Open Access Article
Open Access ArticleToward microfluidic continuous-flow and intelligent downstream processing of biopharmaceuticals
Vikas
Sharma†
 a,
Amirreza
Mottafegh†
a,
Jeong-Un
Joo
a,
Ji-Ho
Kang
a,
Lei
Wang
a,
Amirreza
Mottafegh†
a,
Jeong-Un
Joo
a,
Ji-Ho
Kang
a,
Lei
Wang
 b and
Dong-Pyo
Kim
b and
Dong-Pyo
Kim
 *a
*a
aCenter for Intelligent Microprocess of Pharmaceutical Synthesis, Department of Chemical Engineering, Pohang University of Science and Technology (POSTECH), Pohang 37673, Republic of Korea. E-mail: dpkim@postech.ac.kr
bSchool of Chemistry and Chemical Engineering, Harbin Institute of Technology, Harbin 150001, Heilongjiang, P. R. China
First published on 9th May 2024
Abstract
Biopharmaceuticals have emerged as powerful therapeutic agents, revolutionizing the treatment landscape for various diseases, including cancer, infectious diseases, autoimmune and genetic disorders. These biotherapeutics pave the way for precision medicine with their unique and targeted capabilities. The production of high-quality biologics entails intricate manufacturing processes, including cell culture, fermentation, purification, and formulation, necessitating specialized facilities and expertise. These complex processes are subject to rigorous regulatory oversight to evaluate the safety, efficacy, and quality of biotherapeutics prior to clinical approval. Consequently, these drugs undergo extensive purification unit operations to achieve high purity by effectively removing impurities and contaminants. The field of personalized precision medicine necessitates the development of novel and highly efficient technologies. Microfluidic technology addresses unmet needs by enabling precise and compact separation, allowing rapid, integrated and continuous purification modules. Moreover, the integration of intelligent biomanufacturing systems with miniaturized devices presents an opportunity to significantly enhance the robustness of complex downstream processing of biopharmaceuticals, with the benefits of automation and advanced control. This allows seamless data exchange, real-time monitoring, and synchronization of purification steps, leading to improved process efficiency, data management, and decision-making. Integrating autonomous systems into biopharmaceutical purification ensures adherence to regulatory standards, such as good manufacturing practice (GMP), positioning the industry to effectively address emerging market demands for personalized precision nano-medicines. This perspective review will emphasize on the significance, challenges, and prospects associated with the adoption of continuous, integrated, and intelligent methodologies in small-scale downstream processing for various types of biologics. By utilizing microfluidic technology and intelligent systems, purification processes can be enhanced for increased efficiency, cost-effectiveness, and regulatory compliance, shaping the future of biopharmaceutical production and enabling the development of personalized and targeted therapies.
1. Introduction
The application of recombinant DNA technology has rapidly and significantly transformed the global pharmaceutical sector. Biopharmaceuticals, comprising recombinant proteins, cell-based products, and nucleic acid therapies, exhibit a diverse landscape, where monoclonal antibodies (mAbs) lead in both approvals and sales, while COVID-19 vaccine was among the top-grossing individual products.1 There are active licenses for 443 biopharmaceutical products out of a total of 541 licensed products, since the era of commercial pharmaceutical biotechnology commenced with the approval of the U.S. Food and Drug Administration (FDA) on Eli Lilly's recombinant human insulin in October 1982.2 From 2018 to 2022, regulatory authorities approved 197 products, encompassing mAbs, hormone-related products, gene therapies, and vaccines. Mammalian cell systems, predominantly Chinese hamster ovary (CHO) cells, constitute the dominant platform, contributing to 67% of biopharmaceutical production, while for non-mammalian platforms, E. coli leads with 36 products.1In the US, biotech drugs undergo development following regulations outlined in the Food, Drug & Cosmetic Act and the Public Health Service (PHS) Act. Clinical trials are overseen by Investigational New Drug Applications (INDs). Once ready for market, they require Biologics License Applications, which also cover biosimilars under section 351(k) of the PHS Act.3 The emergence of biosimilars has introduced new dynamics to the market, shifting the emphasis from innovative products to efficient bioprocesses. However, two decades ago, the biologics industry prioritized “quality” with limited drugs and slow regulatory approvals. Since then, platform manufacturing processes have become standard, and critical quality attributes (CQA) for biologics are well-characterized with advanced analytical technologies. “Quality by design (QbD)” is now the norm, emphasizing tracking of process parameters and product attributes across manufacturing. In the competitive biologics sector, cost reduction and speed to market are critical, necessitating a shift beyond quality to cost-effectiveness. Continuous manufacturing offers a clear path for radical cost improvements. Dr. Janet Woodcock's 2007 call for accelerated adoption led to the first continuous pharmaceutical process in 2011 and continuous biopharmaceutical processes in 2012, marking a pivotal step towards efficient and economical manufacturing.4–6
Downstream processing is a pivotal stage in the production of biotherapeutics, representing a significant cost component and being critical for ensuring the quality. It encompasses multiple purification steps to obtain a specific target bio-product with high purity by removing the impurities, and minimizing the risk of undesired immune response from the contaminants. High-quality therapeutic bio-products with intact structure and appropriate modifications ensure the successful production of biopharmaceuticals. Regulatory agencies, such as the FDA and the European Medicines Agency (EMA), have stringent requirements for the quality, safety, and consistency of biotherapeutics.3 Current production strategies require many purification steps and lack efficiency in many aspects. Developing efficient downstream processing methods is critical for streamlining the production of biopharmaceuticals that can reduce production costs and increase accessibility to novel biopharmaceuticals. Furthermore, as the field continues to diversely evolve, rapid optimization of downstream processing methods would be important in realizing the full potential of these therapies. Advancements in science and technology enable the adoption of continuous bioprocessing, and there is a need for further development, including hardware–software interfacing. The implementation of continuous processing can occur at different points in a drug product's lifecycle, such as prior to an IND, during development, or after marketing approval. Continuous-flow microfluidic platforms have shown great potential in serial process intensification of reaction and separations, including purification of biomolecules, with its advantages of mixing efficiency, as well as controlled and tunable setups with high precision and accuracy.7 Achieving a fully continuous, end-to-end bioprocess for biopharmaceutical production still remains a challenge. The development of intensified processes integrated with process analytical technology (PAT) tools facilitates the production of high-quality drugs, providing benefits to both industry and patients.3 Smart biomanufacturing systems streamline the biological processes by enabling the real-time monitoring, precise control and optimization of critical process parameters (CPP). These systems represent the convergence of AI-based systems, automated platforms, and the generation of self-driving biomanufacturing systems, offering a comprehensive solution for the biopharmaceutical industry.8
Hence, this perspective review delves into the pivotal role of microfluidic approaches in the downstream process of biopharmaceuticals throughout microscale unit operations to elevate the efficiency and control of the purification process. However, there is a scarcity of information availability in this field of research, and the existing reviews emphasized on the development of continuous integrated purification processes or specifically discussed the microfluidic unit operations. In turn, this comprehensive review offers a collective approach, exploring the emerging frontier of smart biomanufacturing systems and their potential, and providing insights into the synergistic advancements in these interconnected fields. Our aim is to contribute a holistic perspective that encompasses the evolving landscape of microfluidics, continuous processing, and the integration of intelligent biomanufacturing systems, ultimately paving the way for more efficient and sophisticated biopharmaceutical purification strategies.
2. Current biomanufacturing strategies and associated challenges
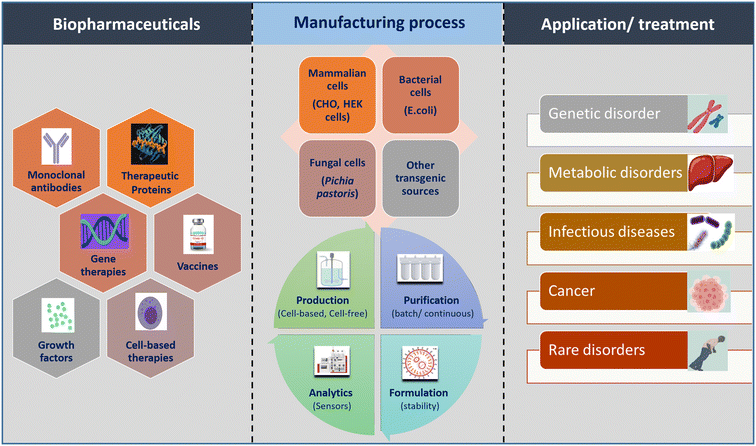
In the realm of biopharmaceutical manufacturing, the promise lies in precisely controlled, miniaturized microfluidic platforms. However, a solid grasp of the foundational unit operations involved in processing crude biotherapeutic extracts is essential. This section meticulously examines the steps and challenges of conventional processing, assessing their adaptability to microfluidic systems. This exploration serves as the bedrock for our subsequent discussion on how microfluidics can revolutionize biopharmaceutical production, particularly for recombinant proteins and mRNA therapeutics.A biopharmaceutical manufacturing process comprises two integral components: upstream processing and downstream processing. The specific steps within the process are contingent upon the type of biopharmaceutical under consideration, categorizing it into cell-based manufacturing and cell-free manufacturing. Fig. 1 provides a brief overview of biopharmaceuticals, illustrating production strategies and applications. The production of cell culture-based therapeutics, including mAbs, therapeutic proteins, and growth hormones, involves culturing of various cellular systems. In the upstream phase, suitable cells (primarily mammalian, bacterial, or yeast cells), cell culture media for nutrient feeds, and bioreactor vessels of varying volumetric scales are selected to provide optimal conditions (temperature, pH, dissolved oxygen, and CO2 levels). These conditions are essential for sustaining cell growth and producing the desired product at high yield.9 In cell-free biotherapeutics production, such as mRNA-based biologics, the enzymatic in vitro transcription (IVT) process replaces the fermentation process. Components like the DNA template, ribonucleotide triphosphates (rNTPs), transcription buffer, and polymerase enzymes are introduced into a bioreactor with specific concentrations, followed by temperature maintenance. The choice of polymerase (T7, SP6, and T3) depends on the DNA template's promoter type for IVT-based mRNA synthesis.10 The absence of cell-derived impurities enhances mRNA manufacturing feasibility and safety.11,12 DNA templates are typically obtained by linearizing purified plasmids or amplifying regions of interest using polymerase chain reaction (PCR). Incorporating a cap analog and poly-A tail at the 5′- and 3′-ends, respectively, boosts mRNA stability and translation efficiency. These steps, co-transcriptional or through additional enzymatic processes post IVT, are being optimized for increased efficiency, mRNA stability, reduced immunogenicity, and enhanced translation efficiency.10
The upstream process generates a complex crude mixture containing the desired therapeutic product. However, extensive downstream purification is necessary to isolate the product and achieve the high purity standards required for a marketable drug, as mandated by regulatory guidelines. The crude mixture undergoes downstream processing involving clarification, capturing, polishing, and formulation steps. Selection of separation methods depends on the product and impurity properties, including charge, molecular weight, solubility, and stability. This process, typically comprising 8–10 steps, contributing to increased production costs and inefficient unit operations, leads to significant loss of the targeted product at each step. For biopharmaceuticals produced from cell culture and fermentation processes, the consideration of cellular location (intracellular or extracellular) is also crucial, where intracellular products necessitate an additional step of cell lysis.13,14
Clarification, the initial purification step for cell-based biopharmaceuticals, segregates the desired product from impurities like cell debris, host cell proteins (HCPs), DNA, and viral particles. Unit operations, including centrifugation, membrane filtration, precipitation, and liquid–liquid extraction, are utilized for this purpose. Controlled centrifugation under low-temperature conditions and specific rotation speeds prevents biomolecule degradation and preserves structural integrity. Membrane filtration serves multiple functions like dialysis, diafiltration (DF), and upconcentration, considering factors such as membrane material, molecular weight cut-off, and crude solution flow direction.15 In protein and enzyme purification, methods like ammonium sulfate and ethanol precipitation are common, often followed by dialysis for impurity removal and buffer exchange.16,17 Aqueous two-phase systems (ATPS) provide an alternative clarification approach, efficiently partitioning the desired product into the aqueous phase while impurities migrate to the other phase. ATPS can be applied directly to the fermentation broth or crude extract, potentially eliminating the need for some centrifugation steps. However, its limited selectivity often hinders ATPS applications in commercial biopharmaceutical processing.18,19
In the capturing step, the unit operation of affinity chromatography employs specially formulated resins with immobilized ligands exhibiting high affinity for the target biomolecule on a solid support. The captured biomolecules can be selectively eluted based on specific binding interactions, facilitating the isolation and concentration of the desired product. For instance, protein A chromatography resins are extensively used in mAb purification due to protein A's high affinity for the Fc region of antibodies.20 In IVT-mRNA capturing, OligodT resins specifically bind to the poly-A tail of the target mRNA sequence. Impurities incapable of binding to affinity resins are washed away, and the desired product is eluted from the resin-packed column using elution buffers, such as salt gradient solutions.10 Although the capturing step significantly reduces undesired molecules, some impurities resembling the desired product may persist in the partially purified solution. Therefore, polishing steps, involving multiple chromatography techniques like ion-exchange chromatography, hydrophobic interaction chromatography, and size-exclusion chromatography, are necessary for the removal of remaining impurities, relying on charge, hydrophobicity, and molecular weight, respectively. The increasing popularity of multi-modal chromatography-based purification is notable, where resins or beads possess multiple features, combining ionic and hydrophobic interactions with the target molecule, thereby reducing the number of purification steps.21 However, the low dynamic binding capacity of resins makes the process inefficient and costly. Advanced ATPS with higher selectivity, utilizing novel phase components such as functionalized polymeric phases and ionic liquid-based phases, have been reported for biomolecule extraction and purification.22 Thus, selective ATPS can be employed for the capturing step of biopharmaceutical purification. Indeed, ATPS hold great potential for selectively purifying biomolecules. However, their current application remains confined to lab-scale processes. By addressing scalability challenges and optimizing their performance, ATPS could become a valuable tool in large-scale biomanufacturing.
While delivering purified products of high quality, factors such as the high expenses associated with individual unit operations, chromatography resins, product degradation, and diminished yields contribute to the elevated cost per dose of biopharmaceuticals. Consequently, research endeavors are directed towards overcoming these challenges and minimizing product costs while upholding quality in downstream processing steps. Continuous manufacturing strategies hold promise to overcome these challenges.
3. Continuous processing of biopharmaceutical manufacturing
Continuous processing of biologics has gained tremendous popularity in recent years due to its high productivity and reduced costs, while ensuring product quality with real-time monitoring.23 Major biologic drug manufacturers are actively involved in developing continuous process platforms for their targeted bio-products. As detailed in Table 1, studies have explored various lab and pilot scale continuous-flow processes for downstream processing of biopharmaceuticals in the macroscale. Individual unit operations (cell disruption, buffer exchange, capturing, etc.) are combined and fully integrated with end-to-end processes encompassing capture, concentration, desalting, and other necessary steps. These techniques employ macrofluidic channels, typically having dimensions in millimeters or centimeters.| Continuous process (macroscale) | Application | Challenges/limitations | Ref. |
|---|---|---|---|
| Focused acoustics based homogenization | Recovery of antibody fragments, cell disruption | Possibility of degradation of biomolecules | 24, 25 |
| Filtration (microfiltration/ultrafiltration) | Buffer exchange and upconcentration | Fouling | 26, 27 |
| Three stage countercurrent diafiltration | Buffer exchange, antibody formulation | Fouling | 28 |
| Precipitation | Upconcentration and solubility based impurity removal | Inefficient mixing and time consuming | 29 |
| ZnCl2 based precipitation | Capturing of mAbs | Low yield (70%) | 30 |
| Precipitation (coiled flow inversion reactor) | Impurity removal from cell culture supernatant, high productivity | Loss of product (10%) | 31 |
| Tabular plug-flow crystallizer | Protein crystallization | — | 32 |
| Integrated precipitation and tangential flow filtration | Upconcentration and washing of antibodies, virus inactivation (antibody purity 97%) | Loss of product (5%) | 33 |
| ATPS | Partitioning of biomolecules | Inefficient phase mixing and time consuming phase separation | 34, 35 |
| ATPS – packed differential contactor (pilot scale) | Extraction of human IgG from CHO cells supernatant | Low recovery yield | 36 |
| Combination of functionalized magnetic nanoparticles and aqueous micellar two-phase system (15 L scale) | Affinity-partitioning of proteins (antibody fragment) from crude extract | Low extraction efficiency (70%) | 37 |
| Coiled flow inverter | Low pH viral inactivation (14.5 min) | — | 38 |
| Chromatography (affinity, ion-exchange, hydrophobic interaction and size-exclusion) | Selective purification of biomolecules, high feasibility for continuous operation | High cost, low dynamic binding capacity, require repeated multiple steps for high purity | 23 |
| Periodic counter-current chromatography | Capturing of proteins (enzymes and antibodies), cost reduction, low buffer consumption | — | 39, 40 |
| Anion and cation exchange chromatography (staggered cycle operation) | Desalting of refolded protein solution, enhance performance of subsequent purification steps | High cost | 41 |
| Multi-column protein A based chromatography | Model based study, effect of particle size on protein capture | Require additional experimental validation | 42 |
| Monolithic anion exchange chromatography (SMB-based) | Purification of cell culture derived influenza virus (vaccine purification) | Require additional steps of subsequent removal of DNA contamination | 43 |
| Twin-column multi-column countercurrent solvent gradient purification (MCSGP) | Isolation of charge isoform of mAbs, removal of multiple impurities | Loss of product (10%) | 44 |
| Integrated multistep lab-scale process | End-to-end monoclonal antibody production | Some steps are semi-continuous | 45 |
| Twin-column counter-current chromatography | mAb capture, high productivity | — | 46 |
| Countercurrent tangential chromatography system | Capture and polishing of mAbs, high productivity | — | 47 |
| Activated carbon and cation exchange resin based integrated process | Purification of mAbs | Low yield (80%) | 48 |
| Combination of inclusion body solubilization and SMB-SEC (lab scale) | High throughput and productivity, low buffer consumption | Semi-continuous | 26 |
In spite of great potential, the path toward continuous integrated bioprocessing remains unclear for the biologics industry due to legacy infrastructure, process integration challenges, vague regulatory guidelines, and a diverging focus toward novel therapies (Rathore et al. 2023).6 The potential benefits of developed continuous downstream processes, coupled with the acknowledged limitations of current methods, underscore the imperative for research and implementation of innovative approaches. In this context, the intrinsic advantages of microfluidic platforms, including efficient mixing, tunable device design and fabrication, precise control and process intensification, have emerged as a promising and innovative approach to solving challenges associated with continuous downstream processing of biopharmaceuticals. This can provide optimized production efficiency by making the manufacturing process more simplified, robust and cost-effective.
4. Harnessing microfluidic technology for enhanced bioprocesses
Microfluidic systems have garnered substantial attention across various domains, encompassing chemical synthesis, materials science, biotechnology, and biopharmaceutical manufacturing, for both upstream and downstream processes.49,50 Distinguished by their reduced channel dimensions (diameter < 1 mm), microfluidic modules display augmented surface-to-volume ratios, resulting in enhanced efficiency of mass and heat transfer as compared to conventional batch modules.51 Furthermore, the strategic placement of microstructures (as passive mixers) within microfluidic channels promotes vortex flow, leading to increased mixing efficiency within a shorter timeframe in comparison to batch-scale modules with significant dead volume.52,53 Consequently, the utilization of microfluidic systems enables the attainment of swift and highly efficient synthesis and separation, surpassing traditional methods, and concurrently ensures enhanced flexibility and reproducibility.54 Additionally, microfluidic systems offer the capability to precisely adjust multiple process variables to achieve optimized conditions.55 In addition to this, microfluidic single-cell analysis offers unparalleled resolution, enabling precise examination of individual cells. It plays a crucial role in upstream drug development by providing insights into cellular responses. Deciphering cellular heterogeneity is a critical challenge, and microfluidics allows manipulation of minute fluid volumes and individual cells. Interrogating individual cells unveils population variations, aiding efficient analysis. Microfluidic platforms integrate functionalities like cell sorting and analysis on a single chip. Microraft arrays (MRAs) enable high-throughput analysis of stem cell–niche interactions and CAR-T cell functions. Laser-induced selective detachment, cloning, and spatiotemporal profiling are also valuable approaches.56–60Efficient processing of small-volume samples, including the preparation or/and analysis of limited quantities of biopharmaceuticals such as recombinant proteins, is a crucial need. Operating at a reduced scale is also imperative for space- and cost-effective screening of protein activity and for evaluating the conditions required for larger-scale manufacture, including expression, purification, and assay conditions.61 Laboratory-scale research has extensively explored microfluidic unit operations for bioprocessing. These operations encompass cell lysis, cell sorting, extraction, partitioning, buffer exchange, and purification of diverse biological products—ranging from enzymes and proteins to antibodies, nucleic acids, and viruses (see Table 2). Our perspective aims to harness the potential of microfluidics and continuous processing to create a miniaturized, integrated downstream processing platform (Fig. 2).
| Microfluidic process | Application | Advantages | Ref. |
|---|---|---|---|
| Microscale cell lysis methods | |||
| NH4Cl mediated micro scale cell lysis device (theoretically milliliter scale) | Erythrocyte lysis and removal, leukocyte isolation | Rapid, ∼100% recovery of leukocytes and reduced duration of exposure to isotonic solution | 64 |
| Nanowire-mediated cell lysis | Bacterial and yeast cell membrane disruption, identification of cells | Non-penetrating approach, contamination free | 65 |
| Ultra-sharp silicon nano-blade arrays for (cell lysis chip) | Intracellular protein extraction | Rapid lysis, simple and cost-effective, does not require additional reagents, low dead volume | 66 |
| Silicon nanospike membrane for cell lysis (electrochemical etching) | Extraction of intracellular proteins and nucleic acids | Rapid and high throughput | 67 |
| Nanostructured barbs with deep reactive ion etching | Accessibility of intracellular proteins | Simple method, chemical free | 68 |
| Multi-turn serpentine microchannel with an attached resistive heater | Release of intracellular nucleic acids and proteins | Portable, rapid and low cost, controlled exposure to heat | 69 |
| Silicon–diamond microcantilever heaters | Thermal lysis of fibroblast and bacterial cells | Uniform temperature distribution, rapid lysis in shorter duration of 15 s (93 °C) | 70 |
| Microscale filtration techniques | |||
| Polydimethylsiloxane (PDMS) tangential flow microfiltration device | Viral separation and concentration | Nearly 100% permeation of viral particles | 77 |
| Polymethylmethacrylate (PMMA) tangential flow microfiltration device (serpentine channels) | Purification of exosomes, removal of proteins | Fast, enhanced efficiency and recovery | 78 |
| Ultrathin nanoporous silicon nitride based tangential flow filtration system | Processing concentrating protein solutions, capturing of extracellular vesicles | Low membrane fouling and possibility of extension to macroscale process | 82 |
| Two membrane ultrafiltration/diafiltration (UF/DF) module (lab scale) | Upconcentration and buffer exchange | Salt reduction to 47%, simple yet simultaneous process | 83 |
| Microscale aqueous two phase system (ATPS) | |||
| Three inlet and single microchannel based ATPS | Partitioning of FITC tagged biomolecules | Low stabilization time, compatible for wide range of biomolecules with differences in molecular weight and pI | 85 |
| Three inlet, serpentine channel with two outlets based ATPS (microscale) | Purification of tagged proteins (glutathione S-transferase) from E. coli lysates | Automated, rapid and high throughput | 86 |
| Combined ATPS and ionic-liquid two phase system | Separation of light sensitive bacteriorhodopsin followed by desalting | Integrated purification and dialysis steps | 87 |
| PDMS-device for ATPS | Extraction of tagged IgG | Reduction in operation time, suitable for process optimization | 88 |
| Co-axial capillary device for co-laminar flow ATPS | Separation of bovine serum albumin (BSA) | Rapid and controlled mass transfer area and time, enhanced recovery | 89 |
| Two module based aqueous two phase extraction | Matrix extraction and analyte pre-concentration for immunoassay | On-chip sample preparation, matrix cleaning, concentration and identification | 90 |
| Y-shaped pressure driven ionic liquid based ATPS | Separation of BSA | High efficiency due to low dynamic viscosity of ionic liquid rich phase | 91 |
| Mixer-settler design based ATPS (microscale to bench scale) | Recovery of human IgG from cell supernatant | Multi-stage, high recovery of antibodies | 92 |
| Glass chip with Y- and Ψ-branched ATPS | α-Amylase extraction | High efficiency, faster equilibrium | 93 |
| ATPS | Purification of membrane proteins from crude cell extract | Faster extraction with high efficiency, minimal emulsification | 94 |
| Glass structured device for ATPS with double interface | BSA recovery | Parallel flow pattern and enhanced recovery | 74 |
| Microscale chromatography modules | |||
| Three chambered bead packed chromatography system | Screening of various multimodal ligands, purification of labelled IgG and BSA | Faster purification along with parallel analysis of multiple conditions | 76 |
| Miniaturized ion exchange, size exclusion and affinity chromatography modules | Purification of recombinant proteins | Highly efficient integrated purification system | 61 |
| PDMS chip packed with methacrylate monolithic polymers (weak anion-exchange) | Investigation of binding capacity for purification of proteins | Rapid separation process | 101 |
| Copolymeric immobilized metal affinity (IMA) adsorbent packed microfluidic device (microgram capacity) | Capturing of histidine tagged recombinant fusion proteins, development of sensing tools | High throughput and in-process monitoring, selective product capture | 106 |
| Ion exchange chromatography with packed polydispersed porous agarose beads (microscale glass chip) | Dynamic binding capacity measurements | 75 | |
| Microfluidic size exclusion chromatography (μSEC) (nano-liter scale) | Isolation of extracellular vesicles from endogenous proteins in biological samples | Automated, integrated and rapid process | 95 |
4.1 Microfluidic modules in biopharmaceutical downstream processing
Fig. 3 demonstrates lab scale microfluidic based unit operations documented for the processing of various biomacromolecules. These reported microfluidic systems investigate different methods of cell lysis, filtration techniques, ATPS and chromatographic separations. Grigorov et al. and Islam et al. described various microscale and milli scale cell lysis methods that are based on chemical, mechanical, electrical, thermal, and laser approaches, specifically designed to release intracellular components of the cell.62,63 The reports indicate that microfluidic reactors substantially decrease the necessary time for lysis, scaling down to the minute level owing to their high mass transfer efficiency, without affecting the structural integrity of the desired components such as DNA or RNA. This approach has also demonstrated cost-effectiveness, characterized by affordable instrument pricing, ease of handling, and high efficiency. A representative microfluidic chemical lysis scheme is illustrated in Fig. 3(a), which has the ability to process milliliters of whole blood for the isolation of leukocytes. The developed microfluidic device lyses and removes erythrocytes from whole blood to achieve nearly 100% recovery of leukocytes that were promptly retuned to physiological conditions.64 Moreover, nano-structure arrays are reported to enable cell lysis without the need for additional reagents or external forces, utilizing approaches such as engineered nanowires, ultra-sharp silicon nano-blades, silicon nanospike membrane and nanoscale barbs.65–68 Compared to other lysis methods, using nanoscale barbs increased the accessibility of total protein and hemoglobin as measured by absorption, from 1.9% and 3.2% to 4.8% and 7.5%, respectively. Microfluidic systems excel in thermal cell lysis, boasting high heat transfer efficiency for precise temperature control, preventing damage to target proteins.69 Privorotskaya et al. utilized silicon-diamond based microcantilever heaters for rapid lysis of bacterial and fibroblast cells.70 A notable study utilized a magnetic field for wireless induction heating in microfluidic channels for the extraction of DNA and RNA from E. coli cells.71 However, microfluidic-based cell lysis, primarily designed for single-cell analysis and diagnostics, faces challenges for long-term continuous operation due to potential clogging issues in small channels.64,72 Moreover, mechanical and thermal lysis methods, while offering continuous operation, suffer from high power consumption and expensive system setups.63 Existing microfluidic devices for cell lysis often function as standalone units, highlighting the need for a comprehensive approach that not only lysates cells to eliminate unwanted substances but also separates target products with minimal contamination. Achieving these objectives requires integration with additional purification techniques like tangential flow filtration, while minimizing cell debris adhesion to enable continuous purification.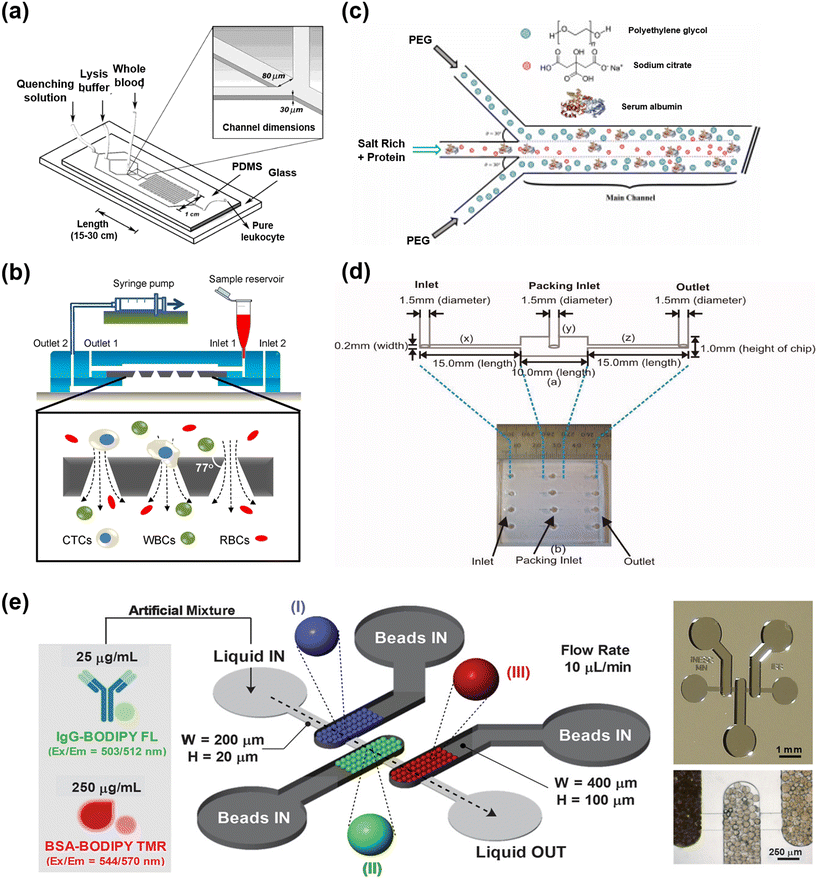 | ||
| Fig. 3 The representative concepts of the microfluidic-based bioprocessing module. (a) Chemical cell lysis: device design and construction to perform rapid lysis of whole blood to obtain pure leukocyte populations, adapted from ref. 64 with permission from American Chemical Society, Copyright 2004; (b) tangential flow filtration (TFF): schematic diagram of the TFF module integrated with a conical-hole filter and experimental setup for capturing circulating tumor cells, adapted from ref. 73 with permission from Springer Nature, Copyright 2014; (c) aqueous two phase system (APTS): schematic diagram of double interface laminar flow in a microfluidic device for the recovery of BSA, adapted from ref. 74 with permission from Elsevier B.V., Copyright 2021; (d) ion-exchange (IEX) chromatography: schematic diagram of a single microfluidic column. Photograph shows the four parallelized columns in a single chip, adapted from ref. 75 with permission from American Institute of Chemical Engineers, Copyright 2009; (e) multimodal chromatography (MMC): schematic illustration of the microfluidic device with three different types of chromatography beads, labelled as I–III (MabSelect SuRe, Capto MMC and MEP HyperCel) packed in series to purify IgG-BODIPY and BSA-BODIPY TMR. Microscopy image of SU-8 mold of microfluidic structure (top of right side) and polydimethylsiloxane (PDMS) structure showing the beads packed well inside the chamber (bottom of right side), adapted from ref. 76 with permission from Wiley-VCH Verlag GmbH & Co. KGaA, Copyright 2019. | ||
Various microfluidic-based membrane filtration processes play a crucial role in the separation and purification of biological products (Table 2). Notably, microfiltration (MF), with a pore size larger than 0.1 μm, was employed for the separation of large biomaterials such as cells, bacteria, and colloids. Additionally, ultrafiltration (UF), with a pore size ranging from 10 nm to 100 nm, is a fundamental unit operation in biopharmaceutical production downstream processes, facilitating protein concentration, virus removal, and buffer exchange.77,78 In a microfluidic system, small molecules like ibuprofen racemate undergo simultaneous enzymatic esterification and chiral-specific separation in a triple-laminar flow of organic, ionic liquid, and aqueous phases as a pseudo-membrane.79 In several studies, TFF was demonstrated as an alternative to dead-end filtration, facilitating the sustained maintenance of high flux by minimizing concentration polarization and cake formation.80 In a study, a modular reactor and in-line TFF microscale system has demonstrated complete substrate conversion, complete enzyme retention and prevention of macromolecule buildup on the membrane.81 A highly efficient size dependent capturing of circulating tumor cells from a blood sample was demonstrated using an integrated microfluidic system equipped with microfilters of conical-shaped holes, as presented in Fig. 3(b).73 Ultrathin nanoporous silicon nitride membranes (NPN) are reported to exhibit high critical flux in concentrated protein solutions, making them ideal for microfluidic TFF, whereas micron-thick membranes perform poorly in this context. The study showcases that NPN with an average 60 nm pore size can process highly concentrated BSA solutions (up to 60 mg mL−1) at 30 μL min−1 without fouling.82 The elevated flow speed in the UF/DF system resulted in minimal concentration effects, requiring frequent recirculation in a loop, increased energy consumption and risking temperature fluctuations. Additionally, the high flow speed and frequent recirculation may induce elevated shear stress on dissolved substances, potentially leading to foaming issues and damage or denaturation of sensitive biomolecules.83 The 3D printed single pass UF/DF module allows continuous concentration of biomolecules and simultaneous reduction of salt buffer, demonstrating a factor of 4.6 protein concentration while reducing salt content to 47%. Despite concentration polarization effects in higher factors, the module's simple design and simultaneous UF/DF capabilities make it economically feasible for small-scale applications.83 Hence, the development of microfluidic or miniaturized SPTFF modules is suggested to offer economic advantages for future downstream processes.
The benefits of precise control in microfluidic platforms and their high mass and heat transfer characteristics have been widely employed in ATPS, as listed in Table 2, for the separation and purification of biomolecules.7 In a microfluidic ATPS, phase components which include polymers (PEG, dextran, etc.), salts (phosphate, sulphate, etc.) or ionic liquids are supplied into the micro-scale inlet channels along with the mixture of biomolecules such as crude extract, in a laminar or serpentine flow. Several efficient extractions have been reported so far, where successfully purification of proteins such as bovine serum albumin (BSA), lipase, α-amylase, bacteriorhodopsin, antibodies and nucleic acids has been demonstrated.84–93 A schematic diagram of the ATPS with double interface laminar flow in a microfluidic device is displayed in Fig. 3(c) for the recovery of BSA. The results suggested that increasing the channel length from 4 cm to 8 cm enhances the BSA recovery from 41.8% to 71.3%.74 However, most of the microfluidic based ATPS were applied for analytical purposes, rather than the continuous purification of targeted bio-products. This method demonstrates rapid extraction efficiency compared to traditional batch-based ATPS, attributed to the high material transfer efficiency between the two phases. As the two phases are inherently well-separated, the need for an additional phase separation process is eliminated. The expeditious and highly efficient purification of membrane proteins, with an extraction efficiency of 90%, was achieved in 7 s through the utilization of a microfluidic continuous-flow ATPS.94 However, maintaining laminar flow poses limitations on increasing the flow rate, necessitating precise flow control due to the substantial viscosity difference between polymeric solutions and salt solutions.7 Hence, to enhance processing capacity and achieve practically feasible continuous purification, it is imperative to develop scalable numbering-up microfluidic systems capable of conducting continuous extraction and phase separation under diverse flow conditions, bypassing the need for a separate phase separation process. Successfully accomplishing this necessitates an in-depth investigation into phase inversion and emulsification phenomena, and the development of precise microfluidic systems capable of inducing mixing without relying on these phenomena.
Chromatography typically serves as the primary purification step to enhance purity and decrease volume, but it entails high costs, constituting a substantial portion of total production costs for therapeutic proteins. Chromatographic process development can be time-consuming and involves large resin quantities. Consequently, there is a growing interest in identifying cost-effective techniques for chromatographic process development without compromising accuracy.95 Leveraging microfluidic devices can facilitate efficient chromatography processes, although numerous challenges remain to transition from mere analysis and diagnosis to practical purification. To achieve continuous purification, the applicability of multicolumn chromatography methods such as SMB and PCC requires evaluation.96 Furthermore, for the swift purification of large volumes, membrane chromatography could be a promising approach.97,98 Meanwhile the development of microfluidic multimodal chromatography strategies is suggested to be considered to further minimize the number of chromatography processes.99 Recent advancements involve the integration of microfluidic systems with automation technology, enabling high-throughput screening for the analysis of various process parameters and product quality.54,100 In various reports (Table 2), several efficient microfluidic chromatographic platforms have been developed for the separation of biomacromolecules which include ion-exchange chromatography,61,101 hydrophobic interaction chromatography102,103 and affinity chromatography.104–106 The investigations showed a noteworthy impact on reagent consumption and sample requirements.76,107 To enhance comprehension, the concept of ion exchange chromatography in microfluidics is depicted in Fig. 3(d). The device possesses a 1 cm length column filled with 70 μm mean diameter porous agarose beads, with the aim of measuring the dynamic binding capacities.75 A multiplex three chambered (8 nL) microfluidic device for multimodal chromatography has been reported as shown in Fig. 3(e), incorporating chromatography ligands with multiple types of interactions into a single chip. The developed system successfully purified IgG from a BSA rich solution in <3 min.76 Microfluidic chromatography is highly considerable for automated flow operations, leading to a substantial increase in purification of the target biomolecule, analysis speed and throughput compared to benchtop methods. This technique has the potential to enhance capabilities such as size fractionation and the removal of high-abundance proteins, steps that are often necessary before on-chip, point-of-care, and mass spectrometric analyses.7 An integrated microscale affinity and size-exclusion chromatographic purification module was developed for the separation of enhanced green fluorescent protein (eGFP) from E. coli lysates. Quantitative measurements indicated an average elution of 650 ± 162 μg eGFP in ∼35 μL of 2 M NaCl.61 The biopharmaceutical industry has recognized the potential of innovative microfluidic designs and their performance. This has led to the adoption of microfluidics for small-scale purification, testing, and data generation, enabling the production of tens to hundreds of thousands of data points per day.100 Furthermore, the uniform distribution of substances within microfluidic channels enhances the reliability of real-time analytical monitoring results and facilitates the application of PAT for biopharmaceutical downstream processes.108 Fouling poses a significant challenge in micro-scale reactors, particularly impacting membrane separation units. Despite ongoing research, fouling remains unsolved, and microfluidics is instrumental in understanding the complex mechanisms governing its progression, offering insights into fundamental interactions and serving as a key tool for dynamic investigation techniques.109
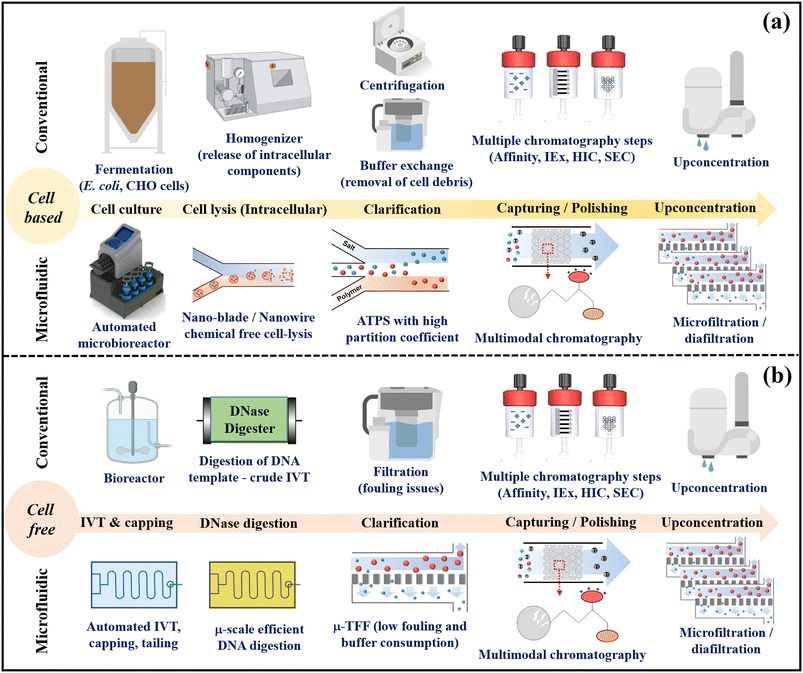
Therefore, microfluidic processes offer a host of advantages and integrated applications. Leveraging these benefits, they can significantly advance continuous downstream processing of biopharmaceuticals, specifically tailored for GMP grade production. Fig. 2 illustrates the conceptual transition from conventional biopharmaceutical downstream unit operations to an integrated microfluidic platform suitable for both cell-based and cell-free biologics production. In cellular-based approaches, automated single-use microfluidic devices include microbioreactors, reagent-free cell lysis units, selective ATPS, mixed-mode chromatography, and TFF units. Microfluidics enables continuous-flow production of bio-products with fewer unit operations. Parallel microscale viral inactivation and removal enhances downstream processing for mammalian cell-based biomanufacturing. In cell-free systems (e.g., mRNA vaccine production), integrated microfluidic devices employ microbioreactors for co-transcriptional mRNA synthesis, capping, poly-A tailing, linked with template digestion units, TFF, mixed-mode chromatography, and upconcentration filtration, achieving continuous-flow purification of mRNA.
These integrated systems offer enhanced control and performance compared to conventional methods. Additionally, the integration of PAT-based sensors at appropriate unit operations, an automated platform, and AI-based optimization ensures the robustness of microfluidic based biomanufacturing. Meanwhile, single use technology enhances the safety and faster production of biologics by reducing the need for sterilization steps, and specific design requirements for a particular manufacturing process. These integrated platforms prove ideal for the production of personalized medicines, particularly for small patient populations. The existing industrial manufacturing infrastructure primarily caters to large-scale production, limiting the production of bio-medicines tailored to individual patients based on their genetic profiles and disorders. In organic synthesis processes using microfluidic platforms, the ‘numbering-up’ method that parallelizes multiple microreactors has been used as an effective approach to increase productivity while maintaining high mass and heat transfer efficiency.110,111 To evenly inject the reactant solution into each microreactor, a baffle disc or a bifurcation type flow distributor was manufactured through 3D printing or lamination of a patterned film and used as a core structure. Using this numbering-up reaction platform, effective scale-up of heterogeneous catalysis, photocatalysis, and ultrafast synthesis was achieved to afford synthetic drugs and their scaffolds.112–115 In a study, the productivity of the letrozole scaffold synthesis using ultrafast synthesis was improved by almost 16-fold from 123.7 g min−1 to 2068.9 g min−1 through the numbering method.114 The successful implementation of the “numbering-up” approach in chemical synthesis can be strategically extended and applied to microfluidic bioprocessing techniques. This approach has the potential to significantly enhance the scalability of these processes.
5. Emergence of intelligent biomanufacturing processes
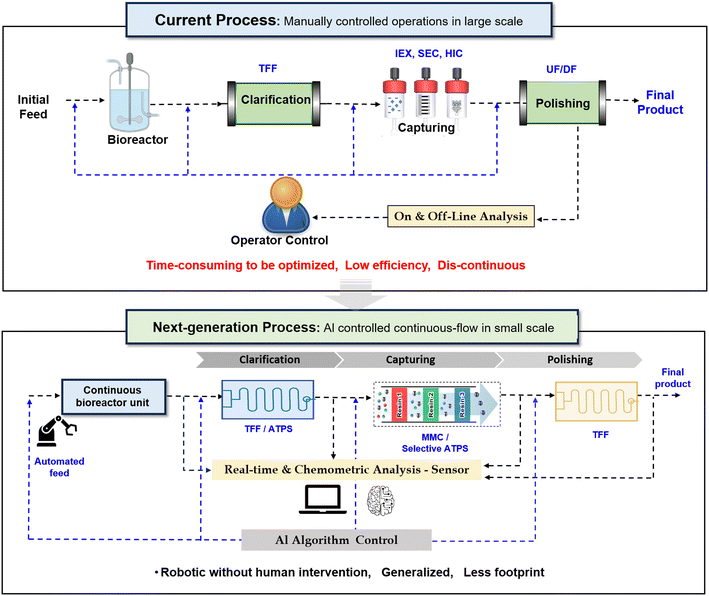
Continuous bioprocessing holds immense promise for the biopharmaceutical industry. However, its true potential lies in seamless integration with intelligent technologies such as artificial intelligence (AI), automation, and real-time PAT. AI processes vast data volumes, revealing complex patterns and adapting parameters in real time. Superior process outcomes and reduced human intervention result from AI's agility. Manual batch processes hinder efficiency and continuity. Labor-intensive sampling, limited analytics, and time-consuming operations persist.Hence, next-generation bioprocessing aims for fully automated, integrated continuous manufacturing with compact, flexible equipment. Advanced PAT tools, multivariate analytics, and adaptive AI control algorithms are essential. Enhanced real-time optimization improves productivity, reduces footprint, and minimizes waste. Automation ensures process robustness and mitigates contamination risks. Key innovations nee rapid at-line analytics, microfluidic technologies (integrated with AI), and single-use components. Intelligent process simulation tools will further revolutionize biopharmaceutical production. The overall schematic comparison of the current and next generation of downstream processing can be seen in Fig. 4.
The development of an intelligent continuous bioprocessing system relies heavily on the implementation of advanced real-time monitoring and PAT. This technology is crucial for improving control mechanisms and data acquisition efficiency in the bioprocessing workflow. In this context, the use of innovative analytical tools becomes paramount. For instance, spectroscopic techniques such as UV, IR, Raman, and online High-Performance Liquid Chromatography (HPLC) are employed for real-time monitoring of biopharmaceutical attributes. These tools provide immediate, accurate data on the bioprocess, enabling swift adjustments to maintain optimal conditions and ensure product quality.116
The integration of AI and automation in upstream bioprocessing has been a focal point of extensive research. This integration has led to the emergence of Bioprocessing 4.0,117–119 a significant advancement that reshapes the field by combining state-of-the-art AI models and automation. In this case, supervised machine learning (ML) models as well as unsupervised models have been successfully applied to upstream bioprocessing showcasing the potential of AI algorithms in this field. As some successful examples, supervised ML models have been used in predicting and influencing the CPP and product CQAs.120 Additionally, the concept of digital twins, also known as the digitalization of bioprocessing, has been employed via supervised ML models leading to creation of a digital replica of the physical bioprocessing system, allowing for real-time monitoring and predictive maintenance, thereby enhancing the efficiency and reliability of the bioprocessing operations.121–124 Supervised ML models have also been utilized as predictive tools for better understanding the underlying factors in the manufacturing of biopharmaceuticals. These models have been applied in various applications, including the manufacturing of mAbs,125,126 enzymes,127 and mammalian cells,128 among others. Aside from the use of supervised ML techniques, a study by Treloar et al. demonstrated the use of deep reinforcement learning as an unsupervised ML method, combined with an automated bioreactor, for controlling microbial co-cultures for the span of around 96 replicate runs.129 The work showed that reinforcement learning effectively maintains target population levels in continuous bioreactors, outperforming traditional control methods. This approach has the potential to optimize microbial community bioprocesses, despite the challenges in assembling such communities for biomanufacturing. The use of continuous integrated microfluidic systems combined with AI and automation can potentially address these challenges. Microfluidic systems can handle small volumes of fluids, allowing for precise control over biological processes and reducing the consumption of expensive reagents.
In the quest to establish intelligent bioprocessing units, it is not only the upstream manufacturing of biopharmaceuticals that needs to evolve as discussed above, but also the downstream processing. The transition towards a more integrated and automated approach in downstream processing is equally crucial for the successful implementation of smart bioprocessing units. To achieve this, several elements need to work in harmony. These include the use of soft sensors, ML algorithms, and automated platforms. Soft sensors, which utilize data-driven or model-based approaches, enable real-time monitoring and control of critical parameters such as concentrations, pH, and conductivities. They provide a bridge between the virtual models and actual processes, enhancing the efficiency and reliability of chromatography and other unit operations. By offering insights into adsorption kinetics, fluid dynamics, and overall process performance, soft sensors facilitate informed decision-making, reduce the necessity for extensive laboratory testing, and significantly contribute to the rapid and cost-effective development of downstream processing units.130 In addition to soft sensors, ML algorithms are also crucial components for constructing smart downstream processing platforms. ML models have vastly been employed to create accurate predictive models for mechanistic modeling131 and parameter optimization132–136 in the chromatography process, for enhancing efficiency and product quality while reducing experimentation time and cost. ML models have also been intensively used for development of membranes for UF136,137 and APTS.138–140
In addition to integrating ML models in downstream processing, the construction of an automated platform capable of smooth coordination with AI algorithms and digitalized processes is of utmost significance. So far, a few studies of fully automated downstream processing platforms have been reported, indicating a lack of comprehensive understanding regarding these platforms. In one study by Winters et al., the authors modified a chromatography system to enable in-line dilution, enhancing the efficiency of automated two-column protein purification.141 This modification allows for the direct loading of a second column from a first column elution, with the pH and ionic strength adjusted for optimal binding enabling the purifications for up to six samples of 1 L volume through two columns without human intervention. In a more extended study by Gomis-Fons et al., as shown in Fig. 5(a), an automated downstream process for the purification and formulation of a recombinant protein at the lab scale in a single chromatography unit was developed.142 This process, which included three bind-and-elute chromatography columns, a flow-through membrane chromatography step, and a final UF-DF step, increased productivity up to 1.09 mg mL−1 h−1 and reproducibility while reducing process time and manual work from almost 2 working days to 1 working day. Another important aspect of the development of an automated small-scale continuous downstream bioprocessing was the buffer management system involving the construction of an automated platform.143 In this work, the authors developed an automated buffer management system for continuous lab-scale bioprocessing, which was able to process 34 and deliver 55 L of buffers, corresponding to 20% of its capacity. The system, integrated with an ÄKTA™ explorer chromatography system and controlled by Orbit software, handled buffer formulation, monitoring, and delivery, demonstrating robust performance and consistency. In another recent study, the authors demonstrated the use of automated liquid handlers (ALHs) to streamline the purification and buffer exchange processes in biopharmaceutical research as shown in Fig. 5(b).144 The authors successfully automated two purification methods, achieving high-quality biologics rapidly without manual intervention, reaching percent recoveries for the three different purified recombinant proteins ranging from 51% to 86%. In addition to the aforementioned studies, Tallvod et al. developed an automatic quality analysis system (QAS) for small-scale biopharmaceutical downstream processes, integrating an ÄKTA Explorer chromatography system and an HPLC system as depicted in Fig. 5(c).145 The QAS was demonstrated in a continuous capture chromatography process, enabling consistent data acquisition without human intervention, thus paving the way for automated process monitoring and control.
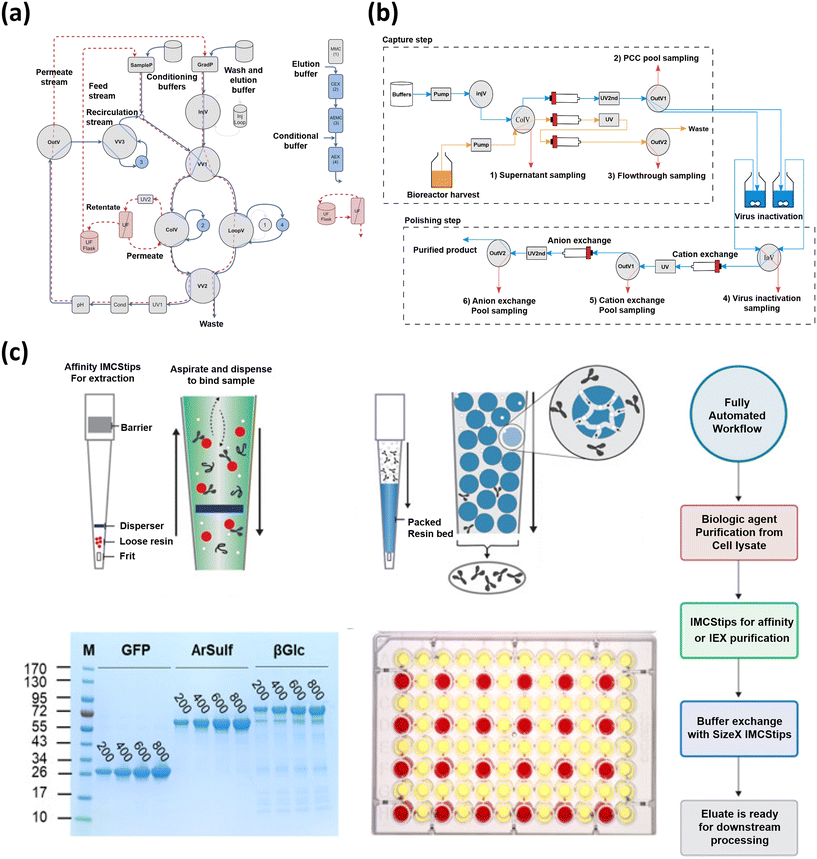 | ||
| Fig. 5 The development and application of automated platforms in downstream processing. (a) Automated downstream process for the purification and formulation of a recombinant protein at the lab scale in a single chromatography unit, adapted from ref. 142 with permission from Elsevier B.V, Copyright 2019. (b) Use of automated liquid handlers (ALHs) to streamline the purification and buffer exchange processes in biopharmaceutical research, adapted from ref. 144 with permission from integrated micro-chromatography systems, Inc. Published by Elsevier Inc., Copyright 2023. (c) Automatic quality analysis system (QAS) for small-scale biopharmaceutical downstream processes, integrating an ÄKTA Explorer chromatography system and an HPLC system, adapted from ref. 145 with permission from Elsevier B.V, Copyright 2023. | ||
Despite the promising advancements in automated downstream processing platforms, several challenges and limitations remain to be addressed. One major challenge is the lack of real-time analysis, which hinders the ability to make immediate adjustments and optimizations during the process. Additionally, the absence of a decision-making policy and limitations in developing an AI-based self-driving system restricts the platform's adaptability and responsiveness to changing conditions. Furthermore, the controlling software's lack of flexibility, being a closed source and difficult to extend, poses challenges for researchers and engineers seeking to customize and improve the system. Lastly, integrating the platform with other independent modules presents challenges in terms of compatibility and seamless communication between different components. Overcoming these challenges will be crucial for the successful implementation and widespread adoption of automated downstream processing platforms in the biopharmaceutical industry, demanding the need for standardization of the systems.
5.1 Toward self-driving downstream processing platforms
Although successful utilization of the previously discussed intelligent system with the use of real-time analytical instruments, ML models, and automated platforms seemed to be promising in further development of bioprocessing in upstream and specifically downstream processing of biopharmaceuticals, the next critical steps to fully harness the potential of intelligent systems are the construction of AI-based self-driving downstream processing platforms and high-throughput experimentation automated platforms. These innovations can generate big data to develop models and share within the community to advance processes. The success of self-driving platforms for organic molecule and material synthesis suggests that similar systems could excel when applied to microfluidic-based continuous downstream processing systems for biopharmaceuticals. Constructing these AI-enabled autonomous platforms and leveraging high-throughput automated experimentation will catalyze breakthroughs in bioprocessing efficiency, precision, and productivity.The recent success of self-driving AI-based platforms in the field of organic synthetic chemistry provides a compelling proof-of-concept and model for the development of similar autonomous systems for biopharmaceutical downstream processing. In organic synthesis, AI-driven robotic platforms have demonstrated capabilities in reaction prediction, optimization, and automation of laboratory procedures.146,147 Of particular relevance is the integration of these platforms with flow and microfluidic systems, enabling real-time monitoring, feedback, and enhanced efficiency compared to batch processes. This parallels the proposed utilization of self-driving units in microfluidic-based continuous bioprocessing platforms, which can precisely control critical parameters like flow rates, mixing, and separation. Such an approach would minimize human intervention and variability while ensuring long-term continuous processing. Additionally, the complex optimization required in organic synthesis, which involves navigating many connected variables, is similar to the complexity in bioprocessing and downstream processing. This underscores the utility of autonomous platforms in both domains. In short, the recent advancements in self-driving organic synthetic platforms provide an optimistic outlook on the potential for similar transformative technologies to be extended to biopharmaceutical downstream processing in the near future.148 The success in organic synthesis highlights the viability of constructing AI-powered autonomous microfluidic platforms to bring enhanced efficiency, consistency, and productivity to biopharmaceutical downstream processing.
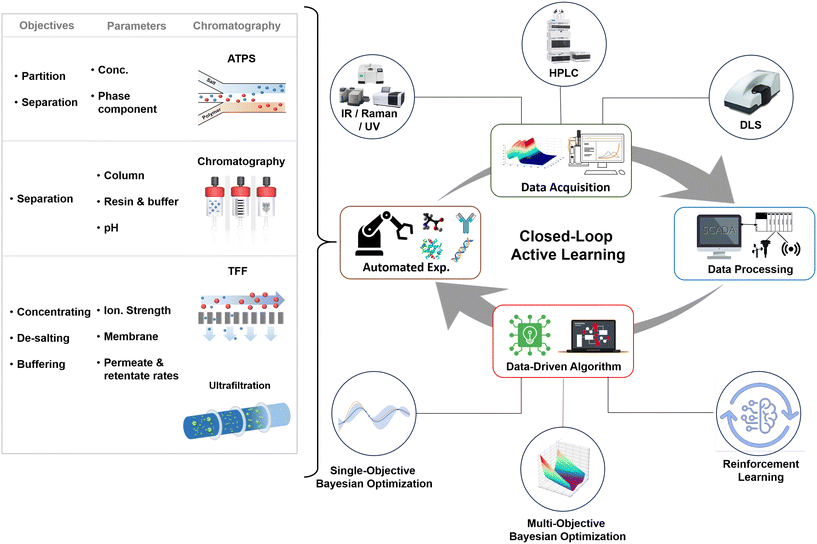
The entire self-driving and self-optimized process can be distilled into four crucial phases. As can be seen in Fig. 6, the process commences with real-time experimentation, where automated exploration of various targets for biopharmaceuticals and downstream processing units takes place. During this stage, the system continually adapts and fine-tunes parameters such as chromatography conditions, filtration rates, and separation techniques to optimize efficiency and product yield. Subsequently, a comprehensive qualification and performance evaluation is undertaken, drawing upon data obtained from real-time analysis such as various spectroscopy techniques (IR, UV-vis, Raman), HPLC, dynamic light scattering (DLS), etc. This includes a variety of sources, including advanced soft sensors149 that computationally estimate critical process variables, and the integration of the internet of things (IoT),150 which facilitates seamless data collection and transmission by interconnecting equipment and devices. The collected data is then rigorously processed through diverse methods, incorporating statistical analysis and data fusion. These techniques cleanse, transform, and extract valuable insights from raw data, serving as the foundation for informed decision-making. In the final phase, AI algorithms come into play as decision-making strategies, aiming to steer the entire microfluidic biopharmaceutical manufacturing process towards an optimal state. These AI algorithms fall into distinct categories, including single-objective151 algorithms that specialize in optimizing specific parameters, multi-objective algorithms152 that balance multiple goals concurrently, and reinforcement learning153 algorithms that introduce adaptability through ongoing interaction and feedback. These AI-driven strategies collectively empower the active learning self-driving self-optimized process, optimizing biopharmaceutical production, and ensuring adaptability to the evolving demands and challenges.
This concept can be applied to various types of microfluidic downstream processing units. In ion exchange chromatography, self-driving AI systems can continuously analyze the binding kinetics of target biomolecules to the ion exchange resin. They can adjust the gradient elution rate based on real-time measurements, optimizing the separation of different proteins with distinct charge properties, integrating a total of 12 different columns and 24 mobile phases that were sequentially operated in a straightforward automated fashion.154 For instance, when purifying mAbs, the AI algorithm can dynamically control the salt concentration in the elution buffer, ensuring that each antibody variant is eluted at the desired point in the chromatogram. In gel filtration chromatography, the AI-driven platform can adapt to variations in sample composition and size. For instance, when purifying VLPs of varying sizes, the AI-assisted system can autonomously select the appropriate column and elution conditions to achieve effective separation. It can also detect changes in sample concentration and adjust the flow rate or sample injection volume in real-time, ensuring that VLPs are consistently purified to meet the required standards. The SEC also can benefit from the discussed methodology by automating the analysis of elution profiles.155 When purifying protein aggregates or viral vectors, the system can identify peak positions and shapes, facilitating the detection of impurities or changes in the product's quality. If anomalies are detected, the AI system can take corrective actions, such as modifying column temperature or flow rate to improve separation or purity.
Aside from chromatography, self-driving membrane-based microfluidic processes can also be considered extremely beneficial in this context. In UF and DF, AI systems monitor the membrane's fouling156 and adjust the transmembrane pressure or flow rate to maintain consistent flux rates and ensure the effective concentration or DF of biomolecules, like mAbs. In TFF also, self-driving processes can bring a high level of precision and automation to the purification of biomolecules.157 A self-driven microfluidic TFF system can continuously adapt and control the filtration parameters in real time to achieve the desired product purity and concentration. This system can monitor the feed flow rate, transmembrane pressure, and filtration flux rate. Based on this data, it can make instant adjustments to the filtration parameters. For instance, if the feed flow rate decreases due to changing characteristics of the feed stream, the AI can increase the transmembrane pressure to maintain a constant flux rate. This ensures that the protein is effectively concentrated without fouling the membrane or risking product loss. Furthermore, advanced AI algorithms, such as recurrent neural networks (RNNs), can predict and mitigate membrane fouling by analyzing historical data and real-time measurements. When the system detects early signs of fouling, it can initiate backflushing or adjust the flow rates to mitigate fouling effects,158 ultimately extending the run time, and improving the overall efficiency of the TFF process. In terms of differential centrifugation, self-driving systems can dynamically adjust the rotor speed, temperature, and centrifugation time to achieve precise fractionation. For instance, when isolating cellular organelles such as mitochondria from a cell lysate, the system can continuously monitor the pellet formation and adjust the centrifugation parameters to ensure optimal organelle recovery. The predictive AI algorithm can analyze real-time data from the centrifuge, detecting the sedimentation rates of different components and adapting the centrifugation conditions accordingly. By doing so, it minimizes the risk of cross-contamination between organelles, ultimately resulting in higher purity and yield of the isolated biomolecules. Additional to the discussed techniques, in ATPS, an AI-driven system can optimize the partitioning of biomolecules between two immiscible aqueous phases. For instance, when isolating a specific enzyme from a cell lysate, the AI can continuously adjust the composition of the two phases, ensuring that the enzyme preferentially partitions into one of them. It can analyze real-time data on the biomolecule's distribution and adapt the phase conditions to maximize the yield and purity of the enzyme, simplifying the purification process and reducing manual intervention.
The closed-loop active learning enables dynamic adaptation to changing constraints and objectives, allowing for optimization over time. With its inherent capability for continuous self-improvement, this framework represents a significant evolutionary leap in bioprocessing. It provides a glimpse into a future characterized by more agile, efficient, and intelligent bio-production processes. The true innovation in constructing this platform lies in the integration of multiple components to achieve a self-driving intelligent system, rather than simply the choice of optimization algorithm (whether classical DoE or AI-based). While advanced algorithms like Bayesian optimization significantly enhance performance, they represent only one element of a breakthrough self-driving platform. This platform's practical success depends on the seamless integration of automated robotic experimentation units, real-time analysis, and robust data processing capabilities.
The future of biopharmaceutical processing lies in a cutting-edge data-sharing platform designed specifically for automated continuous downstream processing. This platform transcends mere data storage; it drives predictive modeling, process development, and performance forecasting. Similar platforms in organic chemistry have revolutionized chemical reaction evaluation.159 While biopharmaceutical predictions may be less complex, this system will fundamentally transform process development, especially for interconnected steps like chromatography. The platform functionality can be broken down into four key stages. First, automated microfluidic experiments: real-time data generation on downstream operations informs bioprocessing trends. Secondly, centralized bioprocessing repository: structured data-sharing foundation. Thirdly, sophisticated ML models: leveraging platform data for accurate outcome prediction.160 Fourthly, empowering researchers and engineers: accelerating process development and modeling complex scenarios. Fig. 7 visualizes the proposed cloud-based platform, illustrating data flow from microfluidic experiments to the centralized dataset, enabling informed decision-making in smart biopharmaceutical downstream processing.
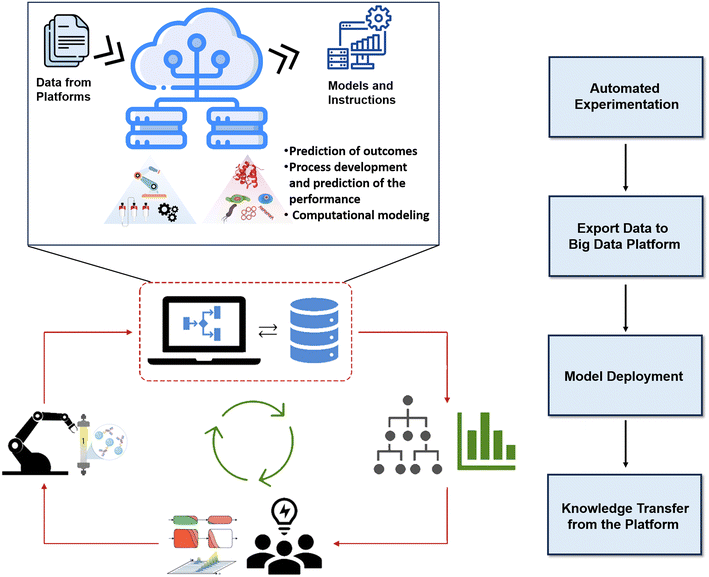 | ||
| Fig. 7 Schematic representation of the proposed cloud-based platform for model deployment and big-data management in a cyber-biophysical biopharmaceutical downstream processing environment. | ||
The future of this field is poised to generate advanced ML models that align with the industry's latest trends. These models, including few-shot learning, address the industry's “big data” challenges, which have traditionally limited the broad applications of ML models in pharmaceutical sciences. Furthermore, the creation of a user-friendly web-based application will democratize access to pre-trained models, making data-driven tools available to a wider range of groups. While the primary focus of self-driving downstream processing units is biopharmaceuticals, the foundational workflows and models developed can be applied in various sectors where formulation is critical, such as agriculture, cosmetics, and paints and coatings. This interdisciplinary potential highlights the broad societal impact of such technological advancements.
Hence, automated, integrated continuous-flow microfluidic platforms would be suitable for various applications, such as faster generation of high-accuracy R&D data to facilitate rapid process optimization through DoE methodologies and development timelines, streamlined clinical trials and manufacturing of emergency-use biopharmaceuticals. This technology has the potential to enable the on-demand production of high quality biologics for pandemics and niche patient populations, particularly those suffering from rare genetic and metabolic disorders. Furthermore, it could pave the way for the application of data-driven autonomous systems in the manufacturing of personalized and precision medicines, ultimately improving cost-effectiveness and accessibility.
6. Concluding remarks
The significance of microfluidic technology in the development of continuous downstream processing for biopharmaceuticals is evident in its ability to enhance control, miniaturization, separation techniques, and process integration while minimizing reagent consumption. The integration of functional microfluidic modules offers a promising avenue for advancing downstream processing in biopharmaceuticals. This combination presents an opportunity for increased robustness, automation, and advanced control, allowing for seamless data exchange, real-time monitoring, and synchronization of purification steps. The resulting improvements in process efficiency, data management, and decision-making contribute to the reliable and streamlined production of high-quality biopharmaceutical products. By emphasizing small-scale continuous, integrated, and autonomous systems, this perspective review underscores the importance, opportunities, and challenges in shaping the future of biopharmaceutical production. Leveraging microfluidic technology and Al-driven autonomous systems not only improves the efficiency and cost-effectiveness of purification processes but also ensures compliance with regulatory standards, paving the way for the development of targeted therapies with enhanced efficacy and fewer side effects. In the context of personalized medicine, the deployment of self-driving downstream processing units and similar autonomous lab technologies can significantly enhance the formulation development process and improve the translation of innovative precision nano-medicines. This provides hope for patients with life-threatening diseases worldwide, underscoring the potential societal benefits of these advancements.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We gratefully acknowledge the support from the National Research Foundation (NRF) of Korea grant funded by the Korean government (NRF-2017R1A3B1023598).References
- G. Walsh and E. Walsh, Nat. Biotechnol., 2022, 40, 1722–1760 CrossRef CAS PubMed.
- I. S. Johnson, Science, 1983, 219, 632–637 CrossRef CAS PubMed.
- A. C. Fisher, M. H. Kamga, C. Agarabi, K. Brorson, S. L. Lee and S. Yoon, Trends Biotechnol., 2019, 37, 253–267 CrossRef CAS PubMed.
- A. Kumar, I. A. Udugama, C. L. Gargalo and K. V. Gernaey, Processes, 2020, 8, 1641 CrossRef.
- NIIMBL National Institute for Innovation in Manufacturing of Biopharmaceuticals, 2022, vol. 1, pp. 1–256.
- A. S. Rathore, G. Thakur and N. Kateja, Biotechnol. Bioeng., 2023, 120, 333–351 CrossRef CAS PubMed.
- F. A. Vicente, I. Plazl, S. P. Ventura and P. Žnidaršič-Plazl, Green Chem., 2020, 22, 4391–4410 RSC.
- C. L. Gargalo, I. Udugama, K. Pontius, P. C. Lopez, R. F. Nielsen, A. Hasanzadeh, S. S. Mansouri, C. Bayer, H. Junicke and K. V. Gernaey, J. Ind. Microbiol. Biotechnol., 2020, 47, 947–964 CrossRef PubMed.
- M. Kesik-Brodacka, Biotechnol. Appl. Biochem., 2018, 65, 306–322 CrossRef CAS PubMed.
- S. S. Rosa, D. M. Prazeres, A. M. Azevedo and M. P. Marques, Vaccine, 2021, 39, 2190–2200 CrossRef CAS PubMed.
- N. Pardi, M. J. Hogan, F. W. Porter and D. Weissman, Nat. Rev. Drug Discovery, 2017, 17, 261–279 CrossRef PubMed.
- S. Pascolo, Expert Rev. Vaccines, 2015, 14, 153–156 CrossRef CAS PubMed.
- B. Owczarek, A. Gerszberg and K. Hnatuszko-Konka, BioMed Res. Int., 2019, 4216060 CAS.
- A. F. Jozala, D. C. Geraldes, L. L. Tundisi, V. D. A. Feitosa, C. A. Breyer, S. L. Cardoso, P. G. Mazzola, L. D. Oliveira-Nascimento, C. D. O. Rangel-Yagui, P. D. O. Magalhães and M. A. D. Oliveira, Braz. J. Microbiol., 2016, 47, 51–63 CrossRef CAS PubMed.
- R. Menzel, I. Pahl, T. Loewe, A. Stuetzer and A. Hauk, Eur. J. Pharm. Sci., 2022, 168, 105982 CrossRef CAS PubMed.
- V. Sharma, G. Pugazhenthi and D. Vasanth, J. Biosci. Bioeng., 2022, 133, 8–16 CrossRef CAS PubMed.
- A. Tscheliessnig, P. Satzer, N. Hammerschmidt, H. Schulz, B. Helk and A. Jungbauer, J. Biotechnol., 2014, 188, 17–28 CrossRef CAS PubMed.
- L. Castro, P. Pereira, M. Freire and A. Pedro, Am. Pharm. Rev., 2019, 1–6 Search PubMed.
- M. Iqbal, Y. Tao, S. Xie, Y. Zhu, D. Chen, X. Wang, L. Huang, D. Peng, A. Sattar, M. A. B. Shabbir and H. I. Hussain, Biol. Proced. Online, 2016, 18, 1–18 CrossRef PubMed.
- A. M. Ramos-de-la-Peña, J. González-Valdez and O. Aguilar, J. Sep. Sci., 2019, 42, 1816–1827 CrossRef PubMed.
- V. Halan, S. Maity, R. Bhambure and A. S. Rathore, Curr. Protein Pept. Sci., 2019, 20, 4–13 CrossRef CAS PubMed.
- F. S. Buarque, A. Carniel, B. D. Ribeiro and M. A. Z. Coelho, Sep. Purif. Technol., 2023, 324, 124539 CrossRef CAS.
- L. Gerstweiler, J. Bi and A. P. Middelberg, Chem. Eng. Sci., 2021, 231, 116272 CrossRef CAS.
- Q. Li, J. P. Aucamp, A. Tang, A. Chatel and M. Hoare, Biotechnol. Bioeng., 2012, 109, 2059–2069 CrossRef CAS PubMed.
- R. J. Carvalho, J. Cabrera-Crespo, M. M. Tanizaki and V. M. Gonçalves, Appl. Microbiol. Biotechnol., 2012, 94, 683–694 CrossRef CAS PubMed.
- M. Wellhoefer, W. Sprinzl, R. Hahn and A. Jungbauer, J. Chromatogr. A, 2013, 1319, 107–117 CrossRef CAS PubMed.
- A. Jungbauer, Trends Biotechnol., 2013, 31, 479–492 CrossRef CAS PubMed.
- M. G. Jabra, C. J. Yehl and A. L. Zydney, Biotechnol. Prog., 2019, 35, e2810 CrossRef PubMed.
- N. Hammerschmidt, A. Tscheliessnig, R. Sommer, B. Helk and A. Jungbauer, Biotechnol. J., 2014, 9, 766–775 CrossRef CAS PubMed.
- G. Dutra, D. Komuczki, A. Jungbauer and P. Satzer, Eng. Life Sci., 2020, 20, 265–274 CrossRef CAS PubMed.
- N. Kateja, H. Agarwal, A. Saraswat, M. Bhat and A. S. Rathore, Biotechnol. J., 2016, 11, 1320–1331 CrossRef CAS PubMed.
- P. Neugebauer and J. G. Khinast, Cryst. Growth Des., 2015, 15, 1089–1095 CrossRef CAS PubMed.
- D. Burgstaller, A. Jungbauer and P. Satzer, Biotechnol. Bioeng., 2019, 116, 1053–1065 CrossRef CAS PubMed.
- F. Ruiz-Ruiz, E. López-Guajardo, P. Vázquez-Villegas, M. E. del Angel-Chong, K. D. P. Nigam, R. C. Willson and M. Rito-Palomares, Chem. Eng. Process., 2019, 142, 107554 CrossRef CAS.
- M. A. Torres-Acosta, K. Mayolo-Deloisa, J. González-Valdez and M. Rito-Palomares, Biotechnol. J., 2019, 14, 1800117 CrossRef PubMed.
- P. A. J. Rosa, A. M. Azevedo, S. Sommerfeld, W. Bäcker and M. R. Aires-Barros, J. Chromatogr., B, 2012, 880, 148–156 CrossRef CAS PubMed.
- I. Fischer, C. C. Hsu, M. Gärtner, C. Müller, T. W. Overton, O. R. Thomas and M. Franzreb, J. Chromatogr. A, 2013, 1305, 7–16 CrossRef CAS PubMed.
- L. David, B. Maiser, M. Lobedann, P. Schwan, M. Lasse, H. Ruppach and G. Schembecker, Biotechnol. Bioeng., 2019, 116, 857–869 CrossRef CAS PubMed.
- R. Godawat, K. Brower, S. Jain, K. Konstantinov, F. Riske and V. Warikoo, Biotechnol. J., 2012, 7, 1496–1508 CrossRef CAS PubMed.
- R. Godawat, K. Konstantinov, M. Rohani and V. Warikoo, J. Biotechnol., 2015, 213, 13–19 CrossRef CAS PubMed.
- N. Walch and A. Jungbauer, Biotechnol. J., 2017, 12, 1700082 CrossRef PubMed.
- D. Baur, J. M. Angelo, S. Chollangi, X. Xu, T. Müller-Späth, N. Zhang, S. Ghose, Z. J. Li and M. Morbidelli, J. Biotechnol., 2018, 285, 64–73 CrossRef CAS PubMed.
- L. M. Fischer, M. W. Wolff and U. Reichl, Vaccine, 2018, 36, 3153–3160 CrossRef CAS PubMed.
- F. Steinebach, N. Ulmer, L. Decker, L. Aumann and M. Morbidelli, J. Chromatogr. A, 2017, 1492, 19–26 CrossRef CAS PubMed.
- F. Steinebach, N. Ulmer, M. Wolf, L. Decker, V. Schneider, R. Wälchli, D. Karst, J. Souquet and M. Morbidelli, Biotechnol. Prog., 2017, 33, 1303–1313 CrossRef CAS PubMed.
- M. Angarita, T. Müller-Späth, D. Baur, R. Lievrouw, G. Lissens and M. Morbidelli, J. Chromatogr. A, 2015, 1389, 85–95 CrossRef CAS PubMed.
- D. Fedorenko, A. K. Dutta, J. Tan, J. Walko, M. Brower, N. D. Pinto, A. L. Zydney and O. Shinkazh, Biotechnol. Bioeng., 2020, 117, 646–653 CrossRef CAS PubMed.
- T. Ichihara, T. Ito, Y. Kurisu, K. Galipeau and C. Gillespie, mAbs, 2018, 10, 325–334 CrossRef CAS PubMed.
- K. S. Elvira, X. C. i Solvas, R. C. Wootton and A. J. Demello, Nat. Chem., 2013, 5, 905–915 CrossRef CAS PubMed.
- A. Enders, A. Grünberger and J. Bahnemann, Mol. Biotechnol., 2022, 1–13 Search PubMed.
- S. Chevalier, J. N. Tourvieille, A. Sommier and C. Pradère, Chem. Eng. J. Adv., 2021, 8, 100166 CrossRef CAS.
- C. Y. Lee, W. T. Wang, C. C. Liu and L. M. Fu, Chem. Eng. J., 2016, 288, 146–160 CrossRef CAS.
- T. Habib, C. Brämer, C. Heuer, J. Ebbecke, S. Beutel and J. Bahnemann, Lab Chip, 2022, 22, 986–993 RSC.
- T. C. Chao and N. Hansmeier, Proteomics, 2013, 13, 467–479 CrossRef CAS PubMed.
- I. Bouhid de Aguiar and K. Schroën, Membranes, 2020, 10, 316 CrossRef PubMed.
- A. D. Gracz, I. A. Williamson, K. C. Roche, M. J. Johnston, F. Wang, Y. Wang, P. J. Attayek, J. Balowski, X. F. Liu, R. J. Laurenza and L. T. Gaynor, Nat. Cell Biol., 2015, 17, 340–349 CrossRef CAS PubMed.
- C. A. LaBelle, R. J. Zhang, S. A. Hunsucker, P. M. Armistead and N. L. Allbritton, Cytometry, Part A, 2023, 103, 208–220 CrossRef CAS PubMed.
- Y. C. Chen, H. W. Baac, K. T. Lee, S. Fouladdel, K. Teichert, J. G. Ok, Y. H. Cheng, P. N. Ingram, A. J. Hart, E. Azizi and L. J. Guo, ACS Nano, 2017, 11, 4660–4668 CrossRef CAS PubMed.
- A. Mocciaro, T. L. Roth, H. M. Bennett, M. Soumillon, A. Shah, J. Hiatt, K. Chapman, A. Marson and G. Lavieu, Commun. Biol., 2018, 1, 41 CrossRef PubMed.
- J. Lee, Z. Liu, P. H. Suzuki, J. F. Ahrens, S. Lai, X. Lu, S. Guan and F. St-Pierre, Sci. Adv., 2020, 6, eabb7438 CrossRef CAS PubMed.
- L. J. Millet, J. D. Lucheon, R. F. Standaert, S. T. Retterer and M. J. Doktycz, Lab Chip, 2015, 15, 1799–1811 RSC.
- E. Grigorov, B. Kirov, M. B. Marinov and V. Galabov, Micromachines, 2021, 12, 498 CrossRef PubMed.
- M. Shehadul Islam, A. Aryasomayajula and P. R. Selvaganapathy, Micromachines, 2017, 8, 83 CrossRef.
- P. Sethu, M. Anahtar, L. L. Moldawer, R. G. Tompkins and M. Toner, Anal. Chem., 2004, 76, 6247–6253 CrossRef CAS PubMed.
- T. Yasui, T. Yanagida, T. Shimada, K. Otsuka, M. Takeuchi, K. Nagashima and Y. Baba, ACS Nano, 2019, 13, 2262–2273 CAS.
- S. S. Yun, S. Y. Yoon, M. K. Song, S. H. Im, S. Kim, J. H. Lee and S. Yang, Lab Chip, 2010, 10, 1442–1446 RSC.
- H. So, K. Lee, Y. H. Seo, N. Murthy and A. P. Pisano, ACS Appl. Mater. Interfaces, 2014, 6, 6993–6997 CrossRef CAS PubMed.
- D. Di Carlo, K. H. Jeong and L. P. Lee, Lab Chip, 2003, 3, 287–291 RSC.
- M. M. Packard, E. K. Wheeler, E. C. Alocilja and M. Shusteff, Diagnostics, 2013, 3, 105–116 CrossRef PubMed.
- N. Privorotskaya, Y. S. Liu, J. Lee, Z. Zeng, J. A. Carlisle, A. Radadia, L. Millet, R. Bashir and W. P. King, Lab Chip, 2010, 10, 1135–1141 RSC.
- S. Baek, J. Min and J. P. Park, Lab Chip, 2010, 10, 909–917 RSC.
- B. P. Mun, S. M. Jung, S. Y. Yoon, S. H. Kim, J. H. Lee and S. Yang, Microfluid. Nanofluid., 2010, 8, 695–701 CrossRef CAS.
- Y. Tang, J. Shi, S. Li, L. Wang, Y. E. Cayre and Y. Chen, Sci. Rep., 2014, 4, 6052 CrossRef CAS PubMed.
- F. Raji and A. Rahbar-Kelishami, Colloids Surf., A, 2021, 624, 126823 CrossRef CAS.
- M. S. Shapiro, S. J. Haswell, G. J. Lye and D. G. Bracewell, Biotechnol. Prog., 2009, 25, 277–285 CrossRef CAS PubMed.
- I. F. Pinto, R. R. Soares, M. R. Aires-Barros, V. Chu, J. P. Conde and A. M. Azevedo, Biotechnol. J., 2019, 14, 1800593 CrossRef CAS PubMed.
- Y. Wang, K. Keller and X. Cheng, Micromachines, 2019, 10, 320 CrossRef PubMed.
- Z. Han, C. Peng, J. Yi, D. Zhang, X. Xiang, X. Peng and L. Qiao, Sens. Actuators, B, 2021, 333, 129563 CrossRef CAS.
- T. H. Yoon, L. Y. Hong and D. P. Kim, Chem. – Asian J., 2011, 6, 1015–1018 CrossRef CAS PubMed.
- P. Agrawal, K. Wilkstein, E. Guinn, M. Mason, C. I. Serrano Martinez and J. Saylae, Org. Process Res. Dev., 2023, 27, 571–591 CrossRef CAS.
- B. O'Sullivan, H. Al-Bahrani, J. Lawrence, M. Campos, A. Cázares, F. Baganz and N. Szita, J. Mol. Catal. B: Enzym., 2012, 77, 1–8 CrossRef.
- K. Lucas, S. D. Ahmad, M. Dehghani, T. Gaborski and J. McGrath, Sep. Purif. Technol., 2020, 251, 117342 CrossRef CAS.
- R. Tan and M. Franzreb, Biotechnol. Bioeng., 2020, 117, 654–661 CrossRef CAS PubMed.
- J. P. Frampton, D. Lai, H. Sriram and S. Takayama, Biomed. Microdevices, 2011, 13, 1043–1051 CrossRef CAS PubMed.
- D. F. C. Silva, A. M. Azevedo, P. Fernandes, V. Chu, J. P. Conde and M. R. Aires-Barros, J. Chromatogr. A, 2017, 1487, 242–247 CrossRef CAS PubMed.
- R. J. Meagher, Y. K. Light and A. K. Singh, Lab Chip, 2008, 8, 527–532 RSC.
- Y. S. Huh, C. M. Jeong, H. N. Chang, S. Y. Lee, W. H. Hong and T. J. Park, Biomicrofluidics, 2010, 4 Search PubMed.
- D. F. C. Silva, A. M. Azevedo, P. Fernandes, V. Chu, J. P. Conde and M. R. Aires-Barros, J. Chromatogr. A, 2012, 1249, 1–7 CrossRef CAS PubMed.
- Y. Huang, T. Meng, T. Guo, W. Li, W. Yan, X. Li and Z. Tong, Microfluid. Nanofluid., 2014, 16, 483–491 CrossRef CAS.
- R. R. G. Soares, P. Novo, A. M. Azevedo, P. Fernandes, M. R. Aires-Barros, V. Chu and J. P. Conde, Lab Chip, 2014, 14, 4284–4294 RSC.
- U. Novak, A. Pohar, I. Plazl and P. Žnidaršič-Plazl, Sep. Purif. Technol., 2012, 97, 172–178 CrossRef CAS.
- E. Espitia-Saloma, P. Vâzquez-Villegas, M. Rito-Palomares and O. Aguilar, Biotechnol. J., 2016, 11, 708–716 CrossRef CAS PubMed.
- U. Novak, M. Lakner, I. Plazl and P. Žnidaršič-Plazl, Microfluid. Nanofluid., 2015, 19, 75–83 CrossRef CAS.
- R. Hu, X. Feng, P. Chen, M. Fu, H. Chen, L. Guo and B. F. Liu, J. Chromatogr. A, 2011, 1218, 171–177 CrossRef CAS PubMed.
- S. Y. Leong, H. B. Ong, H. M. Tay, F. Kong, M. Upadya, L. Gong and H. W. Hou, Small, 2022, 18, 2104470 CrossRef CAS PubMed.
- A. S. Rathore, D. Kumar and N. Kateja, Biotechnol. Lett., 2018, 40, 895–905 CrossRef CAS PubMed.
- R. Ghosh, J. Sep. Sci., 2022, 45, 2024–2033 CrossRef CAS PubMed.
- V. Orr, L. Zhong, M. Moo-Young and C. P. Chou, Biotechnol. Adv., 2013, 31, 450–465 CrossRef CAS PubMed.
- V. Halan, S. Maity, R. Bhambure and A. S. Rathore, Curr. Protein Pept. Sci., 2019, 20, 4–13 CrossRef CAS PubMed.
- T. C. Silva, M. Eppink and M. Ottens, J. Chem. Technol. Biotechnol., 2022, 97, 2365–2375 CrossRef CAS.
- A. S. Chan, M. K. Danquah, D. Agyei, P. G. Hartley and Y. Zhu, Sep. Sci. Technol., 2014, 49, 854–860 CrossRef CAS.
- Y. Li, H. D. Tolley and M. L. Lee, Anal. Chem., 2009, 81, 9416–9424 CrossRef CAS PubMed.
- E. Lietta, A. Pieri, E. Innocenti, R. Pisano, M. Vanni and A. A. Barresi, Pharmaceutics, 2021, 13, 2135 CrossRef CAS PubMed.
- C. Gao, X. Sun and A. T. Woolley, J. Chromatogr. A, 2013, 1291, 92–96 CrossRef CAS PubMed.
- R. R. Soares, A. Pettke, A. Robles-Remacho, S. Zeebaree, S. Ciftci, M. Tampere and N. Madaboosi, Sens. Actuators, B, 2021, 336, 129723 CrossRef CAS.
- A. G. Deshpande, N. J. Darton, K. Yunus, A. C. Fisher and N. K. Slater, New Biotechnol., 2012, 29, 494–501 CrossRef CAS PubMed.
- B. Bao, Z. Wang, D. Thushara, A. Liyanage, S. Gunawardena, Z. Yang and S. Zhao, Separations, 2020, 8, 3 CrossRef.
- M. N. São Pedro, M. E. Klijn, M. H. Eppink and M. Ottens, J. Chem. Technol. Biotechnol., 2022, 97, 2347–2364 CrossRef.
- A. I. Cirillo, G. Tomaiuolo and S. Guido, Micromachines, 2021, 12, 820 CrossRef PubMed.
- Z. Dong, Z. Wen, F. Zhao, S. Kuhn and T. Noël, Chem. Eng. Sci.: X, 2021, 10, 100097 CAS.
- J. C. M. Monbaliu and J. Legros, Lab Chip, 2023, 23, 1349–1357 RSC.
- G. N. Ahn, T. Yu, H. J. Lee, K. W. Gyak, J. H. Kang, D. You and D. P. Kim, Lab Chip, 2019, 19, 3535–3542 RSC.
- S. J. Yim, B. T. Ramanjaneyulu, S. Vidyacharan, Y. D. Yang, I. S. Kang and D. P. Kim, Lab Chip, 2020, 20, 973–978 RSC.
- J. H. Kang, G. N. Ahn, H. Lee, S. J. Yim, S. Lahore, H. J. Lee, H. Kim, J. T. Kim and D. P. Kim, ACS Cent. Sci., 2021, 8, 43–50 CrossRef PubMed.
- S. A. Kawale, D. C. Kang, G. N. Ahn, A. Mottafegh, J. H. Kang, G. S. Na and D. P. Kim, Chem. Eng. J., 2023, 477, 147033 CrossRef CAS.
- A. S. Rathore and D. Sarin, Trends Biotechnol., 2024, 42, 282–292 CrossRef CAS PubMed.
- M. von Stosch, R. M. C. Portela and C. Varsakelis, Curr. Opin. Chem. Eng., 2021, 33, 100692 CrossRef.
- M. Mowbray, T. Savage, C. Wu, Z. Song, B. A. Cho, E. A. Del Rio-Chanona and D. Zhang, Biochem. Eng. J., 2021, 172, 108054 CrossRef CAS.
- J. Smiatek, A. Jung and E. Bluhmki, Trends Biotechnol., 2020, 38, 1141–1153 CrossRef CAS PubMed.
- I. Walsh, M. Myint, T. Nguyen-Khuong, Y. S. Ho, S. K. Ng and M. Lakshmanan, mAbs, 2022, 14, 2013593 CrossRef PubMed.
- H. Helgers, A. Hengelbrock, A. Schmidt, F. L. Vetter, A. Juckers and J. Strube, Processes, 2022, 10, 809 CrossRef CAS.
- Y. Chen, O. Yang, C. Sampat, P. Bhalode, R. Ramachandran and M. Ierapetritou, Processes, 2020, 8, 1088 CrossRef.
- M. Canzoneri, A. De Luca and J. Harttung, Adv. Biochem. Eng./Biotechnol., 2021, 167–184 Search PubMed.
- R. M. Portela, C. Varsakelis, A. Richelle, N. Giannelos, J. Pence, S. Dessoy and M. von Stosch, Digital Twins: Tools and Concepts for Smart Biomanufacturing, 2021, pp. 35–55 Search PubMed.
- M. S. Hong, F. Mohr, C. D. Castro, B. T. Smith, J. M. Wolfrum, S. L. Springs, A. J. Sinskey, R. A. Hart, T. Mistretta and R. D. Braatz, Comput. Chem. Eng., 2023, 179, 108445 CrossRef CAS.
- S. Nikita, G. Thakur, N. G. Jesubalan, A. Kulkarni, V. B. Yezhuvath and A. S. Rathore, Comput. Chem. Eng., 2022, 164, 107896 CrossRef CAS.
- M. Dixit, D. Chhabra and P. Shukla, Bioresour. Technol., 2023, 370, 128467 CrossRef CAS PubMed.
- M. B. Takahashi, J. Leme, C. P. Caricati, A. Tonso, E. G. Fernandez Nunez and J. C. Rocha, Bioprocess Biosyst. Eng., 2015, 38, 1045–1054 CrossRef CAS PubMed.
- N. J. Treloar, A. J. Fedorec, B. Ingalls and C. P. Barnes, PLoS Comput. Biol., 2020, 16, e1007783 CrossRef CAS PubMed.
- A. S. Rathore, S. Nikita and N. G. Jesubalan, Biosens. Bioelectron.: X, 2022, 12, 100263 CAS.
- S. M. Pirrung, L. A. van der Wielen, R. F. Van Beckhoven, E. J. van de Sandt, M. H. Eppink and M. Ottens, Biotechnol. Prog., 2017, 33, 696–707 CrossRef CAS PubMed.
- A. Tiwari, V. Bansode and A. S. Rathore, J. Chromatogr. A, 2022, 1682 Search PubMed.
- M. Vaskevicius, J. Kapociute-Dzikiene and L. Slepikas, Molecules, 2021, 26 Search PubMed.
- K. Ciura, I. Fryca and M. Gromelski, Microchem. J., 2023, 187 Search PubMed.
- X. Y. Zeng and B. Ma, J. Proteome Res., 2021, 20, 3455–3462 CrossRef CAS PubMed.
- H. Xu, J. Lin, D. Zhang and F. Mo, Nat. Commun., 2023, 14, 3095 CrossRef CAS PubMed.
- Y. Zhang, L. Han, L. Zou, M. Zhang and R. Chi, Chem. Eng. Commun., 2021, 208, 1005–1016 CrossRef CAS.
- M. Pirdashti, M. Taheri, E. N. Dragoi and S. Curteanu, Iran. J. Chem. Chem. Eng., 2020, 39, 185–197 Search PubMed.
- Y. Q. Chen, X. D. Liang and G. M. Kontogeorgis, Ind. Eng. Chem. Res., 2023, 62, 11165–11177 CrossRef CAS.
- C. J. Peacock, C. Lamont, D. A. Sheen, V. K. Shen, L. Kreplak and J. P. Frampton, ACS Appl. Mater. Interfaces, 2021, 13, 11449–11460 CrossRef CAS PubMed.
- D. Winters, C. Chu and K. Walker, J. Chromatogr. A, 2015, 1424, 51–58 CrossRef CAS PubMed.
- J. Gomis-Fons, A. Löfgren, N. Andersson, B. Nilsson, L. Berghard and S. Wood, J. Biotechnol., 2019, 301, 45–51 CrossRef CAS PubMed.
- M. Isaksson, J. Gomis-Fons, N. Andersson and B. Nilsson, J. Chromatogr. A, 2023, 1695, 463942 CrossRef CAS PubMed.
- P. A. Kates, J. N. Cook, R. Ghan, H. J. Nguyen, P. Sitasuwan and L. A. Lee, SLAS Technol., 2023, 28, 243–250 CrossRef CAS PubMed.
- S. Tallvod, D. Espinoza, J. Gomis-Fons, N. Andersson and B. Nilsson, J. Chromatogr. A, 2023, 1702, 464085 CrossRef CAS PubMed.
- C. W. Coley, D. A. Thomas III, J. A. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers, H. Gao and R. W. Hicklin, Science, 2019, 365, eaax1566 CrossRef CAS PubMed.
- R. W. Epps, M. S. Bowen, A. A. Volk, K. Abdel-Latif, S. Han, K. G. Reyes, A. Amassian and M. Abolhasani, Adv. Mater., 2020, 32, 2001626 CrossRef CAS PubMed.
- R. J. Hickman, P. Bannigan, Z. Bao, A. Aspuru-Guzik and C. Allen, Matter, 2023, 6, 1071–1081 CrossRef CAS PubMed.
- A. Dürauer, A. Jungbauer and T. Scharl, Biotechnol. Bioeng., 2023, 1–18 Search PubMed.
- H. N. Saha, S. Auddy, S. Pal, S. Kumar, S. Jasu, R. Singh, R. Singh, S. Banerjee, P. Sharan and A. Maity, 8th Annual Industrial Automation and Electromechanical Engineering Conference (IEMECON), IEEE, 2017, pp. 364–369 Search PubMed.
- B. Shahriari, K. Swersky, Z. Wang, R. P. Adams and N. De Freitas, Proc. IEEE, 2015, 104, 148–175 Search PubMed.
- J. A. G. Torres, S. H. Lau, P. Anchuri, J. M. Stevens, J. E. Tabora, J. Li, A. Borovika, R. P. Adams and A. G. Doyle, J. Am. Chem. Soc., 2022, 144, 19999–20007 CrossRef CAS PubMed.
- A. A. Volk, R. W. Epps, D. T. Yonemoto, B. S. Masters, F. N. Castellano, K. G. Reyes and M. Abolhasani, Nat. Commun., 2023, 14, 1403 CrossRef CAS PubMed.
- G. L. Losacco, M. B. Hicks, J. O. DaSilva, H. Wang, M. Potapenko, F. R. Tsay, I. A. H. Ahmad, I. Mangion, D. Guillarme and E. L. Regalado, Anal. Bioanal. Chem., 2022, 414, 3581–3591 CrossRef CAS PubMed.
- D. W. Patrick, D. A. Strand and H. J. Cortes, J. Sep. Sci., 2002, 25, 519–526 CrossRef CAS.
- J. Cen, M. Vukas, G. Barton, J. Kavanagh and H. G. L. Coster, J. Membr. Sci., 2015, 484, 133–139 CrossRef CAS.
- G. Thakur, S. Thori and A. S. Rathore, J. Membr. Sci., 2020, 613, 118492 CrossRef CAS.
- T. Woo, K. Nam, S. Heo, J. Y. Lim, S. Kim and C. Yoo, J. Membr. Sci., 2022, 649, 120400 CrossRef CAS.
- P. Schwaller, T. Gaudin, D. Lanyi, C. Bekas and T. Laino, Chem. Sci., 2018, 9, 6091–6098 RSC.
- Y. Song, T. Wang, P. Cai, K. Mondal and J. P. Sahoo, ACM Comput. Surv., 2023, 55, 1–40 CrossRef.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |