Open Access Article
Open Access ArticlePhytosulfokine peptide library: chemical synthesis and biological evaluation on protoplast regeneration†
Jasper W.
van de Sande
 a,
Dieuwertje E.
Streefkerk
a,
Richard G. H.
Immink
b,
Martijn
Fiers
b and
Bauke
Albada
a,
Dieuwertje E.
Streefkerk
a,
Richard G. H.
Immink
b,
Martijn
Fiers
b and
Bauke
Albada
 *a
*a
aLaboratory of Organic Chemistry, Wageningen University and Research, Stippeneng 4, 6708 WE Wageningen, The Netherlands. E-mail: bauke.albada@wur.nl
bBioscience, Wageningen University and Research, PO Box 16, 6700 AA Wageningen, The Netherlands
First published on 26th March 2024
Abstract
Phytosulfokine (PSK) is a highly conserved plant peptide hormone (PPH) containing two sulfated tyrosine residues. PSK acts as a growth factor in Arabidopsis thaliana plant species and significantly improves the efficiency of cellular proliferation in plant cell culture, a process known as regeneration. However, many important plant species are recalcitrant under exposure to PSK and require e.g., artificially generated PPHs to boost regeneration. Here, we report the design, chemical synthesis, and biological evaluation of the regeneration capacity of a PSK-like peptide library. Our set of thirteen PSK analogues included N-terminal modifications, amino acid substitutions, incorporation of diastereomers, and backbone N-methylation. Insertion of the latter required solution-phase synthesis of the proper building block prior to solid-phase peptide synthesis. The biological activity of the PSK analogues was assessed in the regeneration capacity of Brassica oleracea cells (protoplasts). Most PSK analogues did not affect the regeneration capacity as compared to PSK. For example, when an alanine substitution was introduced and replaced either 2-Ile or 4-Thr, the bio-activity was fully abolished. However, 2-allo-Ile PSK and 4-Nme-Thr PSK showed increased activity in protoplast regeneration when compared with that of the native peptide. These findings indicated that introducing small chemical modifications in the PPH PSK provides opportunities for inducing protoplast regeneration in recalcitrant species.
Introduction
Environmental influences significantly impact plant development and growth. Stress conditions can change plant growth, reduce crop productivity, alter gene expression, or affect cellular metabolism.1,2 In an adaptive response, plant tissue can withstand stress by re-initiation of cell division, or preservation of totipotent cells.3 Analogous to this process, responsive plant tissues or cells can resume cell division when exposed to media enriched with the phytohormone auxin and cytokinin, giving rise to new organs and the potential to grow a full plant that can complete its lifecycle.4 In the field of plant research, farming practices and plant breeding are investigated as methods to reduce plant stress.5–7 Unfortunately, many plant species that are important from an agricultural point of view display poor tolerance to stress conditions, and furthermore, are challenging to improve by efficient breeding and genetic engineering methods due to their recalcitrance in regeneration. Plant hormones are key in the control of totipotency and regeneration capacity and represent signaling molecules that are involved in signal reception, signal transduction, and cellular response. To transmit information intra- and intercellularly between plant cells and their external environment, chemical signaling is used for down- and upregulation of gene expression.8 Various plant hormones have been identified as important regulatory molecules in plant cells and their involvement with plant signaling and molecular trafficking has been established. For example, abscisic acid, auxin, cytokinin, ethylene, and the family of gibberellins have been identified as major plant hormones.9 These hormones play an important role in initiating plant dormancy, cell differentiation, cell division, fruit ripening, and stem elongation, respectively. In addition to these phytohormones, the family of plant peptide hormones (PPHs) was also identified as important in plant signaling.10–13Phytosulfokine (PSK) is a secreted PPH that is bioactive even at nanomolar concentrations.14 It was first isolated from an Asparagus officinalis L. mesophyll cell culture medium and found to promote cell division of asparagus mesophyll cells cultured at low cell density.15 The PPH is a disulfated pentapeptide H-Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln-OH, which is known as PSK-α. In this peptide, both tyrosine residues contain a post-translationally installed sulfate group that is introduced on a PSK precursor by tyrosylprotein sulfotransferase (TPST) using 3′-phosphoadenosine-5′-phosphosulfate (PAPS) as a substrate.16,17 Analysis of PSK analogues revealed the importance of the sulfated residues for its biological activity (i.e., nonsulfated PSK is inactive below 100 nM in stimulating cell proliferation at low density),18 and identified Tyr(SO3H)-Ile-Tyr(SO3H) as the core requirement of PSK to display activity (i.e., the pharmacophore). In the cell, PSK is synthesized from 80–120 amino acid long prepropeptides that are encoded by small gene families and unlike these precursors, the mature PSK peptide is strongly conserved.19,20
The formed mature peptide is recognized by the PSK receptor (PSKR1, Fig. 1A), a transmembrane receptor belonging to the leucine-rich repeat receptor-like kinase (LRR-RLK) family with more than 600 members in Arabidopsis and over 1100 members in rice.21–23 The PSKR1 contains an extracellular domain, located in the cell membrane by a single transmembrane helix, and a C-terminal intracellular kinase domain.24 Binding sites of PSK to the PSKR1 have been found in the plasma membrane of rice suspension cells, asparagus, carrot, maize, tobacco, and tomato plants with photoaffinity labeling and radiolabeling experiments.25–27 Moreover, the crystal structure of the PSKR1 (Fig. 1B) has been resolved. In addition, the formation of the PSK-receptor complex, and allosteric activation by PSK has been discovered, leading to detailed insights in peptide-receptor interaction and receptor conformation in both bound and unbound state.28,29
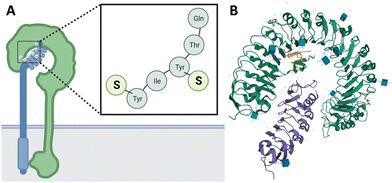 | ||
| Fig. 1 (A) Illustration of phytosulfokine (PSK) being perceived at the plasma membrane by the PSK receptor (PSKR1) and co-receptor SERK1. S represents the sulfate modification on the tyrosine residue. (B) Crystal structure of the plant peptide hormone receptor-coreceptor complex including ligand (PSK-PSKR1-SERK1) in Arabidopsis thaliana (PDB-code: 4z64; receptor in green, co-receptor in purple, PSK in orange). | ||
PSK is universally found in both monocotyledon and dicotyledon plant species and strongly promotes proliferation of plant cells in culture.19 In addition to cell proliferation, PSK also promotes plant cell differentiation, organogenesis, and somatic embryogenesis.30,31 Previously, PSK was tested for its ability to enhance regeneration in cauliflower varieties, showing its variety-dependent potential in shoot regeneration.32,33 In addition to cauliflower, PSK was also tested in sugar beet protoplast regeneration, in which some recalcitrant cultures could be induced to cell division upon application of exogenous PSK peptide.34 So far, characteristics and functions of PSK has been reviewed quite extensively.35–38 PSK have been categorized as a growth factor in Arabidopsis thaliana.39,40 As we now understand, it has many more activities besides growth regulation, such as responses to biotic and abiotic stress and reproductive processes.41,42 In recalcitrant plant species there are still several bottlenecks that need to be solved before PSK can be successfully used as a regenerator peptide. Based on literature, the PSK peptide has potential, but needs optimization for efficient plant regeneration in a broad range of plant varieties, for example Brassica oleracea (red cabbage). Therefore, we used PSK as starting point in search for modified PSK peptides with improved performance in protoplast regeneration of recalcitrant plant species.
Here, we report the chemical synthesis and analysis of the bio-activity of a set of PSK analogues that are not yet present in literature.43–45 To evaluate the PSK-like library of mutated PSK peptides, screening for any induction or enhancement of regeneration as compared to the original PSK peptide was assessed by determining their effect on the regeneration of Brassica oleracea protoplasts. Ultimately, this will lead to new insights into structural features of PSK-like peptides, and the effects of these peptides on cell growth, that improve the regeneration capacity and affects the stress levels of the plant.
Results and discussion
PSK was first discovered by phenotypic analysis and biochemical purification in Asparagus officinalus L. culture medium by Matsubayashi et al.15 The same group was also the first to report the chemical synthesis of PSK-α in 1996.45 In addition to PSK-α, various analogues were reported and structure–activity relationships were established according to mitogenic activities measured by a highly sensitive bioassay method. Results of these studies indicated that the N-terminal tripeptide of PSK acts as the active core.Chemical synthesis of phytosulfokine (PSK)
PSK (1, Fig. 2) was synthesized following standard Fmoc/tBu-based solid-phase peptide synthesis (SPPS). Starting with a Wang resin preloaded with Fmoc-Gln(Trt) (loading: 0.4–0.9 mmol g−1), the peptide chain was elongated with Fmoc-Thr(tBu)-OH using the coupling reagents 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyl-uronium hexafluorophosphate (HBTU) and 1-hydroxy-benzotriazole (HOBt) in the presence of DIPEA. Subsequently, Fmoc-Tyr(SO2ONp)-OH, Fmoc-Ile-OH, and Fmoc-Tyr(SO2ONp)-OH were coupled using similar chemistry. We used commercially available Fmoc-Tyr(SO2ONp)-OH, which contains a neopentyl (Np) protecting group for the sulfated tyrosine residue, as a stable and easy cleavable protecting group to incorporate the sulfated tyrosine residues.46 The Np moiety was removed either by conditions involving unhindered strong nucleophiles in polar aprotic solvent with long reaction times and elevated temperatures,47 or by treatment with an aqueous solution of ammonium acetate (NH4OAc) at 37 °C for at least 12 h,48 resulting in the sulfate monoester.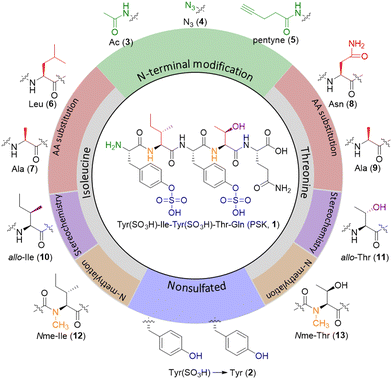 | ||
| Fig. 2 Chemical structure of PSK (1) and its analogues. Nonsulfated PSK (2); N-terminal modified PSK (3–5); amino acid substituted PSK (6–9); diastereomer PSK (10–11); N-methylated PSK (12–13). | ||
After completion of PSK synthesis on solid support, the resin-bound peptide H-Tyr(SO2ONp)-Ile-Tyr(SO2ONp)-Thr(tBu)-Gln(Trt)-OH was cleaved from the solid support and concomitant removal of tert-butyl and trityl protecting groups with an acidic TFA cocktail (95% TFA: 2.5% triisopropylsilane (TIS): 2.5% H2O, reaction time: 1.5–2 h). After acidic cleavage, the crude peptide was precipitated in ice-cold ether and the residue was dissolved in H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN 6
MeCN 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 from which it was lyophilized. The lyophilized product was first purified by reversed-phase (RP) HPLC using acidic buffers. After purification and lyophilization of the PSK-containing fractions, the obtained powder was dissolved in a 1–2 M NH4OAc solution and a minimum amount of acetonitrile as co-solvent, and subsequently heated to 40–45 °C for 40 h. RP-HPLC analysis of the mixture at different time-points showed complete Np-removal after 40 h (Fig. S1, ESI†). The resulting sulfate monoester was purified by preparative RP-HPLC using 10 mM NH4OAc buffers. Purified PSK was analyzed with RP-HPLC coupled to a mass spectrometer (MS), and with nuclear magnetic resonance (NMR; peak assignment was done with standard 2D NMR techniques, Fig. S2, ESI†). In MS, sulfate fragmentation might occur due to small traces of acid in the system (see ESI† for analytical spectra and data). Non-sulfated PSK (PSK-2, Fig. 2) was synthesized analogously, although added steps as result of the sulfated Tyr could be omitted. Compared to the 1H NMR of PSK-1, non-sulfated PSK-2 only shows notable differences in ppm peaks for the protons of the tyrosine residue (Fig. S1, ESI†).
4 from which it was lyophilized. The lyophilized product was first purified by reversed-phase (RP) HPLC using acidic buffers. After purification and lyophilization of the PSK-containing fractions, the obtained powder was dissolved in a 1–2 M NH4OAc solution and a minimum amount of acetonitrile as co-solvent, and subsequently heated to 40–45 °C for 40 h. RP-HPLC analysis of the mixture at different time-points showed complete Np-removal after 40 h (Fig. S1, ESI†). The resulting sulfate monoester was purified by preparative RP-HPLC using 10 mM NH4OAc buffers. Purified PSK was analyzed with RP-HPLC coupled to a mass spectrometer (MS), and with nuclear magnetic resonance (NMR; peak assignment was done with standard 2D NMR techniques, Fig. S2, ESI†). In MS, sulfate fragmentation might occur due to small traces of acid in the system (see ESI† for analytical spectra and data). Non-sulfated PSK (PSK-2, Fig. 2) was synthesized analogously, although added steps as result of the sulfated Tyr could be omitted. Compared to the 1H NMR of PSK-1, non-sulfated PSK-2 only shows notable differences in ppm peaks for the protons of the tyrosine residue (Fig. S1, ESI†).
Design of PSK analogues
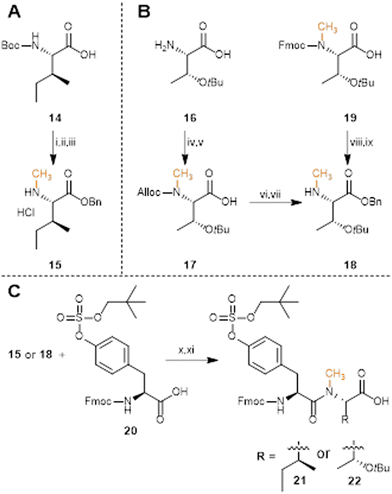
Over 50 PSK analogues have been reported in different studies.43–45 These analogues mainly focused on changes in the tyrosine residues, for example by incorporation of non-proteinogenic aromatic amino acid residues or halogenated phenol amino acids, and showed that the presence of a sulfate group at Tyr-1 or Tyr-3 of the PSK peptide plays an important role in the bio-activity of the PSK peptide. Here we select analogues with different types of small modifications while maintaining the sulfated tyrosine residues intact (Fig. 2).We first installed N-terminal modifications such as an acetyl (Ac-PSK 3), or an azide (N3-PSK 4) or pentynoic amide (pentyne-PSK 5) for further bioconjugation strategies.49 These modifications can be conveniently introduced as final step in SPPS or, for the azide, after peptide purification using a transfer reagent.50 For mutations of the amino acids, Matsubayashi et al.45 reported >95% reduction in activity of a PSK analogue which lacks the δ-CH3-group on position 2 (resulting from the Ile2Val substitution), but also for mutations on position 4 with for example serine. We report four additional mutations at these two sites, namely 2-Leu PSK and 2-Ala PSK (analogues 6 and 7, respectively), and 4-Asn PSK and 4-Ala PSK (analogues 8 and 9, respectively). Next, we selected analogues containing diastereomers of both isoleucine on position 2 and threonine on position 4 by using the allo amino acids. We incorporated 2-allo-Ile PSK 10 and 4-allo-Thr PSK 11, resulting in small changes in spatial orientation of a CH3-group or OH-group at the Cβ atom. These diastereomers can easily be introduced by SPPS using Fmoc-allo-Ile-OH (Bachem) or Fmoc-allo-Thr(tBu)-OH (Bachem). Finally, we selected N-methylated modifications on positions 2 or 4, resulting in 2-Nme-Ile PSK 12 and 4-Nme-Thr PSK 13 in order to prevent hydrogen-bond formation between PSK and the receptor via these peptide bonds. The N-methylated peptides can be synthesized with SPPS, with in solution-phase synthesized building blocks.
Synthesis of mutated PSK peptides
For preparation of the threonine building block, two synthetic routes were explored (Scheme 1B). First, H2N-Thr(tBu)-OH 16 required an orthogonal protecting group strategy to maintain the threonine side chain protected in the final building block.56 Therefore, the N-terminus was first protected with an allyloxycarbonyl (Alloc) protecting group by reacting 16 with allyl chloroformate in the presence of sodium bicarbonate. Afterwards, the carbamate amine was methylated using methyl iodide and sodium hydride to afford Alloc-Nme-Thr(tBu)-OH 17, and the C-terminus was protected with a benzyl group. Finally, the Alloc protecting group was removed from the secondary amine using palladium-catalyzed allyl transfer to thiosalicylic acid as the accepting nucleophile,57,58 and yielded Nme-Thr(tBu)-OBn 18 in 51% yield over 4 steps. We found that the same threonine building block could also be synthesized starting from commercially available Fmoc-Nme-Thr(tBu)-OH 19. In two steps that involved benzylation of the carboxyl moiety and Fmoc-removal with piperidine, Nme-Thr(tBu)-OBn 18 was obtained in 81% yield over 2 steps. Clearly, this last route is preferred, although the former might be more favorable for large amounts of N-methylated amino acid 18. Using building blocks 15 and 18, solution-phase dipeptide synthesis was performed by coupling Fmoc-Tyr(SO2ONp)-OH using HATU and DIPEA in DMF (Scheme 1C). The formed products were hydrogenated with Pd/C and H2 to remove the benzyl protecting group and Fmoc-Tyr(SO2ONp)-Nme-Ile-OH 21 was obtained in 94% yield over 2 steps, whereas Fmoc-Tyr(SO2ONp)-Nme-Thr(tBu)-OH 22 was obtained in 62% yield over 2 steps.
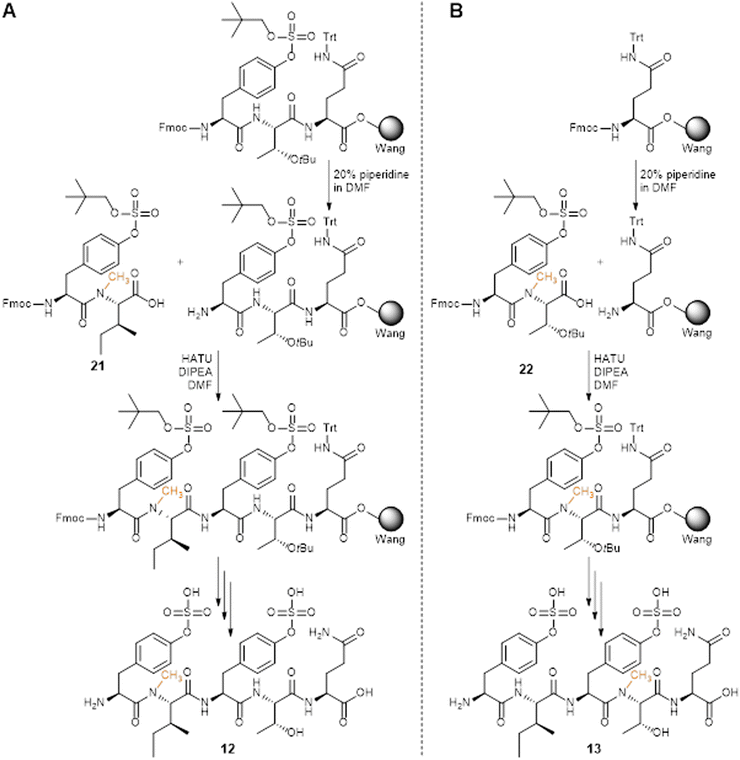
The obtained dipeptide building blocks 21 and 22 were used in SPPS to synthesize 2-Nme-Ile PSK-12 and 4-Nme-Thr PSK-13, respectively (Scheme 2), with HATU as coupling reagent. For 2-Nme-Ile PSK (Scheme 2A), dipeptide 21 was coupled to H2N-Tyr(SO2ONp)-Thr(tBu)-Gln(Trt)-Wang resin to afford the desired pentapeptide. For 4-Nme-Thr PSK (Scheme 2B), dipeptide 22 was coupled to H-Gln(Trt)-Wang resin and was subsequently further elongated with Fmoc-Ile-OH and Fmoc-Tyr(SO2ONp)-OH to afford the desired pentapeptide. Sequential Fmoc-deprotection, acidic cleavage and partial deprotection, initial purification, SO2ONp-deprotection, final purification, and lyophilization of the products afforded pure PSK analogues containing 2-Nme-Ile, in PSK-12, and 4-Nme-Thr, in PSK-13.
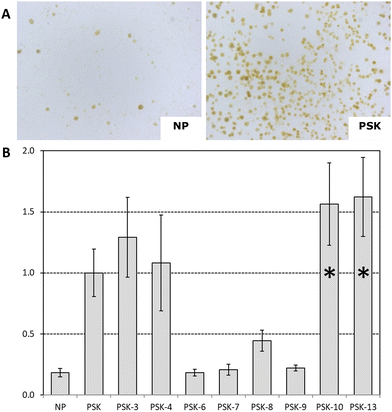
For the PSK analogues, the number of calli formed were counted and compared to the number of calli formed when exposed to the native peptide PSK in the same experiment. We note that PSK-5, PSK-11, and PSK-12 were also tested on red cabbage, but those results were not sufficiently reliable to include in this graph. We found that exposing protoplasts derived from red cabbage to 2-allo-Ile PSK (PSK-10) and 4-Nme-Thr PSK (PSK-13) lead to calli numbers that were significantly higher than those counted for PSK (P < 0.01) (Fig. 3B). Interestingly, treatment of the protoplasts with Ac-PSK (PSK-3) or N3-PSK (PSK-4) resulted in similar calli formation as compared to the native peptide (Fig. 3B), suggesting that the N-terminus tolerates these modifications. Notably, when 2-Ile is substituted for 2-Leu PSK (PSK-6) or 2-Ala (PSK-7), or when 4-Thr is substituted for 4-Ala (PSK-9), calli formation was similar to that in the absence of hormone (Fig. 3B). Higher than background numbers of calli were counted when protoplasts were exposed to Asn-PSK (PSK-8), although not as much as with native PSK. In Brassica oleracea white cabbage protoplasts, similar observations for amino acid substituted analogues PSK-6–9 were found (Fig. S19, ESI†). For 2-Nme-Ile PSK (PSK-12), no increase in formed calli is observed when compared to the absence of peptide (Fig. S19, ESI†), while 4-allo-Thr PSK (PSK-11) resulted in an increased number of calli, although less than with native PSK.
Evaluation of the structures of the synthetic mutants and with respect to the observed protoplast proliferation, structure–activity relationships (SAR) could elucidate essential features for the bio-activity of the PSK peptide. First, the presence of 2-Ile and 4-Thr seems essential for the biological activity. While 4-Asn PSK (PSK-8) is still capable of inducing callus formation, the relative activity is only 30% compared to PSK (PSK-1). N-Terminal modifications Ac-PSK (PSK-3) and N3-PSK (PSK-4) resulted in similar outcomes compared to PSK (PSK-1). This indicates that the pentapeptide pharmacophore still remains bio-active while containing an extended N-terminal modification, which could be useful for peptide labeling. Indeed, the crystal structure of the PSKR-PSK complex indicates the presence of a cavity at the N-terminus, which can be occupied by the respective PSK analogues. Changes in stereochemistry resulted in weaker bio-activity for 4-allo-Thr PSK (PSK-11) but higher activity for 2-allo-Ile PSK (PSK-10) when compared to the parent PPH PSK, with 0.7 and 1.3 relative activity, respectively. Considering the presence of a N-methylated amide backbone, we found that methylation of the Ile amide (PSK-12) does not lead to bio-active compounds, whereas shifting the methyl to the Thr residue, as in 4-Nme-Thr PSK (PSK-13), leads to the most active PSK analogue in our library (>1.5 relative activity compared to PSK).
The resolved crystal structure of the PSK-PSKR1LRR indicates hydrogen bonding and van der Waals packing interactions, where the most prominent contributions arise from the two sulfate moieties.28 Additionally, residues from the inner surface of the PSKR1LRR forms hydrogen bonds with the main chain of PSK. Polar interactions are observed in the amide moiety between Tyr-1 and Ile-2 of PSK with Thr398, between the Thr-4-OH of PSK and Ser370, and between a PSK backbone NH of Thr-4 with Ser372. As the 4-Nme-Thr analogue (PSK-13) does not enable the hydrogen bond between Ser372 and the PSK backbone, the higher biological activity with respect to plant protoplast regeneration shows that this interaction between the PSK analogue and the PSKR is not crucial to elicit a biological response. Changing the stereochemistry of Cβ in 2-Ile (as in PSK-10) resulted in increased bio-activity, which could indicate an increase in polar interaction with Thr398, resulting in stronger receptor activation. In contrast, the reduced bio-activity found for 4-allo-Thr PSK (as in PSK-11) could be directed to weaker Thr-4-OH with Ser370 interactions and thus less receptor activation and weaker bio-activity of the peptide.
Conclusions
We report the synthesis of N-methylated amino acids, their incorporation in PSK analogues, and a set of other substituted PSK-like peptides. We selected a set of peptides including N-terminal modifications, amino acid substitutions, diastereomers and N-methylation. Solution-phase synthesis was combined with solid-phase peptide synthesis and resulted in a set of 13 peptides. For the N-methylated analogues PSK-12 and PSK-13, building blocks 15 and 18 were synthesized with an N-alkylation procedure using methyl iodide. The building blocks were coupled in solution to Fmoc-Tyr(SO2ONp)-OH using HATU as activator to obtain dipeptides 21 and 22. The chemically synthesized PSK analogues were tested in Brassica oleracea protoplast proliferation assays to compare relative activity to the native peptide. Bio-activity of most analogues was reduced or even abolished. Only 2-allo-Ile PSK (PSK-10) and 4-Nme-Thr PSK (PSK-13) displayed a significant increase in bio-activity. These findings indicate that small chemical modifications in PPHs affects bio-activity which arises from weaker or stronger polar interactions in the binding pocket of PSKR1. Potentially, making combinations of PSK-10 and PSK-13 might lead to an even more potent artificial analogue. With these assessments, a bio-active PPH could be designed, synthesized and biologically evaluated in order to improve bio-activity in different recalcitrant crops.Author contributions
BA, RGHI and MF conceived the project, JvdS and DS prepared the chemical compounds, MF performed the biological activity studies and data curation. JvdS and BA wrote the manuscript, DS, RGHI, and MF reviewed and edited the manuscript, MF and RGHI obtained funding for this project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge and thank our industry partners, as well as TKI's Agri & Food, Tuinbouw & Uitgangsmaterialen en Water & Maritiem, for their funding and support under TKI project LWV19054, Hazel van Waijjen for help with the design of the isolation and culture conditions and to many colleagues in our laboratories for helpful discussions during this project.Notes and references
- G. Audil, L. Ajaz Ahmad and W. Noor Ul Islam, Abiotic and Biotic Stress in Plants, IntechOpen, Rijeka, 2019, ch. 1 DOI:10.5772/intechopen.85832.
- N. Suzuki, R. M. Rivero, V. Shulaev, E. Blumwald and R. Mittler, Abiotic and biotic stress combinations, New Phytol., 2014, 203, 32–43 CrossRef PubMed.
- I. K. Vasil and V. Vasil, Totipotency and Embryogenesis in Plant Cell and Tissue Cultures, In Vitro, 1972, 8, 117–127 CrossRef CAS PubMed.
- G. E. Schaller, A. Bishopp and J. J. Kieber, The Yin-Yang of Hormones: Cytokinin and Auxin Interactions in Plant Development, Plant Cell, 2015, 27, 44–63 CrossRef CAS PubMed.
- A. Blum and W. R. Jordan, Breeding crop varieties for stress environments, Crit. Rev. Plant Sci., 1985, 2, 199–238 CrossRef.
- J. Lipiec, C. Doussan, A. Nosalewicz and K. Kondracka, Effect of drought and heat stresses on plant growth and yield: a review, Int. Agrophys., 2013, 27, 463–477 Search PubMed.
- W. Wang, B. Vinocur and A. Altman, Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance, Planta, 2003, 218, 1–14 CrossRef CAS PubMed.
- A. E. Huber and T. L. Bauerle, Long-distance plant signaling pathways in response to multiple stressors: the gap in knowledge, J. Exp. Bot., 2016, 67, 2063–2079 CrossRef CAS PubMed.
- S. Fahad, S. Hussain, A. Matloob, F. A. Khan, A. Khaliq, S. Saud, S. Hassan, D. Shan, F. Khan and N. Ullah, Phytohormones and plant responses to salinity stress: a review, Plant Growth Regul., 2015, 75, 391–404 CrossRef CAS.
- T. Boller, Peptide signalling in plant development and self/non-self perception, Cur. Opin. Cell Biol., 2005, 17, 116–122 CrossRef CAS PubMed.
- Y. Matsubayashi, Posttranslationally Modified Small-Peptide Signals in Plants, Annu. Rev. Plant Biol., 2014, 65, 385–413 CrossRef PubMed.
- N. Czyzewicz, K. Yue, T. Beeckman and I. D. Smet, Message in a bottle: small signalling peptide outputs during growth and development, J. Exp. Bot., 2013, 64, 5281–5296 CrossRef CAS PubMed.
- S. Roy, P. Lundquist, M. Udvardi and W. R. Scheible, Small and Mighty: Peptide hormones in plant biology, Plant Cell, 2018, 30 DOI:10.1105/tpc.118.tt0718.
- H. Hanai, T. Matsuno, M. Yamamoto, Y. Matsubayashi, T. Kobayashi, H. Kamada and Y. Sakagami, A Secreted Peptide Growth Factor, Phytosulfokine, Acting as a Stimulatory Factor of Carrot Somatic Embryo Formation, Plant Cell Physiol., 2000, 41, 27–32 CrossRef CAS PubMed.
- Y. Matsubayashi and Y. Sakagami, Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 7623–7627 CrossRef CAS PubMed.
- R. Komori, Y. Amano, M. Ogawa-Ohnishi and Y. Matsubayashi, Identification of tyrosylprotein sulfotransferase in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15067–15072 CrossRef CAS PubMed.
- M. J. Stone, S. Chuang, X. Hou, M. Shoham and J. Z. Zhu, Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins, N. Biotechnol., 2009, 25, 299–317 CrossRef CAS PubMed.
- M. J. Stone and R. J. Payne, Homogeneous Sulfopeptides and Sulfoproteins: Synthetic Approaches and Applications To Characterize the Effects of Tyrosine Sulfation on Biochemical Function, Acc. Chem. Res., 2015, 48, 2251–2261 CrossRef CAS PubMed.
- H. Yang, Y. Matsubayashi, H. Hanai and Y. Sakagami, Phytosulfokine-α, a Peptide Growth Factor Found in Higher Plants: Its Struc-ture, Functions, Precursor and Receptors, Plant Cell Physiol., 2000, 41, 825–830 CrossRef CAS PubMed.
- Y. Matsubayashi, M. Ogawa, H. Kihara, M. Niwa and Y. Sakagami, Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth, Plant Physiol., 2006, 142, 45–53 CrossRef CAS PubMed.
- S.-H. Shiu and A. B. Bleecker, Plant Receptor-Like Kinase Gene Family: Diversity, Function, and Signaling, Sci. Signaling, 2001, re22 CAS.
- S. A. Morillo and F. E. Tax, Functional analysis of receptor-like kinases in monocots and dicots, Curr. Opin. Plant Biol., 2006, 9, 460–469 CrossRef CAS PubMed.
- X. Gou and J. Li, Paired Receptor and Coreceptor Kinases Perceive Extracellular Signals to Control Plant Development1, Plant Physiol., 2020, 182, 1667–1681 CrossRef CAS PubMed.
- Y. Matsubayashi, M. Ogawa, A. Morita and Y. Sakagami, An LRR Receptor Kinase Involved in Perception of a Peptide Plant Hormone, Phytosulfokine, Science, 2002, 296, 1470–1472 CrossRef CAS PubMed.
- Y. Matsubayashi and Y. Sakagami, Characterization of specific binding sites for a mitogenic sulfated peptide, phytosulfokine-α, in the plasma-membrane fraction derived from Oryza sativa L, Eur. J. Biochem., 1999, 262, 666–671 CrossRef CAS PubMed.
- Y. Matsubayashi, L. Takagi and Y. Sakagami, Phytosulfokine-α, a sulfated pentapeptide, stimulates the proliferation of rice cells by means of specific high- and low-affinity binding
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sites, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13357–13362 CrossRef CAS PubMed.
sites, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13357–13362 CrossRef CAS PubMed. - Y. Matsubayashi and Y. Sakagami, 120- and 160-kDa Receptors for Endogenous Mitogenic Peptide, Phytosulfokine-α, in Rice Plasma Membranes, J. Biol. Chem., 2000, 275, 15520–15525 CrossRef CAS PubMed.
- J. Wang, H. Li, Z. Han, H. Zhang, T. Wang, G. Lin, J. Chang, W. Yang and J. Chai, Allosteric receptor activation by the plant peptide hormone phytosulfokine, Nature, 2015, 525, 265–268 CrossRef CAS PubMed.
- J. V. de Souza, M. Kondal, P. Zaborniak, R. Cairns and A. K. Bronowska, Controlling the Heterodimerisation of the Phytosulfokine Receptor 1 (PSKR1) via Island Loop Modulation, Int. J. Mol. Sci., 2021, 22, 1806 CrossRef CAS PubMed.
- S. Yamakawa, Y. Matsubayashi, Y. Sakagami, H. Kamada and S. Satoh, Promotive Effects of the Peptidyl Plant Growth Factor, Phytosulfokine-α, on the Growth and Chlorophyll Content of Arabidopsis Seedlings under High Night-time Temperature Conditions, Biosci., Biotechnol., Biochem., 1999, 63, 2240–2243 CrossRef CAS PubMed.
- H. Yang, Y. Matsubayashi, K. Nakamura and Y. Sakagami, Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor, Plant Physiol., 2001, 127, 842–851 CrossRef CAS PubMed.
- A. Kiełkowska and A. Adamus, Early studies on the effect of peptide growth factor phytosulfokine-α on Brassica oleracea var. capitata L. protoplasts, Acta Soc. Bot. Pol., 2017, 86, 3558 Search PubMed.
- A. Kiełkowska and A. Adamus, Peptide Growth Factor Phytosulfokine-α Stimulates Cell Divisions and Enhances Regeneration from B. oleracea var. capitata L. Protoplast Culture, J. Plant Growth Regul., 2019, 38, 931–944 CrossRef.
- E. Grzebelus, M. Szklarczyk, J. Greń, K. Śniegowska, M. Jopek, I. Kacińska and K. Mrożek, Phytosulfokine stimulates cell divisions in sugar beet (Beta vulgaris L.) mesophyll protoplast cultures, Plant Growth Regul., 2012, 67, 93–100 CrossRef CAS.
- A. Bahyrycz and D. Konopińska, Plant signalling peptides: some recent developments, J. Pept. Sci., 2007, 13, 787–797 CrossRef CAS PubMed.
- M. Sauter, Phytosulfokine peptide signalling, J. Exp. Bot., 2015, 66, 5161–5169 CrossRef CAS PubMed.
- C. Kaufmann and M. Sauter, Sulfated plant peptide hormones, J. Exp. Bot., 2019, 70, 4267–4277 CrossRef CAS PubMed.
- E. Murphy, S. Smith and I. De Smet, Small Signaling Peptides in Arabidopsis Development: How Cells Communicate Over a Short Distance, Plant Cell, 2012, 24, 3198–3217 CrossRef CAS PubMed.
- N. Stührwohldt, R. I. Dahlke, A. Kutschmar, X. Peng, M.-X. Sun and M. Sauter, Phytosulfokine peptide signaling controls pollen tube growth and funicular pollen tube guidance in Arabidopsis thaliana, Physiol. Plant., 2015, 153, 643–653 CrossRef PubMed.
- N. Stührwohldt, R. I. Dahlke, B. Steffens, A. Johnson and M. Sauter, Phytosulfokine-α Controls Hypocotyl Length and Cell Expansion in Arabidopsis thaliana through Phytosulfokine Receptor 1, PLoS One, 2011, 6, e21054 CrossRef PubMed.
- X. Shen, N. Stührwohldt and C. Lin, The Research Process of PSK Biosynthesis, Signaling Transduction, and Potential Applications in Brassica napus, Plants, 2023, 12, 3075 CrossRef CAS PubMed.
- H. Motose, K. Iwamoto, S. Endo, T. Demura, Y. Sakagami, Y. Matsubayashi, K. L. Moore and H. Fukuda, Involvement of Phytosulfokine in the Attenuation of Stress Response during the Transdifferentiation of Zinnia Mesophyll Cells into Tracheary Elements, Plant Physiol., 2009, 150, 437–447 CrossRef CAS PubMed.
- A. Bahyrycz, Y. Matsubayashi, M. Ogawa, Y. Sakagami and D. Konopińska, Plant peptide hormone phytosulfokine (PSK-α): synthesis of new analogues and their biological evaluation, J. Pept. Sci., 2004, 10, 462–469 CrossRef CAS PubMed.
- A. Bahyrycz, Y. Matsubayashi, M. Ogawa, Y. Sakagami and D. Konopińska, Further analogues of plant peptide hormone phytosulfokine-α (PSK-α) and their biological evaluation, J. Pept. Sci., 2005, 11, 589–592 CrossRef CAS PubMed.
- Y. Matsubayashi, H. Hanai, O. Hara and Y. Sakagami, Active Fragments and Analogs of the Plant Growth Factor, Phytosulfokine: Structure–Activity Relationships, Biochem. Biophys. Res. Commun., 1996, 225, 209–214 CrossRef CAS PubMed.
- J. C. Roberts, H. Gao, A. Gopalsamy, A. Kongsjahju and R. J. Patch, Neopentyl ester protecting groups for arylsulfonic acids, Tetrahedron Lett., 1997, 38, 355–358 CrossRef CAS.
- L. S. Simpson and T. S. Widlanski, A Comprehensive Approach to the Synthesis of Sulfate Esters, JACS Au, 2006, 128, 1605–1610 CrossRef CAS PubMed.
- L. S. Simpson, J. Z. Zhu, T. S. Widlanski and M. J. Stone, Regulation of Chemokine Recognition by Site-Specific Tyrosine Sulfation of Receptor Peptides, Chem. Biol., 2009, 16, 153–161 CrossRef CAS PubMed.
- Y. Matsubayashi, Y. Takahata, A. Morita, K. Atsumi and Y. Sakagami, Preparation and Characterization of Fully Active Biotinylated Analogs of Phytosulfokine-α, Biosci., Biotechnol., Biochem., 1999, 63, 1847–1849 CrossRef CAS PubMed.
- S. Schoffelen, M. B. van Eldijk, B. Rooijakkers, R. Raijmakers, A. J. R. Heck and J. C. M. van Hest, Metal-free and pH-controlled introduction of azides in proteins, Chem. Sci., 2011, 2, 701–705 RSC.
- E. D. Goddard-Borger and R. V. Stick, An Efficient, Inexpensive, and Shelf-Stable Diazotransfer Reagent:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Imidazole-1-sulfonyl Azide Hydrochloride, Org. Lett., 2007, 9, 3797–3800 CrossRef CAS PubMed.
Imidazole-1-sulfonyl Azide Hydrochloride, Org. Lett., 2007, 9, 3797–3800 CrossRef CAS PubMed. - M. B. Hansen, T. H. M. van Gurp, J. C. M. van Hest and D. W. P. M. Löwik, Simple and Efficient Solid-Phase Preparation of Azido-peptides, Org. Lett., 2012, 14, 2330–2333 CrossRef CAS PubMed.
- A. Leggio, E. L. Belsito, G. De Luca, M. L. Di Gioia, V. Leotta, E. Romio, C. Siciliano and A. Liguori, One-pot synthesis of amides from carboxylic acids activated using thionyl chloride, RSC Adv., 2016, 6, 34468–34475 RSC.
- A. V. Malkov, K. Vranková, M. Černý and P. Kočovský, On the Selective N-Methylation of BOC-Protected Amino Acids, J. Org. Chem., 2009, 74, 8425–8427 CrossRef CAS PubMed.
- A. Wunder, M. Rothemund and R. Schobert, Synthesis and anticancer activity of the proposed structure of caldoramide, an N-peptidyltetramate from the cyanobacterium Caldora penicillata, Tetrahedron, 2018, 74, 5138–5142 CrossRef CAS.
- F. Albericio, Orthogonal protecting groups for Nα-amino and C-terminal carboxyl functions in solid-phase peptide synthesis, Pept. Sci., 2000, 55, 123–139 CrossRef CAS.
- D. Fernández-Forner, G. Casals, E. S. Navarro, H. Ryder and F. Albericio, Solid-phase synthesis of 4-aminopiperidine analogues using the Alloc protecting group: an investigation of Alloc removal from secondary amines, Tetrahedron Lett., 2001, 42, 4471–4474 CrossRef.
- J. P. Genêt, E. Blart, M. Savignac, S. Lemeune, S. Lemaire-Audoire and J. M. Bernard, A General and Simple Removal of the Allyloxycarbonyl Protecting Group by Palladium-Catalyzed Reactions Using Nitrogen and Sulfur Nucleophiles, Synlett, 1993, 680–682 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, characterization data, LCMS & NMR spectra. See DOI: https://doi.org/10.1039/d3nj05996k |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |