 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel immunoassay technique using principal component analysis for enhanced detection of emerging viral variants†
Josselyn
Mata Calidonio
a,
Arianna I.
Maddox
b and
Kimberly
Hamad-Schifferli
 *ac
*ac
aDepartment of Engineering, University of Massachusetts Boston, Boston, MA, USA. E-mail: kim.hamad@umb.edu
bDepartment of Biology, University of Massachusetts Boston, Boston, MA, USA
cSchool for the Environment, University of Massachusetts Boston, Boston, MA, USA
First published on 18th July 2024
Abstract
Rapid diagnostics are critical infectious disease tools that are designed to detect a known biomarker using antibodies specific to that biomarker. However, a way to detect unknown disease variants has not yet been achieved in a paper test format. We describe here a route to make an adaptable paper immunoassay that can detect an unknown biomarker, demonstrating it on SARS-CoV-2 variants. The immunoassay repurposes cross reactive antibodies raised against the alpha variant. Gold nanoparticles of two different colors conjugated to two different antibodies create a colorimetric signal, and machine learning of the resulting colorimetric pattern is used to train the assay to discriminate between variants of alpha and Omicron BA.5. By using principal component analysis, the colorimetric test patterns can pick up and discriminate an unknown variant that it has not encountered before, Omicron BA.1. The test has an accuracy of 100% and a potential calculated discriminatory power of 900. We show that it can be used adaptively and that it can be used to pick up emerging variants without the need to raise new antibodies.
Introduction
Infectious diseases are a global health threat that impacts the entire world. Diagnostics are important tools that are at the front line of defense for infectious diseases, enabling decision making for patient treatment and disease surveillance. They are especially critical when a disease is newly emerging, as this is when tools for identification and treatment are not yet in place and stopping the spread is essential. As soon as point of care (POC) tests were widely available for the COVID-19 pandemic, they proved to be vital in disease control because they allowed end users to determine if they were infected. One of the most useful formats for POC diagnostics has been rapid paper tests, such as lateral flow assays (LFAs) and dipstick assays. These are paper strips embedded with gold nanoparticles and antibodies specific to the target. The tests can go from sample to answer within minutes, do not require external instrumentation or power, and can be easily read out by eye or mobile phone cameras. Because of their low cost, they can be manufactured in large numbers and be widely distributed, enabling users to test themselves in a variety of environments.As the infectious disease landscape is constantly evolving, it is urgent that we be ready for the next global pandemic. However, one challenge that has always remained is that we have never come up with a broadly accessible way to diagnose newly emerging infectious diseases.1 Viruses undergo continual mutations across various species, and zoonotic spillovers events, involving the transmission from animals to human, exert a substantial influence on the emergence of new outbreaks. An estimated 60–75% of all emerging human infectious diseases are thought to be zoonotic in origin.2 The heightened frequency of zoonotic spillover events is attributed to increasing levels of globalization, travel, urbanization, and shipping; these occurrences are further exacerbated by climate change, posing a persistent threat.3,4 Any given unknown pathogen might become the next big global pandemic if left unchecked, so the ability to rapidly identify unknowns circulating in the human population is of extreme importance. While complete identification in real time is impossible, information on what virus class it is, vector type (i.e., tick vs. mosquito borne), or what it is most similar to would be valuable in mounting a public health response to contain transmission, treat patients, and allocate resources.
Conventional LFAs require highly specific antibodies, but in order to produce them, the antigen target must first be identified before antibodies can be raised against it, thus preventing the ability to raise antibodies for an unknown. The only tool that can identify pathogens without prior knowledge of the target identity is DNA sequencing. However, sequencing is expensive, as it requires sophisticated equipment, reagents, and personnel, and is performed after PCR registers a positive. As a consequence, sequencing is commonly conducted in a centralized lab, with turnaround times spanning weeks or more. The COVID-19 pandemic spurred the opening of distributed sequencing centers that greatly enhanced the identification of new variants. However, these facilities were disproportionately concentrated in high-income countries, creating a significant disparity in genomic surveillance capabilities between high-income and low- to middle-income countries, primarily attributable to socioeconomic inequalities.5 Eventually, governments ultimately shuttered them because of high associated costs.6 Moreover, sequencing can still miss detection of the target if reference sequence or primers are not well suited to the divergent sequence, or the virus is present at low abundance, or the window in which the virus is present in the patient is short. Many infections by unknowns go undiagnosed. Therefore, we have a major flaw in our strategies to respond to newly emerging infectious diseases, and we urgently need to come up with a different tactic because the emergence of new diseases, strains, variants, and subtypes are now occurring with greater frequency. As it stands, diagnosing unknowns remains a major unmet need in global health.
However, chemists have determined a way to detect unknowns via olfactory arrays. To detect a range of previously unencountered analytes, they have developed novel sensors that act as a “chemical nose” or “chemical tongue”7,8 by employing a selective array rather than a specific binary (yes/no) sensor. In these cases, the signal is read out as a multidimensional pattern, or fingerprint, via principal component analysis (PCA).9 More importantly, chemical olfaction can classify these analytes that have never been encountered by the assay before.10 As selective arrays, they are not constrained to the detection of a limited number of target analytes, but instead can detect hundreds if not thousands of different species in pure form or in complex mixtures. Moreover, the format for olfaction sensors is highly versatile, with many ways to achieve readouts (e.g., electrochemical, fluorescence, colorimetric, nanoplasmonic, and others). It has been demonstrated to be powerful for a variety of analytes, ranging from small molecules, beverages,11 and disease cell types.12 Additionally, the sensor itself does not need to be expensive or difficult to operate, as they have been successfully implemented with colorimetric readouts on paper that can be imaged by a desktop scanner. The development of these sensors has given rise to overarching principles for constructing an olfactory array, where a salient feature is that it possesses some degree of cross-reactivity.13
Recognizing the power of chemical olfaction, we created a selective array in an immunoassay format to result in an adaptive diagnostic that can detect a biomarker that it has not encountered before. We chose to demonstrate it on variants of SARS-CoV-2 because the virus is a compelling model for a pathogen that evolves into new variants. COVID-19 disease evolution has occurred on an exceedingly accelerated timescale. In the last 4 years, we have been faced with more than 35 variants, with intervals between some variants spanning just a few weeks. (e.g., BA.1 and BA.5 in 2022).14,15 Each variant is effectively a new disease, where immunization or vaccination against one does not fully protect against future variants, and the nature of the disease has inflicted grave public health consequences and led to increasing disease burdens. The virus has mostly leveraged mutations on the receptor binding domain (RBD) in the spike protein (S) where it interacts with the angiotensin-converting enzyme 2 (ACE2) receptor on cells. However, nucleocapsid mutations have also been shown to be significant, contributing to increased infectivity, heightened transmission, and, notably, the ability to evade detection in rapid tests.16,17 While there are many available rapid tests that can diagnose SARS-CoV-2 infections, they cannot discriminate between variants.
The selective array also allowed us to leverage antibody cross-reactivity, as specificity is not required. By doing so, antibodies for another target can be repurposed.18 We utilized reagents that were available at a hypothetical time point of 2021–2022 after the alpha variant had emerged, and assumed we would have antibodies for alpha S RBD but not for what was newly emerging, Omicron BA.1 and Omicron BA.5 (timeline, Fig. 1). Starting with a pool of two antibodies from the alpha variant, we constructed a multicolor array and trained it with machine learning to detect the new variant BA.5 based on its colorimetric pattern. Then, using PCA, we show that the assay could pick up an unknown, BA.1, and classify it as something different from the previous variants it has encountered. We demonstrate that a limited set of just two cross-reactive antibodies can be repurposed to identify a newer variant if the array follows basic rules from chemical olfactory arrays. These results illustrate that rethinking how rapid paper tests are used can yield capabilities that go beyond yes/no answers. Furthermore, they show that strategic use of cross-reactive antibodies have the potential to pick up newly emerging pathogenic threats in a point of care format, ultimately aiding emergency preparedness.
 | ||
| Fig. 1 a) Demonstrating a proof-of-concept assay that can detect a variant of SARS-CoV-2 that is previously unencountered by the assay. The assay uses only antibodies raised against alpha (Ab1 and Ab2), and is trained to detect BA.5 by its colorimetric fingerprint; b) the time window of the hypothetical test development (pink) is during the BA.1. and BA.5 outbreaks, and antibodies used are those raised against the earlier variant of alpha; c) phylogenetic tree of main SARS-CoV-2 variants. The scale bar indicates the genetic distance. From Wang et al.;19 d) sequence identity for the variants studied, with overlap in blue, and e) computed sequence identity (orange) and similarity (green) from BLASTp. | ||
Experimental
Reagents
Gold chloride trihydrate (CAS: 16961-25-4), N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) (HEPES) (CAS: 7365-45-9), bis(sulphatophenyl) phenylphosphine dehydrate (BPS) (CAS: 308103-66-4), sodium citrate tribasic trihydrate (CAS: 6132-04-3), tween 20 (CAS: 9005-64-5), and sucrose (CAS: 57-50-1) were all purchased from Sigma-Aldrich. Phosphate buffer saline (PBS) was purchased from Fisher Scientific, and tris-buffered saline (TBS) (10×, pH 7.4) from Boston BioProducts. Thiolated mPEG (5 kDa) was purchased from Nanocs. Casein hydrolysate was purchased from Sigma.From the biological reagents used, the antibodies for the spike receptor binding domain (RBD) were rabbit anti-spike IgG (Ab1) and human anti-spike IgG (Ab2), which were purchased from Sino Biological. Anti-mouse and anti-rabbit IgG antibodies (anti-Fc) for the control line were purchased from Sino Biological. The S RBDs used in the test pertained to the alpha, BA.1, and BA.5 variants, all from Sino Biological.
Nanoparticle (NP) synthesis and conjugation
Blue-colored gold NPs were synthesized according to established methods in the literature.20,21 Gold nanostars (GNS), characterized by their star-shaped morphology and blue color, were generated by combining 1100 μL of Milli-Q (MQ) H2O, 900 μL of 140 mM HEPES (pH 7.4), and 32 μL of 25 mM gold chloride trihydrate. Antibody conjugation to GNS was achieved through physisorption.22 The synthesized GNS underwent centrifugation at 3381g for 12 min to form a pellet, and the supernatant was subsequently removed. The pellet was then resuspended in a solution comprising 140 mM HEPES (pH 7.48) and MQ H2O. Subsequently, Ab1 (10 μg) was added to the solution, and the mixture was incubated for 60 min at room temperature. Following this incubation period, thiolated polyethylene glycol (PEG-SH) (5 kDa) was introduced (5 × 10−10 mol) and allowed to mix with the GNS–Ab1 for 10 min. The PEGylated GNS–Ab1 complex was separated by centrifugation at 2348g for 10 min, and the supernatant containing free PEG-SH and Ab1 was discarded. The remaining pellet was resuspended using approximately 100 μL of 0.01 M PBS buffer.Red-colored gold NPs were synthesized according to established methods in the literature. Preceding the formation of the NP–Ab2 conjugate, 1 mL of red nanoparticles (NPs) underwent centrifugation at 6014g for 10 min to eliminate excess reagents. Following the removal of the supernatant, the NP pellet was reconstituted in a solution containing 140 mM HEPES (pH 7.48) and Ab2 (4.55 μg) and allowed to incubate on an orbital shaker for 60 min. A PEG backfilling step ensued, involving the addition of PEG-SH (1 × 10−9 mol), followed by a 10 min incubation on an orbital shaker. Subsequently, the NP–Ab2 complex underwent centrifugation at 3381g for 10 min and was then resuspended in 50 μL of 0.01 M PBS buffer.
Utilizing cuvettes with a 1 cm path length, optical absorption spectroscopy data were acquired on the spectramax molecular devices plate reader. The nanoparticle analyzer SZ-100 from HORIBA Scientific was employed to determine the hydrodynamic diameter (DH) of both the NPs and the NP conjugates.
Immunoassay construction and running
Dipstick immunoassays were crafted using laser-cut nitrocellulose (Sartorius, Unistart CN140), with an affixed absorbent pad wick (Sigma Aldrich, GB003 Gel Blot paper) serving as a fluid sink, secured by an adhesive backing (DCN Dx, MIBA-050).23 In the control spot (position 4 of the nitrocellulose), 0.4 μg αIgG antibodies were spotted. Test lines, represented by positions 3 and 2 on the nitrocellulose, featured 0.22 μg of Ab1 and 0.176 μg of Ab2, respectively. Each case was tested in triplicate. Given that the immunoassay uses paper as an analytical platform, the cost remains very low per unit, as the reagents and membranes are low cost.24 Upon complete drying of the nitrocellulose strips, a four-step procedure was employed to administer test solutions, where each step took approximately ∼15–20 min to run. Initially, the strips were immersed in individual tubes containing 30 μL of human serum, 12 μL of running buffer (composed of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of 50% sucrose and 1% Tween 20), 3 μL of the GNS–Ab1 conjugate, 5 μL of gold quencher, and the target(s) (S RBD of alpha (0.34 μg), BA.5 (0.5 μg), BA.1 (0.5 μg)). Capillary forces facilitated fluid migration to the absorbent pad. Subsequently, the strips underwent a washing step in tubes containing 25 μL of a 1% casein solution. For the third step, the strips were immersed in a solution containing 30 μL of human serum, 12 μL of running buffer, and 2 μL of the NP–Ab2 conjugate. Finally, a post-wash was performed by placing the strips in 25 μL of the 1% casein solution. The strips were allowed to thoroughly dry before undergoing image analysis.
1 ratio of 50% sucrose and 1% Tween 20), 3 μL of the GNS–Ab1 conjugate, 5 μL of gold quencher, and the target(s) (S RBD of alpha (0.34 μg), BA.5 (0.5 μg), BA.1 (0.5 μg)). Capillary forces facilitated fluid migration to the absorbent pad. Subsequently, the strips underwent a washing step in tubes containing 25 μL of a 1% casein solution. For the third step, the strips were immersed in a solution containing 30 μL of human serum, 12 μL of running buffer, and 2 μL of the NP–Ab2 conjugate. Finally, a post-wash was performed by placing the strips in 25 μL of the 1% casein solution. The strips were allowed to thoroughly dry before undergoing image analysis.
Image analysis
Upon completion of the drying process, all strips were affixed onto a paper sheet and subsequently scanned using a desktop scanner.24 The color analysis of the spots of interest was conducted using the “RGB measure” plugin in ImageJ.25 The resulting RGB colors were in an 8-bit format. The final spot color data for each spot were derived by subtracting the RGB color of the spot of interest (spot 2 and 3) from the background of the strips. The background color, primarily near pure white for all strips, was obtained through spot 4.Pseudocolor/stain deconvolution
To enhance the precision of the LDA model, color deconvolution was applied, as outlined by Ruifrok in 2001.26 This process involved dissecting the color information from the two test areas based on the color details of the immunoprobes (stains). Initially, the red NP–Ab2 and blue GNS–Ab1 conjugates were directly applied to the nitrocellulose, allowing them to fully dry before initiating image analysis. Utilizing the color deconvolution 2 plugin in ImageJ, stain vectors S1, S2, and S3 were established. These vectors were defined according to the optical density for each RGB channel of the red NP–Ab2 (S1), the blue GNS–Ab1 spot (S2), and the cross product of both (S3). Subsequently, the three stain vectors were put together to construct the stain vector matrix. The inverse of this matrix was then employed to determine the quantities of the three stains present at the test spots on the nitrocellulose. The obtained deconvoluted values served as the new input data for the subsequent application of machine learning in the analysis.Machine learning
Linear discriminant analysis (LDA), a supervised learning technique within machine learning, was executed using MATLAB (version R2023a). LDA aims to find the linear combinations of features that best discriminate between different classes in a dataset. The input data for the model consisted of the RnGnBn data, with n = 2, 3 denoting the location of the test spot. Consequently, the model incorporated 6 training features. Each class (control, alpha, and BA.5) was run in triplicates, giving rise to a total of 9 training examples. The machine learning analysis primarily involved the use of confusion matrices and ROC curves.Principal component analysis (PCA)
Principal component analysis (PCA) is a dimensionality reduction technique that identifies the underlying patterns in a dataset by transforming the original variables into a new set of uncorrelated variables called principal components. In the context of the RGB data from spots 2 and 3 of the test strip, PCA was employed in MATLAB (version R2023a) to extract two principal components that captured the maximum variance in the data. In this analysis, the two principal components were utilized to visualize and analyze the clustering of each class. The clustering patterns revealed by PCA show how the data points from different classes are distributed and grouped based on the significant variations within these principal components. This allows for a clear and easy to interpret representation of the inherent structures and relationships within the RGB data, aiding in understanding the patterns that may be indicative of distinct classes or groups.Hierarchical clustering analysis (HCA)
Hierarchical clustering analysis (HCA) was employed to generate dendrograms from RGB data in Origin 2023. HCA allows the identification of inherent structures within datasets by grouping similar data points into clusters, aiding in the visualization of relationships and patterns. In this analysis, the cluster method chosen was based on the group average. This method calculates the average distances between all pairs of data points in different clusters, contributing to a balanced representation of similarities within and between clusters. For the distance metric, the Euclidean distance was selected. The Euclidean distance is a common choice when dealing with multidimensional data, measuring the straight-line distance between two points in a space defined by their RGB values. This metric works well for capturing the geometric relationships between data points, providing a reliable measure of dissimilarity in the RGB space.Results and discussion
Our overarching goal was to make a paper immunoassay that could discriminate between SARS CoV-2 variants repurposing S antibodies for an earlier variant (Fig. 1b). We chose alpha as the earlier variant as it first appeared in the pandemic in late 2020, and sought to demonstrate that alpha antibodies could be reconfigured to detect a later variant. For the variant that occurred later in time, we chose the Omicron variant BA.5 as it was widespread in the pandemic in mid-2022. Therefore, we trained an assay constructed out of alpha antibodies to detect and distinguish S RBD of alpha and also BA.5. Then, we wanted to challenge the assay with an unknown variant that would be new to the assay, and for this we chose Omicron BA.1, which was also circulating in 2022.Measuring the affinity of the antibody pool for the targets
Looking at sequences for the spike protein among variants (ESI† Fig. S1), S RBD of alpha and BA.5 have high similarity, where sequence alignment (BLASTp) shows calculated numbers of 95% identity or higher when considering the S RBD only (Genbank access number YP_009724390.1). Generally, selective arrays generally employ multiple binding agents that possess a range of cross reactivities, where some species are more specific to certain analytes, while others bind more broadly.27 We applied this principle here to our antibody choice so that they would have a range of cross reactivities towards BA.5. One antibody, Ab1, was a rabbit polyclonal antibody for alpha S RBD. The second, Ab2, was a chimeric monoclonal antibody also raised against alpha S RBD, which had a constant domain of the human IgG1 molecule with mouse variable regions.We used microscale thermophoresis (MST) to characterize binding of each of the antibodies for each of the targets, alpha and BA.5 spike RBD proteins (Fig. 2a).28 The MST data was taken using the Monolith instrument and capillaries from NanoTemper. For each reading, ∼10 μL of our sample were added to the premium capillaries. The MST time traces for Ab1 incubated with alpha at different concentrations (Fig. 2b and inset, red) exhibited a systematic change with alpha concentration. The resulting dose response curve based on the MST traces exhibited an upward trend indicative of binding (Fig. 2c, pink), and a fit yielded a dissociation constant KD of 17.1 nM. When Ab1 was incubated with BA.5, the MST traces also exhibited a systematic change (Fig. 2b and inset, blue) and a dose response curve that suggested binding (Fig. 2c, blue) with a KD of 292.3 nM. Thus, Ab1 bound to both targets with reasonable affinity.
MST traces for Ab2 exhibited systematic concentration dependences for alpha (Fig. 2d, red) but not for BA.5 (Fig. 2d, blue). Ab2 dose response curves with alpha exhibited a concentration dependence that yielded KD = 22.8 nM (Fig. 2e, pink), but did not change significantly with BA.5 concentration, and yielded a KD = 12.7 nM (Fig. 2e, blue). Thus, Ab2 bound with different affinities for alpha vs. BA.5. We note that thermophoretic trends for Ab2 binding to the antigens were the opposite direction from Ab1, which can occur in MST due to differences in the thermophoretic properties of the bound complex relative to the free species.29,30 Overall, the MST results showed that the two anti-alpha antibodies exhibit differential binding for each of the targets, and thus the set had potential to be used as a selective array.
Screening pairs of alpha antibodies
For a visible signal to be generated in an immunoassay, a pair of antibodies must be able to bind to the antigen simultaneously to form a sandwich. Even though two given antibodies can bind to a target individually, shared epitopes may prevent sandwich formation for the pair. Therefore, we quantified the immunoassay signals that the antibodies pairs would generate by screening all possible combinations for antibodies immobilized on the nitrocellulose and conjugated to the nanoparticle (NP) (Fig. 3).Gold NPs were synthesized using an aqueous approach using citrate reduction which resulted in ∼30 nm diameter spherical particles that were red in color.31 NPs were conjugated to each of the antibodies by incubation in solution, which allows them to bind to the NP surface by physisorption. While there are conjugation strategies that utilize covalent click chemistry, those tend to result in lower reaction yield, and others have found that physisorption does not compromise the ability of the antibody to bind to a target.32
We then ran every possible antibody pair with each antigen plus no antigen as the negative control (Fig. 3a). Each antibody (Ab1, Ab2) was immobilized on a nitrocellulose strip at the test area. The control antibody (anti-Fc) was immobilized on the control line as a positive control to verify flow. Then, the strip was run with a given NP–Ab conjugate with each of the targets. Given two antibodies which could be either immobilized antibodies or conjugated on the NPs as immunoprobes, this resulted in 2 immobilized antibodies × 2 immunoprobes × 3 antigen cases (alpha, BA.5, negative control) = 12 different strips. Fig. 3b shows test strip images for the pair consisting of immobilized Ab1 and NP–Ab1. The test area intensity varied depending on which antigen was run, with a strong intensity for S RBD from alpha, and a much lower intensity for BA.5. No signal was observed at the test area when no antigen was present (control). RGB intensities of the test areas were quantified by ImageJ25 and plotted as a heat map for the different immobilized antibodies (horizontal) vs. NP–antibody conjugates (vertical) (Fig. 3c). A range of cross reactivities with the antibody pairs was observed. For example, immobilized Ab1 run with NP–Ab1 resulted in a finite signal for BA.5 (14.42 RGB intensity), but when run with NP–Ab2 exhibited nearly no signal (0.55 RGB intensity). On the other hand, all pairs demonstrated signal when run with alpha. Negative controls showed a range of baseline signal below 10 RGB, which could be attributed to non-specific adsorption. This shows that depending on the antibody pair, the antigens produce different signal intensities, illustrating that this set of antibodies had the potential to discriminate between the variants.
Multicolored multiplexed test for alpha and BA.5
Based on the behavior of each of the antibody pairs, we then sought to design a multiplexed test that could be trained on the different antigens. Previously we demonstrated the ability to repurpose antibodies for immunoassays of off-targets for the cases of flaviviruses and filoviruses, where dengue and zika anti-NS1 antibodies could be used to detect yellow fever NS1,18 and Marburg anti-glycoprotein (GP) antibodies to detect Ebola GP.33 For this, we required NP–Ab conjugates of visually distinct colors so they would result in different colors at the test areas.We designed the assay based on how the antibodies paired with each of the antigens (Fig. 3). The multiplexed assay had two test areas with Ab1 and Ab2 immobilized at different locations, and was run with a mixture of blue GNS–Ab1 and red NP–Ab2 so the test areas could yield a range of colors (Fig. 4). Thus, a different colorimetric pattern resulted based on the antigen that was run.
To complement the red NPs, we synthesized star-shaped gold nanoparticles, or gold nanostars (GNS) which were blue in color (Fig. 4c). GNS were synthesized using a reduction of a Goods buffer21 so that their peak SPR was at 708 nm (Fig. 4a, blue line) which was shifted considerably from the SPR of the spherical red NPs (Fig. 4a, red line). DLS showed that the GNS had an average DH of 215.4 ± 44.7 nm. When the NP and GNS antibody conjugates were spotted down on nitrocellulose, the format of the final assay (Fig. 4f), their colors were visually distinguishable.34
Ab1 was conjugated to the blue GNS and Ab2 was conjugated to the red NPs. DLS showed that the GNS exhibited an increase in DH of from 215.4 nm to 246 nm for the GNS–Ab1, and the NPs from 33.6 nm to 56.4 nm for the NP–Ab2 (Fig. 4b), confirming successful conjugation. UV-vis showed a slight peak shift but no major decrease in intensity, thus exhibiting no significant aggregation (Fig. 4a, dotted lines). This also confirmed conjugation to the antibodies and that they were stable in solution.
Machine learning to optimize running conditions to distinguish SARS-CoV-2 variants
We then optimized conditions to result in a test that could successfully discriminate between the alpha and BA.5 variants using the repurposed antibodies for alpha. For this we used supervised machine learning (ML) in an iterative loop to optimize test conditions for improved accuracy, an approach we and others have used before to optimize a multicolor immunoassay (Fig. 4d).35,36 First, the test was run and the R,G, and B intensities of each of the two test areas was quantified by image analysis, resulting in 6 features. Instead of using the RGB values for machine learning, we utilized an approach using stain vectors for the components because the blue GNS are not pure blue, and the red NPs are not pure red. Alternatively, stain vectors that are linear combinations of RGB can better represent the color of the GNS and NPs.26 Stain vectors were defined for both the blue GNS–Ab1 (S2) and red NP–Ab2 (S1) from the color each species spotted on paper individually (Fig. 4f). A third vector S3 orthogonal to both S1 and S2 was defined by the cross product of S1 and S2. Then, the color information for each of the test spots was deconvolved to obtain a value based on S1, S2, and S3.The training model chosen for the training of the test was linear discriminant analysis (LDA). LDA analyses were performed based on the deconvoluted RGB values of the test spots. LDA is advantageous when dealing with multivariate data, such as deconvoluted RGB values of two spots, as it maximizes the separation between class means while minimizing within-class variability. For test optimization, a 5-fold cross-validation was performed, which involves the partitioning of the dataset into subsets, training the model on a subset, and validating its performance on the remaining data. By validating the model on different subsets, cross-validation provides a more robust evaluation of its performance, enhancing the reliability of the LDA analysis on the deconvoluted RGB values of the test spots.
A confusion matrix was used to evaluate the accuracy of the resulting test based on the LDA model, where on-diagonals in the confusion matrix indicated correctly classified results (predicted class = true class) and off diagonals indicated incorrectly classified ones (predicted classes19 true class). Initial conditions resulted in an accuracy of 22.22%. Changing running conditions (washes, buffers, quenching agents) could improve accuracy to 55.56%, 66.67% (ESI† Fig. S2), and then ultimately 100% (Fig. 4h). Using the finalized as the optimal running conditions, if alpha was run, it resulted in signal at both test areas, where the lower area was bluer and the upper one more purple. If BA.5 was run, it resulted in only one spot on the upper test location which was blue in color (Fig. 4e). This shows that the resulting test pattern could discriminate between variants and thus was successfully optimized.
LDA of the top two components showed data clusters based on the variant run (Fig. 4g, ellipses indicating 95% confidence). The cluster for alpha was well separated from both BA.5 and the negative control, with no overlap. This shows that the assay could successfully discriminate between alpha and BA.5 variants using entirely repurposed alpha antibodies without the need for specific antibodies against BA.5. This shows the potential for discriminating SARS-CoV-2 variants in a rapid test format, which to date have not allowed variant identification, and that it can be achieved without antibodies specific for that variant.
Limit of detection
We measured the limit of the detection (LOD) of each of the symmetric antibody pairs (GNS–Ab1/Ab1 and NP–Ab2/Ab2). Strips with immobilized Ab1 and run with GNS–Ab1 were run with varying concentrations of alpha and BA.5 (ESI,† Fig. S3a–d). The LOD calculations were performed by fitting a modified Langmuir curve to the experimental data, which best describes the relationship between signal and concentration.37 The Y-intercept for the LOD was then calculated by multiplying the standard deviation of the signal at 0 nM concentration of the target by three and adding this value to the y-intercept of the calibration curve. Finally, the LOD concentration was determined using the fitted curve equation. The test area RGB intensity was quantified using ImageJ (ESI,† Fig. S3e) and plotted as a function of protein concentration. Blue GNS–Ab1/Ab1 showed a strong dose response curve with alpha, and fit to a Langmuir equation yielding an LOD of 22.2 nM (ESI† S4a), but did not exhibit a dose response with BA.5 that could be fit (ESI,† Fig. S4b). The NP–Ab2/Ab2 pair exhibited a strong dose response and yielded an LOD of 4.17 nM for the area before the hook region (ESI,† Fig. S4c and d, pink). The hook effect, a common issue in immunoassays, occurs when high analyte concentrations produce falsely low readings, limiting the system's dynamic range. This effect causes misleading low readings at high alpha concentrations when using the NP–Ab2/Ab2 conjugate. We measured the LOD by testing smaller concentrations of the alpha spike protein. Moreover, the NP–Ab2/Ab2 pair did not bind to BA.5 (ESI,† Fig. S4c, blue), where it did not show a significant increase in intensity with increasing BA.5 concentration. While MST results indicated that Ab1 could bind to BA.5 and that the NP–Ab1/Ab1 pair could also bind to BA.5, (Fig. 3b), binding was decreased for the GNS–Ab1 conjugate. To compare the limit of detection (LOD) of our system with the target concentration in an infected patient's serum sample, we referenced the expected target concentration range of 2.5 μg mL−1 to 17.5 μg mL−1 from George et al.38 For the GNS–Ab1/Ab1 pair with the alpha protein, our LOD is 0.216 μg mL−1, which is well below the concentration found in an infected patient's sample. Similarly, for the NP–Ab2/Ab2 pair with the alpha protein, our LOD is 0.769 μg mL−1. However, for the BA.5 variant, we were unable to determine a reliable LOD with either antibody pair, precluding a meaningful comparison of LOD versus target concentration for this variant.Challenging the test with an unknown variant
Having demonstrated the ability to discriminate between alpha and BA.5 variants, we then explored whether the test could pick up a variant for which it was not trained, utilizing Omicron BA.1 as an unknown (Fig. 5a). This time we used PCA to evaluate the colorimetric pattern of the assay8,11 as it can classify unknowns into groups with comparable properties, and is a powerful approach for its ability to discriminate between analytes that are highly similar. Here the sequence similarity between the targets was high, where BLASTp results (Fig. S1†) revealed a sequence identity for BA.1 of 96% with BA.5 and 95% with alpha (Fig. 1e).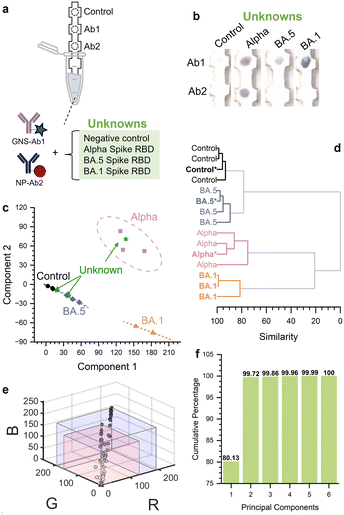 | ||
| Fig. 5 a) Challenging the test with an unknown variant. Strips were run with alpha, BA.5, and then challenged with BA.1 as a new variant; b) resulting strip images run with the unknowns; c) PCA of clustering with 95% confidence ellipses, with unknowns in green and BA.1 unknown in orange triangles; d) dendrogram constructed using HCA; e) RGB plot of test areas showing the range of colors possible for the GNS–Ab1 (blue prism) and NP–Ab2 (pink prism). Each individual dot is the RGB value for a given test result and/or spotted strip (ESI† Fig. S5); f) scree plot of the assay. | ||
Strips run with BA.1 exhibited a different colorimetric pattern compared to alpha, BA.5, and the negative control, where it resulted in a single dark blue spot at the upper test area (Fig. 5b, triplicates, ESI† Fig. S5). To determine whether this pattern was distinguishable from those of the other targets, we used PCA of the stain vectors of the RGB values of the two test areas. BA.1 clustered in an entirely new region that was well separated from alpha, BA.5, and the negative control, with no overlap (Fig. 5c, orange). Thus, the colorimetric pattern was sufficiently distinct to enable discrimination of BA.1 from alpha and BA.5. This shows that the test could identify a new variant on which it was not trained.
The other unknowns were species the assay had encountered before (negative control, alpha, and BA.5) which were also correctly classified, where each of them was properly assigned to their respective clusters (Fig. 5c, green triangles). A zoom in of the data in 3D is shown in ESI,† Fig. S6.
Dendrograms were constructed using hierarchical cluster analysis (HCA) of the RGB data (Fig. 5d).11 Unknowns (bold, marked with *) were located within the clusters of their true classes. Thus, the test correctly classified the unknown variants. This shows that the assay using only antibodies for alpha, could pick up an unknown variant on which it was not trained, and distinguish it from the variants in the original training set.
The 24-spot olfactory array of Suslick et al. could discriminate between a massively large number of analytes.39 In this context, the discriminatory power refers to the ability of the array to distinguish between different analytes based on the unique colorimetric patterns produced by the interaction of the compounds with the array of chemo-responsive dyes. Assuming detectable RGB 8-bit differences of 4 and utilizing their scree plot, they estimated a discriminatory power of 1012.39 To calculate the discriminatory power of our assay, we quantified the number of distinct patterns the multiplexed immunoassay can generate, which is determined by the variability and resolution of the color changes observed, number of test areas available, and the distribution of the variance within the data. Here, the red NPs and blue GNS do not span the entire RGB spectrum, especially when they are spotted onto paper substrates, and thus are expected to yield a narrower range of possible RGB values. To determine the range of possible colors at a test strip, we experimentally measured the RGB values for different concentrations of the red NP–Ab2 and blue GNS–Ab1 on nitrocellulose. We spotted the GNS–Ab1 and NP–Ab2 down on nitrocellulose in aliquots up to the final volume used in an assay run (3 μL for blue GNS–Ab1, 2 μL for red NP–Ab2, ESI† Fig. S7) and measured RGB values of the spots (Fig. 5e). Given that the entire RGB space is not accessible with these two stains, we experimentally determined the number of distinct colors achievable by mixing the NP and GNS at various ratios. Included in the figure are the RGB values for mixtures of red NP–Ab2 and blue GNS–Ab1 spotted down on nitrocellulose as well as all of the RGB values of test areas from the strips for the negative control, alpha, BA.5, and BA.1. The maximum RGB value for each immunoprobe is indicated (blue and pink prisms). Based on the available RGB space, we estimated that there are ∼30 distinguishable colors for a given spot, so for two spots this yields (30)2 = 900 possible combinations. While 8-bit RGB analysis theoretically allows for finer color distinctions, we opted for a more conservative estimate to avoid overstating the assay's discriminatory power. Based on the scree plot showing the cumulative percentage of the variance as a function of the principal components (Fig. 5f), we found that >99% of the information is contained within two components, so that only 2 components are necessary for discriminating the analytes. While this is a rough estimate for the discriminatory power, it is still higher compared to the total number of confirmed SARS-CoV-2 variants, which is currently over 35.40,41
Chemical olfactory arrays can discriminate a large number of closely related species by using a large array of binding agents which possess a range of cross reactivities, where some species will bind broadly to all analytes, and others to only a select few. They typically rely on a single binding event between the analyte and the immobilized sensor molecule to induce a color change, typically employing porphyrins and other organometallic molecules.11,39 However, for paper immunoassays, a signal is generated upon sandwich formation of the antigen with both with NP–Ab and the immobilized Ab. Because there are two binding events, the signal is the result of two different binding affinities. This affords room for differential signals in our 2-spot array, which has only 12 different possible combinations for sandwich formation. Based on the estimated discriminatory power, we believe that an extensive array of antibodies is not necessary in order to discriminate between several variants. Thus, the test can be quite economical, as antibodies are the most expensive component in a lateral flow assay strip.24 Furthermore, using polyclonals for certain antibodies is advantageous, as they can bind to a range of epitopes on the target. Consequently, they have a higher probability of cross-reactivity, a favorable attribute for adaptive immunoassays. While the benefit of monoclonals is attractive due to their high specificity, this actually can be a drawback when trying to make an adaptive assay that can detect an emerging target, and they would need to be combined with the use of polyclonals. Here, because Ab1 and Ab2 are polyclonal, we showed that they exhibited different levels of cross-reactivity sufficient to make an array. Ultimately, determining the number of suitable test areas with unique antibodies and also NP–Ab conjugates is something that requires systematic experimental investigation, where the antibodies are varied and the ability to distinguish unknowns is quantified, and will be subject of future studies.
Conclusions
We demonstrated an approach to make an immunoassay that can detect a variant that was not encountered before by adapting it to behave like a chemical olfactory array. The assay used two different colored nanoparticles in a 2-spot array, and readout of the colorimetric pattern was by PCA as opposed to a binary yes/no answer based on presence of signal. In doing so, the immunoassay could detect the presence of an unknown in a low-cost format.This assay was exclusively built using antibodies raised against the alpha variant, eliminating the necessity for antibodies tailored to each detectable variant. This underscores the ability to repurpose reagents successfully. Furthermore, the assay required only two different antibodies—one that was cross reactive, and one that was less so. Commercial antibodies were deliberately chosen to show that custom antibodies are not required.
While we have demonstrated repurposing antibodies in immunoassays before, here we applied the technique to the SARS-CoV-2 spike protein for variant discrimination. Previously, our studies demonstrated the adaptability of repurposing antibodies from one virus to effectively detect an entirely different virus, e.g., repurposing dengue and Zika antibodies to detect yellow fever NS1, or utilizing dengue antibodies for Zika detection, and even repurposing dengue 3 antibodies to detect dengue serotypes 1–4. Notably, in all of these instances, the targets were already identified, where the test was trained on the targets via supervised learning. In addition, the sequence similarities in the prior works were comparatively lower, approximately ∼55%, ∼75%, and ∼85%, respectively.18,33 Variants of SARS-CoV-2 are more challenging because sequence identity and similarity between them are much higher in comparison (>95%). Taking into consideration these prior examples, this shows that repurposing antibodies can be successful with a large range of sequence similarities. Future work could push the upper and lower bounds on similarities as well as epitope mapping to gain a fuller understanding of the mechanism, and then extend this approach to pick up an unknown disease.
Current approaches in diagnostic development have significant shortcomings, with major bottlenecks in antibody production,42–44 where deployment of a point of care LFA test can take as long as a year. This is far too long to aid in rapid response to new diseases, which are often characterized by large increases in cases within a timeframe of weeks. This problem highlights the need for new ways outside of traditional approaches to get diagnostics for emerging diseases on the ground sooner.45 Ignoring new outbreaks or not scanning for potential ones has dangerous consequences.46 Past experience has shown that public health systems need to be better prepared not just for the next SARS-CoV-2 variant, but for the next virus, pointing to the dire need for widespread disease surveillance.47 While there are surveillance efforts sequencing viruses in animal populations that can potentially induce zoonotic spillover, these spillover events are rare, making it nearly impossible to predict what will cause the next pandemic.48,49 Arguably, our response to new outbreaks has been suboptimal, primarily attributed to our inadequate preparedness for such events.50 This underscores the fact that traditional approaches, i.e., generating a new antibody for every new disease variant once we detect it in circulation, are not rapid enough for an effective response.
The approach here can offer a solution to detect an unknown and classify it in a rapid, widely deployable format. It has the potential to be applied to other pathogens and evolving targets, not just viral but also bacterial targets.1 While it is probably not possible to make a single universal immunoassay that can detect any virus, it seems plausible to make for a broad virus type (e.g., one that encompasses flaviviruses, another for coronaviruses, filoviruses, etc.). Furthermore, the assay can be tailored to target things that could potentially arise in a particular geographic region. It should be of note that the identity of the antibodies used in the assay may result in limitations in their range of cross reactivities and what the ultimate assay can detect and distinguish. Also, traditional challenges faced by paper immunoassays such as non-specific adsorption and sample matrix effects would still be a potential problem.
The protocol is simple to execute and results in a low-cost assay, as it uses colorimetric/visual sensing, where readout does not require sophisticated instrumentation and can be via mobile phone images or desktop scanners. Moreover, the protocol can be easily shared and adapted to whatever reagents an end-user has, eliminating the need for specialized reagents and antibodies that may not be currently accessible. This renders it amenable for wide distribution, unlike sequencing which has intensive requirements for trained personnel, instrumentation, reagents, energy, and lab infrastructure. Additionally, the affinity agents in the immunoassay are not limited exclusively to antibodies; they may encompass peptides, nanobodies, or other species that have reasonable (∼nM) binding affinity.
Data availability
The data supporting this article have been included as part of the ESI.† These include the Matlab script used for the PCA of the unknowns, and data in the form of the RGB values of the assay strips as a Microsoft Excel table.The machine learning (ML) script was used in ref. 35 (Mata Calidonio and Hamad-Schifferli) and is available in the ESI† for that publication.
Author contributions
JMC and AIM executed the experiments, JMC performed the data analysis, KHS supervised the work and administered the project, JMC and KHS wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Biophysical Instrumentation Core at the University of Massachusetts Boston, funded by the Massachusetts Life Sciences Center.Notes and references
- S. A. Walper, G. Lasarte Aragonés, K. E. Sapsford, C. W. Brown, III, C. E. Rowland, J. C. Breger and I. L. Medintz, ACS Sens., 2018, 3, 1894–2024 CrossRef CAS PubMed.
- J. H. Ellwanger and J. A. B. Chies, Genet. Mol. Biol., 2021, 44, e20200355 CrossRef CAS PubMed.
- C. J. Carlson, G. F. Albery, C. Merow, C. H. Trisos, C. M. Zipfel, E. A. Eskew, K. J. Olival, N. Ross and S. Bansal, Nature, 2022, 607, 555–562 CrossRef CAS PubMed.
- A. J. Meadows, N. Stephenson, N. K. Madhav and B. Oppenheim, BMJ Glob. Health, 2023, 8, e012026 CrossRef PubMed.
- A. F. Brito, E. Semenova, G. Dudas, G. W. Hassler, C. C. Kalinich, M. U. G. Kraemer, J. Ho, H. Tegally, G. Githinji and C. N. Agoti, et al. , Nat. Commun., 2022, 13, 7003 CrossRef PubMed.
- F. Jane, BMJ, 2023, 381, p1157 Search PubMed.
- L. Otten, E. Fullam and M. I. Gibson, Mol. BioSyst., 2016, 12, 341–344 RSC.
- J. R. Uzarski and C. M. Mello, Anal. Chem., 2012, 84, 7359–7366 CrossRef CAS PubMed.
- W. J. Peveler, M. Yazdani and V. M. Rotello, ACS Sens., 2016, 1, 1282–1285 CrossRef CAS PubMed.
- S. Rana, A. K. Singla, A. Bajaj, S. G. Elci, O. R. Miranda, R. Mout, B. Yan, F. R. Jirik and V. M. Rotello, ACS Nano, 2012, 6, 8233–8240 CrossRef CAS PubMed.
- C. Zhang and K. S. Suslick, J. Agric. Food Chem., 2007, 55, 237–242 CrossRef CAS PubMed.
- G. Macias, J. R. Sperling, W. J. Peveler, G. A. Burley, S. L. Neale and A. W. Clark, Nanoscale, 2019, 11, 15216–15223 RSC.
- J. R. Sperling, B. Poursat, L. Savage, I. Christie, C. Cuthill, B. Aekbote, K. McGuire, A. S. Karimullah, J. Robbie and W. T. Sloan, et al. , Environ. Sci.: Nano, 2023, 10, 3500–3508 RSC.
- J. Hadfield, C. Megill, S. M. Bell, J. Huddleston, B. Potter, C. Callender, P. Sagulenko, T. Bedford and R. A. Neher, Bioinformatics, 2018, 34, 4121–4123 CrossRef CAS PubMed.
- https://nextstrain.org/ncov/gisaid/global/6m .
- M. Amicone, V. Borges, M. J. Alves, J. Isidro, L. Zé-Zé, S. Duarte, L. Vieira, R. Guiomar, J. P. Gomes and I. Gordo, Evol. Med. Public Health, 2022, 10, 142–155 CrossRef PubMed.
- C. Del Vecchio, B. C. Daniels, G. Brancaccio, A. R. Brazzale, E. Lavezzo, C. Ciavarella, F. Onelia, E. Franchin, L. Manuto and F. Bianca, et al. , Nat. Commun., 2022, 13, 1–16 CrossRef PubMed.
- C. Rodríguez-Quijada, J. Gomez-Marquez and K. Hamad-Schifferli, ACS Nano, 2020, 14, 6626–6635 CrossRef PubMed.
- Q. Wang, S. Iketani, Z. Li, L. Liu, Y. Guo, Y. Huang, A. D. Bowen, M. Liu, M. Wang and J. Yu, et al. , Cell, 2023, 186, 279–286.e278 CrossRef CAS PubMed.
- H. de Puig, J. O. Tam, C.-W. Yen, L. Gehrke and K. Hamad-Schifferli, J. Phys. Chem. C, 2015, 119, 17408–17415 CrossRef CAS PubMed.
- K. Chandra, K. S. B. Culver, S. E. Werner, R. C. Lee and T. W. Odom, Chem. Mater., 2016, 28, 6763–6769 CrossRef CAS.
- G. Ruiz, N. Ryan, K. Rutschke, O. Awotunde and J. D. Driskell, Langmuir, 2019, 35, 10601–10609 CrossRef CAS PubMed.
- D. Hristov, H. Rijal, J. Gomez-Marquez and K. Hamad-Schifferli, Anal. Chem., 2021, 93, 7825–7832 CrossRef CAS PubMed.
- J. O. Tam, H. de Puig, C.-W. Yen, I. Bosch, J. Gómez-Márquez, C. Clavet, K. Hamad-Schifferli and L. Gehrke, J. Immunoassay Immunochem., 2017, 38, 355–377 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- A. C. Ruifrok and D. A. Johnston, Anal. Quant. Cytol., 2001, 23, 291–299 CAS.
- J. R. Askim, M. Mahmoudi and K. S. Suslick, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
- C. J. Wienken, P. Baaske, U. Rothbauer, D. Braun and S. Duhr, Nat. Commun., 2010, 1, 100 CrossRef PubMed.
- M. Jerabek-Willemsen, T. André, R. Wanner, H. M. Roth, S. Duhr, P. Baaske and D. Breitsprecher, J. Mol. Struct., 2014, 1077, 101–113 CrossRef CAS.
- R. Magnez, C. Bailly and X. Thuru, Int. J. Mol. Sci., 2022, 23(14), 7672 CrossRef CAS PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- G. Ruiz, K. Tripathi, S. Okyem and J. D. Driskell, Bioconjugate Chem., 2019, 30, 1182–1191 CrossRef CAS PubMed.
- H. de Puig, I. Bosch, N. Salcedo, C. Clavet, J. J. Collins, K. Hamad-Schifferli and L. Gehrke, Nano Today, 2022, 47, 101669 CrossRef CAS PubMed.
- S. Lee, S. Mehta and D. Erickson, Anal. Chem., 2016, 88, 8359–8363 CrossRef CAS PubMed.
- J. Mata Calidonio and K. Hamad-Schifferli, Sens. Diagn., 2024, 3, 677–687 RSC.
- H.-A. Joung, Z. S. Ballard, J. Wu, D. K. Tseng, H. Teshome, L. Zhang, E. J. Horn, P. M. Arnaboldi, R. J. Dattwyler and O. B. Garner, et al. , ACS Nano, 2020, 14, 229–240 CrossRef CAS PubMed.
- H. de Puig, I. Bosch, M. Carre-Camps and K. Hamad-Schifferli, Bioconjugate Chem., 2017, 28, 230–238 CrossRef CAS PubMed.
- S. George, A. C. Pal, J. Gagnon, S. Timalsina, P. Singh, P. Vydyam, M. Munshi, J. E. Chiu, I. Renard and C. A. Harden, et al. , Kidney360, 2021, 2, 924–936 CrossRef PubMed.
- K. S. Suslick, N. A. Rakow and A. Sen, Tetrahedron, 2004, 60, 11133–11138 CrossRef CAS.
- SARS-CoV-2 Variant Classifications and Definitions, https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html Search PubMed.
- S. Elbe and G. Buckland-Merrett, Glob. Chall., 2017, 1, 33–46 CrossRef PubMed.
- O. Yang, M. Qadan and M. Ierapetritou, J. Pharm. Innov., 2020, 15, 182–200 CrossRef PubMed.
- Monoclonal Antibody Production, National Research Council (US) Committee on Methods of Producing Monoclonal Antibodies, Washington, DC, 1999 Search PubMed.
- K. F. Smith, M. Goldberg, S. Rosenthal, L. Carlson, J. Chen, C. Chen and S. Ramachandran, J. R. Soc., Interface, 2014, 11, 20140950 CrossRef PubMed.
- J. Mata Calidonio and K. Hamad-Schifferli, Biochim. Biophys. Acta, Gen. Subj., 2023, 1867, 130266 CrossRef PubMed.
- D. M. Morens, G. K. Folkers and A. S. Fauci, Nature, 2004, 430, 242–249 CrossRef CAS PubMed.
- M. D. Perkins, C. Dye, M. Balasegaram, C. Bréchot, J.-V. Mombouli, J.-A. Røttingen, M. Tanner and C. C. Boehme, Lancet, 2017, 390, 2211–2214 CrossRef PubMed.
- J. Piret and G. Boivin, Front. Microbiol., 2021, 11, 631736 CrossRef PubMed.
- E. C. Holmes, A. Rambaut and K. G. Andersen, Nature, 2018, 558, 180–182 CrossRef CAS PubMed.
- B. Gates, New Engl. J. Med., 2015, 372, 1381–1384 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00505h |
| This journal is © The Royal Society of Chemistry 2024 |



