. elegans (C.Presl) K.Schum., A. eremochlamys K.Schum., A. flabellata Ridl., A. formosana K.Schum., A. gagnepainii K.Schum., A. galanga (L.) Willd., A. intermedia Gagnep., A. japonica (Thunb.) Miq., A. katsumadae Hayata, A. hainanensis K.Schum., A. malaccensis (Burm.f.) Roscoe, A. mutica Roxb., A. nantoensis F. Y. Lu and Y. W. Kuo, A. nigra (Gaertn.) Burtt, A. nutans (L.) Roscoe, A. officinarum Hance, A. oxyphylla Miq., A. pahangensis Ridl., A. pinnanensis T. L. Wu and S. J. Chen, A. platychilus K.Schum., A. pricei Hayata, A. purpurata (Vieill.) K.Schum., A. rafflesiana Wall. ex Baker, A. sichuanensis Z. Y. Zhu, A. tonkinensis Gagnep., A. zerumbet (Pers.) B. L. Burtt and R. M. Sm, and A. speciosa (J.C.Wendl.) K.Schum.The rhizomes of A. officinarum (Yang Gang), seeds of A. officinarum (Hong Du Gu), seeds of A. katsumadae (Cho Du Gu), and fruits of A. oxyphylla (Ik Ji, bitter cardamon) have been used in traditional medicine to treat indigestion, emesis, and stomach ache.43,44Alpinia species are rich in diarylheptanoids, terpenoids, flavonoids, and phenolic compounds and have shown cytotoxic, anti-inflammatory, antioxidant, and anti-bacterial activities.40,47 In Korea, there have been 95 studies on Alpinia species since 1997, and the most investigated species are A. officinarum (39 cases), A. katsumadae (25 cases), and A. oxyphylla (22 cases). Biological studies have been performed, with analyses of the cytotoxicity, anti-inflammation, and anti-oxidant activity of these species. In the studies performed in Korea, most of the newly found compounds are diarylheptanoids from A. katsumadae and A. officinarum.
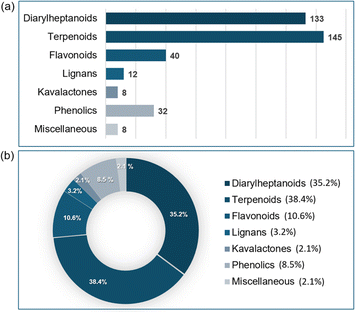
Previous reviews have summarized phytochemistry reports of more than 500 active compounds from the genus Alpinia.40,47,48 However, no reviews have been reported since 2017, though many studies have been performed since then. Here we describe newly isolated and known compounds from the Alpinia species with pharmacological activity reported in 148 references published from 2016 to 2023. This review includes 378 compounds from the Alpinia species, comprising 133 diarylheptanoids, 145 terpenoids, 40 flavonoids, 12 lignans, 8 kavalactones, 32 phenolics, and others (Fig. 1a). The percentage of each class compared with the total number of compounds is shown in Fig. 1b. Moreover, the biological activities of frequently used compounds are reported. A literature search was conducted using online databases (PubMed, SciFinder, and Google scholar), and publications that underwent peer-review and in journals with specific impact factors were considered for this review. Scientific names were validated via the “International Plant Names Index” (https://www.ipni.org/).
2. Phytochemical investigation
2.1. Diarylheptanoids
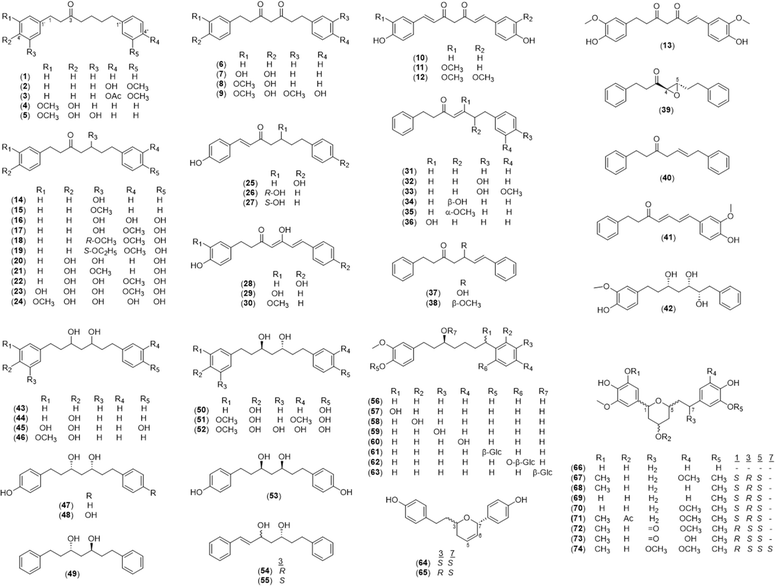
Here, we discuss the structures of diarylheptanoid monomers, dimers, and hybrid conjugates with other structural classes. A total of 133 diarylheptanoids has been identified in Alpinia species since 2016.| No. | Name | No. | Name |
|---|---|---|---|
| 1 | 1,7-Diphenylheptan-3-one25 | 38 | (5S)-5-Methoxy-1,7-diphenylhepta-6-en-3-one22 |
| 2 | 7-(4′′-Hydroxy-3′′-methoxyphenyl)-1-phenylheptan-3-one41 | 39 | trans-(4R,5S)-Epoxy-1,7-diphenylheptan-3-one45 |
| 3 | 7-(4′′-Actetoxy-3′′-methoxy phenyl)-1-phenylheptan-3-one41 | 40 | 1,7-Diphenylhept-5-en-3-one45 |
| 4 | Yakuchinone A49,50 | 41 | (4E,6E)-7-(4′′-Hydroxy-3′′-methoxyphenyl)-1-phenylhepta-4,6-dien-3-one45 |
| 5 | 5′-Hydroxyyakuchinone A50 | 42 | (3S,5S,6S)-1-(4′-Hydroxy-3′-methoxyphenyl)-7-phenylheptane-3,5,6-triol25 |
| 6 | 1,7-Diphenylheptane-3,5-dione25 | 43 | 1,7-Diphenylheptane-3,5-diol25,68 |
| 7 | 1-(3′,4′-Dihydroxyphenyl)-7-phenylheptane-3,5-dione25 | 44 | 1-(4′-Hydroxyphenyl)-7-phenylheptane-3,5-diol25 |
| 8 | 1-(4′-Hydroxy-3′-methoxyphenyl)-7-phenylheptane-3,5-dione25 | 45 | 1-(3′,4′-Dihydroxyphenyl)-7-(4′-hydroxyphenyl)heptane-3,5-diol69 |
| 9 | 1,7-Bis(4-hydroxy-3-methoxyphenyl)heptane-3,5-dione25 | 46 | 1-(4′-Hydroxy-3′-methoxyphenyl)-7-phenylheptane-3,5-diol25 |
| 10 | (1E,6E)-1,7-Bis(4-hydroxyphenyl)hepta-1,6-diene-3,5-dione25 | 47 | (3S,5S)-Alpinikatin65 |
| 11 | (1E,6E)-1-(4′-Hydroxy-3′-methoxyphenyl)-7-(4′′-hydroxyphenyl)hepta-1,6-diene-3,5-dione25 | 48 | (+)-Hannokinol66 |
| 12 | (1E,6E)-1,7-Bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione22,23,25 | 49 | (3S,5R)-1,7-Diphenylheptane-3,5-diol66,68 |
| 13 | (E)-1,7-Bis(4-hydroxy-3-methoxyphenyl)hept-1-ene-3,5-dione25 | 50 | meso-Hannokinol66 |
| 14 | 5-Hydroxy-1,7-diphenylheptan-3-one16,20,41,68,69 | 51 | (3R,5S)-1,7-Bis(4-hydroxy-3-methoxyphenyl)heptane-3,5-diol25 |
| 15 | 5-Methoxy-1,7-diphenylheptan-3-one45 | 52 | Alpinin B69 |
| 16 | 5-Hydroxy-7-(3′′,4′′-dihydroxyphenyl)-1-phenyheptan-3-one41,69 | 53 | (3R,5R)-1,7-Bis(4-hydroxyphenyl)heptane-3,5-diol25 |
| 17 | 5-Hydroxy-7-(4′′-hydroxy-3′′-methoxyphenyl)-1-phenylheptan-3-one16,20,21,25,41,68,69 | 54 | (3R,5S)-trans-3,5-Dihydroxy-1,7-diphenyl-1-heptene66 |
| 18 | (5R)-5-Methoxy-7-(4′′-hydroxy-3′′-methoxyphenyl)-1-phenylheptan-3-one16,21,68 | 55 | (3S,5S)-trans-3,5-Dihydroxy-1,7-diphenyl-1-heptene66 |
| 19 | (5S)-5-Ethoxy-7-(4′′-hydroxy-3′′-methoxyphenyl)-1-phenylheptan-3-one65 | 56 | (3S)-Oxyphyllacinol49 |
| 20 | 5-Hydroxy-1,7-bis(4-hydroxyphenyl)heptan-3-one25 | 57 | (3S)-7-Hydroxyoxyphyllacinol49 |
| 21 | 5-Methoxy-1,7-bis(4-hydroxyphenyl)heptan-3-one25 | 58 | (3S)-2′′-Hydroxyoxyphyllacinol49 |
| 22 | 5-Hydroxy-1-(4′-hydroxyphenyl)-7-(4′′-hydroxy-3′′-methoxyphenyl)heptan-3-one25 | 59 | (3S)-3′′-Hydroxyoxyphyllacinol49 |
| 23 | 5-Hydroxy-1-(3′,4′-dihydroxyphenyl)-7-(4′′-hydroxy-3′′-methoxyphenyl)heptan-3-one25 | 60 | (3S)-4′′-Hydroxyoxyphyllacinol49 |
| 24 | 5-Hydroxy-1-(4′-hydroxy-3′-methoxyphenyl)-7-(3′′,4′′-dihydroxyphenyl)heptan-3-one69 | 61 | (3S)-Oxyphyllacinol-4′-O-β-D-glucopyranoside49 |
| 25 | (E)-1,7-Bis(4-hydroxyphenyl)hept-1-en-3-one22 | 62 | (3S)-2′′-Hydroxyoxyphyllacinol-2′′-O-β-D-glucopyranoside49 |
| 26 | (R)-4′′-Hydroxyashabushiketol65 | 63 | (3S)-Oxyphyllacinol-3-O-β-D-glucopyranoside49 |
| 27 | (S)-4′′-Hydroxyashabushiketol66 | 64 | (3S,7S)-5,6-Dehydro-4′′-de-O-methylcentrolobine65 |
| 28 | 1,2-Dihydro-bis(de-O-methyl)curcumin65 | 65 | (3R,7S)-5,6-Dehydro-4′′-de-O-methylcentrolobine66 |
| 29 | (4Z,6E)-5-Hydroxy-1-(3′,4′-dihydroxyphenyl)-7-phenylhepta-4,6-dien-3-one25 | 66 | Alpinin A69 |
| 30 | (4Z,6E)-5-Hydroxy-1-(4′-hydroxy-3′-methoxyphenyl)-7-phenylhepta-4,6-dien-3-one25,65 | 67 | Coriandralpinin A32 |
| 31 | 1,7-Diphenylhept-4-en-3-one8,16,25,41,45,68,69,96 | 68 | Coriandralpinin B32 |
| 32 | (4E)-7-(4-Hydroxyphenyl)-1-phenylhepten-3-one16,20,98 | 69 | Coriandralpinin C32 |
| 33 | (4E)-7-(4′′-Hydroxy-3′′-methoxyphenyl)-1-phenylhept-4-en-3-one20,21,68,99 | 70 | Coriandralpinin D32 |
| 34 | (4E)-6-Hydroxy-1,7-diphenylhept-4-en-3-one25 | 71 | Coriandralpinin E32 |
| 35 | (6S,4E)-6-Methoxy-1,7-diphenylhept-4-en-3-one25 | 72 | Coriandralpinin F32 |
| 36 | 5-Hydroxy-1,7-diphenylhept-4-en-3-one69 | 73 | Coriandralpinin G32 |
| 37 | 5-Hydroxy-1,7-diphenylhept-6-en-3-one25,68 | 74 | Coriandralpinin H32 |
 | ||
| Fig. 4 Diarylheptanoid conjugates with monoterpenes, sesquiterpenes, butyrovanillones, and flavones isolated from the Alpinia species. | ||
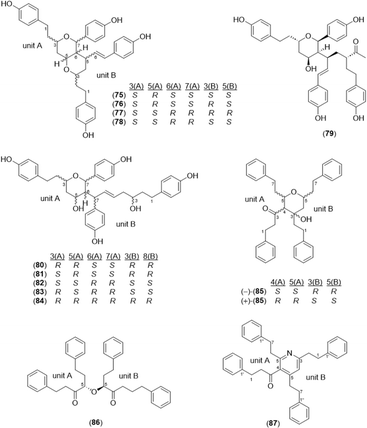
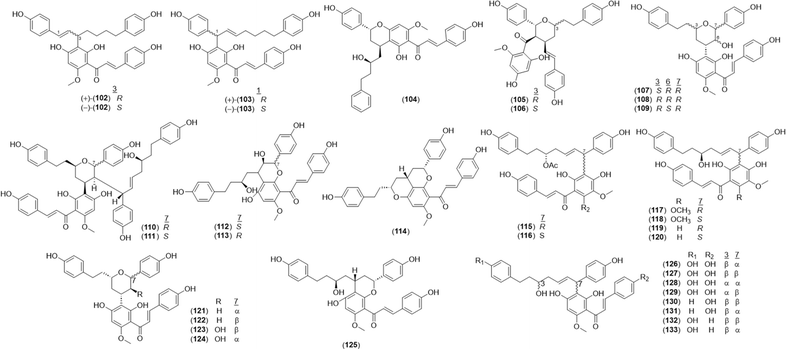
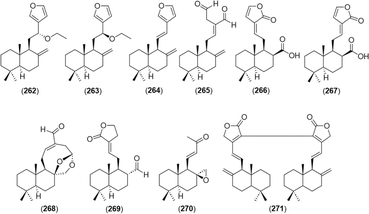
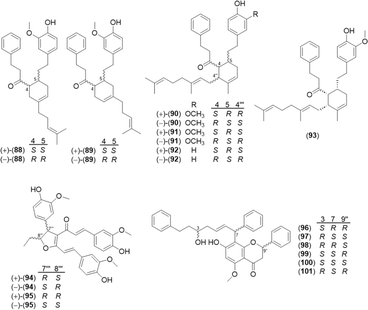
A total of 32 diarylheptanoid-chalcone hybrid conjugates (Fig. 5) was isolated in Alpinia. New structures alphainanins C and D (102 and 103) were isolated from A. katsumadae,22 as was katsumain H (104).82 In addition, 16 new diarylheptanoid–chalcone hybrids, katsumadainols A1–A16 (105–120), were found in A. katsumadae.83 In that same study, 13 known analogs were isolated: calyxin F (121), epi-calyxin F (122), (3S,5S,6S,7R)-6-hydroxycalyxin F (123), (3S,5S,6S,7S)-6-hydroxycalyxin F (124), calyxin L (125), epi-calyxin B (126), calyxin B (127), alpinnanin A (128), alpinnanin B (129), calyxin H (130), epi-calyxin H (131), katsumain C (132), and 7-epi-katsumain C (133).83
2.2. Terpenoids
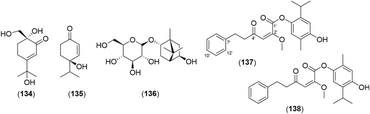
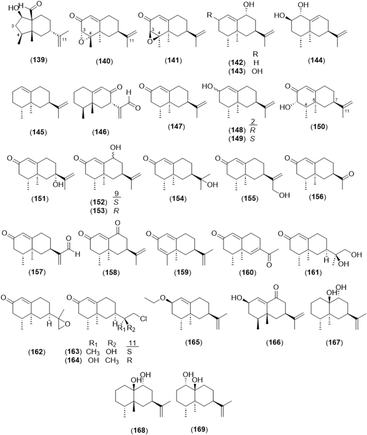
