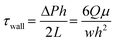Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
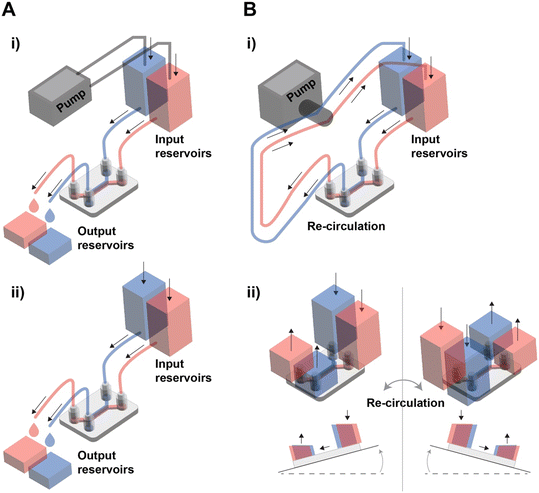
Reconstitution of human tissue barrier function for precision and personalized medicine
Jaehoon
Kim
a,
Taehee
Yoon
ab,
Sungryeong
Lee
 a,
Paul J.
Kim
c and
YongTae
Kim
a,
Paul J.
Kim
c and
YongTae
Kim
 *abde
*abde
aGeorge W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA. E-mail: ytkim@gatech.edu
bParker H. Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332, USA
cDepartment of Psychiatry & Behavioral Sciences, School of Medicine, Emory University, Atlanta, GA 30322, USA
dWallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA
eInstitute for Electronics and Nanotechnology, Georgia Institute of Technology, Atlanta, GA 30332, USA
First published on 19th June 2024
Abstract
Tissue barriers in a body, well known as tissue-to-tissue interfaces represented by endothelium of the blood vessels or epithelium of organs, are essential for maintaining physiological homeostasis by regulating molecular and cellular transports. It is crucial for predicting drug response to understand physiology of tissue barriers through which drugs are absorbed, distributed, metabolized and excreted. Since the FDA Modernization Act 2.0, which prompts the inception of alternative technologies for animal models, tissue barrier chips, one of the applications of organ-on-a-chip or microphysiological system (MPS), have only recently been utilized in the context of drug development. Recent advancements in stem cell technology have brightened the prospects for the application of tissue barrier chips in personalized medicine. In past decade, designing and engineering these microfluidic devices, and demonstrating the ability to reconstitute tissue functions were main focus of this field. However, the field is now advancing to the next level of challenges: validating their utility in drug evaluation and creating personalized models using patient-derived cells. In this review, we briefly introduce key design parameters to develop functional tissue barrier chip, explore the remarkable recent progress in the field of tissue barrier chips and discuss future perspectives on realizing personalized medicine through the utilization of tissue barrier chips.
1. Introduction
A tissue-to-tissue barrier in a body is a biological wall that separates different tissues, controlling the movements of molecules, cells, and other entities across the tissues.1 These barriers are essential for maintaining the physiological homeostasis, protecting the tissues and organs from harmful agents, and regulating the exchange of signaling molecules. The tissue barriers typically form with the endothelium in the blood vessels and the epithelium of the organs. Examples of the tissue barriers in the body include the blood–brain barrier, blood-biliary barrier, the intestinal barrier, the lung barrier, and the skin barrier, among others.In pharmaceutical drug development, understanding the physiology of tissue barriers is crucial for predicting drug response. The absorption of a drug through a tissue barrier is an essential step in its pharmacokinetics.2,3 In the case of oral delivery, the drug is absorbed through the intestinal barrier to enter the blood vessels. For drug administered through intravenous injection, the drug is directly delivered into the blood vessels, but enters into another tissue barriers in the liver and kidney, which are major organs involved in a drug metabolism and excretion. Thus, in addition to minimizing hepatic clearance, attaining high intestinal absorption and low intestinal metabolism are key outcomes in drug discovery. A tissue barrier of tumors containing leaky blood vessel4,5 is one of the tissue barriers and another important biological interface in developing anti-cancer drugs.
Physiologically relevant human tissue barrier models can provide valuable insights into how drugs interact with human tissue barriers,6 aiding in the design and optimization of drug formulations for improved efficacy and safety. These models allow for the study of drug transport, metabolism, and interactions within the context of specific tissue barriers, bringing us closer to more approaches. Traditionally, animal models have played a central role in understanding drug behavior and responses within the body.7 Animal models are expected to be still necessary in preclinical toxicity testing because extensive historical data in support of their utilization, enabling researchers to investigate toxicity across various organs within a single study. Additionally, animal models can capture responses of the entire organism, potentially involving intricate interactions among interconnected organ systems.8 Animal models are known to often unable to align with the outcomes observed in human phase II clinical trials. The substantial failure rates which these phase II/III clinical trials, which can reach as high as 80%, have primarily been attributed to genetic and physiological distinctions between animals and humans, as well as the utilization of overly simplistic in vitro experimental models.9 Ethical issues related to animal studies also have come to the forefront, prompting the inception of alternative technologies permitted by the FDA Modernization Act of 2021 and the Humane Research and Testing Act (HR 1744).10
Organ-on-a-chip or microphysiological system (MPS) is one such alternative to animal models, with tissue barrier applications comprising a significant portion. A tissue barrier chip, one application of an MPS, is a microfluidic cell culture device that aims to replicate the structure and function of human tissue barriers in a controlled micro-environment. These chips are designed to simulate the complex physiological and mechanical properties of specific tissues or organ interfaces within the human body.
Recent advancements in stem cell technology have brightened the prospects for the application of tissue barrier chips in personalized medicine.11,12 The discovery of induced pluripotent stem cells (iPSCs) and adult stem cells (ASCs) enables rapid acquisition of various cells from tissues derived from patients. This breakthrough has facilitated emergence patient derived organoids from patients' iPSCs and ASCs, which demonstrate highly similar structures, cell types, and genetic characteristics to human organs and are thus gaining traction in the field of personalized medicine.13 However, a significant structural limitation arises from the lack of a physiologically relevant tissue-to-tissue interface, an apical in/basal out structure, and the absence of inlets and outlets.14,15 This limitation presents substantial challenges for functional studies. For instance, in the case of skin organoids, simulating drug treatment on the skin surface is not feasible due to the apical side being positioned internally.16 However, the utilization of iPSC- and ASC-derived cells in tissue barrier chips, hinting at promising possibilities for personalized drug tests. Tissue barrier chips are characterized by the presence of distinct apical and basal sides and microchannels connecting each side. This allows for the observation of tissue barrier responses depending on the direction of drug entry, enabling quantitative evaluations of drug penetration and absorption. These advantages, combined with the utilization of iPSC- and ASC-derived cells, are paving the way for remarkable progress in the field of personalized medicine. Hence, the evolution of tissue barrier chips holds tremendous promise for revolutionizing drug development and personalized medicine.
In past decade, designing and engineering these microfluidic devices, and demonstrating the ability to reconstitute tissue functions were main focus of this field.17 However, the field is now advancing to the next level of challenges: validating their utility in drug evaluation and creating personalized models using patient-derived cells. This review discusses key design parameters to develop functional tissue barrier chip. We will also explore the remarkable progress made in the field of tissue barrier chips over the last five years. Additionally, we will discuss future perspectives on realizing personalized medicine through the utilization of tissue barrier chips.
2. Key design parameters to develop functional tissue barrier chip
Creating key biological specifications for organ specific tissue barrier function was covered by excellent reviews which have been already published.9,18–20 This section describes in-depth exploration of the engineering aspects involved in constructing and designing tissue barrier chips.2.1 Compartmentalization
Tissue barriers are specialized structures that separate different body compartments and serve as selective barriers to control the transport of substances and maintain homeostasis. To mimic the tissue barrier, tissue barrier chips contain at least two compartments to provide “inside” and “outside” of the tissue. These compartments are separated by barrier cells such as endothelial or epithelial cells. The barrier cells have the apical and basolateral (basal) side forming polarization. In case of epithelial cell, the apical side faces the lumen meaning the external environment and the basal side faces the internal environment including the underlying connective tissue or blood vessels. For example, in the intestinal epithelium, the apical side of the epithelial cells faces the intestinal lumen and is involved in the absorption of nutrients and water from the digested food equipped with specialized structures, microvilli. In the blood–brain barrier, the apical side of brain endothelial cells faces the lumen of brain microvessel. The basal side faces the brain tissue, providing essential support and stability to brain structures. It's involved in interactions with astrocytes and other supporting cells, contributing to the intricate regulation of the brain microenvironment.21To support the barrier cell lining, tissue barrier chips utilize scaffolds represented by porous membrane and hydrogel (Fig. 1). The most popular design of tissue barrier chip is porous membrane-based tissue barrier chip which is first presented as an organ on a chip22 (Fig. 1A). In this configuration, a chip requires careful consideration of factors such as the membrane's material, porosity, pore size, and thickness. Porous membrane-based chips excel in creating intact two-layered cell structures, making them well-suited for applications in tissues like the lung,22 intestine,23 the blood–brain barrier (BBB),24 blood–cerebrospinal fluid barrier,25 blood–retinal barrier26 and glomerular filtration barriers.27 However, it's important to note that porous membrane chips have limitations in that they culture cells in a 2D format. Another limitation is resulted from material properties of the porous membrane. The commercially available track-etched porous membranes are commonly made of polyester or polycarbonate, which require extracellular matrix (ECM) protein coating for cell adhesion and have higher stiffness than in vivo basement membrane.28 Though a PDMS membrane provides tunable stiffness and facilitates stretch mediated mechanical stimulus to tissue barrier chips, the thickness of the PDMS membrane (∼50 μm) typically thicker than the track-etched membranes (∼10 μm).27,29 Although the porous membrane can be made from electrospun nanofiber or vitrified ECM membrane which have low stiffness, nanofibrous structure, and physiological relevant thickness, these techniques remain lab-scale research.28
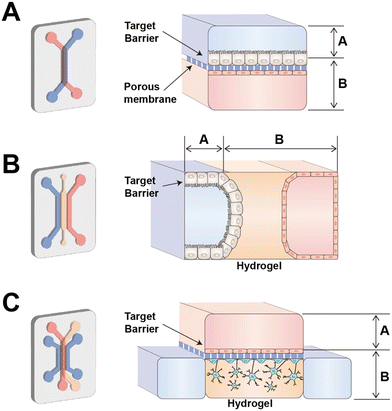 | ||
| Fig. 1 Design of compartments in a tissue barrier chip. (A) Porous membrane-based tissue barrier chip (B) hydrogel based tissue barrier chip (C) hybrid type of tissue barrier chip. | ||
Another popular design is hydrogel-based tissue barrier chip (Fig. 1B) which includes hydrogel channel between two culture media channels. Hydrogel can be pinned by surface tension by structural guidance such as channel height or micropost array. In case of hydrogel-based chip, intestinal and renal proximal tubule epithelial cells were seeded through medium channel and allowed to adhere to the hydrogel scaffold.30,31 When designing a hydrogel-based chip, the choice of hydrogel material is a crucial consideration, as it can impact factors like cell adhesion and migration.32 Hydrogel-based chips are known for their ability to create 3D structures effectively. Within the hydrogel, different cell types can be embedded, allowing for the creation of additional microenvironments.33 Moreover, they are well-suited for studies involving cell migration, angiogenesis, or immune cell transmigration.34 When migrated cells are distributed within the hydrogel, it simplifies imaging and quantification due to their well-defined locations. However, creating an intact two-layer structure such as structure of epithelial cell layer and endothelial cell layer can be challenging due to the thickness of the hydrogel channel.
Within these two categories of organ-on-a-chip configurations, researchers gain access to both the apical side channel, where the epithelial cells are seeded, and the basal side channel, which is situated opposite to the apical channel. For example, to assess drug absorption through the intestine, in a porous membrane-based chip, the drug can be injected into the apical side channel where intestine epithelial cells are seeded to mimic oral administration. The concentration of the drug that is transported through the opposite channel, can be measured.35 To evaluate drug metabolism in the liver, the drug can be injected into the basal side channel to mimic delivery of the drug in blood into the liver. The concentration of metabolic byproducts, which represents liver function and hepatotoxicity, by liver cells can be measured by collecting samples from the apical side channel.36 To simulate drug transport through BBB to brain, the drug can be introduced into the apical side channel where brain endothelial cells were seeded. The extent to which the drug has crossed over to the brain side can then be evaluated by collecting samples from the basal side channel.37
Depending on the context of use, tissue barrier chips need to be designed accordingly. Hybrid designs, combining porous membrane-based and hydrogel-based features, are also possible.38 In such cases, a hydrogel-based chip could be a lower part of the conventional porous membrane-based chip (Fig. 1C). For example, in the development of a BBB chip, a hybrid design was employed to establish contact between 3D-cultured astrocytes and an intact three-layer structure consisting of brain endothelial cells, a porous membrane, and pericytes. This approach was chosen as it allowed for a more physiological morphology and reduced expression of reactive astrocyte markers compared to 2D culture.39 The compartmentalized design of tissue barrier chips empowers functional analysis, highlighting their versatility and strength in various applications.
2.2 Fluid flow
The conventional in vitro human tissue barrier model is known as the Transwell system. Utilizing a porous membrane that separates the upper and lower chambers of Transwell system, cells are typically cultured on the upper side of the membrane, and the lower chamber contains the medium or other substances of interest. It enables polarized cultures of human epithelial and endothelial cells, distinguishing between their apical and basal sides. The tissue barrier models using the Transwell system are designed to study the selective permeability of these barriers, transport of molecules across them, and their responses to different conditions or treatments. However, tissue barrier chips offer a distinctive feature that cannot be achieved with Transwell systems, as they can mimic the fluid flow present in our bodies. In our body, tissue barriers exist in dynamic environments with fluid flow. For example, in the intestine, fluid flow, acting as a mechanical cue, leads to the formation of villi.40,41 Similarly, for endothelial cells, it is known that the expression of transporters, receptors, and other molecules differs under physiological conditions.42 The tissue barrier chip mimic the physiological flow by capturing physiological shear stress range. For example, the shear stress in venous blood vessels in BBB has been known to have range of 1–4 dyn cm−2, and many BBB modeling system have reportedly used this range, which is summarized by Yan et al.43 Also, the shear stress range in the intestinal barrier has been estimated as 0.002–0.4 dyn cm−2, which is recapitulated in various intestine-chip model.29,41,44 The shear stress in a tissue barrier chip can be obtained by:45where Q is the volume flow rate, μ is the absolute fluid viscosity, and W and h are the width and height of the rectangular channel, respectively. ΔP is the pressure difference between the inlet and the outlet of the channel. Thus, pressure controllers and syringe pumps can be easily adopted to capture physiological shear stress level in the microchannel.
Fluid flow in tissue barrier chips serves multiple purposes. It not only facilitates long-term culture by refreshing the medium and providing shear stress but also allows for the modeling of systemic vascular circulation from a pharmacokinetics/pharmacodynamics (PK/PD) perspective. In a static environment, drug processing occurs within limited geometries, relying solely on diffusion which implies limitations in drug transport, adsorption, and cellular responses. However, in a dynamic environment with fluid flow, drugs passing through the tissue barrier are continuously washed out on the opposite side, preventing the saturation of cellular responses.35 Consequently, research akin to animal models, where drugs are administered, and their plasma concentrations are continually monitored, becomes possible.46 Furthermore, when constructing multi-organ-on-a-chip systems, it is even feasible to mimic inter-organ tissue flow rates, advancing the precision of PK/PD studies.47
In tissue barrier chips, there are two types of fluid flow: open flow and closed flow (Fig. 2). Open flow refers to the continuous supply of fresh medium without recirculation, often achieved using an external pump, enabling in vivo-like metabolic agent sampling. During drug tests using the open flow, the fluid input channel carries the drug, and the output channel collects the solution that has passed through the tissue barrier, allowing for real-time measurement of drug permeability.48 Closed flow, on the other hand, involves re-circulation of the medium, typically driven by a peristaltic pump or a tilting instrument using gravity.49–51 Re-circulation is more cost-effective as it maintains a small-medium volume even under similar shear stress conditions. When employing a tilting instrument to establish gravity-driven closed flow, it eliminates the need for tubing and accessories, making high-throughput experiments more straightforward. However, the use of gravity-driven flow by tilting instrument introduces periodic bi-directional flow of which potential impact is not yet fully understood. For example, oscillating bi-directional flow can induce a disruption in endothelial barrier properties.52,53 While various methods have been developed to create uni-directional flow in gravity-driven systems, they have limitations in maintaining a continuous and consistent flow rate.52,54–56
2.3 Compatibility with analysis techniques
Another critical consideration in the design of tissue barrier chips is the compatibility with existing analytical methods. One of the most crucial aspects of tissue barrier functional analysis is permeability measurement. The method for measuring permeability is generally employed from the traditional Transwell system, where molecules applied to the apical side are collected from the basal side, and the concentration difference between the two chambers is used to calculate permeability.57 Therefore, for permeability measurements to be feasible, each compartment of the tissue barrier chip must have accessible ports for collecting solutions. This measurement method can be carried out using fluorescence-conjugated drugs and quantified using a plate reader. Alternatively, for drugs that cannot be conjugated with fluorescence, quantification can be achieved using a mass spectrometer.This method of solution collection has limitations in hydrogel-based chips because drugs and substances need to diffuse through a much thicker hydrogel compared to a porous membrane. Consequently, accurately representing the amount of drug that has passed through by measuring the concentration of the collected solution on the opposite side can be challenging in hydrogel-based chips. In this regard, porous membrane-based chips have a significant advantage. Their thin and intact two-layer cell structure allows for a relatively accurate representation of permeability by measuring the drug concentrations in the input and output channels.58 In hydrogel-based chips, image-based permeability measurement using fluorescence-conjugated molecules is sometimes employed, especially in models where a vascular network is formed within the hydrogel compartment.59 In such cases, selective access to the basal side of blood vessels is limited, so permeability is indirectly measured by observing the intensity profile of fluorescence molecules injected into the apical side as they diffuse through the hydrogel.
Trans-epithelial/endothelial electrical resistance (TEER) measurement is another option to consider when assessing tissue barrier properties. TEER is a method that measures electrical resistance through the paracellular route and is commonly used to assess tissue barrier integrity in the traditional Transwell system.60 To apply this measurement method to tissue barrier chips, various approaches have been developed, such as installing electrodes on the chip's surface61 or integrating electrodes into inlet/outlet ports.39
3. Current efforts for personalized medicine
Personalized medicine refers to a practice of medicine determining or guiding data obtain from an individual patient's cells for the prevention, diagnosis or treatment of disease.17 The concept aims to customize medical decisions, practices, interventions, and therapies to the individual patient, rather than using a “one-size-fits-all” approach. The goal is to maximize the therapeutic benefits while reducing the potential for adverse effects or unnecessary treatments.62 For realization of personalized medicine, it is indispensable to establish physiologically relevant, human-based, patient-based, and individual based pre-clinical model. Since MPSs, termed organ on a chip early, emerged in 2010,22 Neužil et al. first envisioned utilizing patient derived cells could be the groundwork for personalized medicine in 2012.63 In 2022, 10 years later, among many efforts to rapid advances in MPS, undoubtedly, recent progress in processing patient derived cells and stem cell technology has been offering unprecedented opportunities for personalized medicine.64 The advancement of stem cell technology has enabled the acquisition and expansion of patient-derived iPSCs with genetic characteristics of the patient.65 Furthermore, the development of organoid technology has allowed for the rapid and enduring acquisition of adult stem cells specific to certain organs.65,66 The progress in cancer organoid technology has facilitated the mass cultivation of patient tumor cells maintaining the characteristics of cancer stem cells, and the features of patient primary tumors.67,68While there have been various reviews on the prospects of personalized medicine using conventional organ-on-a-chip and MPS,17,69–72 an update is warranted to encompass the diverse array of endeavors driven by the rapid advancements in the field over the past five years. In this section, we aim to focus on recent progress of the tissue barrier chip utilizing patient derived cell and efforts towards clinical translation of tissue barrier chip (Fig. 3). Tissue barrier chip models can be established from a patient's cells, enabling the creation of personalized disease models. This allows researchers and clinicians to study how a specific individual's cells or tissues respond to different treatments, medications, or interventions.
3.1 Patient tumor derived cells
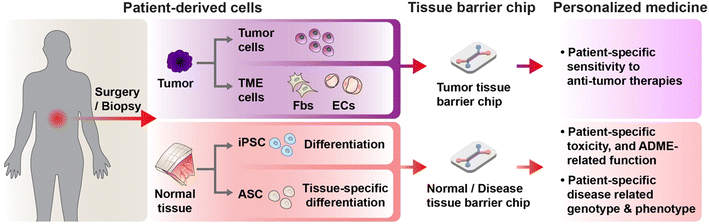
Patient tumor cells obtained from a surgery can be incorporated in the tissue barrier chip to mimic tissue specific tumor model. Leveraging tissue barrier chip, blood-tumor barrier model can be constructed by designing blood vessel compartment and locating tumor compartment outside of the blood vessel's lumen.73 Yi et al. constructed glioblastoma (GBM) with endothelial barrier on a chip by bioprinting.74 The patient-derived GBM cells embedded in decellularized brain tissue ECM were printed and surrounded by ring shaped blood vessel compartment (Fig. 4A). This structure allowed an oxygen gradient generating hypoxic core in the GBM compartment. Concurrent chemoradiation using temozolomide therapies were recapitulated in the chip using three types of patient-derived GBM cells, which exhibited low resistance, high resistance, and aggressive progression under concurrent chemoradiation therapy respectively. The GBM chips showed matched results with the actual clinical results after clinical-mimicking therapy. Furthermore, the GBM chips were utilized to suggest potential drug combination by investigating patient specific sensitivity from the aggressive progression group.Patient specific sensitivity to targeted therapy of tumor cells can be affected by tumor microenvironment (TME). Kim et al. captured how patient-derived brain metastatic non-small cell lung carcinoma cells acquire brain TME-mediated resistance to targeted therapies.75 The brain TME was designed in microchannels by constructing brain endothelial cell-lined channel, the patient-derived tumor cell compartment, and astrocytes-embedded hydrogel compartment separating them (Fig. 4B). The FDA approved drugs for targeted therapy which were predicted from a next generation sequencing were treated to the chip, notably, some of these drugs exhibited limited or even no efficacy under existence of the brain TME. These drug resistance acquisitions were not replicated in the equivalent Transwell co-culture model and were suggested to be mediated by local cytokine accumulation in the chip. The disparity from the Transwell results can be attributed to micro-scale compartments which generate chemical gradients between the compartments affecting cellular interaction mediated by cytokines. Fibroblasts are representative stromal cells in TME.76 Tumor cell mediated fibroblast activation leading to generation of cancer associated fibroblast has been reportedly reconstituted in a microchip.77 Cancer stem cells can reportedly maintain the stemness in a microchip.75,78 Thus, patient derived tumor organoid with TME can be constructed in tissue barrier chip format. Lai et al. constructed endothelial cell lined scaffold, which has nano and micro-pores enable to molecular transport, penetrating patient derived pancreatic tumor organoid compartment14 (Fig. 4C). The patient tumor organoid mixed with human fibroblasts formed densely packed tumor tissues embracing the blood vessel compartment. The co-culture tumor organoid exhibited features of fibrosis such as collagen deposition and increased tissue stiffness by the activated fibroblasts, and as well dramatic increase of resistance to gemcitabine (FDA approved chemotherapeutic drug) when it was injected into the blood vessel lumen compartment.
Not only patient-derived tumor cells but also patient-derived TME cells can be targets for personalized medicine.79–81 A tissue barrier chip can be harness in mechanistic study for TME cells' interaction with normal cells. Lugo-Cintrón et al. studied inter-patient's heterogeneity of the head and neck tumor derived TME fibroblasts and their interaction with a lymphatic vessel82 (Fig. 4D). The lymphatic vessel was constructed in tubular shape in a hydrogel embedding the TME fibroblasts. The TME fibroblasts were found to induce lymphangiogenesis, which reportedly is correlated with a tumor progression, and hyperpermeability. These lymph vessel conditioning of TME fibroblasts showed inter-patient differences in response to IGF-1 blockade. Targeting TME endothelial cells, which means anti-angiogenic treatment, can be considered in pursuing personalized medicine.83–85 Patient derived tumor cells were reported to show organotypic interaction with blood vessels in different organs in microchip.78 The blood vessels model with patient-specific tumor endothelial cells (tECs) and normal endothelial cells (nECs) were replicated86 (Fig. 4E). The tEC vessels displayed increased angiogenic sprouts and disrupted monolayer. Notably, as well inter-patient's heterogeneous responses were detected for five different patients under expose of FDA approved two anti-angiogenic agents, pazopanib and sunitinib.
While various patient-derived tumor cells and TME cells can be isolated and utilized in tissue barrier chips, this approach is primarily limited to personalized cancer medicine. To achieve a broader definition of personalized medicine, which encompasses normal and diseased tissues beyond tumors, the utilization of patient-derived stem cells is necessary.
3.2 Patient derived stem cells
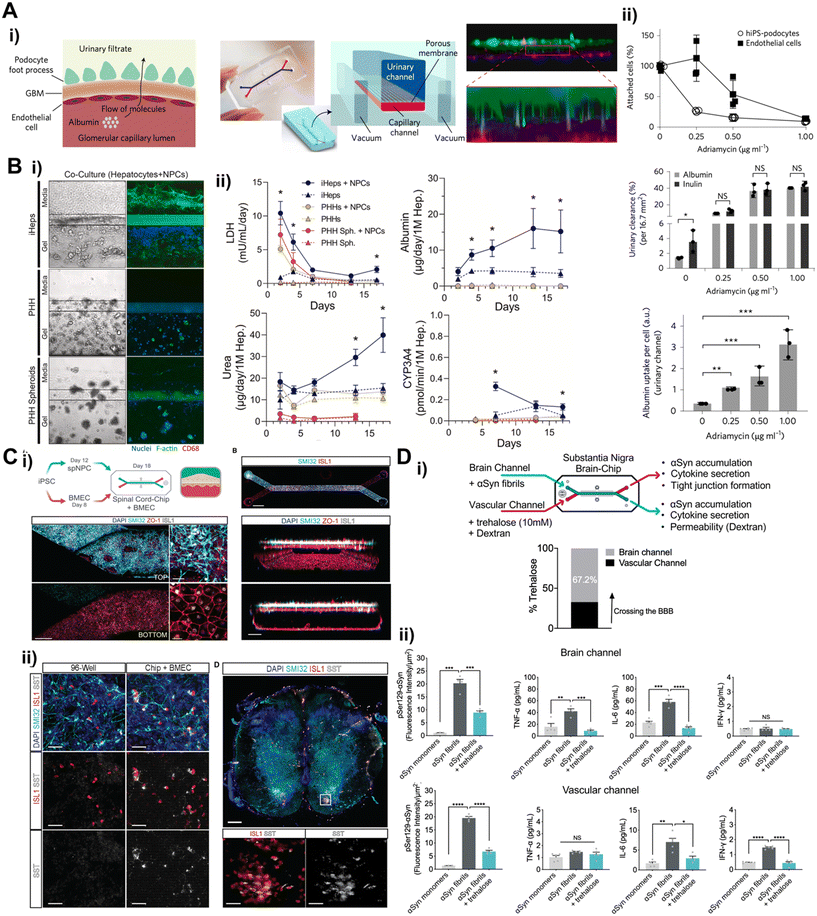
Recent breakthroughs in molecular and nanotechnology, especially the capability to create patient-specific iPSC lines, differentiate these cells into various cell types, and incorporate them within microfluidic chips, have paved the way for the development of tissue barrier chip that faithfully replicate the intricate functions of organs.87 The iPSCs and ASCs can be utilized to generate a variety of cell types while maintaining the genetic background of a single donor, thereby facilitating the establishment of an isogenic ex vivo model. Therefore, the utilization of patient-derived stem cells in tissue barrier chips is essential for advancing towards personalized medicine.The use of iPSCs derived from donors with genetic disorders for large-scale drug screening purposes has already demonstrated promising performance in pre-screening.88 This suggests that incorporating iPSCs into tissue barrier chips holds even greater potential for creating more advanced disease models. Notably, iPSCs offer robust cell expansion for organ research where obtaining primary cells is challenging, such as in the case of the kidney glomerular filtration barrier, liver, and central nervous system (CNS). A kidney glomerular filtration barrier, first place filtering blood into urine, has specialized epithelial cells, podocytes, which are terminally differentiated and no self-renewal cells.89 iPSCs could potentially serve as an unlimited source of podocytes. Musah et al. established method to differentiate iPSC-derived podocytes and utilized the podocytes to model kidney glomerular filtration barrier27 (Fig. 5A). The urinary channel (the iPSC-derived podocyte compartment) and blood channel (glomerular endothelial cell compartment) were separated by porous membrane. The FDA approved cancer drug adriamycin was treated continuously into the blood channel mimicking intravenous administration to test glomerular toxicity. Notably, significant podocyte delamination was observed at the clinically relevant dose. iPSC derived glomerular endothelial cells were also established recently facilitating isogenic glomerular filtration barrier model.90 Hepatotoxicity has significant importance in personalized medicine due to its impact on drug safety and efficacy.91 iPSC-derived hepatocytes (iHeps) were defined92,93 and employed to construct hepatotoxicity model94 where hepatocyte-embedded hydrogel was located adjacent to vascular channel lined with non-parenchymal cells (endothelial cells and monocytes) (Fig. 5B). The iHeps co-cultured with non-parenchymal cells exhibited more robust properties than primary human hepatocyte in terms of prolonged cell viability and stable hepatic function such as albumin/urea secretion, CYP3A4 activity and drug metabolism. The cells in CNS including many types of neurons, needless to say, can be hardly isolated and expanded from a surgery or biopsy. It highlights iPSCs unique solution to obtain human CNS cells. A spinal cord on a chip was constructed by co-culturing iPSC-derived spinal neural progenitor cells (spNPC) with iPSC-derived brain microvascular endothelial cells (iBMEC)95 (Fig. 5C). The iPSC-derived spNPC were successfully differentiated into mature and active spinal cord motor neuron showing spontaneous neuronal physiological function in live calcium transient imaging, which gives promise for modeling blood–spinal cord barrier.
The most emerging field is modeling blood–brain barrier (BBB). Recent breakthrough in simple and rapid differentiation protocol to acquire iBMECs96–98 facilitates mechanistic in vitro studies for drug delivery targeting CNS.24,37,39 The iBMEC was seamlessly applied into BBB on a chip platform.24,37,99–101 To mimic BBB structure, the iBMECs were lined with the underneath of porous membrane in the lower channel, and pericytes and astrocytes, which wrap around blood capillary to form BBB in our body, were cultured on the porous membrane in the upper channel.101 In this model, iBMEC exhibited shear stress responsive BMEC like gene expressions and cellular interactions which led to physiological relevant barrier properties. These barrier properties were more enhanced in developmentally inspired differentiation under hypoxic condition.24
The utilization of the iBMEC has been propagating into disease modeling. Parkinson's disease pathology was recapitulated in a BBB chip containing iBMEC in blood channel99 (Fig. 5D). To mimic synucleinopathy, alpha synuclein pre-formed fibrils were introduced into brain channel containing pericytes, astrocytes, microglia and iPSC-derived dopaminergic neuron. The alpha synuclein fibrils accumulation resulted in microglia activation, astrogliosis, and neuronal loss. The FDA approved drug, trehalose was administered into the blood channel promoted clearance of alpha synuclein displaying about 60% penetration rate. Neuroinflammation has been reportedly a major pathogenetic mechanism underlying neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, multiple sclerosis, and amyotrophic lateral sclerosis.102 The early phases of neuroinflammation were simulated by perfusing TNF-α into either the blood channel lined with iBMEC or the brain channel containing astrocytes, pericytes, microglia, and iPSC-derived neurons.100 This BBB containing brain chip displayed similar transcriptomic profile to the adult cortical tissue than the Transwell model composed with same cells. The key clinical features of neuroinflammation were recapitulated such as BBB disruption, astrocyte and microglial activation, and proinflammatory cytokine secretion.
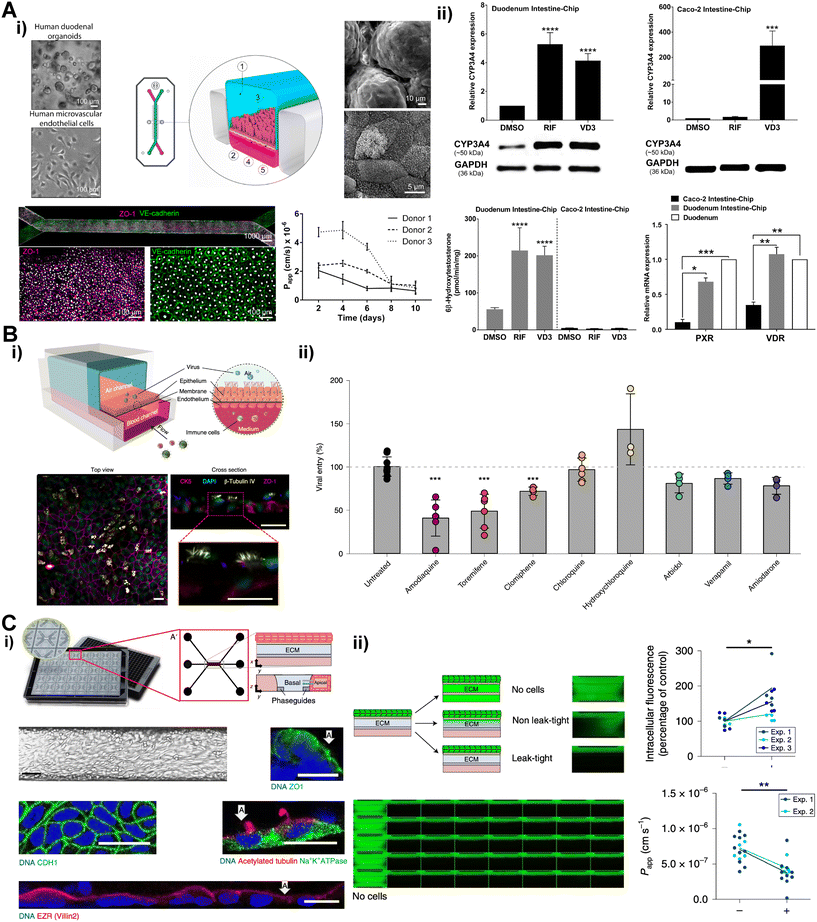
ASCs are facilitating robust expansion of self-renewal organ specific cells such as intestine, lung, liver and kidney tubules. The ASCs from these organs can be easily obtained from a surgery and expanded by organoid culture methods.103–106 The ASC-derived organoids contain organ specific epithelial cells and can be dissociated into single cells and can be seeded into microfluidic chip29,103,107,108 (Fig. 6). Using this technique, biopsy-derived small intestine organoid cells were utilized in constructing small intestine chip.29 The organoid derived intestine cells and intestine microvascular endothelial cells were lined along with each side of porous membrane separating upper and lower channel (Fig. 6A). The intestine epithelium constructed in the chip exhibited villi-like 3D projection and multi-lineage differentiation showed in the organoid culture. In this intestine chip, altered profiles of drug metabolizing enzymes and drug transporters under exposes of two FDA approved drugs, calcitriol and rifampicin, were investigated to identify early risk for drug–drug interaction.109 The alveolus epithelial cells were isolated, expanded in the organoid culture and mono-cultured in the previous lung on a chip.108 This organoid-derived alveolus epithelium chip displayed strong expressions of alveolar type 2 cells markers and prosurfactant proteins, and lower expression of epithelial–mesenchymal transition marker than Transwell model. Human lung bronchial-airway epithelial basal stem cells were also utilized to construct the lung on a chip.110 The lung chips infected with pseudotyped severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were treated with clinically relevant doses of various FDA approved drugs (Fig. 6B). Notably, the anti-malarial drug amodiaquine was found to have inhibitory effects in the entry of the SARS-CoV-2 than other anti-viral drugs. Kidney tubule cells were also successfully expanded over 20 passages in organoid culture and leveraged to construct functional kidney tubule model.103,107 The organoid-derived tubule cells lined in a chip along a hydrogel which separates medium channels. These cells exhibited polarized characteristics of kidney tubules including microvilli formation, tight junction expression, active efflux function of p-glycoprotein 1 (Fig. 6C).
In addition, these stem cells also provide more robust barrier integrity than primary cells and cell lines. For example, the immortalized hCMEC/d3 cell, which is known as human brain endothelial cell line, has typical TEER values of 30–50 Ω cm2in vitro,111 on the other hand, iBMEC provides more well-suited barrier integrity with much higher TEER values as high as estimated in vivo TEER values112 (∼8000 Ω cm2) or even higher. Also, intestine organoids derived epithelial cells showed higher TEER value (1008 Ω cm2) than TEER value (604 Ω cm2) of Caco-2 cells, which are a standard cell line utilized in many intestine modeling studies.113
4. Challenges and future direction
4.1 Inter-donor variation
For realization of personalized medicine using tissue barrier chip, the personalized tissue barrier chip should capture inter-donor variability. The substantial inter-donor variability was confirmed in liver chips using commercial primary hepatocytes from five different donors.114 Metabolic depletion profiles of six drugs (phenacetin, diclofenac, lidocaine, ibuprofen, propranolol, and prednisolone) exhibited substantial inter-donor variation in gene expression levels, drug metabolism, and other hepatocyte functions. The intestine chip constructed from three healthy donors displayed comparable levels in permeabilities however it showed inter-donor variation in sucrase activity and mucin secretion.29These progress in tissue chip performance facilitates to distinguish disease patients from healthy donors. The organoid-derived intestine cells from healthy donors and environmental enteric dysfunction (EED), a chronic inflammation intestine disease, patients were employed in intestine on a chip model.44 A nutrient-deficient medium was applied to mimic EED-associated intestinal injury. In the nutrient-deficient EED model, key features of transcriptome signature of EED patients were recapitulated. The functional characteristics of EED were also reconstituted in the chip including villus blunting, compromised permeability, and lower absorption of metabolites in EED patient intestine chip. These results demonstrate that ASC can maintain patient's pathological characteristics in the tissue barrier chip's disease mimicking environment even in case of non-genetic disease.
Bone marrow is sensitive tissue to drug and radiation related toxicities.115,116 By using hematopoietic stem cells and bone marrow stromal cells isolated from different donors, bone marrow tissue was reconstructed in the opposite channel to the blood vessel channel where endothelial cells attaching to porous membrane.48 The bone marrow chip displayed predicted hepatotoxicity to 5-fluorouracil at clinically relevant concentration where conventional in vitro cultures didn't. The inter-donor variation in the toxicity was consistent between healthy donors. Notably, the bone marrow chip demonstrated its potential in mimicking the phase I studies using AZD2811, small molecule inhibitor in phase II clinical development. To mimic intravenous injection of the phase I studies, clinically relevant dose of AZD2811 was infused through the blood channel over 2 or 48 h. The results showed similar concentration profiles with expectation from patient plasma sample. The bone marrow chip was also utilized cells from patient with genetic disorder, Shwachman-Diamond syndrome (SDS), a genetic BM failure syndrome. Although this study didn't reach to a functional analysis of the bone marrow tissue model. The bone marrow chip from SDS patients was revealed to have defects in hematopoiesis and cell-type specific abnormalities meaning aberrantly blunted maturation similar with their in vivo phenotype.
Thus, leveraging patient derived stem cells in tissue barrier chip enabled ex vivo investigation of genetic disorders. Using iPSC techniques, inter-donor variations in the BBB function were investigated. The variations between healthy donors in permeability were statistically non-significant and in the transcriptomic signature distance were significantly closer to human cerebral cortex.100 However, this model still included primary brain cells such as astrocytes, pericytes, and microglia.
Personalized isogenic iPSC derived BBB chips were constructed by co-culturing iPSC-derived neural progenitor cells from three healthy donors and a Huntington's disease (HD) patient, instead of human primary pericytes and astrocytes.101 As results, the healthy individuals had a consistent permeability meaning insignificant variations, but HD patient exhibited significant increase in the permeability. Notably, this study utilized CRISPR/Cas9 technique to compare BBB models from healthy donor derived iPSCs having CRISPR/Cas9 mediated MCT8 mutation, MCT8 mutation patient derived iPSCs, and patient derived iPSCs in which MCT8 mutation corrected using CRISPR/Cas9. Both MCT8 deficient BBB chips (patient derived and mutation induced) showed disrupted transport of thyroid hormone and the mutation corrected BBB chips exhibited the comparable level of the thyroid hormone transport with BBB chips from healthy donors. These results indicate that tissue barrier chip can precisely discrete the barrier functions constructed from patients among healthy donors.
4.2 Clinically relevant workflow
Since the FDA Modernization Act 2.0, tissue barrier chips have only recently been utilized in the context of drug development. Currently, there is a lack of specific regulations governing their use. To advance towards personalized medicine, it's imperative to establish a framework for employing tissue barrier chips throughout the entire drug evaluation process. According to guidelines proposed by the Innovation and Quality (IQ) Consortium in 2020, several pre-existing pharmaceutical and regulatory practices can be applied, including: i) Good Laboratory Practice (GLP) regulations, ii) International Consortium on Harmonization (ICH) Q2 (R1) guidelines, which emphasize reproducibility even if results differ from traditional validation parameters, iii) instrument qualification, operational qualification, and performance qualification, and iv) good cell culture practice.117In line with these efforts, FDA researchers in 2017 to explore the potential of liver chips in screening for liver toxins. They confirmed that diglycolic acid (DGA), a known liver toxin, exhibited a similar trend in liver toxicity when tested on both the liver chip and conventional well plates.118 The liver chips were also constructed from a rat, dog, and human.119 These multi-species liver chips showed species-specific hepatotoxicities. The animal liver-chips could provide valuable understanding the differences preclinical animal testing between human clinical results. More recently, in 2022, industrial researchers in this field validated the performance of their human liver chip for predicting drug-induced liver injury (DILI),120 aligning with previously discussed liver model guidelines.117,121 Impressively, they used 870 liver-chips to evaluate a blinded set of 27 known hepatotoxic and non-toxic drugs and achieved a sensitivity of 87% and a specificity of 100%.120 The authors devised a liver-chip DILI score based on i) the inhibition of albumin secretion, ii) an increase in alanine aminotransferase (ALT) levels, and iii) morphological scoring of hepatocytes.
In 2023, they also proposed two decision-support frameworks for DILI prediction using the liver-chip.122 The first involves mapping the DILI score to industry-standard Garside DILI severity categories,123 enabling quantitative assessment of severity. The second integrates the liver-chip DILI score with standard animal testing results as an additional screening criterion. The establishment of such standardization frameworks will not only have significant implications for personalized medicine but will also set crucial benchmarks for evaluating its efficacy.
4.3 Standardization and automation
Standardization and automation of tissue barrier chips can lead to the production of high-throughput and replicable data. Experiments involving tissue barrier chips often demand skilled researchers to minimize inter-experiment variation. In this regard, the automation of tissue barrier chip formation is proposed as an alternative to reduce such variation.The current efforts for standardization are primarily focused on aligning the external format with that of the existing standard well plates. This approach has been widely employed51,94,103,124 because it allows for the use of existing multi-pipettes, and the integration into existing automated liquid handling machine. For instance, many designs allocate one well each for the inlet and outlet in 96 or 384 well plates, with connecting channels designed around other well areas. Traditional membrane-based tissue barrier chips allocate inlets and outlets to individual wells in both upper and lower channels, connecting these through channels within the chip (a total of 4 wells per chip unit).125,126 In these designs, the upper channel was modified into open well, linking one well each for the inlet and outlet of the lower channel through a membrane positioned on the opened upper layer, making up a total of 3 wells per chip unit.127 Additionally, hydrogel-based tissue barrier chips have been developed with models incorporating 2 wells each for inlet and outlet of the hydrogel channel and the medium supplying channel, resulting in a 4-well model in 384 well plate format.94,124 There are also versions with two side medium channels, constituting a 6-well model51,128 or 9-well model.31,103
A high-throughput format of tissue barrier chip has been adopted for automated construction of tissue barrier model. In 2021, liver chips containing compartments with iPSC-derived hepatocytes and endothelial cell channels with monocytes were created using a standard liquid handling robot.124 These automated high-throughput liver tissue chips were validated using a well-known hepatotoxin, troglitazone, and were employed to evaluate a library of 159 hepatotoxic compounds. Furthermore, in 2023, the semi-automated high-throughput screening of the inhibitory effects of 1537 protein kinase inhibitors was demonstrated in an angiogenesis model.128 Using an automated liquid handler, they constructed over 4000 endothelial barrier models, exposing them to the respective kinase inhibitors. While reagent preparations and plate transfers were carried out manually, 96.5% (3987) of the 4130 chips passed the quality control of visual inspection. Image-based scoring using Python showed high replicability, with 97.4% of duplicates displaying the same or adjacent scores. This screening identified 53 anti-angiogenic and low-toxicity inhibitors, 44 of which were not previously associated with the angiogenesis pathway. Automation is not limited to liquid handling but also extends to high-content imaging. Automated workflows for liver-chip experiments, from capturing to analyzing morphology, proliferation, and apoptosis endpoints using automated confocal images129 and automated bench-top fluorescence microscopes,118 have been developed.
Automation systems can also be applied to complex multi-organ tissue barrier chips. For instance, a robotic liquid handler, automated culture, perfusion, medium addition, fluidic linking, sample collection, and in situ microscopy imaging were employed to connect eight tissue barrier chips (intestine, liver, kidney, heart, lung, skin, BBB, and brain) over a three-week period.130 This automated system was utilized to demonstrate physiological PK modeling including drug absorption, metabolism and excretion using computationally scaled data correction.47 Multiple fluidically connected tissue barrier chips were employed to predict PK parameters of two well-known drugs, nicotine and cisplatin. For oral administration model, nicotine was infused into the epithelial channel of gut chip and the vascular outflow was collected and transferred the vascular channels of liver chip and kidney chip by robotic liquid handler. For intravenous administration model, cisplatin was infused into the vascular unit chip and transferred into the vascular channel of bone marrow chip, liver chip, and kidney chip. To mimic clinical PK/PD modeling, this system employed partially closed flow, meaning that a portion of the total medium was recirculated, some was collected, and some fresh medium was supplied. This human-relevant, fluidically connected multi-chip PK/PD modeling system facilitated in vitro–in vivo translation to predict human PK parameters using tissue barrier chips.
Although progress is being made towards standardizing the exterior format to align with the standard well plate format, the standardization of the interior format has not yet been achieved. There are many instances of developing multiple tissue barrier models on the same format of chip.22–25,27,48 However, the discussion on standardizing internal parameters —such as 1) the size of the culture area, 2) channel width and height directly related with the volume-to-area ratio (VAR) 3) shear stress, and 4) total volume of culture medium—is still lacking. These parameters primarily affect molecular transport mechanisms such as diffusion profiles, which include nutrient supply, paracrine or autocrine signaling, and drug transport. For example, in channels with VARs, nutrient supply primarily depends on the perfusion of culture medium. However, the physiological level of shear stress limits the use of high flow rates because shear stress is proportional to the square of the flow rate. Paracrine or autocrine signals can be transported by diffusion easily in low VARs forming gradient profiles, but harder in high VARs. For drug efficacy or toxicity evaluation, high VAR channels can provide more sensitive outcomes as the drug can be transported easily by diffusion. Total volume of the culture medium is also important in the molecular transport. Using large amount of medium means large quantity of test molecules in same concentration. For example, in high VAR channels, for physiological relevant shear stress, large or small volume of medium can be re-circulated. Although they have same concentration of target testing drugs, the dose in terms of quantity varies proportionally with the volume. Thus, these internal parameters significantly influence the determination of the culture period and testing time windows, as cells cultured in tissue barrier chips typically undergo stages of barrier formation and maturation, maintaining the barrier properties for a specific period before their function deteriorates.
Another challenge of the current tissue barrier chips is lack of quantitative functional assessment standard. The purpose of the human tissue barrier chip is to create a physiologically relevant tissue barrier model, facilitating more human-relevant disease modeling and drug toxicity assessments. However, the direct assessment of specific tissue barrier function in a human body remains limited due to the lack of quantitative and non-invasive methods, making accurate evaluations of how well these chips reflect human barrier functions challenging. However, the value of human tissue barrier chips is rather emphasized due to this inaccessibility of functional assessment; to evaluate responses of the tissue barrier chip to estimate responses of human tissues. Comparative studies between tissue barrier chips and rodent models exist. For example, Pediaditakis et al.100 demonstrated that the permeability values of their BBB model to 3–70 kDa dextran have similar values with the rat in vivo permeability routinely cited as golden standards131,132 also showed the permeability values of their BBB model to 4–70 kDa dextran have good correlation (R2 = 0.96) with the rat in vivo permeability.132 Musah et al.27 demonstrated that their GFB model has in vivo comparable albumin and inulin clearances (permeability presented as a percent). However, as shown in Table 1, the recent studies of tissue barrier chip utilizing patient derived cells tend to overlook the barrier function assessment, which is essential for quality control, physiological relevance, and future standardization. Although, recent research has indicated that tissue barrier chips can yield results closer to clinical outcomes than mouse models in drug toxicity as mentioned in section 4.2, such as achieving a sensitivity of 87% and a specificity of 100%,120 the ability to perform standardized functional assessment by using personalized cell sources such as iPSC or ASC to form a tissue barrier model that reflects the phenotypical features of each individual is expected as a major advantage of the personalized tissue barrier chip in the future.
| Author | Barrier type | Design | Barrier integrity assessment | Cell sources | Target function and diseases |
|---|---|---|---|---|---|
| GBM: glioblastoma, HUVEC: human umbilical vein endothelial cells, HLEC: human lymphatic endothelial cells, GFB: glomerular filtration barrier, iPSC: induced pluripotent stem cell, BBB: blood–brain barrier HD: Huntington's disease SDS: Shwachman–Diamond syndrome. | |||||
| Yi et al.74 | Tumor blood barrier | Hydrogel based | — | Patient derived GBM, and HUVEC | Tumor drug resistance |
| Kim et al.75 | Tumor blood barrier | Hydrogel based | — | Patient derived brain metastatic lung cancer, human astrocytes, and hCMEC/D3 | Tumor drug resistance |
| Lai et al.14 | Tumor blood barrier | Hydrogel based | Fluorescent dye perfusion – quantified by signal area | Patient derived pancreatic tumor organoid, and HUVEC | Tumor drug resistance |
| Lugo-Cintrón et al.82 | Tumor lymph barrier | Hydrogel based | Permeability–fluorescence conjugated 70 kDa dextran – 1–2 × 10−5 cm s−1 | Patient derived fibroblast in head and neck tumor, and HLECs | Tumor lymphangiogenesis |
| Jiménez-Torres, J. A. et al.86 | Tumor blood barrier | Hydrogel based | — | Patient derived renal cell carcinoma endothelial cells | Tumor angiogenesis |
| Musah et al.27 | Kidney (GFB) | Membrane based | Permeability (named urinary clearance)–fluorescence conjugated inulin and albumin | iPSC derived podocyte and primary glomerular endothelial cells | Drug toxicity |
| in vivo correlation | |||||
| Roye et al.90 | Kidney (GFB) | Membrane based | Permeability (named urinary clearance)–fluorescence conjugated inulin and albumin | iPSC derived podocyte and iPSC derived endothelial cells | Drug toxicity |
| Kato et al.94 | Liver | Hydrogel based | — | iPSC derived hepatocyte, HMEC-1, and THP-1 | Drug metabolism |
| Sances et al.95 | Spinal cord | Membrane based | — | iPSC derived spinal motor neuron and iPSC derived brain microvascular endothelial cell | Vascular–neural interaction |
| Park et al.24 | BBB | Membrane based | TEER, permeability–fluorescence conjugated 3, 10, and 70 kDa dextran – 10−9 (3 kDa)–10−7 (70 kDa) cm s−1 | iPSC derived brain microvascular endothelial cell, primary astrocytes, and pericytes | Therapeutic antibody transport evaluation |
| Choi et al.37 | BBB | Membrane based | Permeability–fluorescence conjugated 10 kDa dextran – 2.5 × 10−8 cm s−1 | iPSC derived brain microvascular endothelial cell, primary astrocytes, and pericytes | Aptamers transport screening |
| Pediaditakis et al.99 | BBB | Membrane based | Permeability–fluorescence conjugated dextran with Cascade blue (3 kDa) – 1–3 × 10−6 cm s−1, and Lucifer yellow (0.5 kDa) – 4–6 × 10−6 cm s−1 | iPSC derived dopaminergic neurons, primary human brain microglia, astrocytes, pericytes, and iPSC derived brain microvascular endothelial cell | BBB in Parkinson's disease |
| Pediaditakis et al.100 | BBB | Membrane based | Permeability–fluorescence conjugated dextran with Cascade blue (3 kDa) – 0.5–1 × 10−6 cm s−1, and 10, 40, 70 kDa dextran (values not available) | iPSC-derived glutamatergic neurons, iPSC-derived GABAergic neurons, primary astrocytes, pericytes, microglia, and iPSC derived brain microvascular endothelial cell | BBB in neuroinflammation |
| in vivo correlation | |||||
| Vatine et al.101 | BBB | Membrane based | Permeability–fluorescence conjugated 3 kDa dextran (1 × 10−7 cm s−1) | iPSC derived brain microvascular cell, iPSC derived neuron from (normal donors or HD patients), primary astrocytes, and pericytes | BBB in Huntington's disease |
| in vivo correlation | |||||
| Gijzen et al.103 | Kidney (tubule) | Hydrogel based | Permeability – image only | Normal kidney tissue ASC derived tubule cells | Tissue barrier formation from kidney ASC |
| Schutgens et al.107 | Kidney (tubule) | Hydrogel based | Permeability – rhodamine 123–2.5 × 10−7–10−6 cm s−1 | Normal kidney tissue derived ASC | Tissue barrier formation from kidney ASC |
| Kasendra et al.29 | Intestine | Membrane based | Permeability – 450 Da Lucifer yellow – 0.5–2 × 10−6 cm s−1 | Normal intestine tissue derived ASC | Tissue barrier formation from intestine ASC, inter-donor variation |
| Kasendra et al.109 | Intestine | Membrane based | Permeability–fluorescence conjugated 3 kDa dextran – 1–5 × 10−6 cm s−1 | Normal intestine tissue derived ASC | Drug–drug interaction in intestine barrier |
| Van Riet et al.108 | Lung | Membrane based | — | Normal lung tissue derived ASC | Lung epithelial-to-mesenchymal transition |
| Si et al.110 | Lung | Membrane based | Permeability – 607 Da Cascade blue – 3–8 × 10−6 cm s−1 | Primary human lung bronchial-airway epithelial basal stem cells and primary pulmonary microvascular endothelial cell | Virus infection |
| Tsamandouras et al.114 | Liver | Membrane based | — | Primary hepatocyte from five different donors | Drug metabolism |
| Bein et al.44 | Small intestine | Membrane based | Permeability – 596 Da Cascade blue (3.82 × 10−7 cm s−1 from healthy donor) | Organoids derived from biopsy of healthy donors or patients | Environmental enteric dysfunction |
| Chou et al.48 | Bone marrow | Membrane based | — | Peripheral blood and leukapheresis product from donors, normal bone marrow stromal tissue from donors, bone marrow mononuclear cells from SDS patients or unaffected donors and HUVEC | Shwachman-Diamond syndrome |
| Eckstrum et al.118 | Liver | Membrane based | — | Primary human hepatocytes from a single donor, and primary liver sinusoidal endothelial cells | Drug metabolism |
| Ewart et al.120 | Liver | Membrane based | — | Primary hepatocytes from three different human donors, primary liver sinusoidal endothelial cells, Kupffer cells, and Stellate cells | Drug metabolism |
| Bircsak et al.124 | Liver | Hydrogel based | — | iPSC derived hepatocyte, HMEC-1, and THP-1 | Drug toxicity and metabolism |
5. Conclusion
The development of the tissue barrier chip appears to be mature, thanks to recent remarkable advancements. However, it is important to note that there isn't a ‘one-size-fits-all’ solution when it comes to tissue barrier chips. To move forward, it is crucial to define the specific contexts of use and design tissue barrier chips accordingly. This means that we can expect ongoing developments and emergence of new designs of tissue barrier chips. To establish these, a fundamental standardization process should precede. In other words, the development of guidelines for assessing drug toxicity and efficacy using tissue barrier chips is imperative to transform them into effective tools for drug evaluation. Within these well-established tissue barrier chips, personalized models using patient-derived tumor cells or patient-derived stem cells should be constructed. Tissue barrier chip developers need to work on increasing throughput and reproducibility, stem cell differentiation techniques should be improved for more relevant differentiation. Regulatory agencies and government authorities should closely monitor advancements in tissue barrier chips, collaborating with researchers to establish suitable workflows, frameworks, and guidelines. This effort would be pivotal in establishing the best practice framework of tissue barrier chips for personalized medicine.Author contributions
J. K.: conceptualization, visualization, writing – original draft, T. Y.: writing – original draft, S. L.: writing – review & editing, P. J. K.: writing – review & editing, Y. K.: conceptualization, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
The authors declare the following competing financial interests: in compliance with the institutional guidelines of the Georgia Institute of Technology, Y. K. discloses his financial interest in Mepsgen Co., Ltd. and Mepsgenus Inc., two biotechnology companies developing microengineered physiological systems and biomimetic nanoparticles for medical applications. The other authors declare no competing interests.Acknowledgements
This work was supported by Mepsgen Co., Ltd. through the research contract (AWD-004440 and #GR00019992) between Mepsgen Co., Ltd. and the Georgia Tech Research Corporation (GTRC).References
- F. Yu, N. D. Selva Kumar, D. Choudhury, L. C. Foo and S. H. Ng, Microfluidic platforms for modeling biological barriers in the circulatory system, Drug Discovery Today, 2018, 23(4), 815–829 CrossRef CAS.
- J. Bicker, G. Alves, A. Falcão and A. Fortuna, Timing in drug absorption and disposition: The past, present, and future of chronopharmacokinetics, Br. J. Pharmacol., 2020, 177(10), 2215–2239 CrossRef CAS.
- S. Chillistone and J. G. Hardman, Factors affecting drug absorption and distribution, Anaesthesia & Intensive Care Medicine., 2017, 18(7), 335–339 Search PubMed.
- A. K. Shenoy and J. Lu, Cancer cells remodel themselves and vasculature to overcome the endothelial barrier, Cancer Lett., 2016, 380(2), 534–544 CrossRef CAS.
- M. W. Dewhirst and T. W. Secomb, Transport of drugs from blood vessels to tumour tissue, Nat. Rev. Cancer, 2017, 17(12), 738–750 CrossRef CAS.
- P. A. Billat, E. Roger, S. Faure and F. Lagarce, Models for drug absorption from the small intestine: where are we and where are we going?, Drug Discovery Today, 2017, 22(5), 761–775 CrossRef CAS.
- M. Sántha, Biologia futura: animal testing in drug development—the past, the present and the future, Biol. Futura, 2020, 71(4), 443–452 CrossRef.
- F. Sewell, J. Edwards, H. Prior and S. Robinson, Opportunities to Apply the 3Rs in Safety Assessment Programs, ILAR J., 2016, 57(2), 234–245 CrossRef CAS.
- A. Gough, A. Soto-Gutierrez, L. Vernetti, M. R. Ebrahimkhani, A. M. Stern and D. L. Taylor, Human biomimetic liver microphysiology systems in drug development and precision medicine, Nat. Rev. Gastroenterol. Hepatol., 2021, 18(4), 252–268 CrossRef.
- J. J. Han, FDA Modernization Act 2.0 allows for alternatives to animal testing, Artif. Organs, 2023, 47(3), 449–450 CrossRef.
- J. M. Ayuso, M. Virumbrales-Muñoz, J. M. Lang and D. J. Beebe, A role for microfluidic systems in precision medicine, Nat. Commun., 2022, 13(1), 3086 CrossRef CAS.
- F. Fanizza, M. Campanile, G. Forloni, C. Giordano and D. Albani, Induced pluripotent stem cell-based organ-on-a-chip as personalized drug screening tools: A focus on neurodegenerative disorders, J. Tissue Eng., 2022, 13, 204173142210953 CrossRef.
- G. C. Napoli, W. D. Figg and C. H. Chau, Functional Drug Screening in the Era of Precision Medicine, Front. Med., 2022, 9, 912641 CrossRef.
- B. F. L. Lai, R. X. Z. Lu, Y. Hu, L. Davenport Huyer, W. Dou and E. Y. Wang, et al., Recapitulating Pancreatic Tumor Microenvironment through Synergistic Use of Patient Organoids and Organ-on-a-Chip Vasculature, Adv. Funct. Mater., 2020, 30(48), 2000545 CrossRef CAS.
- T. Takebe, B. Zhang and M. Radisic, Synergistic Engineering: Organoids Meet Organs-on-a-Chip, Cell Stem Cell, 2017, 21(3), 297–300 CrossRef CAS.
- J. Lee, W. H. Van Der Valk, S. A. Serdy, C. Deakin, J. Kim and A. P. Le, et al., Generation and characterization of hair-bearing skin organoids from human pluripotent stem cells, Nat. Protoc., 2022, 17(5), 1266–1305 CrossRef CAS.
- D. E. Ingber, Human organs-on-chips for disease modelling, drug development and personalized medicine, Nat. Rev. Genet., 2022, 23(8), 467–491 CrossRef CAS.
- B. Cui and S. W. Cho, Blood-brain barrier-on-a-chip for brain disease modeling and drug testing, BMB Rep., 2022, 55(5), 213–219 CrossRef CAS.
- S. Fowler, W. L. K. Chen, D. B. Duignan, A. Gupta, N. Hariparsad and J. R. Kenny, et al., Microphysiological systems for ADME-related applications: current status and recommendations for system development and characterization, Lab Chip, 2020, 20(3), 446–467 RSC.
- N. C. Peterson, P. K. Mahalingaiah, A. Fullerton and M. Di Piazza, Application of microphysiological systems in biopharmaceutical research and development, Lab Chip, 2020, 20(4), 697–708 RSC.
- N. J. Abbott, L. Rönnbäck and E. Hansson, Astrocyte–endothelial interactions at the blood–brain barrier, Nat. Rev. Neurosci., 2006, 7(1), 41–53 CrossRef CAS.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Reconstituting Organ-Level Lung Functions on a Chip, Science, 2010, 328(5986), 1662–1668 CrossRef CAS.
- W. Shin and H. J. Kim, 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert, Nat. Protoc., 2022, 17(3), 910–939 CrossRef CAS.
- T. E. Park, N. Mustafaoglu, A. Herland, R. Hasselkus, R. Mannix and E. A. FitzGerald, et al., Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies, Nat. Commun., 2019, 10(1), 2621 CrossRef.
- Y. Zhou, H. Qiao, F. Xu, W. Zhao, J. Wang and L. Gu, et al., Bioengineering of a human physiologically relevant microfluidic blood–cerebrospinal fluid barrier model, Lab Chip, 2023, 23(13), 3002–3015 RSC.
- H. Ragelle, A. Goncalves, S. Kustermann, D. A. Antonetti and A. Jayagopal, Organ-On-A-Chip Technologies for Advanced Blood–Retinal Barrier Models, J. Ocul. Pharmacol. Ther., 2020, 36(1), 30–41 CrossRef CAS.
- S. Musah, A. Mammoto, T. C. Ferrante, S. S. F. Jeanty, M. Hirano-Kobayashi and T. Mammoto, et al., Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip, Nat. Biomed. Eng., 2017, 1(5), 0069 CrossRef CAS.
- J. Youn and D. S. Kim, Engineering porous membranes mimicking in vivo basement membrane for organ-on-chips applications, Biomicrofluidics, 2022, 16(5), 051301 CrossRef CAS.
- M. Kasendra, A. Tovaglieri, A. Sontheimer-Phelps, S. Jalili-Firoozinezhad, A. Bein and A. Chalkiadaki, et al., Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids, Sci. Rep., 2018, 8(1), 2871 CrossRef.
- A. Nicolas, F. Schavemaker, K. Kosim, D. Kurek, M. Haarmans and M. Bulst, et al., High throughput transepithelial electrical resistance (TEER) measurements on perfused membrane-free epithelia, Lab Chip, 2021, 21(9), 1676–1685 RSC.
- F. Pöschl, T. Höher, S. Pirklbauer, H. Wolinski, L. Lienhart and M. Ressler, et al., Dose and route dependent effects of the mycotoxin deoxynivalenol in a 3D gut-on-a-chip model with flow, Toxicol. In Vitro, 2023, 88, 105563 CrossRef.
- S. R. Caliari and J. A. Burdick, A practical guide to hydrogels for cell culture, Nat. Methods, 2016, 13(5), 405–414 CrossRef CAS.
- L. G. Griffith and M. A. Swartz, Capturing complex 3D tissue physiology in vitro, Nat. Rev. Mol. Cell Biol., 2006, 7(3), 211–224 CrossRef CAS.
- Y. Shin, S. Han, J. S. Jeon, K. Yamamoto, I. K. Zervantonakis and R. Sudo, et al., Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels, Nat. Protoc., 2012, 7(7), 1247–1259 CrossRef CAS.
- M. T. Nelson, M. R. Charbonneau, H. G. Coia, M. J. Castillo, C. Holt and E. S. Greenwood, et al., Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip, Nat. Commun., 2021, 12(1), 2805 CrossRef CAS.
- J. C. Nawroth, D. B. Petropolis, D. V. Manatakis, T. I. Maulana, G. Burchett and K. Schlünder, et al., Modeling alcohol-associated liver disease in a human Liver-Chip, Cell Rep., 2021, 36(3), 109393 CrossRef CAS.
- J. W. Choi, M. Seo, K. Kim, A. R. Kim, H. Lee and H. S. Kim, et al., Aptamer Nanoconstructs Crossing Human Blood–Brain Barrier Discovered via Microphysiological System-Based SELEX Technology, ACS Nano, 2023, 17(9), 8153–8166 CrossRef CAS.
- J. Kim, T. Yoon, P. Kim, M. Bekhbat, S. M. Kang and H. S. Rho, et al., Manufactured tissue-to-tissue barrier chip for modeling the human blood–brain barrier and regulation of cellular trafficking, Lab Chip, 2023, 23(13), 2990–3001 RSC.
- S. I. Ahn, Y. J. Sei, H. J. Park, J. Kim, Y. Ryu and J. J. Choi, et al., Microengineered human blood–brain barrier platform for understanding nanoparticle transport mechanisms, Nat. Commun., 2020, 11(1), 175 CrossRef CAS.
- L. C. Delon, Z. Guo, A. Oszmiana, C. C. Chien, R. Gibson and C. Prestidge, et al., A systematic investigation of the effect of the fluid shear stress on Caco-2 cells towards the optimization of epithelial organ-on-chip models, Biomaterials, 2019, 225, 119521 CrossRef CAS.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow, Lab Chip, 2012, 12(12), 2165 RSC.
- J. M. Tarbell, Shear stress and the endothelial transport barrier, Cardiovasc. Res., 2010, 87(2), 320–330 CrossRef CAS.
- L. Yan, R. A. Moriarty and K. M. Stroka, Recent progress and new challenges in modeling of human pluripotent stem cell-derived blood-brain barrier, Theranostics, 2021, 11(20), 10148–10170 CrossRef CAS.
- A. Bein, C. W. Fadel, B. Swenor, W. Cao, R. K. Powers and D. M. Camacho, et al., Nutritional deficiency in an intestine-on-a-chip recapitulates injury hallmarks associated with environmental enteric dysfunction, Nat. Biomed. Eng., 2022, 6(11), 1236–1247 CrossRef.
- K. Lin, P. P. Hsu, B. P. Chen, S. Yuan, S. Usami and J. Y. J. Shyy, et al., Molecular mechanism of endothelial growth arrest by laminar shear stress, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(17), 9385–9389 CrossRef CAS.
- J. Kühnl, T. P. Tao, K. Brandmair, S. Gerlach, T. Rings and U. Müller-Vieira, et al., Characterization of application scenario-dependent pharmacokinetics and pharmacodynamic properties of permethrin and hyperforin in a dynamic skin and liver multi-organ-chip model, Toxicology, 2021, 448, 152637 CrossRef.
- A. Herland, B. M. Maoz, D. Das, M. R. Somayaji, R. Prantil-Baun and R. Novak, et al., Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips, Nat. Biomed. Eng., 2020, 4(4), 421–436 CrossRef CAS.
- D. B. Chou, V. Frismantas, Y. Milton, R. David, P. Pop-Damkov and D. Ferguson, et al., On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology, Nat. Biomed. Eng., 2020, 4(4), 394–406 CrossRef.
- H. Azizgolshani, J. R. Coppeta, E. M. Vedula, E. E. Marr, B. P. Cain and R. J. Luu, et al., High-throughput organ-on-chip platform with integrated programmable fluid flow and real-time sensing for complex tissue models in drug development workflows, Lab Chip, 2021, 21(8), 1454–1474 RSC.
- Z. Chen, S. He, J. Zilberberg and W. Lee, Pumpless platform for high-throughput dynamic multicellular culture and chemosensitivity evaluation, Lab Chip, 2019, 19(2), 254–261 RSC.
- H. Ehlers, A. Nicolas, F. Schavemaker, J. P. M. Heijmans, M. Bulst and S. J. Trietsch, et al., Vascular inflammation on a chip: A scalable platform for trans-endothelial electrical resistance and immune cell migration, Front. Immunol., 2023, 14, 1118624 CrossRef CAS.
- J. Kim, K. T. Lee, J. S. Lee, J. Shin, B. Cui and K. Yang, et al., Fungal brain infection modelled in a human-neurovascular-unit-on-a-chip with a functional blood–brain barrier, Nat. Biomed. Eng., 2021, 5(8), 830–846 CrossRef.
- Y. J. Sei, S. I. Ahn, T. Virtue, T. Kim and Y. Kim, Detection of frequency-dependent endothelial response to oscillatory shear stress using a microfluidic transcellular monitor, Sci. Rep., 2017, 7(1), 10019 CrossRef.
- Y. I. Wang and M. L. Shuler, UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems, Lab Chip, 2018, 18(17), 2563–2574 RSC.
- Y. Yang, P. Fathi, G. Holland, D. Pan, N. S. Wang and M. B. Esch, Pumpless microfluidic devices for generating healthy and diseased endothelia, Lab Chip, 2019, 19(19), 3212–3219 RSC.
- F. Zhang, D. S. Y. Lin, S. Rajasekar, A. Sotra and B. Zhang, Pump-Less Platform Enables Long-Term Recirculating Perfusion of 3D Printed Tubular Tissues, Adv. Healthcare Mater., 2023, 12(27), 2300423 CrossRef CAS.
- A. Mahringer, J. Delzer and G. Fricker, A fluorescence-based in vitro assay for drug interactions with breast cancer resistance protein (BCRP, ABCG2), Eur. J. Pharm. Biopharm., 2009, 72(3), 605–613 CrossRef CAS.
- R. R. Xiao, B. Jing, L. Yan, J. Li, P. Tu and X. Ai, Constant-rate perfused array chip for high-throughput screening of drug permeability through brain endothelium, Lab Chip, 2022, 22(23), 4481–4492 RSC.
- S. W. L. Lee, M. Campisi, T. Osaki, L. Possenti, C. Mattu and G. Adriani, et al., Modeling Nanocarrier Transport across a 3D In Vitro Human Blood-Brain–Barrier Microvasculature, Adv. Healthcare Mater., 2020, 9(7), 1901486 CrossRef CAS.
- B. Srinivasan, A. R. Kolli, M. B. Esch, H. E. Abaci, M. L. Shuler and J. J. Hickman, TEER Measurement Techniques for In Vitro Barrier Model Systems, SLAS Technol., 2015, 20(2), 107–126 CrossRef CAS.
- O. Y. F. Henry, R. Villenave, M. J. Cronce, W. D. Leineweber, M. A. Benz and D. E. Ingber, Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function, Lab Chip, 2017, 17(13), 2264–2271 RSC.
- J. L. Jameson and D. L. Longo, Precision Medicine — Personalized, Problematic, and Promising, N. Engl. J. Med., 2015, 372(23), 2229–2234 CrossRef CAS.
- P. Neužil, S. Giselbrecht, K. Länge, T. J. Huang and A. Manz, Revisiting lab-on-a-chip technology for drug discovery, Nat. Rev. Drug Discovery, 2012, 11(8), 620–632 CrossRef.
- E. P. Papapetrou, Patient-derived induced pluripotent stem cells in cancer research and precision oncology, Nat. Med., 2016, 22(12), 1392–1401 CrossRef CAS.
- Y. Shi, H. Inoue, J. C. Wu and S. Yamanaka, Induced pluripotent stem cell technology: a decade of progress, Nat. Rev. Drug Discovery, 2017, 16(2), 115–130 CrossRef CAS.
- H. Xu, Y. Jiao, S. Qin, W. Zhao, Q. Chu and K. Wu, Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine, Exp. Hematol. Oncol., 2018, 7(1), 30 CrossRef CAS.
- B. L. LeSavage, R. A. Suhar, N. Broguiere, M. P. Lutolf and S. C. Heilshorn, Next-generation cancer organoids, Nat. Mater., 2022, 21(2), 143–159 CrossRef CAS.
- Z. Zhao, X. Chen, A. M. Dowbaj, A. Sljukic, K. Bratlie and L. Lin, et al., Organoids, Nat. Rev. Methods Primers, 2022, 2(1), 94 CrossRef CAS.
- C. Ma, Y. Peng, H. Li and W. Chen, Organ-on-a-Chip: A New Paradigm for Drug Development, Trends Pharmacol. Sci., 2021, 42(2), 119–133 CrossRef CAS.
- E. Moradi, S. Jalili-Firoozinezhad and M. Solati-Hashjin, Microfluidic organ-on-a-chip models of human liver tissue, Acta Biomater., 2020, 116, 67–83 CrossRef CAS.
- M. Rothbauer, J. M. Rosser, H. Zirath and P. Ertl, Tomorrow today: organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models, Curr. Opin. Biotechnol., 2019, 55, 81–86 CrossRef CAS.
- A. Van Den Berg, C. L. Mummery, R. Passier and A. D. Van Der Meer, Personalised organs-on-chips: functional testing for precision medicine, Lab Chip, 2019, 19(2), 198–205 RSC.
- S. Seo, S. Nah, K. Lee, N. Choi and H. N. Kim, Triculture Model of In Vitro BBB and its Application to Study BBB-Associated Chemosensitivity and Drug Delivery in Glioblastoma, Adv. Funct. Mater., 2022, 32(10), 2106860 CrossRef CAS.
- H. G. Yi, Y. H. Jeong, Y. Kim, Y. J. Choi, H. E. Moon and S. H. Park, et al., A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy, Nat. Biomed. Eng., 2019, 3(7), 509–519 CrossRef CAS.
- H. Kim, J. K. Sa, J. Kim, H. J. Cho, H. J. Oh and D. Choi, et al., Recapitulated Crosstalk between Cerebral Metastatic Lung Cancer Cells and Brain Perivascular Tumor Microenvironment in a Microfluidic Co-Culture Chip, Adv. Sci., 2022, 9(22), 2201785 CrossRef CAS.
- Q. Ping, R. Yan, X. Cheng, W. Wang, Y. Zhong and Z. Hou, et al., Cancer-associated fibroblasts: overview, progress, challenges, and directions, Cancer Gene Ther., 2021, 28(9), 984–999 CrossRef CAS.
- J. Kim, H. Park, H. Kim, Y. Kim, H. J. Oh and S. Chung, Microfluidic one-directional interstitial flow generation from cancer to cancer associated fibroblast, Acta Biomater., 2022, 144, 258–265 CrossRef CAS.
- M. Gerigk, H. Bulstrode, H. H. Shi, F. Tönisen, C. Cerutti and G. Morrison, et al., On-chip perivascular niche supporting stemness of patient-derived glioma cells in a serum-free, flowable culture, Lab Chip, 2021, 21(12), 2343–2358 RSC.
- L. Bejarano, M. J. C. Jordāo and J. A. Joyce, Therapeutic Targeting of the Tumor Microenvironment, Cancer Discovery, 2021, 11(4), 933–959 CrossRef CAS.
- C. Roma-Rodrigues, R. Mendes, P. Baptista and A. Fernandes, Targeting Tumor Microenvironment for Cancer Therapy, Int. J. Mech. Sci., 2019, 20(4), 840 CAS.
- S. Zhong, J. H. Jeong, Z. Chen, Z. Chen and J. L. Luo, Targeting Tumor Microenvironment by Small-Molecule Inhibitors, Transl. Oncol., 2020, 13(1), 57–69 CrossRef.
- K. M. Lugo-Cintrón, J. M. Ayuso, M. Humayun, M. M. Gong, S. C. Kerr and S. M. Ponik, et al., Primary head and neck tumour-derived fibroblasts promote lymphangiogenesis in a lymphatic organotypic co-culture model, EBioMedicine, 2021, 73, 103634 CrossRef.
- H. Choi and A. Moon, Crosstalk between cancer cells and endothelial cells: implications for tumor progression and intervention, Arch. Pharmacal Res., 2018, 41(7), 711–724 CrossRef CAS.
- K. Hida, K. Akiyama, N. Ohga, N. Maishi and Y. Hida, Tumour endothelial cells acquire drug resistance in a tumour microenvironment, J. Biochem., 2013, 153(3), 243–249 CrossRef CAS.
- K. Hida, N. Maishi, Y. Sakurai, Y. Hida and H. Harashima, Heterogeneity of tumor endothelial cells and drug delivery, Adv. Drug Delivery Rev., 2016, 99, 140–147 CrossRef CAS.
- J. A. Jiménez-Torres, M. Virumbrales-Muñoz, K. E. Sung, M. H. Lee, E. J. Abel and D. J. Beebe, Patient-specific organotypic blood vessels as an in vitro model for anti-angiogenic drug response testing in renal cell carcinoma, EBioMedicine, 2019, 42, 408–419 CrossRef.
- V. E. J. M. Palasantzas, I. Tamargo-Rubio, K. Le, J. Slager, C. Wijmenga and I. H. Jonkers, et al., iPSC-derived organ-on-a-chip models for personalized human genetics and pharmacogenomics studies, Trends Genet., 2023, 39(4), 268–284 CrossRef CAS.
- G. Lee, C. N. Ramirez, H. Kim, N. Zeltner, B. Liu and C. Radu, et al., Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression, Nat. Biotechnol., 2012, 30(12), 1244–1248 CrossRef CAS.
- S. J. Shankland, B. S. Freedman and J. W. Pippin, Can podocytes be regenerated in adults?, Curr. Opin. Nephrol. Hypertens., 2017, 26(3), 154–164 CrossRef CAS.
- Y. Roye, R. Bhattacharya, X. Mou, Y. Zhou, M. A. Burt and S. Musah, A Personalized Glomerulus Chip Engineered from Stem Cell-Derived Epithelium and Vascular Endothelium, Micromachines, 2021, 12(8), 967 CrossRef.
- H. Hong and W. Tong, Emerging efforts for discovering new biomarkers of liver disease and hepatotoxicity, Biomarkers Med., 2014, 8(2), 143–146 CrossRef CAS.
- A. Asplund, A. Pradip, M. Van Giezen, A. Aspegren, H. Choukair and M. Rehnström, et al., One Standardized Differentiation Procedure Robustly Generates Homogenous Hepatocyte Cultures Displaying Metabolic Diversity from a Large Panel of Human Pluripotent Stem Cells, Stem Cell Rev. Rep., 2016, 12(1), 90–104 CrossRef CAS.
- J. Lu, S. Einhorn, L. Venkatarangan, M. Miller, D. A. Mann and P. B. Watkins, et al., Morphological and Functional Characterization and Assessment of iPSC-Derived Hepatocytes for In Vitro Toxicity Testing, Toxicol. Sci., 2015, 147(1), 39–54 CrossRef CAS.
- Y. Kato, A. Y. Lim, C. Sakolish, A. Valdiviezo, H. L. Moyer and P. Hewitt, et al., Analysis of reproducibility and robustness of OrganoPlate® 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs, Toxicol. In Vitro, 2022, 85, 105464 CrossRef CAS.
- S. Sances, R. Ho, G. Vatine, D. West, A. Laperle and A. Meyer, et al., Human iPSC-Derived Endothelial Cells and Microengineered Organ-Chip Enhance Neuronal Development, Stem Cell Rep., 2018, 10(4), 1222–1236 CrossRef CAS.
- E. K. Hollmann, A. K. Bailey, A. V. Potharazu, M. D. Neely, A. B. Bowman and E. S. Lippmann, Accelerated differentiation of human induced pluripotent stem cells to blood–brain barrier endothelial cells, Fluids Barriers CNS, 2017, 14(1), 9 CrossRef.
- E. S. Lippmann, A. Al-Ahmad, S. M. Azarin, S. P. Palecek and E. V. Shusta, A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources, Sci. Rep., 2014, 4(1), 4160 CrossRef.
- E. H. Neal, N. A. Marinelli, Y. Shi, P. M. McClatchey, K. M. Balotin and D. R. Gullett, et al., A Simplified, Fully Defined Differentiation Scheme for Producing Blood-Brain Barrier Endothelial Cells from Human iPSCs, Stem Cell Rep., 2019, 12(6), 1380–1388 CrossRef CAS.
- I. Pediaditakis, K. R. Kodella, D. V. Manatakis, C. Y. Le, C. D. Hinojosa and W. Tien-Street, et al., Modeling alpha-synuclein pathology in a human brain-chip to assess blood-brain barrier disruption, Nat. Commun., 2021, 12(1), 5907 CrossRef CAS.
- I. Pediaditakis, K. R. Kodella, D. V. Manatakis, C. Y. Le, S. Barthakur and A. Sorets, et al., A microengineered Brain-Chip to model neuroinflammation in humans, iScience, 2022, 25(8), 104813 CrossRef CAS.
- G. D. Vatine, R. Barrile, M. J. Workman, S. Sances, B. K. Barriga and M. Rahnama, et al., Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications, Cell Stem Cell, 2019, 24(6), 995–1005 CrossRef CAS.
- M. A. Erickson and W. A. Banks, Neuroimmune Axes of the Blood–Brain Barriers and Blood–Brain Interfaces: Bases for Physiological Regulation, Disease States, and Pharmacological Interventions. Dantzer R, editor, Pharmacol. Rev., 2018, 70(2), 278–314 CrossRef CAS.
- L. Gijzen, F. A. Yousef Yengej, F. Schutgens, M. K. Vormann, C. M. E. Ammerlaan and A. Nicolas, et al., Culture and analysis of kidney tubuloids and perfused tubuloid cells-on-a-chip, Nat. Protoc., 2021, 16(4), 2023–2050 CrossRef CAS.
- M. Huch, S. F. Boj and H. Clevers, Lgr5+ liver stem cells, hepatic organoids and regenerative medicine, Regener. Med., 2013, 8(4), 385–387 CrossRef CAS.
- J. R. Rock, M. W. Onaitis, E. L. Rawlins, Y. Lu, C. P. Clark and Y. Xue, et al., Basal cells as stem cells of the mouse trachea and human airway epithelium, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(31), 12771–12775 CrossRef CAS.
- T. Sato, D. E. Stange, M. Ferrante, R. G. J. Vries, J. H. Van Es and S. Van Den Brink, et al., Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett's Epithelium, Gastroenterology, 2011, 141(5), 1762–1772 CrossRef CAS.
- F. Schutgens, M. B. Rookmaaker, T. Margaritis, A. Rios, C. Ammerlaan and J. Jansen, et al., Tubuloids derived from human adult kidney and urine for personalized disease modeling, Nat. Biotechnol., 2019, 37(3), 303–313 CrossRef CAS.
- S. Van Riet, A. Van Schadewijk, P. P. S. J. Khedoe, R. W. A. L. Limpens, M. Bárcena and J. Stolk, et al., Organoid-based expansion of patient-derived primary alveolar type 2 cells for establishment of alveolus epithelial Lung-Chip cultures, Am. J. Physiol., 2022, 322(4), L526–L538 CAS.
- M. Kasendra, R. Luc, J. Yin, D. V. Manatakis, G. Kulkarni and C. Lucchesi, et al., Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model, eLife, 2020, 9, e50135 CrossRef CAS.
- L. Si, H. Bai, M. Rodas, W. Cao, C. Y. Oh and A. Jiang, et al., A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics, Nat. Biomed. Eng., 2021, 5(8), 815–829 CrossRef CAS.
- H. C. Helms, N. J. Abbott, M. Burek, R. Cecchelli, P. O. Couraud and M. A. Deli, et al., In vitro models of the blood–brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use, J. Cereb. Blood Flow Metab., 2016, 36(5), 862–890 CrossRef CAS.
- M. A. Erickson, M. L. Wilson and W. A. Banks, In vitro modeling of blood–brain barrier and interface functions in neuroimmune communication, Fluids Barriers CNS, 2020, 17(1), 26 CrossRef.
- S. Hosic, W. Lake, E. Stas, R. Koppes, D. T. Breault and S. K. Murthy, et al., Cholinergic Activation of Primary Human Derived Intestinal Epithelium Does Not Ameliorate TNF-α Induced Injury, Cell. Mol. Bioeng., 2020, 13(5), 487–505 CrossRef CAS.
- N. Tsamandouras, T. Kostrzewski, C. L. Stokes, L. G. Griffith, D. J. Hughes and M. Cirit, Quantitative Assessment of Population Variability in Hepatic Drug Metabolism Using a Perfused Three-Dimensional Human Liver Microphysiological System, J. Pharmacol. Exp. Ther., 2017, 360(1), 95–105 CrossRef CAS.
- A. Banfi, G. Bianchi, M. Galotto, R. Cancedda and R. Quarto, Bone Marrow Stromal Damage after Chemo/Radiotherapy: Occurrence, Consequences and Possibilities of Treatment, Leuk. Lymphoma, 2001, 42(5), 863–870 CrossRef CAS.
- J. S. Greenberger and M. Epperly, Bone Marrow–Derived Stem Cells and Radiation Response, Semin. Radiat. Oncol., 2009, 19(2), 133–139 CrossRef.
- K. Fabre, B. Berridge, W. R. Proctor, S. Ralston, Y. Will and S. W. Baran, et al., Introduction to a manuscript series on the characterization and use of microphysiological systems (MPS) in pharmaceutical safety and ADME applications, Lab Chip, 2020, 20(6), 1049–1057 RSC.
- K. Eckstrum, A. Striz, M. Ferguson, Y. Zhao, B. Welch and N. Solomotis, et al., Utilization of a model hepatotoxic compound, diglycolic acid, to evaluate liver Organ-Chip performance and in vitro to in vivo concordance, Food Chem. Toxicol., 2020, 146, 111850 CrossRef CAS.
- K. J. Jang, M. A. Otieno, J. Ronxhi, H. K. Lim, L. Ewart and K. R. Kodella, et al., Reproducing human and cross-species drug toxicities using a Liver-Chip, Sci. Transl. Med., 2019, 11(517), eaax5516 CrossRef CAS.
- L. Ewart, A. Apostolou, S. A. Briggs, C. V. Carman, J. T. Chaff and A. R. Heng, et al., Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology, Commun. Med., 2022, 2(1), 154 CrossRef.
- A. R. Baudy, M. A. Otieno, P. Hewitt, J. Gan, A. Roth and D. Keller, et al., Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry, Lab Chip, 2020, 20(2), 215–225 RSC.
- D. Levner and L. Ewart, Integrating Liver-Chip data into pharmaceutical decision-making processes, Expert Opin. Drug Discovery, 2023, 18(12), 1313–1320 CrossRef CAS.
- H. Garside, K. F. Marcoe, J. Chesnut-Speelman, A. J. Foster, D. Muthas and J. Gerry Kenna, et al., Evaluation of the use of imaging parameters for the detection of compound-induced hepatotoxicity in 384-well cultures of HepG2 cells and cryopreserved primary human hepatocytes, Toxicol. In Vitro, 2014, 28(2), 171–181 CrossRef CAS.
- K. M. Bircsak, R. DeBiasio, M. Miedel, A. Alsebahi, R. Reddinger and A. Saleh, et al., A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®, Toxicology, 2021, 450, 152667 CrossRef CAS.
- K. Tan, P. Keegan, M. Rogers, M. Lu, J. R. Gosset and J. Charest, et al., A high-throughput microfluidic microphysiological system (PREDICT-96) to recapitulate hepatocyte function in dynamic, re-circulating flow conditions, Lab Chip, 2019, 19(9), 1556–1566 RSC.
- E. M. Shaughnessey, S. H. Kann, H. Azizgolshani, L. D. Black, J. L. Charest and E. M. Vedula, Evaluation of rapid transepithelial electrical resistance (TEER) measurement as a metric of kidney toxicity in a high-throughput microfluidic culture system, Sci. Rep., 2022, 12(1), 13182 CrossRef CAS.
- H. T. Nguyen, S. L. Rissanen, M. Peltokangas, T. Laakkonen, J. Kettunen and L. Barthod, et al., Highly scalable and standardized organ-on-chip platform with TEER for biological barrier modeling, Tissue Barriers, 2024, 2315702 CrossRef.
- C. Soragni, K. Queiroz, C. P. Ng, A. Stok, T. Olivier and D. Tzagkaraki, et al., Phenotypic screening in Organ-on-a-Chip systems: a 1537 kinase inhibitor library screen on a 3D angiogenesis assay, Angiogenesis, 2024, 27(1), 37–49 CrossRef CAS.
- S. Peel, A. M. Corrigan, B. Ehrhardt, K. J. Jang, P. Caetano-Pinto and M. Boeckeler, et al., Introducing an automated high content confocal imaging approach for Organs-on-Chips, Lab Chip, 2019, 19(3), 410–421 RSC.
- R. Novak, M. Ingram, S. Marquez, D. Das, A. Delahanty and A. Herland, et al., Robotic fluidic coupling and interrogation of multiple vascularized organ chips, Nat. Biomed. Eng., 2020, 4(4), 407–420 CrossRef.
- L. Shi, M. Zeng, Y. Sun and B. M. Fu, Quantification of Blood-Brain Barrier Solute Permeability and Brain Transport by Multiphoton Microscopy, J. Biomech. Eng., 2014, 136(3), 031005 CrossRef.
- W. Yuan, Y. Lv, M. Zeng and B. M. Fu, Non-invasive measurement of solute permeability in cerebral microvessels of the rat, Microvasc. Res., 2009, 77(2), 166–173 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |