Open Access Article
Open Access ArticleSynthesis and bioactivity of a novel surfactin-based lipopeptide for mRNA delivery†
Mohammed S.
Alqahtani
 *ab,
Rabbani
Syed
ab,
Ali S.
Alqahtani
c,
Omer M.
Almarfadi
c,
Monzurul A.
Roni
d and
Satya S.
Sadhu
e
*ab,
Rabbani
Syed
ab,
Ali S.
Alqahtani
c,
Omer M.
Almarfadi
c,
Monzurul A.
Roni
d and
Satya S.
Sadhu
e
aDepartment of Pharmaceutics, College of Pharmacy, King Saud University, Riyadh 11451, Saudi Arabia
bDepartment of Pharmaceutics, Nanomedicine & Biotechnology Research Unit, College of Pharmacy, King Saud University, Riyadh 11451, Saudi Arabia. E-mail: msaalqahtani@ksu.edu.sa
cDepartment of Pharmacognosy, College of Pharmacy, King Saud University, Riyadh 11451, Saudi Arabia
dDepartment of Health Sciences Education and Pathology, University of Illinois College of Medicine, Peoria, IL 61605, USA
eChemistry Department, Northern Michigan University, 1401, Presque, Isle, Marquette, MI 49855, USA
First published on 4th September 2024
Abstract
The effective delivery of messenger ribonucleic acid (mRNA) to specific cell types and target tissues poses a significant challenge in nonviral therapeutic strategies. Lipid-based nanoparticles (LNPs) have emerged as a leading carrier system for delivering mRNA, particularly for infectious diseases, such as COVID-19. This study aimed to describe the synthesis of a novel lipopeptide based on surfactin, a naturally occurring surfactant. Additionally, a series of novel LNPs were rationally designed, based on the modified surfactin, OleSurf, and were formulated and optimized. The physicochemical properties, morphologies, and stabilities of the particles were evaluated. All formulations containing OleSurf produced particles with a diameter <80 nm and an encapsulation efficiency >95%. OleSurf LNPs demonstrated excellent transfection efficiency and luciferase expression with no cytotoxicity, compared to lipofectamine 2000, a known transfection reagent, and were comparable to the DLin-MC3-DMA lipid. OleSurf-based LNPs behaved as efficient mRNA carriers and showed enhanced mRNA-binding capabilities, associated with facilitated intracellular release, endosomal escape, and protection from endonuclease degradation. In addition, OleSurf-LNPs showed a higher mRNA delivery efficiency, a more advantageous biodistribution pattern, and an improved safety profile in vivo. Overall, the novel OleSurf LNPs presented an optimal delivery platform for mRNA therapeutics.
1. Introduction
Lipid nanoparticles (LNPs) have emerged as highly attractive systems for delivering nucleic acid-based drugs.1,2 Historically, cationic liposomes, such as those based on DOTAP and DOTMA, were among the earliest synthetic systems used for nucleic acid delivery. Over time, formulations evolved with the addition of PEGylated lipids and the replacement of traditional cationic lipids with ionizable lipids. New formulations incorporating ionizable lipids, such as Dlin-MC3-DMA, have demonstrated the ability to form lipid nanoparticles (LNPs) that increase mRNA concentration within the particles. Ionizable lipid nanoparticles (iLNPs) typically feature a tertiary amine group that remains uncharged at neutral pH but becomes positively charged at lower pH levels, enhancing endosomal escape. These iLNPs are crucial for encapsulating nucleic acids and disrupting endosomal membranes, which facilitates the release of nucleic acids into the cytosol. Additionally, iLNPs may promote endosomal uptake through interactions with negatively charged cell membranes or plasma proteins that aid in cellular uptake.3 Messenger RNA (mRNA) technology holds significant promise in therapeutic applications.4 However, its molecule, which is relatively large with a length typically ranging from 500 to 5000 nucleotides and carries a negative charge, relies on a delivery vehicle to ensure effective cellular uptake and protection against degradation.5 An example of an FDA-approved RNA-based therapy is Onpattro (Patisiran), an LNP-based small-interfering RNA (siRNA) treatment for hereditary transthyretin amyloidosis (hATTR).6 The main lipid in this LNP was DLin-MC3-DMA [(6Z,9Z,28Z,31Z)-heptatriacont-6,9,28,31-tetraene-19-yl 4-(dimethylamino)butanoate], which is often referred to as MC3. It consists of a dilinoleic acid tail and an ester linker group, making it effective for silencing genes in the liver.7 Additionally, some RNA-based therapeutics have been developed for coronavirus disease 2019 (ref. 8) vaccines, further demonstrating their safety and effectiveness.8 Given the inherent instability of mRNA in vivo and its inability to permeate cell membranes, effective delivery systems are critical to ensure that mRNA reaches the cytoplasm, where it can perform its therapeutic functions.9 To enhance mRNA delivery efficiency, extensive research has been conducted to screen various lipids and refine their properties to further improve their efficiency.3,10 The general structure of lipids can be divided into two main components, namely the head group and the tails. Lipid head groups usually carry positive charges and are primarily composed of amine moieties. The positive charge results in pH-dependent ionization, facilitating endosomal escape. The size and charge density of the headgroups play crucial roles in capturing nucleic acids, providing stability to the lipid nanoparticles (LNPs), and interacting with the cell membrane. Moreover, to reduce accumulation and long-term toxicity, lipids should be readily degraded into nontoxic metabolites after successful intracellular delivery, which is especially crucial for mRNA therapeutics that require repeated dosing.11Over the past 25 years, studies have extensively explored the capacity of numerous cationic lipids to bind mRNA and facilitate its delivery in both cells (in vitro) and in animal models (in vivo).12 Initial attempts to use conventional liposomes were hindered by the limited binding between mRNA and neutral or negatively charged liposomes.13 However, subsequent investigations revealed that the utilization of cationic lipids could significantly improve the rates of association and delivery.13 While the ultimate objective of these investigations is to develop pharmaceutical agents capable of delivering therapeutic mRNA, the initial screening process for identifying promising cationic agents typically occurs via cell culture.3 Consequently, in vitro transfection rates serve as a measure of the delivery efficiency of experimental formulations, and majority of published studies rely solely on cultured cells to evaluate the potential of novel cationic agents for intracellular delivery.14 In addition to the cell type utilized in these studies, factors such as mixing conditions, charge ratio, particle size, zeta potential, and the presence of serum have been widely acknowledged to have a significant impact on the observed transfection rates in such experiments. Previously, mRNA had been formulated using various cationic delivery platforms. Several studies have emphasized the importance of cationic lipids in mRNA transfection.15 The primary advantage of using cationic lipids to complex with anionic RNA molecules is the enhancement of cellular uptake.16 The effectiveness of lipid nanoparticles (LNPs) is due to the synergistic roles of their main components. Helper phospholipids (such as DSPC, DOPE, and DOPC) enhance the stability of the LNP bilayer, which helps prevent leakage of the nucleic acid cargo and facilitates membrane fusion for efficient cellular uptake. Cholesterol, a steroid component, modifies the fluidity and permeability of the bilayer, stabilizing the lipid structure by filling gaps and promoting tighter packing of the lipids. Additionally, the PEG lipid conjugate is crucial for reducing LNP size, protecting them from rapid clearance by the reticuloendothelial system, and preventing protein adsorption.3,17 Collectively, these components make LNPs highly effective as delivery vehicles for vaccines and other therapeutics, especially in large-scale production.18
Biomimetic and bioinspired nanomaterials, such as lipopeptide-based nanoparticles, have gained significant attention in recent years, particularly in the field of drug delivery.19 Lipopeptides exhibit characteristics of both lipids and peptides. They have biological functions, including targeting effects, and leverage lipid properties, such as hydrophobic interactions.20 Lipopeptides constitute a distinct category of a highly potent biosurfactant. These bioactive secondary metabolites have remarkable therapeutic and biotechnological attributes.21 Surfactin, a prominent member of the antimicrobial lipopeptide family, is produced by B. subtilis strain.22 It is a cyclic lipopeptide with amphipathic properties, having a molecular weight of 1036 Da.21 It is chemically defined as a heptapeptide interlinked with a β-hydroxy fatty acid. Owing to its amphipathic nature, surfactin demonstrates a broad spectrum of interactions with the phospholipid bilayer of the target cell and holds promise in various medical applications.21 Surfactin can destabilize cell membranes and compromise their structure through a range of theoretical mechanisms; it has the potential to integrate into lipid bilayers and alter membrane permeability through ion channel formation, phospholipid solubilization, and micelle formation.23 Furthermore, surfactin can spontaneously infiltrate lipid membranes through hydrophobic interactions, leading to changes in the arrangement of hydrocarbon structures and membrane thickness.24 In addition to its antifungal and antibacterial effects, surfactin can hinder fibrin clot formation, trigger the creation of ion channels in lipid bilayer membranes, inhibit cyclic adenosine monophosphate activity, prevent platelet aggregation, and display antiviral and antitumor properties.25 Additionally, surfactin has been used in pharmaceutical formulations to improve the delivery of therapeutic agents.26 Previous studies on mRNA-LNPs had emphasized the importance of developing new cationic lipids for mRNA transfection. Significant disadvantages associated with the frequently used cationic lipids for mRNA delivery include their restricted electrostatic interaction with mRNA, owing to the presence of a single quaternary ammonium head group (in contrast to structures containing multiple amine functionalities), activation of the innate immune system leading to undesirable side effects, and unfavorable biodistribution resulting from plasma protein binding and non-specific tissue distribution.27 Ultimately, these factors contribute to a relatively limited therapeutic window. The purpose of this study was to synthesize a novel modified surfactin (OleSurf) as a primary component of lipid nanoparticles (LNPs). To establish correlations for the LNP performance, we conducted a comparative analysis of OleSurf LNPs. We assessed their effectiveness in transfecting cell lines in vitro, observed their cellular uptake, and evaluated their expression in vivo.
2. Materials and methods
2.1. Materials
Surfactin was purchased from TargetMol (Boston, MA, USA). Lipofectamine 2000 (LP 2000) was from Invitrogen (CA, USA). 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) was obtained from Avanti Polar Lipids (Birmingham, AL, USA). Cholesterol was obtained from MP Biomedicals (Santa Ana, CA, USA). 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG) was purchased from NOF America Corporation (White Plains, NY, USA). Pierce D-Luciferin, Slide-A-Lyzer™ MINI Dialysis Device (MWCO, 10 kDa), RNAse-free, and Quanti-iT Ribogreen RNA reagent were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Firefly luciferase (Fluc) mRNA was purchased from TriLink BioTechnologies (San Diego, CA, USA). Firefly-luciferase mRNA labeled with the fluorophore Cy5 was purchased from APExBIO (Houston, TX, USA). Microfluidic cartridges for the NanoAssemblr were purchased from Precision Nanosystems. The ONE-Glo Luciferase Assay Kit was purchased from Promega. Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), Dlin-MC3-DMA (MC3), sodium hydroxide, citric acid, sodium citrate, and oleylamine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 and LysoBrite Green was purchased from Thermo Fisher Scientific (Cleveland, OH, USA). PAS-Green Stain kit was from Agilent Technologies (Santa Clara, CA, USA). All the other chemicals were of reagent grade.
342 and LysoBrite Green was purchased from Thermo Fisher Scientific (Cleveland, OH, USA). PAS-Green Stain kit was from Agilent Technologies (Santa Clara, CA, USA). All the other chemicals were of reagent grade.
2.2. Synthesis and characterisation of lipopeptide conjugates
Oleylamine was used to modify surfactin via amidation reactions, following a previously reported method with some modifications, and its structure was confirmed by 1H NMR and mass spectrometry.28 Briefly, 20 mg (0.02 mmol) of surfactin was dissolved in methanol and 0.25 g (2 mmol) of oleylamine was mixed with it. After adjusting the pH to 8.0 with 2 M NaOH, 10 mL of dichloromethane (DCM) was added to start the reaction under a nitrogen atmosphere. The reaction mixture was stirred at 25 °C for 24 h. Finally, the reaction mixture was concentrated in an evaporator and purified with silica column chromatography (DCM/MeOH 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). A yellow oil was obtained, which was sealed and stored at 4 °C until further use. 1H-NMR spectra were recorded on a Bruker 300 MHz spectrometer using tetramethylsilane (TMS) as the internal standard, with its residual 1H resonance calibrated at 2.14 ppm. Mass spectra were measured using an AVANT UHPLC system equipped with an Advion expression® S compact mass spectrometer (Advion, Ithaca, NY, USA). OleSurf analysis was performed on this system using a Thermo C18 + UHPLC Column (1.6 μm, 100 × 2.1 mm) at a flow rate of 200 μL min−1. The mobile phases consisted of 5 mM ammonium acetate in water (phase A) and 5 mM ammonium acetate in acetonitrile/water (95/5, v/v) (phase B), with a flow rate of 0.3 mL min−1. Sample elution began with 70% buffer B, followed by a linear gradient to 100% buffer B over 30 minutes. The injection volume of the sample extract was 20 μL. The analysis was monitored at 210 nm and in negative-ion mode over the m/z range from 500 to 1200.
1 v/v). A yellow oil was obtained, which was sealed and stored at 4 °C until further use. 1H-NMR spectra were recorded on a Bruker 300 MHz spectrometer using tetramethylsilane (TMS) as the internal standard, with its residual 1H resonance calibrated at 2.14 ppm. Mass spectra were measured using an AVANT UHPLC system equipped with an Advion expression® S compact mass spectrometer (Advion, Ithaca, NY, USA). OleSurf analysis was performed on this system using a Thermo C18 + UHPLC Column (1.6 μm, 100 × 2.1 mm) at a flow rate of 200 μL min−1. The mobile phases consisted of 5 mM ammonium acetate in water (phase A) and 5 mM ammonium acetate in acetonitrile/water (95/5, v/v) (phase B), with a flow rate of 0.3 mL min−1. Sample elution began with 70% buffer B, followed by a linear gradient to 100% buffer B over 30 minutes. The injection volume of the sample extract was 20 μL. The analysis was monitored at 210 nm and in negative-ion mode over the m/z range from 500 to 1200.
2.3. Lipid nanoparticle (LNP) formulation and characterization
LNPs were formulated via microfluidic mixing using a previously described method with some modifications.29 Briefly, one part of the organic phase was mixed with three parts of the aqueous phase containing mRNA using a microfluidic mixing chip with etched channel depths of 125 μm and a hydrophilic coating (Dolomite Microfluidics, England, UK). The chip features three inlets that converge for immediate mixing, where the aqueous phase containing RNA enters from each side, and the organic phase containing the lipid mixture enters through the center channel. Lipid mixtures were prepared at a total lipid concentration of 1 mM in 200-proof ethanol (Fisher Bioreagents, Pennsylvania, USA) using a molar ratio of 50, 15, 30, and 2 for cationic lipids, DSPC, DMG-PEG, and cholesterol, respectively. The aqueous phase, composed of mRNA in 100 mM citrate buffer at pH 4, was combined with the lipid solution at a weight ratio of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for ionizable lipid to RNA. Before mixing, the lipid solution was heated to 60 °C for one minute and briefly sonicated to ensure uniform solubilization of the lipid components. As the sample exits the microfluidic outlet, it is immediately diluted in 1 mL of phosphate-buffered saline (0.01 M PBS, pH 7.4) and kept in ice. The mRNA-LNPs were purified using a 10 kDa Slide-A-Lyzer™ dialysis cassette (Millipore Sigma, Billerica, MA). The cassettes containing the mRNA-LNPs were left in 1.0 Liter beaker containing 1× PBS, and dialyzed for a minimum of 12 h. After dialysis, the cassettes containing the mRNA-LNPs were kept in a sterile biosafety cabinet and the mRNA-LNPs were removed using a syringe with a needle. Finally, the formulation was further purified using Amicon ultra centrifugal filters (10 kDa, 3000 g, 60 min) and stored at 4 °C. The hydrodynamic radius (nm) and polydispersity index (PDI) of the LNPs were assessed via dynamic light scattering using a Malvern Zetasizer-S 3600 (Malvern Instruments Inc., Southborough, MA, USA). Each sample was measured in triplicate, and the results are expressed as mean ± SD. The efficiency of mRNA encapsulation was determined using a modified version of the Quant-iT RiboGreen RNA reagent, as previously reported.30 Encapsulation efficiency was expressed as % mean of three experiments (±SD).
1 for ionizable lipid to RNA. Before mixing, the lipid solution was heated to 60 °C for one minute and briefly sonicated to ensure uniform solubilization of the lipid components. As the sample exits the microfluidic outlet, it is immediately diluted in 1 mL of phosphate-buffered saline (0.01 M PBS, pH 7.4) and kept in ice. The mRNA-LNPs were purified using a 10 kDa Slide-A-Lyzer™ dialysis cassette (Millipore Sigma, Billerica, MA). The cassettes containing the mRNA-LNPs were left in 1.0 Liter beaker containing 1× PBS, and dialyzed for a minimum of 12 h. After dialysis, the cassettes containing the mRNA-LNPs were kept in a sterile biosafety cabinet and the mRNA-LNPs were removed using a syringe with a needle. Finally, the formulation was further purified using Amicon ultra centrifugal filters (10 kDa, 3000 g, 60 min) and stored at 4 °C. The hydrodynamic radius (nm) and polydispersity index (PDI) of the LNPs were assessed via dynamic light scattering using a Malvern Zetasizer-S 3600 (Malvern Instruments Inc., Southborough, MA, USA). Each sample was measured in triplicate, and the results are expressed as mean ± SD. The efficiency of mRNA encapsulation was determined using a modified version of the Quant-iT RiboGreen RNA reagent, as previously reported.30 Encapsulation efficiency was expressed as % mean of three experiments (±SD).
2.4. Cryo-transmission electron microscopy (cryo-TEM)
Cryo-TEM was performed at 300 kV using the FEI Titan Krios (Thermo Fisher Scientific) equipped with Falcon III and K3 cameras with DED. Three to five microliters of the sample were dispensed on a plasma-cleaned grid (Quantifoil TEM grids) in a Vitrobot chamber and allowed to incubate for 50 s. The chamber was set to 100% humidity at 4 °C. Next, the sample was blotted for 4.5 s and plunged into a propane–ethane mixture (40% ethane and 60% propane) cooled in liquid nitrogen. Subsequently, the frozen grids were observed for defects and were stored in liquid nitrogen. Transmission electron microscopy (JEM-F200 TEM, USA) was used to detect the morphology of the mRNA-encapsulated LNPs. The images obtained were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).2.5. Cell culture
HeLa and HepG2 cell lines were obtained from the American Type Culture Collection (ATCC) (Rockville, MD, USA). Cells were cultured in DMEM supplemented with 10% FBS, 100 mg per mL streptomycin, and 100 U per mL penicillin. The cells were grown in a humidified atmosphere and controlled temperature at 37 °C with 5% CO2. Cells at the exponential growth stage were harvested using 0.25% trypsin–EDTA treatment. In every experiment, the passage number of cells was considered, and after passage 10, the cells were no longer used.2.6. In vitro transfection efficiency
Transfection was performed as described previously.31 Approximately 6000 HeLa cells were seeded in each well of a white-wall clear bottom 96 well plate in 100 μL of complete cell culture medium (DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin). The cells were allowed to adhere overnight in a controlled CO2 incubator at 37 °C and 5% CO2 to ensure approximately 80% confluent cultures. Transfection was carried out using lipofectamine 2000, following the manufacturer's instructions. The LNP formulations encapsulating firefly luciferase-(Fluc mRNA) were diluted in complete cell culture medium so that the total dose was delivered in 100 μL. Briefly, the medium was removed from each well on the day of experiment. The cells were treated with mRNA-LNPs, diluted in complete cell culture medium at the desired doses or in complete medium (control). Thereafter, the 96 well plate containing the treated cells was placed in a controlled CO2 incubator for 24 h. The transfection efficiency was calculated using a luciferase assay. The 1× cell lysis buffer and luciferase reagent were prepared according to the manufacturer's instructions. The 96 well plate containing the cells was removed from the incubator and placed in a biosafety cabinet. Finally, 25 μL of the prepared luciferase assay reagent was added to each well. The plate was placed on a Tecan plate reader to measure the bioluminescence signal in each well. Normalized luciferase expression was quantified by subtracting the bioluminescence values from the untreated or transfected cells without firefly luciferase-encoding mRNA cargo.2.7. Cellular uptake and endosomal escape imaging
For endosomal escape studies, cells were seeded in confocal dishes at a density of 1 × 105 cells and transfected with LNP formulations encapsulating firefly luciferase (Fluc) mRNA. The cells were then incubated for 0.5, 2, and 4 hours. Next, the treated cells were fixed in 4% paraformaldehyde and washed with phosphate-buffered saline (PBS). The cells were then washed thrice with buffer (10× PBS) and incubated for 45 min with LysoBrite Green and Hoechst-33258 for nuclear staining. Subsequently, the cells were washed, and stored in buffer (PBS) until used for imaging. Cells were imaged at ×20 magnification and the fluorescence signals of these cells were analyzed using CLSM (Lecia TCS SP8, Germany).2.8. In vivo bioluminescence imaging of LNP
All animal procedures were approved by the Animal Care and Use Committee of the King Saud University and performed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Female BALB/c mice (6–8 weeks old) were obtained from Charles River Laboratories (Wilmington, MA, USA). The mRNA-LNP solution was diluted with sterile 1× PBS before in vivo administration. Randomized mice received tail injections of 100 μL of luciferase LNPs at a dose of 0.15 mg kg−1. For in vivo imaging, a stock solution of 15 mg per mL D-luciferin potassium salt in sterile 1× PBS was prepared; 9 hours post injection, mice were injected with 200 μL of D-luciferin intraperitoneally. Before imaging, the mice were anesthetized in a chamber with an oxygen flowrate of 2.0 L min−1 and 2.5% isoflurane. The luciferase signal was allowed to stabilize for 15 min before imaging the treated mice. The mice were imaged using an IVIS Spectrum optical imager (PerkinElmer, MA, USA) at an exposure time of 30 s to obtain a full bioluminescence image. Moreover, bioluminescence of organs (kidneys, liver, spleen, heart, and lungs) was obtained at 24 h and was quantified as average radiance (p s−1 cm−2 sr−1). Histopathological study was performed to determine acute liver toxicity. Blood samples were collected 24 h post-injection and sent for biochemical analysis using a Cobas 6000 (Roche Diagnostics, IN, USA). Liver tissue samples were collected and placed in paraffin after being fixed in 4% paraformaldehyde, and glacial acetic acid. 5 μm sections of paraffin-embedded tissues were prepared and stained by Periodic Acid-Schiff (PAS) according to manufacturer's instructions. The area of PAS staining was quantified and carbon tetrachloride (CCl4) was used as a positive control.322.9. Data analysis
All experiments were performed in triplicate, and the results are expressed as mean ± SD. One-way ANOVA (GraphPad Software, La Jolla, CA, USA) was used to compare the effects across different groups, and the results were considered significant at p < 0.05.3. Results and discussion
3.1. Synthesis and preparation of OleSurf LNPs
Surfactin, a cyclic lipopeptide composed of five hydrophobic amino acids and two hydroxy fatty acids, exhibits a slightly negative overall charge due to the carboxyl groups present in aspartic acid and glutamic acid. OleSurf was synthesized as the primary component of lipid nanoparticles (LNPs), achieving a net yield of 1.8 g (72%). The synthesis involved an amidation reaction to conjugate oleylamine with the surfactin acidic amino acids (Fig. 1). Mass spectrometric analysis confirmed the successful synthesis of OleSurf, with a molecular weight consistent with theoretical calculations (Fig. 2). The synthesized OleSurf samples were acidified, and the resulting precipitates were extracted with methanol. These extracts were analyzed by UHPLC/MS. High-resolution mass spectrometry (HRMS) with electrospray ionization (ESI) provided detailed characterization. Surfactin (C53H93N7O13) was detected with principal m/z values of 994.6, 1008.5, 1022.0, and 1036.7, indicating various fatty acid length polymorphisms, with retention times of 17.3, 18.7, and 17.8 minutes. OleSurf (C89H163N9O11) showed a calculated m/z of 1535, which was confirmed as 1535.2. 1H-NMR spectra for surfactins and OleSurf in their ionized forms are shown in (Fig. 3). The spectra reveal three main regions: amide proton resonances (6.5–10 ppm), α-carbon protons (3.5–5.5 ppm), and side-chain protons (0.25–3.0 ppm). Residual water appears at 3.54 ppm. The spectra are sufficiently resolved to display amide proton splittings with coupling constants ranging from 7.0 Hz to 8.2 Hz. Significant differences in relative intensities and chemical shifts are observed in the amide-proton regions, with the presence of some minor impurities also noted. Several studies have indicated that esterification or amidation of the two carboxylic acids of surfactin can result in new derivatives with unique biological and interfacial activities.28,33 In this study, we hypothesized that amidation of aspartic and glutamic acids would eliminate the negative charge of these residues, thereby increasing the contribution of the cationic backbone of surfactin to mRNA complexation. The major drawback of the commonly used cationic lipids is their limited electrostatic interaction with mRNA owing to the presence of a single quaternary ammonium head group.27 However, the surfactin structure contains multiple amine functionalities and had previously been reported to bind RNA.34 Furthermore, incorporating oleylamine lipid chains in addition to the predominantly hydrophobic amino acid residues facilitates the complexation. Previous reports had indicated that including unsaturated fatty acids as lipid tails leads to an increased delivery efficiency of LNP formulations.35 This is potentially due to their lower transition temperatures and their ability to enhance membrane fluidity.36 Indeed, the overall zeta potential of OleSurf LNPs was increased and mRNA encapsulation efficiency was 90.5% with mRNA concentration of 43.25 ng μL−1, which is comparable to that of the common lipid D-Lin-MC3-DMA (MC3). When formulated in LNPs, OleSurf resulted in 85 nm particles having low polydispersity with a slightly positive zeta potential (Table 1). The cryo-TEM image revealed the presence of bleb structures, and these findings, in conjunction with the results from Dynamic Light Scattering (DLS), indicated colloidal stability (Fig. 4). The physicochemical characteristics of LNPs, such as the particle diameter, polydispersity index (PDI), and surface charge (zeta potential), play a significant role in determining their biological effectiveness.38 Previous studies had highlighted the substantial impact of these physicochemical characteristics on the overall success of LNPs in gene delivery.39 Since LNPs can undergo changes in their particle properties over time, during storage, potentially affecting their biological performance, we conducted measurements to track changes in particle size and zeta potential over a seven-day period while keeping the samples refrigerated. Remarkably, no noticeable difference in size, PDI, charge or encapsulation efficiency (EE%) was observed during storage (Table S1†). In the context of LNP-based mRNA formulations, future efforts may be directed towards improving the stability of lipids to prevent mRNA leakage, particularly in the bloodstream following administration.40 Gaining insights from current research that focuses on developing cationic lipids to precisely adjust their surface charge, it is beneficial to explore potential modifications in other LNP constituents, including cholesterol, helper lipids, and PEG. These adjustments can help modify the structure of the LNP shell, leading to enhancements in both colloidal and mRNA stability.35,41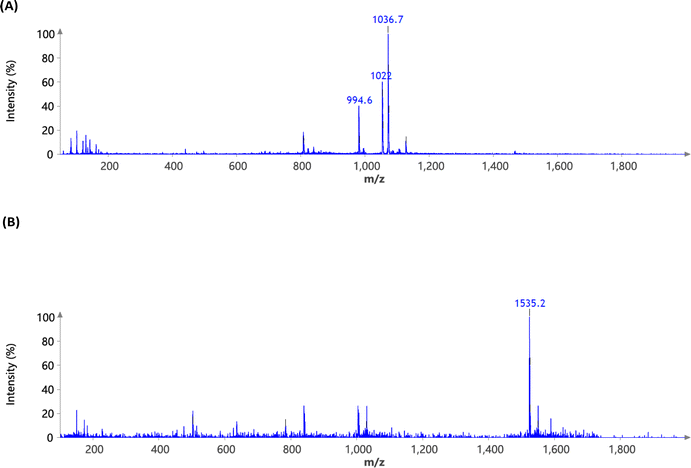 | ||
| Fig. 2 High-resolution mass spectrometry (HRMS) ESI of (A) surfactin (C53H93N7O13) showing the main m/z (M + H) at 994.6, 1008.5, 1022 and 1036.7 (B) OleSurf (C89H163N9O11) calc. 1535 found 1535.2. | ||
| Sample ID | Average particle size (nm) | Polydispersity index (PDI) | Zeta potential37 | Encapsulation efficiency (%) |
|---|---|---|---|---|
| a Data are shown as mean ± SD of three independent experiments. | ||||
| LP2000 | 227.12 ± 11.05 | 0.218 ± 0.103 | −2.05 ± 1.4 | 88.38 ± 4.8 |
| MC3 | 71.61 ± 5.52 | 0.074 ± 0.029 | +5.14 ± 1.6 | 92.35 ± 2.1 |
| OleSurf | 84.91 ± 6.24 | 0.102 ± 0.025 | +1.3 ± 0.7 | 90.51 ± 3.7 |
 | ||
| Fig. 4 Physicochemical characterization of mRNA OleSurf LNPs. (A) Hydrodynamic size measurements and size distribution by dynamic light scattering (DLS). (B) Representative cryoTEM images. | ||
3.2. Cellular transfection efficiency
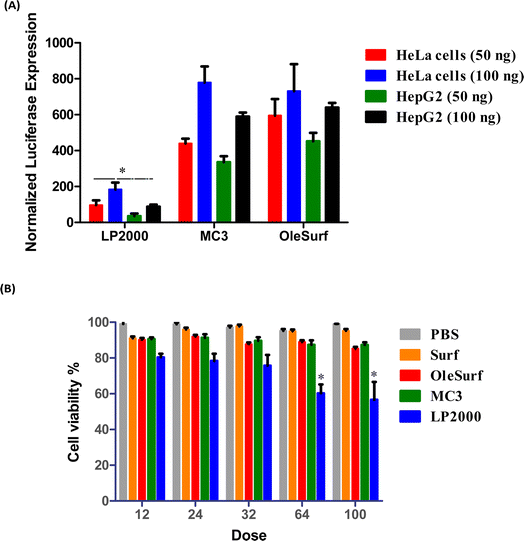
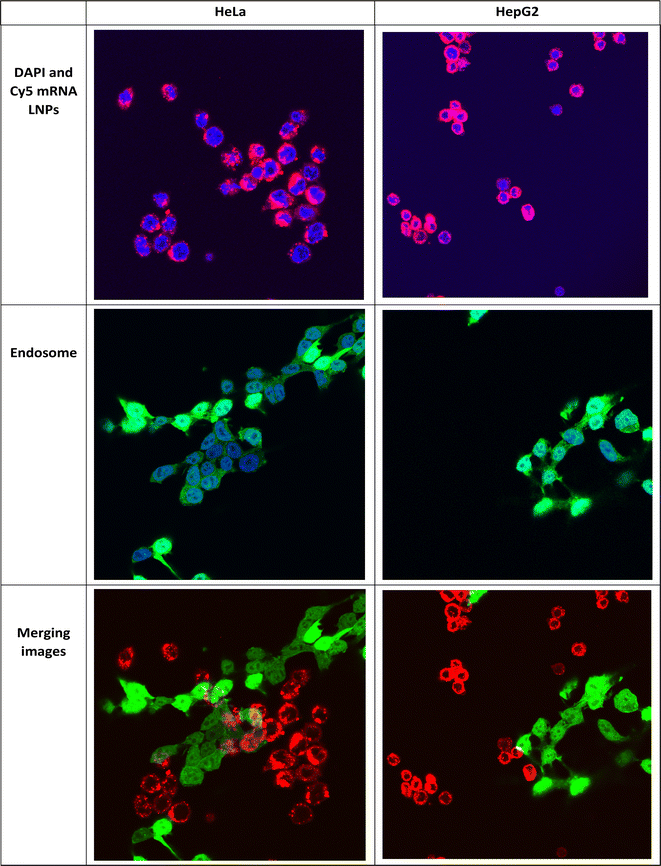
This study was performed using two different cell lines, HeLa and HepG2, which are often used for in vitro transfection studies.40 Luciferase expression was evaluated in vitro as a measure of the functional mRNA expression. Lipofectamine is commonly regarded as a benchmark transfection agent for delivering nucleic acids in vitro.42 Furthermore, as the gold standard, OleSurf LNPs were compared with LNPs containing the FDA-approved lipid D-Lin-MC3-DMA (MC3 LNPs) while encapsulating the same Fluc mRNA components.43,44 LNPs or lipofectamine 2000 (LP2000) were used to treat the cells with either 50 or 100 ng of luciferase mRNA per 6000 cells. OleSurf LNPs exhibited significantly higher luciferase expression than lipofectamine, with an approximately three-fold increase in transfection efficiency (Fig. 5A). Moreover, the trends observed in the transfection with 50 ng of mRNA were also observed at higher doses (100 ng), indicating better transfection and a lack of cytotoxicity. Additionally, cell viability was evaluated 24 h after treatment with different concentrations of Fluc mRNA, and none of the OleSurf LNPs showed a significant decrease in cell viability compared to MC3 LNPs (Fig. 5B). However, we noticed that LP2000 induced cytotoxicity in HepG2 cells, resulting in only 65–70% viability after transfection.Intracellular localization of mRNA is an important factor in successful gene therapy.45 Moreover, cationic lipids in LNPs play a key role in their intracellular uptake and delivery.40 Typically, hydrolytic enzymes and the acidic environment in lysosomes can degrade both the carrier and the mRNA within. Therefore, endosomal escape prior to mRNA degradation is a critical step for the success of mRNA therapy.41 Within the endosome, LNPs can undergo protonation, leading to the fusion of their membrane lipids with the anionic lipids present there. Consequently, their mRNA cargo is released into the cytosol.40,46 We studied the uptake and endosomal escape of OleSurf LNPs loaded with Cy5 mRNA in two different cell lines (Fig. 6). Results showed that the intracellular uptake of Cy5 mRNA-loaded OleSurf LNPs can be seen as red fluorescence around the cell nucleus. Furthermore, after 4 h of incubation, most of the green signal originating from the endosomes did not overlap with the red signal emitted by the Cy5 mRNA, indicating that OleSurf LNPs effectively facilitated the release of mRNA into the cytosol. This observation supported those reported in previous studies, together implying that LNPs deliver mRNAs and release them inside the cells.46,47
3.3. In vivo mRNA delivery
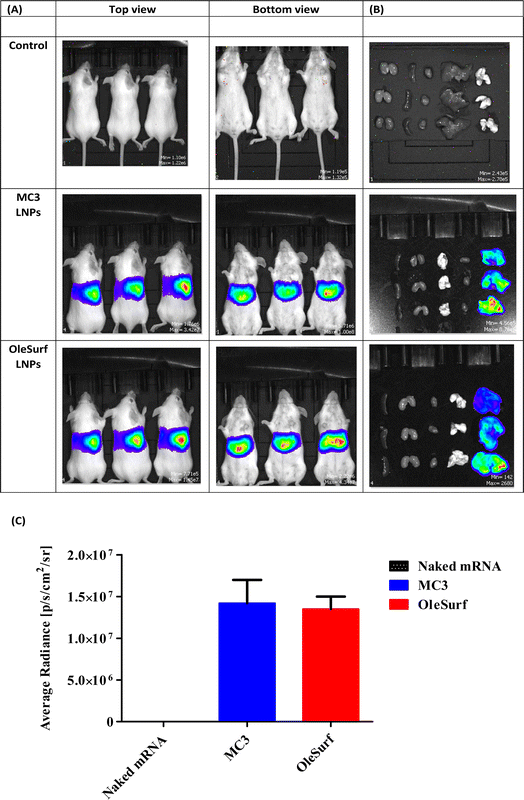
Given the significant in vitro transfection efficiency of OleSurf LNPs, we designed and performed a more detailed in vivo imaging study. MC3 has gained remarkable popularity as an ionizable cationic lipid used in the formulation of LNPs, thereby facilitating mRNA delivery.48 However, achieving high mRNA delivery effectiveness and incorporating biodegradable properties into LNPs are challenging.49 We compared our OleSurf LNPs with the LNPs composed of MC3 lipids, which are commonly used for targeting liver, because previous studies had demonstrated that after systemic administration, LNPs primarily target hepatocytes and accumulate in the liver.7,50 A frequently observed uptake mechanism for LNPs involves their binding to plasma proteins, leading to opsonization and subsequent scavenger receptor-mediated uptake into hepatic Kupffer cells and the sinusoidal endothelium.51Lipopeptides offer distinct advantages, particularly for biodegradability and multifunctionality.52 Biodegradable lipopeptides offer the advantage of rapid clearance from plasma, enhancing their tolerability and safety profile.53 Beyond their role as delivery components, lipopeptides may also exhibit therapeutic effects that work in synergy with mRNA-encoded proteins.54 Recent studies have demonstrated the successful delivery of cholesterol-modified siRNAs using lipopeptide-based materials.55 Amino acids and peptides are intrinsic constituents of apolipoproteins, and several studies have explored the potential of amino acid derivatives in mRNA delivery.10 To the best of our knowledge, none of these systems demonstrated strengths or selectivities comparable to those of conventional lipids. In our study, we observed notable luminescence throughout the entire animal group, particularly in the liver (Fig. 7A and B). This observation indicates the expression of luciferase mRNA loaded into OleSurf LNPs within the liver and no off-target luciferase expression after IV administration. Moreover, no significant difference was observed between MC3 based LNPs and OleSurf LNPs (Fig. 7C). This phenomenon had been reported previously, where LNPs designed for liver targeting were bound to apolipoprotein E (ApoE) in the bloodstream, leading to the uptake of lipoproteins by liver hepatocytes through receptor-mediated endocytosis facilitated by the low-density lipoprotein receptor (LDL-R).50,56
Safety is a crucial consideration before translating LNPs into the clinical stage.40,43 We evaluated whether the OleSurf-based LNPs demonstrated biocompatibility in vivo. We assessed liver toxicity at different doses of OleSurf LNPs. To evaluate liver toxicity, we examined enzyme markers, such as alanine transaminase (ALT) and aspartate aminotransferase57 in the bloodstream, supported by histological analysis. Typically, these enzymes exist in the bloodstream at minimal concentrations, but their levels rise in response to liver damage.58 Following OleSurf LNPs treatment, there was a negligible elevation in liver enzymes at any dose relative to the control, and the enzyme levels remained within the normal range across all treatment groups (Fig. 8A and B). Additionally, histological assessment of the liver samples did not reveal any sign of necrosis, significant bleeding, or other morphological alterations. The utilization of periodic acid-Schiff (PAS) staining revealed the typical histology of hepatocytes and hepatic cords, with the cytoplasm of the majority of hepatocytes displaying a distinct staining pattern following the administration of OleSurf LNPs (Fig. 8C). PAS staining has previously proven to be a reliable method for assessing the differentiation of cells into fully functioning hepatocytes and for evaluating acute hepatotoxicity.59 Following the treatment of OleSurf LNPs, we did not observe any pale patches indicative of glycogen depletion within the parenchyma (Fig. 8D and E). To enhance the relevance of our results, we compared the toxicity of OleSurf LNPs to that of MC3 LNPs as reported in the literature.57 Studies have shown that MC3 LNPs, similar to ALC-0315 LNPs, are effective in delivering RNA to hepatocytes and hepatic stellate cells (HSCs). However, MC3 LNPs exhibit different toxicity profiles compared to ALC-0315. For instance, ALC-0315 LNPs, at high doses, have been associated with increased liver toxicity markers such as ALT. In contrast, MC3 LNPs do not show such effects at similar doses. The observed differences are attributed to variations in lipid structures affecting endosomal escape and liver cell interaction. Specifically, ALC-0315's more pronounced cone-shaped structure enhances endosomal escape but may contribute to higher hepatotoxicity compared to MC3 LNPs. Collectively, the findings indicated that OleSurf LNPs did not induce liver toxicity and have a safety profile similar to that of MC3 LNPs. Nevertheless, conducting long-term toxicity assessments for LNPs is crucial, especially for their frequent use in chronic disease treatment, and future studies are essential to evaluate their long-term safety. Overall, our study confirmed that the LNPs are both safe and effective for mRNA delivery. In future perspective, the approach to formulating OleSurf LNPs will align closely with the central objectives of advanced drug delivery. This involves custom-tailoring the formulation method to encapsulate various macromolecules and a dedicated exploration of other administration routes.
 | ||
| Fig. 8 Evaluation of liver toxicology in mice 24 hours post injection of different LNPs doses (A) serum profile analysis of alanine aminotransferase (ALT) levels. (B) Serum profile analysis of aspartate aminotransferase57 levels. (C & D) Representative periodic acid-Schiff (PAS) stained liver sections of BALB/c mice after intravenous injection of OleSurf LNP at a dose of 0.15 mg kg−1 at 40× and 100× magnification respectively. (E) Quantification of PAS in the hepatic tissues after treatment of carbon tetrachloride (CCl4), phosphate-buffered saline (PBS), and OleSurf LNPs. | ||
4. Conclusions
In summary, we developed a novel LNP platform, based on OleSurf, for mRNA delivery. OleSurf facilitated the efficient encapsulation of mRNA and its transfection in vitro. In addition, OleSurf LNPs protected mRNA molecules from degradation and facilitated their cellular uptake and release into the cytosol. Our study demonstrated that OleSurf LNPs have comparable activity to the clinically approved MC3 LNPs in delivering mRNA. Moreover, LNPs were found to be safe and non-toxic. Overall, the results of this study demonstrated the significant potential of OleSurf LNPs as a highly efficacious delivery system for mRNA-based therapeutics.Conflicts of interest
Dr Mohammed S. Alqahtani is the inventor named on the patent application filed (#18/795![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667) related to this technology.
667) related to this technology.
Acknowledgements
The authors are thankful to the Researchers Supporting Project number (RSP2024R132), King Saud University, Riyadh, Saudi Arabia.References
- M. L. Ibba, G. Ciccone, C. L. Esposito, S. Catuogno and P. H. Giangrande, Advances in mRNA non-viral delivery approaches, Adv. Drug Delivery Rev., 2021, 177, 113930 CrossRef CAS PubMed.
- S. Qin, X. Tang, Y. Chen, K. Chen, N. Fan, W. Xiao, Q. Zheng, G. Li, Y. Teng and M. Wu, mRNA-based therapeutics: powerful and versatile tools to combat diseases, Signal Transduction Targeted Ther., 2022, 7(1), 166 CrossRef CAS PubMed.
- Y. Zhang, C. Sun, C. Wang, K. E. Jankovic and Y. Dong, Lipids and lipid derivatives for RNA delivery, Chem. Rev., 2021, 121(20), 12181–12277 CrossRef CAS PubMed.
- J. C. Kaczmarek, P. S. Kowalski and D. G. Anderson, Advances in the delivery of RNA therapeutics: from concept to clinical reality, Genome Med., 2017, 9, 1–16 CrossRef CAS; U. Sahin, K. Karikó and Ö. Türeci, mRNA-based therapeutics—developing a new class of drugs, Nat. Rev. Drug Discovery, 2014, 13(10), 759–780 CrossRef PubMed.
- Y. Zong, Y. Lin, T. Wei and Q. Cheng, Lipid Nanoparticle (LNP) Enables mRNA Delivery for Cancer Therapy, Adv. Mater., 2023, 2303261 CrossRef CAS PubMed.
- X. Zhang, V. Goel and G. J. Robbie, Pharmacokinetics of Patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis, J. Clin. Pharmacol., 2020, 60(5), 573–585 CrossRef CAS.
- S. C. Semple, A. Akinc, J. Chen, A. P. Sandhu, B. L. Mui, C. K. Cho, D. W. Sah, D. Stebbing, E. J. Crosley and E. Yaworski, Rational design of cationic lipids for siRNA delivery, Nat. Biotechnol., 2010, 28(2), 172–176 CrossRef CAS PubMed.
- Food and Drug Administration, Janssen COVID-19 vaccine emergency use authorization, US Department of Health and Human Services, Food and Drug Administration, Silver Spring, MD, 2021, https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/Janssen-covid-19-vaccine Search PubMed.
- K. A. Hajj, J. R. Melamed, N. Chaudhary, N. G. Lamson, R. L. Ball, S. S. Yerneni and K. A. Whitehead, A potent branched-tail lipid nanoparticle enables multiplexed mRNA delivery and gene editing in vivo, Nano Lett., 2020, 20(7), 5167–5175 CrossRef CAS PubMed; M. Cornebise, E. Narayanan, Y. Xia, E. Acosta, L. Ci, H. Koch, J. Milton, S. Sabnis, T. Salerno and K. E. Benenato, Discovery of a novel amino lipid that improves lipid nanoparticle performance through specific interactions with mRNA, Adv. Funct. Mater., 2022, 32(8), 2106727 CrossRef; L. M. Kranz, M. Diken, H. Haas, S. Kreiter, C. Loquai, K. C. Reuter, M. Meng, D. Fritz, F. Vascotto and H. Hefesha, Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy, Nature, 2016, 534(7607), 396–401 CrossRef PubMed.
- X. Han, H. Zhang, K. Butowska, K. L. Swingle, M.-G. Alameh, D. Weissman and M. J. Mitchell, An ionizable lipid toolbox for RNA delivery, Nat. Commun., 2021, 12(1), 7233 CrossRef CAS PubMed.
- M. A. Maier, M. Jayaraman, S. Matsuda, J. Liu, S. Barros, W. Querbes, Y. K. Tam, S. M. Ansell, V. Kumar and J. Qin, Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics, Mol. Ther., 2013, 21(8), 1570–1578 CrossRef CAS PubMed.
- A. Akinc, A. Zumbuehl, M. Goldberg, E. S. Leshchiner, V. Busini, N. Hossain, S. A. Bacallado, D. N. Nguyen, J. Fuller and R. Alvarez, A combinatorial library of lipid-like materials for delivery of RNAi therapeutics, Nat. Biotechnol., 2008, 26(5), 561–569 CrossRef CAS PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, Lipid nanoparticles – from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement, ACS Nano, 2021, 15(11), 16982–17015 CrossRef CAS PubMed.
- M. Li, Y. Li, S. Li, L. Jia, H. Wang, M. Li, J. Deng, A. Zhu, L. Ma and W. Li, The nano delivery systems and applications of mRNA, Eur. J. Med. Chem., 2022, 227, 113910 CrossRef CAS PubMed.
- M. Maugeri, M. Nawaz, A. Papadimitriou, A. Angerfors, A. Camponeschi, M. Na, M. Hölttä, P. Skantze, S. Johansson and M. Sundqvist, Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells, Nat. Commun., 2019, 10(1), 4333 CrossRef CAS.
- J. A. Kulkarni, M. M. Darjuan, J. E. Mercer, S. Chen, R. Van Der Meel, J. L. Thewalt, Y. Y. C. Tam and P. R. Cullis, On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA, ACS Nano, 2018, 12(5), 4787–4795 CrossRef CAS.
- M. Kim, M. Jeong, S. Hur, Y. Cho, J. Park, H. Jung, Y. Seo, H. Woo, K. Nam and K. Lee, Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver, Sci. Adv., 2021, 7(9), eabf4398 CrossRef CAS PubMed.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. McFadyen and J. Chan, A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates, Mol. Ther., 2018, 26(6), 1509–1519 CrossRef CAS PubMed.
- Y. Dong, K. T. Love, J. R. Dorkin, S. Sirirungruang, Y. Zhang, D. Chen, R. L. Bogorad, H. Yin, Y. Chen and A. J. Vegas, Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(11), 3955–3960 CrossRef CAS PubMed; S. Busatto, S. A. Walker, W. Grayson, A. Pham, M. Tian, N. Nesto, J. Barklund and J. Wolfram, Lipoprotein-based drug delivery, Adv. Drug Delivery Rev., 2020, 159, 377–390 CrossRef; Z. Tai, X. Wang, J. Tian, Y. Gao, L. Zhang, C. Yao, X. Wu, W. Zhang, Q. Zhu and S. Gao, Biodegradable stearylated peptide with internal disulfide bonds for efficient delivery of siRNA in vitro and in vivo, Biomacromolecules, 2015, 16(4), 1119–1130 CrossRef PubMed; Y. Li, Y. Li, X. Wang, R. J. Lee and L. Teng, Fatty acid modified octa-arginine for delivery of siRNA, Int. J. Pharm., 2015, 495(1), 527–535 CrossRef PubMed.
- X. Jing, C. Foged, B. Martin-Bertelsen, A. Yaghmur, K. M. Knapp, M. Malmsten, H. Franzyk and H. M. Nielsen, Delivery of siRNA complexed with palmitoylated α-peptide/β-peptoid cell-penetrating peptidomimetics: membrane interaction and structural characterization of a lipid-based nanocarrier system, Mol. Pharmaceutics, 2016, 13(6), 1739–1749 CrossRef CAS PubMed.
- E. J. Gudiña, V. Rangarajan, R. Sen and L. R. Rodrigues, Potential therapeutic applications of biosurfactants, Trends Pharmacol. Sci., 2013, 34(12), 667–675 CrossRef PubMed.
- J. H. Lee, S. H. Nam, W. T. Seo, H. D. Yun, S. Y. Hong, M. K. Kim and K. M. Cho, The production of surfactin during the fermentation of cheonggukjang by potential probiotic Bacillus subtilis CSY191 and the resultant growth suppression of MCF-7 human breast cancer cells, Food Chem., 2012, 131(4), 1347–1354 CrossRef CAS.
- Y. Han, B. Ding, Z. Zhao, H. Zhang, B. Sun, Y. Zhao, L. Jiang, J. Zhou and Y. Ding, Immune lipoprotein nanostructures inspired relay drug delivery for amplifying antitumor efficiency, Biomaterials, 2018, 185, 205–218 CrossRef CAS PubMed.
- T. Janek, A. Krasowska, A. Radwańska and M. Łukaszewicz, Lipopeptide biosurfactant pseudofactin II induced apoptosis of melanoma A 375 cells by specific interaction with the plasma membrane, PLoS One, 2013, 8(3), e57991 CrossRef CAS PubMed.
- C. Murugan, K. Rayappan, R. Thangam, R. Bhanumathi, K. Shanthi, R. Vivek, R. Thirumurugan, A. Bhattacharyya, S. Sivasubramanian and P. Gunasekaran, Combinatorial nanocarrier based drug delivery approach for amalgamation of anti-tumor agents in breast cancer cells: An improved nanomedicine strategy, Sci. Rep., 2016, 6(1), 34053 CrossRef CAS PubMed; X.-H. Cao, A.-H. Wang, C.-L. Wang, D.-Z. Mao, M.-F. Lu, Y.-Q. Cui and R.-Z. Jiao, Surfactin induces apoptosis in human breast cancer MCF-7 cells through a ROS/JNK-mediated mitochondrial/caspase pathway, Chem.-Biol. Interact., 2010, 183(3), 357–362 CrossRef PubMed; C. Sivapathasekaran, P. Das, S. Mukherjee, J. Saravanakumar, M. Mandal and R. Sen, Marine bacterium derived lipopeptides: characterization and cytotoxic activity against cancer cell lines, Int. J. Pept. Res. Ther., 2010, 16, 215–222 CrossRef.
- F. H. Kural and R. N. Gürsoy, Formulation and Characterization of Surfactin-Containing Self-Microemulsifying Drug Delivery Systems SF-SMEDDS, Hacet. Univ. J. Fac. Pharm., 2011,(2), 171–186 Search PubMed; W. Huang, Y. Lang, A. Hakeem, Y. Lei, L. Gan and X. Yang, Surfactin-based nanoparticles loaded with doxorubicin to overcome multidrug resistance in cancers, Int. J. Nanomed., 2018, 1723–1736 CrossRef CAS.
- H. Lv, S. Zhang, B. Wang, S. Cui and J. Yan, Toxicity of cationic lipids and cationic polymers in gene delivery, J. Controlled Release, 2006, 114(1), 100–109 CrossRef CAS PubMed.
- M. Morikawa, Y. Hirata and T. Imanaka, A study on the structure–function relationship of lipopeptide biosurfactants, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2000, 1488(3), 211–218 CrossRef CAS PubMed.
- J. Kim, A. Jozic and G. Sahay, Naturally derived membrane lipids impact nanoparticle-based messenger RNA delivery, Cell. Mol. Bioeng., 2020, 13, 463–474 CrossRef CAS PubMed.
- R. Aranda IV, S. M. Dineen, R. L. Craig, R. A. Guerrieri and J. M. Robertson, Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 104 concentration range, Anal. Biochem., 2009, 387(1), 122–127 CrossRef PubMed.
- R. El-Mayta, M. S. Padilla, M. M. Billingsley, X. Han and M. J. Mitchell, Testing the In Vitro and In Vivo Efficiency of mRNA-Lipid Nanoparticles Formulated by Microfluidic Mixing, J. Visualized Exp., 2023, 191, e64810 Search PubMed.
- A. H. Baraaj, Histological, biochemical and DNA changes in the liver of male albino rats treated with silver nanoparticles, Nano Biomed. Eng., 2021, 13(1), 20–26 CAS.
- J.-M. Bonmatin, O. Laprévote and F. Peypoux, Diversity among microbial cyclic lipopeptides: iturins and surfactins. Activity-structure relationships to design new bioactive agents, Comb. Chem. High Throughput Screening, 2003, 6(6), 541–556 CrossRef CAS PubMed.
- M. Krishnamurthy, K. Simon, A. M. Orendt and P. A. Beal, Macrocyclic helix-threading peptides for targeting RNA, Angew. Chem., Int. Ed., 2007, 46(37), 7044–7047 CrossRef CAS PubMed; J. Pai, S. Hyun, J. Y. Hyun, S.-H. Park, W.-J. Kim, S.-H. Bae, N.-K. Kim, J. Yu and I. Shin, Screening of pre-miRNA-155 binding peptides for apoptosis inducing activity using peptide microarrays, J. Am. Chem. Soc., 2016, 138(3), 857–867 CrossRef PubMed; B. M. Lunde, C. Moore and G. Varani, RNA-binding proteins: modular design for efficient function, Nat. Rev. Mol. Cell Biol., 2007, 8(6), 479–490 CrossRef PubMed.
- Y. Eygeris, M. Gupta, J. Kim and G. Sahay, Chemistry of lipid nanoparticles for RNA delivery, Acc. Chem. Res., 2021, 55(1), 2–12 CrossRef PubMed.
- R. I. Mahato, Water insoluble and soluble lipids for gene delivery, Adv. Drug Delivery Rev., 2005, 57(5), 699–712 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Understanding biophysicochemical interactions at the nano–bio interface, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS.
- D. Breznan, D. D. Das, C. MacKinnon-Roy, S. Bernatchez, A. Sayari, M. Hill, R. Vincent and P. Kumarathasan, Physicochemical properties can be key determinants of mesoporous silica nanoparticle potency in vitro, ACS Nano, 2018, 12(12), 12062–12079 CrossRef CAS PubMed.
- S. Chen, Y. Y. C. Tam, P. J. Lin, M. M. Sung, Y. K. Tam and P. R. Cullis, Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA, J. Controlled Release, 2016, 235, 236–244 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Lipid nanoparticles for mRNA delivery, Nat. Rev. Mater., 2021, 6(12), 1078–1094 CrossRef CAS PubMed.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith III, S. Hou, A. A. Esposito, T. Ketova and K. Welsher, Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun., 2020, 11(1), 983 CrossRef CAS PubMed.
- F. Cardarelli, L. Digiacomo, C. Marchini, A. Amici, F. Salomone, G. Fiume, A. Rossetta, E. Gratton, D. Pozzi and G. Caracciolo, The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery, Sci. Rep., 2016, 6(1), 25879 CrossRef CAS PubMed.
- R. Ni, R. Feng and Y. Chau, Synthetic approaches for nucleic acid delivery: choosing the right carriers, Life, 2019, 9(3), 59 CrossRef CAS PubMed.
- K. Garber, Alnylam launches era of RNAi drugs, Nat. Biotechnol., 2018, 36(9), 777–779 CrossRef CAS PubMed.
- J. M. Karp and D. Peer, Focus on RNA interference: from nanoformulations to in vivo delivery, Nanotechnology, 2018, 29, 010201 CrossRef PubMed.
- K. A. Whitehead, J. R. Dorkin, A. J. Vegas, P. H. Chang, O. Veiseh, J. Matthews, O. S. Fenton, Y. Zhang, K. T. Olejnik and V. Yesilyurt, Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity, Nat. Commun., 2014, 5(1), 4277 CrossRef CAS PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy, Nano Lett., 2017, 17(3), 1326–1335 CrossRef CAS PubMed.
- M. Jayaraman, S. M. Ansell, B. L. Mui, Y. K. Tam, J. Chen, X. Du, D. Butler, L. Eltepu, S. Matsuda and J. K. Narayanannair, Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo, Angew. Chem., 2012, 124(34), 8657–8661 CrossRef.
- L. Miao, J. Lin, Y. Huang, L. Li, D. Delcassian, Y. Ge, Y. Shi and D. G. Anderson, Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver, Nat. Commun., 2020, 11(1), 2424 CrossRef CAS.
- A. Akinc, W. Querbes, S. De, J. Qin, M. Frank-Kamenetsky, K. N. Jayaprakash, M. Jayaraman, K. G. Rajeev, W. L. Cantley and J. R. Dorkin, Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms, Mol. Ther., 2010, 18(7), 1357–1364 CrossRef CAS PubMed.
- L. Battaglia and M. Gallarate, Lipid nanoparticles: state of the art, new preparation methods and challenges in drug delivery, Expert Opin. Drug Delivery, 2012, 9(5), 497–508 CrossRef CAS PubMed.
- J. G. Tank and R. V. Pandya, Anti-Proliferative activity of surfactins on human cancer cells and their potential use in therapeutics, Peptides, 2022, 170836 CrossRef CAS PubMed; M. Kurpiers, J. D. Wolf, H. Spleis, C. Steinbring, A. M. Jörgensen, B. Matuszczak and A. Bernkop-Schnürch, Lysine-based biodegradable surfactants: increasing the lipophilicity of insulin by hydrophobic ion paring, J. Pharm. Sci., 2021, 110(1), 124–134 CrossRef PubMed.
- S. Crovella, L. C. de Freitas, L. Zupin, F. Fontana, M. Ruscio, E. P. N. Pena, I. O. Pinheiro and T. Calsa Junior, Surfactin bacterial antiviral lipopeptide blocks in vitro replication of SARS-CoV-2, Appl. Microbiol., 2022, 2(3), 680–687 CrossRef.
- Y. Xiong, J. Kong, S. Yi, Q. Tan, E. Bai, N. Ren, Y. Huang, Y. Duan and X. Zhu, Lipopeptide surfactin ameliorates the cell uptake of platensimycin and enhances its therapeutic effect on treatment of MRSA skin infection, J. Antimicrob. Chemother., 2022, 77(10), 2840–2849 CrossRef CAS PubMed.
- M. Yang, H. Jin, J. Chen, L. Ding, K. K. Ng, Q. Lin, J. F. Lovell, Z. Zhang and G. Zheng, Efficient cytosolic delivery of siRNA using HDL-mimicking nanoparticles, Small, 2011, 7(5), 568–573 CrossRef CAS PubMed; K. K. Ng, J. F. Lovell and G. Zheng, Lipoprotein-inspired nanoparticles for cancer theranostics, Acc. Chem. Res., 2011, 44(10), 1105–1113 CrossRef PubMed.
- Y. Guo, W. Yuan, B. Yu, R. Kuai, W. Hu, E. E. Morin, M. T. Garcia-Barrio, J. Zhang, J. J. Moon and A. Schwendeman, Synthetic high-density lipoprotein-mediated targeted delivery of liver X receptors agonist promotes atherosclerosis regression, EBioMedicine, 2018, 28, 225–233 CrossRef PubMed.
- F. Ferraresso, A. W. Strilchuk, L. J. Juang, L. G. Poole, J. P. Luyendyk and C. J. Kastrup, Comparison of DLin-MC3-DMA and ALC-0315 for siRNA delivery to hepatocytes and hepatic stellate cells, Mol. Pharmaceutics, 2022, 19(7), 2175–2182 CrossRef CAS PubMed.
- M. Sedic, J. J. Senn, A. Lynn, M. Laska, M. Smith, S. J. Platz, J. Bolen, S. Hoge, A. Bulychev and E. Jacquinet, Safety evaluation of lipid nanoparticle–formulated modified mRNA in the sprague-dawley rat and cynomolgus monkey, Vet. Pathol., 2018, 55(2), 341–354 CrossRef CAS PubMed.
- J. D. Bancroft and M. Gamble, Theory and Practice of Histological Techniques, Elsevier health sciences, 2008 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00404c |
| This journal is © The Royal Society of Chemistry 2024 |