Upcycling atmospheric CO2 to polyhydroxyalkanoates via sequential chemo-biocatalytic processes†
Manuel
Bruch‡
 ab,
Julian E.
Sanchez-Velandia‡
ab,
Julian E.
Sanchez-Velandia‡
 c,
Jhonatan
Rodríguez-Pereira
c,
Jhonatan
Rodríguez-Pereira
 de,
Michelle
Rich
de,
Michelle
Rich
 b,
Nicole
Pearcy
b,
Nicole
Pearcy
 f,
Tanja
Narančić
f,
Tanja
Narančić
 ab,
Eduardo
Garcia-Verdugo
ab,
Eduardo
Garcia-Verdugo
 c,
Victor
Sans
c,
Victor
Sans
 g,
Kevin
O'Connor
g,
Kevin
O'Connor
 *ab and
Marcileia
Zanatta
*ab and
Marcileia
Zanatta
 *gh
*gh
aSchool of Biomolecular and Biomedical Sciences, University College Dublin, Dublin, Ireland. E-mail: zanatta@uji.es; kevin.oconnor@ucd.ie
bBiOrbic, Bioeconomy SFI Research Centre, Dublin, Ireland
cDepartamento de Quimica Inorganica y Organica, Avda Sos Baynat, s/n, 12071 Castelló de la Plana, Spain
dCenter of Materials and Nanotechnologies, Faculty of Chemical Technology, University of Pardubice, Nam. Cs. Legii 565, 53002 Pardubice, Czech Republic
eCentral European Institute of Technology, Brno University of Technology, Purkynova 123, 61200 Brno, Czech Republic
fBBSRC/EPSRC Synthetic Biology Research Centre (SBRC), University of Nottingham, University Park, Nottingham, NG7 2RD, UK
gInstitute of Advanced Materials (INAM), Universitat Jaume I (UJI), Avda Sos Baynat, s/n, 12071 Castelló de la Plana, Spain
hDepartament de Química Física i Analítica, Universitat Jaume I, Avda Sos Baynat s/n, 12071 Castelló de la Plana, Spain
First published on 17th October 2024
Abstract
The reduction of greenhouse gas emissions and the shift away from petrochemical-derived materials are critical goals in modern industrial development and societal progress. Addressing these intertwined challenges demands innovative and sustainable solutions. Here, we present the first example of synthesizing poly[R-(–)-3-hydroxybutyrate] (PHB) from atmospheric CO2, utilizing a streamlined and integrated process that combines both chemo- and bio-catalytic conditions. Central to our approach is the development of an immobilized catalytic system that efficiently converts atmospheric CO2 into sodium formate, establishing a sustainable carbon source for formatotrophic organisms. Through Adaptive Laboratory Evolution (ALE), we enhanced the growth rate of the bacterium Cupriavidus necator H16, enabling it to utilize formic acid and formate as the sole carbon and energy sources. The evolved strain, C. necator ALE26, achieved a 1.8-fold increase in the maximum growth rate (μmax = 0.25 ± 0.02 h−1), attributed to the loss of the megaplasmid pHG1. Employing the adapted strain, we report the highest PHB production rate in continuous fermentation using C. necator for growth on formate. The development of the different stages (sorption and chemo- and bio-transformation) under compatible conditions that minimize the number of work-up stages demonstrates a major advancement in converting atmospheric CO2 into valuable biopolymers, thus simultaneously contributing to the reduction of greenhouse gases in the atmosphere and to a circular economy of biobased polymers that diminish fossil fuel dependence.
Introduction
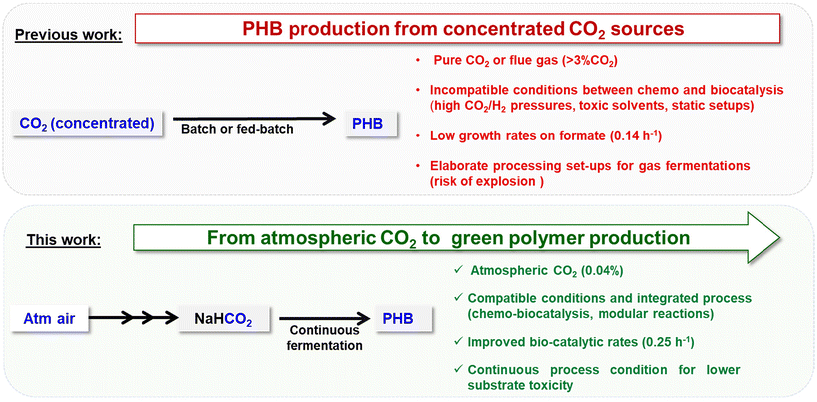
Climate change is one of the major global challenges of the 21st century.1–4 The increasing concentration of carbon dioxide (CO2) in the atmosphere is a major driver of climate change, necessitating innovative solutions for its capture and reuse.5 Currently only 1.3% of the total annual CO2 emission is captured.1,2 For post-combustion carbon capture, two strategies are highlighted in carbon capture, utilisation and storage (CCUS) and direct air capture (DAC). The former aims to remove carbon from flue gas from industrial processes and is generally more energy efficient due to the higher carbon concentrations in these gas streams compared to ambient air. However, it has long been argued that to truly achieve a carbon negative economy, historically emitted carbon needs to be recaptured from the atmosphere, leading to growing interest in DAC technologies.6,7 Efficient direct air capture and integrated conversion (DACC) processes offer a promising opportunity to address these challenges by removing CO2 directly from the atmosphere and converting it into added-value products.8Current state-of-the-art chemical DACC technologies for CO2 hydrogenation reaction primarily focus on the production of C1 compounds such as methanol, methane or formate salts.9–19 In this process, the DAC step is typically performed using amines or hydroxides, which generate carbamate or carbonate species that are subsequently hydrogenated to the desired product.8 In the conversion step, the hydrogenation to formate is particularly interesting due to its potential for further upgrading to higher added-value CO2-derived synthetic products.20 Among the notable catalytic systems for the hydrogenation of carbonate/carbamates to formate, the most employed are iridium (Ir)- and ruthenium (Ru)-based complexes.21–23 Despite significant advances in the DACC technology, several critical challenges remain. One of the primary issues is the difficulty in recycling catalysts and, more significantly, the adsorbents, which are predominantly used in the homogeneous phase. This usage complicates their recovery and reuse, thereby reducing the overall efficiency and sustainability of the process. Additionally, the limited usability of the final products further restricts the practical applications of DAC.8 Thus, it is crucial to design efficient reaction and product separation processes as well as systems for the recovery and reuse of all reaction elements, including catalysts and adsorbents. Additionally, to maximize their impact and reduce the fossil fuel dependence, it is essential to enhance the added value of the products generated from captured CO2, moving beyond simple C1–C2 products to more complex and valuable chemicals.
In this context, biology emerges as a promising avenue for transitioning from C1 to Cn products derived from CO2. Many organisms, including plants and a plethora of bacteria, can utilise CO2 and/or other C1-gases as carbon sources to produce complex molecules, such as sugars, alcohols, lipids, pharmaceutically active ingredients and polymers.24–26 Energy is either derived from light (photosynthesis) or inorganic molecules like molecular hydrogen (H2). Photosynthetic solutions become difficult at large scales when even lighting needs to be provided for efficient production processes. Working with CO2 and H2 directly in microbial fermentations requires elaborate set-ups to avoid the risk of explosions, especially if the organism used is grown aerobically. Furthermore, both CO2 and H2 suffer from low water solubility although much research has been dedicated to tackling this challenge through process engineering.27,28 Thus, alternative strategies for providing carbon and hydrogen to the biological catalyst can improve these processes. Chemical CO2 fixation, therefore, can be a key step in providing easily accessible carbon sources for microbial fermentations.