2.2.2.1 Eremophilane type. The basic skeleton of eremophilane-type sesquiterpenes comprises two six-membered rings and four methyl groups at C-4, 5, and 11.87 The eremophilane-type sesquiterpenoids from the genus Alpinia are presented in Fig. 7 and Table 2. Six newly found eremophilane-type sesquiterpenes were isolated from A. oxyphylla: alpinoxyphyllone C (139); (3S,4S,5R,7R)-eremophila-3,4-epoxy-1(10),11-dien-2-one (140); (3R,4R,5R,7R)-eremophila-3,4-epoxy-1(10),11-dien-2-one (141); (4R,5S,7S,9R)-eremophila-1(10),11-dien-9-ol (142); (2R,4R,5S,7S,9R)-eremophila-1(10),11-dien-2,9-diol (143); and (1R,2R,4R,5S,7R)-eremophila-9,11-dien-1,2-diol (144). In addition, nootkatone and its derivatives (147–155) with a hydroxyl group at various positions are shown. 2-O-Ethyl-β-nootkatol (165), which is a ethoxy substitute of β-nootkatone, was found in A. oxyphylla.28
| No. | Name |
|---|---|
| 139 | Alpinoxyphyllone C24 |
| 140 | (3S,4S,5R,7R)-Eremophila-3,4-epoxy-1(10),11-dien-2-one24 |
| 141 | (3R,4R,5R,7R)-Eremophila-3,4-epoxy-1(10),11-dien-2-one24 |
| 142 | (4R,5S,7S,9R)-Eremophila-1(10),11-dien-9-ol24 |
| 143 | (2R,4R,5S,7S,9R)-Eremophila-1(10),11-dien-2,9-diol24 |
| 144 | (1R,2R,4R,5S,7R)-Eremophila-9,11-dien-1,2-diol24 |
| 145 | Valencene51 |
| 146 | Nigriterpene F28 |
| 147 | Nootkatone3,24,61,62 |
| 148 | α-Nootkatone24 |
| 149 | β-Nootkatone3,24 |
| 150 | 3α-Hydroxynootkatone4 |
| 151 | 7α-Hydroxynootkatone3,24 |
| 152 | 9β-Hydroxynootkatone4,24 |
| 153 | Oxyphyllol B3,4,24 |
| 154 | 11-Hydroxyl-eremophilane-1(10)-en-2-one3,24 |
| 155 | 13-Hydroxynootkatone3,4,7,24 |
| 156 | Diketone3,24 |
| 157 | 12-Al-nootkatone24 |
| 158 | 11(12)-Dien-2,9-dione4,24 |
| 159 | 3,4-Dehydronootkatone4,24 |
| 160 | Oxyphyllanone A3,24 |
| 161 | Nootkatone-11,12-diol4 |
| 162 | Nootkatone-11,12-epoxide4 |
| 163 | (11S)-12-Chloronootkaton-11-ol4,24 |
| 164 | (11R)-12-Chloronootkaton-11-ol24 |
| 165 | 2-O-Ethyl-β-nootkatol28 |
| 166 | Oxyphyllin H3 |
| 167 | Alpinoxyphyllol A28 |
| 168 | Alpinoxyphyllol B28 |
| 169 | Oxyphyllol C24,28 |
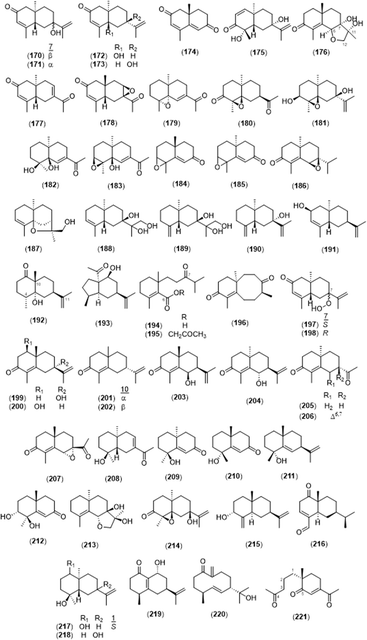
2.2.2.2 Eudesmane type. A total of 52 eudesmane sesquiterpenes was found in the Alpinia species and is summarized in Fig. 8 and Table 3. Oxyphyllins C–G (175–179) were newly isolated from A. oxyphylla,3,88 and oxyphyllin D (176) and oxyphyllol D (213) were eudesmane-12,6-olide structures found in A. oxyphylla.26 Compounds 193 and 196 were rare rearranged eudesmane skeletons with 5/6-fused and 6/8-fused bicyclic backbones, respectively.26 Compounds 194 and 195 were 6,7-secoeudesmane sesquiterpenoids isolated from a plant for the first time, and 221 was a 4,5-secoeudesmane sesquiterpenoid.26 7α-Hydroperoxy eudesma-3,11-diene-2-one (197) and 7β-hydroperoxy eudesma-3,11-diene-2-one (198) were hydrogen peroxide-substituted eudesmanes isolated from the dried fruits of A. oxyphylla.4 In addition, (4S,7S,9R)-14-nor-5(10),11(12)-dien-9-ol-1-one-eudesma (219) was a 14-nor sesquiterpenoid, which is rare in eudesmane sesquiterpenoids.42
| No. | Name |
|---|---|
| 170 | Teucrenone3,4 |
| 171 | 7-epi-Teucrenone3,4,7 |
| 172 | Teacrenone26 |
| 173 | α-Rotunol26,28 |
| 174 | Oxyphyllanene B3,26 |
| 175 | Oxyphyllin C3 |
| 176 | Oxyphyllin D3 |
| 177 | Oxyphyllin E3 |
| 178 | Oxyphyllin F3 |
| 179 | Oxyphyllin G3 |
| 180 | (4R,5R,7R,10R)-12-Noreudesma-4,5-epoxy-3-one26 |
| 181 | (3S,4S,5R,7R,10S)-Eudesma-4,5-epoxy-11-en-3,7-diol26 |
| 182 | (4S,5R,10S)-12-Noreudesma-6-en-4,5-diol-11-one26 |
| 183 | (3S,4S,5R,10R)-12-Noreudesma-3,4-epoxy-6-en-5-ol-11-one26 |
| 184 | (3R,4S,10R)-11,12,13-Trinoreudesma-3,4-epoxy-5-en-7-one26 |
| 185 | (3R,4S,10S)-11,12,13-Trinoreudesma-3,4-epoxy-5-en-7-one26 |
| 186 | (6S,7S,10R)-Eudesma-6,7-epoxy-4-en-3-one26 |
| 187 | (5R,7R,10S,11S)-Eudesma-5,11-epoxy-3-en-12-ol26 |
| 188 | (5S,7R,10S)-Eudesma-3-en-7,11,12-triol26 |
| 189 | (5R,7R,10S)-Eudesma-4(15)-en-7,11,12-triol26 |
| 190 | (5R,7S,10S)-Eudesma-4(15),11-dien-7-ol26 |
| 191 | (2R,5R,7S,10S)-Eudesma-4,11-dien-2-ol26 |
| 192 | (4R,5R,7R,10R)-Eudesma-11-en-5-ol-1-one26 |
| 193 | Epialpiniol26 |
| 194 | Alpinoxyphllaone A26 |
| 195 | Alpinoxyphllaone B26 |
| 196 | Neoxyphyllanene26 |
| 197 | 7α-Hydroperoxyeudesma-3,11-diene-2-one4 |
| 198 | 7β-Hydroperoxyeudesma-3,11-diene-2-one3,4 |
| 199 | (7S,10S)-11-Hydroxyeudesmane-4(5),11(12)-diene-3-one3,26 |
| 200 | Ligucyperonol3,26 |
| 201 | (−)-α-Cyperone26 |
| 202 | (+)-7-epi-α-Cyperone26 |
| 203 | (4aS,7S,8R)-8-Hydroxy-1,4a-dimethyl-7-(prop-1-en-2-yl)-4,4a,5,6,7,8-hexahydronaphthalen-2(3H)-one26 |
| 204 | (6S,7S,10R)-6-Hydroxyeudesmane-4(5),11(12)-diene-3-one3,28 |
| 205 | (10R)-13-Noreudesma-4,6-dien-3,11-dione26 |
| 206 | 7αH-12-Noreudesm-4-ene-3,11-dione26 |
| 207 | Oxyphyllanene C26 |
| 208 | Tephyllone26 |
| 209 | Teuhetenone A1,3,26 |
| 210 | (5E)-(4S,10R)-7-Oxo-trinoreudesm-5-en-4β-ol3,7 |
| 211 | Oxyphyllol A26 |
| 212 | (+)-Oxyphyllenone A26 |
| 213 | Oxyphyllol D26 |
| 214 | Oxyphyllol E26 |
| 215 | Isocyperol26 |
| 216 | 1-Oxo-5α,7αH-eudesm-3-en-15-al26 |
| 217 | Lairdinol A26 |
| 218 | Teucdiol B26 |
| 219 | (4S,7S,9R)-14-Nor-5(10),11(12)-dien-9-ol-1-one-eudesma42 |
| 220 | Litseagermacrane26 |
| 221 | Oxyphyllone A3,26 |
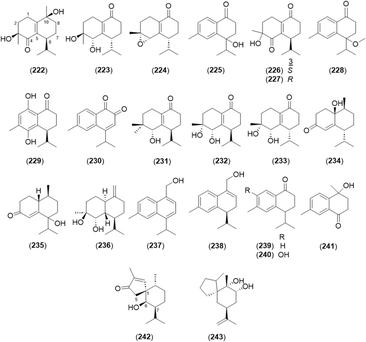
2.2.2.3 Cadinane type. Cadinanes constitute a large family of plant and fungal terpenes with characteristic decaline (cadinane or cadalane) scaffolds that result from the C-1/C-6 and C-5/C-10 cyclization of farnesyl pyrophosphate.89 In total, 22 cardinane-type sesquiterpenoids were isolated from A. oxyphylla, except 4-isopropyl-6-methyl-1-naphthalenemethanol (237) from A. officinarum (Fig. 9 and Table 4).
| No. | Name |
|---|---|
| 222 | Oxyphillin I3 |
| 223 | Oxyphyllin J3 |
| 224 | Oxyphyllone C24 |
| 225 | Oxyphyllone D24,28 |
| 226 | Oxyphyllone E24 |
| 227 | (3R)-Oxyphyllone E24 |
| 228 | Oxyphyllone I24 |
| 229 | Oxyphyllone J24 |
| 230 | Oxyphyllone K24 |
| 231 | (6R,9S,10S)-10-Hydroxyl-15-norcadinane-4(5)-ene-3-one3 |
| 232 | Oxyphyllenodiol A3,24 |
| 233 | Oxyphyllenodiol B7 |
| 234 | (−)-(1R,7S,10R)-1-Hydroxy-11-norcadinan-5-en-4-one28 |
| 235 | Oxyphyllenone H28 |
| 236 | Cadin-10(14)-en-4β,5α-diol24 |
| 237 | 4-Isopropyl-6-methyl-1-naphthalenemethanol73 |
| 238 | (7S)-Calacoren-14-ol24 |
| 239 | 3,4-Dihydro-6-methyl-4-(1-methylethyl)-l(2H)-naphthalenone24 |
| 240 | 2-Hydroxy-14-calamenenone24 |
| 241 | 4-Hydroxy-4,7-dimethyl-1-tetralone24 |
| 242 | Oxyspirone A24 |
| 243 | Oxyspirone B24 |
Oxyspirones A (242) and B (243) were rare spiro[5.6]-bearing rearranged cadinanes.24
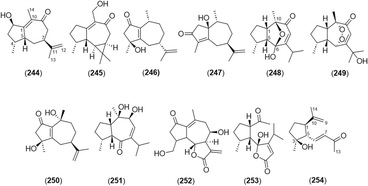
2.2.2.4 Guaiane type. A basic skeleton of guaiane-type sesquiterpene contains a five-membered ring, a seven-membered ring, two methyl groups at C-4 and 10, and an isopropyl group at C-7. A total of 11 guaiane-type sesquiterpenoids was found in this genus (Fig. 10 and Table 5). Among them, alpinenone (248) and 11-hydroxyisohanalpinone (249) have oxygen and peroxide bridges between C-6 and C-10, respectively.3,28,42 Oxyphyllin A (254) was a 7,9-seco-8,12-dinorguaiane-type sesquiterpenoid.3
| No. | Name |
|---|---|
| 244 | Oxyphyllin B3 |
| 245 | Aromadendr-1(10)-ene-9-one3 |
| 246 | (4S,7S,10R)-4-Hydroxyguaiane-1(5),11(12)-diene-2-one3 |
| 247 | (1S,7R,10R)-1-Hydroxyguaiane-4(5),11(12)-diene-3-one3 |
| 248 | Alpinenone3,42 |
| 249 | 11-Hydroxyisohanalpinone28 |
| 250 | Cyperusol A4 42 |
| 251 | [3R-(3α,3aβ,7β,8β,8aβ)]-2,3,3a,7,8,8a-Hexahydro-7,8-dihydroxy-3,8-dimethyl-5-(1-methylethyl)-4(1H)-azulenone42 |
| 252 | 3,4-Dihydrolactucin60 |
| 253 | 6-Hydroxyalpinolide28 |
| 254 | Oxyphyllin A3 |
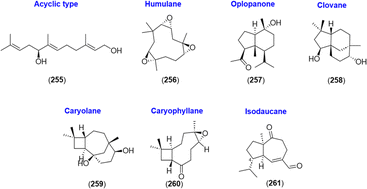
2.2.2.5 Others. Along with the reported subtypes of sesquiterpenoids, acyclic sesquiterpenoid (5S)-5-hydroxy-trans,trans-farnesol (255),66 humulane-type sesquiterpenoid (2S,3S,6R,7R,9S,10S)-humulene triepoxide (256),27 and an oplopanone-type sesquiterpenoid oplopanone (257)24 were isolated from A. katsumadae, A. galanga, and A. oxyphylla, respectively. In addition, four sesquiterpenes were isolated from A. galanga, namely, clovane-2β,9α-diol (258), caryolane-1,9β-diol (259), kobusone (260), and (−)-2-oxoisodauc-5-en-12-al (261).90 Kobusone (260) was a caryophyllene-derived epoxy ketone, and (−)-2-oxoisodauc-5-en-12-al (261) was an isodaucene sesquiterpenoid, which differs from the daucanes in position C-14 and instead attaches to C-4.91 The structures are shown in Fig. 11.
2.3. Flavonoids
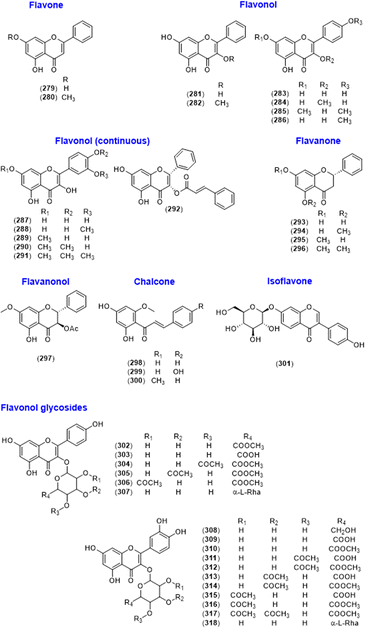
Flavonoids comprise one of the main classes of the genus Alpinia. Several flavonoid subtypes (flavone, flavonol, flavanone, flavanonol, isoflavone, chalcone, and flavonoid glycosides) were found in this genus (Fig. 14 and Table 6). Flavone derivatives chrysin (279)1,2 and tectochrysin (280)2,5,6 are rich in A. oxyphylla. Various flavonol-type structures were also found in the Alpinia genus. Galangin (281) is a major flavonol in A. officinarum and A. calcarata,8–23 and 3-methoxygalangin (282) was also isolated from A. officinarum.10,22 Several kaempferol/quercetin compounds with various functional groups were also found in this genus: kaempferol (283),18,23 3-methoxykaempferol (284),22 7,4′-dimethoxykaempferol (285),32 kaempferide (3,5,7-trihydroxy-4′-methoxyflavone) (286),8,10,11,14,16,18,20,21 quercetin (287),23 isorhamnetin (288),20,21 7-methoxyquercetin (289),32 7,4′-dimethoxyquercetin (290),32 and 7,3′,4′-trimethoxyquercetin (291).32 (2R,3S)-Pinobanksin-3-cinnamate (292) was isolated from A. galanga as a flavonol-type conjugated with a cinnamic acid.100 A flavanone derivative, pinocembrin (293), was found in A. zerumbet,20,53–55 and alpinetin (294) is the main active ingredient in A. katsumadae and A. mutica.56–59| No. | Name |
|---|---|
| 279 | Chrysin1,2 |
| 280 | Tectochrysin2,5,6 |
| 281 | Galangin8–23 |
| 282 | 3-Methoxygalangin10,22 |
| 283 | Kaempferol18,23 |
| 284 | 3-Methoxykaempferol22 |
| 285 | 7,4′-Dimethoxykaempferol32 |
| 286 | Kaempferide, (3,5,7-trihydroxy-4′-methoxyflavone)8,10,11,14,16,18,20,21 |
| 287 | Quercetin23 |
| 288 | Isorhamnetin20,21 |
| 289 | 7-Methoxyquercetin32 |
| 290 | 7,4′-Dimethoxyquercetin32 |
| 291 | 7,3′,4′-Trimethoxyquercetin32 |
| 292 | (2R,3S)-Pinobanksin-3-cinnamate52 |
| 293 | Pinocembrin20,53–55 |
| 294 | Alpinetin56–59 |
| 295 | Pinostrobin54 |
| 296 | 5,7-Dimethoxyflavanone54 |
| 297 | Alpinone 3-acetate64 |
| 298 | Cardamonin67 |
| 299 | Helichrysetin22,54,55 |
| 300 | Flavokawain B22 |
| 301 | Genistein 7-O-β-D-glucoside58,70 |
| 302 | Kaempferol 3-O-β-D-glucuronide-6′′-methyl ester67 |
| 303 | Kaempferol 3-O-β-D-glucuronide67 |
| 304 | Oxyphyllvonide A67,72 |
| 305 | Oxyphyllvonide B67 |
| 306 | Oxyphyllvonide C67 |
| 307 | Kaempferol 3-O-β-D-(6-O-α-L-rhamnopyranosyl)glucopyranoside67 |
| 308 | Quercetin 3-O-β-D-glucoside32,67 |
| 309 | Quercetin 3-O-β-D-glucuronide32,67 |
| 310 | Quercetin 3-O-β-D-glucuronide-6′′-methyl ester67 |
| 311 | Oxyphyllvonide G67 |
| 312 | Oxyphyllvonide D67 |
| 313 | Quercetin 3-O-β-(3′′-O-acetyl-β-D-glucuronide)67 |
| 314 | Oxyphyllvonide E67 |
| 315 | Quercetin 3-O-β-(2′′-O-acetyl-β-D-glucuronide)67 |
| 316 | Oxyphyllvonide F67 |
| 317 | Oxyphyllvonide H67 |
| 318 | Quercetin 3-O-β-D-(6-O-α-L-rhamnopyranosyl)glucopyranoside67 |
Pinostrobin (295) and 5,7-dimethoxyflavanone (296) were obtained by methylating 293 in a structure–activity study.54 Alpinone-3-acetate (297), the acetylated form of 295, was isolated from A. japonica.64 In addition, chalcone derivatives were found in this species including cardamonin (298),22,54,55 helichrysetin (299),22 and flavokawain B (300).58,70 An isoflavone glycoside, genistein 7-O-β-D-glucoside (301), was isolated from A. oxyphylla.67 Flavonol glycosides were isolated from A. coriandriodora, A. oxyphylla, and A. zerumbet: kaempferol 3-O-β-D-glucuronide-6′′-methyl ester (302),67 kaempferol 3-O-β-D-glucuronide (303),72 oxyphyllvonides A–C (304–306),67 kaempferol 3-O-β-D-(6-O-α-L-rhamnopyranosyl)glucopyranoside (307),32,67 quercetin 3-O-β-D-glucoside (308),32,67 quercetin 3-O-β-D-glucuronide (309),67 quercetin 3-O-β-D-glucuronide-6′′-methyl ester (310),67 oxyphyllvonide G (311),67 oxyphyllvonide D (312),67 quercetin 3-O-β-(3′′-O-acetyl-β-D-glucuronide) (313),67 oxyphyllvonide E (314),67 quercetin 3-O-β-(2′′-O-acetyl-β-D-glucuronide) (315),67 oxyphyllvonide F (316),67 oxyphyllvonide H (317),67 and quercetin 6-O-α-L-rhamnosyl-β-D-glucoside (318).67
2.4. Lignans
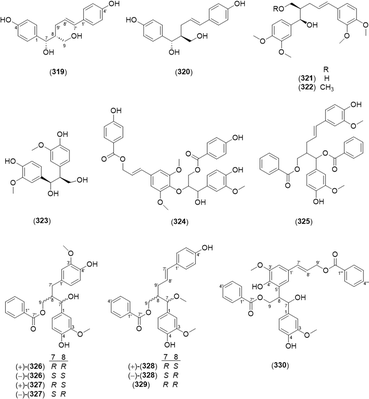
Lignans are compounds in which two phenylpropanoids are connected by the central (β) carbons of each propyl chain.101 Neolignan refers to structures in which the two phenylpropanoids are not linked by a β–β′ bond,102 and the neolignans isolated from the Alpinia species are shown in Fig. 15. Two 8-9′ linked neolignans, galanganol A (319) and galanganol B (320), were isolated from A. galanga.31 Morinol G (321), (1R,2R,4E)-1,5-bis(3,4-dimethoxyphenyl)-2-(methoxymethyl)-pent-4-en-1-ol (322), 1,2-bis(3-methoxy-4-hydroxyphenyl)-1,3-propanediol (323), quique-lignan H (324), and 1,5-bis(3′-methoxyphenyl-4′-hydroxy)-2-[(benzyloxy)methyl]-pent-4-en-1-yl benzoate (325) were previously identified.65 Additionally, 7,8-threo-9-benzoyloxy-3,3′-dimethoxy-8′,9′-dinor-7′,8-neoligane-4,4′,7-triol (326) and 7,8-erythro-9-benzoyloxy-3,3′-dimethoxy-8′,9′-dinor-7′,8-neoligane-4,4′,7-triol (327) have an 8′,9′-dinorneolignan skeleton with a 7′-8 linkage, which is rare in the lignan family. In addition, 7,8-erythro-9-benzoyloxy-3,7-dimethoxy-8,9′-neoligane-4,4′-diol (328) and (−)-7R,8R-9-benzoyloxy-3,7-dimethoxy-8,9′-neoligane-4,4′-diol (329) are 8-9′ linked neolignans. The compound 7,8-erythro-3,3-dimethoxy-9,9′-dibenzoyloxy-5′,8-neoligane-4,4′,7-triol (330) was a racemic mixture, in which two phenylpropanoid units were connected via a linkage from C-8 to C-5′ and possessed a benzoyloxy group at C-9 and C-9′.652.5. Kavalactones
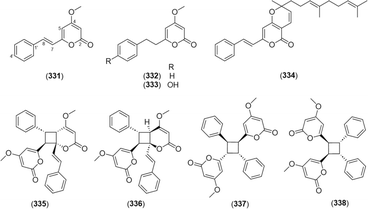
Kavalactones are lactones with an arylethylene-pyrone skeleton.103 Kavalactone monomers 5,6-dehydrokawain (331),66,104–107 7,8-dihydro-5,6-dehydrokawain (332),66,106,107 and 4′-hydroxy-7,8-dihydro-5,6-dehydrokawain (333)103 were found in Alpinia. A kavalactone–sesquiterpene conjugate, malakavalactone (334), was newly found in A. malaccensis.104Asymmetrical cyclobutane dimers of 5,6-dehydrokawain were found in A. zerumbet, namely, aniba dimer A (335),66,103,106 aniba dimer C (336),103,106 6,6′-((1α,2α,3β,4β)-2,4-diphenylcyclobutane-1,3-diyl)bis(4-methoxy-2H-pyran-2-one) (337),103,106 and 6,6′-((1R,2S,3R,4S)-3,4-diphenylcyclobutane-1,2-diyl)bis(4-methoxy-2H-pyran-2-one) (338).66,103,106 The compounds are shown in Fig. 16.
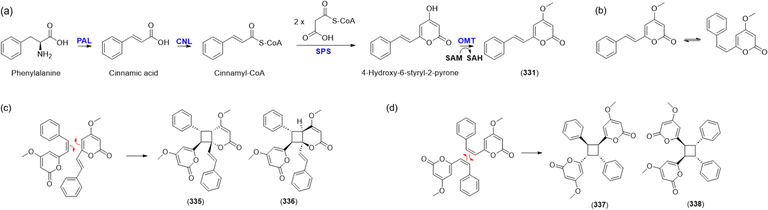
Kavalactones, also known as styrylpyrones, have been found not only in kava root (Piper methysticum), but also in fungi such as Phellinus and Inonotus spp.108,109 Recently, the key enzymes in the biosynthetic pathway of kavalactones were elucidated (Fig. 17a): phenylalanine ammonia-lyase (PAL); cinnamate-CoA ligase (CNL); styrylpyrone synthase (SPS); O-methyltransferase (OMT); S-adenosyl-L-methionine (SAM); and S-adenosyl-L-homocysteine (SAH).86 Kavalactones undergo phyto-isomerization in nature, and oligomeric kavalactones can be produced (Fig. 17b).110 Nishidono et al. proposed a biosynthesis of 335 and 336 by a [2 + 2] photo-cycloaddition from the trans–trans form of 331 (Fig. 17c).106 Dimerization of 337 and 338 from 331 was contrived from a previous study on 5-substituted-2-styryl-4-pyrones (Fig. 17d).110
2.6. Phenolics
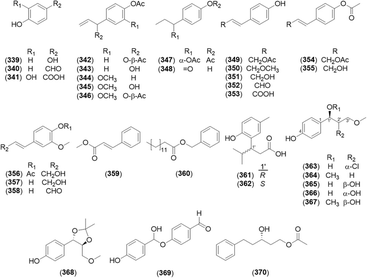
Oxyphylla A was elucidated in 2016,1 and its chiral separation yielded (R)-oxyphylla A (361) and (S)-oxyphylla A (362), with neuroprotective activity against Parkinson's disease.77 Six phenylpropanoids were newly identified from the chemically converted extract of A. galanga: 4-[erytho-2′-chloro-1′-hydroxy-3′-methoxypropyl]phenol (363), 4-(1′,3′-dimethoxypropyl)phenol (364), threo-1-(4-hydroxyphenyl)-3′-methoxypropane-1′,2′-diol (365), erythro-1-(4-hydroxyphenyl)-3′-methoxypropane-1′,2′-diol (366), 4-(threo-2′-hydroxy-1′,3′-dimethoxypropyl)phenol (367), and 4-(threo-5′-(methoxymethyl)-2′,2′-dimethyl-1′,3′-dioxolan-4′-yl)phenol (368).27 The isolated phenolic compounds from the Alpinia species are shown in Fig. 18 and Table 7.| No. | Name |
|---|---|
| 339 | Hydroquinone27 |
| 340 | p-Hydroxybenzaldehyde29–31 |
| 341 | Protocatechuic acid2 |
| 342 | (1′S)-1′-Acetoxychavicol acetate29–31,33–39 |
| 343 | (1′S)-1′-Hydroxychavicol acetate29,30,36,39 |
| 344 | Eugenyl acetate30 |
| 345 | (1′S)-1′-Hydroxyeugenol acetate36 |
| 346 | (1′S)-1′-Acetoxyeugenol acetat28,30,31,36,46 |
| 347 | (1′S)-1′-Acetoxydihydrochavicol acetate36 |
| 348 | 1′-(4-Hydroxyphenyl)-1′-propanone36 |
| 349 | trans-p-Coumaryl acetate29,30,36,48 |
| 350 | trans-p-Coumaryl alcohol γ-O-methyl ether36 |
| 351 | trans-p-Coumaryl alcohol31,36,39 |
| 352 | trans-p-Coumaryl aldehyde30,31,36,39 |
| 353 | trans-p-Coumaric acid63 |
| 354 | trans-p-Coumaryl diacetate29,30,39 |
| 355 | trans-p-Acetoxycinnamoyl alcohol30,31,36,39 |
| 356 | trans-Coniferyl alcohol 4-O-acetate36 |
| 357 | trans-Coniferyl alcohol36 |
| 358 | trans-Coniferyl aldehyde36 |
| 359 | (E)-Methylcinnamate74 |
| 360 | Benzyl myristat76 |
| 361 | (R)-Oxyphylla A1,77 |
| 362 | (S)-Oxyphylla A77 |
| 363 | 4-[erytho-2′-Chloro-1′-hydroxy-3′-methoxypropyl]phenol27 |
| 364 | 4-(1′,3′-Dimethoxypropyl)phenol27 |
| 365 | threo-1-(4-Hydroxyphenyl)-3′-methoxypropane-1′,2′-diol27 |
| 366 | erythro-1-(4-Hydroxyphenyl)-3′-methoxypropane-1′,2′-diol27 |
| 367 | 4-(threo-2′-Hydroxy-1′,3′-dimethoxypropyl)phenol27 |
| 368 | 4-(threo-5′-(Methoxymethyl)-2′,2′-dimethyl-1′,3′-dioxolan-4′′-yl)phenol27 |
| 369 | 4-Hydroxy(4-hydroxyphenyl)methoxybenzaldehyde27 |
| 370 | (S)-1-Acetoxy-5-phenyl-3-pentanol66 |
2.7. Miscellaneous compounds
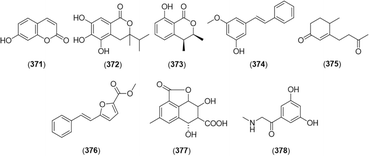
A coumarin structure, umbelliferone (371), was isolated from A. galanga,60 and (3R)-5,6,7-trihydroxy-3-isopropyl-3-methylisochroman-1-one (372) was identified as a novel antioxidant coumarin from A. katsumadae.111,112 In addition, 4-hydroxymelein (373) was an isocoumarin identified in A. galanga.27 Furthermore, 3-methoxy-5-hydroxystilbene (374),22,113 oxyphyllone F (375),3 5-styrylfuran-2-carboxylic acid methyl ester (376),22 3-methyl-6α, 8β-diol-7-carboxylic acid tetralin-11, 9β-olide (377),76 and 1-(3,5-dihydroxyphenyl)-2-(methylamino)ethan-1-one (378)114 were found in Alpinia. Their structures are shown in Fig. 19.3. Pharmacological and toxicological aspects
3.1. Cytotoxicity and anti-tumor effects
Alpinisin A (93), a diarylheptanoid–sesquiterpene conjugate isolated from the rhizomes of A. officinarum, inhibited the growth of human gastric carcinoma (SGC-7901, IC50 = 11 μM), human breast cancer (MCF-7, IC50 = 15 μM), and epidermoid cervical carcinoma (CaSki, IC50 = 15 μM) cell lines.80 Microbial transformation of yakuchinone A (4) using the fungus M. hiemalis KCTC 26779 produced nine metabolites. Among them, compound 63 showed the most selective cytotoxic activities against murine melanoma (B16F1 and B16F10) and human melanoma (A375P) cell lines (IC50 values: 6.1–9.7 μM).49Terpenoid derivatives also exhibited cytotoxic activity. 3,4-Dihydrolactucin (252), a guaiane-type sesquiterpenoid, showed cytotoxicity against MDA-MB-231 and HeLa cells, with IC50 values of 37 and 75 μg mL−1, respectively.60 In addition, a labdane diterpene from A. intermedia, intermedin A (262), at 30 μg mL−1 suppressed the growth of human leukemia cells (HL-60) and prolonged the life of P-388D1 tumor-bearing CDF1 mice by 49% at 20 mg kg−1.92
The cytotoxic and anti-tumor effects of some flavonoids have also been reported. Flavokawain B (300) and alpinetin (294) isolated from A. mutica inhibited the UCK2 enzyme and suppressed the growth of HT-29 cells, with IC50 values of 28 μM and 44 μM, respectively.115 Helichrysetin (299), a chalcone isolated from A. katsumadae, inhibited the growth of human gastric cancer cells (MGC803) with an IC50 value of 16 μM and gastric cancer growth in vivo through mTOR/p70S6K/c-Myc/PDHK1-mediated energy metabolism reprogramming.116 The rates of growth inhibition in an MGC803-xenografted mouse model were 63% (3 mg kg−1), 46% (10 mg kg−1), and 51% (30 mg kg−1). A synergistic cytotoxic effect of tectochrysin (280) with cetuximab (an anti-epidermal growth factor receptor (EGFR) monoclonal antibody) was observed in the human colon cancer cell lines HCT116 and SW480.5 The underlying mechanism involved inhibition of the EGFR pathway, followed by suppression of NF-kB and AP-1 activity. Atwa et al. investigated the combination cancer therapy of galangin (281) and luteolin (100 mg kg−1 each) with doxorubicin in a chemically induced hepatocellular carcinoma (HCC) rat model and observed significant decreases of HCC markers compared to healthy rats.117
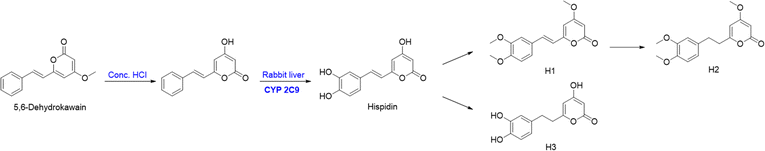
Hispidin and its derivatives H1–H3 were synthesized from 5,6-dehydrokawain (331) by CYP2C9 in the liver microsomes of rabbit.118 The bioconversion pathway of hispidin and H1–H3 are shown in Fig. 20,118 and the IC50 values of hispidin and H1–H3 against A549 cancer cells were 25, 17, 21, and 17 μM, respectively (IC50 of the positive control resveratrol = 23 μM).119
3.2. Anti-bacterial effects
Several diarylheptanoids (2, 3, 14, 17, and 31) isolated from the rhizomes of A. officinarum showed inhibitory activity against Mycobacterium tuberculosis H37Ra and M. bovis BCG.41 Among them, 2 showed the strongest activity against M. buterculosis (IC50 = 4.2 μg mL−1) and M. bovis (IC50 = 4.7 μg mL−1). Additionally, 3,4-dihydrolactucin (252) showed inhibitory activity against Bacillus cereus, B. subtilis, Staphylococcus aureus, and methicillin-resistant S. aureus, with a minimum inhibitory concentration (MIC) of 16–32 μg mL−1.60 Compound 265 induced death of Escherichia coli and S. paratyphi cells at a concentration of 50 μg mL−1.93 The kavalactone derivative malakavalactone (334) from A. malaccensis exhibited weak antibacterial activity (MIC = 60 μM) against four gram-negative bacteria: Enterobacter aerogenes, E. coli, Pseudomonas aeruginosa, and Shigella dysenteriae.104 Compound 378 alleviated the swarming mobility of P. aeruginosa at a minimum concentration of 12.5 μg mL−1 and reduced the virulence of P. aeruginosa.1143.3. Anti-parasitic and insecticidal effects
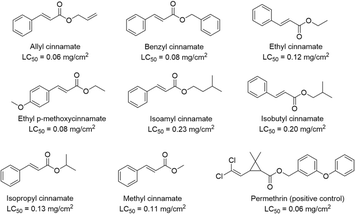
Various phenolic compounds showed anti-parasitic activities. The most active compound was hydroquinone (339), with IC50 values of 0.4 ± 1.4 μg mL−1 (Leishmania major), 3.2 ± 0.1 (Trypanosoma brucei gambiense), and 3.3 ± 0.3 (T. brucei rhodesiense).27 Compound 363 was also confirmed as a significant active component, with IC50 values of 4.2 ± 0.8 (T. brucei gambiense) and 5.6 ± 1.5 (T. brucei rhodesiense). Kang et al.120 observed potent acaricidal activities of the cinnamate derivatives derived from A. galanga oil and conducted a structure–activity relationship (SAR) study of commercial cinnamate derivatives against Haemaphysalis longicornis nymphs (Fig. 21). Allyl cinnamate showed the most potent toxicity (LC50 = 0.06 mg cm−2),120 and the mode of action was inhibition of acetylcholinesterase (AChE) of H. longicornis nymphs.(1′S)-1′-Acetoxychavicol acetate (342, LD50 = 1.6 μg per larva, 24 h) and trans-p-coumaryl diacetate (354, LD50 = 2.4 μg per larva, 24 h) showed insecticidal effects against Spodoptera litura.39,121
3.4. Anti-virus activities
Yoo et al. performed structure-based virtual screening to find inhibitors of neuraminidase, which is a key enzyme of the influenza viruses.25 By virtual screening of 6149 compounds in the drug library, a diarylheptanoid structure was selected as a promising scaffold. Thirty diarylheptanoids were isolated from A. officinarum, and 9, 13, 16, 22, 23, and 51 (10 μM each) showed strong relative inhibitory activity compared with mangiferin (1.9, 0.9, 1.0, 0.9, 2.1, and 0.9, respectively).The viral protein R (Vpr) is found in HIV-1 and 2 and influences G2 cell cycle arrest and apoptosis.122 Monoterpenoids such as 134, 135, and 136 showed dose-dependent anti-Vpr activity in TREx-HeLa-Vpr cells (concentrations: 0.3–5.0 μM) without cytotoxicity.84
3.5. Anti-inflammatory effect
Diarylheptanoids have shown anti-inflammatory activities. In particular, 14, 17, 18, 31, 33, and 37 showed anti-inflammatory activities against lipopolysaccharide (LPS)-induced macrophages (RAW 264.7).68,123 Moreover, 17 and 33 down-regulated the gene expressions of interleukin (IL)-1β, IL-6, and tumor necrosis factor alpha (TNF-α) in LPS-treated HepG2 cells, and the in silico docking simulation revealed that the compounds interact with p38 mitogen-activated protein kinase (MAPK).21 Compound 17 (10 mg kg−1) inhibited right paw edema induced by carrageenan-induced inflammation in a rat model.8 The compound (10 and 20 mg kg−1) also inhibited paw volume, joint diameter, thermal hyperalgesia, and tactile allodynia in mice with Freund's complete adjuvant (FCA)-induced arthritis and oxido-inflammatory markers such as SOD, GSH, MDA, and TNF-α.124 Compound 64 inhibited nitric oxide (NO) release in LPS-induced RAW 264.7 cells (IC50 = 8.3 μM).65 Coriandralpinin C (69) and E (71) isolated from the rhizomes of A. coriandriodora inhibited NO release in LPS-treated RAW 264.7 cells.32 Compounds 12, (−)-94, and (−)-102 significantly inhibited the production of NO, IL-1β, IL-6, and TNF-α via the NF-κB signaling pathway in LPS-treated RAW 264.7 cells.22,23Compound 137, a monoterpenoid isolated from the pericarps of A. zerumbet, inhibited IL-1β-induced production of NO in primary cultured rat hepatocytes (IC50 = 18 μM).85 Eremophilane-type (147, 149, 153, and 158) and cadinane-type (238, 239, and 240) sesquiterpenoids inhibited cytokine release (NO, TNF-α, and IL-6) in LPS-treated BV-2 cells, and compound 158 further suppressed the mRNA levels and protein expressions of TNF-α, IL-6, COX-2, and iNOS.24 Compound 219, a eudesmane-type sesquiterpenoid isolated from the fruits of A. oxyphylla, inhibited PGE2 release in LPS-treated RAW 264.7 cells with an IC50 value of 90 μM (IC50 of hydrocortisone, a positive control = 48 μM).42 Compound 275, an acyclic triterpenoid from the A. katsumadae seed, inhibited the release of iNOS and COX-2 and suppressed the gene expressions of IL-1β, IL-6, and NF-κB in LPS-treated J774 cells, and oral administration of 275 (50 mg kg−1) attenuated paw thickness and volume in a mouse model.97
Tectochrysin (280, 2.5 and 5.0 mg kg−1) alleviated allergic airway inflammation by regulating the Th2 response and oxidative stress in a shrimp tropomyosin-induced mouse asthma model.125 Compounds 281, 283, 284, 286, 287, 288, and 299 suppressed the production of IL-1β, IL-6, and TNF-α.21–23 In addition, 281 (10 mg kg−1) inhibited paw edema in the carrageenan-induced mouse inflammation model.8 Compound 297, a flavanolol from the seeds of A. japonica, mitigated the symptoms of ear edema in a 12-O-tetradecanoylphorbol 13-acetate-induced mouse inflammation model with an ID50 value (50% inhibitory dose) of 176 μg per ear.64 Compound 298 inhibited the IL-1β-induced production of NO in primary cultured rat hepatocytes (IC50 = 10 μM).85
A previous study showed that (+)-326, (−)-326, (+)-327, and (−)-327, neolignans with an 8′,9′-dinorneolignan skeleton, reduced NO production in LPS-induced RAW 264.7 cells with IC50 values of 3.6, 7.6, 6.5, and 5.6 μM, respectively (the IC50 of dexamethasone, a positive control, was 30 nM).65 As the pericarps of A. zerumbet significantly inhibited NO production in IL-1β-treated hepatocytes, the kavalactone derivatives from this plant were subjected to an anti-inflammatory assay, and the IC50 value of each compound on NO suppression was 27 (331), 28 (332), 34 (335), 25 (336), 34 (337), and 26 μM (338).106 (E)-Methylcinnamate (359) alleviated the symptoms of colitis through the MAPK pathway.126 Compound 372 also showed protective effects of H9c2 cells against lipoteichoic acid-induced toxicity through anti-inflammatory and oxidative responses.112
3.6. Anti-oxidant effects
Several compounds from A. oxyphylla exhibited anti-oxidant activity. For example, diarylheptanoids yakuchinone A (4) and 5′-hydroxyyakuchinone A (5) scavenged reactive oxygen species (ROS) in HepG cells.50 Moreover, 13-hydroxynootkatone (155) and nootkatone-11,12-epoxide (162) from A. oxyphylla effectively repaired the damage from tert-butyl hydroperoxide-induced oxidative stress in adipose-derived mesenchymal stem cells in a dose-dependent manner.4 Coriandralpinins A–H (67–74) isolated from A. coriandriodora also exerted ROS inhibition in a dose-dependent manner.32 Compounds 372 (5 mg kg−1) from A. katsumadae and 292 (15 mg kg−1) from A. galanga significantly reduced oxidative stress and improved retinal function in an rd10 mouse model of retinitis pigmentosa.100,1113.7. Neuroprotective effects
The neuroprotective effects of the compounds from Alpinia species have been reported in several studies. Compound 32 has been reported to improve amyloid β (Aβ) 42-induced neuronal damage, apoptosis, and oxidative stress.98 Alpinin A (66, 10 μM) and alpinin B (52, 10 μM) were tested for their effects on the aggregation of α-synuclein (α-syn), a protein mainly expressed in neurons of Parkinson's disease (PD) patients.69,127 The two compounds inhibited α-syn aggregation by 66% and 67%, respectively. Moreover, a diarylheptane–monoterpene conjugate, (−)-alpininoid B (89), inhibited AChE with an IC50 of 2.6 μM.16Qiu et al. observed that the sesquiterpenoids (153, 154, 175, 204, 221, 244, 246, and 247) from the fruits of A. oxyphylla improved the cell viability (≥80%) of a neuroblastoma cell line (SH-SY5Y) in conditions of H2O2-induced oxidation with serial concentrations of the compounds (5, 10, and 20 μM).3 Moreover, 175, 221, 247, and 375 (20 and 100 μM) also reduced the production of ROS.
