Open Access Article
Open Access Article1-Naphthylacetic acid appended amino acids-based hydrogels: probing of the supramolecular catalysis of ester hydrolysis reaction†
Ruchika
Bassan
 a,
Biplab
Mondal
b,
Mayank
Varshney
c and
Subhasish
Roy
a,
Biplab
Mondal
b,
Mayank
Varshney
c and
Subhasish
Roy
 *a
*a
aDepartment of Chemistry, Birla Institute of Technology and Science-Pilani, K K Birla Goa Campus, NH 17B, Zuarinagar, Sancoale, Goa 403726, India. E-mail: subhasishr@goa.bits-pilani.ac.in
bSchool of Biological Sciences, Indian Association for the Cultivation of Science, 2A & 2B, Raja S. C. Mullick Road, Jadavpur, Kolkata-700034, West Bengal, India
cSenior Application Scientist, Characterization Division, Anton Paar India Pvt. Ltd., 582, Phase V, Udyog Vihar Industrial Area, Gurgaon 122016, Haryana, India
First published on 8th May 2024
Abstract
A 1-naphthaleneacetic acid-appended phenylalanine-derivative (Nap-F) forms a stable hydrogel with a minimum gelation concentration (MGC) of 0.7% w/v (21 mM) in phosphate buffer of pH 7.4. Interestingly, Nap-F produces two-component [Nap-F + H = Nap-FH, Nap-F + K = Nap-FK and Nap-F + R = Nap-FR], three-component [Nap-F + H + K = Nap-FH–K, Nap-F + H + R = Nap-FH–R and Nap-F + K + R = Nap-FK–R] and four-component [Nap-F + H + K + R = Nap-FH–K–R] hydrogels in water with all three natural basic amino acids (H = histidine, K = lysine and R = arginine) at various combinations below its MGC. Nap-F-hydrogel forms a nice entangled nanofibrillar network structure as evidenced by field emission scanning electron microscopy (FE-SEM). Interestingly, lysine-based co-assembled two- (Nap-FK), three- (Nap-FH–K and Nap-FK–R) and four-component (Nap-FH–K–R) xerogels exhibit helical nanofibrillar morphology, which was confirmed by circular dichroism spectroscopy, FE-SEM and TEM imaging. However, histidine and arginine-based two-component (Nap-FH and Nap-FR) and three-component (Nap-FH–R) co-assembled xerogels exhibiting straight nanofibrillar morphology. In their co-assembled states, these two-, three- and four-component supramolecular hydrogels show promising esterase-like activity below their MGCs. The enhanced catalytic activity of helical fibers compared to obtained straight fibers (other than lysine-based assembled systems) suggests that the helical fibrillar nanostructure is involved in ordering the esterase-like although all supramolecular assemblies are chemically different from one another.
Introduction
The creation of life on Earth is the biggest mystery. Prebiotic chemistry-based research is attracting substantial attention to understand the complex emergent behaviour of interacting biomolecules, which were responsible for the creation of primitive life on Earth. In this regard, amino acids and peptides are considered building blocks for constructing bioinspired, emerging, smart and functional advanced supramolecular biomaterials due to the ease of their synthesis, amino acid mutation and fantastic self-assembly/co-assembly behavior.1–11 Systems chemistry investigates such emerging behaviour of moleculess that store energy in the form of chemical potential to form various supramolecular assemblies to attain life-like functions.12–16 Simple biomolecular building blocks, which were present when life began on Earth, react to form complex molecules (biopolymers). Such abiotic complex constituents of life can be synthesized and understood by simple chemical reactions. Nature has created enzymes to catalyze chemical reactions with extraordinary efficiency and selectivity. These biocatalysts are the result of millions of years of biochemical evolution. Therefore, they have well-defined three-dimensional nanostructures, which are achieved due to non-covalent interactions. To develop artificial enzymes, covalent modifications are engineered into existing proteins by changing the folding and aggregation behaviour through supramolecular interactions.17 However, the designing and synthesis of artificial enzymes by following a bottom-up approach for the construction of supramolecular nanostructures, which comprise self-assembling amino acid derivatives or short peptides associated via non-covalent interactions, is indeed an elegant approach as the amino acids can mutate to form short peptides/amino acid derivatives, and they can be synthesized with high purity and reproducibility.18 It is believed that amino acids are an important class of biomolecules involved in the origin of primitive life through chemical evolution.19 The study of emerging catalytic behaviour of aqueous supramolecular assemblies containing amino acids and their derivatives is a minimalistic approach to understanding the prebiotic relevancy of the origin-of-life concept. Amino acids and peptide-based hydrogels20 comprise a prebiotic-relevant system that is made of supramolecular nanofibrillar network structure21 can drive various natural chemical transformations, including esterase-like activity. The hydrolysis of an ester bond into an acid and alcohol in the presence of water is performed by the esterase enzyme found in plants, animals and microorganisms.22 Esterase is important in pharmaceutical, food and biofuel industries.23 There is extensive research literature available on the esterase-like activity of short peptides and amino acid derivatives. All these reports can be categorized as (i) template-mediated catalytic activities, (ii) catalytic triad mimicking peptide side chains and (iii) metal-assisted amino acids/peptide assembly as a catalytic substrate.(i) Template-mediated catalytic activity: N-benzoyl L-tyrosine ethyl ester and N-tosyl-L-arginine methyl ester have been hydrolyzed using 29-residue peptides that are made by mimicking the active sites of alpha-chymotrypsin and trypsin enzymes.24 Previously, ester hydrolysis reactions have also been carried out using a polypeptide with 42 amino acid residues that formed a helix-loop-helix motif,25 nonapeptide and heptapeptide-based self-assembled nanofibers,26 dipeptide ser–his27 and green fluorescent protein-tagged hexameric histidine.28 Three different 23-residue-long peptides, such as VK2H (two strands of valine, lysine repeats, and functionalized with histidine), VK2G (two strands of valine, lysine repeats, and functionalized with glycine), and VR2H (two strands of valine, arginine repeats, functionalized with histidine) were synthesized by Ulijn and his co-workers. Among these three peptides, only VK2H has been observed to show efficient esterase-like activity.29 Korendovych and his co-workers engineered a 74-residue-long non-esterase protein, calmodulin, into an esterase protein by introducing a histidine mutation at the proper position.30 Escuder and her co-workers demonstrated imidazole-appended hydrogelators for efficient esterase-like activity.31 Das and his co-workers discovered imidazole-conjugated heptapeptides (a general peptide sequence Im–XLVFFALNH2 where, Im = imidazole, X = lysine (K)/arginine (R)/ornithine (O)/glutamic acid (E), V = valine, F = phenylalanine, A = alanine and L = leucine); for ester hydrolysis reaction where imidazole and lysine-containing peptide nanotubes as well as co-assembled Im–KLVFFAL–NH2 and (2-(4-formylphenoxy)-2-oxoethanesulfonate) exhibited efficient catalysis.32,33 Amphiphilic histidine coupled with stearic acid and its nitrophenol ester have been utilized for transient self-assembled cooperative hydrolysis of ester.34LH–DF–DF (L-histidine–D-phenylalanine–D-phenylalanine)-derived hydrogel has been employed previously for the ester hydrolysis reaction.35 Fmoc–GFFYGHY–OH (glycine–phenylalanine–phenylalanine–tyrosine–glycine–histidine–tyrosine)-based hydrogel has been employed previously for the efficient hydrolysis of p-nitrophenyl acetate along with various tertiary esters.36 Covalent cross-linked through di-tyrosine bond in peptide sequence YYHYY has also been investigated for efficient esterase-like activity.37 The co-assembly of Fmoc–FFH–CONH2 and Fmoc–FFR–CONH2 into nanotubes is also efficient for nitrophenyl acetate hydrolysis.38 14-Amino-acid-long peptides with general formula, NH2–X–SGQQKFQFQFEQQ–CONH2 (where X = glycine/histidine/arginine) have been designed and synthesized for nanofiber preparation, followed by ester hydrolysis.39 Riek and his co-workers dedicated tremendous effort to understanding the ester hydrolysis reaction by a set of 94-peptide libraries and their binary mixtures.40 Shape switchable nanostructures derived from Fmoc–CH–OH have also been utilized for ester hydrolysis.41 Recently, Matson and his co-workers demonstrated S-aroylthiooxime-appended glutamic acid, histidine and two lysine-based isomeric tetrapeptides for the construction of various nanostructures and their ester hydrolysis efficiencies.42 Reches and her co-workers have also recently developed a catalyst based on a non-proteinogenic amino acid L-3,4-dihydroxyphenylalanine (DOPA), for ester hydrolysis reaction.43
(ii) Catalytic triad mimics the peptide side chains: Woolfson and his coworkers developed coiled-coil heptamer-based peptides with 21 mutations to polar amino acids within the barrel structure of preinstalled triads (cysteine–histidine–glutamic acid) on each of helices for the hydrolysis of p-nitrophenyl acetate.44 Guler and his co-workers extensively studied the effect of serine, histidine and aspartic acid in lauryl-appended pentapeptide sequences and effect of these nanostructures on the ester hydrolysis reaction.45
(iii) Metal-assisted amino acid/peptide assembly as catalytic substrate: Ac–IHVHLQI–CONH2–Zn2+–metal–peptide framework for ester hydrolysis reaction has been established previously.46 Korendovych and his co-workers also identified that the heptapeptide, Ac–IHIHIQI–CONH2, can self-assemble in the presence of Zn(II) to form a catalytic amyloid that catalyzes the ester hydrolysis reaction.47 Serpell and his co-workers have discovered amyloid fibrils containing 7 and 11-residue Zn(II)-binding peptides for efficient ester hydrolysis.48 Banerjee and co-workers discovered myristic acid-, ethylene diamine- and histidine-appended amide-based metallo-hydrogel for esterase-like activity.49 Gazit and co-workers successfully invented and studied a phenylalanine-based-Zn(II) complex that forms an amyloid-like structure for ester hydrolysis.50
However, to the best of our knowledge, there is no report available in the literature that shows the tuning of esterase-like activity by 1-naphthylacetic acid-appended-L-phenylalanine–OH (Nap-F) via the formation of template-assisted co-assembled nanostructures with various basic amino acids in water. Interestingly, helical chirality is inherent in nature and chiral environment-assisted selective catalysis is also well known. This motivates us to study the catalytic behaviour of naturally occurring chiral amino acids because it is believed that amino acids are one of the most important classes of biomolecules involved in the origin of primitive life through chemical evolution. An amino acid-based derivative (i.e., Nap-F) was used here as a co-assembly template with all three basic amino acids (histidine, lysine and arginine) in various combinations for the construction of supramolecular nanostructures. Their ester hydrolysis efficiencies were then investigated. Studies of chemically different catalytic supramolecular systems have revealed that helical nanofibrillar network structures have higher catalytic efficiencies compared to straight fibrillar morphologies. In this report, we have discovered the hydrogelation behaviour of 1-naphthaleneacetic acid-appended phenylalanine derivative (Nap-F) in a phosphate buffer of pH 7.4. Interestingly, Nap-F produces two-component (Nap-FH, Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R) and four-component (Nap-FH–K–R) hydrogels below its own MGC in water using all three basic natural amino acids including histidine, lysine and arginine in various combinations. These supramolecular two-, three-, and four-component hydrogels show promising esterase-like activity below their MGCs, as well as below the MGC of the native hydrogel. It has been observed that lysine-based co-assembled systems that are endowed with helical nanofibrillar morphologies can use their helical chiral nanofibers to create a catalytic hotspot for catalyzing the ester hydrolysis reaction. This suggests that the helical chiral nanostructure governs enhanced catalytic activity. It is important to mention that not only the side chains of the amino acids but the assembly template and nanostructure also play crucial roles in achieving the catalytic activity.
Experimental section
The details of synthetic protocols, purifications and chemical characterizations using NMR spectroscopic and mass spectrometric analyses are given in the ESI (Fig. S1–S16†). Detailed experimental methods, protocols and instrumentation are discussed in the ESI.† The hydrogelation behaviour of LNap-F and DNap-F and their co-assembled hydrogels are found same.Results and discussion
It is important to understand the enzymatic activity from a basic building block of life. It is believed that during the formation of the primitive earth, the simplest abiotic biomolecules, such as amino acids, nucleobases, lipids and sugars underwent various chemical reactions to form peptides, proteins, DNA, and RNA followed by the conversion to biotic species through evolution. Indeed, it is significant to understand how various complex molecules evolved from the pool of simple molecules, which is why catalysis by simple building blocks in complex mixtures is relevant for systems chemistry. This motivates us to study the hydrogelation behaviour of 1-naphthaleneacetic acid-conjugated phenylalanine (Nap-F) and hydrogelation behavior of its two-, three- and four-component systems. These systems have designed and developed judiciously by selecting the basic amino acids and Nap-F at various combinations as phenylalanine4,20 is well known for its assembly propensity and basic amino acids are well known for their esterase-like activity in various peptides and proteins.32,33,38,39 No catalytic activity has been identified from the native hydrogel (Nap-F) nor its diluted self-assembled solution. However, the negligible effect of phosphate anion in the phosphate buffer used for the preparation of the Nap-F-hydrogel has been noted.Serendipitously, 1-naphthaleneacetic acid-conjugated phenylalanine (Nap-F) forms two- (Nap-FH, Nap-FK and Nap-FR), three- (Nap-FH–K, Nap-FH–R and Nap-FK–R), and four-component (Nap-FH–K–R) hydrogels below its MGC in pure Milli-Q water. This creates the possibility of studying the catalytic behaviour of co-assembled hydrogels below their MGCs in water as no catalytic activity is observed in their hydrogel states. This can be due to the presence of higher-ordered aggregates that restrict cationic amine groups from taking part in stabilizing the origin of anionic intermediate from the ester hydrolysis reaction. All these hydrogels were thoroughly characterized using Tgel estimation, UV-Vis, FT-IR, and CD spectroscopy, XRD, FE-SEM, TEM, confocal and rheological analyses. The catalytic activity during p-nitrophenyl acetate (PNPA) hydrolysis in water to produce p-nitrophenol (PNP) was investigated using a UV-Vis spectrophotometer. The absorbance of the newly developed peak at 400 nm due to the formation of p-nitrophenolate/p-nitrophenol was recorded under different pHs applied to the systems. The basic amino acid, lysine, bestowed the helical nanofibers to the hydrogels in their co-assembled states; in a helical pitch, lysine and arginine synergistically were protonated by protons abstracted from water molecules to produce hydroxyl ions, which acted as nucleophiles to enhance enzymatic activity. The supramolecular helical co-assembled nanostructures with positively charged amino acids also stabilized the anionic intermediate, which is produced during the ester hydrolysis reaction.
Hydrogelation study
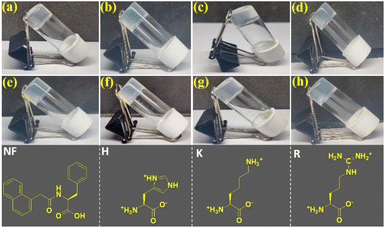
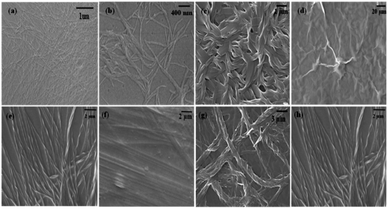
First, 8 mg of 1-naphthaleneacetic acid-appended phenylalanine derivative (Nap-F) was transferred in a glass vial and dissolved in 1 mL of 50 mM phosphate buffer (pH 7.4) with sonication, followed by heating on a hot plate to obtain a clear solution. This hot solution was kept undisturbed at room atmosphere and cooled to room temperature. Hydrogel formation was confirmed by the inverted vial test method (Fig. 1 and Table 1). The minimum gelation concentration (MGC) of the hydrogel, Nap-F, was estimated to be 0.7% w/v. Two-, three-, and four-component hydrogels (Fig. 1b–h) have been prepared using similar protocols.| Hydrogelators | Weight (in mg) ratios for hydrogelation | T gel (°C) of 0.8% w/v hydrogels | MGC (% w/v) | MGC with respect to 10−3 M Nap-F |
|---|---|---|---|---|
| Nap-F | Native gelator | 79 | 0.7 | 24 |
| Nap-FH |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H = 5 H = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
52 ± 0.6 | 0.57 | 15 |
| Nap-FH |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H = 6 H = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
77 ± 1.2 | 0.22 | 18 |
| Nap-FK |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 6 K = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
76 | 0.21 | 18 |
| Nap-FK |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 7 K = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
75 ± 0.6 | 0.258 | 24 |
| Nap-FR |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
73 | 0.257 | 18 |
| Nap-FR |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 7 R = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
74 ± 0.6 | 0.281 | 24 |
| Nap-FH–K |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 6 K = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
73 ± 0.6 | 0.42 | 18 |
| Nap-FH–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
72 | 0.56 | 18 |
| Nap-FK–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
66 ± 1.2 | 0.25 | 18 |
| Nap-FH–K–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 5 R = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
75 ± 0.6 | 0.67 | 15 |
Gel melting temperatures of the hydrogels
To understand the thermal behavior of these hydrogels, each hydrogel at 0.8% w/v concentration were utilized for estimating their gels melting temperatures. The gel melting temperatures are shown in Table 1. The Nap-F hydrogel shows the highest thermal stability compared to all the other hydrogels at 0.8% w/v concentration. However, Nap-FH shows the least thermal stability at a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio. Moreover, at a 6
3 ratio. Moreover, at a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, Nap-FH and Nap-FK show thermal stability that is comparable to that of the Nap-F hydrogel. Gel melting temperatures reduced abruptly when the concentration of the basic amino acids increased, and the concentration of Nap-F reduced, while keeping a fixed (0.8% w/v) hydrogel concentration. The Tgel profile of the native Nap-F hydrogel at various concentrations has been shown in Fig. 2. At three different concentrations with the ratio 6
2 ratio, Nap-FH and Nap-FK show thermal stability that is comparable to that of the Nap-F hydrogel. Gel melting temperatures reduced abruptly when the concentration of the basic amino acids increased, and the concentration of Nap-F reduced, while keeping a fixed (0.8% w/v) hydrogel concentration. The Tgel profile of the native Nap-F hydrogel at various concentrations has been shown in Fig. 2. At three different concentrations with the ratio 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Tgels were measured for Nap-FH–K and Nap-FH–R and are shown in Fig. S17.† The concentration-dependent gel melting profiles are shown in Fig. 2 and S17,† where they suggest an increase in melting points of the hydrogels with an increase in concentrations of the hydrogelators.
1, Tgels were measured for Nap-FH–K and Nap-FH–R and are shown in Fig. S17.† The concentration-dependent gel melting profiles are shown in Fig. 2 and S17,† where they suggest an increase in melting points of the hydrogels with an increase in concentrations of the hydrogelators.
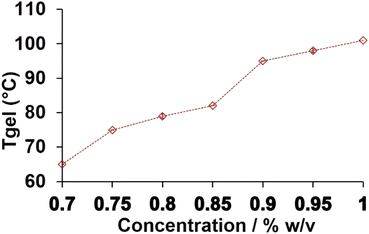 | ||
| Fig. 2 T gel profile of the native Nap-F hydrogel. Error bars are shown inside the symbol denote standard deviation. | ||
UV-Vis spectroscopic study of the hydrogels
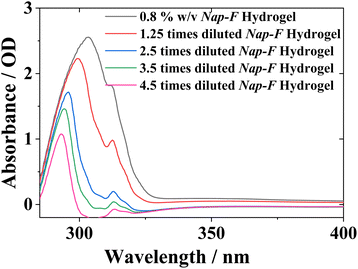
UV-Vis spectroscopic study has been carried out to understand the aggregation behaviour of the hydrogels in their gel states, as well as in their solution states. Hydrogels of 0.8% w/v (see the details of hydrogel preparation procedures) were used for the UV-Vis spectroscopic study. The absorbances were recorded from 250 to 450 nm (Fig. S18†), and the effect of dilution of each of these mono (Fig. 3), two-, three- and four-component hydrogels was investigated. The λmax for Nap-F hydrogel was 303 nm, but it blue shifted to 293 nm (Fig. 3 and S18†) upon dilution. Other than this λmax peak at 303 nm, another shoulder peak was observed at 312 nm. Similar observations were noted for all of these two-component (Nap-FH, Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R) and four-component (Nap-FH–K–R) hydrogels with similar blue shifts upon dilution (Fig. S18†). The blue shifts upon dilution were observed for all the hydrogels due to less scattering from their lower-order aggregates compared to the hydrogels.4 The λmax peak at 303 nm for the native hydrogel Nap-F was red shifted to 315 nm/317 nm for the histidine-based two-component (Nap-FH), three-component (Nap-FH–K and Nap-FH–R) and four-component (Nap-FH–K–R) hydrogels. This could be due to the presence of π–π stacking interactions between the histidine aromatic imidazole side chain unit and phenyl aromatic moiety of the native gelator (Nap-F).Structural studies
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of carboxylic acid from 1712 cm−1 to 1732 cm−1. No carboxylic acid peak, corresponding to the C
O stretching vibration of carboxylic acid from 1712 cm−1 to 1732 cm−1. No carboxylic acid peak, corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, was observed for any of the xerogels; however, a small hump ranging from 1568 cm−1 to 1580 cm−1 was observed for all the xerogels, which corresponded to the presence of deprotonated asymmetric carboxylate groups. The symmetric deprotonated carboxylate group vibration was observed at 1415 cm−1 for all the xerogels, as well as in the bulk state of the native gelator, Nap-F. This suggests that in the bulk state, carboxylic acid and carboxylate ions were present; however, in xerogel states, carboxylate anion formation was observed exclusively. Stretching vibrations ranging from 1537 cm−1 to 1538 cm−1 were observed for the formation of the helical chiral nanofibers in lysine-based two-, three- and four-component assembled systems.56 Peaks at 1635 cm−1 to 1657 cm−1 suggest the
O stretching vibration, was observed for any of the xerogels; however, a small hump ranging from 1568 cm−1 to 1580 cm−1 was observed for all the xerogels, which corresponded to the presence of deprotonated asymmetric carboxylate groups. The symmetric deprotonated carboxylate group vibration was observed at 1415 cm−1 for all the xerogels, as well as in the bulk state of the native gelator, Nap-F. This suggests that in the bulk state, carboxylic acid and carboxylate ions were present; however, in xerogel states, carboxylate anion formation was observed exclusively. Stretching vibrations ranging from 1537 cm−1 to 1538 cm−1 were observed for the formation of the helical chiral nanofibers in lysine-based two-, three- and four-component assembled systems.56 Peaks at 1635 cm−1 to 1657 cm−1 suggest the ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in beta-sheet arrangements, and it could also be due to protonated lysine and arginine amine side chains. N–H bending vibrations peaks from 1530 to 1540 cm−1 region were found absent for the histidine-containing assembled states. Peaks at 3283 cm−1 to 3314 cm−1 were observed, representing N–H stretching vibrations, for all the xerogels.57 Peaks at 1442 cm−1 to 1464 cm−1 were identified as C–N stretching vibrations of the amide bond. Peaks at 3127 cm−1 to 3160 cm−1 and 2623 cm−1 to 2633 cm−1 were identified as the stretching vibration of NH3+ for all the two-, three-, and four-component xerogels.58 FT-IR spectroscopic analyses suggest that due to the presence of anionic carboxylate ions and cationic basic amino acids side chains, molecular packing occurs through electrostatic interactions for the formation of the supramolecular nanofibrillar network structures. FTIR analysis with D2O solvent under ATR mode also shows the
O stretching in beta-sheet arrangements, and it could also be due to protonated lysine and arginine amine side chains. N–H bending vibrations peaks from 1530 to 1540 cm−1 region were found absent for the histidine-containing assembled states. Peaks at 3283 cm−1 to 3314 cm−1 were observed, representing N–H stretching vibrations, for all the xerogels.57 Peaks at 1442 cm−1 to 1464 cm−1 were identified as C–N stretching vibrations of the amide bond. Peaks at 3127 cm−1 to 3160 cm−1 and 2623 cm−1 to 2633 cm−1 were identified as the stretching vibration of NH3+ for all the two-, three-, and four-component xerogels.58 FT-IR spectroscopic analyses suggest that due to the presence of anionic carboxylate ions and cationic basic amino acids side chains, molecular packing occurs through electrostatic interactions for the formation of the supramolecular nanofibrillar network structures. FTIR analysis with D2O solvent under ATR mode also shows the ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration in the beta-sheet arrangements through peaks at 1639 cm−1 for Nap-F, 1649 for cm−1Nap-FH, 1646 for cm−1Nap-FK and 1646 for cm−1Nap-FR in their gel states (Fig. S20†).
O stretching vibration in the beta-sheet arrangements through peaks at 1639 cm−1 for Nap-F, 1649 for cm−1Nap-FH, 1646 for cm−1Nap-FK and 1646 for cm−1Nap-FR in their gel states (Fig. S20†).
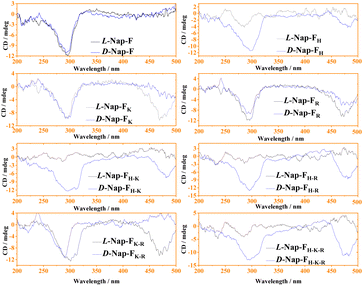
However, two opposite peak patterns (positive CD signals for all D-amino acids and negative signals for all L-amino acids for their mono, two-, three- and four-component systems) were observed at 472 nm (Fig. S21† and 5). A histidine-containing co-assembled two-component system (Nap-FH) shows a weak signal near 472 nm (Fig. 5). However, three and four-component co-assembled systems of histidine (Nap-FH–K, Nap-FH–R, Nap-FH–K–R) show intense CD signals (opposite handedness when L-amino acids are replaced by D-amino acids) at 472 nm, suggesting that lysine, and to some extent, arginine play a crucial role in creating helical chiral nanofibers.59,60 Opposite CD bands have also been observed for the two-component (Nap-FK and Nap-FR) and three-component (Nap-FK–R) hydrogels at 472 nm upon replacement of all used L-amino acids with D-amino acids. This suggests that the helical structure of the nanofibrillar network structures has opposite handedness. This observation is completely supported by our morphological characterizations with FE-SEM imaging, where we observed right-handed helical fibers in all lysine-containing co-assembled systems; all amino acids were used in L-configuration, and the majority of them were left-handed in the co-assembled systems, only when all used amino acids were in D-configuration. Fig. S21† shows similar trends from the CD spectroscopic analyses of all hydrogels in their gel states at 0.8% w/v concentrations and their half-diluted solution states.
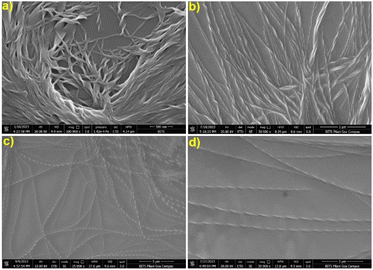
However, below their minimum gelation concentrations at different ratios, two-, three- and four-component supramolecular nanofibrillar assemblies had excellent catalytic efficiency, except Nap-F (Nap-F assemblies were prepared in phosphate buffer, where phosphate ions cause negligible catalysis) and the two-component Nap-FH assemblies. The FE-SEM imaging of all these two-, three-, and four-component supramolecular assemblies was performed using their catalytic solution states, which were drop cast after 50 times dilution (Fig. S23†). Interestingly, all two-, three-, and four-component systems showed nice nanofibrillar network structure in their solution states. Interestingly, the lysine-containing nanofibers are helical in nature. The fibrillar diameters were estimated to be 28.5 ± 0.01 nm, 23.87 ± 0.07 nm, 32.9 ± 0.04 nm, 36.76 ± 0.02 nm and 56.78 ± 0.04 nm for the supramolecular assemblies of Nap-FK, Nap-FH–K, Nap-FH–R, Nap-FK–R and Nap-FH–K–R, respectively. It is interesting to note that the helical fibers were identified using FE-SEM imaging for the two-, three-, and four-component systems with lysine as a common basic amino acid, and helical pitches were estimated to be 157.7 ± 52.6 nm, 178.3 ± 0.1 nm, 249.2 ± 0.1 nm and 258.4 ± 0.2 nm for Nap-FK, Nap-FH–K, Nap-FK–R and Nap-FH–K–R supramolecular assemblies, respectively (Fig. 7). In addition, all the helical fibers were right-handed (Fig. 7 and Table S2†), when all L-amino acids [Nap-F is 1 naphthaleneacetic acid-conjugated L-phenylalanine; H = L-histidine, K = L-lysine and R = L-arginine in two-component (Nap-FK), three-component (Nap-FH–K and Nap-FK–R) and four-component (Nap-FH–K–R) systems] were used for co-assembly. Most of the helical nanofibers are left-handed, while 10 to 20% were right-handed, when all D-amino acids [F as 1-naphthaleneacetic acid-conjugated D-phenylalanine; H = D-histidine, K = D-lysine and R = D-arginine for the two-component (Nap-FK) and three-component (Nap-FK–R)] (Fig. S24 and Table S2†) were used for co-assembly. During the nucleation steps of the supramolecular hydrogel formation, the two-, three-, and four-component hydrogels produced various straight fibrils followed by helical strands, and all strands assembled to construct helical nanofibers network structures; these changes likely happened through electrostatic, π–π stacking, and hydrogen bonding interactions (Fig. S23†). Helical nanofiber formation was also confirmed by a dry-state transmission electron microscopic study (Fig. S25†).
Enzymatic activity
The ester hydrolysis reaction of p-nitrophenyl acetate to p-nitrophenol was investigated on assembled native (Nap-F), two-component (Nap-FH, Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R) and four-component (Nap-FH–K–R) systems. It is interesting to note that none of the hydrogels showed the formation of ester hydrolysis products in their gel states using a UV-Vis spectroscopic study, when p-nitrophenyl acetate in acetonitrile-water solution was added to the hydrogels. Mouldable slices of the hydrogels emerged in a solution of p-nitrophenyl acetate in acetonitrile-water, and no change in colour from colourless to yellow was observed visually or in UV-Vis spectroscopy. The solution of native self-assembled hydrogel (Nap-F) showed a negligible ester hydrolysis response in the UV-Vis spectroscopic study, which could be due to the presence of phosphate ions in the medium. To nullify the effect of the phosphate ions in the hydrolysis, we discovered the two-, three-, and four-component co-assembled systems in simple Milli-Q water at 0.5% w/v concentration (Table 2). Interestingly, the two-component (Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R) and four-component (Nap-FH–K–R) co-assembled systems showed esterase-like activity. They were examined by considering different concentrations of ester as the substrate. Surprisingly we did not observe any catalytic conversion for the Nap-FH system. This suggests the presence of strong supramolecular π–π stacking interactions which do not allow the imidazolium part of the histidine side chain to participate in catalysis. Alternatively, due to the resonance stability of the imidazolium ion, the catalytic anionic intermediate is not stabilized by positive imidazolium ions from histidine. The progress of ester hydrolysis was monitored by UV-Vis spectroscopy until the absorbance reached its saturation. The molar extinction coefficient (ε) of p-nitrophenol (PNP) was estimated from the slope of the linear profile between absorption maxima at 400 nm versus concentrations of p-nitrophenol (Fig. S27†) in water.| Catalytic systems | Hydrogel formation ratios in weight (mg) at 0.8% w/v | Weight (in mg) ratios for catalysis at 0.5% w/v co-assembled systems where no hydrogel formation was observed |
|---|---|---|
| Nap-FH |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H = 5 H = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and Nap-F 3 and Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H = 6 H = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H = 2.5 H = 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (7.5 mM of Nap-F) 2.5 (7.5 mM of Nap-F) |
| Nap-FK |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 6 K = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and Nap-F 2 and Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 7 K = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 2.5 K = 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (7.5 mM of Nap-F) 2.5 (7.5 mM of Nap-F) |
| Nap-FR |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and Nap-F 2 and Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 7 R = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 2.5 R = 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (7.5 mM of Nap-F) 2.5 (7.5 mM of Nap-F) |
| Nap-FH–K |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 6 K = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 3 K = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (9 mM of Nap-F) 1 (9 mM of Nap-F) |
| Nap-FH–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 3 R = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (9 mM of Nap-F) 1 (9 mM of Nap-F) |
| Nap-FK–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 6 R = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 3 R = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (9 mM of Nap-F) 1 (9 mM of Nap-F) |
| Nap-FH–K–R |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 5 R = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Nap-F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H H![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) R = 2 R = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (6 mM of Nap-F) 1 (6 mM of Nap-F) |
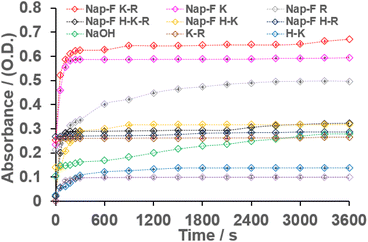
The value of ε was estimated to be 6161 M−1 L−1. The hydrolysis reactions were carried out at 0.5% w/v concentrations for all the two-component (Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R) and four-component (Nap-FH–K–R) co-assembled systems in 1 mL of Milli-Q water (Table 2). Further, 3 μL of 3 mM p-nitrophenyl acetate were added individually to all the assembled systems, and the progress of the hydrolysis reaction was monitored by UV-Vis spectroscopy (Fig. S28†). The absorbance versus time profiles for all the two-component (Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R), and four-component (Nap-FH–K–R) co-assembled systems are shown in Fig. 9 to represent the progress of the hydrolysis reaction. Catalytic studies were performed by taking different concentrations of p-nitrophenyl acetate (substrate), and absorbance values of the hydrolysed product (p-nitrophenol) were recorded at fixed concentrations of co-assembled catalysts.
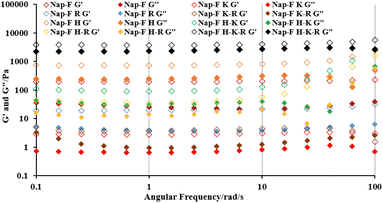
The Michaelis–Menten kinetic parameters (KM) of the ester hydrolysis reactions were calculated using the double reciprocal plots of the initial rate of reaction versus different concentrations of the substrate (Fig. S29†). The hydrolytic rate constant (Kcat) and catalytic efficiency (Kcat/KM) of all the assembled catalytic systems are shown in Table S3.† The highest catalytic efficiency of the lysine containing two-, three- and four-component co-assembled systems with helical nanofibrillar morphologies was determined.
In these co-assembled systems, if the overall concentration (at% w/v) was kept fixed, an increase in the amount of Nap-F increased the catalytic efficiency. Moreover, normal basic amino acid mixtures were employed for catalytic activity at the same concentration (at% w/v) and found that the catalytic activity of these mixed amino acid systems showed one order less catalytic activity compared to the co-assembled lysine-based supramolecular systems. Interestingly, three different crucial factors were identified for enhanced catalytic activity in these amino acid-based co-assembled systems, including the (i) template effect from the native hydrogel system or supramolecular building blocks, (ii) amino acids containing a free amine side chain (lysine in this case, and for histidine and arginine, the protonated side chain is resonance stabilized) and (iii) helical chiral nanostructure environment (although all the studied catalytic systems were chemically different systems). It was also found at room temperature that all the co-assembled systems had much better activity compared to the NaOH solution. The % conversion for the most efficient catalytic system Nap-FK–R was more than 86%. Furthermore, 80 and 75% conversion efficiency were recorded for Nap-FR and Nap-FK co-assembled systems as compared to the conversion by the same concentration of the NaOH solution (70%). The colour of the solution after catalysis under visible light is shown in Fig. 10. An intense yellow colour was observed for lysine-containing assembled systems. The morphological features remained unchanged after catalysis, suggesting the stability of the co-assembled systems under the catalytic medium (Fig. S30†). Interestingly, amino acid derivatives and amino acids can be reused after HPLC purifications, followed by co-assembly for catalysis. A typical ESI-MS analysis of the catalytic solution mixture after catalysis had been performed, which showed masses corresponding to p-nitrophenol and acetic acid. The mass spectrum shows the mass corresponding to the p-nitrophenolate (Fig. S31†) ion, as well as acetic acid with one potassium ion, suggesting the successful formation of the ester hydrolysis product. The pH of the assembled catalytic solutions of the native, two-, three-, and four-component systems before and after catalysis were measured and are shown in Table S4.† LC-MS analyses were carried out under positive mode (see ESI† for LC traces and corresponding mass spectra of all the catalytic systems, Fig. S32–S42 and Table S5†) at lower voltages, including 50 and 70 V, so that the fragmentation of molecular ion peaks could be prevented. The possible ionic species and their corresponding masses are shown in Table S5.† The hydrolysis of PNPA could have happened due to the involvement of the hydroxyl ion as nucleophile40,42,43,62 because an alkoxide ion intermediate was identified in the mass spectrometric analysis (see Table S5 and Fig. S32–S42†). However, the involvement of direct acylation by the amine functional groups of the basic amino acid, arginine cannot be ruled out completely (see Table S5 and Fig. S35†). No acylated fragments or molecular ion peaks were solely identified when the LC-MS study was conducted at lower voltages, including 50 and 70 V, for individual histidine or lysine and neither for most of the catalytic co-assembled systems. The mass spectrometric peak position at m/z 334 corresponded to diacetyl basic amino acids, along with water molecules, in combination with the mass of sodium/potassium/hydrogen. It also corresponds to ionic intermediate species formed when hydroxyl ions act as nucleophiles (see, Table S5†) in the ester hydrolysis reaction. However, it cannot be concluded that this peak at m/z 334 solely comes from the acetylation of arginine and its corresponding co-assembled systems or from the formation of alkoxide ion intermediate due to the involvement of hydroxyl nucleophile in the hydrolysis as the molar mass of Nap-F is 333 g mol−1. The peak at m/z 334 corresponds to [MNap-F + H]+ as confirmed by its isotopic mass distribution in all cases (Fig. S36–S42†). The peak at m/z 356 (Fig. S36–S42†) corresponded to [MNap-F + Na]+. In summary, mechanisms involving hydroxyl ions as nucleophiles, followed by hydrolysis, as well as the acylation of the amine functional groups of the basic amino acid, arginine, followed by hydrolysis are operated during the hydrolysis of PNPA to PNP for the catalytic systems studied for this report. It is also important to note that 1-naphthaleneacetic acid-conjugated phenylalanine (Nap-F) was involved in ordering the catalytic efficiency. In the catalytic experiment, by keeping the amount of two amino acids (lysine and arginine) fixed, the amount of Nap-F was increased, and it enhanced catalytic efficiency (absorbance intensity increased after catalysis corresponding to the peak at 400 nm, Fig. S43 and S44†). This suggests that 1-naphthaleneacetic acid-conjugated phenylalanine (Nap-F) also played a crucial role in ordering the catalytic activity. It can be concluded that Nap-F had a templating effect in making the helical nanofibers with the lysine containing two, three and four-component systems; under these chiral helical environments, catalysis took place efficiently. Moreover, it is interesting to note that there is no difference in the catalytic activity when all the amino acids are used either in L/D configurations.
Effect of pH on the catalytic activity
Indeed, the pH of the medium plays a significant role in the ester hydrolysis of PNPA to PNP as the absorption maxima of the hydrolysed product PNP varies with pH.63 The pH of all catalytic solutions was adjusted to pH 9 using either 1 N NaOH solution or 1 N HCl solution, followed by volume adjustment by Milli-Q water to keep the concentrations of the catalysts fixed (Table S4†). The absorption maxima of PNP at 400 nm were recorded for all the catalytic systems. The calibration plot at pH 9 is shown in Fig. S45.† UV-Vis catalytic responses (Fig. S46†), Lineweaver–Burk plot (Fig. S47†), time-dependent absorption profiles (Fig. S48†) and catalytic comparative table (Table S6†) for hydrolysis are given in the ESI.† Interestingly, at pH 9, the catalytic efficiency (Kcat/KM) trends remained unchanged with respect to the catalytic activity described in water only. As the pH increased for all the catalytic systems to pH 9, Nap-FH also showed notable catalytic activity due to the increase in hydroxyl ions concentration and the imidazole moiety of histidine. It is also interesting to note that at pH 9, lysine-containing catalytic systems showed the formation of the helical nanofibres (Fig. S49†), suggesting a significant contribution of the helical chiral nanofibres in catalysis.Zeta potential study and plausible mechanism for esterase-like activity
The zeta potential of all these catalytic co-assembled solutions was measured. It has been observed that all catalytic samples exhibited average negative zeta potentials (ranging from ≈−20 to −55), suggesting stable assembled solutions. The native hydrogel solution (Nap-F) exists as carboxylate in the phosphate buffer, and thus, it is negative. However, two-component (Nap-FH, Nap-FK and Nap-FR), three-component (Nap-FH–K, Nap-FH–R and Nap-FK–R), and four-component (Nap-FH–K–R) catalytic co-assembled solutions were in Milli Q water, and due to the presence of the basic amino acids, protons were abstracted from water. Thus, in solutions, more hydroxide ions exist that are responsible for the ester hydrolysis reactions. As the basic amino acids abstract protons from water, and the side chain exists in cationic form, cations are stable and delocalized for histidine and arginine side chains due to resonance. However, for lysine, the cationic charge is localized and more exposed for electrostatic interactions with the anionic carboxylate of the 1-naphthaleneacetic acid-appended phenylalanine derivative (Nap-F). The charge also stabilizes the anionic intermediate during catalysis. It has been observed that the helical nanofibrillar morphology participates in catalysis, and for all the lysine-containing systems, the helical nanofibers were discovered serendipitously. This suggests that lysine aliphatic side chains are more flexible in their co-assembled states, compared to histidine and arginine-based co-assembled systems, and thus, in the helical pitch, the lysine side chains are more exposed to the catalytic hotspot, which in turn, exhibits enhanced esterase-like activity.Conclusion
The synthesised amino acid-based two-, three-, and four-component co-assembled systems were utilized for esterase-like activity in water at room atmosphere. This study shows the effect of basic amino acids in building various co-assembled hydrogels that consist of nanofibrillar network structures. On careful observation, it has been inferred that lysine-containing co-assembled supramolecular systems bestow a nanofibrillar network-like structure with helical chirality. Co-assembled systems show right-handed helical fibrillar network structure; however, a majority of left-handed helical fibrillar network structures, along with some right-handedness were observed, when all L-amino acids were replaced by D-amino acids for co-assembly. We have identified three major factors that could be responsible for efficient ester hydrolysis reaction, including the templating effect from native Nap-F hydrogelator for the construction of co-assembled supramolecular nanostructures, helical chirality of the nanofibers, and free basic side chain of lysine. The current study offers a bioinspired, cost-effective and minimalistic approach for efficient amino acid-based supramolecular catalysis in water. These results motivate us to understand the chemistry of the effect of various biomolecules-derived helical nanostructures-based systems in detail for bioinspired functional group transformation reactions under physiological environments. The effect of various nanostructures for the same catalytic chemical system can also be explored in the future.Conflicts of interest
The authors declare no competing financial interest.Author contributions
All authors approved the final version of the manuscript. S. R. is responsible for conceptualization, making graphs and figures from raw data, data interpretation, writing of the manuscript, review, preparation of revised version, editing and supervision. R. B. was responsible for syntheses, purifications, hydrogelations, methodology, investigations, characterizations, catalysis studies and calculations, and writing the early stage draft and formatting. B. M. was responsible for performing the circular dichroism, FTIR-ATR in D2O and estimation of critical aggregation concentrations by concentration-dependent fluorescence experiments. M. V. was responsible for rheological measurements.Acknowledgements
DST-Inspire Faculty Award Research Grant, DST, New Delhi, India (Project no. DST/INSPIRE/04/2017/001084, IFA 17-CH269), is gratefully acknowledged for the financial support. S. R. sincerely thanks the DST-Inspire Faculty Award Research Grant. Research Initiation Grant (BPGC/RIG/2029-20/05-19/04) and Additional Competitive Grant (GOA/ACG/2019-20/Oct/04) funded by BITS-Pilani, K K Birla Goa Campus is highly acknowledged. S. R. sincerely thanks BITS- Pilani, K K Birla Goa Campus for these Research Grants. R. B. is sincerely thanks BITS-Pilani, K K Birla Goa Campus for the institute's Ph.D. research fellowship. We acknowledge the Central Sophisticated Instrumentation Facility (CSIF) and Department of Chemistry of BITS-Pilani, K K Birla Goa Campus for the instrumental facilities. We acknowledge Dr Durga Prasad Muvva and Ms Jyothi Yarram for FE-SEM imaging (CSIF, BITS Goa), Mr Kumar Kanneboina for LC-MS (CSIF, BITS Goa) and Mr Lakshya Raj Khatri for NMR study (CSIF, BITS Goa). We acknowledge all technical experts involved in this work for their help and support.References
- D. Ivnitski, M. Amit, O. Silberbush, Y. Atsmon-Raz, J. Nanda, R. Cohen-Luria, Y. Miller, G. Ashkenasy and N. Ashkenasy, Angew. Chem., Int. Ed., 2016, 55, 9988–9992 CrossRef CAS PubMed.
- S. Bera, B. Xue, P. Rehak, G. Jacoby, W. Ji, L. J. W. Shimon, R. Beck, P. Král, Y. Cao and E. Gazit, ACS Nano, 2020, 14, 1694–1706 CrossRef CAS PubMed.
- D. Ivnitski, M. Amit, B. Rubinov, R. Cohen-Luria, N. Ashkenasy and G. Ashkenasy, Chem. Commun., 2014, 50, 6733–6736 RSC.
- R. Bassan, M. Varshney and S. Roy, ChemistrySelect, 2023, 8, e202203317 CrossRef CAS.
- S. Roy, L. Zheng, O. Silberbush, M. Engel, Y. Atsmon-Raz, Y. Miller, A. Migliore, D. N. Beratan and N. Ashkenasy, J. Phys. Chem. B, 2021, 125, 12741–12752 CrossRef CAS PubMed.
- D. B. Rasale, I. Maity and A. K. Das, Chem. Commun., 2014, 50, 11397–11400 RSC.
- D. B. Rasale, I. Maity, M. Konda and A. K. Das, Chem. Commun., 2013, 49, 4815–4817 RSC.
- M. Amit, S. Roy, Y. Deng, E. Josberger, M. Rolandi and N. Ashkenasy, ACS Appl. Mater. Interfaces, 2018, 10, 1933–1938 CrossRef CAS PubMed.
- D. B. Rasale, M. Konda, S. Biswas and A. K. Das, Chem.–Asian J., 2016, 11, 926–935 CrossRef CAS PubMed.
- P. Singh, S. Misra, A. Das, S. Roy, P. Datta, G. Bhattacharjee, B. Satpati and J. Nanda, ACS Appl. Bio Mater., 2019, 2, 4881–4891 CrossRef CAS PubMed.
- A. K. Das and P. K. Gave, Soft Matter, 2020, 16, 10065–10095 RSC.
- Y. Bai, A. Chotera, O. Taran, C. Liang, G. Ashkenasy and D. G. Lynn, Chem. Soc. Rev., 2018, 47, 5444–5456 RSC.
- B. A. Grzybowski and W. T. S. Huck, Nat. Nanotechnol., 2016, 11, 585–592 CrossRef CAS PubMed.
- N. Giuseppone, Acc. Chem. Res., 2012, 45, 2178–2188 CrossRef CAS PubMed.
- Y. Cao, L. Gabrielli, D. Frezzato and L. J. Prins, Angew. Chem., Int. Ed., 2023, 62, e202215421 CrossRef CAS PubMed.
- R. Merindol and A. Walther, Chem. Soc. Rev., 2017, 46, 5588–5619 RSC.
- D. Qi, C. M. Tann, D. Haring and M. D. Distefano, Chem. Rev., 2001, 101, 3081–3111 CrossRef CAS PubMed.
- I. W. Hamley, Biomacromolecules, 2021, 22, 1835–1855 CrossRef CAS PubMed.
- S. Pizzarello and A. L. Weber, Science, 2004, 303, 1151 CrossRef CAS PubMed.
- S. Roy and A. Banerjee, Soft Matter, 2011, 7, 5300–5308 RSC.
- K. Hawkins, A. K. Patterson, P. A. Clarke and D. K. Smith, J. Am. Chem. Soc., 2020, 142, 4379–4389 CrossRef CAS PubMed.
- Y. Ding, L. Nie, X. C. Yang, Y. Li, Y. Y. Huo, Z. Li, Y. Gao, H. L. Cui, J. Li and X. W. Xu, Front. Microbiol. DOI:10.3389/fmicb.2021.798194.
- K. Sayali, P. Sadichha and S. Surekha, Int. J. Curr. Microbiol. Appl. Sci., 2013, 2, 135–146 Search PubMed.
- M. Z. Atassi and T. Manshouri, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8282–8286 CrossRef PubMed.
- K. S. Broo, H. Nilsson, J. Nilsson, A. Flodberg and L. Baltzer, J. Am. Chem. Soc., 1998, 120, 4063–4068 CrossRef CAS.
- M. O. Guler and S. I. Stupp, J. Am. Chem. Soc., 2007, 129, 12082–12083 CrossRef CAS PubMed.
- M. J. Macdonald, L. D. Lavis, D. Hilvert and S. H. Gellman, Org. Lett., 2016, 18, 3518–3521 CrossRef CAS PubMed.
- L. Schoonen, K. S. Van Esterik, C. Zhang, R. V. Ulijn, R. J. M. Nolte and J. C. M. V. Hest, Sci. Rep., 2017, 7, 14772 CrossRef PubMed.
- C. Zhang, R. Shafi, A. Lampel, D. MacPherson, C. G. Pappas, V. Narang, T. Wang, C. Maldarelli and R. V. Ulijn, Angew. Chem., Int. Ed., 2017, 129, 14703–14707 CrossRef.
- Y. S. Moroz, T. T. Dunston, O. V. Makhlynets, O. V. Moroz, Y. Wu, J. H. Yoon, A. B. Olsen, J. M. McLaughlin, K. L. Mack, P. M. Gosavi, N. A. J. Van Nuland and I. V. Korendovych, J. Am. Chem. Soc., 2015, 137, 14905–14911 CrossRef CAS PubMed.
- N. Singh, M. P. Conte, R. V. Ulijn, J. F. Miravet and B. Escuder, Chem. Commun., 2015, 51, 13213–13216 RSC.
- B. Sarkhel, A. Chatterjee and D. Das, J. Am. Chem. Soc., 2020, 142, 4098–4103 CrossRef CAS PubMed.
- S. Goswami, A. Reja, S. Pal, A. Singh and D. Das, J. Am. Chem. Soc., 2022, 144, 19248–19252 CrossRef CAS PubMed.
- S. Bal, K. Das, S. Ahmed and D. Das, Angew. Chem., Int. Ed., 2019, 58, 244–247 CrossRef CAS PubMed.
- A. M. Garcia, M. Kurbasic, S. Kralj, M. Melchionna and S. Marchesan, Chem. Commun., 2017, 53, 8110–8113 RSC.
- J. Rodon Fores, M. Criado-Gonzalez, A. Chaumont, A. Carvalho, C. Blanck, M. Schmutz, C. A. Serra, F. Boulmedais, P. Schaaf and L. Jierry, Angew. Chem., Int. Ed., 2019, 58, 18817–18822 CrossRef CAS PubMed.
- Y. Tian, L. Yang, X. Peng, W. Qi and M. Wang, Soft Matter, 2023, 19, 3458–3463 RSC.
- Z. Huang, S. Guan, Y. Wang, G. Shi, L. Cao, Y. Gao, Z. Dong, J. Xu, Q. Luo and J. Liu, J. Mater. Chem. B, 2013, 1, 2297–2304 RSC.
- C. Zhang, X. Xue, Q. Luo, Y. Li, K. Yang, X. Zhuang, Y. Jiang, J. Zhang, J. Liu, G. Zou and X. J. Liang, ACS Nano, 2014, 8, 11715–11723 CrossRef CAS PubMed.
- M. P. Friedmann, V. Torbeev, V. Zelenay, A. Sobol, J. Greenwald and R. Riek, PLoS One, 2015, 10, e0143948 CrossRef PubMed.
- A. Singh, J. U. Joo and D. P. Kim, Nanoscale, 2022, 14, 15010–15020 RSC.
- Z. Li, S. Y. Joshi, Y. Wang, S. A. Deshmukh and J. B. Matson, Angew. Chem., Int. Ed., 2023, 62, e202303755 CrossRef CAS PubMed.
- O. Agazani, A. Tulpin and M. Reches, ChemSystemsChem, 2021, 3, e2100005 CrossRef CAS.
- A. J. Burton, A. R. Thomson, W. M. Dawson, R. L. Brady and D. N. Woolfson, Nat. Chem., 2016, 8, 837–844 CrossRef CAS PubMed.
- G. Gulseren, M. A. Khalily, A. B. Tekinay and M. O. Guler, J. Mater. Chem. B, 2016, 4, 4605–4611 RSC.
- M. Lee, T. Wang, O. V. Makhlynets, Y. Wu, N. F. Polizzi, H. Wu, P. M. Gosavi, J. Stöhr, I. V. Korendovych, W. F. Degrado and M. Hong, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6191–6196 CrossRef CAS PubMed.
- C. M. Rufo, Y. S. Moroz, O. V. Moroz, J. Stöhr, T. A. Smith, X. Hu, W. F. Degrado and I. V. Korendovych, Nat. Chem., 2014, 6, 303–309 CrossRef CAS PubMed.
- Z. S. Al-Garawi, B. A. McIntosh, D. Neill-Hall, A. A. Hatimy, S. M. Sweet, M. C. Bagley and L. C. Serpell, Nanoscale, 2017, 9, 10773–10783 RSC.
- K. Gayen, K. Basu, D. Bairagi, V. Castelletto, I. W. Hamley and A. Banerjee, ACS Appl. Bio Mater., 2018, 1, 1717–1724 CrossRef CAS PubMed.
- P. Makam, S. S. R. K. C. Yamijala, K. Tao, L. J. W. Shimon, D. S. Eisenberg, M. R. Sawaya, B. M. Wong and E. Gazit, Nat. Catal., 2019, 2, 977–985 CrossRef CAS PubMed.
- L. C. Serpell, Alzheimer's amyloid fibrils: structure and assembly, Biochim. Biophys. Acta, 2000, 1502, 16–30 CrossRef CAS PubMed.
- I. W. Hamley, A. Dehsorkhi, V. Castelletto, J. Seitsonen, J. Ruokolainen and H. Iatrou, Soft Matter, 2013, 9, 4794–4801 RSC.
- K. Roy, M. Chetia, A. K. Sarkar and S. Chatterjee, RSC Adv., 2020, 10, 42062–42075 RSC.
- P. K. Gavel, N. Kumar, H. S. Parmar and A. K. Das, ACS Appl. Bio Mater., 2020, 3, 3326–3336 CrossRef CAS PubMed.
- J. Madine, E. Jack, P. G. Stockley, S. E. Radford, L. C. Serpell and D. A. Middleton, J. Am. Chem. Soc., 2008, 130, 14990–15001 CrossRef PubMed.
- M. Szefczyk, N. Szulc, M. Gąsior-Głogowska, A. Modrak-Wójcik, A. Bzowska, W. Majstrzyk, M. Taube, M. Kozak, T. Gotszalk, E. Rudzińska-Szostak and Ł. Berlicki, Nanoscale, 2021, 13, 4000–4015 RSC.
- J. Seo, W. Hoffmann, S. Warnke, X. Huang, S. Gewinner, W. Schöllkopf, M. T. Bowers, G. Von Helden and K. Pagel, Nat. Chem., 2017, 9, 39–44 CrossRef CAS PubMed.
- A. Barth, Prog. Biophys, Mol. Biol., 2000, 74, 141–173 CAS.
- S. J. George, A. Ajayaghosh, P. Jonkheijm, A. P. H. J. Schenning and E. W. Meijer, Angew. Chem., Int. Ed., 2004, 43, 3422–3425 CrossRef CAS PubMed.
- P. Xing, H. P. Tham, P. Li, H. Chen, H. Xiang and Y. Zhao, Adv. Sci., 2018, 5, 1700552 CrossRef PubMed.
- K. Dan, R. Pan and S. Ghosh, Langmuir, 2011, 27, 612–617 CrossRef CAS PubMed.
- A. Singh, J. P. Joseph, D. Gupta, C. Miglani, N. A. Mavlankar and A. Pal, Nanoscale, 2021, 13, 13401–13409 RSC.
- Y. Peng, S. Fu, H. Liu and L. A. Lucia, Bioresearch, 2016, 11, 10099–10111 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, materials, methods, instrumentation details, synthetic protocols, hydrogelation, results and discussion on experiments performed using NMR, ESI-MS, FE-SEM, TEM, XRD, FTIR, CD, CMC, UV-Vis and confocal microscopy. See DOI: https://doi.org/10.1039/d4na00268g |
| This journal is © The Royal Society of Chemistry 2024 |