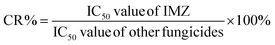Gold nanoparticle-based immunochromatographic assay for rapid detection of imazalil†
Xinxin
Xu
ab,
Aihong
Wu
ab,
Lingling
Guo
ab,
Hua
Kuang
 ab,
Liguang
Xu
ab,
Chuanlai
Xu
ab,
Liguang
Xu
ab,
Chuanlai
Xu
 *ab and
Liqiang
Liu
*ab
*ab and
Liqiang
Liu
*ab
aInternational Joint Research Laboratory for Biointerface and Biodetection, and School of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, P. R. China. E-mail: murel@163.com; xcl@jiangnan.edu.cn
bState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, P. R. China
First published on 6th October 2023
Abstract
Imazalil (IMZ) is a commonly used fungicide for controlling fungus in agriculture, leaving residual IMZ in crops that could be hazardous to human health. In this work, we designed IMZ haptens for mice immunization and prepared sensitive monoclonal antibody (mAb) against IMZ. The subtype of anti-IMZ mAb is IgG2a. It possessed a half inhibition concentration (IC50) of 0.95 ng mL−1 and showed no cross-reactivity against other chemicals in ic-ELISA. Taking advantage of the mAb, we developed a gold nanoparticle-based immunochromatographic assay (GICA) for the rapid detection of IMZ in grapes and tomatoes. The assay gave a visual limit of detection (vLOD) of 25 ng g−1 and cut-off value of 500 ng g−1 in both samples. According to the calibration curves, the calculated LOD were 4.12 ng g−1 and 4.70 ng g−1 in grapes and tomatoes, respectively. The recovery rates of IMZ ranged from 84.7% to 104.4% with variation coefficients (CVs) of 5.7–11.8% in spiked samples, indicating a potent practicability of the GICA. The whole GICA process took 30 min. Therefore, the developed assay can be used for on-site detection and quantitation of IMZ in grape and tomato samples.
New conceptsIMZ haptens were fabricated for mice immunization and prepared sensitive monoclonal antibody (mAb) against IMZ. A gold nanoparticle-based immunochromatographic assay (GICA) for the rapid detection of IMZ in grapes and tomatoes was developed. The assay showed a visual limit of detection (vLOD) of 25 ng g−1 and cut-off value of 500 ng g−1 in both samples, and the calculated LOD were 4.12 ng g−1 and 4.70 ng g−1 in grapes and tomatoes, respectively. |
1. Introduction
Imazalil (IMZ) is a synthetic systemic fungicide, widely used for fungi control on vegetables and fruits in postharvest preservation.1–3 IMZ inhibits demethylation in the biosynthesis of ergosterol in fungal pathogens to achieve its anti-fungi function.4,5 The high effectiveness of IMZ sometimes leads to its over usage, resulting in the emergence of resistant strains and large amounts of residues in the environment.6–9 Several studies have reported that imazalil exposure may induce developmental abnormalities, gut microbiota dysbiosis, colonic inflammation, intestinal barrier disruption, and endocrine system disorder in model animals.10–14 Therefore, IMZ residual on vegetables and fruits poses a potential threat to humans. To limit the extensive use of IMZ and reduce health risks to humans, the maximum residue level (MRL) in vegetables and fruits has been set, ranging from 0.05 to 5 mg kg−1 in China and the EU.Currently, instrumental analysis is the most commonly used method for IMZ monitoring, such as LC-MS/MS,15–18 HPLC coupled with diode array detector,19 and GC-MS/MS.20 Although the above methods can provide specific and precise results, they are expensive and time-consuming and require complicated sample pretreatment and well-trained technicians, limiting the application outside the laboratory. Therefore, the development of rapid detection methods has become attractive and promising. For example, methods based on surface-enhanced Raman spectroscopy (SERS) have been reported for IMZ detection in fruits recently.21–23 Immunoassay is an alternative approach to achieve the rapid detection of analytes, with high throughput. Based on the specific and sensitive binding affinity between the antibody and antigen, immunoassays usually offer great performance for the analysis of agricultural chemicals in real samples, such as insecticides,24,25 herbicides,26,27 acaricide,28 and fungicides.29,30 Enzyme-linked immunosorbent assay (ELISA) is such a method. Watanabe et al. developed a competitive ELISA for the detection of residual IMZ in citrus fruits.31 The method provided half maximal inhibitory concentration (IC50) of 9 pmol mL−1 (2 ng mL−1) and the readout showed good consistency with HPLC analysis. Later, Moreno et al. improve the sensitivity of ELISA (1.6 nM, equal to 0.5 ng mL−1) and applied it to fruit juices.32 Because ELISA always needs repeated washing steps and long-time incubation, it is not suitable for on-site detection.
The gold nanoparticle-based immunochromatographic assay (GICA) is another popular immunoassay in the field of food safety. Compared to ELISA, it is more convenient, portable, and rapid, and meets the requirement of on-site mass sample screening. To the best of our knowledge, there is no GICA reported for the detection of IMZ in food samples. In this paper, we developed a monoclonal antibody (mAb) based GICA and applied it to analyze IMZ residues in grape and tomato samples.
2. Materials and methods
2.1 Chemicals and reagents
IMZ, penconazole, hexaconazole, miconazole, propiconazole, triflumizole, diclobutrazol, and other chemicals for hapten synthesis were purchased from J & K Scientific (Beijing, China). Bovine serum albumin (BSA), ovalbumin (OVA), Freund's complete adjuvant, and Freund's incomplete adjuvant were bought from Sigma Aldrich (Shanghai, China). RPMI 1640 Medium, fetal bovine serum (FBS), HAT, and HT supplements were obtained from Thermo Fisher Scientific (Shanghai, China). HRP-labeled goat anti-mouse IgG was brought from Jackson ImmunoResearch Laboratories (West Grove, USA). N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide (EDC) were brought from East China Chemicals (Wuxi, China).2.2 Preparation of antigens
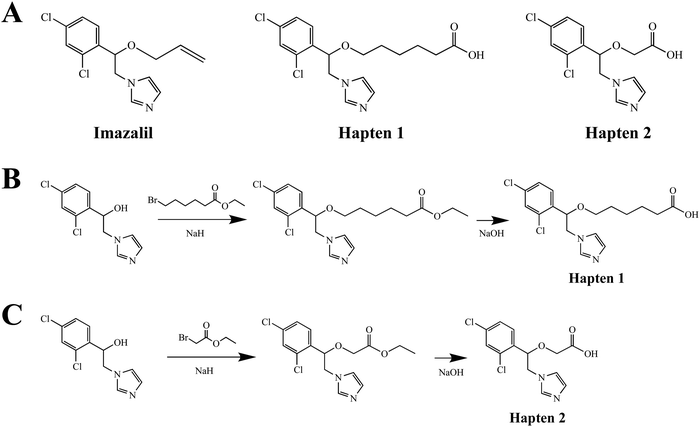 | ||
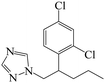
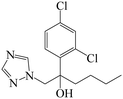
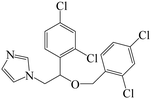
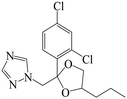
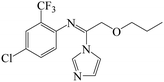
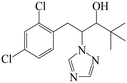
| Fig. 1 (A) The structure of IMZ and two haptens. (B) The synthesis of IMZ hapten 1. (C) The synthesis of IMZ hapten 2. | ||
The synthesis of hapten 2 was similar to that of hapten 1, in which ethyl 2-bromoacetate was used instead of ethyl 6-bromohexanoate at the first step. ESI-MS: calced: C13H12Cl2N2O3 314.02; found: 314.00 [M − H+].
2.3 Mice immunization and antibody production
BALB/c mice (female, 6–8 weeks old) were used for immunization. In the first immunization, mice were subcutaneously injected with 100 μg of immunogens emulsified with Freund's complete adjuvant. Booster immunizations were carried out five times at three-week intervals, during which 50 μg immunogens emulsified with Freund's incomplete adjuvant were administered. One week post booster immunization, the titer and inhibition of antisera were measured by the indirect competitive ELISA (ic-ELISA).The ic-ELISA was performed as follows. The coating antigen was diluted in CBS (15 mM Na2CO3, 34.9 mM NaHCO3, pH 9.6). 96 well microplates were coated with 100 μL of coating antigen and incubated at 37 °C for 2 h. After wash, they were blocked with 200 μL of 0.2% gelatin in CBS at 37 °C for 2 h. After wash, 50 μL of IMZ standard and 50 μL of antisera (or antibody) were added, while PBS was used as blank. The microplates were incubated at 37 °C for 30 min. After wash, 100 μL of HRP-labeled goat anti-mouse IgG was added and incubated for 30 min. The TMB substrate solution was added and reacted for 15 min at 37 °C. The reactions were stopped by the addition of 2 M sulfuric acid. The absorbance at 450 nm was measured using a microplate reader.
The mouse with the highest titer and inhibition was sacrificed and the spleen cells were used for cell fusion. One week later, the cell culture supernatants were analyzed using ic-ELISA. The hybridoma candidates were subcloned using limited dilution for three rounds. The screened cell line was expanded in a cell culture flask and injected into the abdominal cavity of mice. The mice ascites containing anti-IMZ antibody were collected and further purified using saturated ammonium sulfate precipitation methods. The obtained antibody was dialyzed in PBS and stored at 4 °C before use.
2.4 Characterization of anti-IMZ mAb
2.5 Preparation of GNP-mAb
Gold nanoparticles (GNPs), about 15 nm in diameter, were synthesized according to our previous reports.33 GNP solution (10 mL) was adjusted to pH 8.8 using 0.1 M K2CO3 solution; while the mAb 3G5 was diluted to 0.2 mg mL−1 with HEPES buffer. The diluted mAb was then dropwise added to the GNP solution, making the concentration 5 or 10 μg mL−1. The solution was incubated for 45 min at room temperature. Then, 0.5% (w/v) BSA was added and incubated for 2 h. The mixture was centrifuged at 9000g for 45 min at 4 °C. The supernatant was discarded using a pipette; while the pellet was resuspended with 1.2 mL of the basic buffer (PBS containing 0.02% NaN3, 1% BSA, 0.5% PEG 6000, and 5% sucrose) and used as GNP-mAb. The GNP-mAb was stored at 4 °C before use and diluted 10-fold with basic buffer when use.2.6 Preparation of the GICA strip
The GICA strip was prepared as follows (Fig. 2). The nitrocellulose (NC) membrane was pasted on the backing plate, then the NC membrane was sprayed with coating antigen and goat anti-mouse IgG antibody to make the T and C lines.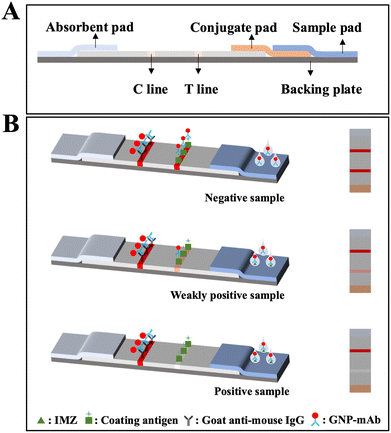 | ||
| Fig. 2 Schematic diagram of the GICA strip. (A) The assembly of the GICA strip. (B) The detection principle of the GICA strip. | ||
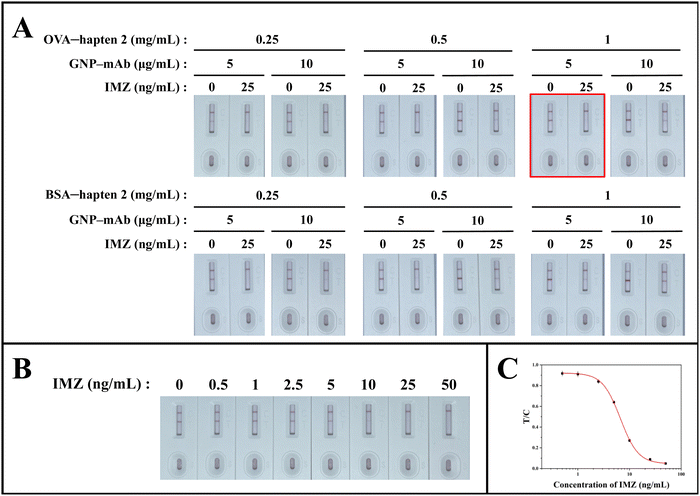
To give the best performance of the GICA strip, the coating antigen and GNP-mAb were optimized. The concentration of goat anti-mouse IgG on the C line was fixed as 0.2 mg mL−1. OVA-hapten 2 or BSA-hapten 2 was sprayed on the T line in concentrations of 0.25, 0.5, and 1 mg mL−1, respectively. As mentioned above, the defined 5 or 10 μg mL−1 of GNP-mAb was tested. Totally 12 groups of the GICA strips were set.
After drying at 37 °C overnight, the absorbent pad, and sample pad were pasted on the backing plate one after another. The assembled plates were introduced to a strip cutter and sliced into 3.0 mm broad strips. The strips were kept in a drying cabinet before use.
The detection principle of the GICA strip was elucidated in Fig. 2B. At first, 50 μL of diluted GNP-mAb solution is mixed with 150 μL of IMZ standard or sample extracts. After 100 μL of above mixture is loaded onto the sample pad, it moves towards the absorbent pad; while the GNP-mAb migrates with the sample. When a negative sample is tested, both T and C lines turn red due to the interactions between GNP-mAb and the coating antigen or anti-mouse IgG. When a weakly positive sample is tested, part of the GNP-mAb prior binds with the analyte in the sample, resulting in a faint T line. When a positive sample is tested, all GNP-mAb binds with the analyte, resulting in a colorless T line. The C line should always be red during the test; otherwise, the test is invalid. It is worthy noted that the strip is incubated for 10 min on a 37 °C thermostatic incubator after the mixture is loaded.
2.7 Pretreatment and IMZ-spiking of food samples
Fresh tomato and grape samples were purchased from the local supermarket and confirmed as IMZ-negative. The samples were pretreated prior to GICA strip analysis. Briefly, fresh samples were finely chopped and homogenized. The homogenates were divided into several groups and spiked with IMZ standard to the final concentrations of 0, 5, 10, 25, 50, 100, 250, and 500 ng g−1. Two grams of IMZ-spiked homogenates were weighed and mixed with 4 mL of 50% methanol in water. After vigorous vortex for 5 min, the mixture was centrifuged at 9000g for 10 min. The pellet was discarded, while appropriate amount of supernatant was collected and diluted three-fold with PBS. The diluted extract (100 μL) was subjected to the GICA strip. Each sample was tested in triplicate.3. Results and discussion
3.1 Characterization of haptens and antigens
Small molecules must be conjugated with carrier proteins to obtain immunogenicity; while the active group, such as the amino group or carboxyl group, is necessary for the conjugation. IMZ has no active group. Based on its structure, we designed two haptens, in which the propylene group of IMZ was replaced by hexanoic acid or acetic acid (Fig. 1A). Hapten 1 and hapten 2 were synthesized as shown in Fig. 1B and C, respectively. Both haptens were characterized by LC-MS (Fig. S1, ESI†). Hapten 1 had a feature at 3.98 min in liquid chromatography and the corresponding m/z was 369 under ESI negative ion mode in the mass spectrum; while Hapten 2 had a feature at 3.14 min with m/z 313. These data suggested that IMZ haptens were successfully synthesized.Subsequently, the haptens were conjugated with carrier proteins, BSA and OVA, as described in Materials and methods. BSA-hapten conjugates were used as the immunogens; while OVA-hapten conjugates were used as the coating antigens. All antigens were characterized using SDS-PAGE (Fig. 3). Compared with the carrier proteins (BSA, 66 kDa; OVA, 45 kDa), the molecular weight of each hapten was slightly larger, indicating that the haptens and proteins were successfully conjugated.
3.2 Characterization of anti-IMZ mAb
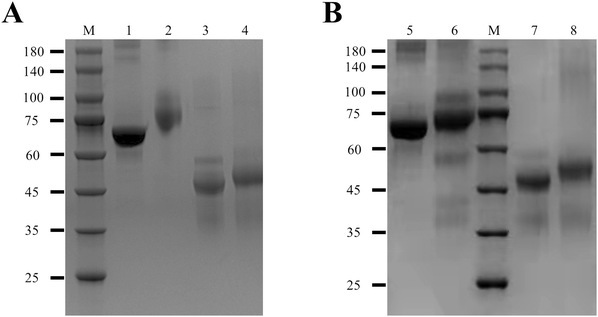
Both BSA-hapten 1 and BSA-hapten 2 were subjected to mice immunization. As shown in Table S1 (ESI†), the antisera from 3rd and 5th immunized mice were analyzed using ic-ELISA, in which the antisera were 9000-fold diluted in PBS. The antisera from mice immunized with BSA-hapten 1 showed high titer but low inhibition in ic-ELISA, possibly due to most antibodies in antisera recognizing the long carbon chain linker of hapten 1. Afterwards BSA-hapten 1 was no longer employed in the mice immunization. BSA-hapten 2-immunized mice antisera had both high titer and great inhibitory effect. Therefore, BSA-hapten 2 and OVA-hapten 2 were chosen as the optimal antigens for anti-IMZ mAb screening.The antisera from No. 7 mouse presented the highest titer and greatest inhibitory effect (Table S1, ESI†). This mouse was chosen for the isolation of monoclonal hybridoma cell line. The screened mAb was designated as 3G5. The subtype of mAb 3G5 was determined as IgG2a (Fig. 4A). To evaluate the sensitivity of the antibody, a series of gradient-diluted IMZ standard solutions (0, 0.04, 0.12, 0.37, 1.11, 3.33, 10, and 30 ng mL−1) were prepared in PBS and tested by ic-ELISA. The logarithm of concentration on the x-axis was plotted against the OD450 value on the y-axis. The calibration curve was fitted using a four-parameter logistic function (Fig. 4B). The curve had an equation of y = 0.08 + 1.58/[1 + (x/0.95)1.10] with R2 = 0.999. The IC50 was calculated as 0.95 ng mL−1 with a linear range of 0.27–3.36 ng mL−1 (IC20–IC80). Several structural analogs of IMZ and other fungicides were analyzed by ic-ELISA. None of these chemicals showed cross-reactivity with mAb 3G5 (Table 1). Collectively, the mAb 3G5 is highly sensitive and specific to IMZ.
3.3 Development of the GICA strip
The GNPs were synthesized in-house and used for the optical signal molecules. The appearance of GNPs was characterized by a transmission electron microscope. As shown in Fig. S2A (ESI†), the GNPs possessed desirable dispersibility and homogeneity, with an average diameter of about 15 nm. In the UV spectroscopic analysis, the GNPs had a ∼520 nm maximum absorption wavelength; while the GNP-mAbs showed a clear peak shift to 525 nm (Fig. S2B, ESI†). These data demonstrated that the GNPs were synthesized successfully and conjugated with mAb.The types and dosages of the coating antigens and GNP-mAbs were the determinants responsible for the performance of GICA strips. We tested series of different combinations of coating antigen and GNP-mAb during the strip optimization, in which goat anti-mouse antibody was set as 0.2 mg mL−1 on the C line. 0 ng mL−1 and 25 ng mL−1 IMZ standard solution were defined as the blank group and test group, respectively (Fig. 5A). Both groups were analyzed using the strips to compare the color intensities of both C and T lines. After the solutions were added, the strips were incubated on a 37 °C thermostatic heater for 10 min. When 1 mg mL−1 of OVA-hapten 2 and 5 g mL−1 of GNP-mAb were combined, the red bands of the C and T lines in the blank group exhibited almost equal findings; while the red bands of the C and T lines were extensively distinguishable in the test group, indicating that the strip was very sensitive to IMZ. None of other combinations simultaneously gave equivalent C and T bands in the blank group and great difference between C and T bands in the test group. As a result, we determined that 1 mg mL−1 of OVA-hapten 2 and 5 μg mL−1 of GNP-mAb provided the optimal performance of the GICA strips in qualitative analysis by naked eyes, offering a superior T/C value.
3.4 Performance of the GICA strip
We wondered if the GICA strips were acceptable for both qualitative and quantitative analysis of IMZ. Under the optimal conditions, we examined a series of gradient-diluted IMZ standard solution (0, 0.5, 1, 2.5, 5, 10, 25, and 50 ng mL−1 in PBS containing 10% methanol) to evaluate the sensitivity of the GICA strip (Fig. 5B). The qualitative result can be estimated by comparing the difference between the C and T lines. Here, we defined the visual limit of detection (vLOD) as the lowest concentration at which the T line differed from the C line, while the cut-off value was the concentration at which the T line completely disappeared. For qualitative analysis, the vLOD and cut-off value of the GICA strip were 5 ng mL−1 and 50 ng mL−1, respectively. For quantitative analysis, the corresponding T/C value of each strip was calculated using a strip scan reader. The calibration curve was established by plotting the logarithm of IMZ concentration on the x-axis and the T/C value on the y-axis and fitted using four-parameter logistic function (Fig. 5C). According to the calibration curve, the equation was y = 0.05 + 0.88/[1 + (x/6.64)2.51] with R2 = 0.999 and the IC50 of the strip for IMZ detection in PBS containing 10% methanol was calculated as 6.64 ng mL−1.3.5 Analysis of IMZ in tomato and grape samples
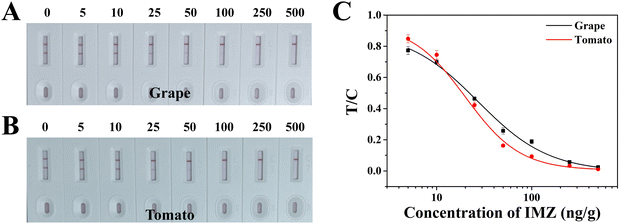
The strip performance for IMZ monitoring in real sample was evaluated. The strips were used to analyze grape and tomato samples that were spiked with IMZ to concentrations of 0, 5, 10, 25, 50, 100, 250, and 500 ng g−1 (Fig. 6). The colorimetric results showed a lighter T line with the increasing concentration of IMZ. In either a tomato or grape sample, the strip provided a vLOD of 25 ng g−1 and cut-off value of 500 ng g−1 (Fig. 6A and B), which were lower than the MRLs in China, the EU, and Japan. As shown in Fig. 6C, the calibration curves were established for quantitative analysis of IMZ. The calculated LODs (cLODs) which are equivalent to IC10 values were 4.12 ng g−1 and 4.70 ng g−1 in grape and tomato samples, respectively.To further verify the strip's accuracy, we reprepared the spiked tomato and grape samples, then pretreated them and analyzed using the strips. As shown in Table 2, the recovery rates of IMZ ranged from 90.3% to 98.2% with a CV of 5.7–11.8% in grapes and 84.7% to 104.4% in tomato with a CV of 5.9–10.6% in tomatoes. Meanwhile, the colorimetric results were stable and consistent. Taken together, we considered that the GICA strip is practical and efficient for IMZ monitoring in grape and tomato samples.
| Samples | Spiked level (ng g−1) | Tested value (ng g−1) mean ± SD | Recovery ± SD (%) | CV (%) | Colorimetric results | ||
|---|---|---|---|---|---|---|---|
| a “ND”, not detected. b “NC”, not calculated. c “—” represented negative result in which similar T and C lines were shown. d “±” represented weakly positive result in which the T line was obvious lighter than the C line. e “+” represented positive result in which the T line was colorless. | |||||||
| Grape | 0 | NDa | NCb | NC | —c | — | — |
| 10 | 9.0 ± 0.5 | 90.3 ± 5.1 | 5.7 | — | — | — | |
| 50 | 49.1 ± 4.4 | 98.2 ± 8.8 | 9.0 | ±d | ± | ± | |
| 250 | 241.3 ± 28.5 | 96.5 ± 11.4 | 11.8 | ± | ± | ± | |
| 500 | ND | NC | NC | +e | + | + | |
| Tomato | 0 | ND | NC | NC | — | — | — |
| 10 | 8.5 ± 0.9 | 84.7 ± 9.0 | 10.6 | — | — | — | |
| 50 | 52.2 ± 3.1 | 104.4 ± 6.2 | 5.9 | ± | ± | ± | |
| 250 | 239.3 ± 17.3 | 95.7 ± 6.9 | 7.2 | ± | ± | ± | |
| 500 | ND | NC | NC | + | + | + | |
3.6 Comparison of the GICA and other methods
As listed in Table 3, several other kinds of analytical methods have been reported for the detection of IMZ in foods. LC-MS/MS analysis has good sensitivities in fruits, wines, and environmental samples due to the low LOD and limit of quantification (LOQ). Even though the development of QuEChERS methodology can reduce the duration of sample pretreatment, LC-MS/MS analysis still requires column packing, washing, and equilibrium, resulting in time-consuming of operation and pollution of organic solvent. The sensitivities of GC-MS analysis were slightly lower than those of LC-MS/MS analysis and it also took more than one hour for the whole detection process. Watanabe et al. developed a direct competitive ELISA for IMZ, in which the LOD was 0.1 ng g−1 in lemon, orange, and grapefruit samples.31 However, this method required twice overnight incubation, limiting its practicability. Compared with above methods, the proposed GICA required less than 30 min to accomplish the testing procedure and result readout for either qualitative or quantitative analysis. The cLOD of GICA was competitive with the other methods and met the IMZ MRL in China and EU (500 ng g−1 in tomatoes and 5000 ng g−1 in grapes). Additionally, the GICA strip is cheap and eco-friendly. These advantages make the GICA strip applicable for the on-site detection of IMZ. Therefore, the GICA strip is a practicable method and tool for the rapid detection of IMZ in foods.| Methods | Samples | LODs | LOQs | Duration of process | Ref. |
|---|---|---|---|---|---|
| a ND, not defined. | |||||
| UPLC-MS/MS | Apple and soil | 0.16–0.18 μg kg−1 | 0.53–0.60 μg kg−1 | More than an hour | 15 |
| UHPLC-MS/MS | Grape, apple, and fruit wine | NDa | 5 μg kg−1 | 16 | |
| UPLC-MS/MS | Banana | ND | 10 μg kg−1 | 17 | |
| UHPLC-MS/MS | Banana, blueberry, golden berry, lime, mango, tamarind, and pineapple | 1.92 μg kg−1 | 6.40 μg kg−1 | 34 | |
| GC-MS/MS | Cucumber, cabbage, apple, and orange | 1.1–2.6 μg kg−1 | 3.5–8.6 μg kg−1 | 20 | |
| GC-MS | Papaya | 7.5 μg kg−1 | 25 μg kg−1 | 35 | |
| GC-SQ/MS | Melons | 10 μg kg−1 | 30 μg kg−1 | 36 | |
| ELISA | Lemon, orange, and grapefruit | 0.1 ng g−1 | ND | >24 h | 31 |
| GICA | Grape and tomato | vLOD: 25 μg kg−1 | ND | Within 30 min | This work |
| cLOD: 4.12–4.70 μg kg−1 | |||||
4. Conclusions
In this study, we designed a IMZ hapten, prepared a sensitive and specific anti-IMZ mAb, and developed the GICA strip for the qualitative and quantitative analysis of IMZ in grape and tomato samples. Under optimal conditions, the GICA strip provided a vLOD of 25 ng g−1 and cut-off value of 500 ng g−1 in both samples. The cLODs of the strip were calculated as 4.12 ng g−1 and 4.70 ng g−1 in grape and tomato samples, respectively. The whole process including sample pretreatment and strip assay can be accomplished within 30 min at the fastest and give satisfactory recovery rates. Collectively, our proposed strip is practicable as a sensitive, rapid and on-site detection tool for IMZ in grape and tomato samples.Ethical approval
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Jiangnan University and approved by the Animal Ethics Committee of Jiangnan University.Conflicts of interest
All the authors declare no competing financial interest.Acknowledgements
This work is financially supported by the National Natural Science Foundation of China (222089658) and Jangsu Agriculture Science and Technology Innovation Fund (JASTIF, CX(21)2036).References
- U. K. Bhatta, Alternative management approaches of citrus diseases caused by Penicillium digitatum (green mold) and Penicillium italicum (blue mold), Front. Plant Sci., 2021, 12, 833328 CrossRef.
- C. Chen, N. Cai, J. Chen and C. Wan, Clove essential oil as an alternative approach to control postharvest blue mold caused by Penicillium italicum in citrus fruit, Biomolecules, 2019, 9(5), 197 CrossRef CAS PubMed.
- R. Li, X. Pan, Y. Tao, D. Jiang, Z. Chen, F. Dong, J. Xu, X. Liu, X. Wu and Y. Zheng, Systematic evaluation of chiral fungicide imazalil and its major metabolite R14821 (Imazalil-M): stability of enantiomers, enantioselective bioactivity, aquatic toxicity, and dissipation in greenhouse vegetables and soil, J. Agric. Food Chem., 2019, 67(41), 11331–11339 CrossRef CAS PubMed.
- M. R. Siegel and N. N. Ragsdale, Antifungal mode of action of imazalil, Pestic. Biochem. Physiol., 1978, 9(1), 48–56 CrossRef CAS.
- X. Sun, J. Wang, D. Feng, Z. Ma and H. Li, PdCYP51B, a new putative sterol 14alpha-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis, Appl. Microbiol. Biotechnol., 2011, 91(4), 1107–1119 CrossRef CAS PubMed.
- A. Erasmus, C. L. Lennox, L. Korsten, K. Lesar and P. H. Fourie, Imazalil resistance in Penicillium digitatum and P. italicum causing citrus postharvest green and blue mould: Impact and options, Postharvest Biol Tec, 2015, 107, 66–76 CrossRef CAS.
- M. de Ramon-Carbonell and P. Sanchez-Torres, Significance of 195 bp-enhancer of PdCYP51B in the acquisition of Penicillium digitatum DMI resistance and increase of fungal virulence, Pestic. Biochem. Physiol., 2020, 165, 104522 CrossRef CAS PubMed.
- A. Ccanccapa, A. Masia, V. Andreu and Y. Pico, Spatio-temporal patterns of pesticide residues in the Turia and Jucar Rivers (Spain), Sci. Total Environ, 2016, 540, 200–210 CrossRef CAS PubMed.
- A. Ccanccapa, A. Masia, A. Navarro-Ortega, Y. Pico and D. Barcelo, Pesticides in the Ebro River basin: Occurrence and risk assessment, Environ. Pollut., 2016, 211, 414–424 CrossRef CAS PubMed.
- S. Huang, M. Huang, S. Tian, Z. Meng, S. Yan, M. Teng, Z. Zhou, J. Diao and W. Zhu, Imazalil and its metabolite imazalil-M caused developmental toxicity in zebrafish (Danio rerio) embryos via cell apoptosis mediated by metabolic disorders, Pestic. Biochem. Physiol., 2022, 184, 105113 CrossRef CAS PubMed.
- Y. Jin, Z. Zhu, Y. Wang, E. Yang, X. Feng and Z. Fu, The fungicide imazalil induces developmental abnormalities and alters locomotor activity during early developmental stages in zebrafish, Chemosphere, 2016, 153, 455–461 CrossRef CAS.
- C. Jin, Z. Zeng, Z. Fu and Y. Jin, Oral imazalil exposure induces gut microbiota dysbiosis and colonic inflammation in mice, Chemosphere, 2016, 160, 349–358 CrossRef CAS.
- C. Jin, T. Luo, Z. Zhu, Z. Pan, J. Yang, W. Wang, Z. Fu and Y. Jin, Imazalil exposure induces gut microbiota dysbiosis and hepatic metabolism disorder in zebrafish, Comp. Biochem. Physiol. C, 2017, 202, 85–93 CAS.
- C. Jin, R. Zhang, Z. Fu and Y. Jin, Maternal exposure to imazalil disrupts the endocrine system in F1 generation mice, Mol. Cell. Endocrinol., 2019, 486, 105–112 CrossRef CAS.
- R. Li, F. Dong, J. Xu, X. Liu, X. Wu, X. Pan, Y. Tao, Z. Chen and Y. Zheng, Enantioseparation of imazalil and monitoring of its enantioselective degradation in apples and soils using ultrahigh-performance liquid chromatography–tandem mass spectrometry, J. Agric Food Chem., 2017, 65(16), 3259–3267 CrossRef CAS.
- R. Li, X. Pan, X. An, K. Wang, F. Dong, J. Xu, X. Liu, X. Wu and Y. Zheng, Monitoring the behavior of imazalil and its metabolite in grapes, apples, and the processing of fruit wine at enantiomeric level, J. Sci. Food Agric., 2021, 101(13), 5478–5486 CrossRef CAS PubMed.
- M. Almutairi, T. Alsaleem, H. Al Herbish, A. A. Al Sayari and A. M. Alowaifeer, LC-MS/MS and GC-MS/MS analysis of pesticide residues in Ecuadorian and Filipino Cavendish bananas imported into Saudi Arabia, Food Addit Contam Part A, 2021, 38(8), 1376–1385 CrossRef CAS PubMed.
- M. E. Casas, A. C. Kretschmann, L. Andernach, T. Opatz and K. Bester, Separation, isolation and stereochemical assignment of imazalil enantiomers and their quantitation in an in vitro toxicity test, J. Chromatogr. A, 2016, 1452, 116–120 CrossRef CAS.
- T. Lv, Y. Zhang, M. E. Casas, P. N. Carvalho, C. A. Arias, K. Bester and H. Brix, Phytoremediation of imazalil and tebuconazole by four emergent wetland plant species in hydroponic medium, Chemosphere, 2016, 148, 459–466 CrossRef CAS.
- S. Wang, M. Li, X. Li, X. Li, X. Li, S. Li, Q. Zhang and H. Li, A functionalized carbon nanotube nanohybrids-based QuEChERS method for detection of pesticide residues in vegetables and fruits, J. Chromatogr. A, 2020, 1631, 461526 CrossRef CAS.
- G. Díaz-Mirón, M. A. Sánchez, D. M. Chemes and R. M. S. Álvarez, SERS spectrum of imazalil. Experimental and quantum-chemical vibrational analysis [J], J. Raman Spectrosc., 2018, 49(4), 638–650 CrossRef.
- H. Zhao, W. Hasi, N. Li, X. Sha, S. Lin and S. Han, In situ analysis of pesticide residues on the surface of agricultural products via surface-enhanced Raman spectroscopy using a flexible Au@Ag–PDMS substrate, New J. Chem., 2019, 43(33), 13075–13082 RSC.
- S. Zhang, M. Liu, J. Chen and J. Zhao, Rapid detection of imazalil residues in navel orange peel using surface-enhanced Raman spectroscopy, Smart Agr, 2021, 3(4), 42 Search PubMed.
- J. Yao, Z. Wang, L. Guo, X. Xu, L. Liu, H. Kuang and C. Xu, Lateral flow immunoassay for the simultaneous detection of fipronil and its metabolites in food samples, Food Chem., 2021, 356, 129710 CrossRef CAS PubMed.
- L. Luo, S. Q. Lin, Z. Y. Wu, H. Wang, Z. J. Chen, H. Deng, Y. D. Shen, W. F. Zhang, H. T. Lei and Z. L. Xu, Nanobody-based fluorescent immunoassay using carbon dots anchored cobalt oxyhydroxide composite for the sensitive detection of fenitrothion, J. Hazard Mater., 2022, 439, 129701 CrossRef CAS PubMed.
- P. Wang, X. Xu, L. Liu, S. Song, H. Kuang, C. Xu and X. Wu, A colloidal gold immunochromatography for the detection of flumioxazin residues in fruits, J. Food Sci., 2022, 87(10), 4538–4547 CrossRef CAS.
- J. Huo, B. Barnych, Z. Li, D. Wan, D. Li, N. Vasylieva, S. Z. Knezevic, O. A. Osipitan, J. E. Scott, J. Zhang and B. D. Hammock, Hapten synthesis, antibody development, and a highly sensitive indirect competitive chemiluminescent enzyme immunoassay for detection of dicamba [J], J. Agric Food Chem., 2019, 67(20), 5711–5719 CrossRef CAS.
- J. Liu, X. Xu, A. Wu, Z. Wang, S. Song, H. Kuang, L. Liu and C. Xu, Development of a gold nanoparticle-based lateral flow immunoassay for the detection of pyridaben, Microchem. J., 2021, 170, 106762 CrossRef CAS.
- J. Liu, X. Xu, A. Wu, Z. Wang, S. Song, H. Kuang, L. Liu and C. Xu, Preparing monoclonal antibodies and developing immunochromatographic assay strips for the determination of propamocarb levels, Food Chem., 2022, 370, 131284 CrossRef CAS PubMed.
- E. Ceballos-Alcantarilla, D. Lopez-Puertollano, C. Agullo, A. Abad-Fuentes, A. Abad-Somovilla and J. V. Mercader, Combined heterologies for monoclonal antibody-based immunoanalysis of fluxapyroxad, Analyst, 2018, 143(23), 5718–5727 RSC.
- E. Watanabe, S. Watanabe, S. Ito, M. Hayashi, T. Watanabe, Y. Yuasa and H. Nakazawa, Development of an enzyme-linked immunosorbent assay for the fungicide imazalil in citrus fruits, J. Agric. Food Chem., 2000, 48(11), 5124–5130 CrossRef CAS PubMed.
- M. J. Moreno, E. Plana, A. Montoya, P. Caputo and J. J. Manclus, Application of a monoclonal-based immunoassay for the determination of imazalil in fruit juices, Food Addit. Contam., 2007, 24(7), 704–712 CrossRef CAS PubMed.
- D. Kong, L. Liu, S. Song, S. Suryoprabowo, A. Li, H. Kuang, L. Wang and C. Xu, A gold nanoparticle-based semi-quantitative and quantitative ultrasensitive paper sensor for the detection of twenty mycotoxins, Nanoscale, 2016, 8(9), 5245–5253 RSC.
- T. Balkan and H. Karaağaçlı, Determination of 301 pesticide residues in tropical fruits imported to Turkey using LC-MS/MS and GC-MS, Food Control, 2023, 147, 109576 CrossRef CAS.
- J. F. de Freitas, M. E. L. R. de Queiroz, A. F. de Oliveira, L. de Paulo Ribeiro, D. V. Salvador, L. D. L. Miranda, R. R. Alves and A. A. Z. Rodrigues, Evaluation of imazalil dissipation/migration in postharvest papaya using low-temperature partition extraction and GC-MS analysis, Food Chem., 2023, 418, 135969 CrossRef CAS PubMed.
- J. da Silva Sousa, R. C. de Castro, G. de Albuquerque Andrade, C. G. Lima, L. K. Lima, M. A. Milhome and R. F. do Nascimento, Evaluation of an analytical methodology using QuEChERS and GC-SQ/MS for the investigation of the level of pesticide residues in Brazilian melons, Food Chem., 2013, 141(3), 2675–2681 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nh00371j |
| This journal is © The Royal Society of Chemistry 2024 |