For decades, the Gram-negative bacterium Cupriavidus necator (C. necator) H16 has been investigated for its capabilities of growing on CO2 as its sole carbon source and as a model organism to produce the biodegradable polyester poly[R-(–)-3-hydroxybutyrate] (PHB), with properties similar to those of polypropylene.29–34 It naturally uses H2 as its energy source in autotrophic growth. Recently, studies in simple batch set-ups have investigated C. necator's PHB production capabilities in true autotrophy on gaseous feedstocks,35,36 focussing on different factors in cell growth and PHB accumulation.37 Alternatively, in a CCUS context, CO2 from flue gas has been used together with in situ electrochemical hydrogen generation to accumulate PHB in the bacterium.38 The same group has developed a two-stage process in which compressed CO2 is first electrochemically reduced to formate which in turn is fed to resting C. necator cells for PHB accumulation.39
In recent years, the use of formic acid and formate salts as an alternative carbon source for a group of bacteria coined “formatotrophs” has been popularised.37,40 However, to date, the integration of DAC technologies and biotechnological product formation remains unexplored. Chemical synthesis of formic acid or formate salts can provide a readily available, water-soluble alternative C1 carbon source for C. necator. Here, we introduce an innovative synthetic route to produce PHB that integrates three key processes: DAC of CO2, chemo-catalytic hydrogenation, and biocatalytic transformation (Fig. 1). To the best of the authors’ knowledge, this is the first example of production of the commercially viable product PHB from captured atmospheric air and also the first example of continuous fermentation for accumulation of PHB from formate. Our primary objective was two-fold: (i) to showcase the conversion of atmospheric CO2 into formate salts under conditions compatible with further bioprocessing steps and (ii) to optimize the growth and polyhydroxybutyrate (PHB) accumulation of C. necator on formic acid/formate in a continuous bioprocess via Adaptive Laboratory Evolution (ALE) and process optimisation. This integrated approach not only demonstrates the feasibility of converting CO2 into useful products but also highlights the potential for sustainable and economically viable processes that can help reduce fossil fuel dependence and mitigate climate change.
Results and discussion
Hydrogenation of atmospheric CO2 to formate
The CO2 molecule, in its gaseous form, exhibits sp hybridisation, resulting in a strong overlap of bonding orbitals that inherently limits its reactivity. However, when subjected to chemical capture processes, CO2 adopts a more reactive configuration, specifically a trigonal planar sp2 hybridisation, as seen in bicarbonate (HCO3−) and carbamates. When formed from CO2, bicarbonate has a reaction enthalpy of −45 kJ mol−1, compared to carbamate (−80 kJ mol−1).41 Thus, bicarbonate is a more accessible and reactive intermediate and can be further transformed into added value products. Therefore, our previous research demonstrated the feasibility of DAC coupled with sequential cyclic carbonates.42 In this study, we chose to utilize water for DAC, rather than organic solvents, due to its environmentally friendly properties and compatibility with subsequent biological processes (Fig. 2A). Tetrabutylammonium hydroxide (TBA·OH) and sodium hydroxide (NaOH) were tested as capture agents for atmospheric CO2 (Fig. 2B). The choice of TBA (tetrabutylammonium) as the preferred cation for CO2 capture is based on our previous work, where it demonstrated superior efficiency compared to other organic salts, such as cholinium, phosphonium, and imidazolium.42 TBA has shown the ability to capture CO2 and convert it into TBAHCO3, which can then be used in the synthesis of industrial chemicals such as PHB. Analysis depicted in Fig. 2C revealed that TBA·OH yielded TBA·HCO3, as evidenced by a signal at 161 ppm in the 13C NMR spectrum, whereas sodium hydroxide yielded carbonate (Na2CO3) species, indicated by a signal at 168 ppm in the 13C NMR spectrum. Furthermore, under these conditions, TBA·OH exhibited an optimal sorption capacity, by fully transforming into TBA·HCO3. Moreover, our findings indicate the scalability of the process to a 100 mL scale.Having successfully captured CO2 into its active form TBA·HCO3, the next objective was to design a catalyst capable of hydrogenating bicarbonate to formate salts to serve as feedstock in the biosynthesis of PHB. Catalytic activity, stability and the ease of separation and reutilisation in subsequent reaction cycles were the key design parameters. Hence, a series of Ru–N-heterocyclic carbenes (NHCs), organometallic complexes supported on macroporous polystyrene-divinylbenzene (PS-DVB) polymeric supports, were synthesised, drawing inspiration from the Ru-MACHO-like shape catalyst, commonly used in the literature.21–23 A one-pot three-step reaction process (Fig. S1–S6†) was employed as a solid-phase synthetic strategy.43 The structure of the resulting organometallic polymeric materials is illustrated in Fig. 3A. These supported Ru–NHC materials were chosen for their potential dual role. They can serve as chemical NHC–metal precursors for metal nanoparticles (MNPs) and act as potential surface ligands for MNPs due to their strong affinity for forming robust bonds with metals.44 This dual functionality can control the generation of MNPs and enhance their effectiveness as catalytic systems by improving their stability, reactivity, and selectivity.45 The formation of NHC complexes and their composition were characterised by different techniques (ESI sections S3–S6 and Table S1†). The Ru–NHC complexes C1 and C5 derived from [RuCl2(CO)2]n exhibited a distinctive band at approximately 1565 cm−1, corresponding to the C–N–C vibration of NHC (Fig. S4†). Additionally, the CO bands linked to the Ru–NHC complex appeared at 2046 cm−1 and 1969 cm−1 and at 2048 cm−1 and 1973 cm−1, for C1 and C5, respectively, confirming the formation of the Ru–NHC complexes. The XPS spectra also confirm their formation (Fig. 3B, left). The catalyst C1 showed two superimposed peaks in the N 1s area (see Fig. S7†). The peak at 399 eV corresponds to a carbenic nitrogen species, while the peak at 402 eV can be attributed to unreacted imidazolium species (Table S1†).46 Despite the strong overlap between the C 1s and Ru 3d spectra, it was also possible to identify the spin–orbit splitting of Ru 3d5/2 and Ru 3d3/2 at 282.1 eV and 286.3 eV, respectively. These peaks correspond to Ru3+, likely in an octahedral geometry as expected for the Ru–NHC complex. Additionally, the Ru–Cl bond was observed, which is consistent with the expected complex (Fig. S7†).47–49 Similar carbene/imidazolium ratios were also observed for C3 obtained employing [RuCl2(p-Cymene)2] instead of [RuCl2(CO)2]n as metal precursors (Fig. S8†).
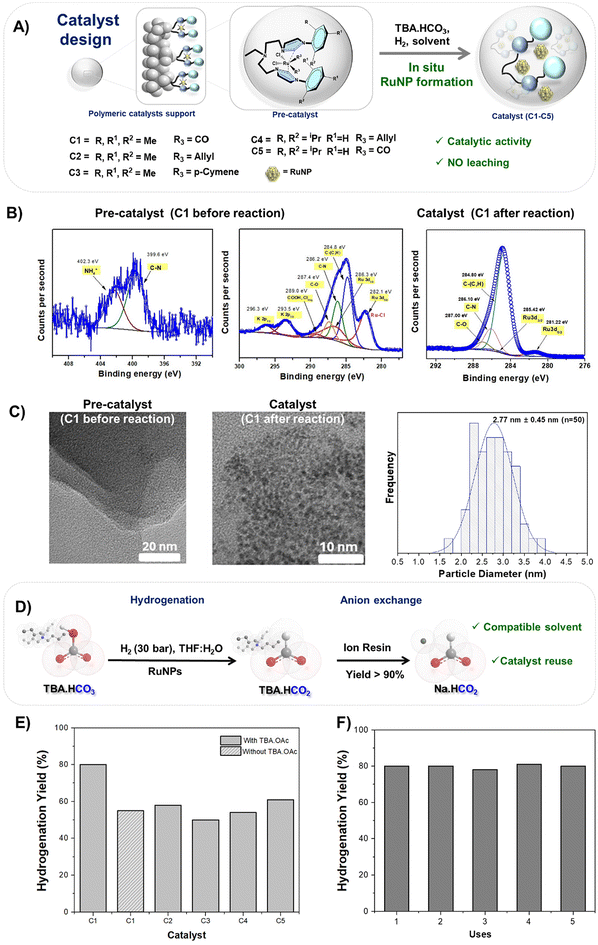 | ||
Fig. 3 Formate generation. (A) General scheme; (B) XPS spectra at the N 1s, C 1s + Ru 3d of C1 before and after the reaction; (C) TEM images of C1 before and after the reaction and histogram of the RuNP size (see Fig. S10–S15†); and (D) scheme of the production of NaHCO2. Cations are not drawn at scale for clarity. (E) Hydrogenation reaction using different catalysts. (F) Recycling experiments of the C1 catalyst. Reaction conditions: TBA·HCO3 (1 mmol); TBA·OAc (0–0.5 mmol), 30 mg pre-catalyst, 30 bar H2, 100 °C, THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 mL H2O (1 mL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL), and 24 h. 1 mL), and 24 h. | ||
After the reaction, the carbon species remain unchanged for compound C1 (Fig. 3B, right). However, the Ru doublet exhibited a shift towards a lower binding energy, with peaks centered at 281.2 eV and 285.4 eV, suggesting a change in its chemical environment, indicating an interaction between Ru and O (Ru–O) bonds. Additionally, the presence of two peaks centered at 462.5 eV and 485 eV, related to Ru 3p3/2 and Ru 3p1/2, respectively, clearly suggests the formation of RuO2 nanoparticles after the reaction. This observation aligns well with previous literature research.47–50
These systems underwent testing in the hydrogenation reaction of TBA·HCO3 to produce TBA·formate (TBA·HCO2) (Fig. 3D–E). Notably, C1 exhibited superior activity (78%) to the rest of the catalysts. TEM images of this catalyst after the reaction revealed the in situ formation of small and homogeneous Ru nanoparticles (RuNPs) with sizes of 2.77 nm ± 0.45 nm (Fig. 3C). Other catalysts displayed larger particle sizes and occasional cluster formation (Fig. S10–S15†). The different results obtained confirm the importance of the electronic parameters and steric environment of the NHC ligands in leading to stable and active RuNP systems. These factors are crucial for optimizing the formation and performance of Ru nanoparticles, thereby enhancing the efficiency of the catalytic process. Indeed, when similar hydrogenation tests were conducted using either [RuCl2(CO)2]n or RuCl3 as metal precursors absorbed onto the support ligand used for the preparation of C1 but without forming NHC complexes, a significant reduction in reaction activity was observed (Fig. S16†). Furthermore, this was accompanied by both metal leaching and the formation of larger nanoparticles (Fig. S16 and S17†). These results underscore the importance of the Ru–NHC precursor in controlling the formation of RuNPs, stabilizing them to prevent aggregation and leaching and thereby enhancing their catalytic efficiency.
A test evaluating catalyst reuse was conducted, revealing that the catalyst (C1) maintained consistent catalytic activity even after undergoing five reaction cycles (Fig. 3F and Fig. S9†). The quantities of the base and bicarbonate were adjusted to ensure equivalence with respect to the Ru equivalents present in the reaction, thus ensuring comparability across all cases. Additionally, the supernatant obtained after drying was analyzed to verify the presence of Ru. The results indicated a minimal degree of leaching (<0.05% relative to Ru amount in the fresh catalyst). The results demonstrate the material's robustness and stability after multiple cycles under identical reaction conditions, thereby confirming the recyclability of the Ru catalyst within the supported heterogeneous material.
It is worth noting that C1 maintained good activity when used in the absence and in the presence of TBA acetate (TBA·OAc), achieving yields of 78% in its presence and 55% in its absence. This reduction in yield can be elucidated by the buffer effect exerted by TBA·OAc. This buffer shifts the equilibrium towards bicarbonate formation, thereby favouring the hydrogenation process towards formate production.51–55 In the absence of this buffer salt, as formate starts to accumulate, the medium becomes more acidic, thereby shifting the equilibrium towards the formation of free CO2, impeding further reaction progression. This methodology was chosen for scaling up production to a larger laboratory scale (10 mL tested) to generate pure formate solutions for the subsequent biocatalytic transformations. The carbon source for formatotrophs and all possible impurities accompanying them should be compatible with the enzymes and microorganisms needed for microbial fermentation, both in terms of toxicity and osmoregulation. For these reasons, two different carbon sources were considered (TBA·HCO2 and NaHCO2) for their conversion into PHB. NaHCO2 was easily obtained from TBA·HCO2 through a cation exchange reaction using an ion exchange resin column, achieving a yield exceeding 90% without exhaustive optimization (Fig. 3D and S19†). In addition, TBA was further reused to regenerate TBA·OH through an anion exchange resin, as detailed in the experimental procedure. It is important to highlight that this approach represents a closed-loop system, ensuring the complete recycling of the materials used. Once it was demonstrated that both sources of formic acid can be produced from CO2 captured from the air, we focused on optimizing a bioprocess that employs them as the sole carbon and energy source for the growth and polyhydroxybutyrate (PHB) accumulation in Cupriavidus necator H16. This continuous bioprocess was optimized using Adaptive Laboratory Evolution (ALE).
Adaptive laboratory evolution
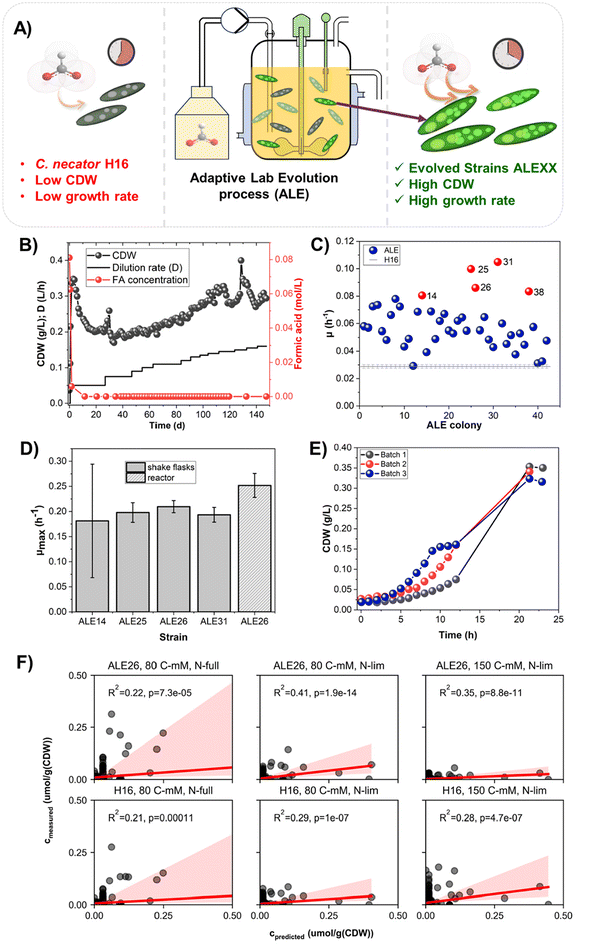
ALE experiments to maximise the growth of C. necator H16 on formic acid with the aim of increasing the maximum growth rate (μmax) of the organism were conducted (Fig. 4A) The mass balance equations around the biomass and growth-limiting substrate in continuous (chemostat) fermentation were calculated. The direct link between the dilution rate (D) and the growth rate (μ) makes the chemostat a great tool to target the growth rate as the optimisation target for ALE campaigns.In this study, C. necator H16 was subjected to continuous fermentation over almost five months with stepwise increasing dilution rates. In prior batch fermentations, the maximum growth rate of the strain in 1 L batch cultivations on 80 mM free formic acid as the sole carbon source was determined as 0.14 ± 0.01 h−1 (Fig. S20†). Throughout the chemostat fermentation, biomass levels were stable over the varying dilution rates and no residual formic acid was detected in the reactor (Fig. 4B, online data shown in Fig. S21†). As a general trend, after the initial establishment of the chemostat, it could be observed that the biomass increased with the increase in the dilution rate. To test the formic acid tolerance of the bacterium, the respective concentration was doubled temporarily after 129 days resulting in a spike in biomass. It normalised again after reverting to the desired 80 mM formic acid in the feed, demonstrating the stability of the process. The highest dilution rate used in the ALE experiment was 0.16 h−1 after which the evolution campaign was terminated.
Screening of ALE variants
From an endpoint sample taken from the ALE experiment, 42 isolated single colonies were selected randomly for growth screening in a BioLector I against a triplicate culture of the wild-type (WT) strain. The μmax of each strain was determined and compared to that of the WT triplicate (Fig. 4C). The five best performing colonies (C. necator ALE14, C. necator ALE25, C. necator ALE26, C. necator ALE31, and C. necator ALE38) were subsequently analysed in shake flask cultures.Growth profiles of C. necator ALE25, ALE26 and ALE31 in flasks (Fig. S22†) were very similar whilst precultures for ALE38 did not grow and ALE14 showed long lag phases. The observed maximum growth rates are shown in Fig. 4D. C. necator ALE26 was selected for further experiments. When grown in a 1 L batch fermentation, growth profiles of C. necator ALE26 differed slightly across repeats (Fig. 4E). However, the final biomass titres and maximal observed growth rates across repeats are very similar (Fig. 4D). The observed μmax for C. necator ALE26 on 80 mM formic acid was 0.25 ± 0.02 h−1, signifying a 1.8-fold increase compared to what was observed for the WT strain in this study and a 1.4-fold improvement compared to the literature.29
Strain characterisation
Genome sequencing of C. necator ALE26 and comparison with the genome of the WT strain revealed that the main difference between the strains originated from the absence of the megaplasmid pHG1 in the evolved strain (see ESI, section S10 and Fig. S23†).This pHG1-loss is interestingly parallel to the findings by Calvey et al., although here a full loss of the plasmid was found, whereas their study reported large partial deletions.56 The rationalisation for the deletions given in their work holds true in this case as well. While pHG1 harbours a second copy of the CBB operon, otherwise also present on chromosome 2, this second copy lacks the LysR-type transcriptional regulator gene cbbR.57 Other genetic elements responsible for hydrogen metabolism, degradation of aromatic compounds and plasmid maintenance and transmission do not play a vital role in the cultivation conditions applied,58 although many of the respective genes have been shown to be constitutively expressed across several growth conditions, even in those where they do not seem to have an active metabolic role.59,60 Both strains exhibit very similar maximum growth rates on formic acid despite the different approaches for laboratory evolution. Strikingly, however, the highest growth rate documented in their work (0.245 ± 0.011 h−1) was observed for a deletion mutant of the transcriptional regulator PhcA (ΔphcA). The gene in their adapted strain was disrupted by a frameshift mutation and was identified as the major contributor to the improved growth rates alongside the (partial) deletion of pHG1. This gene corresponds to the locus E6A55_00465 in the genome sequence used for reference in this work.61 The same mutation could not be observed in this study, where the gene appears to be undisrupted.
For further functional analysis of the newly evolved strain, both this and the WT strain were grown in continuous culture under three different growth conditions: on 80 mM formic acid at a carbon/nitrogen ratio (C/N ratio, [mol/mol]) of (i) 5.3 mol/mol or (ii) 50 mol/mol as well as (iii) 150 mM formic acid at C/N = 50 mol/mol, all at a dilution rate of 0.09 h−1. Under every condition, a large number of proteins was observed only in one of the two strains (Fig. S24†). Many of these proteins correlate with proteins found on pHG1, yet there are additional chromosomal genes that only see the functional expression in one or the other organism. Regulatory shifts across cellular processes like quorum sensing, for which annotated proteins appear to be consistently upregulated across growth conditions, indicate a wide-ranging reorganisation of the gene expression in the evolved strain.
To analyse general trends of proteome utilisation between the strains, resource balance analysis (RBA) was employed.62,63 In the mathematical model, the metabolic cost associated with the protein expression is minimised by reducing the expression to a minimum necessary to support that growth rate. Our hypothesis was that if C. necator ALE26 outperforms the WT strain in terms of growth the improved growth rate of C. necator ALE26 should be reflected to some extent in the allocation of resources, i.e. the protein expression. Therefore, the proteome profile of the strain should correlate more closely with an optimised prediction provided by the RBA module. Proteomic prediction was performed for proteins contained in the model as well as in the proteomic data sets, predicted and measured levels were then correlated and the squared Pearson correlation coefficient was calculated (Fig. 4F).59 For each tested growth condition, the correlation is higher for the evolved strain than for the WT strain, indicating a protein distribution closer to a predicted optimum. Notably, this correlation is the greatest for the growth condition on 80 mM formic acid and under nitrogen limiting (i.e. PHB accumulating) conditions. Here, the overall nutrient availability is the lowest out of the three tested conditions (low carbon and nitrogen), leading to the predicted μmax being the closest to the actual growth rate (as determined by the dilution rate). This feeds into our hypothesis of resource conservation following a deletion of unnecessarily expressed genes (in the form of pHG1) enabling increased growth. While the strain presented here shows a similar growth rate to the ΔphcA-strain mentioned above without the loss or disruption of the eponymous gene, it should be noted that the growth rate determined by Calvey et al. was taken from a flask experiment. The one demonstrated here stems from fermentation in a stirred tank reactor. Growth rates tend to be higher in these more controlled systems. Therefore, a genomic deletion of the transcriptional regulator might further enhance the growth performance of C. necator ALE26 in bioreactors.
Polyhydroxybutyrate production
The accumulation of PHB in continuous fermentation was optimised with C. necator ALE26 in a biotechnological context (Fig. 5A). The parameters optimised in continuous fermentation were C/N ratio, dilution rate and absolute carbon concentration. Growth profiles for fermentations altering these parameters can be found in the ESI (Fig. S25–S28† for cultivation of C. necator H16).C/N ratios of 25 mol/mol and lower resulted in a very low PHB content (Fig. 5A). A C/N ratio of 50 mol/mol and higher resulted in bacterial cells accumulating approximately 15% of CDW as PHB, with higher C/N ratios showing similar PHB contents. Nevertheless, the maximum space time yield (STY) was achieved at 50 mol/mol with a drop in productivity at higher ratios. This correlates with a higher total biomass at lower C/N ratios (Fig. S30†). A low C/N ratio showing the best productivity falls in line with experiments that have shown that the ratio between carbon and the growth-limiting nutrient is not arbitrary as a minimal residual nutrient concentration is required for cellular maintenance processes.64–66
The results from varying the dilution rate suggest a decreasing trend in PHB accumulation with increasing dilution rates, as the percentage of PHB of the CDW dropped from around 21.37 ± 4.99% at 0.050 h−1 to 9.47 ± 0.05% at 0.165 h−1 (Fig. 5C). However, when factoring in the time component via the dilution rate, it became apparent that the STY did not change drastically for dilution rates between 0.07 h−1 and 0.165 h−1, reaching its maximum at 3.9 ± 0.2 mg L−1 h−1 at D = 0.09 h−1. This follows the proposed partial coupling between growth- and PHB accumulation rates by Grousseau et al.65 which explains why the chemostat is ideally performed at medium dilution rates rather than close to the μmax of the organism.
The higher formic acid concentration did not correlate with a higher PHB content per cell (Fig. 5D). In fact, the PHB content dropped slightly at the higher substrate concentrations. Overall, the PHB content of C. necator ALE26 was significantly higher than in the WT strain, reaching up to 20.3 ± 0.5% (Fig. S29†). However, the productivity (STY) in both cases increased with higher formic acid concentrations as the active biomass increased. Again, the evolved strain outperformed the WT strain with a maximum productivity of 8.67 ± 1.26 mg L−1 h−1 at a formic acid feed concentration of 150 mM. This translates to a carbon yield of 0.03 mol(CPHB)/mol(Cformic acid).
The conservation of resources from losing pHG1, as discussed earlier, could also explain the discrepancy seen in PHB accumulation between C. necator H16 and ALE26 (Fig. S29†). As fewer nutrients need to be dedicated to the protein expression or cell division, more can be allocated towards carbon storage in the adapted strain. It has been shown before that under PHA producing conditions, the cellular composition changes, with proportionally less protein expression during nutrient limitation64 which reflects the shifting of resources necessary to accumulate the polymer. This shift might be made easier by the lack of the additional metabolic burden exhibited by the megaplasmid.
Overall, the STY of PHB from formic acid achieved in this work is on par with or surpasses those documented so far whilst being more robust over time than a fed-batch fermentation.35,36,39 The comparatively low polymer yields might be improved with further process optimisation and more complex setups such as multiphase fermentations and medium recirculation methods.
Conversion of atmospheric CO2 into PHP via formate to bridge air-capturing CO2 to producing PHB, the primary product, TBA·HCO2, was tested for the growth of C. necator ALE26 in the BioLector. The organism was cultivated on varying concentrations of TBA·HCO2 as the sole carbon source as well as on free formic acid and sodium gluconate supplemented with TBA·HCO2 in varying concentrations (Fig. S30†). Growth was observed for conditions in which TBA·HCO2 was the sole carbon source as well as when it was added with formic acid or sodium gluconate, albeit at very slow rates.
While C. necator ALE26 grew on low concentrations of TBA·HCO2 and accumulated up to 29.51% (gPHB gCDW−1) in shake flasks (Fig. S31†), its growth rate was too low to permit growth in continuous fermentations. With the development of the ion-exchange method to convert TBA·HCO2 into sodium formate, the latter can be used for continuous fermentation whilst avoiding the toxicity of the TBA cation. The overall productivity of PHB on sodium formate was similar to that of the free acid (Fig. 5E). Thus, fermentation was carried out on 160 mM sodium formate as the sole carbon source and with optimised conditions for PHB accumulation (D = 0.09 h−1 and C/N = 50 mol/mol, Fig. 5F). Samples for PHB measurement were taken under full and nitrogen-limited conditions for comparison with the growth on free formic acid. A final PHB content of 18.73 ± 5.59% was achieved, giving a STY of 6.34 ± 1.75 mg L−1 h−1 (Fig. 5E).
The synthesis of PHB from CO2 waste streams is the subject of numerous studies, but the use of CO2 or CO2-derived compounds as primary reactants remains relatively uncommon. Table S6† highlights the most relevant studies where pure CO2 or CO2 from flue gas has been employed in the production of PHB under diverse biological conditions. In phototrophic processes with the flue gas containing 3–6% of CO2, STYs of maximum 3.02 mg L−1 h−1 have been reported (entries 1 and 2).38,67 The highest productivity to date described for PHB accumulation from CO2-derived formate was found in a two-stage system employing non-growing C. necator H16 cells in a biotransformation (entry 3).39 With this system, Dinges et al. achieved up to 8.4 mg L−1 h−1 (per OD600). However, while their experiments gave valuable insights into theoretical PHB yields from formate, biotransformations such as the one described generally become unstable over time due to cell lysis or enzyme degradation. While yields lower than those reported for pure CO2 as the carbon source (up to 12.54 mg L−1 h−1),68,69 those studies using diluted sources of CO2 demonstrate that these waste gases may still be usable as a direct feedstock for polymer accumulation.
Our results, where CO2 was directly captured from the air (0.04% of CO2), show that the PHB STY (6.34 ± 1.75 mg L−1 h−1, entry 4) is comparable to results where diluted CO2 or formate was utilized. At the same time, to the best of the author's knowledge, this is the first time continuous fermentation for accumulation of PHB from formate has been described, which can be run for long periods of time without the risk of instabilities or the requirement for repeated stopping and starting of the process. This underscores the potential of our integrated biochemical and catalytic process to efficiently produce PHB, a chemical of significant industrial interest.
Conclusion
This study is a proof-of-concept of a chemo-biocatalytic process to upcycle captured atmospheric CO2via formate salts to polyhydroxybutyrate. The direct capture process employing ionic liquids under aqueous conditions, and novel immobilised nanostructured catalysts, represents a simple, yet efficient means that will open up new possibilities for transforming CO2 into valuable products. This non-conventional feedstock was employed to produce a biodegradable polymer polyhydroxybutyrate. This process integrates CO2 capture and polypropylene-like polymer production completely decoupled from fossil-feedstock, and the end product opens multiple applications and new end-of-life management options. The power of adaptive laboratory evolution to yield a strain with an improved growth rate due to the optimised resource efficiency, coupled with bioprocess development showcased here can also serve as a basis to further exploit existing microbial biocatalysts’ anabolic reactions, or metabolism extended by new-to-nature synthesis pathways to produce a plethora of valuable products.This first proof of concept of an interdisciplinary pipeline for atmospheric CO2 fixation and utilisation to generate biodegradable polymers (Fig. 6) will inspire new avenues to simultaneously help tackle climate change and generate circular economy approaches for the plastic industry.
 | ||
| Fig. 6 Schematic representation of a pipeline for atmospheric CO2 capture and transformation in biodegradable polymers. | ||
Experimental
Direct air capture
The samples for CO2 capture were prepared using a solution of 1 mol L−1 of TBA·OH in H2O (0.5–100 mL). The sorption experiments were performed by bubbling the gas (compressed air) in vial glass tubes with a septum at 25 °C for 48 h.42 For the CO2 sorption quantification, we have previously established this NMR methodology for CO2 quantification in ILs.70,71 Typically, 13C NMR inverse gated 1H decoupled spectra were acquired using an inversion recovery experiment (zgig) with a relaxation delay of 60 seconds. The experiments were performed with 64 transients with 64k data points were collected.Catalyst preparation
Ru(CO)2Cl2 was synthesised according to a previously reported method (see section S3 in the ESI†).72Hydrogenation reaction
Catalytic reactions were performed in a stainless-steel batch reactor of 30 mL with a pressure regulator. Typically, 1 mmol of TBA·HCO3, 0.5 mmol of TBA·OAc and a mixture of the solvent (in almost all of the cases 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of water/THF) were put together along with the desired amount of the catalyst. Then, the reactor was fully pressured at 5, 30 or 50 H2 bar based on each specific condition. Initially, the reactions were carried out at 100 °C; however, in some cases, the effects of the temperature were also studied. Quantification of the amount of formate (yield) was performed with 1H NMR with the signal at ∼8.3 ppm and referenced with the signals of the TBA cation at ∼0.8 ppm (18 protons) (see Fig. S18 and S19 in the ESI†). For the reuse, the catalyst was removed from the reaction mixture by centrifugation and the residual was washed with dichloromethane and methanol (3 × 10 mL) and dried overnight at 50 °C (vacuum). The liquid phase was separated to further cation exchange. The remaining catalyst was analyzed by using ICP-MS to estimate the Ru amount (in ppm). The catalyst was also tested as used without any pretreatment, and the results were compared with the washed and dried material.
1 of water/THF) were put together along with the desired amount of the catalyst. Then, the reactor was fully pressured at 5, 30 or 50 H2 bar based on each specific condition. Initially, the reactions were carried out at 100 °C; however, in some cases, the effects of the temperature were also studied. Quantification of the amount of formate (yield) was performed with 1H NMR with the signal at ∼8.3 ppm and referenced with the signals of the TBA cation at ∼0.8 ppm (18 protons) (see Fig. S18 and S19 in the ESI†). For the reuse, the catalyst was removed from the reaction mixture by centrifugation and the residual was washed with dichloromethane and methanol (3 × 10 mL) and dried overnight at 50 °C (vacuum). The liquid phase was separated to further cation exchange. The remaining catalyst was analyzed by using ICP-MS to estimate the Ru amount (in ppm). The catalyst was also tested as used without any pretreatment, and the results were compared with the washed and dried material.
Cation and anion exchange procedure
The cation exchange from TBA·HCO2 to NaHCO2 was conducted using an ion exchange resin (Amberlite IR-120 in H+ form). Aqueous TBA·HCO2 solution (1 M) was gradually passed through a resin-filled column. The eluate was then collected in a vial containing an equimolar amount of NaOH solution, resulting in the formation of Na HCO2. The solvent was evaporated under reduced pressure using a rotary evaporator and dried for 8 hours at 60 °C under vacuum. The TBA was further reused to produce TBA·OH an Amberlyst A26 (OH− form), according to a previously described procedure.42Cultivation in bioreactors
All bioreactor cultivations were carried out in stirred tank vessels of the F0-Baby model by Bionet (Fuente Álamo (Murcia), Spain) at a working volume of 1 L. The process was controlled and monitored using the company's ROSITA software.Cultivation in a BioLector
For condition testing and screening, C. necator was cultivated in a BioLector 1 (Beckman Coulter, Brea, California, USA). Cultivations were performed in 1 mL of the total culture volume in 48-well flower plates with oxygen and pH sensors (MTP-48-BOH 1, Beckman Coulter). The temperature in the incubation chamber was controlled at 30 °C, humidity at 70–80%, and shaking was set to 1000 rpm. The plate was inoculated from two-step 3 mL J-MM precultures, similar to the preparation for Cultivation in Baffled Shake Flasks. Cultivations were performed in J-MM with the required amounts of carbon and nitrogen added for each experiment. Blanks were run in triplicate of the uninoculated medium. The ALE experiment and subsequent screening are described in the ESI in sections S2.5 and S2.6–S2.9.† Analytical procedures accompanying the fermentations are described in the ESI in section S2.10.†Proteomics analysis
For proteomics analysis of C. necator strains under different growth conditions, biological duplicate samples were collected and further split into two technical replicates each (sample preparation is detailed in the ESI, S2.11.1†). Samples were handed to the Proteomics Core facilities of University College Dublin. In these facilities, samples were run in a Thermo Scientific Q Exactive workflow with the Thermo Scientific Ultimate 3000 RSLCnano chromatography system (electrospray ionisation liquid chromatography-mass spectrometry (ESI LC-MS), using a quadrupole with high resolution high mass accuracy (HRAM) by Orbitrap). Peaks were analysed in these facilities using the MaxQuant software. Data analysis and statistics were performed in Perseus (Max Planck Institute of Biochemistry, München, Germany), GraphPad PRISM and Python, as outlined in the ESI, S2.11.2 and S2.11.3.†Author contributions
VS, KO, MZ, and TN: conceptualisation; MB, JESV, and EGV: methodology; MB and NP: software programming; MB, MZ, JESV and JRP: investigation; MB, JESV, and MZ: writing – original draft; all authors: writing – review and editing; MZ, VS, KO, TN, and EGV: supervision; MZ, VS, KO, and EGV: project administration; MZ, VS, EGV, and KO: funding acquisition.Data availability
The data supporting this article are available on Zenodo (https://doi.org/10.5281/zenodo.13366128), the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE74 partner repository with the dataset identifier PXD056335, and are also included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the projects (PID2021-124695OB-C22), MICINN/AEI/10.13039/501100011033 and TED2021-130288B-I00 funded by MCIN/AEI/10.13039/501100011033 and the Unión Europea NextGenerationEU/PRTR. Generalitat Valenciana is gratefully acknowledged for funding for GenT (CIDEGENT 2018/036). The project that gave rise to these results received the support of a fellowship LCF/BQ/PR24/12050016 from the “la Caixa” Foundation (ID 100010434). This work was further supported by the Science Foundation Ireland BiOrbic Research Centre (SFI reference number: 16/RC/3889) through the Centre for Doctoral Training Atoms-to-Products – an Integrated Approach to Sustainable Chemistry (BiOrbic) under the grant number 18/EPSRC-CDT/3582. TN is a recipient of the Ad Astra Fellowship at UCD. The authors acknowledge the Ministry of Education, Youth and Sports of the Czech Republic, for the financial support of XPS measurements using the CEMNAT infrastructure (project LM 2023037).References
- IPCC, Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, 2021 Search PubMed.
- IPCC, Climate Change 2022: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, 2022 Search PubMed.
- IPCC, Climate Change 2022: Mitigation of Climate Change. Contribution of Working Group III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, 2022 Search PubMed.
- G. Althor, J. E. M. Watson and R. A. Fuller, Sci. Rep., 2016, 6, 20281 CrossRef CAS.
- P. Friedlingstein, M. O'Sullivan, M. W. Jones, R. M. Andrew, L. Gregor, J. Hauck, C. Le Quéré, I. T. Luijkx, A. Olsen, G. P. Peters, W. Peters, J. Pongratz, C. Schwingshackl, S. Sitch, J. G. Canadell, P. Ciais, R. B. Jackson, S. R. Alin, R. Alkama, A. Arneth, V. K. Arora, N. R. Bates, M. Becker, N. Bellouin, H. C. Bittig, L. Bopp, F. Chevallier, L. P. Chini, M. Cronin, W. Evans, S. Falk, R. A. Feely, T. Gasser, M. Gehlen, T. Gkritzalis, L. Gloege, G. Grassi, N. Gruber, Ö. Gürses, I. Harris, M. Hefner, R. A. Houghton, G. C. Hurtt, Y. Iida, T. Ilyina, A. K. Jain, A. Jersild, K. Kadono, E. Kato, D. Kennedy, K. Klein Goldewijk, J. Knauer, J. I. Korsbakken, P. Landschützer, N. Lefèvre, K. Lindsay, J. Liu, Z. Liu, G. Marland, N. Mayot, M. J. McGrath, N. Metzl, N. M. Monacci, D. R. Munro, S. I. Nakaoka, Y. Niwa, K. O'Brien, T. Ono, P. I. Palmer, N. Pan, D. Pierrot, K. Pocock, B. Poulter, L. Resplandy, E. Robertson, C. Rödenbeck, C. Rodriguez, T. M. Rosan, J. Schwinger, R. Séférian, J. D. Shutler, I. Skjelvan, T. Steinhoff, Q. Sun, A. J. Sutton, C. Sweeney, S. Takao, T. Tanhua, P. P. Tans, X. Tian, H. Tian, B. Tilbrook, H. Tsujino, F. Tubiello, G. R. van der Werf, A. P. Walker, R. Wanninkhof, C. Whitehead, A. Willstrand Wranne, R. Wright, W. Yuan, C. Yue, X. Yue, S. Zaehle, J. Zeng and B. Zheng, Earth Syst. Sci. Data, 2022, 14, 4811–4900 CrossRef.
- T. M. Gür, Prog. Energy Combust. Sci., 2022, 89, 100965 CrossRef.
- H. Li, M. E. Zick, T. Trisukhon, M. Signorile, X. Liu, H. Eastmond, S. Sharma, T. L. Spreng, J. Taylor, J. W. Gittins, C. Farrow, S. A. Lim, V. Crocellà, P. J. Milner and A. C. Forse, Nature, 2024, 630, 654–659 CrossRef CAS.
- M. Zanatta, ACS Mater. Au, 2023, 3, 576–583 CrossRef CAS.
- A. Weilhard, S. P. Argent and V. Sans, Nat. Commun., 2021, 12, 231 CrossRef CAS.
- S. Moret, P. J. Dyson and G. Laurenczy, Nat. Commun., 2014, 5, 4017 CrossRef CAS.
- E. González, C. Marchant, C. Sepúlveda, R. García, I. T. Ghampson, N. Escalona and J. L. García-Fierro, Appl. Catal., B, 2018, 224, 368–375 CrossRef.
- A. Bahuguna and Y. Sasson, ChemSusChem, 2021, 14, 1258–1283 CrossRef CAS.
- C. Hao, S. Wang, M. Li, L. Kang and X. Ma, Catal. Today, 2011, 160, 184–190 CrossRef CAS.
- X. Meng, Z. Ju, S. Zhang, X. Liang, N. Von Solms, X. Zhang and X. Zhang, Green Chem., 2019, 21, 3456–3463 RSC.
- W. Hui, X. Wang, X.-N. Li, H.-J. Wang, X.-M. He and X.-Y. Xu, New J. Chem., 2021, 45, 10741–10748 RSC.
- J. Kothandaraman, A. Goeppert, M. Czaun, G. A. Olah and G. K. S. Prakash, J. Am. Chem. Soc., 2016, 138, 778–781 CrossRef CAS.
- R. Sen, A. Goeppert, S. Kar and G. K. S. Prakash, J. Am. Chem. Soc., 2020, 142, 4544–4549 CrossRef CAS.
- R. Sen, C. J. Koch, V. Galvan, N. Entesari, A. Goeppert and G. K. S. Prakash, J. CO2 Util., 2021, 54, 101762 CrossRef CAS.
- S. Kar, A. Goeppert and G. K. S. Prakash, ChemSusChem, 2019, 12, 3172–3177 CrossRef CAS.
- D. Mellmann, P. Sponholz, H. Junge and M. Beller, Chem. Soc. Rev., 2016, 45, 3954–3988 RSC.
- R. Bhardwaj, A. Kumar and J. Choudhury, Chem. Commun., 2022, 58, 11531–11534 RSC.
- D. Wei, H. Junge and M. Beller, Chem. Sci., 2021, 12, 6020–6024 RSC.
- C. Guan, Y. Pan, E. P. L. Ang, J. Hu, C. Yao, M.-H. Huang, H. Li, Z. Lai and K.-W. Huang, Green Chem., 2018, 20, 4201–4205 RSC.
- A. B. Hooper and A. A. DiSpirito, in Encyclopedia of Biological Chemistry (Second Edition), ed. W. J. Lennarz and M. D. Lane, Academic Press, Waltham, 2013, pp. 486–492. DOI:10.1016/B978-0-12-378630-2.00219-X.
- H. Salehizadeh, N. Yan and R. Farnood, Chem. Eng. J., 2020, 390, 124584 CrossRef CAS.
- N. S. Kruyer and P. Peralta-Yahya, Biochemistry, 2020, 59, 731–732 CrossRef CAS.
- F. Joó, ChemSusChem, 2008, 1, 805–808 CrossRef.
- S. F. Tolesorkhi, F. Esmaeilzadeh and M. Riazi, Pet. Res., 2018, 3, 370–380 Search PubMed.
- S. Grunwald, A. Mottet, E. Grousseau, J. K. Plassmeier, M. K. Popović, J.-L. Uribelarrea, N. Gorret, S. E. Guillouet and A. Sinskey, Microb. Biotechnol., 2015, 8, 155–163 CrossRef CAS.
- M. Beeby, M. Cho, J. Stubbe and G. J. Jensen, J. Bacteriol., 2012, 194, 1092–1099 CrossRef CAS.
- L. Garcia-Gonzalez, M. S. I. Mozumder, M. Dubreuil, E. I. P. Volcke and H. De Wever, Catal. Today, 2015, 257, 237–245 CrossRef CAS.
- H. G. Schlegel, R. Lafferty and I. Krauss, Arch. Mikrobiol., 1970, 71, 283–294 CAS.
- A. Steinbüchel and B. Füchtenbusch, Trends Biotechnol., 1998, 16, 419–427 CrossRef.
- J. Lu, C. J. Brigham, S. Li and A. J. Sinskey, in Biotechnology for Biofuel Production and Optimization, ed. C. A. Eckert and C. T. Trinh, Elsevier, Amsterdam, 2016, ch. 12, pp. 325–351. DOI:10.1016/B978-0-444-63475-7.00012-1.
- R. Tang, C. Weng, X. Peng and Y. Han, Metab. Eng., 2020, 61, 11–23 CrossRef CAS.
- S. Kim, Y. J. Jang, G. Gong, S.-M. Lee, Y. Um, K. H. Kim and J. K. Ko, Microb. Cell Fact., 2022, 21, 231 CrossRef CAS.
- A. Satanowski and A. Bar-Even, EMBO Rep., 2020, 21, e50273 CrossRef CAS.
- A. Langsdorf, J. P. Schütz, R. Ulber, M. Stöckl and D. Holtmann, J. CO2 Util., 2024, 83, 102800 CrossRef CAS.
- I. Dinges, I. Depentori, L. Gans, D. Holtmann, S. R. Waldvogel and M. Stöckl, ChemSusChem, 2024, 17, e202301721 CrossRef CAS.
- K. Müller, K. Brooks and T. Autrey, Energy Fuels, 2017, 31, 12603–12611 CrossRef.
- D. J. Heldebrant, J. Kothandaraman, N. M. Dowell and L. Brickett, Chem. Sci., 2022, 13, 6445–6456 RSC.
- M. Zanatta, E. García-Verdugo and V. Sans, ACS Sustainable Chem. Eng., 2023, 11, 9613–9619 CrossRef CAS.
- V. Sans, F. Gelat, M. I. Burguete, E. Garcia-Verdugo and S. V. Luis, Catal. Today, 2012, 196, 137–147 CrossRef CAS.
- M. Ghosh and S. Khan, ACS Catal., 2023, 13, 9313–9325 CrossRef CAS.
- M. Koy, P. Bellotti, M. Das and F. Glorius, Nat. Catal., 2021, 4, 352–363 CrossRef CAS.
- A. Palazzolo, T. Naret, M. Daniel-Bertrand, D.-A. Buisson, S. Tricard, P. Lesot, Y. Coppel, B. Chaudret, S. Feuillastre and G. Pieters, Angew. Chem., Int. Ed., 2020, 59, 20879–20884 CrossRef CAS.
- S. M. Thalluri, J. Rodriguez-Pereira, R. Zazpe, B. Bawab, E. Kolíbalová, L. Jelinek and J. M. Macak, Small, 2023, 19, 2300974 CrossRef CAS.
- H. Wang, X. Li, Q. Ruan and J. Tang, Nanoscale, 2020, 12, 12329–12335 RSC.
- A. F. Pedersen, M. Escudero-Escribano, B. Sebok, A. Bodin, E. Paoli, R. Frydendal, D. Friebel, I. E. L. Stephens, J. Rossmeisl, I. Chorkendorff and A. Nilsson, J. Phys. Chem. B, 2018, 122, 878–887 CrossRef CAS.
- M. Lin, L.-X. Dai, J. Gu, L.-Q. Kang, Y.-H. Wang, R. Si, Z.-Q. Zhao, W.-C. Liu, X. Fu, L.-D. Sun, Y.-W. Zhang and C.-H. Yan, RSC Adv., 2017, 7, 33078–33085 RSC.
- A. Weilhard, M. I. Qadir, V. Sans and J. Dupont, ACS Catal., 2018, 8, 1628–1634 CrossRef CAS.
- L. Piccirilli, B. Rabell, R. Padilla, A. Riisager, S. Das and M. Nielsen, J. Am. Chem. Soc., 2023, 145, 5655–5663 CrossRef CAS.
- W. D. G. Gonçalves, M. Zanatta, N. M. Simon, L. M. Rutzen, D. A. Walsh and J. Dupont, ChemSusChem, 2019, 12, 4170–4175 CrossRef.
- N. M. Simon, M. Zanatta, F. P. dos Santos, M. C. Corvo, E. J. Cabrita and J. Dupont, ChemSusChem, 2017, 10, 4927–4933 CrossRef CAS.
- Y. Yasaka, C. Wakai, N. Matubayasi and M. Nakahara, J. Phys. Chem. A, 2010, 114, 3510–3515 CrossRef CAS.
- C. H. Calvey, V. Sànchez I Nogué, A. M. White, C. M. Kneucker, S. P. Woodworth, H. M. Alt, C. A. Eckert and C. W. Johnson, Metab. Eng., 2023, 75, 78–90 CrossRef CAS.
- A. Pohlmann, W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Pötter, E. Schwartz, A. Strittmatter, I. Voß, G. Gottschalk, A. Steinbüchel, B. Friedrich and B. Bowien, Nat. Biotechnol., 2006, 24, 1257–1262 CrossRef.
- E. Schwartz, A. Henne, R. Cramm, T. Eitinger, B. Friedrich and G. Gottschalk, J. Mol. Biol., 2003, 332, 369–383 CrossRef CAS.
- M. Jahn, N. Crang, M. Janasch, A. Hober, B. Forsström, K. Kimler, A. Mattausch, Q. Chen, J. Asplund-Samuelsson and E. P. Hudson, eLife, 2021, 10, e69019 CrossRef CAS.
- N. Pearcy, M. Garavaglia, T. Millat, J. P. Gilbert, Y. Song, H. Hartman, C. Woods, C. Tomi-Andrino, R. Reddy Bommareddy, B.-K. Cho, D. A. Fell, M. Poolman, J. R. King, K. Winzer, J. Twycross and N. P. Minton, PLoS Comput. Biol., 2022, 18, e1010106 CrossRef CAS.
- G. T. Little, M. Ehsaan, C. Arenas-López, K. Jawed, K. Winzer, K. Kovacs and N. P. Minton, Microbiol. Resour. Announce., 2019, 8 DOI:10.1128/mra.00814-19.
- A. Goelzer, V. Fromion and G. Scorletti, Automatica, 2011, 47, 1210–1218 CrossRef.
- A. Goelzer, J. Muntel, V. Chubukov, M. Jules, E. Prestel, R. Nölker, M. Mariadassou, S. Aymerich, M. Hecker, P. Noirot, D. Becher and V. Fromion, Metab. Eng., 2015, 32, 232–243 CrossRef CAS.
- M. Zinn, R. Durner, H. Zinn, Q. Ren, T. Egli and B. Witholt, Biotechnol. J., 2011, 6, 1240–1252 CrossRef CAS.
- E. Grousseau, E. Blanchet, S. Déléris, M. G. E. Albuquerque, E. Paul and J.-L. Uribelarrea, Bioresour. Technol., 2013, 148, 30–38 CrossRef CAS.
- E. Grousseau, E. Blanchet, S. Déléris, M. G. E. Albuquerque, E. Paul and J.-L. Uribelarrea, Bioresour. Technol., 2014, 153, 206–215 CrossRef CAS.
- H. Roh, J. S. Lee, H. I. Choi, Y. J. Sung, S. Y. Choi, H. M. Woo and S. J. Sim, Bioresour. Technol., 2021, 327, 124789 CrossRef CAS.
- S. Cestellos-Blanco, S. Friedline, K. B. Sander, A. J. Abel, J. M. Kim, D. S. Clark, A. P. Arkin and P. Yang, Front. Microbiol., 2021, 12, 700010 CrossRef.
- N. Taga, K. Tanaka and A. Ishizaki, Biotechnol. Bioeng., 1997, 53, 529–533 CrossRef CAS.
- M. Zanatta, M. Lopes, E. J. Cabrita, C. E. S. Bernardes and M. C. Corvo, J. CO2 Util., 2020, 41, 101225–101225 CrossRef CAS.
- M. Zanatta, N. M. Simon, F. P. dos Santos, M. C. Corvo, E. J. Cabrita and J. Dupont, Angew. Chem., Int. Ed., 2019, 58, 382–385 CrossRef CAS.
- P. A. Anderson, G. B. Deacon, K. H. Haarmann, F. R. Keene, T. J. Meyer, D. A. Reitsma, B. W. Skelton, G. F. Strouse and N. C. Thomas, Inorg. Chem., 1995, 34, 6145–6157 CrossRef CAS.
- H. Li, P. H. Opgenorth, D. G. Wernick, S. Rogers, T.-Y. Wu, W. Higashide, P. Malati, Y.-X. Huo, K. M. Cho and J. C. Liao, Science, 2012, 335, 1596–1596 CrossRef CAS.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. García-Seisdedos, S. Hewapathirana, S. Kamatchinathan, D. J. Kundu, A. Prakash, A. Frericks-Zipper, M. Eisenacher, M. Walzer, S. Wang, A. Brazma and J. A. Vizcaíno, Nucleic Acids Res., 2022, 50(D1), D543–D552 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc04228j |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2024 |