Open Access Article
Open Access ArticleMetabolomic approaches to dissect dysregulated metabolism in the progression of pre-diabetes to T2DM†
Wenrui
Ji‡
,
Xiaomin
Xie‡
 *,
Guirong
Bai
,
Yanting
He
,
Ling
Li
,
Li
Zhang
and
Dan
Qiang
*,
Guirong
Bai
,
Yanting
He
,
Ling
Li
,
Li
Zhang
and
Dan
Qiang
Department of Endocrinology, The First People's Hospital of Yinchuan, Yinchuan, People's Republic of China. E-mail: xxm2324@126.com
First published on 30th April 2024
Abstract
Many individuals with pre-diabetes eventually develop diabetes. Therefore, profiling of prediabetic metabolic disorders may be an effective targeted preventive measure. We aimed to elucidate the metabolic mechanism of progression of pre-diabetes to type 2 diabetes mellitus (T2DM) from a metabolic perspective. Four sets of plasma samples (20 subjects per group) collected according to fasting blood glucose (FBG) concentration were subjected to metabolomic analysis. An integrative approach of metabolome and WGCNA was employed to explore candidate metabolites. Compared with the healthy group (FBG < 5.6 mmol L−1), 113 metabolites were differentially expressed in the early stage of pre-diabetes (5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1), 237 in the late stage of pre-diabetes (6.1 mmol L−1 ⩽ FBG < 7.0 mmol L−1), and 245 in the T2DM group (FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 7.0 mmol L−1). A total of 27 differentially expressed metabolites (DEMs) were shared in all comparisons. Among them, L-norleucine was downregulated, whereas ethionamide, oxidized glutathione, 5-methylcytosine, and alpha-D-glucopyranoside beta-D-fructofuranosyl were increased with the rising levels of FBG. Surprisingly, 15 (11 lyso-phosphatidylcholines, L-norleucine, oxidized glutathione, arachidonic acid, and 5-oxoproline) of the 27 DEMs were ferroptosis-associated metabolites. WGCNA clustered all metabolites into 8 modules and the pathway enrichment analysis of DEMs showed a significant annotation to the insulin resistance-related pathway. Integrated analysis of DEMs, ROC and WGCNA modules determined 12 potential biomarkers for pre-diabetes and T2DM, including L-norleucine, 8 of which were L-arginine or its metabolites. L-Norleucine and L-arginine could serve as biomarkers for pre-diabetes. The inventory of metabolites provided by our plasma metabolome offers insights into T2DM physiology metabolism.
7.0 mmol L−1). A total of 27 differentially expressed metabolites (DEMs) were shared in all comparisons. Among them, L-norleucine was downregulated, whereas ethionamide, oxidized glutathione, 5-methylcytosine, and alpha-D-glucopyranoside beta-D-fructofuranosyl were increased with the rising levels of FBG. Surprisingly, 15 (11 lyso-phosphatidylcholines, L-norleucine, oxidized glutathione, arachidonic acid, and 5-oxoproline) of the 27 DEMs were ferroptosis-associated metabolites. WGCNA clustered all metabolites into 8 modules and the pathway enrichment analysis of DEMs showed a significant annotation to the insulin resistance-related pathway. Integrated analysis of DEMs, ROC and WGCNA modules determined 12 potential biomarkers for pre-diabetes and T2DM, including L-norleucine, 8 of which were L-arginine or its metabolites. L-Norleucine and L-arginine could serve as biomarkers for pre-diabetes. The inventory of metabolites provided by our plasma metabolome offers insights into T2DM physiology metabolism.
Introduction
Type 2 diabetes mellitus (T2DM) is a form of noninsulin-dependent diabetes mellitus with a chronic and complex metabolic condition. The burden of T2DM is much higher than that of other types of diabetes mellitus, accounting for over 90% of the incidence of diabetes1. According to the International Diabetes Federation, the current number of people living with diabetes worldwide is 382 million, which is expected to exceed 592 million by 2035.2 In addition, there is evidence that during the COVID-19 pandemic, patients with T2DM have dramatically increased health risks, such as depression, possibly due to the increased risk of death and morbidity from COVID-19 infection3. This aggravates the weight of T2DM attracting the attention of scholars. However, despite promising results in suppressing the diabetes epidemic, the understanding of T2DM still lacks valid evidence that can benefit the general public.Pre-diabetes is a transitional stage in the progression from a healthy state to diabetes, characterized by elevated blood sugar levels that are higher than normal but below the diagnostic thresholds for diabetes.4 Pre-diabetes is a reversible stage, therefore people with pre-diabetes have an opportunity to regain health5. Therefore, interrupting the progression of pre-diabetes may be an effective targeted preventive measure for T2DM. However, current diagnostic methods show various insufficiencies for the early diagnosis of T2DM. Therefore, a biomarker that can predict the risk of prediabetes progression is urgently needed. In the pre-diabetes period, insulin resistance already occurs with the increase of fasting blood glucose (FBG),6 but it is difficult to find the pathological molecular mechanism of T2DM by detecting FBG alone. Numerous studies have shown that the differential expression of metabolites analysed by metabolomics is helpful in elucidating the molecular mechanism of T2DM. For example, metabolic biomarkers based on the circulating nuclear magnetic resonance raised T2DM risk prediction.7 Plasma phosphatidylinositol and sphingomyelin were capable of discriminating healthy individuals and T2DM patients.8 Long et al. suggested that the levels of alanine, glutamate, and palmitic acid (C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) may be biomarkers for the progression of pre-diabetes to T2DM.9 Zhu et al. also proved that a serological metabolite panel can distinguish asymptomatic populations at risk of T2DM based on global and targeted mass spectrometry (MS)10. Although the value of metabolomics in biomarker identification of T2DM has now been repeatedly validated and applied, metabolomics studies based on FBG gradients to distinguish cohorts of subjects to uncover biomarkers for prediabetes have not been reported. Furthermore, due to the incredible diversity of metabolite network structures, it is challenging and innovative to understand how dysregulation in metabolites occurs during progression to T2DM in pre-diabetic patients.
0) may be biomarkers for the progression of pre-diabetes to T2DM.9 Zhu et al. also proved that a serological metabolite panel can distinguish asymptomatic populations at risk of T2DM based on global and targeted mass spectrometry (MS)10. Although the value of metabolomics in biomarker identification of T2DM has now been repeatedly validated and applied, metabolomics studies based on FBG gradients to distinguish cohorts of subjects to uncover biomarkers for prediabetes have not been reported. Furthermore, due to the incredible diversity of metabolite network structures, it is challenging and innovative to understand how dysregulation in metabolites occurs during progression to T2DM in pre-diabetic patients.
In this study, we sought to analyse plasma metabolites in subjects with different glucose metabolism states and identify differential metabolites based on changes in FBG concentrations. Bioinformatics analysis was subsequently performed with a view to identifying pre-diabetes and T2DM-related key metabolites.
Results
Study cohort and the overall metabolome
Clinical characteristics of the study subjects are shown in Table 1. Compared with the healthy control group A, FBG, HbA1c, and fasting insulin were significantly higher in groups B and C of pre-diabetes patients and further significantly elevated in group D of T2DM patients (Table 1). Additionally, creatinine was only significantly upregulated in patients of the D group compared to the healthy group A (Table 1). Other parameters exhibited no significant difference among groups. These results indicated that the studied cohort has been separated by the parameters of T2DM. We next profiled the metabolome of these four groups (20 samples per group).| A | B | C | D | |
|---|---|---|---|---|
| *Kruskal–Wallis test. #One-way ANOVA with Tukey's test. *P < 0.05; **P < 0.01; ***P < 0.001; ##P < 0.01; ###P < 0.001. The significance mark represents the results of A vs. B, C, D. | ||||
| Age, years | 45.15 ± 6.47 | 41.45 ± 10.06 | 47.15 ± 6.37 | 43.80 ± 5.50 |
| Male sex | 10 (50%) | 14 (70%) | 9 (45%) | 11 (55%) |
| BMI, kg m−2 | 23.17 ± 2.79 | 24.36 ± 3.29 | 24.10 ± 3.11 | 24.08 ± 2.03 |
| Waist circumference, cm | 78.75 ± 9.68 | 83.95 ± 10.77 | 80.65 ± 8.71 | 83.80 ± 6.94 |
| FBG, mmol L−1 | 4.88 ± 0.40 | 5.75 ± 0.12* | 6.42 ± 0.21*** | 11.14 ± 2.73*** |
| Mean systolic BP, mmHg | 119.15 ± 8.67 | 118.75 ± 9.12 | 119.35 ± 7.91 | 118.70 ± 9.63 |
| Mean diastolic BP, mmHg | 76.15 ± 7.25 | 75.85 ± 7.88 | 74.25 ± 6.62 | 74.00 ± 6.15 |
| Biochemical parameters | ||||
| HbA1c, ng mL−1 | 164.52 ± 25.81 | 191.31 ± 20.79## | 226.88 ± 19.55### | 251.24 ± 22.20### |
| Triglycerides, mmol L−1 | 1.47(0.98, 1.88) | 1.79(1.19, 2.10) | 2.40(1.29, 3.21) | 2.69(1.29, 2.69) |
| Total cholesterol, mmol L−1 | 4.80(4.15, 5.40) | 4.49(4.04, 5.30) | 5.77(4.75, 6.09) | 5.39(4.56, 6.14) |
| LDL-cholesterol, mmol L−1 | 2.62 ± 0.69 | 2.60 ± 0.67 | 3.37 ± 1.44 | 3.14 ± 0.83 |
| HDL-cholesterol, mmol L−1 | 1.35 ± 0.25 | 1.30 ± 0.28 | 1.32 ± 0.21 | 1.29 ± 0.23 |
| ALT, U L−1 | 19.50 ± 7.98 | 21.74 ± 13.68 | 26.44 ± 14.36 | 28.35 ± 17.81 |
| AST, U L−1 | 25.53 ± 3.68 | 24.84 ± 6.36 | 26.12 ± 8.66 | 25.18 ± 7.55 |
| Urea, mmol L−1 | 4.95 ± 1.02 | 4.72 ± 1.25 | 4.42 ± 1.07 | 4.65 ± 1.28 |
| Uric acid, μmol L−1 | 313.02 ± 86.24 | 332.57 ± 87.15 | 323.68 ± 102.2 | 279.23 ± 62.62 |
| Creatinine, μmol L−1 | 65.41 ± 9.29 | 64.50 ± 10.56 | 62.49 ± 14.62 | 55.42 ± 10.35* |
| Fasting insulin, mIU L−1 | 4.84 ± 0.49 | 5.40 ± 0.61## | 5.98 ± 0.53### | 6.78 ± 0.44### |
To measure the stability during instrumental detection, we evaluated the stability of the QC samples. The total ion current (TIC) map of the mixed QC sample (the spectrum obtained by adding the intensities of all ions in the mass spectrum at each time point and then continuously delineating the spectrum) is shown in ESI,† Fig. S1A. The multi-peak map of MRM metabolite detection of multi-substance extraction is shown in ESI,† Fig. S1B. The results of the TIC overlap map of the QC sample showed that the curve overlap of total ion currents detected by metabolites was high (Fig. S1C, ESI†), that is, the retention time and peak intensity were consistent, indicating that the signal stability of MS was better when the same sample was detected at different time points.
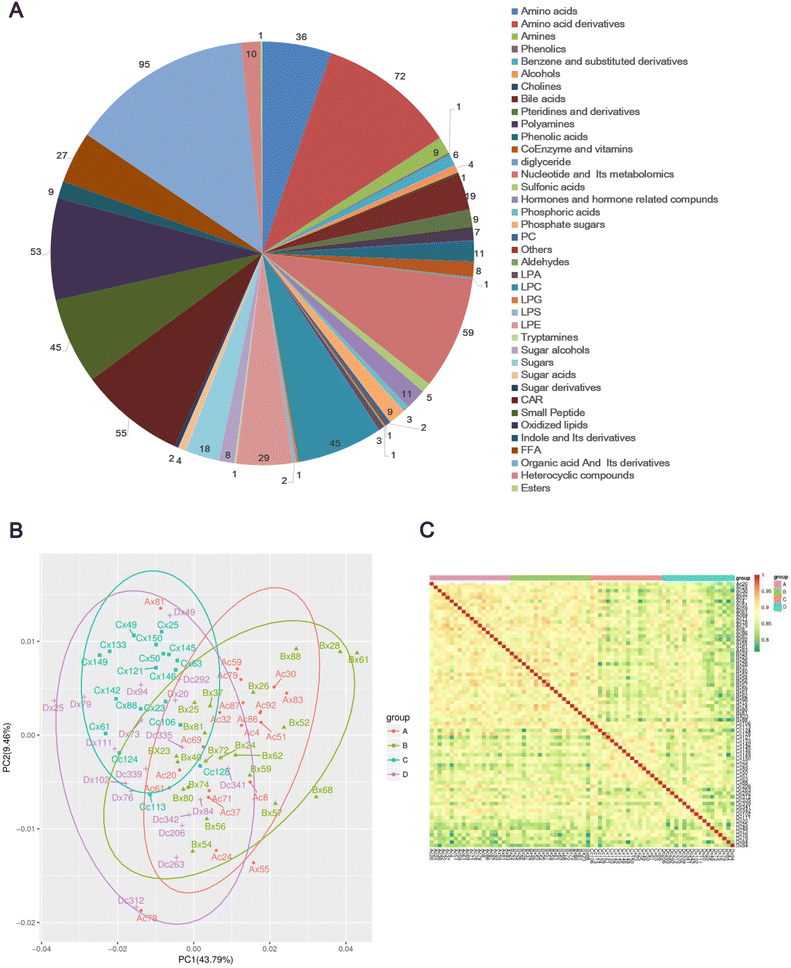
A total of 683 metabolites were detected composed of 39 metabolites from secondary metabolism, including 95 organic acids and their derivatives, 72 amino acid derivatives, and 59 nucleotides and their metabolomics (Fig. 1A and Table S2, ESI†). The principal components analysis (PCA) plot of metabolome explained 53.25% of the variation in the data (PCA1: 43.79% and PCA2: 9.46%) (Fig. 1B), indicating significant differences in overall metabolism between groups of samples. Subsequently, the biological repeatability between samples within the group was assessed by correlation analysis between samples using the Spearman rank correlation. As shown in Fig. 1C, the correlation coefficients of samples within groups were higher than those between groups, suggesting that the repeatability of samples within each group was satisfactory, and the differential metabolites obtained based on these samples will be reliable.
Multivariate analysis using OPLS-DA
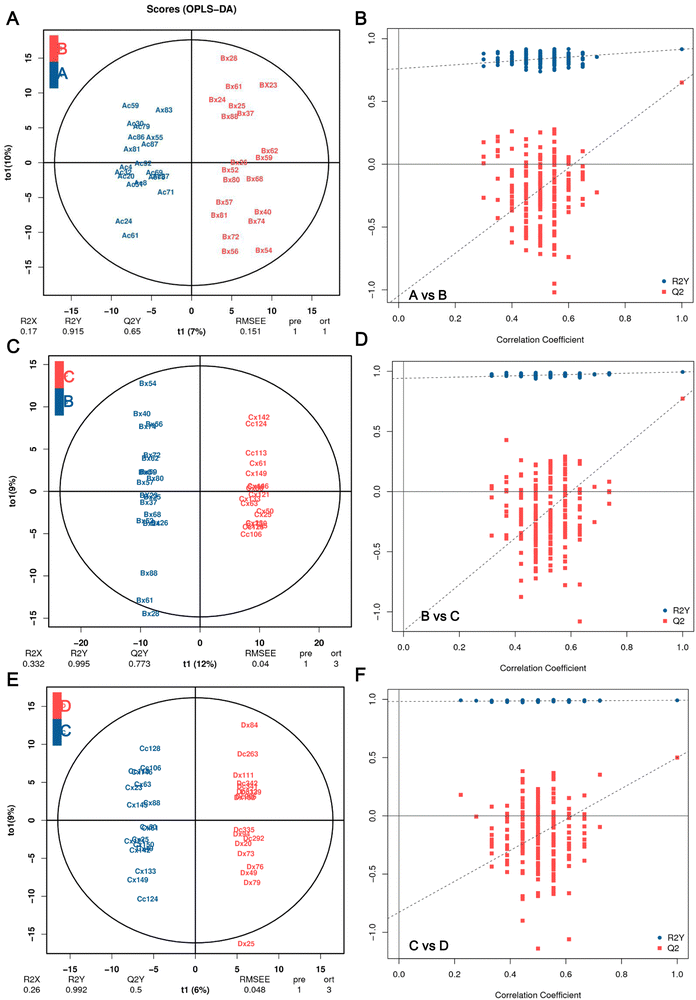
The OPLS-DA models were constructed and validated to compare the metabolic changes. The OPLS-DA score plot revealed a clear and separate clustering between healthy control (A group, FBG < 5.6 mmol L−1) and pre-diabetes patients (B group, 5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1), and these were reflected at Q2Y values exceeding 0.5, as shown in Fig. 2A, and Q2Y was 0.65 (A vs. B). These OPLS-DA models passed the validation of the permutation test (200 times) (Fig. 2B). A separate clustering between Group B pre-diabetes (5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1) and Group C pre-diabetes (6.1 mmol L−1 ⩽ FBG < 7.0 mmol L−1) was also revealed by the OPLS-DA score plot, as reflected at a Q2Y value of 0.773 (B vs. C), which also passed the validation of the permutation test (Fig. 2C and D). However, the Q2Y value was 0.339 when comparing C vs. D (group D, FBG![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 7.0 mmol L−1) (data not shown). We subsequently found that the exclusion of four samples, Cx87, Cc158, Dx31, and Dc1Q26, resulted in Q2Y values of 0.5 when comparing C vs. D in OPLS-DA analysis (Fig. 2E) and this model also passed the validation of the permutation test (Fig. 2F), indicating that this model can effectively screen differential metabolites. Therefore, all analyses in this study are based on the results obtained after excluding these 4 samples. Therefore, these results implicated that these models fit the data very well and have a good predictive ability. More importantly, the VIP values obtained based on OPLA-DA analysis can be used for screening DEMs.
7.0 mmol L−1) (data not shown). We subsequently found that the exclusion of four samples, Cx87, Cc158, Dx31, and Dc1Q26, resulted in Q2Y values of 0.5 when comparing C vs. D in OPLS-DA analysis (Fig. 2E) and this model also passed the validation of the permutation test (Fig. 2F), indicating that this model can effectively screen differential metabolites. Therefore, all analyses in this study are based on the results obtained after excluding these 4 samples. Therefore, these results implicated that these models fit the data very well and have a good predictive ability. More importantly, the VIP values obtained based on OPLA-DA analysis can be used for screening DEMs.
Identification of DEMs
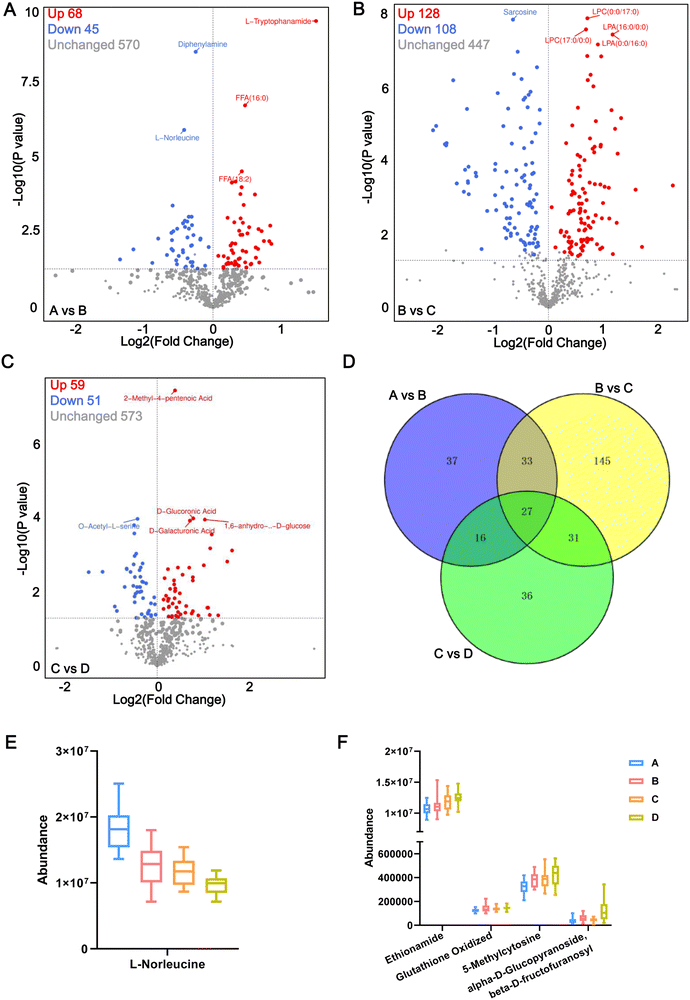
In this study, we considered metabolites with a fold change >1, P < 0.05 and VIP > 1.0 to be representative of DEMs. Under such conditions, compared with the healthy control group A, a total of 113 metabolites were differentially expressed in the early stage of pre-diabetes patients (A vs. B), including 68 upregulated DEMs and 45 downregulated DEMs in pre-diabetes patients (5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1) (Fig. 3A). They are potential markers of early onset of pre-diabetes in patients with FBG changes, including L-tryptophanamide and L-norleucine. Among them, L-norleucine was also the only DEM shared between all comparison groups, suggesting its critical position in the progression of diabetes (Fig. S2A, ESI†). Compared with the initial stage of pre-diabetes onset (Group B), 108 metabolites were upregulated and 128 were downregulated in Group C, indicating that these 236 DEMs changed with FBG concentration in the progression of pre-diabetes (Fig. 3B; B vs. C). Subsequently, the level of another 110 DEMs changed significantly with further increases in FBG concentration (Fig. 3C; C vs. D). Moreover, there were 27 metabolites shared by the above three comparison groups (Fig. 3D) and we visualized the abundance of these 27 metabolites in 4 patterns using box plots. Interestingly, we found that only L-norleucine abundance was successively decreased along with the FBG concentration growth (Fig. 3E and Table S2, ESI†), whereas the abundance of ethionamide, oxidized glutathione (GSSG), 5-methylcytosine, and alpha-D-glucopyranoside beta-D-fructofuranosyl increased with the growth of FBG concentration (Fig. 3F, and Table S2, ESI†). Thus, these five metabolites may be important drivers of pre-diabetes progression. The remaining 22 overlapped DEMs showed incomprehensible trends (Fig. S2B–S2D, ESI†).In addition, compared with T2DM patients of group D, a total of 113 DEMs (including 68 upregulated and 45 downregulated) were identified in the pre-diabetes group under ADA criteria (B vs. D), and 110 DEMs (including 59 upregulated and 51 downregulated) were identified in the pre-diabetes group under CDS criteria (C vs. D) (Table S3, ESI†). A total of 51 DEMs were shared in B vs. D and C vs. D (Fig. S2A, ESI†), serving as biomarkers for differentiating pre-diabetes and T2DM. Details of each DEM for the six comparison groups are presented in Table S4 (ESI†).
The DEMs of the A vs. B, B vs. C, and C vs. D comparison groups were classified, and the results showed that a total of 75 amino acids and their metabolomics, 114 lipids, 31 carboxylic acids and their derivatives (including 16 sugars, 7 phospholipids, and 3 sugar alcohols), 34 organic acid and their derivatives, etc. were obtained (Table S5, ESI†). Of these 16 sugars, the abundance of D-glucose, D-mannose, 1,6-anhydro-β-D-glucose, D-tagatose, and D-allose were significantly increased in the A vs. B comparison group. There was no difference in the B vs. C comparison group, but a significant increase was again observed in the C vs. D comparison group. However, alpha-D-glucopyranoside beta-D-fructofuranosyl, D-trehalose, lactose, lactulose, and maltose were significantly increased in the A vs. B comparison group which then decreased in the B vs. C comparison group and showed no difference in the C vs. D comparison group. These abnormal metabolisms of sugars associated with glycolysis suggest that glycolysis gradually enters a state of high throughput or even overload with the increase of FBG concentration. Of these organic acids and their derivatives, there was no difference in the abundance of α-ketoglutaric acid, methylmalonic acid, and citramalic acid in the A vs. B comparison group, but significantly increased in the B vs. C comparison group and were significantly decreased in the C vs. D comparison group; however, the abundance of pyruvate was significantly upregulated in the B vs. C comparison group but showed no difference between A vs. B and C vs. D comparison groups. These results imply that the TCA cycle is overloaded when FBG concentrations enter the range of the C group (FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 6.1 mmol L−1).
6.1 mmol L−1).
Ferroptosis may contribute to pre-diabetes progression
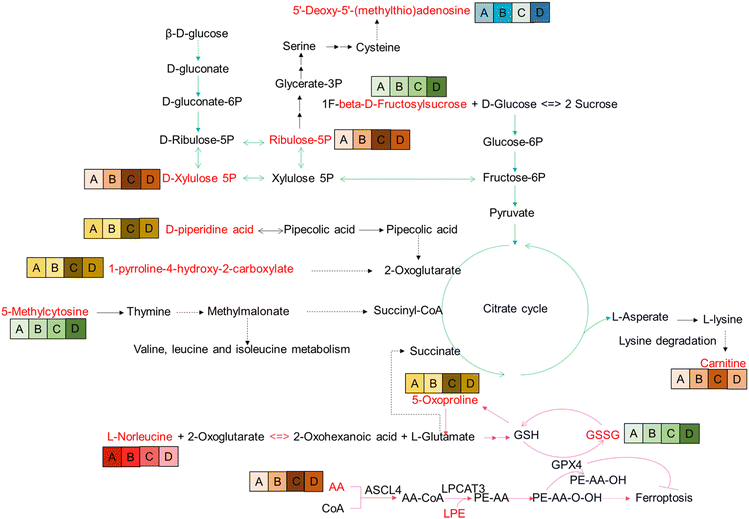
The location of these shared DEMs in the metabolic network is visualized as a flow chart (Fig. 4). Interestingly, of these 27 shared DEMs, there were 11 lyso-phosphatidylcholines (LPCs) which serve as a substrate for lipid peroxidation closely associated with ferroptosis. The expression patterns of the 11 LPCs were the same among all groups (Fig. S2C, ESI†). Four of the remaining 15 DEMs were also involved in the ferroptosis pathway, which were GSSG, arachidonic acid (AA), L-norleucine, and 5-oxoproline. Among them, the abundance of GSSG (A < B < C < D), AA (A < B < D < C) and L-norleucine (A > B > C > D) was positively, positively, and negatively correlated with FBG concentration, respectively (Fig. 4). The remaining eight metabolites were all associated with glycolysis metabolism or the tricarboxylic acid (TCA) cycle. These results suggest that ferroptosis and glycolytic disturbances may be intrinsic drivers of pre-diabetes progression.WGCNA cluster
All 683 metabolites were included in the WGCNA analysis. The similarity between metabolites was calculated according to the adjacency, the coefficient of dissimilarity between metabolites was deduced, and the systematic clustering tree between metabolites was obtained. As shown in Fig. S3 (ESI†), the metabolite profiles of the same group showed high biological reproducibility and clustered into the same branch, but the clustering of DEMs did not have a clear distinction between groups A and B. Moreover, in the present study, WGCNA clustered all DEM features into 8 different modules ranging from 39 to 175 in size, as identified by the hierarchical clustering tree, among which the gray module is one particular module (Fig. 5A). The heatmap showed the correlation (coefficient and p-value) between 8 modules and groups. A most positive correlation between the yellow module and C group of 0.68 (p-value = 2 × 10−11) was found; a positive correlation between the turquoise module and B group was 0.51 (p-value = 3 × 10−6), and a positive correlation between the blue module and D group of 0.48 (p-value = 1 × 10−5), and a positive correlation between the green module and D group of 0.45 (p-value = 4 × 10−5) were uncovered (Fig. 5B).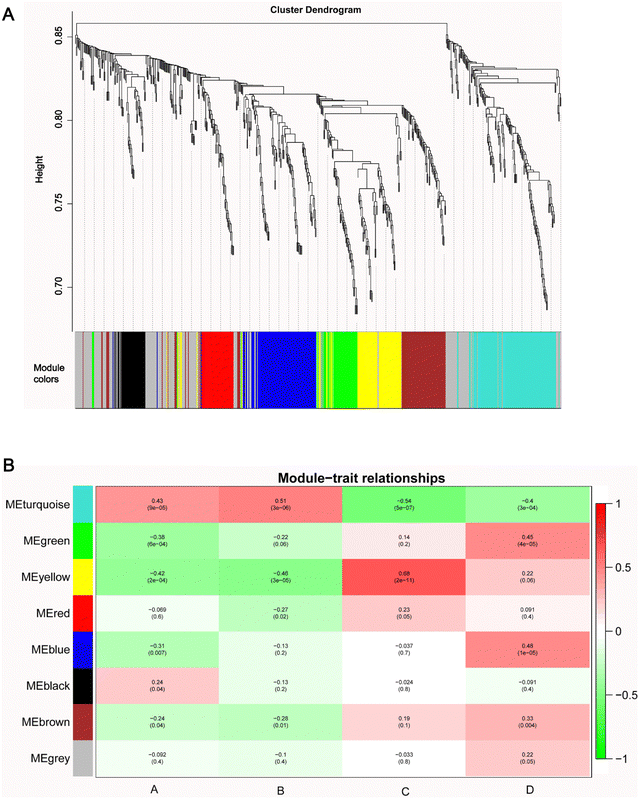 | ||
| Fig. 5 WGCNA network. (A) The clustering dendrogram identifying WGCNA modules. (B) Heatmap of modules–trait relationships. | ||
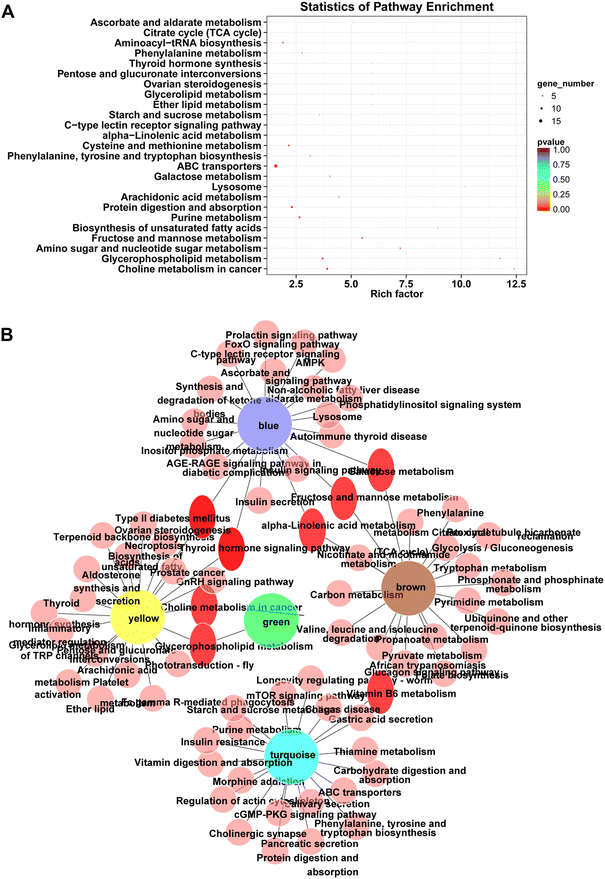
Next, the pathway enrichment analysis of metabolites in the eight modules was performed, and the results showed a significant annotation to insulin resistance-related pathways, such as thyroid hormone synthesis, starch and sucrose metabolism, galactose metabolism, fructose and mannose metabolism, and glycerophospholipid metabolism (Fig. 6A). Moreover, among the top 20 enrichment pathways of each module, the type 2 diabetes mellitus pathway and the thyroid hormone signaling pathway were shared by blue and yellow modules, whereas the insulin resistance pathway was unique to the turquoise module (Fig. 6B).
Assessment of diagnostic potential and predictive ability of DEMs
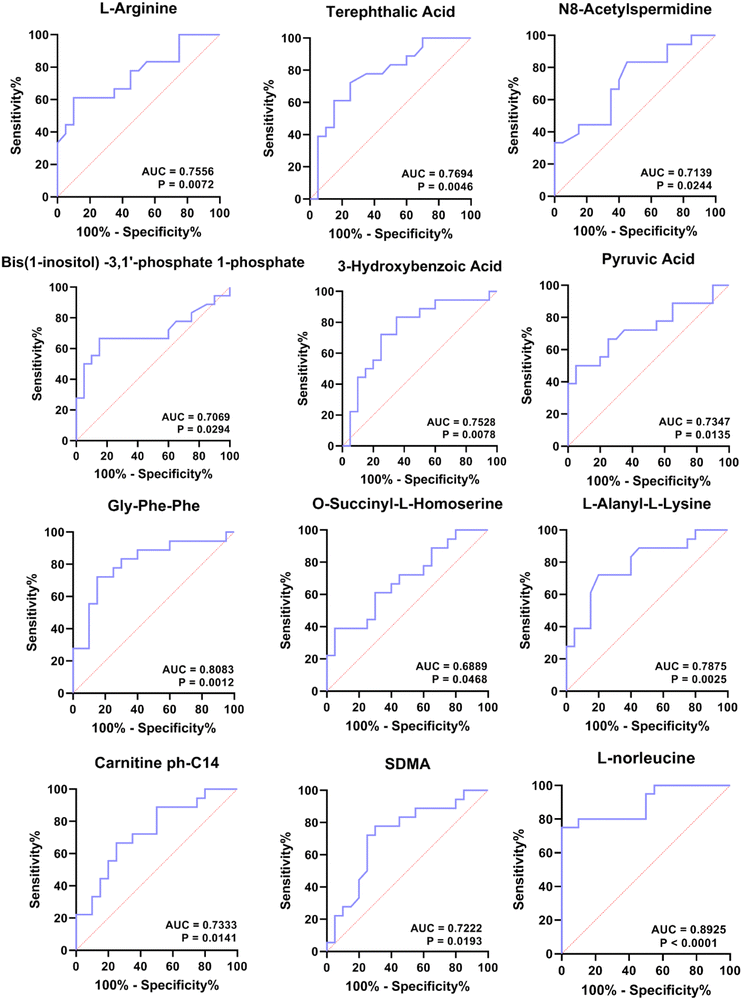
Finally, we sought to find metabolites that could be used to distinguish patients in the four periods. Since groups B, C, and D were significantly associated with the turquoise, yellow and blue modules, respectively, we focused on these three modules. To explore metabolites that can be used as potential biomarkers for diabetes, we intersected DEMs and metabolites in turquoise, yellow and blue modules to screen out candidate biomarkers in three stages of diabetes (Table 2). The results suggested that the 14 metabolites that could be used as disease markers in each group include amino acids and their derivatives, organic acids and their derivatives, and lyso-phosphatidylethanolamine (LPE), etc. (Table 2). Among them, ROC curve analysis was performed for individual metabolites to further determine the diagnostic potential of each metabolite (Fig. 7). The area under the curve (AUC) was used to evaluate the diagnostic performance of the above 14 candidate metabolites. Except for the unreliable biological predictive value of DL-2-methylglutamic acid and LPE (AUC of 0.5, data not shown), the AUC values of the remaining 11 metabolites were all greater than 0.7, such as AUC = 0.7556 (95% CI – 0.5986 to 0.9125, P = 0.0072) for L-arginine, AUC = 0.7694 (95% CI – 0.6168 to 0.9220, P = 0.0046) for terephthalic acid, AUC = 0.7139 (95% CI – 0.5492 to 0.8786, P = 0.0244) for N8-acetylspermidine, AUC = 0.7069 (95% CI – 0.5275 to 0.8864, P = 0.0294) for bis(1-inositol) −3,1′-phosphate 1-phosphate, AUC = 0.7528 (95% CI – 0.5925 to 0.9130, P = 0.0078) for 3-hydroxybenzoic acid, AUC = 0.6889 (95% CI – 0.5200 to 0.8578, P = 0.0468) for O-succinyl-L-homoserine, AUC = 0.8083 (95% CI – 0.6639 to 0.9527, P = 0.0012) for Gly-Phe-Phe, AUC = 0.7347 (95% CI – 0.5691 to 0.9003, P = 0.0135) for pyruvic acid, AUC = 0.7875 (95% CI – 0.6406 to 0.9344, P = 0.0025) for L-alanyl-L-lysine, AUC = 0.7333 (95% CI – 0.5737 to 0.8930, P = 0.0141) for carnitine ph-C14, and AUC = 0.7222 (95% CI – 0.5557 to 0.8887, P = 0.0193) for SDMA (Fig. 7). For ROC analysis, except for SDMA, which predicts for group D (AUC for C vs. D), the other 10 DEMs predict for group B or C (Table 2), and the results indicate that these 10 metabolites exhibit potential values in diagnosing pre-diabetes. Moreover, we also performed ROC analysis on L-norleucine, with AUC = 0.8925 (95% CI – 0.7901 to 0.9949, P < 0.0001), suggesting that the biological value of L-norleucine for differentiating pre-diabetes has high credibility (Fig. 7). Therefore, ROC analysis determined 11 potential biomarkers for pre-diabetes and one, specifically SDMA, for differentiating pre-diabetes and T2DM.| Biomarker | Group | Module | DEMs | Class II | A vs. B | B vs. C | C vs. D |
|---|---|---|---|---|---|---|---|
| B | B vs. C | Turquoise | L-Arginine | Amino acid derivatives | Unchanged | Down | Unchanged |
| B | B vs. C | Turquoise | Terephthalic acid | Phenolic acids | Unchanged | Down | Unchanged |
| B | B vs. C | turquoise | N8-Acetylspermidine | Amino acid derivatives | Unchanged | Down | Unchanged |
| B | B vs. C | Turquoise | Bis(1-inositol)-3,1′-phosphate 1-phosphate | Alcohols | Unchanged | Down | Unchanged |
| B | B vs. C | Turquoise | 3-Hydroxybenzoic acid | Organic acid and its derivatives | Unchanged | Down | Unchanged |
| B | B vs. C | Turquoise | O-Succinyl-L-homoserine | Amino acid derivatives | Unchanged | Down | Unchanged |
| B | B vs. C | Turquoise | Gly-Phe-Phe | Small peptide | Unchanged | down | Unchanged |
| B | B vs. C | Turquoise | DL-2-Methylglutamic acid | Amino acids | Down | Up | Unchanged |
| B, C, D | Turquoise | L-Norleucine | Amino acids | Down | Down | down | |
| C | B vs. C | Yellow | Pyruvic acid | Organic acid and its derivatives | Unchanged | Up | Unchanged |
| C | B vs. C | Yellow | L-Alanyl-L-lysine | Amino acid derivatives | unchanged | Up | Unchanged |
| C | B vs. C | Yellow | Carnitine ph-C14 | CAR | Unchanged | Up | Unchanged |
| D | C vs. D | Green | LPE(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
LPE | Unchanged | Unchanged | Up |
| D | C vs. D | Blue | SDMA | Organic acid and its derivatives | Unchanged | Unchanged | Up |
Discussion
The pandemic of T2DM has seriously threatened public health. Early prevention and screening for pre-diabetes and T2DM are important to reduce the health burden worldwide. At present, there are many metabolomic studies on patients with T2DM and its complications,11–13 but there are few studies on pre-diabetes in the context of T2DM. In the present study, we performed metabolome and WGCNA analysis on pre-diabetic patients without any intervention and patients initially diagnosed with T2DM. This preliminarily characterized metabolites associated with the pre-diabetic progression, and we revealed changes in profiles dominated by ferroptosis and glycolysis metabolism.In this study, we found that L-norleucine was the only metabolite shared in all the comparison groups (A vs. B, A vs. C, A vs. D, B vs. C, B vs. D, and C vs. D), and L-norleucine abundance was negatively correlated with T2DM progression, as L-norleucine expression diminished with rising FBG (the expression of L-norleucine was ranked A > B > C > D in the four groups), indicating that the lower the expression of L-norleucine, the more severe the insulin resistance. This phenomenon may be partly attributed to the insulin secretion-regulating function of L-norleucine. Actually, L-norleucine and L-leucine could stimulate insulin release in the presence of a suitable activator of glutamate dehydrogenase.14 Early studies on mice confirmed that dietary leucine reversed high-fat diet-induced insulin resistance, obesity, and inflammation.15 These studies support our conclusion that a low expression of L-norleucine contributes to the insulin resistance involved in T2DM progression. Therefore, changes in the level of L-norleucine may also be critical to the progression of T2DM and could be a reliable biomarker of pre-diabetes. Moreover, L-norleucine is an isomer of leucine which is a branched-chain amino acid (BCAA) (contains three amino acids, leucine, isoleucine, and valine) that is linked to insulin resistance in multiple studies.16 A study of plasma samples from obese and insulin resistant people found that the main components associated with insulin resistance were BCAAs, aromatic amino acids, carnitine, etc.17 However, the role of BCAAs in insulin resistance is still controversial. On the one hand, it is believed that BCAAs have a strong positive correlation with insulin resistance in the T2DM progression,18,19 but on the other hand, BCAA supplementation did not worsen insulin resistance.20 We speculate that this may be due to the unpredictable differential effects of different leucine isomers mixed with different amino acids. Unfortunately, the current study of L-norleucine in T2DM is insufficient, so there is a lack of strong evidence to support our conclusions; therefore, we will continue to study the significance of L-norleucine abundance changes in T2DM in the future. In addition, among the 75 amino acids and their metabolomics, the number of BCAAs and their derivatives was as high as 12, four of which were already significantly altered in concentration at FBG > 5.6, and the remaining eight showed different patterns with increasing FBG (Table S5, ESI†). In summary, BCAAs are consistently abnormal during the rise in FBG concentration, thus they are involved in the progression of pre-diabetes.
Surprisingly, among 27 shared DEMs, a total of 15 DEMs (11 LPCs, GSSG, AA, L-norleucine, and 5-Oxoproline) were involved in ferroptosis. Ferroptosis is a novel non-apoptotic death form, characterized by iron-dependent, membrane damage from lipid peroxidation. The dysregulation of ferroptosis is highly associated with a variety of diseases, including diabetes-related diseases and the induction of ferroptosis is also proposed as a potential strategy for them. For example, the suppression of pancreatic iron deposition and pancreatic β cells ferroptosis by quercetin significantly alleviates T2DM.21 Inhibition of ferroptosis by cryptochlorogenic acid protects β-cells’ function from diabetes.22 Ferroptosis and ferritinophagy also contribute to the occurrence and development of diabetes complications,23 including diabetes myocardial ischemia/reperfusion,24 diabetes-induced endothelial dysfunction,25 diabetic nephropathy,26,27 and diabetic atherosclerosis.28 Therefore, ferroptosis is involved in the occurrence and development of T2DM. Here, we superficially analyzed the mechanism of ferroptosis-mediated T2DM in conjunction with the identified-DEMs. First, the LPC lipid peroxidation pathway is driven by AA as a substrate. Abnormalities in AA and LPC initiate ferroptosis leading to pre-diabetes progression. Second, the GSH/GSSG pathway involves L-norleucine as the source of occurrence. Abnormal metabolism of L-norleucine, 5-oxoproline, and GSSG in this pathway initiates ferroptosis leading to pre-diabetes progression. Third, the ROS-glycolysis pathway is implicated. ROS, a byproduct of ferroptosis, not only enhances the occurrence of ferroptosis but also impairs insulin synthesis and secretion.29 In addition, ROS acts as a signaling molecule to assist in the cellular disposal of glucose uptake.30 Fourth, there is aberrant metabolism observed in the TCA cycle starting with 5-methylcytosine and ending with carnitine. The TCA cycle is based on glycolysis as the material basis, with L-glutamate/succinate to hook up the ferroptosis pathway. Therefore, abnormalities of 5-methylcytosine and carnitine as well as glycolysis-related metabolites (ribulose-5P, D-xylulose 5P, D-piperidine acid, and 1-pyrroline-4-hydroxy-2-carboxylate) are suggestive of disturbed glucose metabolism, which may be intrinsic to the progression of pre-diabetes. Taken together, we recommend treating diabetic patients with ferroptosis blockers and controlling pre-diabetic patients with dietary restrictions such as L-norleucine and AA.
In this study, a total of 12 biomarkers were predicted (Fig. 7). Interestingly, 7 of these metabolites are arginine or arginine-related urea cycle metabolites, including L-arginine, N8-acetylspermidine, O-succinyl-L-homoserine, Gly-Phe-Phe, terephthalic acid, 3-hydroxybenzoic acid, and bis(1-inositol)-3,1′-phosphate 1-phosphate. Arginine is a multifaceted amino acid because it can act as a precursor for many biologically active substances, such as nitric oxide (NO), agmatine, and proline.31 Emerging evidence showed that arginine is involved in diabetic progression by (1) synthesizing NO through nitric oxide synthase (NOS) to regulate diabetes-related pathways.32 Induced NOS-derived NO plays a central role in the regulation of several biochemical pathways including glucose and lipid metabolism and energy metabolism under inflammatory conditions.33,34 When the body is deficient in L-arginine, the iNOS/L-arginine pathway induces increased NO production, leading to aggravated inflammation and insulin resistance, ultimately contributing to the progression of diabetes.35,36 (2) Arginine is a secretagogue that stimulates the secretion of anabolic hormones, including insulin and glucagon, somatostatin, and adrenal catecholamine in a direct or indirect manner,37–39 which are all hormones associated with the progression of diabetes. These results suggest that abnormal arginine metabolism may be an important marker for the development of T2DM. ROC results in the present study supported the conclusion that arginine can be used as a predictive biomarker for pre-diabetes.
TCA cycle defects were also found in this study due to the energy production overload from glycolysis. In this study, we found that pyruvate did not change in the A vs. B comparison group but was significantly upregulated in the B vs. C comparison group, suggesting that when FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 6.1 mmol L−1, glycolysis overload produced excessive pyruvate. Pyruvate is an intermediate between glycolysis and TCA cycle. Excess pyruvate requires the consumption of large amounts of oxaloacetic acid to maintain the normal functioning of the TCA cycle, which is accompanied by rapid production and consumption of intermediates. We found that the abundance of TCA intermediates such as α-ketoglutaric acid, methylmalonic acid, and citramalic acid accumulated significantly at FBG
6.1 mmol L−1, glycolysis overload produced excessive pyruvate. Pyruvate is an intermediate between glycolysis and TCA cycle. Excess pyruvate requires the consumption of large amounts of oxaloacetic acid to maintain the normal functioning of the TCA cycle, which is accompanied by rapid production and consumption of intermediates. We found that the abundance of TCA intermediates such as α-ketoglutaric acid, methylmalonic acid, and citramalic acid accumulated significantly at FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 6.1 mmol L−1. The results of some studies are similar to ours. For example, mitochondrial pyruvate transporter protein activity is reduced in diabetic rats;40 pyruvate and malate are increased simultaneously in the TCA cycle after glucose loading;41 excess pyruvate in pre-diabetic Zucker (fa/fa) rats can lead to increased TCA cycle and mitochondrial overload.42 Therefore, we conclude that when FBG is elevated to 6.1, glycolytic overload produces excess pyruvate causing subsequent TCA cycle overload; however, when a diagnosis of T2DM is confirmed, that is FBG
6.1 mmol L−1. The results of some studies are similar to ours. For example, mitochondrial pyruvate transporter protein activity is reduced in diabetic rats;40 pyruvate and malate are increased simultaneously in the TCA cycle after glucose loading;41 excess pyruvate in pre-diabetic Zucker (fa/fa) rats can lead to increased TCA cycle and mitochondrial overload.42 Therefore, we conclude that when FBG is elevated to 6.1, glycolytic overload produces excess pyruvate causing subsequent TCA cycle overload; however, when a diagnosis of T2DM is confirmed, that is FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 7.0 mmol L−1, injury occurs after TCA cycle overload, and the TCA cycle exhibits a low-flow state.
7.0 mmol L−1, injury occurs after TCA cycle overload, and the TCA cycle exhibits a low-flow state.
Among the 324 DEMs, lipids were the most abundant species with 114, 51 of which were significantly altered at FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 5.6 mmol L−1 (A vs. B) (Table S5, ESI†), suggesting that abnormal blood glucose can cause lipid metabolism disorder. For example, the abundance of AA (an essential fatty acid) and FFA (20
5.6 mmol L−1 (A vs. B) (Table S5, ESI†), suggesting that abnormal blood glucose can cause lipid metabolism disorder. For example, the abundance of AA (an essential fatty acid) and FFA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) was significantly elevated at FBG
4) was significantly elevated at FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 5.6 mmol L−1 (A vs. B). Consistent with our results, studies have demonstrated that blood glucose overload can lead to glycolytic overload, resulting in an abnormal increase in lipid metabolites.43 Abnormal lipid accumulation, such as lysophosphatidic acid, can exert negative feedback to further enhance hepatic insulin resistance.44 Moreover, the main mode of fatty acid catabolism is β-oxidation, wherein fatty acid is activated to lipid acyl coenzyme A and then transported to mitochondria via carnitine; therefore, carnitine is considered an important marker for the evaluation of metabolic disorders such as mitochondrial dysfunction and insulin resistance.45,46 In the present study, a partial imbalance in carnitine levels (such as carnitine C24
5.6 mmol L−1 (A vs. B). Consistent with our results, studies have demonstrated that blood glucose overload can lead to glycolytic overload, resulting in an abnormal increase in lipid metabolites.43 Abnormal lipid accumulation, such as lysophosphatidic acid, can exert negative feedback to further enhance hepatic insulin resistance.44 Moreover, the main mode of fatty acid catabolism is β-oxidation, wherein fatty acid is activated to lipid acyl coenzyme A and then transported to mitochondria via carnitine; therefore, carnitine is considered an important marker for the evaluation of metabolic disorders such as mitochondrial dysfunction and insulin resistance.45,46 In the present study, a partial imbalance in carnitine levels (such as carnitine C24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and DL-carnitine) was already present at FBG
2 and DL-carnitine) was already present at FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 5.6 mmol L−1. We hypothesize that the body's fatty acid requirements have been disrupted, and there is an overload in the transport of lipids through carnitine. In short, in the early stage of pre-diabetes (FBG
5.6 mmol L−1. We hypothesize that the body's fatty acid requirements have been disrupted, and there is an overload in the transport of lipids through carnitine. In short, in the early stage of pre-diabetes (FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 5.6 mmol L−1), increased glycolytic load initiates abnormalities in lipid metabolism, leading to further deterioration of diabetes.
5.6 mmol L−1), increased glycolytic load initiates abnormalities in lipid metabolism, leading to further deterioration of diabetes.
Experimental section
Subjects
This is a cross-sectional study. All the subjects (n = 80) in this study were from The First People's Hospital of Yinchuan.Inclusion criteria: all pre-diabetes and T2DM patients were newly diagnosed and did not receive any treatment. Enrolled subjects met the 2019 US American Diabetes Association (ADA) diagnostic criteria for pre-diabetes and diabetes. According to the criteria of the Chinese Diabetes Society (CDS) about FBG, pre-diabetes was defined as 6.1 mmol L−1 ⩽ FBG < 7.0 mmol L−1, whereas according to the criteria of the ADA about FBG, pre-diabetes was defined as 5.6 mmol L−1 ⩽ FBG < 6.9 mmol L−1. Due to the difference in the diagnostic criteria between China and America, we want to explore the abnormal metabolic changes that occur in Chinese T2DM patients when their FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 5.6 mmol L−1, to provide a theoretical basis for the early prevention and control of diabetes in China. Therefore, subjects were divided into four groups according to FBG values: group A (n = 20), FBG < 5.6 mmol L−1, healthy people with normal glucose tolerance; group B (n = 20), 5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1, the early stage of pre-diabetes patients; group C (n = 20), 6.1 mmol L−1 ⩽ FBG < 7.0 mmol L−1, the late stage of pre-diabetes patients; group D (n = 20), FBG
5.6 mmol L−1, to provide a theoretical basis for the early prevention and control of diabetes in China. Therefore, subjects were divided into four groups according to FBG values: group A (n = 20), FBG < 5.6 mmol L−1, healthy people with normal glucose tolerance; group B (n = 20), 5.6 mmol L−1 ⩽ FBG < 6.1 mmol L−1, the early stage of pre-diabetes patients; group C (n = 20), 6.1 mmol L−1 ⩽ FBG < 7.0 mmol L−1, the late stage of pre-diabetes patients; group D (n = 20), FBG ![[greater than or equal, slant]](https://www.rsc.org/images/entities/char_2a7e.gif) 7.0 mmol L−1, T2DM patient. Details of each subject are shown in Table S1 (ESI†).
7.0 mmol L−1, T2DM patient. Details of each subject are shown in Table S1 (ESI†).
Exclusion criteria: Type 1 diabetes and other special types of diabetes; individuals with diabetes-related acute or chronic complications; various secondary hypertensive conditions; those with chronic kidney or liver diseases, or cancer; absence of anemia and a history of severe cardiovascular, cerebrovascular diseases, or tumors; recent lack of subjective meal management in daily life; no recent blood donation or blood product transfusion events; no history of drug abuse or other psychotropic medications; females who are pregnant or in the lactation period, and so forth.
All subjects provided signed informed consent. The study was approved by the First People's Hospital of Yinchuan Ethics Committee. All experiments were conducted in strict accordance with the relevant laws in China and followed the institutional guidelines of The First People's Hospital of Yinchuan.
Sample preparation
All subjects fasted overnight for at least 12 h before blood collection. The collected venous blood samples were allowed to stand at room temperature for 1 h for coagulation and stratification47 and then centrifuged at 2500g for 4 min. The supernatant was retained and centrifuged again for 10 min, and finally the supernatant was collected and stored at −80 °C for the subsequent extraction study.For metabolite extraction, 1 mL of 80% methanol internal standard extractant was added to the plasma samples, followed by vortexing for 2 min, quick freezing in liquid nitrogen for 5 min, thawing in an ice bath for 5 min, and repeated freezing and thawing three times. Finally, the plasma sample was centrifuged at 12 000 rpm for 10 min at 4 °C, and 200 μL of the supernatant was collected for subsequent liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis (20 samples per group).
UPLC conditions
The ultra performance liquid chromatography (UPLC; ExionLC AD, https://sciex.com.cn/) system equipped with a C18 column (Waters ACQUITY UPLC HSS T3, 1.8 μm, 2.1 mm × 100 mm) was used for molecular separation. The column temperature was 40 °C. Liquid phase A consisted of 0.1% formic acid aqueous solution, and liquid phase B consisted of a 0.1% formic acid acetonitrile aqueous solution. The liquid phase gradient was as follows: 0–11 min, liquid phase B maintained at 5%; 11–12 min, liquid phase B maintained at 90%; 12–14 min, liquid phase B maintained at 5%. Flow rate was 0.4 mL min−1 over 14 min, and the injection volume was 2 μL.ESI-Q TRAP-MS/MS
After separation by UPLC, linear ion trap (LIT) and triple quadrupole scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Qtrap), QTRAP® LC-MS/MS System (Applied Biosystems 6500+ Quadrupole Trap, https://sciex.com.cn/), equipped with an ESI Turbo Ion-Spray interface, operating in positive and negative ion mode and controlled by Analyst 1.6.3 software (Sciex). The ESI source operation parameters were as follows: for electrospray ionization, gas temperature, 500 °C; ion spray voltage for positive and negative ion mode was, respectively, 5500 V and −4500 V; ion source gases I and II were, respectively, 55 psi and 60 psi; curtain gas, 25 psi; collision-activated dissociation, high. Instrument tuning and mass calibration were performed using 10 and 100 μmol L−1 polypropylene glycol solutions in triple quadrupole and LIT modes, respectively. A specific set of multiple reaction monitoring (MRM) transitions was monitored for each period according to the metabolites eluted within this period. An internal standard, L-2-chlorophenylalanine (106151-100 mg, J&K Scientific, China), was used for data quality control (QC) and normalization.Disregarding background noise, signals with peak heights below 4000 were not picked up. Data were normalized by dividing each metabolite for each sample by the total peak area for that sample. Based on the self-built target standard database, identification analysis of molecules was carried out according to the retention time of the detected molecules, the information of the parent ion pair and the MS2 data. Each molecule was quantified using a targeted multiple reaction monitoring approach based on the peak area of the chromatographic peak on the MultiQuant software. Based on the metabolite abundance obtained above, PCA and OPLS-DA were performed for each sample using the R (v 3.3.2) package. Fold changes of molecules were determined between the different groups using the t test to assess whether differences between the groups were statistically significant. The screening criterion for differentially expressed metabolites (DEMs) was fold change >1, P < 0.05 and variable importance in the projection (VIP) > 1. GO and KEGG enrichment analysis was carried out for the DEMs using the R package of ClusterProfiler. UPLC-MS/MS and bioinformatics analysis were performed by Beijing Biomarker Technology (Beijing, China). Raw metabolite data are available in the Metabolights Database at https://www.ebi.ac.uk/metabolights/MTBLS8644.
Weighted gene co-expression network analysis (WGCNA)
We performed WGCNA on all the metabolites using an R package of WGCNA. Different colors were used to represent different metabolites when visualizing co-expression modules, among which gray defaulted to metabolites that cannot be classified in any module. Correlation analysis was performed between modules and each group of samples, and between modules and clinical phenotype, to screen biomarkers for T2DM.Statistical analysis
For clinical data of study cohort, statistical analysis was performed using Graphpad Prism 9.00. Normal distribution was determined by the Shapiro-Wilk test, and the Brown–Forsythe test was used for the homogeneity of variances. The one-way ANOVA with Tukey's test was used when the data were normality and homogeneity of variance, otherwise, the Kruskal–Wallis test followed by the Dunn's test for nonparametric data was used. P < 0.05 was considered significant.Conclusions
In conclusion, we reveal metabolite changes from healthy individuals to T2DM patients as the FBG concentration increases. Among them, the level of L-norleucine was significantly downregulated in the pre-diabetes group and T2DM group, with a correlation of FBG concentration. Moreover, ROC analysis and WGCNA together determined 11 biomarkers for pre-diabetes, 7 of which were L-arginine or its metabolites, and SDMA for predicted T2DM. L-Norleucine and L-arginine have great potential to differentiate healthy people from those with pre-diabetes. Hopefully, our plasma metabolite inventory could possibly help physicians understand the metabolism alteration in the progression of T2DM so as to predict and even diagnose T2DM early.Data availability
Raw metabolite data are available in the Metabolights Database at https://www.ebi.ac.uk/metabolights/MTBLS8644.Author contributions
J. W. R. and X. X. M. contributed equally in this study. J. W. R. and X. X. M. came up with the conception and revised the final manuscript. Q. D., J. W. R., and X. X. M. wrote the draft manuscript and participated in execution. H. Y. T., Q. D., J. W. R., and X. X. M. completed acquisition of data. L. L., H. Y. T., Q. D., J. W. R., and X. X. M. were in charge of analysis and interpretation. Z. L., L. L., H. Y. T., Q. D., J. W. R., and X. X. M. took part in the literature search. Z. L., L. L., H. Y. T., Q. D., and J. W. R. gave suggestions for revision. X. X. M. acquired the funding. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Not applicable.References
- Z. Xu, J. Geng, S. Zhang, K. Zhang, L. Yang, J. Li and J. Li, JMIR mHealth uHealth, 2020, 8, e19869 CrossRef PubMed.
- L. Guariguata, D. Whiting, C. Weil and N. Unwin, Diabetes Res. Clin. Pract., 2011, 94, 322–332 CrossRef PubMed.
- L. E. Joensen, K. P. Madsen, L. Holm, K. A. Nielsen, M. H. Rod, A. A. Petersen, N. H. Rod and I. Willaing, Diabetic Med.: J. Br. Diabetic Assoc., 2020, 37, 1146–1154 CrossRef CAS PubMed.
- R. M. M. Khan, Z. J. Y. Chua, J. C. Tan, Y. Yang, Z. Liao and Y. Zhao, Medicina, 2019, 55, 546 CrossRef PubMed.
- J. Beulens, F. Rutters, L. Ryden, O. Schnell, L. Mellbin, H. E. Hart and R. C. Vos, Eur. J. Prev. Cardiol., 2019, 26, 47–54 CrossRef PubMed.
- J. J. Matthews, E. Dolan, P. A. Swinton, L. Santos, G. G. Artioli, M. D. Turner, K. J. Elliott-Sale and C. Sale, Syst. Rev., 2020, 9, 282 CrossRef PubMed.
- F. Bragg, E. Trichia, D. Aguilar-Ramirez, J. Besevic, S. Lewington and J. Emberson, BMC Med., 2022, 20, 159 CrossRef CAS PubMed.
- C. Zhu, Q. L. Liang, P. Hu, Y. M. Wang and G. A. Luo, Talanta, 2011, 85, 1711–1720 CrossRef CAS PubMed.
- J. Long, Z. Yang, L. Wang, Y. Han, C. Peng, C. Yan and D. Yan, BMC Endocr. Disord., 2020, 20, 174 CrossRef CAS PubMed.
- L. Zhu, Q. Huang, X. Li, B. Jin, Y. Ding, C. J. Chou, K. J. Su, Y. Zhang, X. Chen, K. Y. Hwa, S. Thyparambil, W. Liao, Z. Han, R. Mortensen, Y. Jin, Z. Li, J. Schilling, Z. Li, K. G. Sylvester, X. Sun and X. B. Ling, Front Mol. Biosci., 2022, 9, 841209 CrossRef CAS PubMed.
- Z. Huang, L. Klaric, J. Krasauskaite, S. McLachlan, M. W. J. Strachan, J. F. Wilson and J. F. Price, Cardiovasc. Diabetol., 2022, 21, 62 CrossRef PubMed.
- Z. Yang, W. Dan, Y. Li, X. Zhou, T. Liu, C. Shi, R. Li, Y. Zhang, J. Zhang, J. Yan, X. Zhu, Y. Li, M. Gong, C. Wang, C. Yuan, Y. Cui and X. Wu, Biomed. Pharmacother. Biomed. Pharmacother., 2022, 146, 112495 CrossRef CAS PubMed.
- L. Michalkova, S. Hornik, J. Sykora, L. Habartova, V. Setnicka and B. Bunganic, J. Proteome Res., 2021, 20, 1744–1753 CrossRef CAS PubMed.
- A. Sener, F. Malaisse-Lagae and W. J. Malaisse, J. Biol. Chem., 1983, 258, 6693–6694 CrossRef CAS PubMed.
- Y. Macotela, B. Emanuelli, A. M. Bang, D. O. Espinoza, J. Boucher, K. Beebe, W. Gall and C. R. Kahn, PLoS One, 2011, 6, e21187 CrossRef CAS PubMed.
- C. Nie, T. He, W. Zhang, G. Zhang and X. Ma, Int. J. Mol. Sci., 2018, 19, 954 CrossRef PubMed.
- C. B. Newgard, Cell Metab., 2012, 15, 606–614 CrossRef CAS PubMed.
- M. Guasch-Ferre, A. Hruby, E. Toledo, C. B. Clish, M. A. Martinez-Gonzalez, J. Salas-Salvado and F. B. Hu, Diabetes Care, 2016, 39, 833–846 CrossRef CAS PubMed.
- I. Owei, N. Umekwe, F. Stentz, J. Wan and S. Dagogo-Jack, Metabolism: Clin. Exp., 2019, 98, 76–83 CrossRef CAS PubMed.
- J. Lee, A. Vijayakumar, P. J. White, Y. Xu, O. Ilkayeva, C. J. Lynch, C. B. Newgard and B. B. Kahn, Endocrinology, 2021, 162, bqab062 CrossRef PubMed.
- D. Li, C. Jiang, G. Mei, Y. Zhao, L. Chen, J. Liu, Y. Tang, C. Gao and P. Yao, Nutrients, 2020, 12, 2954 CrossRef CAS PubMed.
- Y. Zhou, Diabetes, Metab. Syndr. Obes., 2020, 13, 1921–1931 CrossRef CAS PubMed.
- J. He, Z. Li, P. Xia, A. Shi, X. FuChen, J. Zhang and P. Yu, Mol. Metab., 2022, 60, 101470 CrossRef CAS PubMed.
- W. Li, W. Li, Y. Leng, Y. Xiong and Z. Xia, DNA Cell Biol., 2020, 39, 210–225 CrossRef CAS PubMed.
- E. F. Luo, H. X. Li, Y. H. Qin, Y. Qiao, G. L. Yan, Y. Y. Yao, L. Q. Li, J. T. Hou, C. C. Tang and D. Wang, World J. Diabetes, 2021, 12, 124–137 CrossRef PubMed.
- S. Li, L. Zheng, J. Zhang, X. Liu and Z. Wu, Free Radic. Biol. Med., 2021, 162, 435–449 CrossRef CAS PubMed.
- X. Feng, S. Wang, Z. Sun, H. Dong, H. Yu, M. Huang and X. Gao, Front Endocrinol., 2021, 12, 626390 CrossRef PubMed.
- Z. Meng, H. Liang, J. Zhao, J. Gao, C. Liu, X. Ma, J. Liu, B. Liang, X. Jiao, J. Cao and Y. Wang, Life Sci., 2021, 284, 119935 CrossRef CAS PubMed.
- W. Sha, F. Hu, Y. Xi, Y. Chu and S. Bu, J. Diabetes Res., 2021, 2021, 9999612 Search PubMed.
- K. Rehman and M. S. H. Akash, J. Cell. Biochem., 2017, 118, 3577–3585 CrossRef CAS PubMed.
- T. Wu, C. Wang, L. Ding, Y. Shen, H. Cui, M. Wang and H. Wang, Mediators Inflammation, 2016, 2016, 9618795 Search PubMed.
- S. Anavi and O. Tirosh, Free Radical Biol. Med., 2020, 146, 16–35 CrossRef CAS PubMed.
- I. L. A. A. Marti and W. Reith, Cellular and molecular life sciences: CMLS, 2021, 78, 5303–5324 CrossRef PubMed.
- S. Hu, M. Han, A. Rezaei, D. Li, G. Wu and X. Ma, Curr. Protein Pept. Sci., 2017, 18, 599–608 CrossRef CAS PubMed.
- L. D. Monti, E. Galluccio, V. Villa, B. Fontana, S. Spadoni and P. M. Piatti, Eur. J. Nutr., 2018, 57, 2805–2817 CrossRef CAS PubMed.
- J. Cho, Y. Horikawa, M. Enya, J. Takeda, Y. Imai, Y. Imai, H. Handa and T. Imai, Commun. Biol., 2020, 3, 497 CrossRef CAS PubMed.
- P. Newsholme, L. Brennan, B. Rubi and P. Maechler, Clin. Sci., 2005, 108, 185–194 CrossRef CAS PubMed.
- N. Honzawa, K. Fujimoto, M. Kobayashi, D. Kohno, O. Kikuchi, H. Yokota-Hashimoto, E. Wada, Y. Ikeuchi, Y. Tabei, G. W. Dorn II, K. Utsunomiya, R. Nishimura and T. Kitamura, Int. J. Mol. Sci., 2022, 23, 4003 CrossRef CAS PubMed.
- J. Cordoba-Chacon, M. D. Gahete, A. I. Pozo-Salas, J. P. Castano, R. D. Kineman and R. M. Luque, Endocrinology, 2013, 154, 2393–2398 CrossRef CAS PubMed.
- A. Kielducka, G. Paradies and S. Papa, J. Bioenerg. Biomembr., 1981, 13, 123–132 CrossRef CAS PubMed.
- O. Shaham, R. Wei, T. J. Wang, C. Ricciardi, G. D. Lewis, R. S. Vasan, S. A. Carr, R. Thadhani, R. E. Gerszten and V. K. Mootha, Mol. Syst. Biol., 2008, 4, 214 CrossRef PubMed.
- E. S. Jin, B. H. Park, A. D. Sherry and C. R. Malloy, Diabetes, 2007, 56, 777–785 CrossRef CAS PubMed.
- J. Lund, D. M. Ouwens, M. Wettergreen, S. S. Bakke, G. H. Thoresen and V. Aas, Cells, 2019, 8, 1101 CrossRef CAS PubMed.
- C. A. Nagle, J. An, M. Shiota, T. P. Torres, G. W. Cline, Z. X. Liu, S. Wang, R. L. Catlin, G. I. Shulman, C. B. Newgard and R. A. Coleman, J. Biol. Chem., 2007, 282, 14807–14815 CrossRef CAS PubMed.
- N. Longo, M. Frigeni and M. Pasquali, Biochim Biophys Acta, 2016, 1863, 2422–2435 CrossRef CAS PubMed.
- R. L. Mynatt, Diabetes/Metab. Res. Rev., 2009, 25(Suppl 1), S45–S49 CAS.
- A. M. Kauppi, A. Edin, I. Ziegler, P. Mölling, A. Sjöstedt, Å. Gylfe, K. Strålin and A. Johansson, PloS One, 2016, 11, e0147670 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3mo00130j |
| ‡ Wenrui Ji and Xiaomin Xie contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |