Open Access Article
Open Access ArticleA comprehensive review on ginger-derived exosome-like nanoparticles as feasible therapeutic nano-agents against diseases
Faegheh
Bahri
a,
Mahna
Mansoori
 b,
Shayan
Vafaei
b,
Shayan
Vafaei
 b,
Saba
Fooladi
c,
Yousof
Mir
d,
Mehrnaz
Mehrabani
e,
Yaser
Hozhabri
d,
Mohammad Hadi
Nematollahi
b,
Saba
Fooladi
c,
Yousof
Mir
d,
Mehrnaz
Mehrabani
e,
Yaser
Hozhabri
d,
Mohammad Hadi
Nematollahi
 *f and
Siavash
Iravani
*f and
Siavash
Iravani
 *g
*g
aHerbal, and Traditional Medicines Research Center, School of Pharmacy, Kerman University of Medical Sciences, Kerman, Iran
bDepartment of Anatomical Sciences, Afzalipour School of Medicine, Kerman University of Medical Sciences, Kerman, Iran
cYale Cardiovascular Research Center, Section of Cardiovascular Medicine, Department of Internal Medicine, Yale School of Medicine, New Haven, CT 06511, USA
dApplied Cellular and Molecular Research Center, Kerman University of Medical Sciences, Kerman, Iran
ePhysiology Research Center, Institute of Neuropharmacology, Kerman University of Medical Sciences, Kerman, Iran
fNeuroscience Research Center, Institute of Neuropharmacology, Kerman University of Medical Sciences, Kerman, Iran. E-mail: mh.nematollahi@yahoo.com
gIndependent Researcher, W Nazar ST, Boostan Ave, Isfahan, Iran. E-mail: siavashira@gmail.com
First published on 20th January 2024
Abstract
Plant-derived exosome-like nanoparticles (PDENs) are lipid-membrane nanovesicles derived from different edible plant species (e.g., ginger, grape, carrot, and lemon), showing therapeutic applications (e.g., anticancer, anti-inflammatory, and antidiabetic activities). In this context, ginger-derived exosome-like nanoparticles (GDENs) derived from ginger (Zingiber officinale Roscoe) have shown versatile biomedical potentials, including anticancer and anti-inflammatory effects. These nanoparticles with salient advantages of a suitable internalization rate, low immunogenicity/toxicity, stability in the gastrointestinal tract, and the ability to pass the blood–brain barrier can be considered potential candidates for drug delivery and cancer therapy. However, challenges still exist regarding their separation/isolation processes, optimal preparation conditions, and industrialization/commercialization, as well as clinical translation studies and systematic in vitro/in vivo analyses. GDENs are considered an accessible, low-cost drug, or even drug carriers, for different diseases owing to their potential applications such as bioactive molecules, therapeutics mRNAs, enzymes, and other cargoes. In this review, our primary objective is to delve into the most recent breakthroughs in the field of GDENs. We have extensively examined their intricate structures, essential components, various isolation techniques, and the wide range of biomedical applications they offer. In addition, we have discussed the significant challenges that researchers face when working with GDENs and shed light on the future prospects of this innovative technology.
1. Introduction
Herbal plants and their derivatives are popular medications for several diseases such as pulmonary fibrosis,1 asthma,2 hepatitis,3 parasitic infections, bacterial infections, viral infections,4–6 tendinitis,7 burn injury,8 cancer,9,10 inflammation, and oxidative stress.11Zingiber officinale Roscoe, commonly known as ginger, is a member of the Zingiberaceae family and Zingiber genus, which is used as a food flavoring and a herbal medicine worldwide, such as in Iran, China, and India. Traditionally, its rhizomes (ginger root) have been used worldwide, especially in Asian countries.12–14 Scientists have identified that ginger has many bioactive constituents. For instance, it is rich in phenolic compounds, including gingerols, shogaols, and paradols; terpene compounds, including zingiberene and curcumin; polysaccharides; lipids; organic acids; and raw fibers. Apparently, the bioactivity and probably the therapeutic effects of ginger come from these compounds, and all of these together lead to the widespread use of ginger in research and medicine, which is further discussed herein.15,16Traditional health care providers used ginger rhizomes that were either fresh, dried, or processed by stir-frying.17,18 Chemical analysis showed that ginger consists of carbohydrates (60–70%), proteins (9%), water (9–12%), fatty oil (3–6%), crude fiber (3–6%), volatile oil (2–3%), and some minerals (such as calcium, iron, magnesium, phosphorus, potassium, sodium, and zinc). Phenolic and terpene compounds are thought to be responsible for the pharmacological activity of ginger, found in volatile oil and non-volatile components.19,20 Ginger is rich in phenols including gingerols, 6-shogaols, and 6-paradols, terpene compounds including zingiberene, and curcumin. For more information please refer to Table 1.15 Li et al. demonstrated that ginger components are stable at 4 °C for one day and at −20 °C for one month. These components, when administered to animals, decrease very fast in the body (after 3 hours) and are metabolized by the liver and kidneys.21 The U.S. FDA in 2016 classified ginger as “Generally Recognized as Safe” and the German Commission Monographs describes it as not having any studied after-effects or pharmacological interactions.22,23 Active ginger components used as therapeutic agents in various pathological conditions are summarized in Table 1.
| Group | Compounds | Chemical structure | Therapeutic effect |
|---|---|---|---|
| Monoterpene | Borneol |
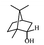
|
Reduces pain and inflammation in Mice24 |
| Phellandrene |
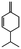
|
Reduces pain and tumorigenesis in mice25 | |
| Sesquiterpene | α-Zingiberene |
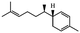
|
Subcutaneous implants of α-zingiberene suppress angiogenesis due to inflammation and increase collagen depo in mice26 |
| Zerumbone |
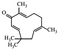
|
Shows anti-cancer effects27 | |
| Phenols | Gingerols |
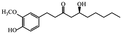
|
Show anticancer effects28 |
| Shogaols |
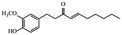
|
Ameliorate aging signs and degenerative diseases29 | |
| Paradols |
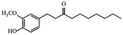
|
Show anticancer effect by affecting signaling pathways30 | |
| Zingerone |
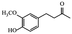
|
Different therapeutic properties31 | |
| Heptanoid | Curcumin |
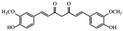
|
Different pharmacological effects32 |
| Gingerenone |
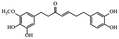
|
Reduces obesity and its inflammation in fat mice33 |
Due to the high antioxidant components in the ginger rhizome, its anti-inflammatory and antioxidant effects have been reported in past studies.34,35 For instance, Ji et al. have shown the protective effects of ginger against ionizing radiation in human mesenchymal stem cells (hMSCs) by a reduction in reactive oxygen species (ROS) production related to Nrf2 and its downstream genes heme oxygenase-1 (HO-1) and NAD(P)H quinone oxidoreductase 1 (NQO-1) expressions.36 Morvaridzadeh et al. in a meta-analysis in 2020 indicated that ginger can reduce inflammatory markers including C-reactive protein (CRP), high sensitivity C-reactive protein (hs-CRP), and tumor necrosis factor-alpha (TNF-α) levels.37 The beneficial effects of ginger in reducing neuro-inflammation by inhibiting nitric oxide, IL-1β, IL-6, and TNF-α mRNA levels and lipopolysaccharide-activated BV2 microglia production,38 and its effects in reducing anxiety symptoms in mice are proven.39 The pain relief effects of ginger in some clinical trials have also been shown previously. It is assumed that the pain relief action of ginger is probably related to anti-inflammatory and anti-oxidant effects.40 In the study by Kashefi et al., it was proved that ginger can relieve dysmenorrhea pain in young women.41 Likewise, studies have suggested that ginger is probably effective in the cure of rheumatoid arthritis by FoxP3 enhancement and reduction of RORγt and T-bet gene expression.42 In 2014, Bartels et al. designed a meta-analysis to show the safety and efficiency of ginger for clinical use in osteoarthritis. Accordingly, they demonstrated that ginger is efficient in treating osteoarthritis pain and disability in comparison to placebo, and it has no reported adverse side effects so it can be considered as safe and efficient in osteoarthritis patients.43
Considering the anti-oxidant and anti-inflammation properties of ginger, it seems that it has potent therapeutic effects against cancers. The effects of ginger on cancer were found in previous in vitro/in vivo research in addition to clinical studies. It works through impression on some molecular pathways including NF-κB, STAT3, Rb, MAPK, PI3K, Akt, ERK, cIAP1, cyclin A, cyclin-dependent kinase (Cdk), cathepsin D, and caspase-3/7, displaying antitumor, antiproliferative, and anti-invasive properties on cancers, specifically gastrointestinal (GI) ones.44 Yousef et al. demonstrated that ginger can decrease lipid peroxidation and scavenge free radical formation resulting in the suppression of carcinogenesis in the liver.45 The remarkable thing besides the anti-cancer properties of ginger is its effect on nausea and vomiting reduction after chemotherapy.46 It has been concluded that ginger's antiemetic effects are due to serotonin receptor inhibition in the GI and central nervous systems.47 Some clinical trials have also shown that ginger can reduce nausea and vomiting in different conditions such as post-anesthesia, during pregnancy, and motion sickness.48,49
There have been a lot of studies about the therapeutic effects of ginger in various conditions apart from those mentioned above. These include cardiovascular problems50 and respiratory diseases,51 for example, asthma.52 Li et al. in a study in 2017 suggested that ginger can protect from ischemia/reperfusion-induced harm in intestinal mucosa to affect ROS and p38 activity.53 Also, ginger's anti-microbial and anti-bacterial effects have been accepted.54 Some studies confirmed by in silico analysis that ginger has a positive influence on influenza-like symptoms and can inhibit Influenza H1N1 Neuraminidase protein.55,56
2. Different formulations of ginger
As mentioned above, besides the flavoring application of ginger, it has been used in folk medicine for therapeutic purposes.57 Ginger has been used to treat some diseases traditionally, including GI problems, infections, inflammation and inflammatory diseases, colds, fever, pain, etc.,58,59 and different formulations have been used. Traditionally, the fresh rhizome and the dried product have been used as a spice and efficient drug.44,60 Nowadays, ginger is used alone in different formulations or with other products as a treatment. It can be in the form of pills, tablets, or capsules in addition to its fresh and dried rhizome.60 Some studies use its powder for therapeutic procedures.22 It is also used topically, especially in the form of oil for pain relief.61 Its leaves and flowers are rich in antioxidant compounds and have been used in studies chiefly for antioxidant activities.62 Nowadays nano-lipid vectors based on ginger are used as an efficient drug delivery system in studies.63 In this regard, 6-shogaol-loaded micelles showed more efficiency than free 6-shogaol in hepatoprotective and anti-tumor activities.64Secretomes, which are secreted from different cells, can contain growth factors, cytokines, proteins, DNA, RNA, and extracellular vesicles (EVs).65 EVs are defined as nanoparticles (NPs) with a lipid bilayer membrane, carrying variable molecules including proteins, lipids, and nucleic acids.66 The EV contents are protected from damage in the extracellular space due to this bilayer lipid membrane.67 They can be ordered into three major groups by their size: exosomes (30–100 nm), ectosomes (100–1000 nm), and apoptotic bodies (1–5 μm).68,69 Plant-derived exosome-like NPs (PDENs) are found in extracts from edible herbs like ginger, lemon, etc.70 They have the same characteristics as mammalian exosomes, including morphology, density, electric charge, size, and contents.71 They are also non-toxic and have no detectable immunogenicity for use in mammalian therapeutic applications, their uptake in the target region is efficient and their capacity for drug transport and delivery is strong.72 PDENs can be used for biomolecule transportation and have therapeutic value.73 Ginger-derived exosome-like NPs (GDENs) and therapeutic strategies based on them are useful in the treatment and prevention of various diseases.74Table 2 shows different formulations of ginger. This study provides relevant information from past research on the beneficial effects of GDENs on various diseases.
| Ginger leaf | Ginger rhizome | Torch ginger (ginger flower) | Carrier-based delivery |
|---|---|---|---|
| Fresh75 | Topical ointment | Gummy Jelly76 | Vesicles-based77 |
| Extract62 | Jelly candies78 | Topical ointment79 | Micelles-based80 |
| Fresh81 | Extracts82,83 | Plant Derived NPs (PDNs)84 | |
| Powder85 | Solid–lipid nanoparticle86 | ||
| Dried87 | Phytosome88 | ||
| Syrup, Capsules89 | Conjugated to nanoparticle90 | ||
| Extract91 | |||
| Essential oil92 | |||
| Aromatherapy, Nasal spray93,94 |
3. Ginger-derived exosome-like NPs (GDENs)
Around 15 years before the discovery of exosomes derived from mammalian cells, in 1967, evidence of exosome-like NPs was found in plants; in an ultrastructural study on the growth and embryogenesis of carrot cells, a fusion of multi-vesicular bodies (MVBs) with plasmalemma was observed, which resulted in the release of the contents of the MVBs, which were small vesicles, into the extracellular space.95 These plasmalemma-related membranous structures were also frequently observed in published micrographs of lower and higher plants, which were either ignored or considered artifacts and unfortunately, they have not been studied until recently.96,97 PDENs are lipid-membrane nanovesicles that are derived from different sections (i.e., fruits, roots, seeds, leaves) of various edible plant species.98,99 PDENs are structurally similar to extracellular vesicles, which are released by cells into the extracellular space after merging MVBs with the plasma membrane; however, due to the mechanical isolation and homogenization methods of plant materials in the case of vesicles that are derived from ground tissue of plants or juices, the extracellular nature of these vesicles has not been confirmed and there is a mixture of intracellular and extracellular vesicles.72,100–102 Because specific markers of mammalian cell exosomes have not been identified in these vesicles, they are called exosome-like NPs.103 Early evidence has shown that plants produce PDENs in response to fungal infections and pathogen attacks. It was further found that PDENs can enter mammalian cells, cause changes in them and contribute to inter-kingdom communication.104–106 PDENs are non-toxic, environmentally friendly particles that are also economically viable to produce. Because of their natural source, the immune system cannot recognize them, and they are safer than mammalian exosomes since they do not have common pathogens with humans, as seen in zoonotic diseases.73,107,108 Ginger, the rhizome of Zingiber officinale, is one of the edible plants71,98 that have been shown to produce large amounts of GDENs; in a study by Zhang et al., 48.5 ± 4.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg of NPs were obtained per kilogram of ginger. In another study, from each 100 grams of ginger, about 380 mg of NPs were obtained.98 Concerning their large-scale production, GDENs can be used in medicine, for the treatment of diseases, as well as for the development of nanocarriers. To describe GDENs, it is necessary to determine their characteristics. Examination of the size and surface charge of GDENs by dynamic light scattering (DLS) showed that these particles have an approximate size and surface charge of 100–300 nm and −20 to −40 mV, respectively.72,98,109,110 However, ambient pH changes lead to altered particle size and surface charge.98,109 The analysis of GDENs via transmission or scanning electron microscopy also indicated that they are morphologically spherical or cup-shaped, consist of lipid bilayer membranes, and are uniformly arranged.84,98,109,111
mg of NPs were obtained per kilogram of ginger. In another study, from each 100 grams of ginger, about 380 mg of NPs were obtained.98 Concerning their large-scale production, GDENs can be used in medicine, for the treatment of diseases, as well as for the development of nanocarriers. To describe GDENs, it is necessary to determine their characteristics. Examination of the size and surface charge of GDENs by dynamic light scattering (DLS) showed that these particles have an approximate size and surface charge of 100–300 nm and −20 to −40 mV, respectively.72,98,109,110 However, ambient pH changes lead to altered particle size and surface charge.98,109 The analysis of GDENs via transmission or scanning electron microscopy also indicated that they are morphologically spherical or cup-shaped, consist of lipid bilayer membranes, and are uniformly arranged.84,98,109,111
3.1. Biogenesis
As was mentioned earlier, PDENs are lipid-membrane nanovesicles that are released by cells into the extracellular space.98 Two routes have been proposed for the generation and biogenesis of exosome-like NPs in plants. The first pathway is almost similar to the formation and secretion of extracellular vesicles in mammalian cells; MVBs containing intraluminal vesicles (ILVs) fuse with the plasmalemma and their contents are released into the extracellular space. It has already been confirmed that this mechanism is involved in papilla formation and defense mechanisms of plants.112–114 This pathway depends on the function of the multi-protein complex called ESCRT (endosomal sorting complex required for transport), which acts in a ubiquitin-dependent mechanism.113,114 ESCRT consists of 5 complexes including ESCRT-0, -I, -II, -III, VPS4-VTa1, and accessory proteins.115 Since ESCRT0 has not been identified in plants, TOM-like (TOL) proteins have been suggested as alternatives in plants that bind to ubiquinone proteins and are involved in vacuolar protein sorting.114,116The next biogenesis mechanism that has been proposed as an alternative to ESCRT is EXPO (exocyst-positive organelle). EXPO is a double-membrane non-spherical cell structure that fuses with the plasmalemma to release single-membrane extracellular EVs outside the cell. EXPO is structurally similar to the autophagosome but is not co-localized with markers of autophagosome formation. The components of the exocyst complex Exo70E2 are a way to describe and characterize EVs that are derived from EXPO.114,117,118
3.2. Isolation of GDENs
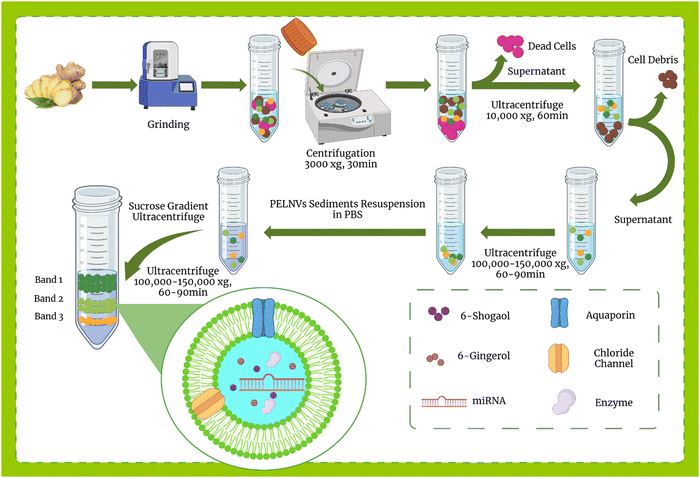
Exosome isolation methods from mammalian cells are well-defined, however, in plants, a specific and uniform isolation method has not yet been introduced. Some of the methods used to isolate mammalian exosomes, such as ultrafiltration, chromatography, PEG-based precipitation, and affinity capture on antibody-coupled magnetic beads, have been reported in plants.114,119 Here, the methods of isolation of exosomes from ginger are discussed.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–150
000–150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g) for 1 hour. This step causes NPs to settle in the form of pellets, then the pellet is suspended in PBS and, as an optional step, can be sonicated. The resulting suspension is usually contaminated with nucleic acids and aggregated proteins, so auxiliary methods such as sucrose step gradient should be used for further purification. For this purpose, the suspension was subjected to a sucrose gradient (8%, 15%, 30%, 45%, and 60%) and centrifuged at more than 100
000 × g) for 1 hour. This step causes NPs to settle in the form of pellets, then the pellet is suspended in PBS and, as an optional step, can be sonicated. The resulting suspension is usually contaminated with nucleic acids and aggregated proteins, so auxiliary methods such as sucrose step gradient should be used for further purification. For this purpose, the suspension was subjected to a sucrose gradient (8%, 15%, 30%, 45%, and 60%) and centrifuged at more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–150
000–150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for about 2 hours. Eventually, the nano-particles floated in different bands of sucrose. All processes were performed at 4 °C. The overview of plant exosome-like nanovesicles (PELNVs) extraction stages (here for ginger) is presented in Fig. 1. After grinding and multiple separation steps with different centrifugation speeds, GDENs can be isolated.120,121 According to the literature, the bands between the layer of 8/30 and 30/45% are considered as GDENs (Fig. 2).72 As has been shown in Fig. 1, GDENs contain active compounds such as 6-gingerol, 6-shogaol,122 transmembrane proteins like aquaporins, chloride channels,123 miRNA,124 and enzymes.125
000 × g for about 2 hours. Eventually, the nano-particles floated in different bands of sucrose. All processes were performed at 4 °C. The overview of plant exosome-like nanovesicles (PELNVs) extraction stages (here for ginger) is presented in Fig. 1. After grinding and multiple separation steps with different centrifugation speeds, GDENs can be isolated.120,121 According to the literature, the bands between the layer of 8/30 and 30/45% are considered as GDENs (Fig. 2).72 As has been shown in Fig. 1, GDENs contain active compounds such as 6-gingerol, 6-shogaol,122 transmembrane proteins like aquaporins, chloride channels,123 miRNA,124 and enzymes.125
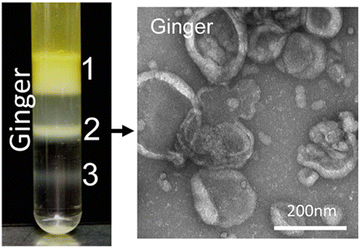 | ||
| Fig. 2 Three populations of GDENs with different contents and lipid compositions can be separated via sucrose gradient (8%, 15%, 30%, 45%, and 60%) ultracentrifugation. TEM images demonstrate the vesicular structure of band 2 of GDENs. Reproduced with permission from ref. 98. Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
3.3. Composition and content of GDENs
PELNs as extracellular phospholipid bilayer vesicles contain various cargoes, such as miRNA, mRNA, DNA, proteins, and active molecules toward mammalian cells, and regulate cellular activities. PELNs can also be used as drug carriers to increase drug stability and cellular uptake in vivo.128 The lipidomic composition of EVs produced by plants has shown that one of the most common lipidic compounds in plant-derived extracellular vesicles is phosphatidic acid (PA), which has recently come to be recognized as an essential lipid agent in intercellular communication and with crucial properties in drug transport.129 MicroRNAs (miRNAs), a group of short non-coding RNAs that are crucial for many physiological and pathological processes like cell division, cell death, metabolism, and immunological responses, are abundant in PDENs. For instance, it was shown that mi168a and miR159 are present in plant-derived EVs that can participate in the regulation of mammalian gene expression.130 Plant miR159 has been proven to be an interesting cross-kingdom inhibitor of breast cancer growth.131 Some studies have also shown that PELNs such as GDENs can be used as an efficient delivery system for nucleic acids and small molecule (siRNAs, miRNAs, and DNA) therapeutics. In some studies, deep sequencing analysis has demonstrated that GDENs contain 125 miRNAs.72 Ju et al. reported that 124 miRNAs found in grape exosome-like NPs can target and control human gene expression by binding to 3′ untranslated regions (UTR).132 PDENs contain different exosome-like particles with different properties. Zhang et al. isolated and identified three populations of NPs, namely, GDENs 1, GDENs 2, and GDENs 3, from the edible plant ginger.133 This study demonstrated that the GDENs population had low protein content. However, GDENs 2 indicated a higher bioactive content such as 6-gingerol and 6-shogaol as compared to others. Interestingly, GDENs 1 and GDENs 2 could tolerate freeze/thaw cycles and were very stable at room temperature as compared to GDENs 3, from which it was concluded that each has a different lipid composition.72Tables 3 and 4 show lipid composition and content characteristics and comparable criteria between GDENs in several studies, respectively. Table 5 indicates the comparison of edible ginger-derived NPs as a cargo vehicle for drugs or siRNA delivery in different studies.| Type | Method of preparation or isolation | Lipid composition/bioactive content | Characterization of lipid/protein/miRNA/bioactive/shape | Function | Target | Size (nm) | Size characterized by | Zeta potential (mV) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| GDEN: ginger-derived exosome-like NPs, GDNV: ginger-derived nanovectors, PELN: plant-derived exosome-like NPs, EV: extracellular vesicles, DGDG: digalactosyldiacylglycerol, MGDG: monogalactosyldiacylglycerol, PC: phosphatidylcholine, PA: phosphatidic acid, PI: phosphatidyl inositol, DAG: diacylglycerol, HPLC: high-performance liquid chromatography, AFM: atomic force microscopy, TEM: transmission electron microscopy, ATM: atomic force microscopy, DLS: dynamic light scattering, UPLC-MS/MS: ultra-performance liquid chromatography-tandem mass spectrometry, CAC: colitis-related cancer. | |||||||||
| GDEN | Differential ultracentrifugation | DGDG > PA > MGDG/Shogaols are much higher in GDN than in GDEN2 | Triple quadrupole tandem mass spectrometer/BCA assay/HPLC analysis/AFM | Mediate the activation of Nrf2, resulting in an increase in the expression of a group of liver detoxification and antioxidant genes, and inhibiting the production of reactive oxygen species, which partially helps to protect the liver. | C57BL/6j mice | 386.6 | DLS | −24.6 | 109 |
| GDEN2 | Differential ultracentrifugation | PA > DGDG > MGDG | Triple quadrupole tandem mass spectrometer, TLC analysis, BCA assay/HPLC analysis/AFM | Mediate the activation of Nrf2, resulting in an increase in the expression of a group of liver detoxification and antioxidant genes, and inhibiting the production of reactive oxygen species, which partially helps to protect the liver. | C57BL/6j mice | 294.1 | DLS | −29.7 | 109 |
| GDEN 1 | Differential ultracentrifugation | DGDG > MGDG > PA> PI | Triple quadrupole tandem mass spectrometer/UPLC-MS/MS/RNA deep sequencing/TEM/AFM | Oral administration of GDNPs 2 increased the survival and proliferation of IECs and reduced the proinflammatory cytokines (TNF-α, IL-6, and IL-1β), and increased the anti-inflammatory cytokines (IL-10 and IL-22) in colitis models suggesting that the higher content of 6-gingerol and 6-shogaol in GDNPs 2 might play a role in their anti-inflammatory activities | DSS-induced colitis mouse mode | 292.5 | DLS | −11.3 | 72 |
| GDEN2 | Differential ultracentrifugation | PA> DGDG > MGDG > PC | Triple quadrupole tandem mass spectrometer/UPLC-MS/MS/RNA deep sequencing/TEM/AFM | Not effective | Dextran sulfate sodium (DSS)-induced acute colitis with ulceration | 231.6 | DLS | −12.9 | 72 |
| GDEN3 | Differential ultracentrifugation | PA> d DGDG > MGDG > PC | Triple quadrupole tandem mass spectrometer | Not examined due to instability | — | 219.6 | DLS | −2.1 | 72 |
| GELN | Differential ultracentrifugation | PA > PC> PE> DGDG, MGDG, DAG | Triple quadrupole tandem mass spectrometer/HPLC | GELNs are selectively taken up by the periodontal pathogen Porphyromonas gingivalis in a GELN phosphatidic acid (PA)-dependent manner via interactions with hemin-binding protein 35 (HBP35) on the surface of P. gingivalis to prevent or treat chronic periodontitis | Porphyromonas gingivalis | 204 | DLS | — | 134 |
| GDNP | Differential ultracentrifugation | PA > DGDG > MGDG | Triple quadrupole tandem mass spectrometer/TEM | Prevent HFD-induced obesity and insulin resistance by protecting the Foxa2 from Akt-1-mediated phosphorylation | High-fat diet-fed mice | 250 ± 72/1 × 1012 GDNP/g | NTA | −220 ± 131 | 74 |
| GELN | Differential ultracentrifugation | PA > MGDG > DGDG > PC | Triple quadrupole tandem mass spectrometer/TEM | GELNs can cross-talk with gut microbiota and are preferentially taken up by Lactobacillus rhamnosus (LGG). GELN mdo-miR7267-3p-mediated targeting of the LGG monooxygenase ycnE yields increased indole-3-carboxaldehyde (I3A). GELN RNAs or I3A, a ligand for aryl hydrocarbon receptor (AHR), are sufficient to induce the production of IL-22, which can ameliorate mouse colitis via IL-22-dependent mechanisms. | Mouse colitis | 206.8 | DLS | — | 135 |
| Type/name | Method of preparation or isolation | Content | Characterization of lipid, protein, miRNA, and bioactive content by | Function | Target | Size (nm) | Size characterized by | Zeta potential (mV) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| ENP: edible NPs, PELN: plant-derived exosome-like NPs, TPC: total polyphenolic content, TLC: thin layer chromatography, AFM: atomic force microscopy, TEM: transmission electron microscopy, AFM: atomic force microscopy, SEM: scanning electron microscopy DLS: dynamic light scattering, PEG: polyethylene glycol, CAC: colitis-related cancer. | |||||||||
| ENPs | PEG6000 precipitated | Small RNA, bioactive content such as polyphenolic | TLC analysis, SDS-PAGE analysis, ATR-FTIR technique | Antioxidant activity | In vitro antioxidant activity assay on RAW macrophages, DPPH assay, Total polyphenolic content estimation | 8% PEG (365), 10% PEG (304), 12%PEG (263), 15%PEG (252) | DLS | 8% PEG (−25.5), 10% PEG (−25.1), 12%PEG (−21.4), 15%PEG (−21.2) | 101 |
| Ginger ENPs | Differential ultra-centrifugation | Small RNA, bioactive content such as polyphenolic | TLC analysis, TPC, SDS-PAGE analysis, ATR-FTIR technique | Antioxidant activity | In vitro antioxidant activity assay on RAW macrophages, DPPH assay, Total polyphenolic content estimation | 403 | DLS | −25.7 | 101 |
| GDEN1 | Differential ultra-centrifugation | 6-Gingerol (0.56 μg mg−1) and 6-shogaol (0.22 μg mg−1)/some cytosolic and membrane proteins | Triple quadrupole tandem mass spectrometer/UPLC-MS/MS/RNA deep sequencing/AFM/TEM | Not effective | Dextran sulfate sodium (DSS)-induced acute colitis with ulceration | 292.5 | DLS | −11.3 | 72 |
| GDEN2 | Differential ultra-centrifugation | 6-Gingerol (5.68 μg mg−1) and 6-shogaol (2.95 μg mg−1)/125 different miRNAs/some cytosolic and membrane proteins | Triple quadrupole tandem mass spectrometer/UPLC-MS/MS/RNA deep sequencing/AFM/TEM | Oral administration of GDNPs 2 increased the survival and proliferation of IECs and reduced the proinflammatory cytokines (TNF-α, IL-6 and IL-1β), and increased the anti-inflammatory cytokines (IL-10 and IL-22) in colitis models suggesting that the higher content of 6-gingerol and 6-shogaol in GDNPs 2 might play a role in their anti-inflammatory activities | DSS-induced acute colitis with ulceration | 292.5 | DLS | −12.9 | 72 |
| GDEN | Differential ultracentrifugation | Shogaols (much higher than GDEN2) | Triple quadrupole tandem mass spectrometer/BCA assay/HPLC analysis/AFM | Mediate the activation of Nrf2, resulting in an increase in the expression of a group of liver detoxification and antioxidant genes, and inhibiting the production of reactive oxygen species, which partially helps to protect the liver. | C57BL/6j mice/hepatocytes | 386.6 | DLS | −24.6 | 109 |
| GDEN2 | Differential ultracentrifugation | Shogaols (much lower than GDN) | Triple quadrupole tandem mass spectrometer/BCA assay/HPLC analysis/AFM | Mediate the activation of Nrf2, resulting in an increase in the expression of a group of liver detoxification and antioxidant genes, and inhibiting the production of reactive oxygen species, which partially helps to protect the liver. | C57BL/6j mice/hepatocytes | 294.1 | DLS | −29.7 | 109 |
| GDEN | Differential ultracentrifugation | Total RNAs (30.0 ± 10.8 ng/1010 vesicles)/total proteins (2.8 ± 0.3 μg/1010 vesicles)/total lipids (32 ± 2.9 μg/1010) | Thin-layer chromatography (TLC)/RNA extraction/SEM | Block NLRP3 inflammasome assembly and activation | Bone marrow-derived macrophages (BMDMs) from C57BL/6J mice | 130/2 × 1011 vesicles per g | NTA | — | 111 |
| GDEN | Polyethylene glycol (PEG)-based precipitation method | Six miRNAs (miR-5077, miR-6300, miR156a, miR-169, miR-5059 and miR-166 m) | In silico analysis | Antiviral bioactive as an effective therapeutic modality against COVID-19 | Target ORF1ab, ORF3a, and ORF8 genes | — | — | — | 126 |
| GELN | Differential ultracentrifugation | 30 miRNA was determined and miRNAs aly-miR159a-3p, gma-miR166u, and gma-miR166p was selected for inhibitory effect | Triple quadrupole tandem mass spectrometer/HPLC | GELNs are selectively taken up by the periodontal pathogen Porphyromonas gingivalis in a GELN phosphatidic acid (PA) dependent manner via interactions with hemin-binding protein 35 (HBP35) on the surface of P. gingivalis to prevent or treat chronic periodontitis | Porphyromonas gingivalis | 204 | DLS | — | 134 |
| GELN | Differential ultracentrifugation | 106 miRNA was isolated including ath-miR167a, gma-miR396e | Deep RNA sequencing | GELNs can cross-talk with gut microbiota and are preferentially taken up by Lactobacillus rhamnosus (LGG). GELN mdo-miR7267-3p-mediated targeting of the LGG monooxygenase ycnE yields increased indole-3-carboxaldehyde (I3A). GELN RNAs or I3A, a ligand for aryl hydrocarbon receptor (AHR), are sufficient to induce the production of IL-22, which can ameliorate mouse colitis via IL-22-dependent mechanisms. | Mouse colitis | 206.8 | DLS | — | 135 |
| GELN | Commercial exosome isolation kits | miRNAs | High-throughput sequencing followed by qPCR | Counteract lipopolysaccharide (LPS)-induced inflammation by downregulating NF-κβ, IL-6, IL-8, and TNF-α expression | Caco-2 cells | 156 ± 36 | DLS | −26.6 ± 5 | 124 |
| GDNP | Differential ultracentrifugation | miR-375 or antisense-miR375 | miRNA PCR microarray/TEM | GDNP to HFD mice improves host glucose tolerance and insulin response via regulating AhR expression by GDNP-induced miR-375 | High-fat diet (HFD) fed mice/Human with Type 2 diabetes/murine colon cell line (MC-38) | ∼200 | DLS | — | 136 |
| PDEN | PEG 6000 isolation/total exosome isolation reagent | Antioxidant content | Uptake assay/antioxidant assay/NTA | PDENs from ginger were taken up by human Wharton's jelly mesenchymal stem cells faster (after 30 minutes) than human dermal fibroblast (21 hours). DPPH assays revealed that ginger PDENs have antioxidant activity, indicating the presence of some nano-molecules in those particles | Human dermal fibroblast (HDF) and human Wharton's jelly mesenchymal (hWj MSC) stem cells/DPPH assay | 132.81/371.61 | DLS | — | 137 |
| GDEN | Differential ultracentrifugation | Proteins, lipids, and miRNAs (Higher RNA content/100 mg GDENs was found compared to Grape, Grapefruit, and carrot) | TLC | GDENs induce the expression of the anti-oxidation gene, heme oxygenase-1 (HO-1) and the anti-inflammatory cytokine, IL-10; | In vitro model (RAW 264.7 macrophage)/in vivo model (transgenic mice) | 100–1000 | DLS | N/A | 98 |
| GDEV | Differential ultracentrifugation | 6-Gingerol, 8-gingerol, and 10-gingerol | HPLC/TEM | 6-Gingerol, 8-gingerol, and 10-gingerol in GDEVs were 10.21-fold, 22.69-fold, and 32.36-fold of those in ginger slices, respectively. | In situ intestinal absorption model in rats | 70.09 ± 19.24 | DLS | −27.70 ± 12.20 | 138 |
| Type | Method of preparation or isolation | Composition | Synthetic content/surface ligand | Characterization of NPs | Function | Target | Size (nm) | Size characterized by | Charge (zeta potential measurements) (mV) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| FA-GDLV: folic acid-coupled, ginger nanoparticle-derived lipid vectors, GDLV: ginger-derived lipid vectors, NTA: nanoparticle tracking analysis, TEM: transmission electron microscopy, DLS: dynamic light scattering, AFM: atomic force microscopy. | ||||||||||
| GDENs | Differential ultracentrifugation | — | Survivin siRNA delivery/Folic acid conjugated on surface | BCA assay, NTA, Negative staining TEM | Inhibition of tumor growth on a xenograft model by intravenous administration | In vitro (HEK293 cells, Raw. 264.7 macrophage and KB cells)/in vivo (KB cell xenograft mice mode) | 124.5 | DLS | — | 139 |
| FA-GDLVs | Differential ultracentrifugation | — | Dmt1 siRNA/Acid folic conjugated GDENs | — | Reduces pre-existing iron overload in the hereditary hemochromatosis mouse model | Hepcidin KO (Hamp−/−) mice on the C57BL/6 genetic background | — | — | — | 140 |
| GDLVs | Differential ultracentrifugation | Phosphatidic acid/monogalactosyldiacylglycerol/di galactosyl diacylglycerol/phosphatidylcholine | Doxorubicin loaded by layer ginger-derived lipid vector (LbL-GDLV) | Triple quadrupole Mass spectrometer, TEM | Inhibited tumor growth by targeting P selectin and demonstrated much better therapeutic efficiency than free Dox | Xenograft tumor growth model e with Luc-HT-29 cells or HCT-116 | 219. ± 2.2 | DLS | −47.2 ± 1.1 | 141 |
| GDENs | Differential ultracentrifugation | Phosphatidic acid/monogalactosyldiacylglycerol/di galactosyl diacylglycerol/phosphatidylcholine phosphatidic acid (∼47% of total lipids), di galactosyl diacylglycerol (∼15% of total lipids), and monogalactosyldiacylglycerol (∼27% of total lipids) | Doxorubicin/Acid folic conjugated GDENs | Triple quadrupole mass spectrometer, TEM, AFM | Enhanced the chemotherapeutic inhibition of tumor growth in a pH-dependent drug-release profile than commercially available liposomal-Dox | Colon-26 tumors in vivo | 188 | DLS | −15.5 | 133 |
| GDENs | Thin film hydration | phosphatidic acids (41.9% of total lipids), di galactosyl diacylglycerol (27.4%), and monogalactosyldiacylglycerol (18.9%) | siRNA against CD98 | Triple quadrupole mass spectrometer | Reduced expression of CD98 | In vitro (o RAW 264.7 macrophage-like cells and colon-26 colonic cancer cells)-In vivo | 189.5 | DLS | 18.1 | 84 |
4. GDENs in biomedicine
4.1. Liver disease
Liver diseases account for more than 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths yearly and the incidence is rapidly accelerating. A significant relationship has been found between liver fat, edibles, gut microbiota, and liver damage. Therapeutic alternatives for treatment are extremely limited, especially for alcoholic liver disease (ALD).142 Many natural NPs are present in human food, being absorbed each day via the gut, and they can be transported from the gut to the liver, thus biologically affecting the liver. Research has indicated that ginger exerts a hepatoprotective impact against carbon tetrachloride, and ethanol, as well as hepatotoxicity induced by acetaminophen.143 An in vitro study on the anti-inflammatory impacts of ginger has mainly focused on shogaols, the dehydration products of gingerols.144 Numerous and diverse biological insults are received by the liver daily. To maintain liver homeostasis and avoid harm caused by absorbed endotoxin, it is vital to induce cellular protective enzymes such as carcinogen-detoxifying and antioxidant enzymes. The gene expression of various cytoprotective enzymes is transcriptionally controlled by nuclear factor erythroid-related factor 2 (Nrf2), which can effectively protect the liver from insult.145 It was shown that after the oral administration of 50 mg/day of fluorescent-labeled GDENs in C57BL/6j mice aged 6 to 8 weeks, the tissue distribution of GDENs was predominantly detected in liver cells after 12 h of oral administration. Afterward, 100 μg mL−1 of GDEN 1 and 2 bands (4 hours) were used to treat the primary hepatocytes, resulting in a remarkable induction of Nrf2 expression, reduction of ROS production, and finally the protection of mice against liver damage by alcohol. They detected shogaol containing GDENs, a dehydrated analog of gingerol, which activated Nrf2 in a TLR4/TRIF-dependent way, resulting in the expression of liver antioxidant/detoxification genes such as NQO-1 and HO-1. Since Nrf2 plays an important role in the modulation of different cellular mechanisms such as the proliferation of hepatocytes during liver regeneration, drug metabolism, and inflammation, it opens a new approach to investigating the role of GDENs in such relevant molecular and cellular processes. This result suggests that in addition to acting as a new agent for protecting the liver against harm, GDENs can also provide a basis for investigating the interspecies communication mechanism via NPs that are consumed daily in diverse varieties of edible plants.109
000 deaths yearly and the incidence is rapidly accelerating. A significant relationship has been found between liver fat, edibles, gut microbiota, and liver damage. Therapeutic alternatives for treatment are extremely limited, especially for alcoholic liver disease (ALD).142 Many natural NPs are present in human food, being absorbed each day via the gut, and they can be transported from the gut to the liver, thus biologically affecting the liver. Research has indicated that ginger exerts a hepatoprotective impact against carbon tetrachloride, and ethanol, as well as hepatotoxicity induced by acetaminophen.143 An in vitro study on the anti-inflammatory impacts of ginger has mainly focused on shogaols, the dehydration products of gingerols.144 Numerous and diverse biological insults are received by the liver daily. To maintain liver homeostasis and avoid harm caused by absorbed endotoxin, it is vital to induce cellular protective enzymes such as carcinogen-detoxifying and antioxidant enzymes. The gene expression of various cytoprotective enzymes is transcriptionally controlled by nuclear factor erythroid-related factor 2 (Nrf2), which can effectively protect the liver from insult.145 It was shown that after the oral administration of 50 mg/day of fluorescent-labeled GDENs in C57BL/6j mice aged 6 to 8 weeks, the tissue distribution of GDENs was predominantly detected in liver cells after 12 h of oral administration. Afterward, 100 μg mL−1 of GDEN 1 and 2 bands (4 hours) were used to treat the primary hepatocytes, resulting in a remarkable induction of Nrf2 expression, reduction of ROS production, and finally the protection of mice against liver damage by alcohol. They detected shogaol containing GDENs, a dehydrated analog of gingerol, which activated Nrf2 in a TLR4/TRIF-dependent way, resulting in the expression of liver antioxidant/detoxification genes such as NQO-1 and HO-1. Since Nrf2 plays an important role in the modulation of different cellular mechanisms such as the proliferation of hepatocytes during liver regeneration, drug metabolism, and inflammation, it opens a new approach to investigating the role of GDENs in such relevant molecular and cellular processes. This result suggests that in addition to acting as a new agent for protecting the liver against harm, GDENs can also provide a basis for investigating the interspecies communication mechanism via NPs that are consumed daily in diverse varieties of edible plants.109
Another study investigated the effects of GDENs in protecting the liver from liver damage induced by alcohol, as well as in treating intestinal colitis. Bakr et al.'s study investigated the protective activity of 120 mg kg−1 GDENs against daily acetaminophen toxicity of male rats for 3 months and evaluated the biochemical and histological aspects. The result of this study indicated that pre-treatment with GDENs improved oxidative stress, biochemical markers, and the histopathological structures of the kidney and liver. Interestingly, the protective effect of GDENs was better than the ginger extract.146 After oral administration, acetaminophen (APAP) is quickly absorbed and metabolized through the liver. The production of N-acetyl-benzoquinone imine (NAPQI) during the metabolism of APAP causes liver toxicity. At toxic doses of APAP, NAPQI binds with cellular protein, and the level of glutathione in hepatocytes is reduced. Glutathione depletion will cause damage to the cells via the production of ROS, including the activation of stress proteins, injury of the cell membrane, nitric oxide, and mitochondrial oxidative stress.147,148
4.2. Inflammation and oxidative stress
The nucleotide-binding domain and leucine-rich repeat-containing family, the pyrin domain-containing 3 (NLRP3) inflammasome, is a cytoplasmic multiprotein complex consisting of the NLRP3 sensor, the apoptotic adapter spike protein containing the caspase recruitment domain (ASC), and the effector caspase 1 (Casp1).149,150 The innate immune system's activation of NLRP3 inflammation is a key function, and recent research has shown that excessive inflammatory activity causes or contributes to the pathogenesis of a variety of diseases, including inflammatory diseases such as cryopyrin-associated auto-inflammatory syndrome (CAPS),151 gout,152 neurodegenerative diseases,153 and metabolic diseases (type 2 diabetes,154,155 atherosclerosis156,157). Thus, NLRP3 inflammation-targeting may be able to slow or stop the progression of the disease, making it a promising therapeutic approach for many complex disorders.111The incubation of GDENs diet with bone marrow-derived macrophages (BMDM) from C57BL/6J mice inhibited the downstream activation of inflammasome pathways such as the production of interleukin (IL)-1, IL-18, and caspase-1 autocleavage, as well as the death of the pyroptotic cell. In more detail, Chen et al. revealed that the treatment of C57BL/6J mice with GDENs (2–10 g per mice) blocked the NLRP3 inflammasome activation and accumulation and prevented the downstream activation of the NLRP3 inflammasome pathway, such as IL-18 secretion, caspase-1 pyroptosis, and autophagy. Hence, GDENs are considered a promising novel approach for inhibiting the NLRP3 inflammasome.111
Research has also shown that GDENs decrease severe inflammation in mice after being orally administered by reducing the levels of pro-inflammatory cytokine expressions (e.g., IL-6, IL-1β, and TNF-α) and elevating the levels of expression of anti-inflammatory cytokines (e.g., IL-10 and IL-22).146,158 Several cases have shown the effects of GDENs on periodontitis, a severe inflammation of the periodontium, which causes gingival swelling, periodontal ligament destruction, chronic pain, and alveolar bone and tooth loss.128,159 GDENs showed an antibacterial impact on Porphyromonas gingivalis (P. gingivalis).134P. gingivalis, which is a common Gram-negative anaerobic oral bacteria, is a main pathogen in severe periodontitis, which is an inflammation related to dysbiotic host reactions.160 Periodontal and periodontitis pathogens are also related to severe systemic problems such as type 2 diabetes, cardiovascular disease, as well as adverse pregnancy outcomes.161P. gingivalis generates several virulent factors that can colonize oral surfaces, destroy periodontal tissues, induce harmful immune responses, and grow a hemin-rich and peptide-inflammatory microenvironment. These factors involve fimbrial adhesions with components FimA and Mfa1, specific gengipain proteases, lipopolysaccharides, lysine (Kgp) and arginine (Rgp), and land hemin transport systems.162–164 In this regard, GDENs change the composition of gut microbiome and the physiology of the host. In this regard, GDENs are preferentially taken up by Lactobacillaceae in a GDENs lipid-dependent manner and contain microRNAs that target various genes in Lactobacillus rhamnosus. It has been shown that the mdo-miR7267-3p-mediated targeting of the LGG monooxygenase ycnE yields increased indole-3-carboxaldehyde (I3A). I3A could induce the production of IL-22, which in turn ameliorated mouse colitis via IL-22-dependent mechanisms. Fig. 3 illustrates the details of the mechanism of protection from colitis by GDENs.135
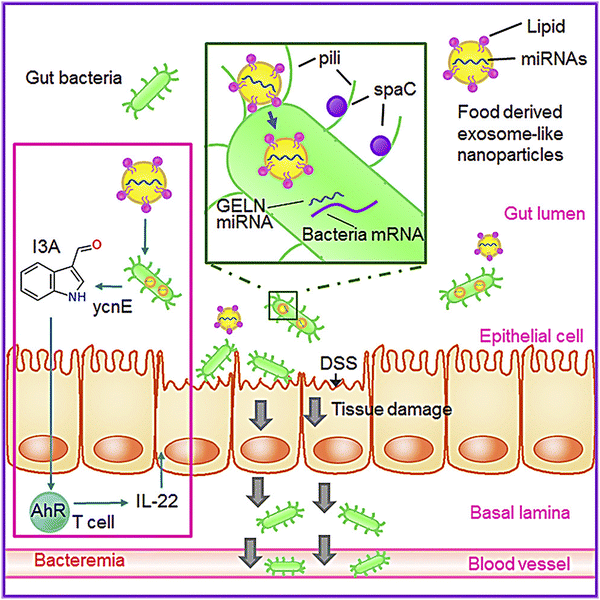 | ||
| Fig. 3 GDENs are taken up by bacteria in a lipid-dependent manner. GDENs containing miRNA affect bacteria (Lactobacillus rhamnosus) gene expression to produce I3A. GDENs-mediated I3A alterations affect IL-22 production, resulting in notably enhanced gut barrier function to alleviate colitis. Reproduced with permission from ref. 135. Copyright 2018 Elsevier Inc. | ||
Sundaram et al. evaluated GDENs to antagonize the virulence factors of P. gingivalis and inhibit pathogenicity in a mouse model with chronic periodontitis. Their data showed that GDENs were drawn selectively through P. gingivalis, thereby reducing the organism's pathogenicity. Pathogenic mechanisms under the influence of GDENs are adhesion, entry, proliferation, and growth in host cells, and as a result, reduce the severity in the periodontal disease mouse model.134 Moreover, promising antibacterial strategies with a combination of Pd–Pt nanosheets and natural GDENs have been proposed. The biomimetic nano-platform (GDENs-Pd–Pt) has prolonged blood circulation without immune clearance, as well as accumulation at infection sites. More interestingly, EV-Pd–Pt can enter the bacteria in an EV lipid-dependent manner. The combination of Pd and Pt in nanosheets enhances synergistic bacteria eradication via electrodynamic therapy (EDT) and also adds photothermal properties. These nanoparticles after entering the bacteria ROS generation trigger via EDT exhibited bactericidal effects in in vitro and in vivo models. In addition to the photothermal properties of Pt–Pd, photoacoustic imaging-guiding can be achieved. A schematic representation of EV-Pd–Pt nanoparticles by conjugating carboxylic group-functionalized Pd–Pt nanoparticles on the surface of GDENs with abundant amino groups can be seen in Fig. 4.165
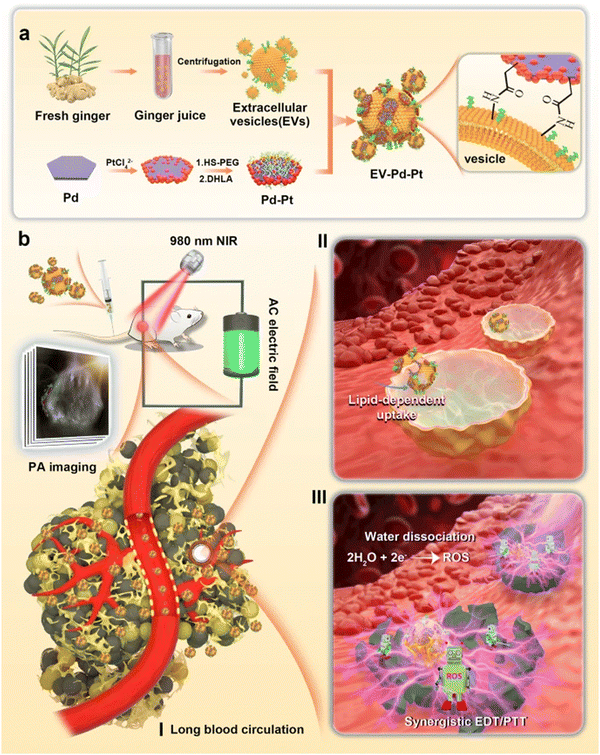 | ||
| Fig. 4 (a) Schematic representation of GDENs-Pd–Pt nanosheet preparation by conjugation of carboxylic groups of Pd–Pt nanoparticles to surface amino groups of GDENs. (b) Bactericidal activity of the GDENs-Pd–Pt nanosheet in an in vivo model guided by the photoacoustic effect of nanosheets. Long blood circulation, lipid uptake-dependent manner, and efficient accumulation at the infection site are introduced in this system. Reproduced with permission from ref. 165. Copyright 2022 Springer Nature, under the terms of the Creative Commons CC BY license. | ||
6-Gingerol as a bioactive compound encapsulated in GDENs, which exhibits anti-inflammation activities in in vitro and in vivo sepsis models. It also diminished inflammatory cytokine IL-18 levels in colon tissues and the serum of mice that already had sepsis. In addition, 6-gingerol suppresses MAPK signaling pathways and diminishes macrophage pyroptosis by decreasing the production of HMGB1, IL-18, and caspase-1-p20 in response to treatment with lipopolysaccharides (LPS) and ATP. 6-Gingerol prevents the production of proteins related to pyroptosis such as IL-1, NLRP3, and caspase-1, which helps to alleviate sepsis. Interestingly, 6-gingerol reduces sepsis by stimulating the Nrf2 pathway through a different method.166
Likewise, 6-shogaol targets the Nrf2 gene expression of HO-1 and metallothionein 1 (MT1), and the aldo-keto reductase family 1 member B10 (AKR1B10) increases ferritin light chain (FTL) as well as glutamyl transferase-like function in human colon cancer cells (GGTLA4). Following the 6-shogaol treatment of HCT-116 cells, the intracellular ratio of GSH/GSSG was first reduced and then it increased more than the basal level. The earlier reduction is due to GSH conjugation to 6-shogaol, and the later increase in the content of GSH may be due to the induction of the antioxidant enzymes of phase II, which are controlled via the antioxidant-response element (ARE) by the Keap1/Nrf2 signaling pathway.167
4.3. Gastrointestinal diseases
Inflammatory bowel disease (IBD) is a general expression describing a condition with severe and relapsing immune responses and GI tract inflammation that results in chronic diarrhea and stomach cramps.168 The GDENs technology is simple to create in large-scale manufacturing and might be a viable therapeutic method for IBD and CAC (colitis-associated cancer) prevention and treatment. In a mouse model with IBD and CAC, the oral administration of GDENs improved intestinal mucosa, decreased severe and acute inflammation, and diminished CAC, implying that GDENs can inhibit tumor growth and chronic colitis. It has been shown that GDENs decrease the production of pro-inflammatory cytokines (IL-6, IL-1, and TNF-) while elevating the anti-inflammatory cytokines expression (IL-10 and IL-22), indicating that such ENs inhibit intestine-impairing agents while promoting intestine-curing agents.169For the treatment of IBD, regular exposure to anti-TNF-α antibodies received via clinical injection induces adverse side effects. Targeted colonic delivery of anti-TNF-α antibodies via the oral channel is significantly important; however, it persists as a major problem. In this regard, Mao et al. reported a biomimetic nanocomposite consisting of a mineral framework and a ginger-derived exosome. Large mesoporous silicon nanoparticles (LMSN) were developed for loading with the antibodies (infliximab) at elevated levels up to 61.3 wt%. Exosome-like nanovesicles were separated from ginger with a high production level (17.5 mg kg−1). Afterward, ultrasound was utilized for coating GDENs on LMSN to obtain a biomimetic nanocomposite of LMSN and GDENs. As was expected, GDENs and LMSN were effective in orally delivering the infliximab with GI tract stability, increased permeability of the intestinal epithelium, and targeted delivery to the colon. Surprisingly, GDENs could also provide an anti-inflammatory impact by inhibiting the NLRP3 inflammasome. To sum up, this work presented a new method for drug delivery via a therapeutic agent (exosomes extracted from plants).170 In another study, it was observed that administering GDENs to model mice orally decreased CAC as well as acute and severe inflammation by enhancing the anti-inflammatory cytokine expression and increasing intestinal mucosa, suggesting that GDENs can inhibit tumor growth and chronic colitis.169 Furthermore, recent studies have shown that ginger-derived lipid nano-factors can be loaded with a therapeutic agent (doxorubicin) as a new drug delivery method for the treatment of colon cancer. Further findings indicated that GDENs efficiently occupied colon cancer cells. GDENs were able to load doxorubicin with high efficiency. They found that modified GDENs combined with folic acid-targeting ligands and intravenous injection of DOX-FA-GDENs in FVB/NJ and C57BL/6 mice with colon cancer-26 could help provide targeted chemotherapy drug delivery to tumors through blood vessels.133
In one study, it was observed that GDEN2 from the family of GDENs in mouse models of colitis increased intestinal repair and reduced acute colitis, preventing the development of chronic colitis. In this study, colitis was induced by dextran sodium sulfate in C57BL/6 or FVB/NJ female mice aged 6 to 8 weeks.133 To evaluate the anti-inflammatory effects of GDNPs in the large intestine, the effects of GDNPs 1 and GDNPs 2 were investigated in a rat with acute colitis induced by dextran sodium sulfate (DSS) with ulceration.171 After reaching the large intestine, GDENs 2 were equally absorbed via macrophages and intestinal epithelial cells (IECs) in animals with colitis. Functional comparative analysis of two populations of GDNP with acceptable biophysical features (GDNPs 1 and GDNPs 2) indicated that the oral administration of GDNPs 2 (0.3 mg per mouse), but not GDNPs 1, decreased acute DSS-induced inflammation and interestingly, oral GDNPs 2 enhanced the IEC proliferation and survival, reduced the expression of pro-inflammatory cytokines (IL-6, IL-1b, and TNF a), and improved the expression of anti-inflammatory cytokines (IL-10 and IL-22) in induced colitis, showing that GDNPs 2 can inhibit agents that impair the intestines while increasing agents that cure them. The analysis of differentially expressed genes after orally administering GDNPs indicated that 2 molecular targets of GDNPs play a potential role in alleviating severe colitis. The majority of these molecular targets involve the proteins expressed in the cell cytoplasm, mitochondria, membrane, or nucleus of the intestinal mucosa. For instance, GDNPs 2 enhanced the expression level of a negative regulator of reactive oxygen species (NRROS), which is assumed to restrict ROS production via phage cells during the inflammatory response, thus soothing the inflammation. The greater dose of 6-shogaol and 6-gingerol in GDNPs 2 may contribute to their anti-inflammatory activities.172
GDENs 2 (0.1 mg ml−1) remodel intestinal mucosa in wound healing models. Interestingly, GDENs 2 therapy of wounded intestinal mucosa restores average doses of anti- and pro-inflammatory cytokines, MPO activity, as well as IEC amplification-apoptosis balance in the mucosa.173 In addition, treating the injured intestinal mucosa with GDNPs 2 restored average doses of anti- and pro-inflammatory cytokines, IEC proliferation-apoptotic balance, and MPO activity in the intestinal mucosa. As the GDNPs 2 molecular targets at the end of the recovery step were primarily membrane/cytoplasmic proteins in intestinal mucosa, the treatment with GDNPs 2 considerably enhanced the expression of such proteins such as carbonic anhydrase 1 (CAR1) (∼14-fold) that was found on the surface of intestinal enterocytes and was a major cecal antigen involved in the development of IBD in the murine models.173,174 Interestingly, the treatment with GDNPs 2 focused on the expression of some proteins involved in the CAC development. Recently, research has demonstrated that cGMP signaling significantly regulates tissue homeostasis in the GI tract, and the activation of cGMP-dependent protein kinase hinders the signaling of the T cell transcription factor (TCF) in colon cancer cells by the inhibition of b-catenin and activation of forkhead boxO4 (FOXO4).175,176 Interestingly, treatment via GDNPs 2 leads to an increase in PKG expression, which may somehow contribute to the therapeutic effectiveness of GDNPs 2. This justification can be further approved by reports that showed that therapeutic activation of cGMP/PKG is a possible technique to prohibit and heal colon cancer.72,176
A recently developed siRNA delivery vehicle based on ginger-derived lipids demonstrated that GDENs can encapsulate siRNA-CD98 and that orally administered GDENs loaded with a very low dose of siRNA-CD98 selectively and efficiently suppressed colonic CD98 gene expression. siRNA-CD98/GDENs can change the siRNA delivery method from manufactured NPs to naturally occurring plant-derived NPs, which could form the basis of a safe siRNA delivery system for the treatment of colitis.84 Certain groups of ginger-derived NPs may also effectively reduce the risk of colitis, which are probably the most important pharmacologically active compounds in these ginger NPs, gingerols, and shogaols.177
An in vivo study showed that the oral administration of ENs-6-shogaol significantly reduced the symptoms of colitis and accelerated the healing of colitis ulcers in mice by regulating the expression level of inflammatory factors such as TNFα, IL-6, IL-1β, iNOS, and COX2, as well as anti-inflammatory agents such as Nrf-2 and HO-1. This nanoparticle system is considered a new and promising therapeutic approach for the treatment of IBD.177 By suppressing PI3K/Akt, 6-shogaol prevents TNF-induced barrier disruption by decreasing the production of the strong binding protein of channel-forming claudin-2. Furthermore, 6-shogaol suppresses TNFα-induced NF-κB signaling, maintaining the strong binding protein of claudin-1 at the binding site and eventually contributing to intestinal inflammatory barrier malfunction. 6-Shogaol protects human HT-29/B6 colon cell components against TNF-induced barrier disorders via modulating the production and assembly of strong binding proteins, according to studies. 6-Shogaol also protects Caco-2 cells against TNF-induced barrier disturbance, demonstrating that this protective effect is not limited to an in vitro model.178
4.4. Cancers
Cancer is considered a result of genetic and epigenetic modifications.179,180 Apoptosis and necrosis cascades are tightly regulated by several factors. Among these factors, Bcl-2 as an anti-apoptotic, and Bax protein as a pro-apoptotic, in addition to p53 protein (apoptosis inhibitor), have been well studied. The ratio of these proteins is considered the main indicator of the regulation of the apoptotic process because the ratio of Bax/Bcl-2 proteins increases during apoptosis.181,182 Apoptotic activity as a protective action of GDENs against the 2,3,7,8-tetracholorodibenzo-p-dioxin (TCDD) in colon cancer in male rats was evaluated and the results showed that GDENs controlled the ratio of these proteins and the expression of Bcl-2, Bax, and p53 genes, reduced the amount of necrosis/apoptosis, and inhibited the initiation of colon cancer.183 In another study, GDENs modified by folic acid could be adsorbed by colon cancer cells more efficiently. Modified folic acid-conjugated GDENs deliberately transferred a therapeutic agent (doxorubicin) to colon 26 tumors in vivo and thus increased the effect of chemotherapy to inhibit tumor growth as compared to the free drug.133 It is well understood that 6-gingerol and 6-shogaol, which are present in GDNEs, are important in the prevention of cancer cell development.166 In various human cancer cell lines, including colorectal cancer, leukotriene A4 hydrolase (LTA4H) is known to be overexpressed. 6-Gingerol is cytotoxic to HCT116 colon cancer cells and has been demonstrated to impede the action of LTA4H. Together with 6-gingerol, 6-shogaol has been shown to have impacts on various cancer signaling pathways. By inhibiting the activity of AKT, mTOR, fork transcription factors (FKHR), and glycogen synthase kinase-3 (GSK-3) in human A549 lung cancer cells, 6-shogaol inhibits the survival of the AKT/mTOR signaling pathway.184 Besides, 6-shogaol induces apoptosis by producing free radicals and releasing mitochondrial-related apoptotic molecules such as cytochrome C via caspases 3 and 9 via the p53 pathway.185 In MDA-MB-231 breast cancer cells, 6-shogaol inhibits another survival signaling pathway. 6-Shogaol blocks STAT3 and MAPK signaling pathways while JNK, p38 MAPK, and ERK are activated via ROS. Bcl-2, Bcl-xL, and survivin, which regulate tumor cell survival, proliferation, invasion, and metastasis, are modulated by 6-shogaol, leading to significant apoptosis.186 Also, 6-shogaol was found to trigger autophagy in human lung cancer cells by inhibiting the AKT/mTOR pathway.184 In addition, it was demonstrated that 6-gingerol decreases the expression of multidrug resistance-associated protein (MRP1) and glutathione-S-transferase (GSTπ) in human prostate cancer cells, which has an anti-proliferative impact.1874.5. Diabetes
In one study, 6-shogaol and 6-gingerol were shown to slow the development of diabetes and inhibit the formation of advanced glycated end products (AGEs) by trapping methylglyoxal (MGO), an AGE precursor. Furthermore, 6-gingerol decreases plasma glucose and insulin levels in obese rats.188 By activating Nrf2, 6-gingerol suppresses carboxymethyl lysine (CML), which is known as an AGE marker.189 6-Shogaol enhances glucose usage in 3T3-L1 adipocytes and C2C12 myotubes by enhancing AMPK phosphorylation. Enhancing glucagon-like peptide 1, 6-gingerol improves glucose tolerance and increases glucose-stimulated insulin production in type 2 diabetic rats (GLP-1).190 Furthermore, 6-gingerol treatment activates glycogen synthase 1 and enhances the expression of the glucose transporter cell type 4 (GLUT4) cell membrane, resulting in an increase in glycogen storage in skeletal muscle.189 In addition, taking 6-gingerol can help individuals with type 2 diabetes to decrease their fasting plasma glucose, glycosylated hemoglobin A (HbA1C), insulin, TG, and TC levels (DM2). As a result, treatment with 6-gingerol, a major component of GDENs, enhances insulin sensitivity in mice with metabolic syndrome, resulting in improved energy metabolism.154.6. Brain diseases
Because of the presence of the blood–brain barrier in the CNS, the brain is one of the most challenging organs for drug delivery. The BBB controls the transport of components into and out of the brain.191 However, nanoscale drug carriers such as PDENs, can pass through the BBB. For instance, the anti-glioma properties of ginseng-derived exosome-like nanoparticles have been proven by Kim, et al.192 The lipid constitution of GDENs showed 42% phosphatidic acid (PA), 27% di-galactosyl diacylglycerol (DGDG), and 19% monogalactosyldiacylglycerol (MGDG).193 As mentioned before, ginger exosomes contain many active components, including 6-gingerol and 6-shogaol, which possess proven anti-oxidative, anti-cancer, and anti-inflammatory properties.170 PA has important roles in the human nervous system functions; for instance, diacylglycerol kinase beta (DGKβ), an enzyme contributing to the production of phosphatidic acid, takes part in the conservation of neural networks participating in memory processes and also in enhancing the hippocampal long-term potentiation.194 PA is also a lipid messenger; mTOR and phosphatidylinositol 4 phosphate (PI4P)-5 kinase are two suggested downstream molecules195,196 and the role of mTOR in dendrite morphology and (PI4P)-5 kinase-related spine configuration has been shown.197–200 On the other hand, NLPR3 inflammasomes are being activated inappropriately in a variety of diseases, including neurodegenerative ones such as Alzheimer's disease201,202 and multiple sclerosis,203 and in mice models, inappropriate activity of NLRP3 inflammasomes plays a role in migraine pathogenicity.204,205 In 2019 Chen et al. showed that GDENs inhibited NLRP3 inflammasome activation111 due to the bad prognosis of the neurodegenerative diseases and their prevalence; targeting the NLRP3 activation pathway in these diseases would be a functional field of research.The expression of indole-3-carboxaldehyde (I3A) has been shown to increase interleukin-22 production taking part in the enhancement of the intestinal mucosal barrier. Interestingly, ginger exosome-like nanoparticles containing mdo-miR7267-3p can increase I3A expression by acting on the monooxygenase of normal intestinal flora and balancing the microbiota, preventing dysbiosis. Several studies have proven that the gut flora is altered in autism spectrum disorders (ASD) and it can be concluded that GDENs have potential therapeutic effects on ASD.135,206–208 Moreover, the intestinal barrier is also more permeable in anxiety and depression and with the effects of ginger nanoparticle-derived mdo-miR7267-3p on the intestinal barrier, new drug designs can be used for these disorders.209 HO-1 activity is detected in the preconditioned-induced preservation of the ischemic brain.210 HO-1 expression can also be increased in intestinal cells by GDENs to make the gut-brain axis but further studies need to take place for GDENs' role in neuro-inflammatory diseases.98
GDENs are shown to deliver siRNA for cancer suppression but their effect on neuroblastoma is not clearly defined.211 Teng et al. demonstrated that ginger exosomes can improve intestinal flora,135 while a varied microbiota profile in the guts of patients with Alzheimer's Disease was seen,212 as well as in patients suffering from Parkinson's Disease.213 Besides, a connection has been shown between altered intestinal flora and autism spectrum disorders through the gut-brain axis in a meta-analysis;214 it has been proven that Nrf2 activity can ameliorate the consequences of head trauma in mice.215 It has also been suggested that the liver can suffer less harm from alcohol usage via ginger-derived NP protection through the Nrf2 pathway, but future studies are needed for Nrf2 pathway activation in the ischemic brain by ginger-derived NPs.216 Furthermore, in PC12 pheochromocytoma neurons in rats, the Nrf2 pathway was activated, NQO-1 and HO-1 the phase II antioxidants were increased, and free radicals were lowered by the neuroprotective effects of 6-shogaol.217 It is worthwhile to mention that neuritogenesis was also increased, suggesting the effects of 6-shogaol on the memory and aging of the brain.218 To sum up, GDENs are expected to bring new insights into neuroscientific issues but more studies are needed.
5. Sustainability, biodegradability, and biocompatibility
In the context of materials chemistry, sustainability is a concept that embodies many green chemistry fundamentals.219 These concepts also include using less hazardous precursors for the development of nanomaterials, using water as a solvent whenever possible, as well as consuming fewer reagents, minimizing synthetic steps, reducing waste and byproducts, keeping reaction temperatures near room temperature, and cost-effectiveness.219,220Conventional differential velocity centrifugation with ultracentrifugation is a popular approach for the purification of edible nanoparticles, which has significant limitations in terms of its cost-effectiveness. From a sustainability point of view, the efficacy of the polyethylene glycol-6000-based purification technique is introduced as an alternative to the expensive ultracentrifugation process. The results have indicated that PEG-generated NPs have similar size, zeta potential, and biochemical components as compared to those of NPs generated by ultracentrifugation. Thus, the PEG technique described in this study will offer a more affordable alternative for purifying GDENs that can be utilized in medicinal formulations as a dietary supplement.101 Another study introduced a novel strategy for the high-yield purification of GDENs under low pH settings without compromising the primary bioactive components. Accordingly, the results demonstrated that accomplishment of PEG participation in low pH conditions increases the recovery of GDENs 4–5 times.127
On the other hand, the nanocarrier systems should essentially remain stable during the therapy and be eliminated from the body once their therapeutic function has been completed; thus, the biodegradability of the nanocarriers has great importance and should be investigated extensively.221 In research investigating the effectiveness of doxorubicin-loaded ginger-derived nano-vectors (GDNVs) on the treatment of colon cancer, the results of zeta potential and size distribution analysis proposed that these nanovectors are remarkably stable for 25 days if stored at 4 °C. This stability is a key characteristic in therapeutic delivery applications and suggests that ginger-derived lipids can be rearranged into stable NPs and produced in vast quantities.133
GDENs demonstrated their efficiency in inflammatory bowel disease treatment, so their stability, especially when delivered orally, is of great importance. Accordingly, researchers studied the stability of GDENs in the stomach- and intestine-like solutions and the results indicated that the GDENs’ size decreased slightly as compared to that of PBS. Besides, the pH value changed the GDENs’ zeta potential in the stomach- and intestine-like solutions; an acidic stomach-like environment made GDENs’ zeta potential weakly positive, while GDENs had a negative charge in neutral pH solution and intestine-like solutions. The aforementioned results proposed that GDENs are firm in solutions resembling stomach and intestinal fluids and the alternation of GDENs' zeta potential corresponds to their inherent characteristics. Thus, orally administered GDENs remain intact during the transition through the GI tract and target the colon effectively.72 Furthermore, another research studying GDENs stability in biofluids revealed that the size and zeta potential were altered slightly in biological fluids, suggesting that the ginger nanocarriers would be stable in the environments of the stomach and intestine. They also firmly assert that the zeta potential in both PBS and simulated intestinal fluid is negative, while negligibly positive in the simulated gastric fluid, which is due to the influence of the medium's pH on the zeta potential of GDENs, and this is why we observe a slight positive zeta potential in simulated gastric fluid with acidic pH.221
Regarding GDENs’ biocompatibility, a research project studied the effects of GDENs on the cell viability of Colon-26 and RAW 346 264.7 cell lines. This study indicated that cell treatment with up to 100 mg ml−1 GDEN for a day does not affect cell viability. This study examined the effects of GDENs on the integrity of the Caco2-BBE monolayers barrier function using electric cell-substrate impedance sensing experiments, and the results revealed no significant changes in the integrity of the barrier function of Caco2-BBE monolayers. The propidium iodide (PI)/Annexin V staining also indicated no significant increase in the amount of Colon-26 or RAW 264.7 apoptotic cells in the presence of less than 100 mg ml−1 GDENs. On the other hand, the in vivo toxicity assessment of GDENs demonstrated no considerable alternation in the activity of colonic myeloperoxidase or stimulation of pro-inflammatory cytokines in mice treated with ginger NPs. In addition, histological examination of the heart, liver, spleen, kidney, and lung in mice gavaged with GDENs suggested no morphological or pathological alternation in comparison with the control group. Accordingly, the obtained results suggest that GDENs are non-toxic in vitro and in vivo.72 Another in vivo study also indicated that surface decoration and reconstruction of GDENs could be a safe approach for disease treatment. In this study, Zhang et al. loaded nano-lipids, which were extracted from ginger, with siRNA-CD98 and orally administered to the mice. The complete blood count results presented no significant change in the number of red blood cells, white blood cells, and hemoglobin in mice treated with NPs compared to the control group. In addition, the results of renal and liver function tests including total bilirubin, blood urea nitrogen, alanine aminotransferase, and total protein showed that reconstructed ginger-derived nano-lipids were non-toxic for treated mice.84
The surface of the ginger nano-lipid was covered with folic acid and then loaded with doxorubicin as an anti-cancer medication. The results proposed that modified GDENs with folic acid target tumors more efficiently in mice with induced colon cancer as compared to non-modified ones loaded with doxorubicin. Furthermore, hematoxylin and eosin staining of tissues in folic acid/GDEN/doxorubicin-treated mice indicated no evidence of cellular or tissue damage in the liver, heart, kidney, spleen, or lung of mice in this group.133 Zhang et al. demonstrated that intestinal homeostasis was sustained by GDENs, while the gene expressions of HO-1 and IL-10 anti-inflammatory cytokines, as well as IL-6 and TNF-α pro-inflammatory cytokines, are upregulated in the presence of GDENs. These findings suggest that the oral administration of GDENs has no effect on cell viability and it is unable to cause in vivo local or systemic side effects.72 Altogether, an ample body of studies investigating the effects of GDENs on disease treatment reported ginger-derived NPs as a non-toxic and safe drug-delivery system that has the potential to be utilized in clinical practice in the future.
6. Scalable production: strategies for GDENs production and potential challenges
Low drug doses can be delivered to specific cell types and tissues using artificially synthesized NPs to decrease systemic side effects of medications and GDENs are among these synthetic NPs. Successfully synthesized NPs should meet two important criteria. First, the in vivo toxicity of these NPs should be examined and their safety should be confirmed before clinical application. Second, they should be producible on large scales at low cost.72 Natural NPs meet the aforementioned criteria as they are non-toxic and can be produced on an industrial scale at an affordable price.109 The latest investigations have characterized exosome-like NPs that were extracted from plants utilizing eco-friendly protocols.98 The rhizome of Z. officinale is an extensively used natural product, which is not only consumed as a spice but also serves as a treatment for various digestive tract disorders including colic, diarrhea, and dyspepsia.222–225Compared to synthetic nanocarriers, plant-derived extracellular vesicles (plant-derived EVs) including ginger-derived vesicles have several benefits as therapeutic agents. These include increased internalization rates, lower immunogenicity and bio-toxicity, stability in the GI tract, and the capability to pass the blood–brain barrier. Furthermore, they can also be manufactured on large scales with reasonable prices and deliver a variety of medications to the target region properly.70 However, despite plant-derived EVs’ benefits in terms of therapeutic efficacy, they lack established, efficient, convenient, and affordable guidelines for their separation and application.226
At present, different strategies have been introduced for plant-derived EVs including ginger-derived nanovesicle isolation; however, none of these strategies have gained desirable results in terms of their applicability, extraction pace and yield, and purity.226 Ultracentrifugation followed by sucrose gradient density centrifugation is a prevalent strategy for the isolation of plant-derived EV among all other isolation methods.70 Although ultracentrifugation provides plant-derived EVs with high purity, it is a time-consuming procedure that is highly dependent on instruments. In addition, this method has a low extraction efficacy and the obtained plant-derived EVs are sometimes aggregated. To address these limitations, scientists suggest taking advantage of high-density iodosanol pads for the enhancement of plant-derived EV extraction while minimizing the aggravation of protein pollution.227 This modified protocol is a simple operation approach that has a higher extraction yield and purity; however, the requirement of costly ultra-high-speed centrifuges and the vesicles’ strong precipitation aggregation force restrict its application in industries. PEG precipitation has similar vesicle extraction steps to ultracentrifugation in terms of eliminating impurities. The zeta potential of the obtained vesicle is similar, while the average diameter of extracellular vesicles formed using this technique is slightly smaller than that obtained by ultracentrifugation.101 PEG precipitation seems to be an efficient and straightforward strategy for GDEN extraction, which can be used for large-scale and economic production of GDENs; however, its application in GDENs production is constrained by its low purity and high cost.228 Size exclusion chromatography is another plant-derived extracellular vesicle extraction approach that is rarely used in laboratories but it provides more uniform vesicles with a significantly increased extraction rate per unit mass of plants as compared to the aforementioned methods. The noteworthy challenge restricting the usage of size exclusion chromatography for plant-derived vesicle extraction is its long vesicle separation procedure. Thus, size exclusion chromatography is not a suitable vesicle extraction approach for GDNP mass production due to its time-consuming and inconvenient procedure, as well as difficulty in the separation of large-size impurities.229 There are also convenient commercially available plant-derived EV separation kits that not only have low separation efficacy but also separate a limited number of vesicles each time, so they are not suitable candidates for large-scale GDNP production.226 Briefly, the goal of producing plant-derived EVs in large quantities is to find a more practical and cost-effective extraction technique, which also preserves extraction quality. Thus, given the benefits and drawbacks of each technique, it seems that combining the aforementioned separation techniques will lead to the extraction of ginger-derived extracellular vesicles with higher quality.
7. Conclusion
The salient advantages of PDENs such as good stability in the GI tract, excellent rate of internalization, low immunogenicity, biodegradability, and biocompatibility/low toxicity make these NPs promising candidates for cancer therapy and the targeted drug delivery of small molecular chemicals/therapeutic agents or nucleic acid drugs. Among them, GDENs have been widely explored because of their fascinating properties such as low toxicity and immunogenicity. In this context, surface modification utilizing nanoscale biomaterials can help to tune the targeting properties of GDENs, paving the way for designing next-generation delivery nanosystems for the treatment of cancers and other chronic diseases. PDENs with the capability of targeting intestinal cells and enduring activity in colitis should be further explored, especially for the induction of apoptosis in tumors. Besides, since these NPs can accumulate in the liver, they can be deployed to provide protective effects for the alcohol-stimulated liver. Several studies have been focused on their therapeutic effects for combating microbial and viral infections, especially in the case of coronaviruses. On the other hand, because of their potential for entering the brain through the blood–brain barrier, future explorations should be conducted on their inhibitory effects against brain tumors/cancers or other neurological diseases/disorders. However, there is a lack of established and affordable guidelines for their separation/isolation, as well as limited clinical trials and clinical translation studies. Remarkably, the systematic in vitro/in vivo toxicological studies along with feasible studies for large-scale production of these NPs are crucial challenges, especially for their future clinical applications.Author contributions
Faegheh Bahri: preforming the experiment; analyzing the data. Mahna Mansoori: data collection and writing the draft manuscript. Shayan Favaei: data collection and writing the draft manuscript. Saba Fouladi: data collection and writing the draft manuscript. Yousof Mir: figure preparation. Mehrnaz Mehrabani: critically revising the manuscript. Yaser Hozhabri: data collection and writing the draft manuscript. Mohammad Hadi Nematollahi: supervising; designing the project; editing and revising the manuscript. Siavash Iravani: supervising; designing the project; editing and revising the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by grant No. 401000945 from the Neuroscience Research Center, Institute of Neuropharmacology, Kerman University of Medical Sciences and the authors greatly acknowledge the support for the financial support. The protocol was approved by the ethics committee of Kerman University of Medical Sciences, Kerman, Iran (IR.KMU.REC.1401.567).References
- H. R. Poursalehi, M. S. Fekri, F. S. Far, A. Mandegari, A. Izadi, R. Mahmoodi, H. Nematollahi, F. Porgholamhosein, V. Ghorani and M. S. Fekri, Avicenna J. Phytomed., 2018, 8, 263 CAS.
- M. A. Rajizadeh, M. H. Nematollahi, E. Jafari, M. A. Bejeshk, M. Mehrabani, M. S. Razeghinia and H. Najafipour, OpenNano, 2023, 100129 CrossRef.
- Z. Sarhadynejad, F. Sharififar, A. Pardakhty, M.-H. Nematollahi, S. Sattaie-Mokhtari and A. Mandegary, J. Ethnopharmacol., 2016, 190, 387–395 CrossRef PubMed.
- K. Amiri, S. Nasibi, M. Mehrabani, M. H. Nematollahi and M. F. Harandi, Exp. Parasitol., 2019, 199, 111–115 CrossRef PubMed.
- M. Raeiszadeh, A. Pardakhty, F. Sharififar, M. Mehrabani and M. Mehrabani, Iran. J. Pharm. Res., 2018, 17, 804 CAS.
- C. A. Demeke, A. E. Woldeyohanins and Z. D. Kifle, Metabolism Open, 2021, 12, 100141 CrossRef CAS PubMed.
- M. M. Oloumi, D. Vosough, A. Derakhshanfar and M. H. Nematollahi, J. Equine Veterinary Sci., 2011, 31, 470–474 CrossRef.
- N. Ebrahimpour, M. Mehrabani, M. Iranpour, Z. Kordestani, M. Mehrabani, M. H. Nematollahi, A. Asadipour, M. Raeiszadeh and M. Mehrbani, J. Ethnopharmacol., 2020, 252, 112570 CrossRef CAS PubMed.
- H. Harandi, S. K. Falahati-Pour, M. Mahmoodi, S. Faramarz, H. Maleki, F. B. Nasab, H. Shiri, S. Fooladi and M. H. Nematollahi, Mol. Biol. Rep., 2022, 1–9 Search PubMed.
- M. L. Sargazi, K. B. Juybari, M. E. Tarzi, A. Amirkhosravi, M. H. Nematollahi, S. Mirzamohammdi, M. Mehrbani, M. Mehrabani and M. Mehrabani, Mol. Biol. Rep., 2021, 48, 6413–6421 CrossRef CAS PubMed.
- A. Bastin, A. Sadeghi, M. H. Nematollahi, M. Abolhassani, A. Mohammadi and H. Akbari, J. Cell. Physiol., 2021, 236, 2790–2799 CrossRef CAS PubMed.
- T. Volqvartz, A. L. Vestergaard, S. Aagaard, M. Andreasen, I. Lesnikova, N. Uldbjerg, A. Larsen and P. Bor, BMC Complementary Altern. Med., 2019, 19, 5 CrossRef PubMed.
- A. E. Kate and P. P. Sutar, J. Food Sci., 2020, 85, 432–441 CrossRef CAS PubMed.
- S. M. Mohammad and H. K. Hamed, J. Med. Plants Res., 2012, 6, 4255–4258 Search PubMed.
- Q.-Q. Mao, X.-Y. Xu, S.-Y. Cao, R.-Y. Gan, H. Corke, T. Beta and H.-B. Li, Foods, 2019, 8, 185 CrossRef CAS PubMed.
- R. Indiarto and E. Subroto, Angeline and Selly, Food Res., 2021, 5, 497–505 Search PubMed.
- M. H. Shahrajabian, W. Sun and Q. Cheng, Acta Agric. Scand., Sect. B, 2019, 69, 546–556 CAS.
- M. Zhang, R. Zhao, D. Wang, L. Wang, Q. Zhang, S. Wei, F. Lu, W. Peng and C. Wu, Phytother. Res., 2021, 35, 711–742 CrossRef CAS PubMed.
- V. S. Govindarajan and D. W. Connell, Crit. Rev. Food Sci. Nutr., 1983, 17, 189–258 CrossRef PubMed.
- M. S. Baliga, R. Haniadka, M. M. Pereira, J. J. D’Souza, P. L. Pallaty, H. P. Bhat and S. Popuri, Crit. Rev. Food Sci. Nutr., 2011, 51, 499–523 CrossRef CAS PubMed.
- S. Tong, J. Li and Y.-M. Wen, Reference Module in Biomedical Sciences, Elsevier, 2019 DOI:10.1016/B978-0-12-801238-3.11319-4.
- F. Rayati, F. Hajmanouchehri and E. Najafi, Dent. Res. J., 2017, 14, 1–7 CrossRef PubMed.
- 1999, 130, 459.
- J. R. G. D. S. Almeida, G. R. Souza, J. C. Silva, S. R. G. D. L. Saraiva, R. G. D. O. Júnior, J. D. S. S. Quintans, R. D. S. S. Barreto, L. R. Bonjardim, S. C. D. H. Cavalcanti and L. J. Q. Junior, Sci. World J., 2013, 2013, 808460 Search PubMed.
- F. R. Pinheiro-Neto, E. M. Lopes, B. T. Acha, L. D. S. Gomes, W. A. Dias, A. C. D. Reis Filho, B. S. Leal, D. Rodrigues, J. D. N. Silva, D. Dittz, P. M. P. Ferreira and F. R. C. Almeida, Toxicol. Appl. Pharmacol., 2021, 418, 115497 CrossRef CAS PubMed.
- B. A. Ferreira, R. F. Silva, F. B. R. de Moura, C. T. Narduchi, S. R. Deconte, P. Sartorelli, T. C. Tomiosso, J. H. G. Lago and F. A. Araújo, Nat. Prod. Res., 2021, 1–5, DOI:10.1080/14786419.2021.2019729.
- S. Girisa, B. Shabnam, J. Monisha, L. Fan, C. E. Halim, F. Arfuso, K. S. Ahn, G. Sethi and A. B. Kunnumakkara, Molecules, 2019, 24(4), 734 CrossRef CAS PubMed.
- R. M. T. de Lima, A. C. Dos Reis, A. P. M. de Menezes, J. V. O. Santos, J. Filho, J. R. O. Ferreira, M. de Alencar, A. da Mata, I. N. Khan, A. Islam, S. J. Uddin, E. S. Ali, M. T. Islam, S. Tripathi, S. K. Mishra, M. S. Mubarak and A. A. C. Melo-Cavalcante, Phytother. Res., 2018, 32, 1885–1907 CrossRef CAS PubMed.
- N. F. N. Mohd Sahardi and S. Makpol, J. Evidence-Based Complementary Altern. Med., 2019, 2019, 5054395 Search PubMed.
- A. Almatroudi, M. A. Alsahli, F. Alrumaihi, K. S. Allemailem and A. H. Rahmani, Curr. Pharm. Biotechnol., 2019, 20, 5–16 CAS.
- B. Ahmad, M. U. Rehman, I. Amin, A. Arif, S. Rasool, S. A. Bhat, I. Afzal, I. Hussain, S. Bilal and M. Mir, Sci. World J., 2015, 2015, 816364 Search PubMed.
- M. U. Akbar, K. Rehman, K. M. Zia, M. I. Qadir, M. S. H. Akash and M. Ibrahim, Crit. Rev. Eukaryotic Gene Expression, 2018, 28, 17–24 CrossRef PubMed.
- S. Suk, G. T. Kwon, E. Lee, W. J. Jang, H. Yang, J. H. Kim, N. R. Thimmegowda, M. Y. Chung, J. Y. Kwon, S. Yang, J. K. Kim, J. H. Y. Park and K. W. Lee, Mol. Nutr. Food Res., 2017, 61(10) DOI:10.1002/mnfr.201700139.
- M. Feldmann and S. R. Maini, Immunol. Rev., 2008, 223, 7–19 CrossRef CAS PubMed.
- B. Ahmad, M. U. Rehman, I. Amin, A. Arif, S. Rasool, S. A. Bhat, I. Afzal, I. Hussain, S. Bilal and M. U. R. Mir, Sci. World J., 2015, 2015, 816364 Search PubMed.
- K. Ji, L. Fang, H. Zhao, Q. Li, Y. Shi, C. Xu, Y. Wang, L. Du, J. Wang and Q. Liu, Oxid. Med. Cell. Longevity, 2017, 2017, 1480294 Search PubMed.
- M. Morvaridzadeh, S. Fazelian, S. Agah, M. Khazdouz, M. Rahimlou, F. Agh, E. Potter, S. Heshmati and J. Heshmati, Cytokine, 2020, 135, 155224 CrossRef CAS PubMed.
- S.-C. Ho, K.-S. Chang and C.-C. Lin, Food Chem., 2013, 141, 3183–3191 CrossRef CAS PubMed.
- F. Fadaki, M. Modaresi and I. Sajjadian, Indian J. Pharm. Educ. Res., 2017, 51(3), S159–S162 CrossRef CAS.
- M. Rondanelli, F. Fossari, V. Vecchio, C. Gasparri, G. Peroni, D. Spadaccini, A. Riva, G. Petrangolini, G. Iannello, M. Nichetti, V. Infantino and S. Perna, Phytother. Res., 2020, 34(11), 2843–2856 CrossRef PubMed.
- F. Kashefi, M. Khajehei, M. Tabatabaeichehr, M. Alavinia and J. Asili, Pain Manage. Nursing, 2014, 15, 826–833 CrossRef PubMed.
- N. Aryaeian, F. Shahram, M. Mahmoudi, H. Tavakoli, B. Yousefi, T. Arablou and S. Jafari Karegar, Gene, 2019, 698, 179–185 CrossRef CAS PubMed.
- E. M. Bartels, V. N. Folmer, H. Bliddal, R. D. Altman, C. Juhl, S. Tarp, W. Zhang and R. Christensen, Osteoarthritis Cartilage, 2015, 23, 13–21 CrossRef CAS PubMed.
- S. Prasad and A. K. Tyagi, Gastroenterology Res. Practice, 2015, 2015, 142979 CrossRef PubMed.
- Y. A. Yusof, N. Ahmad, S. Das, S. Sulaiman and N. A. Murad, Afr. J. Tradit., Complementary Altern. Med., 2008, 6, 87–93 Search PubMed.
- W. P. Chang and Y. X. Peng, Cancer Nursing, 2019, 42, E14–E23 CrossRef PubMed.
- J. Walstab, D. Krüger, T. Stark, T. Hofmann, I. E. Demir, G. O. Ceyhan, B. Feistel, M. Schemann and B. Niesler, Neurogastroenterol. Motil., 2013, 25, 439–e302 CrossRef CAS PubMed.
- J. J. Stewart, M. J. Wood, C. D. Wood and M. E. Mims, Pharmacology, 1991, 42, 111–120 CrossRef CAS PubMed.
- F. Sharifzadeh, M. Kashanian, J. Koohpayehzadeh, F. Rezaian, N. Sheikhansari and N. Eshraghi, J. Matern.-Fetal Neonat. Med., 2018, 31, 2509–2514 CrossRef CAS PubMed.
- A. J. Akinyemi, G. R. Thome, V. M. Morsch, N. Stefanello, J. F. Goularte, A. Belló-Klein, G. Oboh and M. R. C. Schetinger, J. Funct. Foods, 2015, 17, 792–801 CrossRef CAS.
- E. A. Townsend, M. E. Siviski, Y. Zhang, C. Xu, B. Hoonjan and C. W. Emala, Am. J. Respir. Cell Mol. Biol., 2013, 48, 157–163 CrossRef CAS PubMed.
- Z. Li, Z. Liu, V. V. S. Uddandrao, P. Ponnusamy, S. Balakrishnan, P. Brahmanaidu, S. Vadivukkarasi and S. Ganapathy, J. Environ. Pathol., Toxicol. Oncol., 2019, 38, 41–50 CrossRef PubMed.
- Y. Li, B. Xu, M. Xu, D. Chen, Y. Xiong, M. Lian, Y. Sun, Z. Tang, L. Wang, C. Jiang and Y. Lin, Pharmacol. Res., 2017, 119, 137–148 CrossRef CAS PubMed.
- B. K. Tan and J. Vanitha, Curr. Med. Chem., 2004, 11, 1423–1430 CrossRef CAS PubMed.
- N. Vijendra Kumar, P. S. Murthy, J. R. Manjunatha and B. K. Bettadaiah, Food Chem., 2014, 159, 451–457 CrossRef CAS PubMed.
- M. Sahoo, L. Jena, S. N. Rath and S. Kumar, Genomics Bioinf., 2016, 14, 96–103 Search PubMed.
- I. P. Kaur, P. K. Deol, K. K. Kondepudi and M. Bishnoi, Curr. Pharm. Des., 2016, 22, 4160–4172 CrossRef CAS PubMed.
- B. H. Ali, G. Blunden, M. O. Tanira and A. Nemmar, Food Chem. Toxicol., 2008, 46, 409–420 CrossRef CAS PubMed.
- R. Haniadka, E. Saldanha, V. Sunita, P. L. Palatty, R. Fayad and M. S. Baliga, Food Funct., 2013, 4, 845–855 RSC.
- M. Zhang, R. Zhao, D. Wang, L. Wang, Q. Zhang, S. Wei, F. Lu, W. Peng and C. Wu, Phytother. Res., 2021, 35, 711–742 CrossRef CAS PubMed.
- N. Sritoomma, W. Moyle, M. Cooke and S. O'Dwyer, Complementary Ther. Med., 2014, 22, 26–33 CrossRef PubMed.
- G. H. Park, J. H. Park, H. M. Song, H. J. Eo, M. K. Kim, J. W. Lee, M. H. Lee, K.-H. Cho, J. R. Lee and H. J. Cho, BMC Complementary Altern. Med., 2014, 14, 1–8 CrossRef PubMed.
- H. Quach, T. V. Le, T. T. Nguyen, P. Nguyen, C. K. Nguyen and L. H. Dang, Pharmaceutics, 2022, 14(8), 1654 CrossRef CAS PubMed.
- H. Zhang, Q. Wang, C. Sun, Y. Zhu, Q. Yang, Q. Wei, J. Chen, W. Deng, M. Adu-Frimpong, J. Yu and X. Xu, Pharmaceutics, 2019, 11(3), 107 CrossRef PubMed.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789–804 CrossRef CAS PubMed.
- D. W. Greening, R. Xu, H. Ji, B. J. Tauro and R. J. Simpson, Methods Mol. Biol., 2015, 1295, 179–209 CrossRef CAS PubMed.
- H. Kalra, C. G. Adda, M. Liem, C. S. Ang, A. Mechler, R. J. Simpson, M. D. Hulett and S. Mathivanan, Proteomics, 2013, 13, 3354–3364 CrossRef CAS PubMed.
- K. Rilla, A. M. Mustonen, U. T. Arasu, K. Härkönen, J. Matilainen and P. Nieminen, Matrix Biol., 2019, 75–76, 201–219 CrossRef CAS PubMed.
- H. Alqurashi, I. Ortega Asencio and D. W. Lambert, Tissue Eng., Part B, 2020, 530–538 Search PubMed.
- C. Yang, M. Zhang and D. Merlin, J. Mater. Chem. B, 2018, 6, 1312–1321 RSC.
- M. Zhang, E. Viennois, C. Xu and D. Merlin, Tissue Barriers, 2016, 4, e1134415 CrossRef PubMed.
- M. Zhang, E. Viennois, M. Prasad, Y. Zhang, L. Wang, Z. Zhang, M. K. Han, B. Xiao, C. Xu and S. Srinivasan, Biomaterials, 2016, 101, 321–340 CrossRef CAS PubMed.
- Q. Wang, X. Zhuang, J. Mu, Z.-B. Deng, H. Jiang, L. Zhang, X. Xiang, B. Wang, J. Yan and D. Miller, Nat. Commun., 2013, 4, 1–13 Search PubMed.
- A. Kumar, K. Sundaram, Y. Teng, J. Mu, M. K. Sriwastva, L. Zhang, J. L. Hood, J. Yan, X. Zhang, J. W. Park, M. L. Merchant and H. G. Zhang, Theranostics, 2022, 12, 1388–1403 CrossRef CAS PubMed.
- M. H. Shahrajabian, W. Sun and Q. Cheng, Acta Agric. Scand., Sect. B, 2019, 69, 546–556 CAS.
- K. Kanpairo, Burapha Sci. J., 2018, 23, 944–958 Search PubMed.
- M. Yavari, M. R. Jaafari, F. Mirzavi, G. Mosayebi, A. Ghazavi and A. Ganji, Iran. J. Basic Med. Sci., 2022, 25, 890 Search PubMed.
- K. M. Suman, A. Gupta, D. Vaidya and R. Kaushik, Pharm. Innov. J., 2021, 10, 608–613 Search PubMed.
- A. F. Pulungan, D. D. O. Sitepu and D. M. Sinaga, J. Penelitian Farmasi Herbal, 2018, 1, 1–5 CrossRef.
- L. Zhen, Q. Wei, Q. Wang, H. Zhang, M. Adu-Frimpong, C. Kesse Firempong, X. Xu and J. Yu, Pharm. Dev. Technol., 2020, 25, 1–8 CrossRef CAS PubMed.
- J. San Chang, K. C. Wang, C. F. Yeh, D. E. Shieh and L. C. Chiang, J. Ethnopharmacol., 2013, 145, 146–151 CrossRef PubMed.
- S. J. T. Lachumy, S. Sasidharan, V. Sumathy and Z. Zuraini, Asian Pac. J. Trop. Med., 2010, 3, 769–774 CrossRef.
- A. Binti Anzian, S. Rashidah, S. Nazamid, C. W. N. S. Binti Che and W. Sapawi, Food Appl. Biosci. J., 2017, 5, 32–49 Search PubMed.
- M. Zhang, X. Wang, M. K. Han, J. F. Collins and D. Merlin, Nanomedicine, 2017, 12, 1927–1943 CrossRef CAS PubMed.
- D. Azhir, A. Zakeri and A. Kargare-Rezapour, Eur. J. Exp. Biol., 2012, 2, 2090–2092 Search PubMed.
- N. Ratcharin, P. Wongtrakul and R. Indranupakorn, 2012.
- E. Jayashree, R. Visvanathan and J. Zachariah, J. Food Sci. Technol., 2014, 51, 3190–3198 CrossRef CAS PubMed.
- J. Wattanathorn, N. Palachai, W. Thukham-Mee and S. Muchimapura, Oxidative Med. Cellular Longevity, 2020, 2020, 3096826 Search PubMed.
- S. Jafarnejad, S. A. Keshavarz, S. Mahbubi, S. Saremi, A. Arab, S. Abbasi and K. Djafarian, J. Funct. Foods, 2017, 29, 127–134 CrossRef CAS.
- A. H. Alkhathlan, H. A. Al-Abdulkarim, M. Khan, M. Khan, M. Alkholief, A. Alshamsan, A. Almomen, N. Albekairi, H. Z. Alkhathlan and M. R. H. Siddiqui, Materials, 2021, 14, 3368 CrossRef CAS PubMed.
- R. Lantz, G. Chen, M. Sarihan, A. Solyom, S. Jolad and B. Timmermann, Phytomedicine, 2007, 14, 123–128 CrossRef CAS PubMed.
- S. Munda, S. Dutta, S. Haldar and M. Lal, J. Essential Oil Bearing Plants, 2018, 21, 994–1002 CrossRef CAS.
- A. S. Keyhanmehr, S. Kolouri, G. Heydarirad, B. Mofid and S. H. Mosavat, Complementary Ther. Clin. Practice, 2018, 31, 175–180 CrossRef PubMed.
- E. Casula, M. L. Manca, M. Perra, J. L. Pedraz, T. B. Lopez-Mendez, A. Lozano, E. Calvo, M. Zaru and M. Manconi, Antioxidants, 2021, 10, 1109 CrossRef CAS PubMed.
- W. Halperin and W. A. Jensen, J. Ultrastruct. Res., 1967, 18, 428–443 CrossRef CAS PubMed.
- R. Marchant and A. W. Robards, Ann. Bot., 1968, 32, 457–471 CrossRef.
- B. D. Rutter and R. W. Innes, Plant Physiol., 2016, 173, 728–741 CrossRef PubMed.
- J. Mu, X. Zhuang, Q. Wang, H. Jiang, Z. B. Deng, B. Wang, L. Zhang, S. Kakar, Y. Jun and D. Miller, Mol. Nutr. Food Res., 2014, 58, 1561–1573 CrossRef CAS PubMed.
- Q. Wang, Y. Ren, J. Mu, N. K. Egilmez, X. Zhuang, Z. Deng, L. Zhang, J. Yan, D. Miller and H.-G. Zhang, Cancer Res., 2015, 75, 2520–2529 CrossRef CAS PubMed.
- M. De Palma, A. Ambrosone, A. Leone, P. Del Gaudio, M. Ruocco, L. Turiák, R. Bokka, I. Fiume, M. Tucci and G. Pocsfalvi, Plants, 2020, 9, 1777 CrossRef CAS PubMed.
- S. P. Kalarikkal, D. Prasad, R. Kasiappan, S. R. Chaudhari and G. M. Sundaram, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- G. Van Niel, G. d'Angelo and G. Raposo, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS PubMed.
- J. Xiao, S. Feng, X. Wang, K. Long, Y. Luo, Y. Wang, J. Ma, Q. Tang, L. Jin and X. Li, PeerJ, 2018, 6, e5186 CrossRef PubMed.
- M. Regente, M. Pinedo, H. San Clemente, T. Balliau, E. Jamet and L. de la Canal, J. Exp. Bot., 2017, 68, 5485–5495 CrossRef CAS PubMed.
- Q. Cai, L. Qiao, M. Wang, B. He, F. M. Lin, J. Palmquist, S. D. Huang and H. Jin, Science, 2018, 360, 1126–1129 CrossRef CAS PubMed.
- J. H. Lee, Y. G. Kim, P. Choi, J. Ham, J. G. Park and J. Lee, Front. Cell. Infect. Microbiol., 2018, 8, 299 CrossRef PubMed.
- X. Zhuang, Y. Teng, A. Samykutty, J. Mu, Z. Deng, L. Zhang, P. Cao, Y. Rong, J. Yan, D. Miller and H.-G. Zhang, Mol. Ther., 2016, 24, 96–105 CrossRef CAS PubMed.
- H. A. Dad, T.-W. Gu, A.-Q. Zhu, L.-Q. Huang and L.-H. Peng, Mol. Ther., 2021, 29, 13–31 CrossRef CAS PubMed.
- X. Zhuang, Z.-B. Deng, J. Mu, L. Zhang, J. Yan, D. Miller, W. Feng, C. J. McClain and H.-G. Zhang, J. Extracell. Vesicles, 2015, 4, 28713 CrossRef PubMed.
- R. Markam and A. Bajpai, React. Funct. Polym., 2020, 149, 104520 CrossRef CAS.
- X. Chen, Y. Zhou and J. Yu, Mol. Pharmaceutics, 2019, 16, 2690–2699 CrossRef CAS PubMed.
- M. E. Nielsen, A. Feechan, H. Böhlenius, T. Ueda and H. Thordal-Christensen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11443–11448 CrossRef CAS PubMed.
- Y. Ding, J. Wang, J. Wang, Y.-D. Stierhof, D. G. Robinson and L. Jiang, Trends Plant Sci., 2012, 17, 606–615 CrossRef CAS PubMed.
- M. Alfieri, A. Leone and A. Ambrosone, Pharmaceutics, 2021, 13, 498 CrossRef CAS PubMed.
- A. L. Schuh, M. Hanna, K. Quinney, L. Wang, A. Sarkeshik, J. R. Yates III and A. Audhya, Biochem. J., 2015, 466, 625 CrossRef CAS PubMed.
- F. C. Reyes, R. Buono and M. S. Otegui, Curr. Opin. Plant Biol., 2011, 14, 666–673 CrossRef CAS PubMed.
- J. Wang, Y. Ding, J. Wang, S. Hillmer, Y. Miao, S. W. Lo, X. Wang, D. G. Robinson and L. Jiang, The Plant Cell, 2010, 22, 4009–4030 CrossRef CAS PubMed.
- Y. Ding, J. Wang, J. H. Chun Lai, V. H. Ling Chan, X. Wang, Y. Cai, X. Tan, Y. Bao, J. Xia and D. G. Robinson, Mol. Biol. Cell, 2014, 25, 412–426 CrossRef PubMed.
- S. J. Gould and G. Raposo, J. Extracell. Vesicles, 2013, 2, 20389 CrossRef PubMed.
- P. Pérez-Bermúdez, J. Blesa, J. M. Soriano and A. Marcilla, Eur. J. Pharm. Sci., 2017, 98, 40–50 CrossRef PubMed.
- G. Pocsfalvi, L. Turiák, A. Ambrosone, P. del Gaudio, G. Puska, I. Fiume, T. Silvestre and K. Vékey, J. Plant Physiol., 2018, 229, 111–121 CrossRef CAS PubMed.
- S. Raimondo, F. Naselli, S. Fontana, F. Monteleone, A. Lo Dico, L. Saieva, G. Zito, A. Flugy, M. Manno, M. A. Di Bella, G. De Leo and R. Alessandro, Oncotarget, 2015, 6(23), 19514–19527 CrossRef PubMed.
- M. Zhang, E. Viennois, M. Prasad, Y. Zhang, L. Wang, Z. Zhang, M. K. Han, B. Xiao, C. Xu, S. Srinivasan and D. Merlin, Biomaterials, 2016, 101, 321–340 CrossRef CAS PubMed.
- L. Yin, L. Yan, Q. Yu, J. Wang, C. Liu, L. Wang and L. Zheng, J. Agric. Food Chem., 2022, 70, 4725–4734 CrossRef CAS PubMed.
- Y. Zhang, Y. Liu, H. Liu and W. H. Tang, Cell Biosci., 2019, 9, 19 CrossRef PubMed.
- S. P. Kalarikkal and G. M. Sundaram, Toxicol. Appl. Pharmacol., 2021, 414, 115425 CrossRef CAS PubMed.
- A. P. Suresh, S. P. Kalarikkal, B. Pullareddy and G. M. Sundaram, ACS Omega, 2021, 6, 17635–17641 CrossRef CAS PubMed.
- Z. Zhang, Y. Yu, G. Zhu, L. Zeng, S. Xu, H. Cheng, Z. Ouyang, J. Chen, J. L. Pathak and L. Wu, Front. Immunol., 2022, 13, 896745 CrossRef CAS PubMed.
- S. Iravani and R. S. Varma, ACS Sustainable Chem. Eng., 2019, 7, 8055–8069 CrossRef CAS.
- T. Karamanidou and A. Tsouknidas, Int. J. Mol. Sci., 2021, 23, 191 CrossRef PubMed.
- A. R. Chin, M. Y. Fong, G. Somlo, J. Wu, P. Swiderski, X. Wu and S. E. Wang, Cell Res., 2016, 26, 217–228 CrossRef CAS PubMed.
- S. Ju, J. Mu, T. Dokland, X. Zhuang, Q. Wang, H. Jiang, X. Xiang, Z.-B. Deng, B. Wang and L. Zhang, Mol. Ther., 2013, 21, 1345–1357 CrossRef CAS PubMed.
- M. Zhang, B. Xiao, H. Wang, M. K. Han, Z. Zhang, E. Viennois, C. Xu and D. Merlin, Mol. Ther., 2016, 24, 1783–1796 CrossRef CAS PubMed.
- K. Sundaram, D. P. Miller, A. Kumar, Y. Teng, M. Sayed, J. Mu, C. Lei, M. K. Sriwastva, L. Zhang and J. Yan, iScience, 2019, 21, 308–327 CrossRef CAS PubMed.
- Y. Teng, Y. Ren, M. Sayed, X. Hu, C. Lei, A. Kumar, E. Hutchins, J. Mu, Z. Deng, C. Luo, K. Sundaram, M. K. Sriwastva, L. Zhang, M. Hsieh, R. Reiman, B. Haribabu, J. Yan, V. R. Jala, D. M. Miller, K. Van Keuren-Jensen, M. L. Merchant, C. J. McClain, J. W. Park, N. K. Egilmez and H.-G. Zhang, Cell Host Microbe, 2018, 24, 637–652.e638 CrossRef CAS PubMed.
- A. Kumar, Y. Ren, K. Sundaram, J. Mu, M. K. Sriwastva, G. W. Dryden, C. Lei, L. Zhang, J. Yan and X. Zhang, Theranostics, 2021, 11, 4061 CrossRef CAS PubMed.
- D. Ratnadewi, C. H. Widjaja, A. Barlian, R. M. Amsar, I. D. Ana, A. C. Hidajah, H. B. Notobroto and T. D. K. Wungu, Hayati J. Biosci., 2023, 30, 182–192 CrossRef.
- F. Man, C. Meng, Y. Liu, Y. Wang, Y. Zhou, J. Ma and R. Lu, AAPS PharmSciTech, 2021, 22, 1–10 CrossRef PubMed.
- J. Yan, S. Hou, Y. Yu, Y. Qiao, T. Xiao, Y. Mei, Z. Zhang, B. Wang, C.-C. Huang and C.-H. Lin, Colloids Surf., B, 2018, 171, 241–249 CrossRef CAS PubMed.
- X. Wang, M. Zhang, R. R. Woloshun, Y. Yu, J. K. Lee, S. R. Flores, D. Merlin and J. F. Collins, Nutrients, 2021, 13, 1686 CrossRef CAS PubMed.
- M. Zhang, C. Yang, X. Yan, J. Sung, P. Garg and D. Merlin, Adv. Ther., 2019, 2, 1900129 CrossRef CAS PubMed.
- P. D. Subudhi, C. Bihari, S. K. Sarin and S. Baweja, Nanotheranostics, 2022, 6, 365 CrossRef PubMed.
- O. K. Yemitan and M. C. Izegbu, Phytother. Res., 2006, 20, 997–1002 CrossRef PubMed.
- S. Raimondo, F. Naselli, S. Fontana, F. Monteleone, A. L. Dico, L. Saieva, G. Zito, A. Flugy, M. Manno and M. A. Di Bella, Oncotarget, 2015, 6, 19514 CrossRef PubMed.
- D. A. Johnson, S. Amirahmadi, C. Ward, Z. Fabry and J. A. Johnson, Toxicol. Sci, 2010, 114, 237–246 CrossRef CAS PubMed.
- A. F. Bakr, S. S. Abdelgayed, O. S. El-Tawil and A. M. Bakeer, Pak. Vet. J., 2019, 39, 479–486 CrossRef CAS.
- B. L. Woolbright and H. Jaeschke, J. Hepatol., 2017, 66, 836–848 CrossRef CAS PubMed.
- M. J. Hodgman and A. R. Garrard, Crit. Care Clin., 2012, 28, 499–516 CrossRef PubMed.
- F. Martinon, K. Burns and J. Tschopp, Mol. Cell, 2002, 10, 417–426 CrossRef CAS PubMed.
- M. Lamkanfi and V. M. Dixit, Annu. Rev. Cell Dev. Biol., 2012, 28, 137–161 CrossRef CAS PubMed.
- S. L. Masters, A. Simon, I. Aksentijevich and D. L. Kastner, Annu. Rev. Immunol., 2009, 27, 621 CrossRef CAS PubMed.
- F. Martinon, V. Pétrilli, A. Mayor, A. Tardivel and J. Tschopp, Nature, 2006, 440, 237–241 CrossRef CAS PubMed.
- P. J. Shaw, J. R. Lukens, S. Burns, H. Chi, M. A. McGargill and T.-D. Kanneganti, J. Immunol., 2010, 184, 4610–4614 CrossRef CAS PubMed.
- H. Wen, D. Gris, Y. Lei, S. Jha, L. Zhang, M. T.-H. Huang, W. J. Brickey and J. P. Ting, Nat. Immunol., 2011, 12, 408–415 CrossRef CAS PubMed.
- B. Vandanmagsar, Y.-H. Youm, A. Ravussin, J. E. Galgani, K. Stadler, R. L. Mynatt, E. Ravussin, J. M. Stephens and V. D. Dixit, Nat. Med., 2011, 17, 179–188 CrossRef CAS PubMed.
- P. Duewell, H. Kono, K. J. Rayner, C. M. Sirois, G. Vladimer, F. G. Bauernfeind, G. S. Abela, L. Franchi, G. Nuñez and M. Schnurr, Nature, 2010, 464, 1357–1361 CrossRef CAS PubMed.
- K. Rajamäki, J. Lappalainen, K. Öörni, E. Välimäki, S. Matikainen, P. T. Kovanen and K. K. Eklund, PLoS One, 2010, 5, e11765 CrossRef PubMed.
- M. M. Soliman and A. M. S. Elfeky, Natl. J. Physiol., Pharm. Pharmacol., 1970, 6, 349 CrossRef.
- M. Sanz, A. M. Del Castillo, S. Jepsen, J. R. Gonzalez-Juanatey, F. D’Aiuto, P. Bouchard, I. Chapple, T. Dietrich, I. Gotsman and F. Graziani, J. Clin. Periodontol., 2020, 47, 268–288 CrossRef PubMed.
- I. Olsen, J. D. Lambris and G. Hajishengallis, J. Oral Microbiol., 2017, 9, 1340085 CrossRef PubMed.
- L. S. Liu, N. Gkranias, B. Farias, D. Spratt and N. Donos, Clin. Oral Investig., 2018, 22, 2743–2762 CrossRef PubMed.
- C. Zenobia and G. Hajishengallis, Virulence, 2015, 6, 236–243 CrossRef CAS PubMed.
- R. J. Lamont, H. Koo and G. Hajishengallis, Nat. Rev. Microbiol., 2018, 16, 745–759 CrossRef CAS PubMed.
- K. Bao, G. N. Belibasakis, T. Thurnheer, J. Aduse-Opoku, M. A. Curtis and N. Bostanci, BMC Microbiol., 2014, 14, 1–8 CrossRef PubMed.
- Z. Qiao, K. Zhang, J. Liu, D. Cheng, B. Yu, N. Zhao and F.-J. Xu, Nat. Commun., 2022, 13, 7164 CrossRef CAS PubMed.
- V. Menon, M. Elgharib, R. El-awady and E. Saleh, Food Biosci., 2021, 41, 100934 CrossRef CAS.
- H. Chen, J. Fu, H. Chen, Y. Hu, D. N. Soroka, J. R. Prigge, E. E. Schmidt, F. Yan, M. B. Major and X. Chen, Chem. Res. Toxicol., 2014, 27, 1575–1585 Search PubMed.
- M. Saleh and G. Trinchieri, Nat. Rev. Immunol., 2011, 11, 9–20 CrossRef CAS PubMed.
- M. Zhang, J. F. Collins and D. Merlin, Journal, 2016, 12, 3035–3037 Search PubMed.
- Y. Mao, M. Han, C. Chen, X. Wang, J. Han, Y. Gao and S. Wang, Nanoscale, 2021, 13, 20157–20169 RSC.
- E. Viennois, B. Xiao, S. Ayyadurai, L. Wang, P. G. Wang, Q. Zhang, Y. Chen and D. Merlin, Lab. Invest., 2014, 94, 950–965 CrossRef CAS PubMed.
- R. Noubade, K. Wong, N. Ota, S. Rutz, C. Eidenschenk, P. A. Valdez, J. Ding, I. Peng, A. Sebrell and P. Caplazi, Nature, 2014, 509, 235–239 CrossRef CAS PubMed.
- S. Parkkila, A. Parkkila, T. Juvonen and H. Rajaniemi, Gut, 1994, 35, 646–650 CrossRef CAS PubMed.
- J. L. Coombes, K. R. Siddiqui, C. V. Arancibia-Cárcamo, J. Hall, C.-M. Sun, Y. Belkaid and F. Powrie, J. Exp. Med., 2007, 204, 1757–1764 CrossRef CAS PubMed.
- I.-K. Kwon, R. Wang, M. Thangaraju, H. Shuang, K. Liu, R. Dashwood, N. Dulin, V. Ganapathy and D. D. Browning, Oncogene, 2010, 29, 3423–3434 CrossRef CAS PubMed.
- D. D. Browning, I.-K. Kwon and R. Wang, Future Med. Chem., 2010, 2, 65–80 CrossRef CAS PubMed.
- M. Zhang, C. Xu, D. Liu, M. K. Han, L. Wang and D. Merlin, J. Crohn's Colitis, 2018, 12, 217–229 CrossRef PubMed.
- J. Luettig, R. Rosenthal, I. F. M. Lee, S. M. Krug and J. D. Schulzke, Mol. Nutr. Food Res., 2016, 60, 2576–2586 CrossRef CAS PubMed.
- P. Paydar, G. Asadikaram, H. Z. Nejad, H. Akbari, M. Abolhassani, V. Moazed, M. H. Nematollahi, G. Ebrahimi and H. Fallah, J. Cell. Biochem., 2019, 120, 13726–13736 CrossRef CAS PubMed.
- C. Whibley, P. D. Pharoah and M. Hollstein, Nat. Rev. Cancer, 2009, 9, 95–107 CrossRef CAS PubMed.
- I. Karmakar, S. Haldar, M. Chakraborty, K. Chaudhury, S. Dewanjee and P. K. Haldar, Pharm. Biol., 2016, 54, 503–508 CrossRef CAS PubMed.
- A. Mohammadpour-Gharehbagh, D. Jahantigh, M. Eskandari, M. H. Sadegh, M. H. Nematollahi, M. Rezaei, A. Rasouli, F. Eskandari, M. Z. Heydarabad and B. Teimoori, Apoptosis, 2019, 24, 301–311 CrossRef CAS PubMed.
- S. B. Abdu, F. Abdu and W. K. B. Khalil, Int. J. Pharm., 2017, 13, 946–957 CAS.
- J.-Y. Hung, Y.-L. Hsu, C.-T. Li, Y.-C. Ko, W.-C. Ni, M.-S. Huang and P.-L. Kuo, J. Agric. Food Chem., 2009, 57, 9809–9816 CrossRef CAS PubMed.
- M. H. Pan, M. C. Hsieh, J. M. Kuo, C. S. Lai, H. Wu, S. Sang and C. T. Ho, Mol. Nutr. Food Res., 2008, 52, 527–537 CrossRef CAS PubMed.
- S. M. Kim, C. Kim, H. Bae, J. H. Lee, S. H. Baek, D. Nam, W. S. Chung, B. S. Shim, S. G. Lee and S. H. Kim, Mol. Carcinog., 2015, 54, 1132–1146 CrossRef CAS PubMed.
- C.-M. Liu, C.-L. Kao, Y.-T. Tseng, Y.-C. Lo and C.-Y. Chen, Molecules, 2017, 22, 1477 CrossRef PubMed.
- Y. Zhu, Y. Zhao, P. Wang, M. Ahmedna and S. Sang, Chem. Res. Toxicol., 2015, 28, 1842–1849 Search PubMed.
- M. B. Samad, M. Mohsin, N. A. Bin, B. A. Razu, M. T. Hossain, S. Mahzabeen, N. Unnoor, I. A. Muna, F. Akhter and A. U. Kabir, BMC Complementary Altern. Med., 2017, 17, 1–13 Search PubMed.
- C.-K. Wei, Y.-H. Tsai, M. Korinek, P.-H. Hung, M. El-Shazly, Y.-B. Cheng, Y.-C. Wu, T.-J. Hsieh and F.-R. Chang, Int. J. Mol. Sci., 2017, 18, 168 Search PubMed.
- A. Poustforoosh, M. H. Nematollahi, H. Hashemipour and A. Pardakhty, J. Controlled Release, 2022, 343, 777–797 CrossRef CAS PubMed.
- J. Kim, Y. Zhu, S. Chen, D. Wang, S. Zhang, J. Xia, S. Li, Q. Qiu, H. Lee and J. Wang, J. Nanobiotechnol., 2023, 21, 253 CrossRef CAS PubMed.
- M. Zhang, E. Viennois, M. Prasad, Y. Zhang, L. Wang, Z. Zhang, M. K. Han, B. Xiao, C. Xu, S. Srinivasan and D. Merlin, Biomaterials, 2016, 101, 321–340 CrossRef CAS PubMed.
- Y. Shirai, T. Kouzuki, K. Kakefuda, S. Moriguchi, A. Oyagi, K. Horie, S.-Y. Morita, M. Shimazawa, K. Fukunaga, J. Takeda, N. Saito and H. Hara, PLoS One, 2010, 5, e11602 CrossRef PubMed.
- A. Avila-Flores, T. Santos, E. Rincón and I. Mérida, J. Biol. Chem., 2005, 280, 10091–10099 CrossRef CAS PubMed.
- C. Stace, M. Manifava, C. Delon, J. Coadwell, S. Cockcroft and N. T. Ktistakis, Adv. Enzyme Regul., 2008, 48, 55–72 CrossRef CAS PubMed.
- J. Jaworski and M. Sheng, Mol. Neurobiol., 2006, 34, 205–219 CrossRef CAS PubMed.
- V. Kumar, M. X. Zhang, M. W. Swank, J. Kunz and G. Y. Wu, J. Neurosci., 2005, 25, 11288–11299 CrossRef CAS PubMed.
- N. Takei, N. Inamura, M. Kawamura, H. Namba, K. Hara, K. Yonezawa and H. Nawa, J. Neurosci., 2004, 24, 9760–9769 CrossRef CAS PubMed.
- T. Takenawa and S. Suetsugu, Nat. Rev. Mol. Cell Biol., 2007, 8, 37–48 CrossRef CAS PubMed.
- A. Halle, V. Hornung, G. C. Petzold, C. R. Stewart, B. G. Monks, T. Reinheckel, K. A. Fitzgerald, E. Latz, K. J. Moore and D. T. Golenbock, Nat. Immunol., 2008, 9, 857–865 CrossRef CAS PubMed.
- M. T. Heneka, M. P. Kummer, A. Stutz, A. Delekate, S. Schwartz, A. Vieira-Saecker, A. Griep, D. Axt, A. Remus, T. C. Tzeng, E. Gelpi, A. Halle, M. Korte, E. Latz and D. T. Golenbock, Nature, 2013, 493, 674–678 CrossRef CAS PubMed.
- P. J. Shaw, J. R. Lukens, S. Burns, H. Chi, M. A. McGargill and T. D. Kanneganti, J. Immunol., 2010, 184, 4610–4614 CrossRef CAS PubMed.
- W. He, T. Long, Q. Pan, S. Zhang, Y. Zhang, D. Zhang, G. Qin, L. Chen and J. Zhou, J. Neuroinflammation, 2019, 16, 78 CrossRef PubMed.
- Y. Wang, Z. Shan, L. Zhang, S. Fan, Y. Zhou, L. Hu, Y. Wang, W. Li and Z. Xiao, J. Headache Pain, 2022, 23, 75 CrossRef CAS PubMed.
- M. Fiorentino, A. Sapone, S. Senger, S. S. Camhi, S. M. Kadzielski, T. M. Buie, D. L. Kelly, N. Cascella and A. Fasano, Mol. Autism, 2016, 7, 49 CrossRef PubMed.
- D. L. Coury, P. Ashwood, A. Fasano, G. Fuchs, M. Geraghty, A. Kaul, G. Mawe, P. Patterson and N. E. Jones, Pediatrics, 2012, 130, S160–S168 CrossRef PubMed.
- M. De Angelis, R. Francavilla, M. Piccolo, A. De Giacomo and M. Gobbetti, Gut Microb., 2015, 6, 207–213 CrossRef CAS PubMed.
- B. R. Stevens, R. Goel, K. Seungbum, E. M. Richards, R. C. Holbert, C. J. Pepine and M. K. Raizada, Gut, 2018, 67, 1555 CrossRef CAS PubMed.
- E. Zeynalov, Z. A. Shah, R.-C. Li and S. Doré, Neurobiol. Dis., 2009, 35, 264–269 CrossRef CAS PubMed.
- Z. Li, H. Wang, H. Yin, C. Bennett, H.-G. Zhang and P. Guo, Sci. Rep., 2018, 8, 14644 CrossRef PubMed.
- Z.-Q. Zhuang, L.-L. Shen, W.-W. Li, X. Fu, F. Zeng, L. Gui, Y. Lü, M. Cai, C. Zhu, Y.-L. Tan, P. Zheng, H.-Y. Li, J. Zhu, H.-D. Zhou, X.-L. Bu and Y.-J. Wang, J. Alzheimer's Dis., 2018, 63, 1337–1346 CAS.
- F. Scheperjans, V. Aho, P. A. Pereira, K. Koskinen, L. Paulin, E. Pekkonen, E. Haapaniemi, S. Kaakkola, J. Eerola-Rautio and M. Pohja, Mov. Disord, 2015, 30, 350–358 CrossRef PubMed.
- M. Xu, X. Xu, J. Li and F. Li, Front. Psychiatry, 2019, 10, 473 CrossRef PubMed.
- W. Jin, H. Wang, W. Yan, L. Zhu, Z. Hu, Y. Ding and K. Tang, J. Neurotrauma, 2009, 26, 131–139 CrossRef PubMed.
- X. Zhuang, Z. B. Deng, J. Mu, L. Zhang, J. Yan, D. Miller, W. Feng, C. J. McClain and H. G. Zhang, J. Extracell Vesicles, 2015, 4, 28713 CrossRef PubMed.
- S. L. S. Seow, S. L. Hong, G. S. Lee, S. N. A. Malek and V. Sabaratnam, BMC Complementary Altern. Med., 2017, 17, 334 CrossRef PubMed.
- W.-H. Hsu, N.-K. Huang, Y.-J. Shiao, C.-K. Lu, Y.-M. Chao, Y.-J. Huang, C.-H. Yeh and Y.-L. Lin, Phytomedicine, 2021, 87, 153576 CrossRef CAS PubMed.
- P. T. Anastas and M. M. Kirchhoff, Acc. Chem. Res., 2002, 35, 686–694 CrossRef CAS PubMed.
- J. A. Dahl, B. L. Maddux and J. E. Hutchison, Chem. Rev., 2007, 107, 2228–2269 CrossRef CAS PubMed.
- R. Markam, J. Bajpai and A. Bajpai, J. Drug Delivery Sci. Technol., 2019, 50, 355–364 CrossRef CAS.
- M. S. Butt and M. T. Sultan, Crit. Rev. Food Sci. Nutr., 2011, 51, 383–393 CrossRef CAS PubMed.
- S. Prasad and B. Aggarwal, J. Clin. Cell. Immunol., 2014, 5, 10.4172 Search PubMed.
- P. Karna, S. Chagani, S. R. Gundala, P. C. Rida, G. Asif, V. Sharma, M. V. Gupta and R. Aneja, Br. J. Nutr., 2012, 107, 473–484 CrossRef CAS PubMed.
- M. Brahmbhatt, S. R. Gundala, G. Asif, S. A. Shamsi and R. Aneja, Nutr. Cancer, 2013, 65, 263–272 CrossRef CAS PubMed.
- Y. Wang, J. Wang, J. Ma, Y. Zhou and R. Lu, Pharmaceuticals, 2022, 15, 708 CrossRef CAS PubMed.
- P. Duong, A. Chung, L. Bouchareychas and R. L. Raffai, PLoS One, 2019, 14, e0215324 CrossRef CAS PubMed.
- M. C. Deregibus, F. Figliolini, S. D'antico, P. M. Manzini, C. Pasquino, M. De Lena, C. Tetta, M. F. Brizzi and G. Camussi, Int. J. Mol. Med., 2016, 38, 1359–1366 CrossRef CAS PubMed.
- J. Y. You, S. J. Kang and W. J. Rhee, Bioactive Mater., 2021, 6, 4321–4332 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |