Open Access Article
Open Access ArticleSelf-actuated microfluidic chiplet for two-stage multiplex nucleic acid amplification assay†
Felix
Ansah
ab,
Marziyeh
Hajialyani
a,
Fatemeh
Ahmadi
 a,
Yuming
Gu
a,
Yuming
Gu
 a,
Ergün Alperay
Tarım
a,
Ergün Alperay
Tarım
 a,
Michael G.
Mauk
a,
Gordon A.
Awandare
b and
Haim H.
Bau
a,
Michael G.
Mauk
a,
Gordon A.
Awandare
b and
Haim H.
Bau
 *a
*a
aDepartment of Mechanical Engineering and Applied Mechanics, University of Pennsylvania, 233 Towne Building, Philadelphia, Pennsylvania 19104, USA. E-mail: bau@seas.upenn.edu; Fax: +1 215 573 6334
bWest African Centre for Cell Biology of Infectious Pathogens (WACCBIP), College of Basic and Applied Sciences, University of Ghana, Legon, Ghana
First published on 22nd October 2024
Abstract
Effective diagnosis of comorbidities and infectious diseases that present similar symptoms requires point-of-need assays capable of co-detecting and differentiating among multiple co-endemic pathogens to enable timely, precision medicine and effective control measures. We previously developed a two-stage isothermal amplification assay dubbed Penn-RAMP to address this need. Penn-RAMP's first stage comprises a recombinase polymerase amplification (RPA), which amplifies all targets of interest in a single reaction chamber for a short duration. The RPA amplicons are then aliquoted into multiple loop-mediated isothermal amplification (LAMP) reaction chambers, each customized with pre-dried primers to amplify a single target or a group of targets. To enable Penn-RAMP at the point of need, we describe here a self-actuated Penn-RAMP chiplet that accommodates the Penn-RAMP assay. Our chiplet employs temperature-controlled phase change valves and capillary valves to self-aliquot first-stage amplicons into multiple (five) second-stage reaction chambers and to seal these chambers. The functionality of our device is demonstrated by co-detecting plant pathogens. The analytical performance of our chiplet is comparable to that of the benchtop Penn-RAMP assay and surpasses that of standalone LAMP assays. Our self-actuated chiplet can be operated standalone with purified nucleic acids or as the downstream amplification module of a sample preparation cassette.
Introduction
Effective, rapid diagnosis of comorbidities and infectious diseases that present similar symptoms requires point-of-need assays capable of detecting and differentiating among multiple co-endemic pathogens to enable timely precision medicine and effective control measures.1,2 This need is further aggravated by the impact of climate change on microbe evolution, spread into and adaptation in new regions of the world, infecting previously unexposed hosts. Such point-of-need diagnostics are needed for human health to enable timely and effective therapies and on the farm to detect animal and plant diseases to facilitate effective control measures, assuring food security and avoiding staggering economic losses.Standard, cell-based diagnostic methods comprising culturing and microscopy are labor-intensive, have long turnaround times, and often do not provide strain-specific detection.3,4 Tests based on antigen–antibody interactions, such as serology and rapid diagnostic tests, usually have suboptimal sensitivity and poor specificity and, in the case of host antibodies, may detect past infections rather than active ones.5,6 Furthermore, such tests require lengthy development time compared to nucleic acid-based tests that can be implemented soon after the pathogen's nucleic acid sequence has been identified.
Nucleic acid-based amplification tests (NAATs) detect active infection with high sensitivity and specificity7 and can be adapted rapidly to address new threats. In addition, NAATs can detect drug resistance genotypes to enable precision medicine.8,9 Among the available NAATs, polymerase chain reaction (PCR) is widely used in clinical pathology labs and is considered the gold standard.10 However, widespread adaptation of PCR at the point of need faces significant challenges, especially in resource-poor settings, due to the lack of technical expertise and the need for costly infrastructure and laborious sample preparation.11,12
Recently, isothermal amplification assays such as recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP) have emerged as attractive alternatives to PCR.10,13,14 These methods do not require thermal cycling and thus can be carried out with simple incubators that consume less energy than PCR.10,14–16 Isothermal assays produce many more amplicons than PCR, enabling faster tests, often ranging from 10 to 30 min, and simpler detection methods.17 They are tolerant to temperature variations and contaminants, thus providing powerful, field-adaptable, user-friendly methods for diverse point-of-need applications, ranging from disease surveillance, clinical diagnostics, animal and plant disease control, and food-quality control to environmental sample monitoring, among others.10,16,18
Most point-of-need NAATs are either single-plex or capable of co-detecting just a couple of targets. Although it is possible to achieve high-level multiplexing by aliquoting a single sample into multiple reaction chambers, each specialized for a single target, increasing the number of targets will require a proportional large sample volume.19 Where sample volume is constrained, high-level multiplexing will inevitably result in sample dilution and loss of sensitivity.
To address the challenge of high-sensitivity, co-detection of multiple co-endemic targets, we previously described a multiplexed two-stage, isothermal amplification assay (Penn-RAMP)20 comprising a first-stage RPA reaction and a second-stage LAMP reaction. The first stage of Penn-RAMP includes forward and backward primers for all targets of interest and amplifies all these targets, if present in the sample, at approximately ∼42 °C for a short time, enough to produce multiple amplicons and retain high sensitivity upon dilution but not sufficient to produce a detectable signal. The first-stage amplicons are then aliquoted into multiple LAMP reaction chambers, each customized (with pre-stored, lyophilized primers) to amplify a single target or a group of targets at ∼65 °C, enabling co-detection of as many as 16 different targets. Penn-RAMP can also operate as a single-plex assay in a single pot to provide higher sensitivity than standalone RPA and standalone LAMP.20–22
This paper describes a novel, simple Penn-RAMP-chiplet to accommodate our Penn-RAMP assay. Our Penn-RAMP chiplet utilizes phase change valves, capillary imbibition, capillary valves for flow control, and phase change material for evaporation barriers during incubation and to entomb the reaction products after the test's conclusion. Our system operation is solely controlled by temperature variations. We operated our chiplet with a custom incubator that provided temperature control and included a USB camera to monitor, in real-time, fluorescent emission from the second-stage reaction chambers.
To verify our Penn-RAMP chiplet's functionality, we used it to amplify and detect tomato brown rugose fruit virus (ToBRFV), tobacco mosaic virus (TMV), and genomic DNA from tomato extract (internal positive control). Unless detected early, ToBRFV and TMV cause severe damage to vegetable crops worldwide. The analytical performance of our on-chip Penn-RAMP assay was comparable to benchtop Penn-RAMP and surpassed standalone LAMP. Our multiplex RAMP chiplet has the potential to significantly expand the use of nucleic acid-based assays at the point of need for applications such as disease surveillance, precision medicine, and ensuring food safety and security.
Methods
Microfluidic chiplet architecture and fabrication
Our 3D-printed chiplet measures 29 mm (length) × 24 mm (width) × 14 mm (height) (Fig. 1A and B). Our chiplet was designed with SOLIDWORKS™ software (2023) and fabricated with Formlabs™ 3B+ Low Force Stereolithography (LFS) resin-based 3D printer (Formlabs) (Fig. 1C). For flow control optimization, we used chiplets fabricated with a clear, transparent photopolymer resin (Formlabs, RS-F2-GPCL-04) that allowed flow visualization. Chiplets used to amplify nucleic acids were printed with black resin (Formlabs, RS-F2-GPBK-04) to minimize background fluorescence emission (Fig. S1†). The thermal conductivity of the SLA resin is estimated as ∼0.3 W m−1 K−1.23 The 3D-printed chiplets were post-processed following the manufacturer's instructions. Briefly, the chiplet was washed in 2-propanol for 30 minutes and then cured at 60 °C for 15 minutes in the Form Cure UV unit. Following post-processing, our chiplet was incubated with 2% polyethylene glycol (PEG) 3350 at room temperature for 30 minutes to improve surface hydrophilicity and biocompatibility.The chiplet (Fig. 1) comprises a sample introduction well that doubles as an RPA reaction chamber (1). The bottom of the entry well (1) is connected to a distribution manifold (2) that leads to five (more are possible) LAMP reaction chambers (3). The LAMP reaction chambers connect at their distal ends to exit wells (6) through two capillary valves (4 and 5). A composite biocompatible phase change material (7 + 8) forms the bottom of the RPA chamber (1), functioning as a valve that prevents fluid from leaving the RPA chamber (1) during RPA incubation. To prevent premature valve (7 + 8) opening, the valve is made of two different phase change materials (PCMs): PureTemp 58 (8), which melts at 58 °C, is placed around the circumference of the opening, and PureTemp 42 (7) with 42 °C melting temperature is placed in the center. The exit wells (6) house the phase change material PureTemp 58 (9). The floor of these wells is located a sufficient distance above the chiplet's (heated) bottom to delay the melting of the PCM (9) until after the LAMP reaction chambers have been filled. The heating time is estimated based on the thermal diffusion time h2α−1, where h is the distance from the heated surface and α ∼ 4.5 × 10−7 m2 s−1 is the resin's thermal diffusivity. Alternatively, the melting time of valve (9) can be delayed by using a higher melting temperature PCM.
The capillary valves (4) prevent liquid from escaping from any of the LAMP reaction chambers' exits during 2nd stage aliquoting. This ensures that all the LAMP reaction chambers are filled before the 2nd stage LAMP incubation. We measured the capillary valve bridging pressure by measuring an upstream water column's critical height (hC) when any capillary valve was compromised. These experiments were carried out with 0.05% phenol red aqueous solution mixed with 0.1% Tween 20, which has a surface tension like that of the LAMP reaction mix.
The LAMP reaction chambers were left roofless during design and 3D printing. Each LAMP chamber, except one (negative, no primer control), was specialized to detect a target by inserting a target-specific LAMP primer set (Table S1†) through its open top. The primers were then dried at 50 °C for 20 min, and the LAMP reaction chambers were sealed with a transparent PCR tape (ThermalSeal RT2RR film). The chiplets with dried primers were stored under room conditions and typically used within 12 hours after primer drying. Based on prior work,24 we anticipate that the primers can be preserved for months, if not years, without refrigeration.
The non-primer, negative control guards against the eventuality that the 1st stage amplification produces enough amplicons to generate a signal. We did not include a non-template control since such control requires a separate flow path and is not part of the self-actuated chiplet.
The SLA resin used for the chiplet fabrication and the phase change materials are bio-compatible and do not inhibit RPA and LAMP amplification25,26 and partially wet aqueous solutions without and with 2% PEG coating (Fig. S5†).
Temperature control
For incubation, our chiplet was placed in our homemade processor (Fig. S1†) and heated at its bottom with a closed-loop PID-controlled heater equipped with a copper plate for temperature uniformity (Fig. S1†). We equipped a chiplet with thermocouples (K type, OMEGA, # TFIR-24S-50) in the RPA chamber, LAMP chambers, and exit wells to examine temperature variations with time. All experiments were carried out in triplicates at a room temperature of ∼22 °C. Since our processor is closed, the air temperature in the processor was about 10 °C higher than the room temperature. The placement of the camera and its LED illumination did not affect the heating. We acquired thermal images of the chiplet (Fig. S6†) during incubation with an infrared thermal camera (FLIR E8) to examine temperature uniformity.We designed our chiplet such that the bottom of the RPA chamber, the LAMP chambers, and the exit wells are located at different heights from the heated bottom of the chiplet to provide a time delay during the heating process. The floor (bottom) of the RPA chamber (1), LAMP chambers (3), and exit wells (6) were, respectively, 6.5 mm, 0.7 mm, and 7 mm from the bottom of the chiplet (Fig. 1A). The melting time of the various PCM valves was controlled by their position and melting temperatures.
Assay
To test our chiplet operation, we developed a Penn-RAMP assay for the co-detection of the two tomato viruses: tomato brown rugose fruit virus (ToBRFV) and tobacco mosaic virus (TMV) and genomic DNA from tomato extract (internal positive control). The sequences of LAMP primer sets for ToBRFV, TMV, and tomato gDNA are available in Table S1.† The specificity of these primers has been previously determined.27,28The Penn-RAMP assay comprises a first-stage RPA reaction and a second-stage LAMP reaction. To design this assay, we first optimized the individual LAMP assays for each target.
Benchtop LAMP assay
The LAMP assay (20 μL) comprised 1X Isothermal Mastermix (ISO-001, OptiGene); 0.2 μM of each outer primer (F3 and B3); 1.0 μM of each inner primer (FIB and BIP); 0. 4 μM of each loop primer (LF and LB); 2.5 μM EvaGreen® dye; and 1 μL of template DNA. Serially-diluted gBlock gene fragments of ToBRFV and TMV with concentrations ranging from 5 to 5 × 108 per microliter were used as templates. The reaction was carried out with Bio-Rad CFX96 Real-Time PCR system at 63 °C for 45 minutes with fluorescence intensity reading acquired every minute. The specificity of the amplicons was analyzed using melting curves.Benchtop multiplexed Penn-RAMP assay
The 1st stage of Penn-RAMP is RPA. The RPA reaction was carried out at a volume of 10 μL laden with 240 nM of each F3 and B3 outer LAMP primers for ToBRFV, TMV, and tomato gDNA (internal positive control) (Table S1†), 1X rehydrated TwistAmp Basic mix (TwistDx Limited, UK), 14 mM Mg(CH3COO)2, and 1 μL of each template DNA (Table S2†) at various concentrations. The RPA reaction was incubated with Bio-Rad CFX96 Real-Time PCR system at 38 °C for 10 minutes. One microliter of the 1st stage products was added to each of the five tubes containing the LAMP reaction mix. Each of the four tubes contained target-specific LAMP primers. The fifth tube contained no primers and served as a negative (non-primer) control to verify that the 1st stage RPA produced insufficient amplicons for a detectable signal. All assays were run at least in triplicates using ToBRFV and TMV templates with concentrations ranging from 5 to 5 × 108 copies per reaction.Chiplet-based Penn-RAMP
The RPA reaction was carried out in the chiplet's entry well (Fig. 1(1)). A 10 μL volume of 240 nM of each F3 and B3 outer LAMP primers for ToBRFV, TMV, and tomato gDNA (internal positive control) (Table S1†), 1X rehydrated TwistAmp Basic mix (TwistDx Limited, UK), 14 mM Mg(CH3COO)2, and 1 μL of each template DNA at various concentrations were added into the RPA reaction chamber. The RPA chamber was sealed with a transparent PCR tape, and the chiplet was placed in our portable, homemade processor that houses a thin film heater (Fig. S1†) and incubated at 38 °C for 10 minutes. Temperature control was achieved with a closed-loop microcontroller (Raspberry Pi 4 mode B).After the RPA process, a 200 μL LAMP reaction mix was added to the RPA well. The LAMP reaction mix consisted of 1X Isothermal Mastermix (ISO-001, Optigene) and 2.5 μM EvaGreen® dye (Biotium). No primers were included in the LAMP reaction mix. Two of the LAMP reaction chambers stored dry primers for TMV to demonstrate reproducibility, the 3rd LAMP reaction chamber pre-stored primers for ToBRFV, the 4th LAMP reaction chamber stored primers for cytochrome oxidase gene of tomato gDNA (positive control), and the 5th reaction chamber did not contain any primers and served as negative control to alert the user against the eventuality of RPA products producing detectable signal.
Then, the heater's temperature was increased to 70 °C for 45 minutes. As a result, the phase change valve (7 + 8) melted, and the molten PCM floated to the top of well 1, forming an evaporation barrier. The RPA products mixed with the LAMP reaction mix self-aliquoted into the five reaction chambers by the combined action of gravity and capillarity (Video S2 and Section S3†). The distal capillary valves (4) halted the flow, assuring that all the LAMP reaction chambers were filled. After a short delay, the PCM (9) melted, penetrated the distal ends of the LAMP reaction chambers (3), and halted at the capillary valves (5), sealing the exits of the LAMP reaction chambers. After LAMP incubation, after the chiplet cooled to room temperature, the PCMs (7 + 8) and (9) solidified, entombing the reaction products in the chiplet.
Our homemade processor houses a Dino-Lite fluorescence USB microscope (AM4113T-GFBW) at ∼30× magnification and approximately 60 mm working distance (Fig. S1†). The USB microscope was connected to a computer. Fluorescence images were collected from all LAMP reaction chambers once a minute. These images were then processed with our custom MATLAB code to produce amplification curves (averaged emission intensity for each LAMP reaction chamber as a function of time) and threshold times for each curve.
Results and discussion
Many infectious diseases in humans, animals, and plants exhibit similar clinical symptoms, making it difficult to differentiate among them based solely on clinical presentation. Multiplex assays that can co-detect co-endemic pathogens from a single sample provide a comprehensive diagnostic solution that differentiates among causative agents and facilitates precise diagnosis, effective therapy, and appropriate control measures. High-level multiplexing in a single pot is challenging because of the need to use and detect multi-color probes. Furthermore, LAMP requires many primers that may lead to false positives. In microfluidics, multiplexing is often achieved by aliquoting a sample into multiple reaction chambers, each specialized to amplify a specific target. Often, the sample volume is constrained. For example, a finger prick typically contains 10–40 μL of whole blood per drop.29 Hence, such an approach requires sample dilution and inevitable loss of sensitivity.To address these challenges, we have developed a two-stage amplification method dubbed Penn-RAMP that can co-amplify and co-detect multiple nucleic acids in a single sample.20–22 Penn-RAMP's 1st stage is RPA, and its 2nd stage is LAMP. The RPA stage includes forward (F3) and backward (B3) primers for all the targets of interest to amplify any present in the sample sufficiently to avoid loss of sensitivity upon aliquoting 1st stage products into 2nd stage reaction but not enough to produce any detectable signal. This approach also allows for nested amplification, wherein fractions of the same 1st stage amplicon are specifically amplified in the 2nd stage. To accommodate the use of our Penn-RAMP assay at the point of need, we describe here a self-actuated chiplet capable of aliquoting Penn-RAMP 1st stage products into multiple 2nd stage target-specific reaction chambers.
Chiplet self-aliquots 1st stage reaction products into 2nd stage reaction chambers
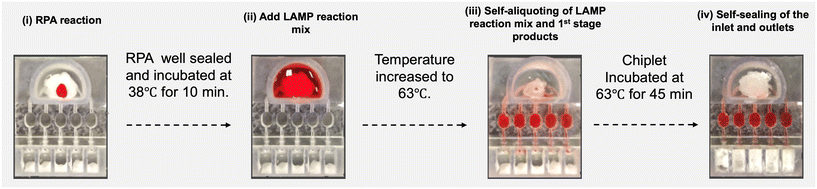
To examine our flow control and valves' integrity, we fabricated chiplets with a clear, transparent resin that allows us to image liquid flow inside the chiplet. Flow control experiments (video S1 and S2 in ESI†) were carried out with phenol red dye solution mixed with 0.1% Tween 20 to mimic the physiochemical properties of our LAMP reaction mix. 10 μL of red dye was placed in the RPA reaction chamber (Fig. 1(1)) on top of the low-temperature composite valve (7 + 8). The well was then sealed with a PCR tape and incubated at 38 °C for 10 minutes (Fig. 2(i)). The valve integrity was maintained during this process. There was no apparent penetration of liquid into the composite PCM material.Next, we added 200 μL of dyed liquid to mimic the addition of LAMP buffer (Fig. 2(ii)) and increased the temperature of the LAMP chambers to 63 °C. With this increase in temperature, the phase change valve (7 + 8) melted and floated to the surface of the liquid, clearing the opening to the manifold located at the bottom of the RPA reaction chamber (2). The liquid was then self-aliquoted into the five LAMP reaction chambers by gravity and capillarity (Fig. 2(iii) and Video S2†). The resins are partially wetting. The contact angles of the clear and black resins, in the absence of coating, are, respectively, 67.9° ± 2.8° and 50.2° ± 2.1°. Our PEG reduces the contact angles slightly to 58.5° ± 2.5° and 47.6° ± 0.9° (Fig. S5†).
A mathematical model of this imbibition process has been presented in Section S3 (Fig. S2 and S3†). During this process, the LAMP reaction chambers' exits remained open, allowing air to escape. When the liquid arrived at the capillary valves (4) that comprise abrupt increases in the conduits' cross-sections (4), the contact line was pinned, and the flow halted (Fig. S2†). This flow halting facilitates the complete filling of all the LAMP reaction chambers. The capillary valve is sufficiently robust to withstand a pressure difference of 104 ± 9 Pa, which is more than enough for our purposes (Fig. S4†).
The phase change material in the exit wells (9) was placed at a greater distance from the heater and had a higher melting temperature than the PCM valve (7 + 8) and melted only after a time delay. This time delay was enough for all the LAMP reaction chambers to completely fill but short enough to avoid excessive evaporation. When the phase change material (9) melted, it flowed into the exits of the LAMP reaction chambers. It got pinned at the capillary valves (5), sealing the LAMP chambers' exits and providing evaporation barriers. Capillary valves (5) prevented the phase change material from further penetrating the reaction chambers and displacing the reaction mix. After LAMP incubation (∼45 min), the chiplet cooled to room temperature. The phase change materials at the inlet (7 + 8) and exits (9) solidified and entombed the reaction products, preventing carry-over contamination (Fig. 2(iv)).
Temperature distribution in the chiplet
Our processor includes a closed-loop PID-controlled heater to provide desired incubation temperatures for the RPA and LAMP amplification processes. Heating also controls the PCM valves. We measured (Fig. S6†) the temperature at the bottom of the chiplet, at the bottom of the RPA chamber, the LAMP chambers, and exit wells as functions of time during the ramp-up to and during RPA incubation (Fig. S6A†), ramp-up to and during LAMP incubation (Fig. S6B†), and after the power was turned off (Fig. S6C†).The RPA chamber achieved the steady state temperature of 38 °C within 3 min and remained at this temperature (±0.5 °C) during the 10 min incubation. At this temperature, the low-temperature composite valve (7 + 8) remained solid, preserving the integrity of the RPA chamber. We did not observe any penetration of the RPA mix into the solid composite PCM valve (7 + 8). After incubation, we recovered 8.6 ± 0.1 μL of the 10 μL solution we initially inserted in the RPA chamber with a pipette (Fig. S7†). This volume loss can be attributed to incomplete recovery and evaporation during RPA incubation.
Following the RPA incubation, 200 μL dyed liquid, mimicking LAMP buffer, was added to the RPA chamber. The PID controller set the heater temperature to 70 °C (Fig. S6B†). When the RPA chamber temperature reached 42 °C, the low-temperature PCM (TrueTemp42, component (7) at the center of the valve) melted, resulting in self-aliquoting of the liquid into the LAMP chambers to initiate the 2nd stage LAMP reaction at 63 °C (Fig. S6B†). When the temperature at the exit well (6) reaches 58 °C after an additional 5 ± 1 min, the TrueTemp58 (9) melted to seal the LAMP chamber's exits, preventing further evaporation (Fig. S6B†). During the 45-min LAMP incubation, the LAMP reaction chambers maintained a steady temperature at 63 ± 0.5 °C.
At the end of the 2nd stage LAMP incubation, the heater was turned off, allowing for the TrueTemp58 PCM (9) at the exits and inlet (8) to solidify within <3 min (Fig. S6C†). The chiplet can then be disposed.
The temperature uniformity across the RPA chamber, LAMP chambers, and exits were confirmed with images obtained with an infrared thermal camera (Fig. S6D†).
Chiplet successfully detects tomato viruses
To verify that our chiplet can, indeed, co-detect multiple pathogens in a single test, we processed contrived samples comprised of nucleic acids (Table S2†) associated with ToBRFV and TMV spiked with tomato genomic DNA extract (internal positive control) with our self-actuated chiplet. Tomato viruses cause unusual color patterns in tomato leaves and fruit, impair plant growth, reduce yield and quality, induce plant death, and may spread to other plants such as peppers (ToBRFV) and solanaceous plants (TMV).30 Rapid, specific, inexpensive detection of tomato viruses is critical to enable effective control measures, prevent infection spread, and minimize the economic damage that can be caused to tomato production.We carried out Penn-RAMP experiments with our self-actuated chiplet (Fig. 3). The 10 μL RPA assay included forward (F3) and backward (B3) LAMP primers for ToBRFV, TMV, and Tomato DNA and various combinations of targets (103 copies of each target). Four of the LAMP reaction chambers of each chiplet pre-stored LAMP primer sets for individual targets as indicated (Fig. 3A–D). The fifth LAMP chamber served as a negative control and contained no primers. The LAMP reactions were incubated for 45 min at 63 °C. The fluorescence emission from all the LAMP reaction chambers was recorded once every minute and transmitted to a computer for further processing. The LAMP reaction chamber fluoresced when their specific target was present and remained dark in the absence of their specific target (Fig. 3A–D). The negative control remained dark for all tests, and the positive control fluoresced in all tests (Fig. 3A–D). The results were confirmed with the corresponding amplification curves (Fig. S8†). We observed no false positives within the 45-minute incubation time and no crosstalk among the reaction chambers. In summary, our self-actuated chiplet enabled automated processing of the two-stage Penn-RAMP, detecting the presence of ToBRFV gBlock (103 copies with threshold time of 28 min), TMV gBlock (103 copies with threshold time of 32 min), and tomato DNA (with threshold time 23 min) without any false positives and false negatives.
 | ||
| Fig. 3 On-chip Penn-RAMP assay for tomato viruses. Second-stage reaction fluorescent images as functions of time for samples comprised of (A) ToBRFV, TMV, and tomato DNA; (B) only tomato DNA; (C) TMV and tomato DNA; and (D) ToBRFV and tomato DNA. The chambers labeled positive control, TMV and ToBRFV pre-stored, respectively, lyophilized LAMP primers for tomato cytochrome oxidase gene, TMV, and ToBRFV. The negative control chamber had no LAMP primers. The plus (+) and minus (−) symbols indicate, respectively, presence and absence of target DNA in the sample. 103 copies of each target were used per RAMP assay. The chiplet was incubated with our home-made processor (Fig. S1†). | ||
High analytical performance of the chiplet
We first performed benchtop experiments with individual LAMP assays with dilution series of ToBRFV and TMV, using previously reported primers (Table S1†). The LAMP assays detected all replicates down to 500 copies/reaction volume of ToBRFV and TMV (Table S3†). The average threshold times for reaction with 500 target copies were, respectively, 14.5 ± 1.8 min and 13.5 ± 0.1 min for ToBRFV and TMV (Table S3†). At lower concentration (50 target copies/reaction), the percentage of positives detected was, respectively, 33.3% (2/6) and 50.0% (3/6) of ToBRFV and TMV. We did not observe any false positives within 45 minutes incubation.Second, we performed benchtop Penn-RAMP experiments with ToBRFV and TMV samples at various concentrations spiked with tomato genomic DNA extract. The 1st stage, 10 min RPA, employed the LAMP outer primers F3 and B3 for ToBRFV, TMV and tomato gDNA in a single pot. The RPA amplicons then served as templates for target-specific LAMP assays, each with a primer set for a single target: ToBRFV, TMV, and positive control (tomato gDNA) and negative (no primer) control. One microliter of RPA products was added to each 20 μL of LAMP reaction mix. The benchtop Penn-RAMP assay detected all replicates down to 5 copies/reaction volume for both ToBRFV and TMV. The average threshold times (defined by the 2nd stage LAMP reaction) for 5 copies/reaction were, respectively, 9.1 ± 1.3 min and 17.2 ± 0.7 min for ToBRFV and TMV (Table S3†). None of the reactions presented false positives within 45 minutes of incubation time.
We then compared the performance of our benchtop LAMP and benchtop Penn-RAMP with our self-actuated Penn-RAMP chiplet tests using the same concentrations of the serially diluted templates of ToBRFV and TMV spiked with tomato gDNA extract. Like the benchtop Penn-RAMP, our on-chip Penn-RAMP assay detected as few as 5 target copies per reaction with 100% sensitivity (3/3) for both ToBRFV and TMV (Table S3†). No false positives were observed after 45 minutes of incubation. Our results show that our on-chip Penn-RAMP assay has a limit of detection comparable to that of the benchtop Penn-RAMP assay and at least 10-fold better than the benchtop standalone LAMP assay. However, we observed relatively high threshold times for both ToBRFV and TMV compared to our benchtop Penn RAMP (Table S3†). The threshold time was measured from the instant when the power to the heater was stepped up to increase the incubation temperature from 38 °C to 63 °C. The larger threshold time of the chiplet compared to the benchtop is likely due, in part, to the less sophisticated thermal control of our homemade incubator and the time needed to melt the PCM barrier (7 + 8).
To assess variability among the LAMP reaction chambers of our chiplet, we functionalized all five 2nd stage reaction chambers with LAMP primers for ToBRFV and processed a sample with 103 ToBRFV template copies per reaction. The fluorescence intensity of all reaction chambers increased with time (Fig. 4A). Based on the monitored images, our custom MATLAB software program produces amplification curves (fluorescent emission intensity as a function of time, Fig. 4B). The threshold times were within 1 minute apart from each other (29.1 ± 1.1 min) (Fig. 4C). This suggests that the chiplet's threshold times enable quasi-quantification of target concentration. We then determined the threshold time as a function of the log of the target concentration (Fig. 4D). The threshold times for ToBRFV (R2 = 0.95) and TMV (R2 = 0.95) exhibited nearly linear dependence on the log of target concentration, enabling one to estimate the target concentration from the threshold time.
Conclusions and outlook
Diverse co-endemic, human, animal, and plant infectious agents induce similar (non-specific) symptoms but require diverse control strategies. Enactment of effective treatments, control measures, and public health policies requires rapid, precise identification of causative pathogens. This need becomes more urgent with climate change and the migration of vectors and pathogens to new regions lacking prior exposure and defense mechanisms against invading pathogens. Multiplexed molecular assays are needed to meet this challenge. Conventional multiplexed molecular assays typically require laboratory facilities, sophisticated equipment, and trained personnel. Such facilities are often unavailable in resource-poor settings or located large distances from the site of need, which imposes significant and costly delays between sample collection and pathogen identification.To address these needs, we developed a two-stage isothermal amplification assay dubbed Penn-RAMP.20 Penn-RAMP's first stage comprises a recombinase polymerase amplification RPA that amplifies all targets of interest in a single reaction well for a short time. RPA amplicons are then aliquoted into multiple LAMP reaction chambers, each specialized to detect a single target or a group of targets. This approach enables us to use spatial multiplexing without loss of sensitivity. Even a single-plex Penn-RAMP offers higher sensitivity than a standalone LAMP.21,22
To enable the use of our Penn-RAMP assay by minimally trained personnel without a need for pipetting and multiple unit operations, we designed, fabricated, and tested a self-actuated Penn-RAMP chiplet that employs temperature-controlled phase change valves and capillary valves to aliquot first-stage amplicons into five-second stage reaction chambers, and to seal these chambers. A normally closed phase change valve enables the distribution of 1st stage products to 2nd stage reaction chambers by opening just in time when the 1st stage incubation temperature increases to the 2nd stage incubation temperature. Passive capillary valves ensure that all 2nd stage reaction chambers are filled and prevent phase change materials from invading 2nd stage reaction chambers. Furthermore, the phase change materials provide evaporation barriers during incubation and seal amplicons at the process conclusion. Our Penn-RAMP chiplet mates with a simple homemade processor that provides thermal control and imaging.
The functionality of our device was demonstrated by co-detecting two tomato viruses, ToBRFV and TMV, spiked with tomato DNA extract (housekeeping gene). Our on-chip multiplex assay showed no cross-talk among the reaction chambers. The performance of the Penn-RAMP chiplet was comparable to that of benchtop assays, detecting as few as 5 target copies per reaction with 100% sensitivity and surpassed that of the benchtop LAMP assay with the detection limit of 50 target copies per reaction volume. The 2nd stage threshold time depended nearly linearly on the log target concentration, suggesting that the Penn-RAMP chiplet can provide quasi-quantification. Our chiplet can be stand-alone with purified nucleic acids or interface with our sample preparation slider cassette.26
Future improvements of the Penn-RAMP chiplet may include lyophilizing RPA and LAMP reagents and storing them refrigeration-free inside the chiplet, and increasing the level of multiplexing by increasing the number of 2nd stage reaction chambers to enable co-detection of multiple pathogens. Previously, we have demonstrated Penn-RAMP's ability to co-detect as many as 16 targets.20 To reduce the cost of operation, it is possible to replace the fluorescent dye with colorimetric dye, which would eliminate the need for a UV source and a camera. Temperature control can be provided with an exothermic reaction, such as used in meals ready to eat (MRE) and controlling temperature with phase change materials.13 These modifications would enable electricity-free, instrument-free operation, making the entire system disposable.
Here, we have used our Penn-RAMP chiplet to co-detect plant pathogens. The same chiplet can be used for numerous other applications that could benefit from rapid detection such as animal and human pathogens and food contaminants.
Data availability
The data that support the findings of this study are available on request from the corresponding author [H. H. Bau].Author contributions
Conceptualization, microfluidic chip design, and experiment design: H. H. Bau, F. Ansah. Chip fabrication, methodology, data curation, and analysis: F. Ansah, M. G. Mauk, Y. Gu. Assay development and benchtop experiment: M. Hajialyani, F. Amadi. Imbibition modeling: E. A. Tarim. Writing: F. Ansah, H. H. Bau. Mentoring for F. Ansah: G. A. Awandare, H. H. Bau. All the co-authors read and approved this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported, in part, by NIH/DHHS grant R33HD101937 (chiplet development) and a grant from Wageningen Research, Wageningen, Netherlands (plant assay development), to the University of Pennsylvania. Felix Ansah and Gordon Awandare are partly supported by a grant from the Science for Africa Foundation to the Developing Excellence in Leadership, Training and Science in Africa (DELTAS Africa) Programme [DEL-22-014] with support from Wellcome Trust and the UK Foreign, Commonwealth & Development Office.References
- P. Van de Vuurst and L. E. Escobar, Infectious Diseases of Poverty, 2023, 12, 51 CrossRef PubMed.
- C. Caminade, K. M. McIntyre and A. E. Jones, Ann. N. Y. Acad. Sci., 2019, 1436, 157–173 CrossRef PubMed.
- S. A. Hudu, A. S. Alshrari, A. Syahida and Z. Sekawi, J. Clin. Diagn. Res., 2016, 10, DE01 CAS.
- A. Hematian, N. Sadeghifard, R. Mohebi, M. Taherikalani, A. Nasrolahi, M. Amraei and S. Ghafourian, Osong Public Health Res. Perspect., 2016, 7, 77–82 CrossRef PubMed.
- V. M. Corman, V. C. Haage, T. Bleicker, M. L. Schmidt, B. Mühlemann, M. Zuchowski, W. K. Jo, P. Tscheak, E. Möncke-Buchner and M. A. Müller, Lancet Microbe, 2021, 2, e311–e319 CrossRef CAS PubMed.
- A. B. Bhatti, F. Ali and S. A. Satti, Cureus, 2015, 7, e396 Search PubMed.
- H. Deng, A. Jayawardena, J. Chan, S. M. Tan, T. Alan and P. Kwan, Sci. Rep., 2021, 11, 15176 CrossRef CAS PubMed.
- K. Malpartida-Cardenas, J. Rodriguez-Manzano, L.-S. Yu, M. J. Delves, C. Nguon, K. Chotivanich, J. Baum and P. Georgiou, Anal. Chem., 2018, 90, 11972–11980 CrossRef CAS PubMed.
- J. de Korne-Elenbaas, A. Pol, J. Vet, M. Dierdorp, A. P. van Dam and S. M. Bruisten, Sex. Transm. Dis., 2020, 47, 238–242 CrossRef CAS PubMed.
- B. B. Oliveira, B. Veigas and P. V. Baptista, Front. Sens., 2021, 2, 752600 CrossRef.
- S. Chakraborty, Sens. Diagn., 2024, 3, 536–561 RSC.
- M. Teymouri, S. Mollazadeh, H. Mortazavi, Z. N. Ghale-Noie, V. Keyvani, F. Aghababaei, M. R. Hamblin, G. Abbaszadeh-Goudarzi, H. Pourghadamyari and S. M. R. Hashemian, Pathol., Res. Pract., 2021, 221, 153443 CrossRef CAS PubMed.
- R. J. Li, M. G. Mauk, Y. Seok and H. H. Bau, Analyst, 2021, 146, 4212–4218 RSC.
- H. H. Bau, C. Liu, M. Mauk and J. Song, Expert Rev. Mol. Diagn., 2017, 17, 949–951 CrossRef CAS PubMed.
- M. G. Mauk, J. Song, C. Liu and H. H. Bau, Biosensors, 2018, 8, 17 CrossRef PubMed.
- P. Srivastava and D. Prasad, 3 Biotech, 2023, 13, 200 CrossRef PubMed.
- M. El-Tholoth and H. H. Bau, Viruses, 2024, 16, 1248 CrossRef CAS PubMed.
- A. A. Sayad, F. Ibrahim, S. M. Uddin, K. X. Pei, M. S. Mohktar, M. Madou and K. L. Thong, Sens. Actuators, B, 2016, 227, 600–609 CrossRef CAS.
- F. Marks, J. Liu, A. B. Soura, N. Gasmelseed, D. J. Operario, B. Grundy, J. Wieser, J. Gratz, C. G. Meyer and J. Im, Clin. Infect. Dis., 2021, 73, 1338–1345 CrossRef PubMed.
- J. Song, C. Liu, M. G. Mauk, S. C. Rankin, J. B. Lok, R. M. Greenberg and H. H. Bau, Clin. Chem., 2017, 63, 714–722 CrossRef CAS PubMed.
- J. Song, M. El-Tholoth, Y. Li, J. Graham-Wooten, Y. Liang, J. Li, W. Li, S. R. Weiss, R. G. Collman and H. H. Bau, Anal. Chem., 2021, 93, 13063–13071 CrossRef CAS PubMed.
- M. El-Tholoth, E. Anis and H. H. Bau, Analyst, 2021, 146, 1311–1319 RSC.
- A. Oval-Trujillo, A. Rodríguez, G. Pérez-Artieda, Y. Dung and P. Alegría, SN Appl. Sci., 2022, 4, 205 CrossRef CAS.
- J. Song, C. Liu, M. G. Mauk, J. Peng, T. Schoenfeld and H. H. Bau, Anal. Chem., 2018, 90, 1209–1216 CrossRef CAS PubMed.
- M. El-Tholoth, H. Bai, M. G. Mauk, L. Saif and H. H. Bau, Lab Chip, 2021, 21, 1118–1130 RSC.
- Y. Seok, Q. Yin, R. Li, M. G. Mauk, H. Bai and H. H. Bau, Sens. Actuators, B, 2022, 369, 132353 CrossRef CAS PubMed.
- Eindrapport Final report: On-site plantpathogen detection and barcode sequencing for improving plant health and phytosanitary control, https://www.glastuinbouwnederland.nl/content/glastuinbouwnederland/docs/themas/Plantgezondheid/2023_Eindverslag-End_Report_PPS_On-site_en_Barcoding.pdf, (accessed June 2024).
- J. Tomlinson, M. Dickinson and N. Boonham, Phytopathology, 2010, 100, 143–149 CrossRef CAS PubMed.
- M. M. Bond and R. R. Richards-Kortum, Am. J. Clin. Pathol., 2015, 144, 885–894 CrossRef CAS PubMed.
- S. Zhang, J. S. Griffiths, G. Marchand, M. A. Bernards and A. Wang, Mol. Plant Pathol., 2022, 23, 1262–1277 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00752b |
| This journal is © The Royal Society of Chemistry 2024 |