The protective effects of (R)-oxyphylla A (361) and (S)-oxyphylla A (362) against PD have been reported in several studies. Zhou et al. demonstrated elevated α-syn degradation by 361in vitro (10, 30, and 100 μM) and in vivo (30 mg kg−1).128 The compound induced the PKA/Akt/mTOR pathway and reduced α-syn accumulation. Li et al. also reported that 361 (5, 10, and 20 mg kg−1) protected against neuron loss and improved neuro-behavioral changes of PD mice via Nrf2 signal transduction. The efficacy of 361 at 20 mg kg−1 for the survival of neurons was similar to that of rasagiline (a positive control).1 Both 361 and 362 at low doses (1.3, 2.5, and 5.0 μM) exerted mild neuroprotective activities in the MPTP-induced PD zebrafish model, whereas high doses (5, 13, and 25 μM) did not show comparable toxicities in zebrafish larvae.77 Additionally, 361 and 362 (10 and 20 mg kg−1) inhibited the expression levels of amyloid precursor protein (APP) and Aβ protein and attenuated cognitive decline in SAMP8 mice via antioxidative effects through the Akt-GSK3β and Nrf2-Keap1-HO-1 pathways.129
Ischemic stroke is caused by reduced blood supply to the brain, and the current therapy is thrombolytic treatment with a limited time window (4.5 to 6 h after the incident). Therefore, the need for neuroprotection in stroke treatment is urgent. A previous study showed that (+)-85 (alpinidinoid A) alleviated oxygen-glucose deprivation and reoxygenation (OGD/R) damage in primary cortical neurons by inducing the PI3K/AKT/mTOR signaling pathway.78p-Coumaric acid (353) activated the BDNF/TrkB/AKT signaling pathway to proliferate neural stem cells in vitro.63 In addition, the compound (50, 100, and 200 mg kg−1) improved neuronal proliferation and spatial/memory functions in a transient middle cerebral artery occlusion rat model.
As neuroprotective effects of A. galanga have been reported in several studies, the aqueous stability and gastro-intestinal/blood–brain barrier (GI/BBB) permeability of the main compounds in the extract were investigated.31 The phenolics (340, 342, 346, 351, 352, and 355) showed good permeability, which contributes to the neuroprotective effects of A. galanga. Four compounds with neuroprotective activity in A. oxyphylla were tested for BBB permeability,2 and the order of permeability was 341, 147, 279, and 280.
3.8. Anti-diabetic effects
Insulin resistance (IR) indicates decreased responses of insulin-target tissues to insulin simulation,130 and it causes several metabolic disorders such as nonalcoholic fatty liver disease and type 2 diabetes.131 Compounds 14 and 31 alleviated oxidative stress and glucose metabolism by regulating the PI3K/AKT-Nrf2-GSK3β pathway, improving the IR of HepG2 cells.96,132 Li et al. reported that 33 improves IR through the PI3K/AKT and TNF-α signaling pathways.99 Among the diarylheptanoid dimers isolated from A. katsumadae, katsumadainols C1–C4 (75–78) and C7–C10 (81–84) showed significant stimulation of glucagon-like peptide-1 receptor (GLP-1) secretion. Additionally, 75–78 significantly inhibited glycogen phosphorylase (GPa) (IC50 values of 18–31 μM), and 75–79 inhibited α-glucosidase (IC50 values of 6.9–18 μM) and protein tyrosine phosphatase 1B (PTP1B, IC50 values of 36 to 80 μM).71 He et al. revealed that the A. katsumadae EtOH extract (200 mg kg−1) reduced blood glucose level in a db/db mouse model; the authors identified 29 diarylheptanoid-chalcone hybrids (105–133) from A. katsumadae to test α-glucosidase and PTP1B inhibition.83 All the isolated compounds exhibited α-glucosidase inhibition with IC50 values of 2.9–30 μM, and 105–107, 109–111, 115–118, 125–129, and 131 showed PTP1B selective inhibition with IC50 values ranging from 22 to 97 μM.Sesquiterpenoids isolated from the fruits of A. oxyphylla were also tested for anti-diabetic activity.28 The results showed that 173 and (6R)-235 (20–100 μM) stimulated GLP-1 by 151–1242% and 421–1090%, respectively, via Ca2+/CaMKII and PKA pathways. Compound 346 (2.5, 5.0, and 10 μM) from A. galanga improved glucose-stimulated insulin secretion and inhibited α-glucosidase activity in rat pancreatic β-cells.30 Moreover, 335 (135 μM) and 338 (25 μM) increased the viability of human umbilical vein endothelial cells damaged by high glucose by 83% and 75%, respectively.103
3.9. Osteoblast-protective effects
Mesenchymal stem cells differentiate into osteoblasts via the Smad- and Runt-related transcription factor 2 (RUNX2) pathways by osteogenic factors, e.g., bone morphogenetic protein (BMP). Apoptosis and migration/differentiation of osteoblasts are important mechanisms by which osteoblasts maintain bone physiology and mitigate bone-related diseases.74 Pinocembrin (293) isolated from the leaves of A. zerumbet improved osteoblast differentiation of pre-osteoblasts (MC3T3-E1) by inducing BMP-2 expression via endoplasmic reticulum (ER) stress and mineralization through the Smad and RUNX2 pathways.53 (E)-Methylcinnamate (359) inhibited the migration of pre-osteoblasts and induced their differentiation.74 In addition, 5,6-dehydrokawain (331) and 7,8-dihydro-5,6-dehydrokawain (332) stimulated alkaline phosphatase activity and mineralization in MC3T3-E1 cells.107 As 331 improved osteoblastogenesis of MC3T3-E1, semi-synthesis was performed for 5,6-dehydrokawain analogs (Fig. 22). The synthesized compounds (E)-6-(4-ethylstyryl)-4-methoxy-2H-pyran-2-one and (E)-6-(4-butylstyryl)-4-methoxy-2H-pyran-2-one significantly stimulated osteoblastic expression of RUNX2 and Osterix genes and showed preventive effects on osteoblasts.133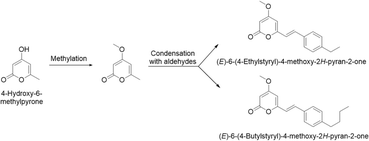 | ||
| Fig. 22 Semi-synthesis of (E)-6-(4-ethylstyryl)-4-methoxy-2H-pyran-2-one and (E)-6-(4-butylstyryl)-4-methoxy-2H-pyran-2-one. | ||
3.10. Miscellaneous activities
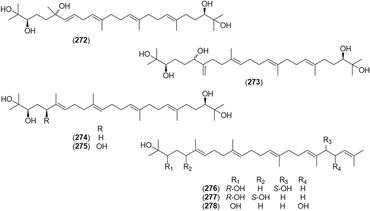
Compounds 14, 15, 31, 39, and 41 (10–80 μM), isolated from A. officinarum, induced differentiation of 3T3-L1 preadipocytes in a dose-dependent manner, which may help protect against obesity.45 Compounds 276 and 277, acyclic triterpenoids isolated from A. katsumadae, suppressed the mRNA and protein levels of proprotein convertase subtilisin/kexin type 9 (PCSK9), which regulates low-density lipoprotein cholesterol (LDL-C) levels in the blood.66Melanogenesis is the production of melanin, which protects the skin from UV damage. However, excessive melanin production leads to dermatological disorders. Melanogenesis inhibitory activities (IC50) of compounds from A. galanga were investigated using theophylline-stimulated murine B16 melanoma 4A5 cells:90257 (IC50 = 18 μM), 258 (IC50 = 9.4 μM), 259 (IC50 = 30 μM), 260 (IC50 = 2.9 μM), 268 (IC50 = 4.4 μM), 269 (IC50 = 8.6 μM), and 270 (IC50 = 4.6 μM). These activities were more potent than that of arbutin (174 μM), a positive control.
Tectochrysin (280, 0–500 μM) extended the lifespan of Caenorhabditis elegans by up to 21% and protected worms against Aβ1-42-induced toxicity by regulating FOXO/DAF-16 and HSF-1.134 Flavonoid glycosides (301, 303, 309, 311, 313, 315, 317, and 318) isolated from A. oxyphylla exhibited anti-renal fibrosis activity at concentrations of 10, 20, and 40 μM in TGF-β1-induced kidney proximal tubular cells, a reno-protective activity.67
4. Compounds with various activities
4.1. Nootkatone
Nootkatone (147, 0.02 and 0.2 mg kg−1) showed protective effects in an Alzheimer's disease (AD) mouse model by decreasing malondialdehyde, Aβ, and AChE and dramatically enhanced the performance of the mice in the Y-maze and Morris water maze tests.62 Nootkatone (5 and 10 mg kg−1) also improved learning and memory impairment in mice, and this may be related with attenuated inflammatory cytokines (IL-1β, IL-6, TNF-α, and NF-κB p65) in the hippocampus.135 Moreover, 147 showed modest BBB permeability in human brain microvascular endothelial cells co-cultured with astrocytes with passive diffusion, which supports the compounds as a good candidate for CNS-related illnesses.2 In addition, administration of nootkatone (10 μM) with schisandrin (50 μM) showed a synergistic neuroprotective activity in Aβ1-42-induced differentiated PC12 cells.136 The underlying mechanism involves the PI3K/AKT/Gsk-3β/mTOR pathway and decreased pro-inflammatory cytokines including NF-κB, IKK, IL-1β, IL-6, and TNF-α.Compound 147 (5, 10, and 20 mg kg−1) showed anti-oxidant and anti-inflammatory activities through NOX4, NF-κB, and Nrf2/HO-1 pathways in kidneys of mice exposed to CCl4.137 It (10 mg kg−1) also protected an obstructive nephropathy mouse model from oxidative stress and the inflammatory response, reduced cell apoptosis, and attenuated renal fibrosis.138
Another study showed that 147 (25 and 50 mg kg−1) improved body weight, liver weight/index elevation, glycolipid metabolism, glucose tolerance, and hepatic lipid accumulation in high fat diet-induced metabolic-associated fatty liver disease (MAFLD) mice.61 Additionally, 147 down-regulated NAG-1 induction and cyclin D and showed cytotoxic activities in colorectal cancer cells (HCT-116 and SW480).139 It reduced NO production in LPS-treated RAW 264.7 cells and exhibited anti-inflammatory activity.51
4.2. Galangin
Galangin (258) is a natural flavonoid mainly isolated from A. officinarum and A. galanga, and it has been reported to show cytotoxic and anti-tumor effects. For example, it prevented the growth of renal cell lines (789-0 and Caki-1) by controlling epithelial/mesenchymal markers (e.g., E-cadherin, N-cadherin, and vimentin) and suppressing cell invasion.9 It also inhibited the proliferation of human laryngeal cancer cells through p38 and AKT/NF-κB/mTOR pathways,140 and activated apoptosis of bladder cancer cells by promoting the p53 signaling pathway.141 Galangin inhibited a human gastric carcinoma cell line (MGC 803) by regulating the STAT3/ROS axis.142 Co-treatment of 258 (30 μM) and berberine (90 μM) suppressed the growth of esophageal carcinoma cells (ECa9706) by modulating anti-apoptotic proteins Bcl-2, Mcl-1, and XIAP and the pro-apoptotic protein Bax.143 Compound 258 (2, 5, and 10 μM) also mitigated the drug resistance of cisplatin (2 μM) in human lung cancer cells by dose-dependent suppression of cell proliferation and apoptosis induction.144 This combination therapy (10 mg kg−1 of 258 and 5 mg kg−1 of cisplatin) also attenuated tumor growth in a mouse xenograft model to a greater extent compared with that of 258 or cisplatin treatment alone.One study reported that 258 shows beneficial effects with cardioprotective properties. It (15 mg kg−1) attenuated cardiac myofibril damage and infarction size, improved cardiac function, and inhibited mitochondrial injury for myocardial ischemic reperfusion injury in a mouse model.145 In hypertensive rats, 258 (30 and 60 mg kg−1) attenuated hypertension, cardiorenal damage, and oxidative stress by modulating the expression of AT1R, TGF-β1, and Collgen I (Col-I) protein in the heart and AT1R/Nox-4 and Nrf2/HO-1 protein in renal tissue.146 The compound (1 mg kg−1) also showed preventive effects against isoproterenol-induced myocardial fibrosis in male albino Wistar rats.17
Another report found that 258 showed osteoprotective effects in LPS-stimulated bone marrow-derived dendritic cells and promoted osteogenic differentiation through activation of AKT/mTOR signaling.147 It also improved the osteogenic differentiation in human amniotic mesenchymal stromal cells by regulating the JAK2/STAT3 signaling pathway.148 In addition, 258 regulated lipid metabolism by reducing lipid accumulation in HepG2 liver cells and improving lipid destruction by increasing the expressions of beclin1, LC3-II/LC3-I, Atg3, AMPKα1, and phosphorylated AMPKα1 proteins.12 Administration of 258 (4, 8, and 16 mg kg−1) also reduced hyperlipidemia in a streptozotocin-induced hyperglycemia rat model.15
Compound 258 has shown protective effects against hepatic fibrogenesis by downregulating the levels of α-smooth muscle actin and Col-I in LX-2 cells.13 It (100 mg kg−1) alleviated the virulence of S. aureus in a mouse model with S. aureus-induced pneumonia.149 The antioxidant activity of 258 was confirmed against DPPH, with an IC50 value of 4.2 μM.14
4.3. Alpinetin
As a main active flavonoid from A. katsumadae, alpinetin (271) has been reported to have cytotoxicity in several cell lines. For example, 271 induced tumor regression via inhibition of the NF-κB signaling pathway in breast cancer cells.57 It also inhibited uridine-cytidine kinase 2 enzyme (UCK2), which is related with uncontrolled cell proliferation, at the concentration of 50 μM.58 Compound 271 also attenuated the viability of the SKOV3 ovarian cancer cell line by inhibiting the STAT3 signaling pathway.150271 showed preventive effects against cancer cachexia caused by chemotherapy.151 It reduced myotube atrophy in carcinoma-conditioned murine myoblasts (25–100 μM) and markedly mitigated the losses of body weight and skeletal muscle in the mouse model (25 and 50 mg kg−1).Compound 271 has been shown to have anti-inflammatory effects in several studies. It down-regulated acute pancreatitis-induced acute lung injury (ALI) by promoting aquaporin-1 and decreasing TNF-α expression in a HPMVEC cell line (0.01, 0.1, 1.0, and 10 μg mL−1) and in a rat model (40, 80, 160, and 320 μg mL−1).56 Two research teams revealed that the progression of dextran sulfate sodium (DSS)-induced colitis in mice was relieved by 271 (25, 50, and 100 mg kg−1) through the toll-like receptor 4 (TLR4)/NF-kB/NOD-like receptor protein 3 (NLRP3) signaling and the Nrf2/HO-1 signaling pathways.152,153 It (13, 25, and 50 mg kg−1) also improved LPS/D-galactosamine-induced liver injury in the mouse model by recruiting the NF-κB and Nrf2 signaling pathways.59 In a septic mouse model, the inflammatory symptoms and the release of proinflammatory cytokines were alleviated by 271 (50 mg kg−1).154 Compound 271 (50 mg kg−1) also enhanced Nrf2-mediated redox homeostasis, inhibited macrophage infiltration, and decelerated atherosclerotic plaque development in an ApoE−/− mouse model.155
4.4. Cardamonin
Cardamonin (276) is a chalcone mainly found in A. katsumadae and A. conchigera and was demonstrated to exhibit anti-inflammatory and anti-oxidant activities in several studies. Treatment with 276 (50 and 100 mg kg−1) significantly relieved acetaminophen (APAP)-induced hepatoxicity in APAP-stimulated mice by suppressing high mobility group box 1 (HMGB1), TLR4, and NLRP3.156 Wang et al. also observed that 276 (2.5 mg kg−1) significantly reduced IL-1β secretion and caspase-1 activity by inhibiting NLRP3 inflammasome activation in a monosodium urate-induced gouty arthritis rat model.157 Oral administration of 276 (15, 30, and 60 mg kg−1) in a DSS- and 2,4,6-trinitrobenzene sulfonic acid-induced colitis mouse model mitigated the symptoms of the disease, and the underlying mechanism involves AhR/Nrf2/NQO1 and NLRP3 inflammasome pathways.158 It (1 mg kg−1) also exhibited critical anti-inflammatory effects for lung injury through the regulation of the TLR2,4-MyD88 and mTOR pathways in a mouse model.159 Additionally, 276 (25, 50, and 100 μM) inhibited the expression of COX-2, iNOS, NO, TNF-α, and IL-6 in rat nucleus pulposus cells, and it (20 mg kg−1) showed protective effects in a puncture-induced intervertebral disc degeneration (IVDD) mouse model through the Nrf2/HO-1 signaling pathway.160 In addition, it (0.8–200 μM) exhibited neuroprotective effects by exerting anti-oxidant and anti-inflammatory activities in LPS-treated BV-2 microglial cells.161 The mechanism of action may involve Nrf2/Keap1, NF-κB, and antioxidant-related enzymes (e.g., SOD, CAT, and GSSH). The application of doxorubicin (DOX) is limited in cancer treatment due to its cardiotoxicity. Notably, 276 (20, 40, and 80 mg kg−1) exerted cardioprotective activity by suppressing oxidative stress and inflammatory response through the Nrf2 signaling pathway in a DOX-treated mouse model.162Compound 276 exerted cytotoxic effects on HepG2 cells, with an IC50 value of 15 μM after 72 h of treatment.163 It induced pro-apoptotic proteins (e.g. FADD, FAS, TRIAL, and HIF-1) and downregulated anti-apoptotic proteins like heat shock proteins (HSP) 60, 27, and 70. Moreover, 276 (0–20 μM) sustained cell proliferation and stimulated apoptosis of pancreatic cancer (PC) cells (PANC-1 and SW1990) by regulating the FOXO3a-FOXM1 axis and improved chemosensitivity of PC cells to gemcitabine, a chemotherapy drug.164276 (1 and 5 mg kg−1) also improved immune responses on WEHI-3 cell-generated leukemia mice by fortifying the phagocytic ability of macrophages and reducing the populations of CD3 (T cells), CD11b (monocytes), and Mac-3 (macrophages).165
Another study showed that 276 (10 mg kg−1) exerted an anti-nociceptive effect by up-regulating the serotonin 1A receptor (5-HT1A) in the central nervous system (CNS) of a neuropathic pain mouse model.166 Moreover, 276 showed selective inhibitory activity (IC50 = 454 nM) against transient receptor potential ankyrin 1 (TRPA1), a receptor involved in pain.167 In addition, it suppressed the cytopathic property induced by the human coronavirus (HCoV-OC43) in human lung cells (MRC-5), with an IC50 value of 3.6 μM, and showed anti-virus activity through the p38 MAPK pathway.168
4.5. 1′S-1′-Acetoxychavicol acetate
1′S-1′-Acetoxychavicol acetate (342) can be easily found in A. galanga and exhibits various biological activities. In particular, 342 inhibited the growth of colorectal adenocarcinoma cells (SW480, IC50 = 80 μM) by regulating apoptosis and the G0/G1 cell cycle check point with significant DNA damage.33 Phuah et al. revealed that miR-629 is a key microRNA that enhances sensitivity of cancer cells toward 342.169 Moreover, overexpression of Ras suppressor-1 (RSU1), which is regulated by miR-629, improved the cytotoxicity of 342. Sok et al. demonstrated the autophagy-inducing ability of 342 in A549 and SK-LU-1 with IC50 values of 29 and 25 μM, respectively.170 Compound 342 exhibited cytotoxic activity by regulating the human epidermal growth factor receptor 2 (HER2) signaling pathway in breast cancer cells (MCF7, IC50 = 12 μM) and HER2-overexpressed MCF7 cells (IC50 = 5.9 μM).37Another study showed that 342 significantly inhibited larval growth of S. frugiperda by 34% at the concentration of 20 mg L−1 and showed cytotoxicity in ovarian tissue of S. frugiperda (IC50 = 1.9 μM).35 It also showed anti-microbial activity against methicillin-resistant S. aureus (MIC = 0.5 mg mL−1).29,121342 showed anti-tuberculosis activity with an MIC of 0.2 μg mL−1 (M. tuberculosis H37Ra ATCC 25177) and 0.7 μg mL−1 (M. tuberculosis H37Rv ATCC 27294).38
While protein degradation by proteasomes is important to maintain protein homeostasis, the age-related reduction of proteasome activity leads to neurodegenerative processes. One study showed that 342 (0.02% of the diet) improved the spatial and memory performance of mice in the Y maze and Morris water maze tests and increased the serum concentrations of β-hydroxybutyric acid and palmitic acid, which help maintain cognitive function.171342 also improved proteasome activity in neuronally differentiated murine pheochromocytoma (PC12) cells by stimulating the cAMP/PKA pathway.172 It also showed good GI/BBB permeability in the artificial membrane permeability assay and thus has potential for the treatment for neurodegenerative diseases.31
One study showed that 342 (5 mg kg−1) inhibited the releases of proinflammatory cytokines, such as IL-6 and TNF-α, and also relieved lung inflammation in the LPS-challenged mouse model by regulating NF-κB and MAP kinases.173 In addition, it (0.03 and 0.05% of food intake) acted as a TRPA1 agonist and stimulated lipolysis of adipose tissue in a diet-induced obesity mouse model.174 In a drug metabolism study of 342, the compound showed moderate inhibition against CYP1A2 (IC50 = 4.5 μM), CYP2D6 (IC50 = 7.5 μM), and CYP3A4 (IC50 = 9.5 μM). Drug–drug interactions were observed when 342 was co-administered with drugs metabolized by CYP1A2, CYP2D6, or CYP3A4 enzymes.34
5. Conclusions
The family Zingiberaceae is composed of approximately 47 genera and 1400 species. Alpinia is a largest genus in Zingiberaceae, followed by Globba, Amomum, and Zingiber.175 Chemical profiles of Alpinia genus demonstrated that diarylheptanoid is the most abundant structure; terpenoids and flavonoids are commonly isolated compounds in the genus Amomum, Hedychium, and Zingiber.175–177 As the largest genus in the family, Alpinia has been investigated for potential pharmacological activities, and evidence has indicated Alpinia as a promising treatment for various diseases, including neurodegenerative disorders, cancers, and metabolic diseases.In this review, the compounds isolated in Alpinia species were categorized and the characteristics related to structures were discussed. Compounds usually found in Alpinia included diarylheptanoids, terpenoids, flavonoids, lignans, kavalactones, and phenolic compounds. Diarylheptanoid–chalcone conjugates are unique structures exclusively found in the genus Alpinia.178,179 The sesquiterpenoids found in this genus were mainly eremophilane, eudesmane, cadinane, and guaiane types. Moreover, the pharmacological properties of the compounds from Alpinia genus included cytotoxic/anti-tumor, anti-bacterial, anti-parasitic/insecticidal, anti-virus, anti-inflammatory, anti-oxidant, neuroprotective, anti-diabetic, and osteoblast-protective activities. Phenylpropanoid from the shikimic acid pathway including cinnamate derivatives showed anti-parasitic and insecticidal effects.
This article provides newly updated structural information of the unique substances isolated from various species of the genus Alpinia and biological activities from the compounds. As the life expectancy of people is increasing, these findings will help researchers find treatments for metabolic syndromes including obesity, type-2 diabetes, and cardiovascular diseases, and for neurodegenerative diseases such as AD and PD.
6. Author contributions
Conceptualization, A. R. H. and E. K. S.; formal analysis, I. Y. and A. R. H.; investigation, D. P., H. L., H. K., and Y. L.; writing-original draft preparation, I. Y. and A. R. H.; writing-review and editing, I. Y., J. W. N., and E. K. S.; visualization, I. Y.; supervision, E. K. S.; project administration, E. K. S.; funding acquisition, I. Y. and E. K. S. All authors have read and agreed to the published version of the manuscript.7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2023-00243759) and by an NRF grant funded by the Korean government (MSIT) (No. 2021R1A2C1003350).9. Notes and references
- G. Li, Z. Zhang, Q. Quan, R. Jiang, S. S. Szeto, S. Yuan, W.-T. Wong, H. H. Lam, S. M.-Y. Lee and I. K. Chu, J. Proteome Res., 2016, 15(8), 2595–2606 CrossRef CAS PubMed.
- T. Xiao, M. Pan, Y. Wang, Y. Huang, M. Tsunoda, Y. Zhang, R. Wang, W. Hu, H. Yang and L.-S. Li, J. Pharm. Biomed. Anal., 2023, 235, 115637 CrossRef CAS PubMed.
- C. Qiu, L. Mu, J. Wang, R. Tang, B. Hou, W. Hu, R. Zhang and X. Chen, Phytochemistry, 2023, 211, 113680 CrossRef CAS PubMed.
- P. Thapa, Y. J. Lee, T. T. Nguyen, D. Piao, H. Lee, S. Han, Y. J. Lee, A.-R. Han, H. Choi and J.-H. Jeong, Molecules, 2021, 26(6), 1762 CrossRef CAS PubMed.
- M. H. Park, J. E. Hong, C. J. Hwang, M. Choi, J. S. Choi, Y. J. An, D. J. Son and J. T. Hong, Arch. Pharmacal Res., 2016, 39, 721–729 CrossRef CAS PubMed.
- B. He, F. Xu, T. Yan, F. Xiao, B. Wu, Y. Wang, K. Bi and Y. Jia, Eur. J. Pharmacol., 2019, 842, 365–372 CrossRef CAS PubMed.
- L. Zuo, J. Li, L. Xue, Q. Jia, Z. Li, M. Zhang, M. Zhao, M. Wang, J. Kang and S. Du, Curr. Drug Metab., 2021, 22(1), 70–82 CAS.
- V. S. Honmore, A. D. Kandhare, P. P. Kadam, V. M. Khedkar, D. Sarkar, S. L. Bodhankar, A. A. Zanwar, S. R. Rojatkar and A. D. Natu, Int. Immunopharmacol., 2016, 33, 8–17 CrossRef CAS PubMed.
- J. Cao, H. Wang, F. Chen, J. Fang, A. Xu, W. Xi, S. Zhang, G. Wu and Z. Wang, Mol. Med. Rep., 2016, 13(5), 4238–4244 CrossRef CAS PubMed.
- K. Lin, Y. Wang, J. Gong, Y. Tan, T. Deng and N. Wei, Pharm. Biol., 2020, 58(1), 854–862 CrossRef CAS PubMed.
- R. Ou, L. Lin, M. Zhao and Z. Xie, Int. J. Biol. Macromol., 2020, 162, 1526–1535 CrossRef CAS PubMed.
- X. Zhang, Y. Deng, J. Xiang, H. Liu, J. Zhang, J. Liao, K. Chen, B. Liu, J. Liu and Y. Pu, Drug Des., Dev. Ther., 2020, 3393–3405 CrossRef CAS PubMed.
- Y. Xiong, H. Lu and H. Xu, Biol. Pharm. Bull., 2020, 43(11), 1634–1642 CrossRef CAS PubMed.
- L. Fang, H. Zhang, J. Zhou, Y. Geng and X. Wang, J. Anal. Methods Chem., 2018, 3158293 Search PubMed.
- A. A. Aloud, V. Chinnadurai, C. Govindasamy, M. A. Alsaif and K. S. Al-Numair, Pharm. Biol., 2018, 56(1), 302–308 CrossRef CAS PubMed.
- J. S. Lee, J. H. Kim, Y. K. Han, J. Y. Ma, Y. H. Kim, W. Li and S. Y. Yang, Int. J. Biol. Macromol., 2018, 120, 2442–2447 CrossRef CAS PubMed.
- R. Thangaiyan, S. Arjunan, K. Govindasamy, H. A. Khan, A. S. Alhomida and N. R. Prasad, Front. Pharmacol, 2020, 11, 585163 CrossRef CAS PubMed.
- J.-J. Lee, J.-H. Lee, N.-H. Yim, J.-H. Han and J. Y. Ma, Sci. Rep., 2017, 7(1), 8207 CrossRef PubMed.
- T. Erusappan, S. Paramasivam and S. P. Ekambaram, J. Ethnopharmacol., 2022, 288, 114975 CrossRef CAS PubMed.
- D. Wu, D. Ge, Y. Dai, Y. Chen, Q. Fu and Y. Jin, J. Sep. Sci., 2023, 2300156 CrossRef CAS PubMed.
- A. A. Elgazar, N. M. Selim, N. M. Abdel-Hamid, M. A. El-Magd and H. M. El Hefnawy, Phytother. Res., 2018, 32(7), 1273–1288 CrossRef CAS PubMed.
- K.-L. Ji, M.-Z. Wu, C.-Y. Huang, P.-C. GongPan, P. Sun, Y.-L. Sun, J. Li, C.-F. Xiao, Y.-K. Xu and Q.-F. Fan, J. Agric. Food Chem., 2022, 70(13), 3989–3999 CrossRef CAS PubMed.
- C.-Y. Li, S.-E. Cheng, S.-H. Wang, J.-Y. Wu, C.-W. Hsieh, H.-K. Tsou and M.-S. Tsai, Chin. J. Physiol., 2021, 64(1), 32 CrossRef CAS PubMed.
- J. Dong, M. Zhou, D.-b. Pan, Q.-y. Qin, T. Li, X.-s. Yao, H.-b. Li and Y. Yu, Food Funct., 2023, 14(21), 9755–9766 RSC.
- G. Yoo, J. H. Park and S. H. Kim, Bioorg. Chem., 2021, 107, 104526 CrossRef CAS PubMed.
- J. Dong, M. Zhou, Q. Qin, T. Li, X. Yao, J. Geng and Y. Yu, Bioorg. Chem., 2023, 134, 106431 CrossRef CAS PubMed.
- M. I. Sulistyowaty, N. H. Uyen, K. Suganuma, B.-Y. A. Chitama, K. Yahata, O. Kaneko, S. Sugimoto, Y. Yamano, S. Kawakami and H. Otsuka, Molecules, 2021, 26(6), 1756 CrossRef CAS PubMed.
- C. Cui, S.-L. Wu, J.-J. Chen, P. Gongpan, M. Guan and C.-A. Geng, J. Agric. Food Chem., 2023, 71(43), 16148–16159 CrossRef CAS PubMed.
- M. N. A. M. Taib, N. Anuar, K. M. Hanafiah, A. A. K. Al-Shammary, M. Saaid and K. Awang, Trop. Life Sci. Res., 2020, 31(1), 159 CrossRef PubMed.
- D. Lee, S.-R. Son, Y. Qi, K. S. Kang and D. S. Jang, Plants, 2023, 12(3), 579 CrossRef CAS PubMed.
- A. Simon, K. S. Nghiem, N. Gampe, Z. Garádi, I. Boldizsár, A. Backlund, A. Darcsi, A. N. Nedves and E. Riethmüller, Pharmaceutics, 2022, 14(9), 1967 CrossRef CAS PubMed.
- X.-L. Cheng, H.-X. Li, J. Chen, P. Wu, J.-H. Xue, Z.-Y. Zhou, N.-H. Xia and X.-Y. Wei, Nat. Prod. Bioprospect., 2021, 11, 63–72 CrossRef CAS PubMed.
- R. Baradwaj, M. Rao and T. S. Kumar, Biomed. Pharmacother., 2017, 91, 485–493 CrossRef CAS PubMed.
- A. M. Haque, K. H. Leong, Y. L. Lo, K. Awang and N. H. Nagoor, Phytomedicine, 2017, 31, 1–9 CrossRef PubMed.
- T. Ruttanaphan, G. de Sousa, A. Pengsook, W. Pluempanupat, H.-I. Huditz, V. Bullangpoti and G. Le Goff, Insects, 2020, 11(10), 686 CrossRef PubMed.
- Y. Manse, K. Ninomiya, R. Nishi, I. Kamei, Y. Katsuyama, T. Imagawa, S. Chaipech, O. Muraoka and T. Morikawa, Bioorg. Med. Chem., 2016, 24(23), 6215–6224 CrossRef CAS PubMed.
- N. Pradubyat, A. Giannoudis, T. Elmetwali, P. Mahalapbutr, C. Palmieri, C. Mitrpant and W. Ketchart, Planta Med., 2022, 88(02), 163–178 CrossRef CAS PubMed.
- S. Warit, K. Rukseree, T. Prammananan, P. Hongmanee, P. Billamas, S. Jaitrong, A. Chaiprasert, B. U. Jaki, G. F. Pauli and S. G. Franzblau, Sci. Pharm., 2017, 85(3), 32 CrossRef PubMed.
- A. Pengsook, A. Puangsomchit, T. Yooboon, V. Bullangpoti and W. Pluempanupat, Nat. Prod. Res., 2021, 35(23), 5261–5265 CrossRef CAS PubMed.
- X.-N. Ma, C.-L. Xie, Z. Miao, Q. Yang and X.-W. Yang, RSC Adv., 2017, 7(23), 14114–14144 RSC.
- V. S. Honmore, S. R. Rojatkar, L. U. Nawale, M. A. Arkile, V. M. Khedkar, A. D. Natu and D. Sarkar, Nat. Prod. Res., 2016, 30(24), 2825–2830 CrossRef CAS PubMed.
- W. Bai, T. Wang, X. Yang, Z. Wang, H. Li and J. Geng, Nat. Prod. Res., 2023, 22, 1–7 Search PubMed.
- Textbook Compilation Committee of Traditional Korean Medicine, Herbal Medicine, Yeonglimsa, 1991 Search PubMed.
- Textbook Compilation Committee of Korean Pharmacognosy, Pharmacognosy, Dongmyeongsa, 2001 Search PubMed.
- X. Zhang, X. Zhang, Y. Wang, F. Chen, Y. Li, Y. Li, Y. Tan, J. Gong, X. Zhong and H. Li, Nat. Prod. Res., 2018, 32(5), 529–535 CrossRef CAS PubMed.
- H. S. J. Chellammal, A. Veerachamy, D. Ramachandran, S. B. Gummadi, M. M. Manan and N. R. Yellu, Biomed. Pharmacother., 2019, 109, 1454–1461 CrossRef PubMed.
- W.-J. Zhang, J.-G. Luo, L.-Y. Kong and W. J. Trad, Chin. Med., 2016, 2(1), 26–41 Search PubMed.
- H. T. Van, T. D. Thang, T. N. Luu and V. D. Doan, RSC Adv., 2021, 11(60), 37767–37783 RSC.
- C. Huo, F. Han, Y. Xiao, H. J. Kim and I.-S. Lee, Int. J. Mol. Sci., 2022, 23(7), 3992 CrossRef CAS PubMed.
- C. L. Park, J. H. Kim, J.-S. Jeon, J.-h. Lee, K. Zhang, S. Guo, D.-h. Lee, E. M. Gao, R. H. Son and Y.-M. Kim, Antioxidants, 2022, 11(5), 1032 CrossRef CAS PubMed.
- F. K. Busayo, J.-L. Yang, X.-P. Ding, Y.-L. Wang, C.-J. Gai, F. Wu, H.-F. Dai, W.-L. Mei and H.-Q. Chen, Nat. Prod. Res., 2023, 5, 1–6 Search PubMed.
- B. Sur, S. Kang, M. Kim and S. Oh, Inflammation, 2019, 42, 928–936 CrossRef CAS PubMed.
- N. Natsume, T. Yonezawa, J.-T. Woo and T. Teruya, Cytotechnology, 2021, 73, 307–317 CrossRef CAS PubMed.
- Y. Okuno, S. Marumoto and M. Miyazawa, Nat. Prod. Res., 2019, 33(6), 862–865 CrossRef CAS PubMed.
- J. Liu, Y. Lu, B. Si, A. Tong, Y. Lu and L. Lv, Foods, 2023, 12(12), 2326 CrossRef CAS PubMed.
- X. Liang, B. Zhang, Q. Chen, J. Zhang, B. Lei, B. Li, Y. Wei, R. Zhai, Z. Liang and S. He, Drug Des., Dev. Ther., 2016, 841–850 CAS.
- T. Zhang, S. Guo, X. Zhu, J. Qiu, G. Deng and C. Qiu, J. Cell. Mol. Med., 2020, 24(15), 8430–8440 CrossRef CAS PubMed.
- I. Malami, A. B. Abdul, R. Abdullah, N. K. Bt Kassim, P. Waziri and I. Christopher Etti, Molecules, 2016, 21(4), 417 CrossRef PubMed.
- T.-G. Liu, K.-H. Sha, L.-G. Zhang, X.-X. Liu, F. Yang and J.-Y. Cheng, Microb. Pathog., 2019, 126, 239–244 CrossRef CAS PubMed.
- T. Taechowisan, T. Chuen-Im and W. S. Phutdhawong, Pak. J. Biol. Sci., 2022, 25(10), 922–928 CrossRef CAS PubMed.
- Z. Yong, H. Zibao, Z. Zhi, M. Ning, W. Ruiqi, C. Mimi, H. Xiaowen, D. Lin, X. Zhixuan and L. Qiang, Front. Pharmacol, 2022, 13, 909280 CrossRef CAS PubMed.
- B. He, F. Xu, F. Xiao, T. Yan, B. Wu, K. Bi and Y. Jia, Metab. Brain Dis., 2018, 33, 251–259 CrossRef CAS PubMed.
- Y. He, S. Chen, B. Tsoi, S. Qi, B. Gu, Z. Wang, C. Peng and J. Shen, Front. Cell Dev. Biol., 2021, 8, 577790 CrossRef PubMed.
- T. Kakegawa, A. Miyazaki and K. Yasukawa, J. Nat. Med., 2016, 70, 653–660 CrossRef CAS PubMed.
- Y. Zhang, Y.-Y. Yu, F. Peng, W.-T. Duan, C.-H. Wu, H.-T. Li, X.-F. Zhang and Y.-S. Shi, J. Agric. Food Chem., 2021, 69(32), 9229–9237 CrossRef CAS PubMed.
- C.-Y. An, M.-G. Son and Y.-W. Chin, ACS Omega, 2023, 8(36), 32804–32816 CrossRef CAS PubMed.
- Y.-T. Zhu, H.-B. Fang, X.-N. Liu, Y.-M. Yan, W.-S. Feng, Y.-X. Cheng and Y.-Z. Wang, Phytochemistry, 2023, 215, 113849 CrossRef CAS PubMed.
- G. Zhang, L. Zhao, J. Zhu, Y. Feng and X. Wu, Biomed. Chromatogr., 2018, 32(2), e4094 CrossRef PubMed.
- G. Fu, W. Zhang, D. Du, Y. P. Ng, F. C. Ip, R. Tong and N. Y. Ip, J. Agric. Food Chem., 2017, 65(31), 6608–6614 CrossRef CAS PubMed.
- Y. C. Hseu, Y. C. Huang, V. Thiyagarajan, D. C. Mathew, K. Y. Lin, S. C. Chen, J. Y. Liu, L. S. Hsu, M. L. Li and H. L. Yang, J. Cell. Physiol., 2019, 234(10), 17514–17526 CrossRef CAS PubMed.
- X.-F. He, S.-L. Wu, J.-J. Chen, J. Hu, X.-Y. Huang, T.-Z. Li, X.-M. Zhang, Y.-Q. Guo and C.-A. Geng, Bioorg. Chem., 2022, 120, 105653 CrossRef CAS PubMed.
- N. Taira, B. C. Q. Nguyen and S. Tawata, Molecules, 2017, 22(1), 132 CrossRef PubMed.
- Q.-Y. Zou, H.-F. Wu, Y.-L. Tang and D.-Z. Chen, Nat. Prod. Res., 2016, 30(1), 1–6 CrossRef CAS PubMed.
- K.-R. Park, H. Lee, M. Cho and H.-M. Yun, Int. J. Mol. Sci., 2020, 21(10), 3700 CrossRef CAS PubMed.
- Y. Tezuka, M. S. Ali, A. H. Banskota and S. Kadota, Tetrahedron Lett., 2000, 41(31), 5903–5907 CrossRef CAS.
- P. K. Sharma, S. Fuloria, M. Ali, A. Singh, S. P. Kushwaha, V. K. Sharma, V. Subramaniyan and N. K. Fuloria, Pak. J. Biol. Sci., 2021, 34(4), 1397–1401 CAS.
- Y. Chen, G. Li, H. C. H. Law, H. Chen and S. M.-Y. Lee, J. Agric. Food Chem., 2020, 68(40), 11170–11181 CrossRef CAS PubMed.
- H. Liu, X. Wang, Q. Shi, L. Li, Q. Zhang, Z.-L. Wu, X.-J. Huang, Q.-W. Zhang, W.-C. Ye and Y. Wang, ACS Omega, 2020, 5(17), 10167–10175 CrossRef CAS PubMed.
- H. Liu, Z.-L. Wu, X.-J. Huang, Y. Peng, X. Huang, L. Shi, Y. Wang and W.-C. Ye, J. Nat. Prod., 2018, 81(1), 162–170 CrossRef CAS PubMed.
- N. Wei, Z. Zhou, Q. Wei, Y. Wang, J. Jiang, J. Zhang, L. Wu, S. Dai and Y. Li, Nat. Prod. Res., 2016, 30(20), 2344–2349 CrossRef CAS PubMed.
- X.-B. Wang, C.-S. Yang, J.-G. Luo, C. Zhang, J. Luo, M.-H. Yang and L.-Y. Kong, Fitoterapia, 2017, 119, 121–129 CrossRef CAS PubMed.
- J.-W. Nam and Y.-S. Lee, Molecules, 2017, 22(10), 1750 CrossRef PubMed.
- X.-F. He, J.-J. Chen, T.-Z. Li, J. Hu, X.-M. Zhang and C.-A. Geng, Bioorg. Chem., 2021, 108, 104683 CrossRef CAS PubMed.
- N. Nam Hoang, T. Kodama, N. Nwet Win, Prema, K. Minh Do, I. Abe and H. Morita, Chem. Biodiversity, 2021, 18(10), e2100401 CrossRef CAS PubMed.
- Y. Nishidono, Y. Iwama, S. Shirako, T. Ishii, T. Okuyama, M. Nishizawa and K. Tanaka, Nat. Prod. Res., 2023, 37(21), 3694–3701 CrossRef CAS PubMed.
- T. Pluskal, M. P. Torrens-Spence, T. R. Fallon, A. De Abreu, C. H. Shi and J.-K. Weng, Nat. Plants, 2019, 5(8), 867–878 CrossRef PubMed.
- L. Wu, Z. Liao, C. Liu, H. Jia and J. Sun, Chem. Biodiversity, 2016, 13(6), 645–671 CrossRef CAS PubMed.
- Q.-X. Wu, Y.-P. Shi and Z.-J. Jia, Nat. Prod. Rep., 2006, 23(5), 699–734 RSC.
- L. Ding, H. Görls and C. Hertweck, Magn. Reson. Chem., 2021, 59(1), 34–42 CrossRef CAS PubMed.
- Y. Manse, K. Ninomiya, R. Nishi, Y. Hashimoto, S. Chaipech, O. Muraoka and T. Morikawa, Molecules, 2017, 22(12), 2279 CrossRef PubMed.
- E. L. Ghisalberti, Phytochemistry, 1994, 37(3), 597–623 CrossRef CAS.
- L.-G. Chen, P.-J. Su, P.-W. Tsai, L.-L. Yang and C.-C. Wang, Planta Med., 2017, 83(1–2), 151–157 CAS.
- I. Chakrabartty, A. Vijayasekhar and L. Rangan, Nat. Prod. Res., 2021, 35(6), 1000–1004 CrossRef CAS PubMed.
- Y. K. Loo, K. H. Leong, Y. Sivasothy, H. Ibrahim and K. Awang, Chem. Biodiversity, 2019, 16, e1900032 CrossRef PubMed.
- H.-J. Jang, S.-J. Lee, S. Lee, K. Jung, S. W. Lee and M.-C. Rho, Molecules, 2017, 22(10), 1611 CrossRef PubMed.
- X.-g. Zhang, A.-x. Liu, Y.-x. Zhang, M.-y. Zhou, X.-y. Li, M.-h. Fu, Y.-p. Pan, J. Xu and J.-q. Zhang, J. Ethnopharmacol., 2022, 295, 115397 CrossRef CAS PubMed.
- H. J. Lim, S. G. Bak, H. J. Lim, S. W. Lee, S. Lee, S.-K. Ku, S.-I. Park, S.-J. Lee and M.-C. Rho, Molecules, 2020, 25(15), 3345 CrossRef CAS PubMed.
- X. Huang, G. Tang, Y. Liao, X. Zhuang, X. Dong, H. Liu, X.-J. Huang, W.-C. Ye, Y. Wang and L. Shi, Biol. Pharm. Bull., 2016, 39(12), 1961–1967 CrossRef CAS PubMed.
- X. Li, H. Wen, Y. Zhang, A. Liu, X. Zhang, M. Fu, Y. Pan, J. Xu and J. Zhang, Front. Pharmacol, 2022, 13, 956812 CrossRef CAS PubMed.
- Y. Li, T. Li, J.-Z. Li and Q.-S. Wu, Cutaneous Ocul. Toxicol., 2017, 36(3), 273–277 CrossRef CAS PubMed.
- R. D. Haworth, J. Chem. Soc., 1942, 448–456 RSC.
- O. R. Gottlieb and A. Da Rocha, Phytochemistry, 1972, 11(5), 1861–1863 CrossRef CAS.
- H. You, M. He, D. Pan, G. Fang, Y. Chen, X. Zhang, X. Shen and N. Zhang, Nat. Prod. Res., 2022, 36(22), 5740–5746 CrossRef CAS PubMed.
- T. Juwitaningsih, L. D. Juliawaty and Y. M. Syah, Nat. Prod. Commun., 2016, 11(9), 1297–1298 CrossRef PubMed.
- I. Malami, A. Muhammad, I. B. Abubakar, I. C. Etti, P. M. Waziri, R. M. Abubakar and H. E. Mshelia, Nat. Prod. Res., 2018, 32(24), 2964–2967 CrossRef CAS PubMed.
- Y. Nishidono, R. Okada, Y. Iwama, T. Okuyama, M. Nishizawa and K. Tanaka, Fitoterapia, 2020, 140, 104444 CrossRef CAS PubMed.
- M. Kumagai, T. Mishima, A. Watanabe, T. Harada, I. Yoshida, K. Fujita, M. Watai, S. Tawata, K. Nishikawa and Y. Morimoto, Biosci., Biotechnol., Biochem., 2016, 80(7), 1425–1432 CrossRef CAS PubMed.
- I.-K. Lee and B.-S. Yun, J. Antibiot., 2011, 64(5), 349–359 CrossRef CAS PubMed.
- R. B. Soares, R. J. Dinis-Oliveira and N. G. Oliveira, J. Clin. Med., 2022, 11(14), 4039 CrossRef CAS PubMed.
- Z. Stiplošek, M. Šindler-Kulyk, K. Jakopčić, A. Višnjevac and B. KojiĆ-ProdiĆ, J. Heterocycl. Chem., 2002, 39(1), 37–44 CrossRef.
- S.-L. Hu and C.-P. Zheng, Cutaneous Ocul. Toxicol., 2018, 37(3), 245–251 CrossRef CAS PubMed.
- Z. Liu, L. Xie, T. Bian, G. Qi and Z. Wang, Anatolian J. Cardiol., 2018, 19(3), 198 CAS.
- K. S. Kumar, S.-H. Wang, Y.-H. Tseng, N.-W. Tsao, Y.-H. Kuo and S.-Y. Wang, Phytomedicine, 2018, 50, 223–230 CrossRef CAS PubMed.
- D. Lakshmanan, A. Harikrishnan, K. Jyoti, M. Idul Ali and K. Jeevaratnam, J. Appl. Microbiol., 2020, 128(5), 1355–1365 CrossRef CAS PubMed.
- I. Malami, A. B. Abdul, R. Abdullah, N. K. B. Kassim, R. Rosli, S. K. Yeap, P. Waziri, I. C. Etti and M. B. Bello, PLoS One, 2017, 12(1), e0170233 CrossRef PubMed.
- P. Wang, J.-m. Jin, X.-h. Liang, M.-z. Yu, C. Yang, F. Huang, H. Wu, B.-b. Zhang, X.-y. Fei and Z.-t. Wang, Acta Pharmacol. Sin., 2022, 43(6), 1581–1593 CrossRef CAS PubMed.
- G. Atwa, G. Omran, A. Abd Elbaky and T. Okda, Wspolczesna Onkol., 2021, 25(3), 174–184 CrossRef CAS PubMed.
- B. C. Q. Nguyen, N. Taira and S. Tawata, Drug Des. Discovery, 2014, 8(6), 238–244 Search PubMed.
- B. C. Q. Nguyen, N. Taira, H. Maruta and S. Tawata, Phytother. Res., 2016, 30(1), 120–127 CrossRef CAS PubMed.
- M.-S. Kang, J.-H. Park and H.-S. Lee, Exp. Appl. Acarol., 2022, 86(2), 313–326 CrossRef CAS PubMed.
- R. Datta, A. Kaur, I. Saraf, I. P. Singh and S. Kaur, Ecotoxicol. Environ. Saf., 2019, 168, 324–329 CrossRef CAS PubMed.
- J. L. Andersen, E. Le Rouzic and V. Planelles, Exp. Mol. Pathol., 2008, 85(1), 2–10 CrossRef CAS PubMed.
- Y. She, Q. Zheng, X. Xiao, X. Wu and Y. Feng, Anal. Bioanal. Chem., 2016, 408, 3185–3201 CrossRef CAS PubMed.
- V. S. Honmore, A. D. Kandhare, P. P. Kadam, V. M. Khedkar, A. D. Natu, S. R. Rojatkar and S. L. Bodhankar, J. Ethnopharmacol., 2019, 229, 233–245 CrossRef CAS PubMed.
- L. Fang, Y. Yan, Z. Xu, Z. He, S. Zhou, X. Jiang, F. Wu, X. Yuan, T. Zhang and D. Yu, Eur. J. Pharmacol., 2021, 902, 174100 CrossRef CAS PubMed.
- E. Lilin, W. Li, Y. Hu, L. Deng, J. Yao and X. Zhou, Acta Biochim. Biophys. Sin., 2023, 55(11), 1806 CrossRef CAS PubMed.
- M. J. Benskey, R. G. Perez and F. P. Manfredsson, J. Neurochem., 2016, 137(3), 331–359 CrossRef CAS PubMed.
- H. Zhou, S. Li, C. Li, X. Yang, H. Li, H. Zhong, J.-H. Lu and S. M.-Y. Lee, Aging Dis., 2020, 11(3), 559 CrossRef PubMed.
- Y. Bian, Y. Chen, X. Wang, G. Cui, C. O. L. Ung, J.-H. Lu, W. Cong, B. Tang and S. M.-Y. Lee, J. Adv. Res., 2021, 34, 1–12 CrossRef CAS PubMed.
- G. Daryabor, M. Atashzar, D. Kabelitz, S. Meri and K. Kalantar, Front. Immunol., 2020, 11, 1582 CrossRef CAS PubMed.
- A. M. Freeman, and N. Pennings, Insulin resistance, StatPearls, 2018 Search PubMed.
- X. Zhang, Y. Zhang, M. Zhou, Y. Xie, X. Dong, F. Bai and J. Zhang, Front. Pharmacol, 2022, 12, 792977 CrossRef PubMed.
- M. Kumagai, K. Nishikawa, T. Mishima, I. Yoshida, M. Ide, K. Koizumi, M. Nakamura and Y. Morimoto, Bioorg. Med. Chem. Lett., 2017, 27(11), 2401–2406 CrossRef CAS PubMed.
- M. Lu, L. Tan, X.-G. Zhou, Z.-L. Yang, Q. Zhu, J.-N. Chen, H.-R. Luo and G.-S. Wu, Biogerontology, 2020, 21, 669–682 CrossRef CAS PubMed.
- Y. Wang, M. Wang, M. Xu, T. Li, K. Fan, T. Yan, F. Xiao, K. Bi and Y. Jia, Int. Immunopharmacol., 2018, 62, 77–85 CrossRef CAS PubMed.
- Y. Qi, X. Cheng, G. Gong, T. Yan, Y. Du, B. Wu, K. Bi and Y. Jia, Food Funct., 2020, 11(3), 2427–2438 RSC.
- C. Dai, M. Liu, Q. Zhang, S. Das Gupta, S. Tang and J. Shen, Antioxidants, 2023, 12(2), 370 CrossRef CAS PubMed.
- C.-M. Chen, C.-Y. Lin, Y.-P. Chung, C.-H. Liu, K.-T. Huang, S.-S. Guan, C.-T. Wu and S.-H. Liu, Nutrients, 2021, 13(11), 3921 CrossRef CAS PubMed.
- E. Yoo, J. Lee, P. Lertpatipanpong, J. Ryu, C.-T. Kim, E.-Y. Park and S. J. Baek, BMC Cancer, 2020, 20(1), 1–12 CrossRef PubMed.
- H.-X. Wang and C. Tang, Oncol. Rep., 2017, 38(2), 703–714 CrossRef CAS PubMed.
- X. Long, L. Chen, J. Yang, T. Dong, Q. Cheng, W. Wang, Y. Zou, Y. Su, W. Dai and B. Chen, Anti-Cancer Agents Med. Chem., 2023, 23(7), 847–857 CrossRef CAS PubMed.
- X. Liang, P. Wang, C. Yang, F. Huang, H. Wu, H. Shi and X. Wu, Front. Pharmacol, 2021, 12, 646628 CrossRef CAS PubMed.
- K. Ren, W. Zhang, G. Wu, J. Ren, H. Lu, Z. Li and X. Han, Biomed. Pharmacother., 2016, 84, 1748–1759 CrossRef CAS PubMed.
- S. Yu, L.-s. Gong, N.-f. Li, Y.-f. Pan and L. Zhang, Biomed. Pharmacother., 2018, 97, 213–224 CrossRef CAS PubMed.
- T. Yang, H. Liu, C. Yang, H. Mo, X. Wang, X. Song, L. Jiang, P. Deng, R. Chen and P. Wu, Drug Des., Dev. Ther., 2023, 2495–2511 CAS.
- N. Chaihongsa, P. Maneesai, W. Sangartit, S. Rattanakanokchai, P. Potue, J. Khamseekaew, S. Bunbupha and P. Pakdeechote, Biomed. Pharmacother., 2022, 152, 113231 CrossRef CAS PubMed.
- H.-Y. Song, W. S. Kim, J. M. Han, H. S. Seo, S.-T. Lim and E.-B. Byun, J. Nutr. Biochem., 2021, 87, 108524 CrossRef CAS PubMed.
- Y. Xing, M.-S. Zhang, J.-H. Xiao and R.-M. Liu, Eur. J. Pharmacol., 2022, 935, 175326 CrossRef CAS PubMed.
- Y. Jin, P. Yang, L. Wang, Z. Gao, J. Lv, Z. Cui, T. Wang, D. Wang and L. Wang, J. Cell. Mol. Med., 2022, 26(3), 828–839 CrossRef CAS PubMed.
- X. Zhao, X. Guo, J. Shen and D. Hua, Mol. Med. Rep., 2018, 18(4), 4030–4036 CAS.
- Y. Zhang, Y. Zhang, Y. Li, L. Zhang and S. Yu, Front. Pharmacol, 2021, 12, 687491 CrossRef CAS PubMed.
- X. He, Z. Wei, J. Wang, J. Kou, W. Liu, Y. Fu and Z. Yang, Sci. Rep., 2016, 6(1), 28370 CrossRef CAS PubMed.
- Y. Tan and C. Zheng, Am. J. Med. Sci., 2018, 355(4), 377–386 CrossRef PubMed.
- Y. Liu, K. Wang, Q. Feng, Y. Zhang, C. Wang, Q. Liu, X. Liu, X. Wang, W. Gao and X. Bai, J. Immunol. Res., 2021, 9998517 CAS.
- D. Dong, Y. Zhang, H. He, Y. Zhu and H. Ou, FASEB J., 2022, 36(4), e22261 CrossRef CAS PubMed.
- Q. Xu, Y. Fan, J. J. Loor, Y. Liang, X. Sun, H. Jia, C. Zhao and C. Xu, Front. Pharmacol, 2020, 11, 601716 CrossRef CAS PubMed.
- C.-C. Wang, J.-W. Lu, Y.-J. Peng, C.-H. Lee, H.-S. Lee, Y.-H. Chu, C.-J. Huang, Y.-J. Ho, F.-C. Liu and C.-C. Wu, Medicina, 2021, 57(9), 898 CrossRef PubMed.
- K. Wang, Q. Lv, Y.-m. Miao, S.-m. Qiao, Y. Dai and Z.-f. Wei, Biochem. Pharmacol., 2018, 155, 494–509 CrossRef CAS PubMed.
- W. Lee, D. Hahn, H. Sim, S. Choo, S. Lee, T. Lee and J.-S. Bae, Fitoterapia, 2020, 146, 104724 CrossRef CAS PubMed.
- C. Xie, H. Ma, Y. Shi, J. Li, H. Wu, B. Wang, Z. Shao, C. Huang, J. Chen and L. Sun, Food Funct., 2021, 12(6), 2703–2714 RSC.
- K. Barber, P. Mendonca, J. A. Evans and K. F. Soliman, Int. J. Mol. Sci., 2023, 24(13), 10872 CrossRef CAS PubMed.
- W. Qi, W. Boliang, T. Xiaoxi, F. Guoqiang, X. Jianbo and W. Gang, Biomed. Pharmacother., 2020, 122, 109547 CrossRef PubMed.
- N. A. Badroon, N. Abdul Majid and M. A. Alshawsh, Nutrients, 2020, 12(6), 1757 CrossRef CAS PubMed.
- H. Sun, N. Zhang, Y. Jin and H. Xu, Dose-Response, 2021, 19(4), 15593258211042163 CrossRef CAS PubMed.
- N. C. Liao, Y. L. Shih, M. T. Ho, T. J. Lu, C. H. Lee, S. F. Peng, S. J. Leu and J. G. Chung, Environ. Toxicol., 2020, 35(4), 457–467 CrossRef CAS PubMed.
- N. K. Kaswan, N. A. B. Mohammed Izham, T. A. S. Tengku Mohamad, M. R. Sulaiman and E. K. Perimal, Molecules, 2021, 26(12), 3677 CrossRef CAS PubMed.
- S. Wang, C. Zhai, Y. Zhang, Y. Yu, Y. Zhang, L. Ma, S. Li and Y. Qiao, Molecules, 2016, 21(9), 1145 CrossRef PubMed.
- Y.-H. Jin, J. S. Min and S. Kwon, Nutrients, 2023, 15(6), 1335 CrossRef CAS PubMed.
- N. H. Phuah, M. N. Azmi, K. Awang and N. H. Nagoor, OncoTargets Ther., 2017, 1695–1705 CrossRef CAS PubMed.
- S. P. Sok, N. M. Arshad, M. N. Azmi, K. Awang, B. Ozpolat and N. Hasima Nagoor, PLoS One, 2017, 12(2), e0171329 CrossRef PubMed.
- A. Kojima-Yuasa, T. Yamamoto, K. Yaku, S. Hirota, S. Takenaka, K. Kawabe and I. Matsui-Yuasa, Chem.-Biol. Interact., 2016, 257, 101–109 CrossRef CAS PubMed.
- K. Yaku, I. Matsui-Yuasa and A. Kojima-Yuasa, Planta Med., 2018, 84(03), 153–159 CrossRef CAS PubMed.
- G. H. Ong, D. Ori, T. Kawasaki and T. Kawai, Genes Cells, 2022, 27(7), 482–492 CrossRef CAS PubMed.
- T. Ikeya, Y. Terada, Y. Morimitsu, K. Kubota, K. Ito and T. Watanabe, Biosci., Biotechnol., Biochem., 2021, 85(10), 2191–2194 CrossRef PubMed.
- R. Hartati, A. G. Suganda and I. Fidrianny, Procedia Chem., 2014, 13, 150–163 CrossRef CAS.
- R. Cai, X. Yue, Y. Wang, Y. Yang, D. Sun, H. Li and L. Chen, J. Ethnopharmacol., 2021, 281, 114563 CrossRef CAS PubMed.
- M. Deng, X. Yun, S. Ren, Z. Qing and F. Luo, Molecules, 2022, 27(9), 2826 CrossRef CAS PubMed.
- D.-J. Sun, L.-J. Zhu, Y.-Q. Zhao, Y.-Q. Zhen, L. Zhang, C.-C. Lin and L.-X. Chen, Fitoterapia, 2020, 142, 104490 CrossRef CAS PubMed.
- S. Kadota, Y. Tezuka, J. Prasain, M. Shawkat Ali and A. H. Banskota, Curr. Top. Med. Chem., 2003, 3(2), 203–225 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |