Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic device for the high-throughput and selective encapsulation of single target cells
Masahiko
Nakamura
 ,
Masahiro
Matsumoto
,
Masahiro
Matsumoto
 ,
Tatsumi
Ito
,
Isao
Hidaka
,
Hirokazu
Tatsuta
and
Yoichi
Katsumoto
,
Tatsumi
Ito
,
Isao
Hidaka
,
Hirokazu
Tatsuta
and
Yoichi
Katsumoto
 *
*
Life Science Technology Research & Development Dept., Application Technology Research & Development Div., Technology Development Laboratories, Sony Corporation, Tokyo, Japan. E-mail: Yoichi.Katsumoto@sony.com; Fax: +81 3 5803 4790; Tel: +81 3 5803 4791
First published on 9th May 2024
Abstract
Droplet-based microfluidic technologies for encapsulating single cells have rapidly evolved into powerful tools for single-cell analysis. In conventional passive single-cell encapsulation techniques, because cells arrive randomly at the droplet generation section, to encapsulate only a single cell with high precision, the average number of cells per droplet has to be decreased by reducing the average frequency at which cells arrive relative to the droplet generation rate. Therefore, the encapsulation efficiency for a given droplet generation rate is very low. Additionally, cell sorting operations are required prior to the encapsulation of target cells for specific cell type analysis. To address these challenges, we developed a cell encapsulation technology with a cell sorting function using a microfluidic chip. The microfluidic chip is equipped with an optical detection section to detect the optical information of cells and a sorting section to encapsulate cells into droplets by controlling a piezo element, enabling active encapsulation of only the single target cells. For a particle population including both targeted and non-targeted particles arriving at an average frequency of up to 6000 particles per s, with an average number of particles per droplet of 0.45, our device maintained a high purity above 97.9% for the single-target-particle droplets and achieved an outstanding throughput, encapsulating up to 2900 single target particles per s. The proposed encapsulation technology surpasses the encapsulation efficiency of conventional techniques, provides high efficiency and flexibility for single-cell research, and shows excellent potential for various applications in single-cell analysis.
Introduction
Single-cell analysis, which is the high-resolution analysis of cell populations at the single-cell level, has become one of the most important approaches for understanding complex biological systems.1–7 Droplet-based microfluidic technologies have developed rapidly in the field of single-cell analysis such as single-cell RNA sequencing8–11 owing to their ability to isolate cells within droplets. With its high encapsulation capacity, droplet-based technology enables a higher throughput than well-based techniques,12,13 rendering scaling-up the number of analyzed cells feasible,13 and facilitating the precise assessment of cellular heterogeneity as well as the identification of rare cells.14Previous studies have shown that stable water-in-oil droplets can be generated using microfluidic geometries such as the T-junction,15,16 flow-focusing,16,17 and co-flow16,18 by applying aqueous solutions as the dispersed phase and oils containing surfactants as the continuous phase. In a T-junction, the dispersed phase is supplied by the main channel and the continuous phase is introduced through the side channel orthogonal to the main channel. In flow-focusing, the continuous phase is supplied from two side channels arranged vertically with respect to the central channel to squeeze the dispersed phase flow. In co-flow techniques, the inner dispersed phase and the outer continuous phase flow in the same direction through coaxial channels. Droplets are formed continuously at or downstream of the junction in different breakup modes, as determined by the balance of inertia, viscosity, and interfacial tension, which vary with the flow rate, channel diameter, and material properties of the fluid system.19
Loading a single cell into a droplet in such passive droplet generators uses a cell suspension as the dispersed phase. The arrival of cells at the droplet generation section is random and the number of cells encapsulated in each droplet follows a Poisson distribution.20,21 In single-cell analysis, it is desirable for only one cell to be encapsulated in each droplet. Therefore, the probability of encapsulating multiple cells in the same droplet should be minimized. To achieve the high-purity encapsulation of single cells (purity is defined as the ratio of the number of droplets containing a single cell to the number of droplets containing any cell), it is necessary to reduce the average number of cells per droplet, by reducing the average frequency at which cells arrive per unit time (event rate) at the droplet generation section. However, this also leads to a larger number of droplets that do not contain any cells. This makes maximizing the encapsulation rate obtained using droplet microfluidic technology challenging.
Droplet generation instruments such as Chromium (10xGenomics), Nadia (dolomite), and InDrop system (1CellBio) are available as commercial platforms for droplet-based single-cell analysis, but none of them have cell sorting functions. Cell sorting operations such as with cell sorters are required prior to cell encapsulation when analyzing specific phenotypic cells.22
To address these challenges, we present a cell-encapsulation technology with cell-sorting functions using a microfluidic chip. This technology enables the encapsulation of single target cells into droplets with high purity and throughput.
System implementation
Principles of selective encapsulation
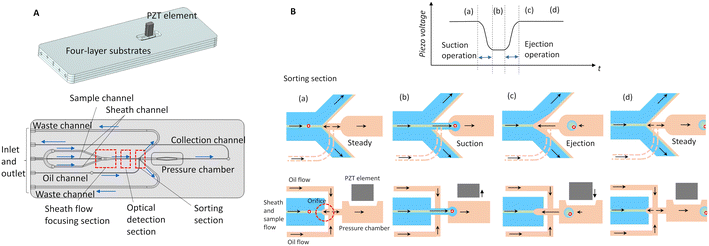
A microfluidic chip was used to encapsulate cells selectively, as shown in Fig. 1A. The chip was fabricated by bonding four layers of pre-grooved plastic substrate composed of a cyclic olefin copolymer (TOPAS, Polyplastics Corporation). A piezo element was fixed to the substrate surface for the sorting operation, which will be described later. The microchannel was composed of three sections for sheath flow focusing, optical detection, and sorting. During operation, the cell suspension is loaded from the sample inlet and merged with the sheath fluid in the sheath-flow-focusing section. The cross-sectional area of the channel in this section decreases in the downstream direction and the thickness of the sample flow is reduced to approximately the diameter of the cells. Consequently, the cells are aligned in a single file in the flow direction and introduced into the optical detection section. In this section, multiple excitation lasers irradiate the cells through the chip, and forward-scattered light (FSC), backward-scattered light (BSC), and fluorescence can be observed. The sample flow containing the cells passes through the optical detection section and reaches the sorting section. The sorting section is equipped with waste channels on both sides and a collection channel in the center, which is connected through an orifice. An oil immiscible with the aqueous sheath liquid is loaded into the orifice in the vertical direction and a flow divided into the upstream and downstream directions is formed in the orifice. The upstream flow prevents the sample from entering through the orifice steadily and the downstream flow enters the collection channel and fills it with oil. The collection channel is equipped with a pressure chamber and a piezo element is connected to it through a thin upper wall. By controlling the voltage applied to the piezo element, rapid pressure control can be achieved in the channel, enabling the sorting of target cells into the collection channel.A basic sorting operation can be performed as shown in Fig. 1B. In the initial state, a driving voltage is applied to the piezo element to stretch it and reduce the volume of the pressure chamber. When the target cell is detected by the optical detection section and reaches the sorting section, a single-step waveform is applied to the piezo element. This causes the piezo element to shrink and a negative pressure is generated inside the pressure chamber. The target cell, sheath fluid, and sample fluid are then sucked and encapsulated in the collection channel filled with oil. After suction, the voltage is released and the piezo element is stretched again to provide the original pressure field. When pressure is restored, the fluid is ejected into the sorting section to generate an ejection flow. As a result, a water-in-oil droplet containing the target cell, sheath fluid, and sample fluid is formed in the collection channel. If the target cell is not detected, then pressure control is not performed and the cells flow into either of the two adjacent waste channels. In this series of sorting operations, a holding time for maintaining a constant displacement (voltage) of the piezo element can be provided between the suction operation when the piezo element shrinks and the ejection operation when the piezo element is stretched to recover pressure. When the holding time is long, the suction of the sheath fluid and sample fluid increase as a result of the longer duration of the suction flow. Therefore, by increasing the holding time during the sorting operation, the volume of the dispersed phase sucked into the collection channel can be increased, resulting in the generation of larger droplets.
To study the response of the flow in the sorting section to piezo actuation, numerical simulations were performed using COMSOL Multiphysics®. Fig. 2 presents the simulation results for the displacement pattern of the piezo element and the flow generated inside the orifice. As shown in Fig. 2A, structural analysis was applied to the structure around the pressure chamber, and partial and temporal deformation was considered. Specifically, the sorting operation was simulated by applying time displacement to a structure resembling the piezo element attached to the upper wall of the pressure chamber. In this study, all fluids were analyzed as having the properties of water. Fig. 2B presents the temporal variation of the mean flow velocity in the mainstream direction inside the orifice when the displacement pattern of the piezo element is applied to the basic sorting operation (basic pulse). The flow velocity U′ and time t′ in the graph were non-dimensionalized using the absolute value of the mean flow velocity in the mainstream direction inside the orifice at a steady-state with U0 as the representative velocity and the orifice diameter D0 as the representative length. When the basic pulse is applied, after the piezo element shrinks, the flow rapidly accelerates in the suction direction and the fluid is sucked into the collection channel. Next, when the piezo element stretches and the pressure-recovery operation is performed, the flow in the suction direction decelerates and a flow in the ejection direction of similar magnitude to that of the suction flow is generated. The labels B1 and B2 indicate the distribution of the mainstream flow velocity in the sorting section at the time when the flow velocity reaches its maximum in the suction direction and ejection direction, respectively. When the ejection flow is generated, the target cells cannot be sucked into the collection channel, even if they reach the sorting section. In particular, when the event rate of the target cell is high, the yield decreases owing to the restriction of the fluid response.
To suppress the ejection flow and minimize the decrease in yield, an offset pulse is applied, which adds a step-like displacement pattern with a reduced displacement (drive voltage) to the piezo element after the pressure-recovery operation (Fig. 2C). As shown in the simulation results for the temporal variation in the orifice flow velocity and flow velocity distribution at the same time as B2, the generation of the ejection flow is suppressed. This enables the minimization of the settling time of the fluid fields in the sorting section. However, because the displacement of the piezo element remains during the sorting operation, the number of continuous sorting operations is limited by restrictions on the maximum displacement of the piezo element. The sorting pulses applied to the piezo element are optimally controlled by an original algorithm to maximize yield with an offset pulse being applied when the event rate is high.
System for selective single-cell encapsulation
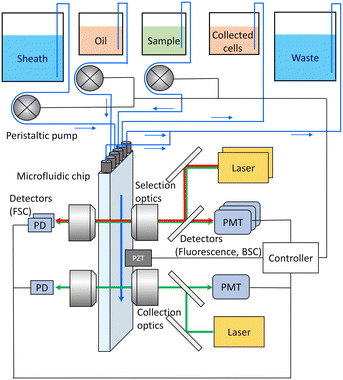
Our experimental setup is presented in Fig. 3. The system consisted of two optical systems, namely a selection optical system and collection optical system, as well as a fluid system, device control system, and signal processing system. The system was equipped with an interchangeable microfluidic chip connected to the fluid system through tubes. The system was also equipped with three peristaltic tube pumps for controlling the sheath, sample, and oil fluids. These pumps supplied the corresponding fluids to the chip. The sample fluid supplied flowed through the channels inside the microfluidic chip and reached the selection optical system. The selection optical system was designed to enable the selection of fluorescently labeled cells within the microfluidic channel. This system was equipped with two lasers (488 nm: OBIS 488LS, 637 nm: OBIS 637LX, COHERENT) as excitation light sources for fluorescent labels. It measured the intensity of FSC, BSC, and fluorescence emitted from the fluorescent labels at each excitation wavelength. Target cells encapsulated in the sorting section were transported downstream to the collection optical system. The collection optical system was equipped with a laser (488 nm: OBIS 488LS, COHERENT) as the excitation light source. It measured the intensity of FSC emitted mainly from droplets and fluorescence emitted from labeled cells within the droplets. Scattering and fluorescence signals from the collection optical system were used to determine the presence or absence of fluorescently labeled cells within the droplets. The signal information obtained from the optical systems was used to control the piezo element attached to the microfluidic chip surface. Additionally, sorting parameters such as the delay time from the detection of target cells to the application of the sorting pulse and the driving voltage applied to the piezo element were adjusted.Materials and methods
Sorting and encapsulation
Sorting experiments were performed using polystyrene particles and human peripheral blood mononuclear cells (PBMCs) as samples. In the sorting experiments using polystyrene particles, phosphate-buffered saline (PBS, 10010031, Thermo Fisher Scientific) was used as the sheath fluid. In the sorting experiments using PBMCs, a cell buffer of PBS containing 0.04% BSA (130-091-376, Miltenyi Biotec) was used as the sheath fluid. Additionally, 008-FluoroSurfactant (RT008-2%500, RAN Biotechnologies) was used as the oil fluid in both experiments. Each liquid was supplied at a constant flow rate using corresponding peristaltic pumps: the sheath flow rate was 10 mL min−1, the sample flow rate was 45 μL min−1, and the oil flow rate was 170 μL min−1. In the steady state without encapsulation, the oil supplied was distributed at a rate of 70 μL min−1 to the orifice and 100 μL min−1 to the collection channel. During encapsulation, a displacement of approximately 1 μm was applied to the piezo element, causing a flow-rate change equivalent to one droplet volume at the orifice. The capillary number Cad indicating the conditions for droplet generation for dispersed phase (sheath and sample) was calculated using the simulation value of the average cross-sectional flow velocity at the orifice during suction and found to be 0.74. The capillary number Cac for the continuous phase (oil) was calculated using the average cross-sectional flow velocity in the oil-supply channel and found to be 0.16. The gates used for selection detection were set to include events based on the intensity of each signal emitted by the target particles (FSC, BSC, and fluorescence signals).Calculation of effective yield and purity for fluorescent polystyrene particles
The effective yield and purity of the microfluidic device were evaluated using a sample consisting of a mixture of 5 μm-diameter polystyrene particles (G0500, excitation: 468 nm; emission: 508 nm, Thermo Fisher Scientific) as target particles and 3 μm-diameter fluorescent polystyrene particles (R0300, excitation: 542 nm; emission: 612 nm, Thermo Fisher Scientific) with different fluorescence properties as non-target particles. The effective yield was defined as the ratio of the number of target particles encapsulated as single particles to the number of target particles detected in the selection optical system. Purity was defined as the ratio of the number of droplets containing only single target particles to the number of droplets containing any particles. The plurality of encapsulated fluorescent polystyrene particles was imaged twice consecutively using a fluorescence microscope (ECLIPSE Ti, Nikon) equipped with a lens (Plan Fluor 4×/NA0.13, Nikon) with an optical filter (excitation: 389/38-25; emission: 483/32-25) for the fluorescence observation of target particles and an optical filter (excitation: 531/40-25; emission: 593/40-25) for the fluorescence observation of non-target particles. The effective yield was calculated by multiplying the ratio of the number of particles counted in the collection optical system to the number of particles detected in the selection optical system by the ratio of the number of particles encapsulating only single target particles to the number of particles in the images captured by the fluorescence microscope. The purity was calculated as the ratio of the number of droplets containing only a single target particle to the number of droplets containing any particles in the images captured by the fluorescence microscope. During fluorescence microscopy imaging, weak bright-field light was applied to capture fluorescent particles and droplets simultaneously.Preparation of PBMC samples
PBMCs derived from healthy individuals (CTL-UP1: Uncharacterised PBMC, Cellular Technology Limited) were cultured in ImmunoCult™-XF T Cell Expansion Medium (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 981, STEMCELL Technologies) supplemented with 12.5 μL mL−1 of ImmunoCult™ Human CD3/CD28 T Cell Activator (10
981, STEMCELL Technologies) supplemented with 12.5 μL mL−1 of ImmunoCult™ Human CD3/CD28 T Cell Activator (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991, STEMCELL Technologies), 20 ng mL−1 of Human Recombinant IL-7 (78053.3, STEMCELL Technologies), 20 ng mL−1 of Human Recombinant IL-15 (7831.3, STEMCELL Technologies), and 0.5% Human Plasma (Cellular Technology Limited).
991, STEMCELL Technologies), 20 ng mL−1 of Human Recombinant IL-7 (78053.3, STEMCELL Technologies), 20 ng mL−1 of Human Recombinant IL-15 (7831.3, STEMCELL Technologies), and 0.5% Human Plasma (Cellular Technology Limited).
Viability assay using Calcein AM
Cell viability after encapsulation was evaluated using PBMCs. The PBMCs (1 × 106 cells per ml in 1 mL) were stained with 2 μL of 50 μM Calcein AM (C1430, Thermo Fisher Scientific) for 20 min, and then centrifuged and replaced with 0.04% BSA PBS. The viability of the PBMCs prior to encapsulation was quantified using an automated cell counter (NucleoCounter ® NC-202, ChemoMetec). The concentration of PBMCs was adjusted to 2.6 × 106 cells per ml. The encapsulated PBMCs were imaged using a fluorescence microscope (ECLIPSE Ti, Nikon) equipped with a lens (Nikon, Plan Fluor 4×/NA0.13) using an optical filter (excitation: 457/50; emission: 520/28). The live and dead cells were counted based on the size of the bright dots in the fluorescence images, and viability (live cell count/cell count) was calculated.Evaluation of lysis efficiency using Calcein AM
Homemade 1× ThermoPol reaction buffer and 0.3% NP-40 (Thermo Fisher Scientific) were added to the sheath fluid as a cell lysis buffer to evaluate cell lysis efficiency after encapsulation. Live and dead cells were counted, and the lysis efficiency (number of dead cells/number of cells) was calculated using the same evaluation method used for the viability assay.Results
Encapsulation rate
The encapsulation speed (minimum time interval after particle encapsulation before pressure recovery and encapsulation of the next particle) of the encapsulation system was evaluated using 5 μm polystyrene particles (G0500, excitation: 468 nm, emission: 508 nm, Thermo Fisher Scientific). When the time interval between arriving particles is below a certain value, the disturbance of the fluid field generated by the encapsulation of leading particles does not fully settle and the following particles cannot be encapsulated. To evaluate this time interval, two consecutive encapsulation operations were performed, as shown in Fig. 4A: first a pseudo-encapsulation operation that only generated a droplet, then true encapsulation of the detected particles. The recovery performance was evaluated for various time intervals Δt between these two operations. The waveforms applied to the piezo element were evaluated using both basic and offset pulses.The relationship between the time interval Δt and recovery is presented in Fig. 4B. For each Δt, the two consecutive encapsulation operations were performed 1000 times. Here, the ratio of the actual number of droplets counted based on the FSC signal of the collection optical system to the total number of particle-encapsulation and pseudo-encapsulation operations is defined as the droplet recovery. The ratio of the actual number of particles counted based on the fluorescence signal of the collection optical system to the number of particle-encapsulation operations is defined as the particle recovery. When the basic pulse was applied, both the droplet recovery and particle recovery increased rapidly at Δt = 165 μs; the recoveries reached almost 100%. At Δt < 165 μs, particle encapsulation failed: number of particles and droplets equivalent to the number of particle-encapsulation operations were not detected in the collection optical system. When the offset pulse was applied, again, both recoveries were almost 100%, even at Δt = 75 μs. When the offset pulse was applied, the fluid field settled more rapidly after pseudo-encapsulation, and particle encapsulation was successful even for the short interval of Δt = 75 μs. The response speed of the sorting device was limited by the settling time of the fluid field after encapsulation. However, our evaluation indicates that by designing the driving waveform applied to the piezo element based on the results of numerical simulations, the settling time can be minimized.
Droplet size
Next, by using the same polystyrene particles described above, the droplet size was evaluated by changing only the holding time between the suction and ejection operations, while the driving time and voltage amplitude of the piezo element in the suction and ejection operations remained constant, under conditions where the average event detection rate of particles arriving at the sorting section randomly was 1900 s−1. The relationship between the holding time and droplet size is presented in Fig. 5. Here, the holding time ratio is defined as the ratio between the holding time and total driving time of the piezo element in the suction and ejection operations. At each holding time ratio, a plurality of droplets (n = 249 to 432) was imaged using a microscope (VHX-1000, KEYENCE), and the mean droplet diameter and coefficient of variation (CV, defined as the ratio of the standard deviation to the mean) were calculated. The droplet size increased with holding time. At a holding time ratio of zero, a mean droplet diameter of 56 μm was obtained and at a holding time ratio of 0.56, a mean droplet diameter of 71 μm was obtained. This change corresponds to an approximately two-fold change in volume. The holding time ratio and the droplet volume calculated from the droplet diameter have a nearly linear relationship. This suggest that the dispersed phase sucked from the orifice into the collection channel was transported downstream, maintaining its liquid column shape and flow velocity during the holding time; then, droplets were formed. The CV was 5.1% ± 0.4% under all evaluated holding time ratios. Increasing the piezo displacement (the amplitude of the driving voltage applied to the piezo element) also changed the droplet size. However, as the piezo displacement increased, the continuous number of offset pulses was limited by the maximum displacement of the piezo element, decreasing the yield. From the perspective of yield, therefore, it is preferable to change the droplet size based on the holding time. These results indicate that in our system, the droplet size can be adjusted by setting the parameters of the driving waveform.Effective yield and purity
The effective yield and purity were evaluated using a sample mixed with the same polystyrene particles described above (used as target particles) and 3 μm fluorescent polystyrene particles with different fluorescence characteristics (used as non-target particles). When increasing the sample concentration, the average event detection rate increased from 2000 to 6000 s−1. Approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles were sorted in each experiment (n = 3). Fig. 6A and B present parts of the fluorescence images of the droplets obtained at average event detection rates of 2000 and 6000 s−1, respectively, with a target concentration of 48.7%. Almost all droplets contained only a single target fluorescent particle. The effective yield was defined as the ratio of the number of target particles encapsulated as single particles to the number of target particles detected in the selection optical system, and purity was defined as the ratio of the number of droplets containing only single target particles to the number of droplets containing any particles. The results for the effective yield and purity are presented in Fig. 7 and listed in Table 1. At a target concentration of 48.7% and average event detection rate of 2000 s−1, an effective yield of 86.9% ± 0.4% and purity of 99.9% ± 0.1% were obtained. An effective yield of 66.0% ± 0.5% and purity of 99.0% ± 0.3% were obtained at an average event detection rate of 6000 s−1. As shown in Fig. 7, the effective yield decreased with an increase in the event detection rate and target concentration. This trend can be attributed to the restrictions of the continuous response of the fluid as the event rate of the target particles (target rate = target concentration × event rate) increases. An effective yield of 56.7% ± 1.0% was maintained even at a high target detection rate with a target concentration of 96.9% and average event rate of 5300 s−1 by using the designed piezo driving waveform to maximize the response speed of the fluid. Purity was maintained at a high level (>97.9%) for all event detection rates. A breakdown of the number of particles and droplets used to calculate purity is presented in Table 1.
000 particles were sorted in each experiment (n = 3). Fig. 6A and B present parts of the fluorescence images of the droplets obtained at average event detection rates of 2000 and 6000 s−1, respectively, with a target concentration of 48.7%. Almost all droplets contained only a single target fluorescent particle. The effective yield was defined as the ratio of the number of target particles encapsulated as single particles to the number of target particles detected in the selection optical system, and purity was defined as the ratio of the number of droplets containing only single target particles to the number of droplets containing any particles. The results for the effective yield and purity are presented in Fig. 7 and listed in Table 1. At a target concentration of 48.7% and average event detection rate of 2000 s−1, an effective yield of 86.9% ± 0.4% and purity of 99.9% ± 0.1% were obtained. An effective yield of 66.0% ± 0.5% and purity of 99.0% ± 0.3% were obtained at an average event detection rate of 6000 s−1. As shown in Fig. 7, the effective yield decreased with an increase in the event detection rate and target concentration. This trend can be attributed to the restrictions of the continuous response of the fluid as the event rate of the target particles (target rate = target concentration × event rate) increases. An effective yield of 56.7% ± 1.0% was maintained even at a high target detection rate with a target concentration of 96.9% and average event rate of 5300 s−1 by using the designed piezo driving waveform to maximize the response speed of the fluid. Purity was maintained at a high level (>97.9%) for all event detection rates. A breakdown of the number of particles and droplets used to calculate purity is presented in Table 1.
| Target population [%] | Event detection rate [s−1] | Number of droplets containing single target particles | Number of droplets containing multiple particles | Purity [%] |
|---|---|---|---|---|
| 18.2 | 1938 | 402 | 2.0 | 99.5 |
| 3829 | 482 | 3.0 | 99.3 | |
| 6036 | 334 | 4.0 | 99.0 | |
| 48.7 | 2074 | 492 | 0.3 | 99.9 |
| 4280 | 561 | 3.0 | 99.5 | |
| 5959 | 547 | 5.7 | 99.0 | |
| 96.9 | 1946 | 416 | 0.3 | 99.9 |
| 3978 | 689 | 3.7 | 99.4 | |
| 5345 | 663 | 11.7 | 98.3 |
Cell viability and lysis efficiency
PBMCs were used to evaluate cell viability after encapsulation. The cells were stained with Calcein AM, gate setting of the sorting system was set to live cells only, and a plurality of encapsulated cells was imaged using fluorescence microscopy to evaluate viability. Live cells can be recognized as bright small dots, as shown in Fig. 8, because calcein emits fluorescence inside the cells. In contrast, dead cells are recognized as large bright dots because calcein diffuses through the droplets. Multiple fluorescence images were captured and the viability of 4739 cells was evaluated based on the size of the bright dots. The viability was evaluated as 96.4%, indicating that the cells encapsulated by the experimental system maintained high viability. Next, a cell lysis buffer containing a surfactant was added to the sheath fluid and the lysis efficiency of the cells after encapsulation was evaluated. To encapsulate only live cells, cell encapsulation was performed with the same gate setting as when the viability evaluation was performed and cell lysis efficiency was evaluated based on the size of the bright dots in the fluorescence images. For the 4234 cells recognized in the fluorescence images, all bright dots were diffuse and the cell lysis efficiency was evaluated as 100%. The intensity distributions of the scattering and fluorescence signals of the cells obtained from the selection optical system differed between live and lysed dead cells. Because the gate setting only permitted live cells, the cell lysis buffer contained in the sheath fluid did not affect the cells in the sample fluid at the time of selection detection, where the sheath and sample flow formed a laminar flow. Instead, the cells were mixed with cell lysis buffer and lysed within the droplets at the time of encapsulation in the sorting section.Discussion
In this paper, we proposed a microfluidic device that selectively encapsulates single cells with high throughput. In conventional passive devices, to achieve high purity (more than 95%), the average number of cells per droplet λ must be maintained at less than 0.1. In other words, because λ is equivalent to the ratio of the average event detection rate to the droplet generation rate, the event detection rate must be set to less than one-tenth of the droplet generation rate, resulting in very low efficiency of single-cell encapsulation. To improve the efficiency, a particle-ordering technique utilizing Dean flow generated within a spiral microchannel has been developed for passive devices. When the event and droplet-generation rates are nearly equal, a high encapsulation efficiency of 79.2% and a high purity of 96.2% have been reported.23,24 However, the particle-ordering performance is sensitive to parameters such as sample concentration, sample flow rate, and channel dimensions. Moreover, the particle spacing and the droplet-generation rate of the downstream droplet-generation device must be adjusted, restricting usability. By contrast our proposed device can acquire the optical and arrival information of cells using a selection optical system located upstream of the sorting section. Therefore, it can actively encapsulate single target cells. Experimental results show that our droplet generation device, which has a response time of 75 μs, can maintain a high purity (>97.9%), under a wide range of event rates: the average event-detection rate ranged from 2000 to 6000 s−1 (corresponding to λ = 0.15 to 0.45, calculated as the ratio of the event rate to the reciprocal of the response time). Active encapsulation technology can provide flexibility in supply conditions, such as sample and target concentration. Under the experimental conditions with the highest target rate (target concentration of 96.9% and event detection rate of 5300 s−1), an effective yield of 56.7% was obtained. This indicates that our system can encapsulate 2900 cells s−1 and generate more than 10 million droplets containing single cells through continuous operation for 1 h. High throughput is beneficial for the immune repertoire analysis of T and B cells with extremely diverse receptors, large-scale CRISPR screening of cells edited with diverse CRISPR gRNA libraries, and exploration of novel cell types and rare cells from diverse cell populations.Additionally, the optical settings and sensitivity of our sorting system are similar to those of conventional cell sorters; hence, the gating settings used in standard flow cytometry can be used. The use of a multi-parameter sorting system with multiple scattering and fluorescence signals enables the more efficient study of cellular heterogeneity for subpopulations by focusing on specific cell types. For example, it enables the removal of dead cells and analysis of only live cells, as well as the encapsulation and analysis of T cells or B cells from PBMCs containing diverse cell types.
The high viability of cells encapsulated in microchannels can be maintained even after encapsulation based on the physical advantages of a closed cell sorter that does not generate the high pressure or impact associated with droplet flight.25 In addition, the flow sucked into the short orifice along with the cells does not develop, and cells smaller than 10 μm are minimally affected by wall shear stress. In this study, the cells inside droplets were lysed by mixing the cell lysis buffer with the sheath fluid. This indicates that by introducing reagents into the sheath fluid, it is possible for specific cells to react after encapsulation. Based on these findings, our method can be applied to a wide range of single-cell analyses. For example, by taking advantage of the system's high cell viability, it may be applicable to single-cell culture and live cell imaging. The variability in droplet size in our system enables the adaptation of droplet sizes according to the cell type and culture duration with reduced reagent consumption, which is an advantage of single-cell culture. By using sheath fluid containing drugs, it may be possible to analyze drug responses because drugs are mixed with the cells contained in the sample fluid when encapsulated. By using sheath fluid containing antigens, capture beads for secreted molecules, and fluorescent antibodies binding to secreted molecules, it may be possible to identify droplets containing cells with high secretion productivity by aggregating secreted molecules on the beads within the droplets. Although this is possible with existing methods,26–29 our method may be superior in terms of throughput.
Our selective encapsulation system enables cell sorting immediately before encapsulation within a single device. Compared to cell sorting using a separate cell sorter, cell viability can be improved and cell loss can be reduced. It is also possible to simplify complex workflows using multiple instruments, thereby leading to more efficient experiments. Additionally, active encapsulation using our sorting system can overcome the poor encapsulation efficiency of passive devices and achieve an outstanding throughput. With progress in single-cell analysis, the number of cells analyzed has increased annually, reaching 105 cells by 2020. It has also been reported that there is a strong correlation between the number of cell types and number of cells analyzed,13 suggesting that the number of cell analyses is expected to continue to increase. The proposed technology may bring high efficiency to single-cell research, enabling larger-scale analyses with a greater number of analyzed cells.
Conclusions
In this paper, we reported a single-cell encapsulation technology with a sorting function and demonstrated its high encapsulation performance. The proposed method enables high-throughput and high-purity encapsulation by actively encapsulating only single target cells based on the optical information of cells obtained from a selection optical system upstream of the sorting section within the microfluidic chip. Furthermore, a sorting function for cells, variability in droplet size, and ability to mix reagents within droplets enable flexible adaptation to various application requirements. We believe that the proposed device can support advanced single-cell studies. Additionally, we expect that our device will facilitate potential applications that have not yet been foreseen.Author contributions
Conceptualization: M. M. and H. T.; formal analysis: M. N.; investigation: M. N. and M. M.; methodology: M. N., M. M., T. I., and I. H.; project administration: H. T.; software: I. H.; supervision: Y. K.; visualization: M. N.; writing – original draft: M. N., M. M., and Y. K.; writing – review and editing: M. N., M. M., T. I., H. T., and Y. K.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We thank Alexia Devulder for her contribution to the evaluation of cell viability and lysis efficiency, Gakuji Hashimoto for his contribution to designing the mechanical and optical setup of the experimental system, and Tsutomu Maruyama for his contribution to designing the electrical setup of the experimental system.Notes and references
- T. Hayashi, H. Ozaki, Y. Sasagawa, M. Umeda, H. Danno and I. Nikaido, Nat. Commun., 2018, 9, 619 CrossRef PubMed.
- K. E. Yost, A. T. Satpathy, D. K. Wells, Y. Qi, C. Wang, R. Kageyama, K. L. McNamara, J. M. Granja, K. Y. Sarin, R. A. Brown, R. K. Gupta, C. Curtis, S. L. Bucktrout, M. M. Davis, A. L. S. Chang and H. Y. Chang, Nat. Med., 2019, 25, 1251–1259 CrossRef CAS PubMed.
- T. D. Wu, S. Madireddi, P. E. de Almeida, R. Banchereau, Y. J. Chen, A. S. Chitre, E. Y. Chiang, H. Iftikhar, W. E. O'Gorman, A. Au-Yeung, C. Takahashi, L. D. Goldstein, C. Poon, S. Keerthivasan, D. E. de Almeida Nagata, X. Du, H. M. Lee, K. L. Banta, S. Mariathasan, M. Das Thakur, M. A. Huseni, M. Ballinger, I. Estay, P. Caplazi, Z. Modrusan, L. Delamarre, I. Mellman, R. Bourgon and J. L. Grogan, Nature, 2020, 579, 274–278 CrossRef CAS PubMed.
- Z. Zhang, D. Xiong, X. Wang, H. Liu and T. Wang, Nat. Methods, 2021, 18, 92–99 CrossRef CAS PubMed.
- Z. Bai, S. Woodhouse, Z. Zhao, R. Arya, K. Govek, D. Kim, S. Lundh, A. Baysoy, H. Sun, Y. Deng, Y. Xiao, D. M. Barrett, R. M. Myers, S. A. Grupp, C. H. June, R. Fan, P. G. Camara and J. J. Melenhorst, Sci. Adv., 2022, 8, eabj2820 CrossRef CAS PubMed.
- K. A. Jagadeesh, K. K. Dey, D. T. Montoro, R. Mohan, S. Gazal, J. M. Engreitz, R. J. Xavier, A. L. Price and A. Regev, Nat. Genet., 2022, 54, 1479–1492 CrossRef CAS PubMed.
- J. Cheng, G. Lin, T. Wang, Y. Wang, W. Guo, J. Liao, P. Yang, J. Chen, X. Shao, X. Lu, L. Zhu, Y. Wang and X. Fan, Adv. Sci., 2023, 10, e2204484 CrossRef PubMed.
- K. Matula, F. Rivello and W. T. S. Huck, Adv. Biosyst., 2020, 4, e1900188 CrossRef PubMed.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev and S. A. McCarroll, Cell, 2015, 161, 1202–1214 CrossRef CAS PubMed.
- A. M. Klein, L. Mazutis, I. Akartuna, N. Tallapragada, A. Veres, V. Li, L. Peshkin, D. A. Weitz and M. W. Kirschner, Cell, 2015, 161, 1187–1201 CrossRef CAS PubMed.
- G. X. Zheng, J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, T. D. Wheeler, G. P. McDermott, J. Zhu, M. T. Gregory, J. Shuga, L. Montesclaros, J. G. Underwood, D. A. Masquelier, S. Y. Nishimura, M. Schnall-Levin, P. W. Wyatt, C. M. Hindson, R. Bharadwaj, A. Wong, K. D. Ness, L. W. Beppu, H. J. Deeg, C. McFarland, K. R. Loeb, W. J. Valente, N. G. Ericson, E. A. Stevens, J. P. Radich, T. S. Mikkelsen, B. J. Hindson and J. H. Bielas, Nat. Commun., 2017, 8, 14049 CrossRef CAS PubMed.
- L. Valihrach, P. Androvic and M. Kubista, Int. J. Mol. Sci., 2018, 19, 807 CrossRef PubMed.
- V. Svensson, E. da Veiga Beltrame and L. Pachter, Database, 2020, 2020, baaa073 CrossRef PubMed.
- J. Ding, X. Adiconis, S. K. Simmons, M. S. Kowalczyk, C. C. Hession, N. D. Marjanovic, T. K. Hughes, M. H. Wadsworth, T. Burks, L. T. Nguyen, J. Y. H. Kwon, B. Barak, W. Ge, A. J. Kedaigle, S. Carroll, S. Li, N. Hacohen, O. Rozenblatt-Rosen, A. K. Shalek, A. C. Villani, A. Regev and J. Z. Levin, Nat. Biotechnol., 2020, 38, 737–746 CrossRef CAS PubMed.
- T. Thorsen, R. W. Roberts, F. H. Arnold and S. R. Quake, Phys. Rev. Lett., 2001, 86, 4163–4166 CrossRef CAS PubMed.
- L. Xiang, F. Kaspar, A. Schallmey and I. Constantinou, Biosensors, 2021, 11 Search PubMed.
- S. L. Anna, N. Bontoux and H. A. Stone, Appl. Phys. Lett., 2003, 82, 364–366 CrossRef CAS.
- C. Cramer, P. Fischer and E. J. Windhab, Chem. Eng. Sci., 2004, 59, 3045–3058 CrossRef CAS.
- P. Zhu and L. Wang, Lab Chip, 2016, 17, 34–75 RSC.
- S. Moon, E. Ceyhan, U. A. Gurkan and U. Demirci, PLoS One, 2011, 6, e21580 CrossRef CAS PubMed.
- M. Najah, A. D. Griffiths and M. Ryckelynck, Anal. Chem., 2012, 84, 1202–1209 CrossRef CAS PubMed.
- D. Kim, K. B. Chung and T. G. Kim, J. Dermatol. Sci., 2020, 99, 74–81 CrossRef CAS PubMed.
- E. W. Kemna, R. M. Schoeman, F. Wolbers, I. Vermes, D. A. Weitz and A. van den Berg, Lab Chip, 2012, 12, 2881–2887 RSC.
- T. Tang, H. Zhao, S. Shen, L. Yang and C. T. Lim, Microsyst. Nanoeng., 2024, 10, 3 CrossRef CAS PubMed.
- M. Matsumoto, S. Tashiro, T. Ito, K. Takahashi, G. Hashimoto, J. Kajihara, Y. Miyahara, H. Shiku and Y. Katsumoto, Mol. Ther.--Methods Clin. Dev., 2023, 30, 367–376 CrossRef CAS PubMed.
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Nat. Protoc., 2013, 8, 870–891 CrossRef CAS PubMed.
- E. Brouzes, M. Medkova, N. Savenelli, D. Marran, M. Twardowski, J. B. Hutchison, J. M. Rothberg, D. R. Link, N. Perrimon and M. L. Samuels, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14195–14200 CrossRef CAS PubMed.
- H. N. Joensson and H. Andersson Svahn, Angew. Chem., Int. Ed., 2012, 51, 12176–12192 CrossRef CAS PubMed.
- A. Rakszewska, J. Tel, V. Chokkalingam and W. T. S. Huck, NPG Asia Mater., 2014, 6, e133 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |