Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Two-stage tuberculosis diagnostics: combining centrifugal microfluidics to detect TB infection and Inh and Rif resistance at the point of care with subsequent antibiotic resistance profiling by targeted NGS†
Judith
Schlanderer
 a,
Harald
Hoffmann
b,
Jan
Lüddecke
ac,
Andrey
Golubov
d,
Wolfgang
Grasse
e,
Elisabeth V.
Kindler
e,
Thomas A.
Kohl
a,
Harald
Hoffmann
b,
Jan
Lüddecke
ac,
Andrey
Golubov
d,
Wolfgang
Grasse
e,
Elisabeth V.
Kindler
e,
Thomas A.
Kohl
 f,
Matthias
Merker
f,
Christoph
Metzger
e,
Vanessa
Mohr
f,
Stefan
Niemann
f,
Claudia
Pilloni
d,
Sara
Plesnik
d,
Bijendra
Raya
g,
Bhawana
Shresta
g,
Christian
Utpatel
f,
Roland
Zengerle
ac,
Markus
Beutler
d and
Nils
Paust
f,
Matthias
Merker
f,
Christoph
Metzger
e,
Vanessa
Mohr
f,
Stefan
Niemann
f,
Claudia
Pilloni
d,
Sara
Plesnik
d,
Bijendra
Raya
g,
Bhawana
Shresta
g,
Christian
Utpatel
f,
Roland
Zengerle
ac,
Markus
Beutler
d and
Nils
Paust
 *ac
*ac
aHahn-Schickard, 79110 Freiburg, Germany. E-mail: Nils.Paust@Hahn-Schickard.de; Tel: +49 761 203 73245
bSYNLAB Gauting SYNLAB Human Genetics Munich, 82131 Gauting, Germany
cLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, 79110 Freiburg, Germany
dWHO supranational Tuberculosis Reference Laboratory, IML red, 82131 Gauting, Germany
egerbion GmbH & Co KG, 70806 Kornwestheim, Germany
fMolecular and Experimental Mycobacteriology, Forschungszentrum Borstel, 23845 Borstel, Germany
gGerman Nepal Tuberculosis Project (GENETUP), Nepal Anti-Tuberculosis Association (NATA), Kalimati, Nepal
First published on 16th November 2023
Abstract
Globally, tuberculosis (TB) remains the deadliest bacterial infectious disease, and spreading antibiotic resistances is the biggest challenge for combatting the disease. Rapid and comprehensive diagnostics including drug susceptibility testing (DST) would assure early treatment, reduction of morbidity and the interruption of transmission chains. To date, rapid genetic resistance testing addresses only one to four drug groups while complete DST is done phenotypically and takes several weeks. To overcome these limitations, we developed a two-stage workflow for rapid TB diagnostics including DST from a single sputum sample that can be completed within three days. The first stage is qPCR detection of M. tuberculosis complex (MTBC) including antibiotic resistance testing against the first-line antibiotics, isoniazid (Inh) and rifampicin (Rif). The test is automated by centrifugal microfluidics and designed for point of care (PoC). Furthermore, enriched MTBC DNA is provided in a detachable sample tube to enable the second stage: if the PCR detects MTBC and resistance to either Inh or Rif, the MTBC DNA is shipped to specialized facilities and analyzed by targeted next generation sequencing (tNGS) to assess the complete resistance profile. Proof-of-concept testing of the PoC test revealed an analytical sensitivity of 44.2 CFU ml−1, a diagnostic sensitivity of 96%, and a diagnostic specificity of 100% for MTBC detection. Coupled tNGS successfully provided resistance profiles, demonstrated for samples from 17 patients. To the best of our knowledge, the presented combination of PoC qPCR with tNGS allows for the fastest comprehensive TB diagnostics comprising decentralized pathogen detection with subsequent resistance profiling in a facility specialized in tNGS.
Introduction
Tuberculosis (TB), caused by bacteria of the M. tuberculosis complex (MTBC), remains one of the deadliest infectious diseases worldwide. It can affect any part of the human body, leading to chronic inflammation and organ destruction. Despite a slow decline in TB incidences and mortality rates over the past decade, the Covid-19 pandemic and other global crises have reversed these trends,1 which is considered dramatic and highly alarming by experts.2–4 Patients with multi-drug resistant TB (MDR-TB) defined by resistance to isoniazid (Inh) and rifampicin (Rif) pose the greatest challenge to TB control as their treatment is extremely costly and their chances of being cured range below 60%.1,5 As a regimen of four active antibiotics is recommended to prevent resistance development and treatment failure, fast identification of TB and determination of antibiotic resistances are key for effective treatment and rapid interruption of transmission chains.6The Xpert MTB/RIF system (Cepheid, USA) is widely used for TB diagnostics and has contributed to an increase of global TB notification by over 30%.7 However, it only detects Rif resistance and not Inh resistance. As Inh-mono resistance is much more prevalent than MDR-TB,8 undetected Inh resistance poses a significant risk of treatment failure and the development of further resistances.9 Several newer WHO-endorsed molecular tests from, for example, Abbott, Becton Dickinson, Roche, or Hain-Bruker detect both Rif and Inh resistance markers, but are only suitable for larger laboratory hubs.10 Fully automated PoC solutions with detection of MTBC and Rif and Inh resistances are still under development.11–13 A lab-on-a-film platform that integrates sample preparation and detection of MTBC and Rif and Inh resistance markers was demonstrated by Kukhtin et al.14 Its long time to result of 8 h and a complex instrumental setup, however, challenge its use as a PoC test.
Several WHO-endorsed reflex test solutions like Xpert MTB/XDR or the line probe assays from HAIN or Nipro are capable of detecting antibiotic drug resistances toward Inh, fluoroquinolones, and second-line injectable drugs. However, they either demand multiple samples or a positive culture. In addition, detectable antibiotic drugs are limited and only mutations in hot spots of the target gene are covered, allowing other mutations to gain an advantage and spread unnoticed.15–17 The only widespread tool for comprehensive DST so far is culture-based,18 which is a time-consuming analysis and needs a complex laboratory infrastructure. We address the shortcomings of TB diagnostics by combining two main innovations in the field: PoC MTBC and Rif and Inh resistance detection automated by centrifugal microfluidics and subsequent targeted next generation sequencing (tNGS)-based resistance profiling employed directly from the sample from a patient. The two analysis technologies are interfaced by providing a genetic material from the PoC test that is transferred to a facility capable of doing the tNGS analysis.
The PoC test employs centrifugal microfluidics. Automated centrifugal microfluidic sample preparation of DNA from clinical sputum samples was shown previously.19,20 With this approach, detection of MTBC and resistance markers must subsequently be performed in manual steps and with an additional PCR cycler. This limits its application at the PoC, as a laboratory infrastructure is required. Here, the previously demonstrated centrifugal microfluidic purification of DNA from clinical sputum samples for subsequent molecular TB diagnostics19,20 is further developed to a fully integrated sample-to-answer analysis. Automated PoC sample-to-answer analysis for pathogen detection is in principle not new to the field. It has been demonstrated for plasma and respiratory samples of 200 μl (ref. 21 and 22) or nasal swabs.23 However, large volumes of 3 ml liquefied sputum24 and sputum as a complex sample matrix25 pose particular challenges to highly integrated PoC diagnostics that have so far not been overcome. The presented fully integrated PoC system is enabled by a newly developed enrichment and lysis method for MTBC within a filter, a new waste management system on centrifugal microfluidic platforms, and the integrated provision of enriched MTBC DNA for downstream tNGS.
The detection of resistance-associated mutations in MTBC strains by whole genome sequencing (WGS) enables comprehensive genetic antibiotic resistance profiling.26,27 The technology has been widely validated with genomic DNA extracted from positive MTBC cultures.28,29 As the culturing step adds considerable turnaround time, efforts have been made to sequence directly from a patient's primary material such as sputum. Selective washing to enrich intact MTBC cells is an interesting option, but currently does not reliably yield full genomes.30 Other approaches include tNGS by either selective enrichment or targeted amplification.30,31 However, approaches are still evolving, with selective enrichment facing difficulties in achieving comprehensive target coverage,32 and proposed amplification panels being limited in the genomic regions to be analysed.33 Therefore, we aim to establish a tNGS amplification approach encompassing the full panel of genomic regions known or suspected to be involved in antibiotic resistance of MTBC strains.
This paper describes the technical implementation of our proposed two-stage diagnostic workflow. We present results for MTBC detection, a proof of principle for the assessment of genetic resistances to Rif and Inh, and the demonstration of downstream susceptibility testing by tNGS from the provided MTBC DNA.
In summary, the system integration approach provides the following main novelties in comparison to the existing state-of-the-art technologies:
• A new waste management method in centrifugal microfluidics enables concentrating bacteria, washing the bacteria and lysing the bacteria from 3 ml liquefied sputum on less than one half of a disk footprint.
• A newly developed enrichment and lysis method that captures bacteria in the filter and employs thermal lysis.
• A sequencing workflow that does not need a bacterial culture and can be performed with MTBC DNA gained in a lab-on-a-chip cartridge.
Two-stage TB diagnostic workflow
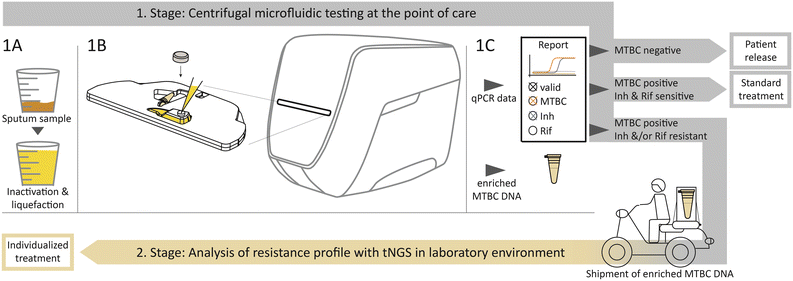
The two-stage diagnostic workflow algorithm is schematically visualized in Fig. 1. The required equipment for the first stage as PoC solution includes a Rhonda player, a portable, electric power operated benchtop device and test cartridges that are further detailed in Fig. S1 in the ESI.† Immediate initial inactivation and liquefaction of the sputum sample after collection guarantees biologically safe handling (Fig. 1-1A). After manual transfer of a 3 ml sample into the cartridge, the inlet is closed with a cap. This provides a completely closed and airtight system to ensure safe handling of pathogens inside the cartridge. The cartridge is automatically retracted into the device (Fig. 1-1B).The outputs provided are the results of the qPCR and from a detachable sample tube with enriched MTBC DNA (Fig. 1-1C). According to the qPCR result, three decision strands are predefined. The first strand defines patient release if no MTBC is detected. Secondly, if MTBC is detected and none of the resistance markers indicate a resistance, the standard antibiotic treatment can be initiated immediately. The third strand should be pursued if MTBC and either one or both of the resistance markers for Inh or Rif are detected. The detachable sample tube with enriched MTBC DNA is intended for shipment to a specialized laboratory with sequencing equipment, as shown on the bottom in Fig. 1. At the specialized facility, tNGS can be performed to reveal the comprehensive resistance profile. The necessary information regarding an individualized antibiotic treatment can then be returned to the physician.
Materials & methods
Microfluidic cartridge design and fabrication
Device
An adapted Rhonda player36 was used, specified by Hahn-Schickard with support by Spindiag and manufactured by DIALUNOX. The fluidic protocol is enabled by rotation up to 70 Hz and contact heating through the sealing foil up to 120 °C by local and individually controllable heating zones. The arrangement of the heating zones in respect to the cartridge is shown in Fig. S1 in the ESI.†qPCR for resistance detection
Colorimetric multiplex qPCR detects gene mutations associated with antibiotic resistance using an endogenous mycobacterial gene sequence control from the wild type and a single nucleotide polymorphism (SNP) sequence in separate fluorescent channels of the same reaction chamber. If a mutation is present, both sequences are amplified equally, resulting in low ΔCT values between the fluorescent channels. Unspecific amplification may occur without a mutation, leading to higher or no CT values in the mutation channel. Larger ΔCT values confirm no mutation is present. A detailed decision scheme of this method is given in Table S1 in the ESI.† This results in a positive detection of specific SNPs preventing false positives due to silent mutations. The assay was validated with a liquid real time qPCR reagent on an AriaMx Instrument (Agilent).WGS and tNGS workflow
For WGS, we prepared libraries following an adapted Illumina Nextera XT protocol37 starting from DNA extracted from H37Rv spiked in sputum samples. Libraries were then run on the Illumina NextSeq 500 or NextSeq 2000 platform in a 2 × 150 bp paired end run according to the manufacturer's specifications.For tNGS, we employed a custom AmpliSeq for the Illumina panel covering altogether 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369 bp from 150 genes or upstream regions implicated in resistance with altogether 856 amplicons. The target regions used for the panel design are listed in dataset S1 in the ESI.† We employed the AmpliSeq panel according to the manufacturer's instructions, using 10 ng and 20 cycles for the DNA solutions extracted from patients' sputum samples.
369 bp from 150 genes or upstream regions implicated in resistance with altogether 856 amplicons. The target regions used for the panel design are listed in dataset S1 in the ESI.† We employed the AmpliSeq panel according to the manufacturer's instructions, using 10 ng and 20 cycles for the DNA solutions extracted from patients' sputum samples.
WGS and tNGS data analysis
For determining the enrichment enabled by sputum processing with the cartridge, WGS data were classified using Kraken38 with default settings.For the detection of resistance mutations in the samples from patients, tNGS data were processed with the MTBseq analysis pipeline.39
Sample collection and description
All sputum samples were stored at −20 °C until analysis.Ethical considerations
Ethical clearance is provided by the Nepal Health Research Council and the Ethical committee of the medical faculty of the University of Munich (project ID 17-761) for the collection and use of clinical sputum samples and metadata from patients.Results
Microfluidic implementation
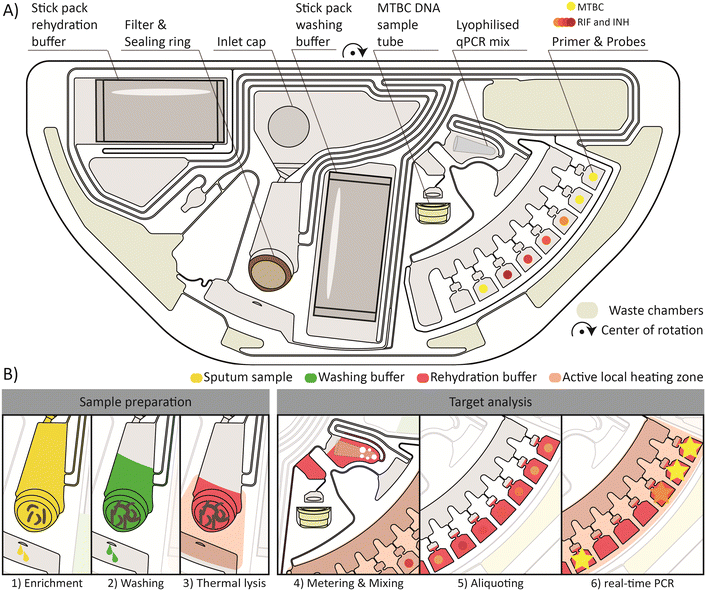
The first-stage PoC solution for testing in peripheral and low-resource settings is based on centrifugal microfluidics.41–43 The portable benchtop device processes the microfluidic cartridges required for the analysis of samples from individual patients.The layout of the centrifugal microfluidic cartridge with its main fluidic steps is depicted in Fig. 2. A more detailed description of the fluidic operation processes, including the frequency and temperature protocol, can be found in the ESI† Fig. S2. The entire workflow from sample preparation to qPCR detection was successfully integrated on a semi-circular cartridge. Consequently, two cartridges can be processed at the same time on the turntable of the device. In Fig. 2A, the cartridge with all necessary components for the functional workflow is shown.
A brief overview of the main steps required for the automated sample preparation and qPCR detection is provided in Fig. 2B. The centerpiece for the sample preparation unit is the integrated filter stack. The first step is the enrichment of MTBC in the filter, as shown in Fig. 2B1. The initial sample filtration takes 25 min at a maximum frequency of 70 Hz. Meanwhile, the buffers, pre-stored in the stick packs,34 are released. 2.5 ml of the sample filtrate is distributed by an overflow mechanism to the four radial, azimuthally distributed outer chambers for waste storage (outer waste chambers). The waste chambers (outer and inner waste chambers) are shown in yellow-gray in Fig. 2A. For the remaining 500 μl of sample filtrate, a new waste management method based on vapor pressure assisted pumping is applied. In brief, the heating zone below the inner waste chamber is heated, and the air inside the chamber expands and is displaced by air bubbles through the liquid in the collection chamber to the periphery of the fluidic network. During the subsequent cooling of the heating zone, negative pressure is created by the contraction of the trapped air and the liquid is drawn into the inner waste chamber. This process can be repeated sequentially. However, from the second pumping cycle onwards, an additional vapor pressure is generated, which significantly increases the pressure differences and thus the transferable liquid volumes. The detailed illustration is shown in the ESI† Fig. S2e and h.
Then, a 600 μl washing buffer is loaded onto the filter by thermo-pneumatic pumping as further detailed by Schlenker et al.44 The washing buffer is perfused through the filter to remove contaminating residues of the sputum and the liquefaction reagent from the filter which could inhibit the subsequent qPCR (Fig. 2B2). After the removal of the washing buffer filtrate to the inner waste chamber, again by vapor pressure assisted pumping, a 500 μl rehydration buffer is loaded onto the filter. A thermal lysis for 3 min is performed by activating the heating zone underneath the filter to a set point of 100 °C (Fig. 2B3). The MTBC DNA is released by the lysis and subsequently flushed through the filter at 70 Hz. This step completes the sample preparation from the sputum sample, making the DNA available for subsequent analyses inside and outside of the cartridge. To access the enriched MTBC DNA, a detachable sample tube is mounted to the cartridge, which is loaded with 70 μl MTBC DNA solution for the subsequent external tNGS analyses. Another 150 μl is further transferred into a chamber where the lyophilized qPCR mix is pre-stored. Combined bubble and shake-mode mixing45,46 is performed to rehydrate and homogenize the qPCR mix with the MTBC DNA solution (Fig. 2B4). Primers and probes are pre-stored in a dry state in seven different reaction chambers. The qPCR mixture is aliquoted into the reaction chambers, 20 μl each (Fig. 2B5). Finally, the qPCR is performed by thermal cycling (45 cycles) and real-time fluorescence readout under rotation at 5 Hz (Fig. 2B6). Geometric multiplexing is combined with colorimetric multiplexing by four different fluorescence detection channels. The first two and the last reaction chamber are functionalized with primers and probes for the detection of MTBC, an internal control (IC), Mycobacterium avium complex (MAC) and nontuberculous mycobacteria (NTM). The chambers in between contain dried primer and probe mixes for the detection of resistance genes against Rif and Inh. After the completion of the microfluidic process, the detachable sample tube filled with MTBC DNA can be removed and used for subsequent analysis by tNGS.
In total, 84 successful cartridge runs were performed for the proof-of concept study: 34 to assess the analytic sensitivity, 25 plus 10 to evaluate the diagnostic sensitivity and specificity, respectively, and 15 to study the antibiotic resistance testing. In all, 21 enriched MTBC DNA solutions extracted from the samples from patients were further analyzed with tNGS.
The current protocol for the PoC test takes 180 min. The filtration and qPCR steps contribute most to the overall runtime, requiring 137 min together. Optimizing the qPCR assay from 90 min to less than 30 min has major potential for saving time, which was shown to be possible with the applied Rhonda player.47 Yet, comparatively long filtration times were applied to ensure that the various sputum samples with different properties are filtered. We hypothesize that a large-scale study with many different sputum samples could optimize and reduce the filtration time from 47 min to less than 25 min. Six cartridge runs were evaluated fluidically to demonstrate reproducible functionality. To avoid contamination during sample preparation (Fig. 2B1–3 – Sample preparation), it is essential that all filtrates of the samples or washing buffer are removed into the waste chambers before the next step begins. This is the case for 100% of the evaluated cartridges after the enrichment and the washing step. In this way, robust sample processing against liquid carryover into the DNA solution can be ensured. On average, 6.75 ± 0.46 reaction chambers are filled after metering, mixing and aliquoting (Fig. 2B5). A detailed analysis with more data is given in the ESI† Fig. S3.
Analytical sensitivity for MTB detection on the cartridge
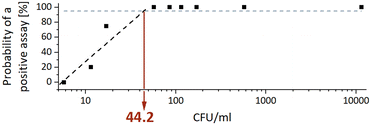
The analytical sensitivity was determined by a total of 34 sputum samples ranging from 5.7 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 475 CFU ml−1 of MTBC strain H37Rv. Concentrations below 50 CFU ml−1 were tested in at least four replicate and the corresponding CT values are given in the ESI† Table S2. The analytical sensitivity, expressed as the MTBC concentration that yields a positive result in 95% of the runs (limit of detection – LoD95), was calculated by interpolating a semi-logarithmic curve using the first four data points. Fig. 3 shows that the analytical sensitivity was 44.2 CFU ml−1.
475 CFU ml−1 of MTBC strain H37Rv. Concentrations below 50 CFU ml−1 were tested in at least four replicate and the corresponding CT values are given in the ESI† Table S2. The analytical sensitivity, expressed as the MTBC concentration that yields a positive result in 95% of the runs (limit of detection – LoD95), was calculated by interpolating a semi-logarithmic curve using the first four data points. Fig. 3 shows that the analytical sensitivity was 44.2 CFU ml−1.
Diagnostic sensitivity & specificity for MTB detection on the cartridge
The diagnostic sensitivity and specificity for MTBC detection on the cartridge are shown in Table 1. In total, 24 out of 25 positive samples from patients were identified as MTBC positive, which results in a diagnostic sensitivity (the proportion of patients correctly identified as positive) of 96%. Accordingly, this results in a 4% probability that a positive patient will not be identified as positive. The specificity (proportion of patients correctly identified as negative) was determined to be 100%; 10 out of 10 negative tested sputum samples were identified as negative.| Pre-classification | |||
|---|---|---|---|
| MTBC positive | MTBC negative | ||
| Cartridge | MTBC positive | 96% (24/25) | 0% (0/10) |
| MTBC negative | 4% (1/25) | 100% (10/10) | |
Resistance detection on the cartridge
The detection of resistances to Rif and Inh was shown for five different bacterial strains: one wild-type strain with no pre-classified resistance, one strain with resistance to Inh only (Inh-mono), and three MDR-TB strains with resistances to both Rif and Inh. The five strains contained three different mutations associated with Rif resistance and one mutation associated with Inh resistance. Results are given in Table 2. Each strain was tested in triplicate. As expected, no mutations were detected with the wild-type strain by the test.| Bacterial strain | Rifampicin resistance | Isoniazid resistance | ||
|---|---|---|---|---|
| Mutation | Mutation detected | Mutation | Mutation detected | |
| Wild-type – sensitive to antibiotics; Inh-mono – resistant to isoniazid; multi-drug-resistant (MDR) – resistant to isoniazid and rifampicin.Mutations are denoted in the Mycobacterium tuberculosis complex numbering system. | ||||
| Wild-type | None | No | None | No |
| Inh-mono | None | No | S315T | 2/3 |
| MDR#1 | H445Y | 3/3 | S315T | 2/3 |
| MDR#2 | D435V | 3/3 | S315T | 1/3 |
| MDR#3 | S450L | 3/3 | S315T | 3/3 |
Resistance to Rif resulting from the different mutations of H445Y (MDR#1), D435V (MDR#2) and S450L (MDR#3) was detected in all three cartridges tested (3/3) for each strain. The Inh resistance resulting from the S315T mutation, present in the Inh-mono, MDR#1, MDR#2 and MDR#3 bacterial strains, was detected at a percentage of 67%. In detail, two out of three cartridges for the Inh-mono and MDR#1 bacterial strains, one out of three for MDR#2 and three of three for MDR#3 detect the Inh resistance.
Targeted NGS
We compared the sputum samples spiked with H37Rv with respect to the relative enrichment of MTBC DNA processed either by direct DNA extraction or by the cartridge. The extracted DNA solutions were sequenced by WGS and classified with Kraken38 as shown in Table S3 in the ESI.† The results demonstrate a relative enrichment of MTBC DNA compared to the background human DNA by the cartridge for the two concentrations tested.Regarding resistance profiling, 21 DNA solutions from patients provided by the cartridge were sequenced using a dedicated custom AmpliSeq for the Illumina tNGS panel, and the sequence data were analyzed using the MTBseq pipeline.
The success of the tNGS as inferred from the achieved coverage of the target regions with sequence information correlates with the CT values determined by the cartridge MTBC qPCR detection as shown in Table S4 in the ESI.† Out of the 21 samples from patients, four failed to achieve a coverage of at least 10 reads and a 75% allele frequency of at least 95% of the target regions. The failed samples all featured MTBC CT values of 29 or higher. From the analytical sensitivity data set, a correlation of ∼11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CFU ml−1 H37Rv spiked in sputum with a CT value of 28 was determined (ESI† Table S2). This suggests that the samples with a low coverage consistently correlated with a bacterial load of less than 10
000 CFU ml−1 H37Rv spiked in sputum with a CT value of 28 was determined (ESI† Table S2). This suggests that the samples with a low coverage consistently correlated with a bacterial load of less than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CFU ml−1, which may be too low for successful sequencing. The samples with a failed tNGS status (Table S4†) from the coverage analysis were excluded from the following resistance profiling analyses.
000 CFU ml−1, which may be too low for successful sequencing. The samples with a failed tNGS status (Table S4†) from the coverage analysis were excluded from the following resistance profiling analyses.
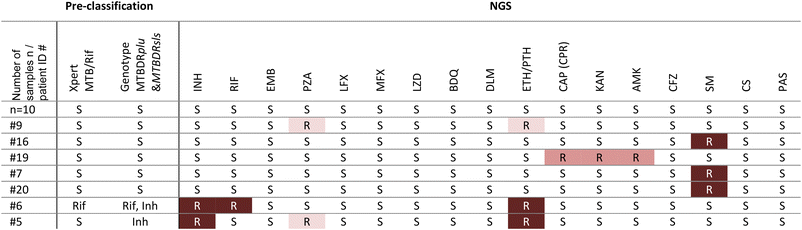
From the analysis of the sequence data, we inferred resistance profiles for 17 samples from patients, depicted in Table 3. The results show full concordance with the pre-classification results from the Xpert MTB/Rif and the Genotype MTBDRplus for the resistances to Rif and Inh.
Discussion
The presented workflow aims to support doctors' decisions for antibiotic treatment in settings with limited resources. To date, rapid genetic resistance testing at the PoC covers too few antibiotic drug groups or takes too long because phenotypic bacterial culture is required. The two-stage workflow we've introduced combines a centrifugal microfluidic PoC test for decentralized detection of MTBC and Rif and Inh resistance with the subsequent comprehensive resistance profiling by tNGS. The implementation of this algorithm could be an important step in fighting the spread of TB and preventing harm to patients caused by drug resistant MTBC.The recommended diagnostic algorithm offers several major advantages. Firstly, it provides a user-friendly PoC test, eliminating the need to send samples to external laboratories. It does not require complex steps at the PoC and thus there is no need for extensive infrastructure, trained personnel, installations, and investments. Intuitive PoC tests have been shown to increase the TB notification numbers by about 30% and thus have a significant impact.7 Our approach not only minimizes delays by the shipment of samples, but also enables the detection of both TB and the two most critical resistances to Rif and Inh. Patient case #5 in Table 3 illustrates the importance of testing for Inh resistance. Inh-mono resistances account for about 11% of all TB cases8 and thus it is of major importance to be detected as quickly as possible. Such cases with Inh resistance fall through existing testing algorithms available at the PoC. Detecting Inh resistance at the PoC not only is favorable for treatment success, but also has been identified as the most cost-effective strategy that has the greatest impact on preventing MDR transmissions and deaths.6 Enriched, unamplified MTBC DNA is provided by the cartridge for reflex testing such as tNGS. In the past, workarounds using the Xpert MTB/Rif Ultra cartridges were considered for further use of the extracted DNA for downstream testing.48,49
This involves puncturing of the cartridges with a syringe to recover the amplification product. In addition to the risk of injury from syringe handling, the DNA used from Xpert cartridges is already amplified, posing a serious risk of bias and high error rates in downstream analyses. However, these approaches highlight the need for two-stage analyses from a single sample. Our workflow allows for the selective use of tNGS for a comprehensive resistance profile analysis from the enriched MTBC DNA provided by the cartridge. This allows antibiotic treatment to be adjusted accordingly in a timely manner. We expect this to result in cost savings as NGS can be reserved for important samples and increases the chances of successful treatment. The centralized implementation of tNGS as a second step is proposed in dedicated facilities equipped to handle the sequencing process. By concentrating investments in sequencers and trained personnel, this approach ensures a favorable price per sample and efficient turnaround time due to higher throughput. The CT values determined in the cartridge qPCR can likely serve as reliable indicators of downstream tNGS success, optimizing the overall efficiency of the coupled PoC cartridge plus tNGS analysis. The entire diagnostic workflow is based on a single sample, eliminating the necessity for time consuming bacterial cultures and reducing the number of visits patients need to make to the healthcare facilities. Overall, significant time savings can be achieved, because no time-consuming bacterial culture is required for DST or NGS resistance profiling.
The presented study demonstrates the technical feasibility of the two-stage diagnostic workflow. In the following, the current performance of the system is critically discussed.
The biochemical and fluidic functionality of the cartridge is demonstrated with the data set from the analytical sensitivity analysis and provides consistent results. The analytical sensitivity of 44.2 CFU ml−1 of the presented system is intermediate between the Xpert MTB/Rif Ultra (3.1 CFU ml−1 in sputum) and the FluoroType MTB (52.1 CFU ml−1 in sputum), which was previously evaluated using a comparable experimental method.40 The analytical sensitivity can still be improved. So far, we have used a volume of 500 μl buffer for MTBC lysis and the uptake of the released DNA. This volume could be reduced to 200 μl to yield a higher concentration of target DNA in the lysate and thus increase the probability of MTBC detection at low concentrations. Further optimization of the PCR on the Rhonda system would probably lower the LoD as well. The diagnostic performance characteristics of the cartridge were comparable to those of the Xpert MTB/Rif Ultra and the FluoroType MTB systems.50,51 Specifically, the diagnostic sensitivity of the cartridge (96%; n = 25) is in the same range as the results of the Xpert MTB/Rif Ultra (97.8%; n = 91) and the FluoroType MTB (100%; n = 43) for smear-positive specimens from patients. The same is found for the diagnostic specificity with 100% (n = 10) for the cartridge, 98.7% (n = 77) for Xpert MTB/Rif Ultra, and 98.9% (n = 912) for FluoroType MTB.
With 180 min for the PoC test, the current time to result can still be optimized. From our perspective, the measures discussed above for time reduction to less than 60 min are straightforward and should be accomplishable with medium resources during product development. While the qPCR worked well for the MTBC detection and the endogenous mycobacterial control (wild type) of the resistance assay, the detection of resistance associated SNP was partially affected by the harsh conditions of the liquefaction reagent. In particular, the detection of Inh resistance markers still needs to provide satisfying results and should be improved in future studies. A qPCR system in combination with melting curves is a promising approach to overcome this challenge in the future. The feasibility of sequencing the enriched MTBC DNA after cartridge processing was demonstrated. We could successfully analyze 17 out of 21 samples from patients, with a clear indication of limited DNA input materials in the remaining four samples. For samples with a bacterial load of <10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CFU ml−1, our tNGS method did not provide adequate quality to generate resistance profiles. Further work is needed to optimize the AmpliSeq panel performance to cover samples with lower bacterial loads, to achieve a 100% target region coverage, and to evaluate the tNGS resistance profiles.
000 CFU ml−1, our tNGS method did not provide adequate quality to generate resistance profiles. Further work is needed to optimize the AmpliSeq panel performance to cover samples with lower bacterial loads, to achieve a 100% target region coverage, and to evaluate the tNGS resistance profiles.
In summary, this proof-of-concept study aims to highlight the societal need for and technical feasibility of improved diagnostic algorithms to combat TB. It is the first overall implementation of a diagnostic algorithm for TB with predefined decision and action strands for clinicians from a rapid initial PoC TB diagnosis to comprehensive resistance analysis by tNGS. By enabling fast and accurate diagnosis and individualized treatment, this approach has the potential to substantially improve TB treatment success rates and reduce transmission. Based on this proof-of-principle study, product development can be initiated. In the future, a full assessment study on a larger scale needs to refine the performance parameters. The technical focus would be to improve the run time of the microfluidic PoC test, to increase the robustness of qPCR detection of Rif and Inh resistance markers, and the introduction of quality control into the manufacturing process. To even further accelerate the time to identification of the MTBC resistance profile, PoC sequencing and the automation of all required preparation steps, e.g., PoC library preparation,46 could be investigated.
Author contributions
Judith Schlanderer: conceptualization (microfluidic cartridge), formal analysis, investigation, methodology, project administration, visualization, and writing – original draft. Harald Hoffmann: conceptualization, funding acquisition, resources, supervision, and writing – review & editing. Jan Lüddecke: conceptualization, investigation, supervision, and writing – review & editing. Andrey Golubov: investigation. Wolfgang Grasse: conceptualization (qPCR), investigation, methodology (qPCR), and writing – review & editing. Elisabeth V. Kindler: investigation (qPCR). Thomas Kohl: conceptualization (tNGS), formal analysis, methodology (tNGS), and writing – review & editing. Matthias Merker: methodology (tNGS). Christoph Metzger: resources and supervision. Vanessa Mohr: investigation (tNGS). Stefan Niemann: resources and supervision. Claudia Pilloni: investigation. Sara Plesnik: investigation. Bijendra Raya: investigation. Bhawana Shresta: investigation. Christian Utpatel: formal analysis (tNGS). Roland Zengerle: resources and supervision. Markus Beutler: conceptualization, investigation, formal analysis, methodology, project administration, and writing – review & editing. Nils Paust: conceptualization, funding acquisition, resources, supervision, and writing – review & editing.Conflicts of interest
We declare that the three unit operation of the integrated workflow is partially protected by patents. The rehydration of the pre-stored reagents is protected by DE102013220257 and family. The switching after the rehydration is patented by DE102016207845 and family. The aliquoting is protected by DE102008003979 and family. All patents are owned by Hahn-Schickard. On the first two, Nils Paust is a co-inventor.Acknowledgements
The authors thank the team at Hahn-Schickard Stuttgart for injection molding of the cartridges and the Hahn-Schickard Lab-on-a-Chip Foundry service in Freiburg for supporting the manufacturing of the cartridges. Financial support by the Federal Ministry of Education and Research, Germany within the projects TB-SeqDisK (16GW0153-158) and ASTANA (13N15803) is gratefully acknowledged.References
- Global tuberculosis report 2022, World Health Organization, 2022.
- Stop TB Partnership sounds the alarm on the TB funding crisis as TB deaths increase for a second year in a row and TB incidence rises for the first time in 20 years, https://www.stoptb.org/news/stop-tb-partnership-sounds-alarm-tb-funding-crisis-tb-deaths-increase-second-year-row-and-tb, Stop TB Partnership, 22 February 2023.
- Tuberculosis deaths rise for the first time in more than a decade due to the COVID-19 pandemic, https://www.who.int/news/item/14-10-2021-tuberculosis-deaths-rise-for-the-first-time-in-more-than-a-decade-due-to-the-covid-19-pandemic, World Health Organization, 8 February 2023.
- Global tuberculosis cases increase for the first time in 20 years, https://www.theguardian.com/global-development/2022/oct/28/global-tuberculosis-tb-cases-increase, The Guardian, 28 October 2022.
- R. Diel, R. Loddenkemper and J.-P. Zellweger, et al., Old ideas to innovate tuberculosis control: preventive treatment to achieve elimination, Eur. Respir. J., 2013, 42, 785–801 CrossRef.
- O. Oxlade, D. Falzon and D. Menzies, The impact and cost-effectiveness of strategies to detect drug-resistant tuberculosis, Eur. Respir. J., 2012, 39, 626–634 CrossRef CAS PubMed.
- G. L. Calligaro, L. S. Zijenah and J. G. Peter, et al., Effect of new tuberculosis diagnostic technologies on community-based intensified case finding: a multicentre randomised controlled trial, Lancet Infect. Dis., 2017, 17, 441–450 CrossRef PubMed.
- Global tuberculosis report 2020, World Health Organization, Geneva, 2020.
- N. Dookie, S. Rambaran and N. Padayatchi, et al., Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care, J. Antimicrob. Chemother., 2018, 73, 1138–1151 CrossRef CAS.
- M. de Vos, L. Scott and A. David, et al., Comparative Analytical Evaluation of Four Centralized Platforms for the Detection of Mycobacterium tuberculosis Complex and Resistance to Rifampicin and Isoniazid, J. Clin. Microbiol., 2021, 59, 3 CrossRef.
- A. Paul, N. Dutta and D. Moschou, et al., Advanced integrative sensing technologies for detection of drug-resistant tuberculosis in point-of-care settings, Sens. Int., 2020, 1, 100036 CrossRef.
- Z. Wang, Y. Wang and L. Lin, et al., A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis, Biosens. Bioelectron., 2022, 195, 113663 CrossRef CAS PubMed.
- A. Molloy, J. Harrison and J. S. McGrath, et al., Microfluidics as a Novel Technique for Tuberculosis: From Diagnostics to Drug Discovery, Microorganisms, 2021, 9, 2330 CrossRef CAS.
- A. V. Kukhtin, R. Norville and A. Bueno, et al., A Benchtop Automated Sputum-to-Genotype System Using a Lab-on-a-Film Assembly for Detection of Multidrug-Resistant Mycobacterium tuberculosis, Anal. Chem., 2020, 92, 5311–5318 CrossRef CAS.
- N. A. Makhado, E. Matabane and M. Faccin, et al., Outbreak of multidrug-resistant tuberculosis in South Africa undetected by WHO-endorsed commercial tests: an observational study, Lancet Infect. Dis., 2018, 18, 1350–1359 CrossRef PubMed.
- G. R. Pereira, M. S. Barbosa and N. J. D. Dias, et al., Impact of introduction of Xpert MTB/RIF test on tuberculosis (TB) diagnosis in a city with high TB incidence in Brazil, PLoS One, 2018, 13, e0193988 CrossRef.
- A. Bainomugisa, C. Gilpin and C. Coulter, et al., New Xpert MTB/XDR: added value and future in the field, Eur. Respir. J., 2020, 56, 2003616 CrossRef.
- WHO consolidated guidelines on tuberculosis: Rapid diagnostics for tuberculosis detection; Module 3: Diagnosis, 2021 update, World Health Organization, Geneva, Switzerland, 2021 Search PubMed.
- A. R. Homann, L. Niebling and S. Zehnle, et al., A microfluidic cartridge for fast and accurate diagnosis of Mycobacterium tuberculosis infections on standard laboratory equipment, Lab Chip, 2021, 21, 1540–1548 RSC.
- M. Beutler, A. R. Homann and M. Mihalic, et al., Rapid Tuberculosis Diagnostics Including Molecular First- and Second-Line Resistance Testing Based on a Novel Microfluidic DNA Extraction Cartridge, J. Mol. Diagn., 2021, 23, 643–650 CrossRef CAS.
- G. Czilwik, T. Messinger and O. Strohmeier, et al., Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation, Lab Chip, 2015, 15, 3749–3759 RSC.
- M. Rombach, S. Hin and M. Specht, et al., RespiDisk: a point-of-care platform for fully automated detection of respiratory tract infection pathogens in clinical samples, Analyst, 2020, 145, 7040–7047 RSC.
- Spindiag, Rhonda SARS-CoV-2 disk – Spindiag, https://www.spindiag.de/rhonda-sars-cov-2-disk/, (2023, accessed 18 March 2023).
- A. Niemz, T. M. Ferguson and D. S. Boyle, Point-of-care nucleic acid testing for infectious diseases, Trends Biotechnol., 2011, 29, 240–250 CrossRef CAS PubMed.
- J. Sturgess, A. J. Palfrey and L. Reid, Rheological properties of sputum, Rheol. Acta, 1971, 10, 36–43, DOI:10.1007/bf01972474.
- R. E. Colman, A. Mace and M. Seifert, et al., Whole-genome and targeted sequencing of drug-resistant Mycobacterium tuberculosis on the iSeq100 and MiSeq: A performance, ease-of-use, and cost evaluation, PLoS Med., 2019, 16, e1002794 CrossRef CAS.
- The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide, World Health Organization, 2018 Search PubMed.
- S. M. Gygli, P. M. Keller and M. Ballif, et al., Whole-Genome Sequencing for Drug Resistance Profile Prediction in Mycobacterium tuberculosis, Antimicrob. Agents Chemother., 2019, 63, 4 CrossRef PubMed.
- M. R. Farhat, R. Sultana and O. Iartchouk, et al., Genetic Determinants of Drug Resistance in Mycobacterium tuberculosis and Their Diagnostic Value, Am. J. Respir. Crit. Care Med., 2016, 194, 621–630 CrossRef CAS.
- A. A. Votintseva, P. Bradley and L. Pankhurst, et al., Same-Day Diagnostic and Surveillance Data for Tuberculosis via Whole-Genome Sequencing of Direct Respiratory Samples, J. Clin. Microbiol., 2017, 55, 1285–1298 CrossRef CAS.
- M. Barbosa-Amezcua, B. Cuevas-Córdoba and C. Fresno, et al., Rapid Identification of Drug Resistance and Phylogeny in M. tuberculosis, Directly from Sputum Samples, Microbiol. Spectrum, 2022, 10, e0125222 CrossRef PubMed.
- A. C. Brown, J. M. Bryant and K. Einer-Jensen, et al., Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples, J. Clin. Microbiol., 2015, 53, 2230–2237 CrossRef CAS PubMed.
- S. Feuerriegel, T. A. Kohl and C. Utpatel, et al., Rapid genomic first- and second-line drug resistance prediction from clinical Mycobacterium tuberculosis specimens using Deeplex-MycTB, Eur. Respir. J., 2021, 57, 7 CrossRef PubMed.
- T. van Oordt, Y. Barb and J. Smetana, et al., Miniature stick-packaging--an industrial technology for pre-storage and release of reagents in lab-on-a-chip systems, Lab Chip, 2013, 13, 2888–2892 RSC.
- M. Rombach, D. Kosse and B. Faltin, et al., Real–time stability testing of air–dried primers and fluorogenic hydrolysis probes stabilized by trehalose and xanthan, BioTechniques, 2014, 57, 151–155 CrossRef CAS PubMed.
- Spindiag, Rhonda player – Spindiag, https://www.spindiag.de/rhonda-player/, (2022, accessed 22 February 2023).
- M. Baym, S. Kryazhimskiy and T. D. Lieberman, et al., Inexpensive multiplexed library preparation for megabase-sized genomes, PLoS One, 2015, 10, e0128036 CrossRef.
- S. L. Salzberg and D. E. Wood, Releasing the Kraken, Front. bioinform., 2021, 1, 808003 CrossRef.
- T. A. Kohl, C. Utpatel and V. Schleusener, et al., MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates, PeerJ, 2018, 6, e5895 CrossRef.
- M. Beutler, S. Plesnik and M. Mihalic, et al., A pre-clinical validation plan to evaluate analytical sensitivities of molecular diagnostics such as BD MAX MDR-TB, Xpert MTB/Rif Ultra and FluoroType MTB, PLoS One, 2020, 15, e0227215 CrossRef CAS PubMed.
- R. Gorkin, J. Park and J. Siegrist, et al., Centrifugal microfluidics for biomedical applications, Lab Chip, 2010, 10, 1758–1773 RSC.
- V. Sunkara, S. Kumar and J. Del Sabaté Río, et al., Lab-on-a-Disc for Point-of-Care Infection Diagnostics, Acc. Chem. Res., 2021, 54, 3643–3655 CrossRef CAS.
- O. Strohmeier, M. Keller and F. Schwemmer, et al., Centrifugal microfluidic platforms: advanced unit operations and applications, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- F. Schlenker, P. Juelg and J. Lüddecke, et al., Nanobead handling on a centrifugal microfluidic LabDisk for automated extraction of cell-free circulating DNA with high recovery rates, Analyst, 2023, 148, 932–941 RSC.
- S. Hin, N. Paust and M. Keller, et al., Temperature change rate actuated bubble mixing for homogeneous rehydration of dry pre-stored reagents in centrifugal microfluidics, Lab Chip, 2018, 18, 362–370 RSC.
- J. F. Hess, M. Kotrová and S. Calabrese, et al., Automation of Amplicon-Based Library Preparation for Next-Generation Sequencing by Centrifugal Microfluidics, Anal. Chem., 2020, 92, 12833–12841 CrossRef CAS.
- Spindiag, Spindiag erhält EU-Marktzulassung für Rhonda Corona PCR-Schnelltest – Spindiag, https://www.spindiag.de/spindiag-erhalt-eu-marktzulassung-fur-rhonda-corona-pcr-schnelltest/, (2020, accessed 23 February 2023).
- R. Venter, B. Derendinger and M. de Vos, et al., Mycobacterial genomic DNA from used Xpert MTB/RIF cartridges can be utilised for accurate second-line genotypic drug susceptibility testing and spoligotyping, Sci. Rep., 2017, 7, 14854 CrossRef.
- R. Venter, S. Minnies and B. Derendinger, et al., Extract from used Xpert MTB/RIF Ultra cartridges is useful for accurate second-line drug-resistant tuberculosis diagnosis with minimal rpoB-amplicon cross-contamination risk, Sci. Rep., 2020, 10, 2633 CrossRef CAS.
- S. Chakravorty, A. M. Simmons and M. Rowneki, et al., The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing, MBio, 2017, 8, 4 CrossRef PubMed.
- S. Hofmann-Thiel and H. Hoffmann, Evaluation of Fluorotype MTB for detection of Mycobacterium tuberculosis complex DNA in clinical specimens from a low-incidence country, BMC Infect. Dis., 2014, 14, 59, DOI:10.1186/1471-2334-14-59.
Footnote |
| † Electronic supplementary information (ESI) available: Dataset S1. See DOI: https://doi.org/10.1039/d3lc00783a |
| This journal is © The Royal Society of Chemistry 2024 |