Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Characterizing induced pluripotent stem cells and derived cardiomyocytes: insights from nano scale mass measurements and mechanical properties†
Irene C.
Turnbull
 *a and
Angelo
Gaitas
*bc
*a and
Angelo
Gaitas
*bc
aCardiovascular Research Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. E-mail: irene.turnbull@mssm.edu
bThe Estelle and Daniel Maggin Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. E-mail: angelo.gaitas@mssm.edu
cBioMedical Engineering & Imaging Institute, Leon and Norma Hess Center for Science and Medicine, New York, NY 10029, USA
First published on 28th November 2023
Abstract
Our study reveals that the nano-mechanical measures of elasticity and cell mass change significantly through induced pluripotent stem cell (iPSC) differentiation to cardiomyocytes, providing a reliable method to evaluate such processes. The findings support the importance of identifying these properties, and highlight the potential of AFM for comprehensive characterization of iPSC at the nanoscale.
This research employs atomic force microscopy (AFM)1–3 to investigate single-cell characteristics of human induced pluripotent stem cells (iPSCs) and iPSCs-derived cardiomyocytes (iPSC-CMs) at the nanoscale. iPSCs, reprogrammed from mature cells like human skin cells, can be specialized into various types, including cardiomyocytes (CMs).4 The promise of iPSC-CMs extends to numerous areas such as cell therapy, drug testing, and investigating aspects of cardiac disease.5–8 They are increasingly recognized for their potential in drug evaluation, with particular interest for their use to predict subject-specific drug effects, thereby paving the way for personalized medicine.9–15 iPSC-CMs hold promise for heart tissue repair in regenerative medicine.16 Progress in these applications of hiPSC-CMs hinges on the ability to accurately characterize these cells at the single-cell level. Understanding their maturation status by adding new phenotypic markers is key.
Cell mass is an integral parameter which provides fundamental insights into cellular health, growth, and proliferative potential.17 Mass measurements at the single-cell level enable precise quantification of cellular growth rates, biomass production, and metabolic activities.18–29 Changes in cell mass are reflective of shifts in intracellular processes, including protein synthesis, cell division, and differentiation, thereby offering a valuable tool for monitoring these key cellular events.19–29 Therefore, measuring single-cell mass is crucial for characterizing cell populations, evaluating the impacts of external stimuli, and understanding cell cycle dynamics and disease processes at an unprecedented resolution. Similarly, single-cell elasticity is of significant importance as it is intrinsically linked to the cell's functionality and state of differentiation.30 The elasticity of a cell, or its stiffness, is a critical physical property that is affected by cytoskeletal organization and intracellular force generation mechanisms, often reflective of cellular behavior and biological processes such as proliferation, migration, and differentiation.31 Thus, understanding these mechanical properties at a single-cell level provides invaluable insights into cell physiology and pathology, helping to elucidate the underpinnings of cellular transformations and their role in tissue function and disease states.19–29,32
In this study, we employed two AFM techniques to evaluate the elasticity and mass of individual iPSCs and iPSC-CM.32–37 AFM has emerged as a key tool in biomechanics for measuring single-cell elasticity, allowing for nanoscale resolution in mapping the elastic properties of cells, thereby providing valuable insights into cell health, behavior, and response to various stimuli.38–44 Utilizing the innovative fluidic-AFM technique we measured single cell mass, the technique allows for the measurement of multiple individual cells using fluidic pressure to attached the live cells in vitro.45–47 We report for the first time that elasticity and mass measurements can discern cells pre- and post-differentiation. Thus, AFM can provide a detailed single-cell analysis of cell elasticity and cell mass to offer insights into iPSC differentiation.48
For cell source we used a well-validated healthy iPSC cell line (SKiPS-31.3).49 iPSCs were grown in StemFlex media on 6-well plates coated with hESC-qualified Matrigel in 5% CO2 incubator at 37 °C. iPSCs were differentiated into CMs by following a monolayer-based differentiation protocol with minor modifications.50–52 When iPSCs reached about 80% confluency, StemFlex media was replaced with basal medium (RPMI 1640 media plus 2% B27 supplement minus Insulin), and 10 μM CHIR99021 for 24 hours. After 24 hours, the media was replaced with basal medium without CHIR99021. Then, after 48 hours (day 3), the medium was replaced with basal medium plus 5 μM IWR-1. After 48 hours (day 5), the media was replaced with basal medium. On day 7, the medium was replaced with RPMI 1640 media plus 2% B27 supplement. Thereafter, the media was exchanged every 2 days. Starting on days 7–10 post-CHIR treatment, iPSC-CMs begin to display spontaneous contractions.
For elasticity measurements of the iPSCs on day 0, 70–80% confluent iPSCs were treated with ReLeSR (STEMCELL technologies; 1 ml per well of a 6-well plate), after 1 minute the ReLeSR was removed and the cells were maintained for 5–8 minutes in the incubator (5% CO2, 37 °C) while intermittently monitoring their detachment by visualization under the microscope. Then the cells were resuspended in Stemflex medium (1 ml per well of a 6-well plate) and the cells were plated onto a Matrigel coated glass-bottom cell culture dish. Stemflex medium was exchanged the following day. On day 3 after replating, the medium was exchanged and the cells were used for elasticity analysis. For the iPSC-CMs, the cells were maintained in culture for at least 141 days. Of note, during this time in culture replating was necessary when the cell monolayer acquired a mesh like morphology and were at risk of detaching from the plate. Five-seven days before the AFM elasticity measurements, the iPSC-CMs were treated with 0.25% trypsin. After 5–10 minutes the cells were resuspended in RPMI 1640 media plus 2% B27 supplement with 10% FBS (2 ml per 1 ml of trypsin). After centrifugation (300 g × 5 minutes) the supernatant was removed and the cells were resuspended in RPMI 1640 media plus 2% B27 supplement with 10% FBS and plated on Matrigel coated glass-bottom cell culture dish. After 24 hours, the media was replaced with RPMI 1640 media plus 2% B27 supplement, and every other day thereafter until AFM measurements were performed.
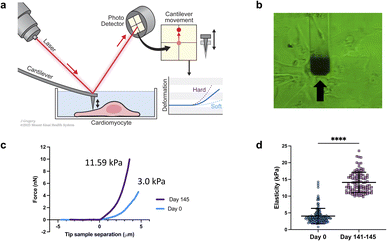
Elastic modulus of the cells was measured using a FlexAFM (Nanosurf AG, Switzerland), with inbuilt temperature controller and an inverted Axio Observer (Carl Zeiss) microscope. Nanosensors qp-SCONT-10 probes with a spring constant of ∼0.01 N m−1, and tip height of 8 μm were used for measurements (Fig. 1a and b). Elasticity measurements were performed on various iPSCs on day zero and on day 141–145 of differentiation. We measured multiple sites on each cell. Total number of sampled sites were: n = 150 for day zero and n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) =
=![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 for day 141–145. The elastic modulus, which was determined from the force curves through the application of the Hertz model, was computed using the Automated Nanomechanical Analysis (ANA) software provided by Nanosurf.
104 for day 141–145. The elastic modulus, which was determined from the force curves through the application of the Hertz model, was computed using the Automated Nanomechanical Analysis (ANA) software provided by Nanosurf.
We conducted a comparison of iPSC-CM at two different time points: 0 days of differentiation and 141–145 days from differentiation. The results revealed that the older/differentiated cells exhibited an average modulus of elasticity of 14.1 ± 4 kPa (average ± standard deviation), whereas the cells at 0 days of differentiation had a modulus of 3 ± 2.3 kPa (Fig. 1c and d). During the differentiation and maturation process of iPSCs,53 the mechanical properties (stiffness/elasticity), undergo changes influenced by the organization and composition of the cytoskeleton, this includes enhanced alignment and organization of the cytoskeletal components—actin filaments, intermediate filaments, and microtubules.53–55 Immature cells have a less organized cytoskeleton and fewer myofibrils, resulting in lower stiffness. However, as cells mature, they develop more myofibrils, composed of actin and myosin proteins, which contribute to higher stiffness. The observed structural and functional alterations mirror those found in native cardiomyocyte development, which is characterized by the emergence of a more robust contractile apparatus and alterations in cellular stiffness. Our findings of increased stiffness following iPSC differentiation and maturation are consistent with prior reports from other research groups studying iPSCs.53–55 Notably, this study represents, to our knowledge, the first instance of a direct comparative analysis between iPSCs and their cardiomyocyte counterparts (iPSC-CMs).
For nano-gram mass measurements of iPSCs on day 0, when the iPSCs reached about 80% confluency, which is the point when the iPSC-CM differentiation starts (see above), the iPSCs were treated with ReLeSR; after 1 minute the ReLeSR was removed and the cells were maintained for 5–8 minutes in the incubator (5% CO2, 37 °C) while intermittently monitoring their detachment by visualization under the microscope. Then the cells were resuspended in Stemflex medium, with gentle pipetting for the cells to be singularized, and transferred to a microcentrifuge tube and prepared immediately for fluidic-AFM assessment. A glass bottom dish (without Matrigel coating) was filled with Stemflex culture medium, then a 100 μl aliquot from the cell suspension in the microcentrifuge tube was dispensed onto the center of the dish. The cells remained singularized and floating, rendering them accessible for capture by the microfluidic cantilever. For the iPSC-CMs, the cells were maintained in culture for 173 days; with replating as needed. On the day of the mass measurement, the iPSC-CMs were treated with 0.25% trypsin. After 5–10 minutes the cells were resuspended in RPMI 1640 media plus 2% B27 supplement with 10% FBS (2 ml per 1 ml of trypsin). After centrifugation (300 g × 5 minutes) the supernatant was removed and the cells were resuspended in RPMI 1640 media plus 2% B27 supplement with gentle pipetting while assuring that the cells were singularized, passed through a 100 μm cell strainer, and transferred to a microcentrifuge tube and prepared immediately for fluidic-AFM assessment. A glass bottom dish (without Matrigel coating) was filled with RPMI 1640 media plus 2% B27 supplement culture medium, then a 100 μl aliquot from the cell suspension of iPSC-CMs in the microcentrifuge tube was dispensed onto the center of the dish. The cells remained singularized and floating, rendering them accessible for capture by the microfluidic cantilever.
The experimental framework for measuring mass uses a microfluidic cantilever from Cytosurg AG, Switzerland. This cantilever includes a 4-micron aperture at the tip (Fig. 2a). The Sader method is used to determine the thermal resonance frequency. A pump provided by Cytosurge is connected to the cantilever, capable of exerting pressure from 800 mbar to 1000 mbar. The method, described in our previous work,46 involves moving the cantilever to position the tip of the fluidic probe above the cell of interest, then applying negative pressure to generate a suction force that attaches the cell to the opening of the fluidic probe. The measurement process involves recording the cantilever's resonance frequency while the cell is adhered to it; following cell detachment (achieved by applying positive pressure) the resonance frequency is subsequently re-measured in the absence of any attached cell (ESI videos 1 and 2†).
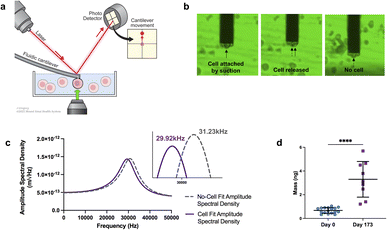 | ||
| Fig. 2 Mass analysis. (a) Schematic of mass measurements with fluidic-AFM, illustrating an iPSC-CM captured by the microfluidic cantilever. Not to scale. (b) Screen shots illustrate the sequence of events during the cell mass measurement process. The three static images show the cell attached (captured by the cantilever), the immediate moment that the cell is released and no longer attached to the cantilever, lastly the empty cantilever with no cell. Refer to ESI videos 1 and 2† to see the sequence in real-time. (c) Representative graph of the shift in resonance frequency post cell attachment (purple line) in comparison to the free/empty cantilever (gray dash line). Utilizing the frequencies and applying eqn (1), the calculated mass is approximately 5.69 ng. (d) Dot plot of cell mass measurements at day 0 and day 173 of differentiation. N = 17 for day 0, and N = 9 for day 173; bar and error bars represent the mean ± SD; ****P < 0.0001, unpaired t-test. | ||
After cell detachment using positive pressure, the fluidic AFM cantilever is used to measure subsequent cells. This method allows the reuse of the same microfluidic cantilever for several measurements. An example of a microfluidic cantilever with a 4 μm aperture submerged in media with a captured iPSC-CM cell is shown in Fig. 2b. The entire measurement for each cell is completed in less than three minutes. A limitation in this application is the risk for clogging of the microfluidic probe, this can be minimized by careful selection according to cell size of the aperture tip and timing of negative pressures applied.
In our experiments, we measured cells at day 0, and at day 173 from the start of differentiation. We used an equation described in ref. 45, 46, 56 and 57 that allows for mass calculation based on the frequency peak changes (eqn (1)):
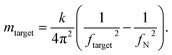 | (1) |
While we anticipate that the general trend of increased stiffness and mass upon differentiation will hold true across various cell types, the absolute values and variability might differ. In a heterogeneous cell population, a larger variability in mechanical properties and cell mass is expected. The strength of our method lies in its ability to perform single-cell analyses; even in heterogeneous population, the mechanical properties and mass can be individually assessed.
In conclusion, for the tested iPSC cell line, we found that mass measurements and mechanical properties can serve as critical morphological markers for monitoring the differentiation of iPSC to iPSC-CMs. Our investigation showed a notable difference in cell mass and elasticity values pre- and post-differentiation, demonstrating our ability to distinguish between these stages in this specific cell line. While our findings are promising, further studies are required to determine if this ability extends universally to other cell lines and types. Our findings underscore the potential of AFM as a comprehensive tool for single-cell characterization at the nanoscale level. Both techniques can be used for a more detailed monitoring of differentiation and maturation of iPSC.33–35
The integrative and non-invasive nature of mass measurements offers great promise especially when combined with other AFM techniques. Fluidic AFM probes, for instance, not only perform mass measurements, but also can be used to extract genomic content of individual cells, thereby providing insights into the nuances of iPSC differentiation enhancing our understanding of iPSC differentiation processes.58 However, it is important to note that the eqn (1) employed in our study serves as an approximation. Its application, especially in a liquid medium, is not entirely accurate because it assumes there is no damping and operates under the assumption of a point mass.59 Improvements can be achieved by using more accurate mathematical modelling to derive mass and employing different cantilever materials and designs.60 Incorporating active vibration strategies, instead of the thermal tune method,61,62 can increase the amplitude of oscillation and in turn reduce the minimum detectable mass. Techniques such as photo-thermal modulation (where thermally induced stress is applied with a laser near the cantilever's fixed end),63,64 piezoelectric actuation,65–67 thermomechanical actuation,68–70 and magnetic excitation71–73 are potential avenues that can be explored to further improve the resolution of these measurements in liquids.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation under award number 226930, the National Institute of General Medical Sciences under award number R44GM146477 and the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number R21HL165298; the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation. The authors gratefully acknowledge Jill K. Gregory, MFA, CMI, Associate Director of Instructional Technology, Icahn School of Medicine at Mount Sinai for illustrations 1a and 2a.References
- D. Borin, I. Pecorari, B. Pena and O. Sbaizero, in Seminars in Cell & Developmental Biology, Elsevier, pp. , pp. 4–12 Search PubMed.
- J. Liu, N. Sun, M. A. Bruce, J. C. Wu and M. J. Butte, Atomic force mechanobiology of pluripotent stem cell-derived cardiomyocytes, PLoS One, 2012, 7, e37559 CrossRef CAS PubMed.
- N. Nagarajan, V. Vyas, B. D. Huey and P. Zorlutuna, Modulation of the contractility of micropatterned myocardial cells with nanoscale forces using atomic force microscopy, Nanobiomedicine, 2016, 3, 1849543516675348 CrossRef PubMed.
- C. L. Mummery, et al., Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes, Circ. Res., 2012, 111, 344–358 CrossRef CAS PubMed.
- D. A. Robinton and G. Q. Daley, The promise of induced pluripotent stem cells in research and therapy, Nature, 2012, 481, 295 CrossRef CAS PubMed.
- T. Shimizu, et al., Fabrication of pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique and temperature-responsive cell culture surfaces, Circ. Res., 2002, 90, e40–e48 CrossRef CAS PubMed.
- I.-H. Park, et al., Disease-specific induced pluripotent stem cells, Cell, 2008, 134, 877–886 CrossRef CAS PubMed.
- M. Bellin, et al., Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome, EMBO J., 2013, 32, 3161–3175 CrossRef CAS PubMed.
- Q. Zhao, et al., Cardiotoxicity evaluation using human embryonic stem cells and induced pluripotent stem cell-derived cardiomyocytes, Stem Cell Res. Ther., 2017, 8, 54 CrossRef PubMed.
- B. J. van Meer, L. G. Tertoolen and C. L. Mummery, Concise review: measuring physiological responses of human pluripotent stem cell derived cardiomyocytes to drugs and disease, Stem Cells, 2016, 34, 2008–2015 CrossRef CAS.
- S. H. Tan and L. Ye, Maturation of pluripotent stem cell-derived cardiomyocytes: a critical step for drug development and cell therapy, J. Cardiovasc. Transl. Res., 2018, 11, 375–392 CrossRef PubMed.
- M. Schmitz, Measurement of Cardiomyocyte Contractions, 2012 Search PubMed.
- H. Laverty, et al., How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines?, Br. J. Pharmacol., 2011, 163, 675–693 CrossRef CAS PubMed.
- W. Redfern, et al., Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development, Cardiovasc. Res., 2003, 58, 32–45 CrossRef CAS.
- F. Stillitano, et al., Modeling susceptibility to drug-induced long QT with a panel of subject-specific induced pluripotent stem cells, Elife, 2017, 6, e19406 CrossRef PubMed.
- H. Abou-Saleh, et al., The march of pluripotent stem cells in cardiovascular regenerative medicine, Stem Cell Res. Ther., 2018, 9, 1–31 CrossRef.
- N. Cermak, et al., High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays, Nat. Biotechnol., 2016, 34, 1052–1059 CrossRef CAS PubMed.
- K. Park, et al., Measurement of adherent cell mass and growth, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20691–20696 CrossRef CAS.
- C. Cadart, L. Venkova, P. Recho, M. C. Lagomarsino and M. Piel, The physics of cell-size regulation across timescales, Nat. Phys., 2019, 15, 993–1004 Search PubMed.
- A. A. Amodeo and J. M. Skotheim, Cell-size control, Cold Spring Harbor Perspect. Biol., 2016, 8, a019083 Search PubMed.
- M. B. Ginzberg, R. Kafri and M. Kirschner, On being the right (cell) size, Science, 2015, 348, 1245075 CrossRef.
- K. N. Pollizzi, A. T. Waickman, C. H. Patel, I. H. Sun and J. D. Powell, Cellular size as a means of tracking mTOR activity and cell fate of CD4+ T cells upon antigen recognition, PLoS One, 2015, 10, e0121710 CrossRef.
- A. C. Lloyd, The regulation of cell size, Cell, 2013, 154, 1194–1205 CrossRef CAS.
- J. J. Turner, J. C. Ewald and J. M. Skotheim, Cell size control in yeast, Curr. Biol., 2012, 22, R350–R359 CrossRef CAS.
- J. Yang, et al., Cell size and growth rate are major determinants of replicative lifespan, Cell Cycle, 2011, 10, 144–155 CrossRef CAS.
- A. Tzur, R. Kafri, V. S. LeBleu, G. Lahav and M. W. Kirschner, Cell growth and size homeostasis in proliferating animal cells, Science, 2009, 325, 167–171 CrossRef CAS PubMed.
- S. Di Talia, J. M. Skotheim, J. M. Bean, E. D. Siggia and F. R. Cross, The effects of molecular noise and size control on variability in the budding yeast cell cycle, Nature, 2007, 448, 947–951 CrossRef CAS PubMed.
- P. Echave, I. J. Conlon and A. C. Lloyd, Cell size regulation in mammalian cells, Cell Cycle, 2007, 6, 218–224 CrossRef CAS.
- T. T. Su and P. H. O'Farrell, Size control: cell proliferation does not equal growth, Curr. Biol., 1998, 8, R687–R689 CrossRef PubMed.
- S. E. Cross, Y.-S. Jin, J. Rao and J. K. Gimzewski, in Nano-enabled Medical Applications, Jenny Stanford Publishing, 2020, pp. 547–566 Search PubMed.
- S. Suresh, Biomechanics and biophysics of cancer cells, Acta Biomater., 2007, 3, 413–438 CrossRef PubMed.
- K. D. Costa, Single-cell elastography: probing for disease with the atomic force microscope, Dis. Markers, 2004, 19, 139–154 CrossRef PubMed.
- S. Vučković, et al., Characterization of cardiac metabolism in iPSC-derived cardiomyocytes: lessons from maturation and disease modeling, Stem Cell Res. Ther., 2022, 13, 332 CrossRef PubMed.
- P. Wu, G. Deng, X. Sai, H. Guo, H. Huang and P. Zhu, Maturation strategies and limitations of induced pluripotent stem cell-derived cardiomyocytes, Biosci. Rep., 2021, 41(6), BSR20200833 CrossRef CAS PubMed.
- G. Maroli and T. Braun, The long and winding road of cardiomyocyte maturation, Cardiovasc. Res., 2021, 117, 712–726 CrossRef CAS.
- A. Gaitas, R. Malhotra, T. Li, T. Herron and J. Jalife, A device for rapid and quantitative measurement of cardiac myocyte contractility, Rev. Sci. Instrum., 2015, 86, 034302 CrossRef.
- A. Gaitas, R. Malhotra and K. Pienta, A method to measure cellular adhesion utilizing a polymer micro-cantilever, Appl. Phys. Lett., 2013, 103, 123702 CrossRef PubMed.
- M. Radmacher, Studying the mechanics of cellular processes by atomic force microscopy, Methods Cell Biol., 2007, 83, 347–372 CAS.
- Y. F. Dufrêne, et al., Imaging modes of atomic force microscopy for application in molecular and cell biology, Nat. Nanotechnol., 2017, 12, 295–307 CrossRef.
- Y. Liu, C. Zhou, Z. Su, Q. Chang, Y. Qiu, J. Bei, A. Gaitas, J. Xiao, A. Drelich, K. Khanipov, Y. Jin, G. Golovko, T. B. Saito and B. Gong, Endothelial exosome plays a functional role during rickettsial infection, mBio, 2021, 12(3), e00769 CAS.
- C. Zhou, et al., Exosomally targeting microRNA23a ameliorates microvascular endothelial barrier dysfunction following rickettsial infection, Front. Immunol., 2022, 13, 904679 CrossRef CAS.
- X. He, et al., Exchange protein directly activated by cAMP plays a critical role in regulation of vascular fibrinolysis, Life Sci., 2019, 221, 1–12 CrossRef CAS.
- K. W. C. Lai, A. Gaitas, N. Xi, R. Yang and C. K. M. Fung in 11th IEEE International Conference on Nanotechnology, IEEE, 2011, pp. 1066–1069 Search PubMed.
- C. Mondal, M. J. Gacha-Garay, K. A. Larkin, R. C. Adikes, J. S. Di Martino, C. C. Chien, M. Fraser, I. Eni-Aganga, E. Agullo-Pascual., K. Cialowicz, U. Ozbek, A. Naba, A. Gaitas, T. M. Fu, S. Upadhyayula, E. Betzig, D. Q. Matus, B. L. Martin and J. J. Bravo-Cordero, A proliferative to invasive switch is mediated by srGAP1 downregulation through the activation of TGF-β2 signaling, Cell Rep., 2022, 40(12), 111358 CrossRef CAS PubMed.
- D. Ossola, P. Dörig, J. Vörös, T. Zambelli and M. Vassalli, Serial weighting of micro-objects with resonant microchanneled cantilevers, Nanotechnology, 2016, 27, 415502 CrossRef PubMed.
- C.-C. Chien, J. Jiang, B. Gong, T. Li and A. Gaitas, AFM microfluidic cantilevers as weight sensors for live single cell mass measurements, Meas. Sci. Technol., 2022, 33, 095009 CrossRef.
- Y. Qiu, et al., Extending applications of AFM to fluidic AFM in single living cell studies, J. Cell. Physiol., 2022, 237, 3222–3238 CrossRef CAS PubMed.
- A. J. Ribeiro, et al., Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12705–12710 CrossRef CAS PubMed.
- E. Galende, I. Karakikes, L. Edelmann, R. J. Desnick, T. Kerenyi, G. Khoueiry, J. Lafferty, J. T. McGinn, M. Brodman, V. Fuster, R. J. Hajjar and K. Polgar, Amniotic fluid cells are more efficiently reprogrammed to pluripotency than adult cells, Cell. Reprogram., 2010, 12(2), 117–125 CrossRef CAS PubMed.
- X. Lian, et al., Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1848–E1857 CrossRef CAS.
- J. G. Oh, J. Dave, C. Kho and F. Stillitano, Generation of Ventricular-Like HiPSC-Derived Cardiomyocytes and High-Quality Cell Preparations for Calcium Handling Characterization, J. Visualized Exp., 2020, e60135 Search PubMed.
- I. C. Turnbull, W. Zhu, F. Stillitano, C.-C. Chien and A. Gaitas, A micromachined force sensing apparatus and method for human engineered cardiac tissue and induced pluripotent stem cell characterization, Sens. Actuators, A, 2021, 112874 CrossRef CAS.
- C. L. Keefer and J. P. Desai, Mechanical phenotyping of stem cells, Theriogenology, 2011, 75, 1426–1430 CrossRef.
- S. Pagliari, et al., YAP–TEAD1 control of cytoskeleton dynamics and intracellular tension guides human pluripotent stem cell mesoderm specification, Cell Death Differ., 2021, 28, 1193–1207 CrossRef CAS.
- K. E. Hammerick, et al., Elastic properties of induced pluripotent stem cells, Tissue Eng., Part A, 2011, 17, 495–502 CrossRef CAS PubMed.
- J. Cleveland, S. Manne, D. Bocek and P. Hansma, A nondestructive method for determining the spring constant of cantilevers for scanning force microscopy, Rev. Sci. Instrum., 1993, 64, 403–405 CrossRef CAS.
- S. Kim, K. D. Kihm and T. Thundat, Fluidic applications for atomic force microscopy (AFM) with microcantilever sensors, Exp. Fluids, 2010, 48, 721–736 CrossRef CAS.
- W. Chen, et al., Live-seq enables temporal transcriptomic recording of single cells, Nature, 2022, 608, 733–740 CrossRef CAS PubMed.
- D. Kim and M. Gharib, Experimental study of three-dimensional vortex structures in translating and rotating plates, Exp. Fluids, 2010, 49, 329–339 CrossRef.
- C.-C. Chien, J. Jiang, B. Gong, T. Li and A. Gaitas, AFM Microfluidic Cantilevers as Weight Sensors for Live Single Cell Mass Measurements, Measurement Science and Technology, 2022 Search PubMed.
- J. L. Hutter and J. Bechhoefer, Calibration of atomic-force microscope tips, Rev. Sci. Instrum., 1993, 64, 1868–1873 CrossRef CAS.
- H.-J. Butt and M. Jaschke, Calculation of thermal noise in atomic force microscopy, Nanotechnology, 1995, 6, 1 CrossRef.
- G. C. Ratcliff, D. A. Erie and R. Superfine, Photothermal modulation for oscillating mode atomic force microscopy in solution, Appl. Phys. Lett., 1998, 72, 1911–1913 CrossRef CAS.
- A. Labuda, et al., Photothermal excitation for improved cantilever drive performance in tapping mode atomic force microscopy, Microsc. Anal., 2014, 21–25 Search PubMed.
- B. Rogers, L. Manning, T. Sulchek and J. Adams, Improving tapping mode atomic force microscopy with piezoelectric cantilevers, Ultramicroscopy, 2004, 100, 267–276 CrossRef CAS PubMed.
- C. A. Putman, K. O. Van der Werf, B. G. De Grooth, N. F. Van Hulst and J. Greve, Tapping mode atomic force microscopy in liquid, Appl. Phys. Lett., 1994, 64, 2454–2456 CrossRef CAS.
- P. Hansma, et al., Tapping mode atomic force microscopy in liquids, Appl. Phys. Lett., 1994, 64, 1738–1740 CrossRef CAS.
- J. Lee and W. P. King, Microcantilever actuation via periodic internal heating, Rev. Sci. Instrum., 2007, 78, 126102 CrossRef PubMed.
- B. Mitra and A. Gaitas, Thermally actuated tapping mode atomic force microscopy with polymer microcantilevers, Rev. Sci. Instrum., 2009, 80, 23703 CrossRef PubMed.
- R. Pedrak, et al., Micromachined atomic force microscopy sensor with integrated piezoresistive sensor and thermal bimorph actuator for high-speed tapping-mode atomic force microscopy phase-imaging in higher eigenmodes, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.-Process., Meas., Phenom., 2003, 21, 3102–3107 CrossRef CAS.
- E. L. Florin, M. Radmacher, B. Fleck and H. E. Gaub, Atomic force microscope with magnetic force modulation, Rev. Sci. Instrum., 1994, 65, 639–643 CrossRef CAS.
- W. Han, S. Lindsay and T. Jing, A magnetically driven oscillating probe microscope for operation in liquids, Appl. Phys. Lett., 1996, 69, 4111–4113 CrossRef CAS.
- I. Revenko and R. Proksch, Magnetic and acoustic tapping mode microscopy of liquid phase phospholipid bilayers and DNA molecules, J. Appl. Phys., 2000, 87, 526–533 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00727h |
| This journal is © The Royal Society of Chemistry 2024 |