Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Increasing the diversity of nylonases for poly(ester amide) degradation†
Jan
de Witt
 a,
Maike-Elisa
Ostheller
b,
Kenneth
Jensen
a,
Maike-Elisa
Ostheller
b,
Kenneth
Jensen
 c,
Christian A. M. R.
van Slagmaat
c,
Christian A. M. R.
van Slagmaat
 d,
Tino
Polen
d,
Tino
Polen
 a,
Gunnar
Seide
b,
Stephan
Thies
a,
Gunnar
Seide
b,
Stephan
Thies
 a,
Benedikt
Wynands
a,
Benedikt
Wynands
 a and
Nick
Wierckx
a and
Nick
Wierckx
 *a
*a
aInstitute of Bio- and Geosciences IBG-1: Biotechnology, Forschungszentrum Jülich, Jülich, Germany. E-mail: n.wierckx@fz-juelich.de
bAachen-Maastricht Institute for Biobased Materials (AMIBM), Maastricht University, Brightlands Chemelot Campus, Urmonderbaan 22, 6167 RD Geleen, The Netherlands
cNovonesis A/S, Biologiens Vej 2, Kgs. Lyngby DK-2800, Denmark
dB4Plastics BV, IQ-parklaan 2A, 3650, Dilsen-Stokkem, Belgium
First published on 6th August 2024
Abstract
Global production of synthetic polyamides (PA), or nylons, is increasing while recycling rates are currently below 5% contributing to the global plastics crisis. Enzymatic depolymerization is a powerful strategy to overcome the drawbacks of mechanical and chemical recycling and has the potential to increase PA recycling rates. However, enzymatic depolymerization of PA is currently limited to a small group of nylonases (NylC) that exhibit low activities making them unsuitable for efficient enzymatic recycling. In this study, we extend the diversity of nylonases, namely NylC1, NylC2, and NylC3 by library screenings and in silico analysis. Three novel nylonases were identified that showed varying sequence identities ranging from 84 to 32% compared to the previously characterized NylCp2 from Paenarthrobacter ureafaciens. Activity of these nylonase candidates towards cyclic PA-oligomers was confirmed via the detection of soluble degradation products. These nylonases were also active on synthesized poly(ester amides) (PEA), and this activity was synergistically increased by combination with the leaf and branch compost cutinase LCC resulting in the hydrolysis of approximately 1% of the total polymer. Overall, our discoveries greatly increase the sequence space of NylC enzymes for future enzyme engineering strategies to boost their activities, and they show the potential of PEA for tuning the biodegradability of performance polymers. Thereby, this study leads the path for developing efficient enzymatic PA and PEA depolymerization processes, revealing significant insights into combining the contrary parameters of performance and biodegradability of polymers.
1 Introduction
Synthetic polyamides (PA), or nylons, are characterized by their durability and high tensile strength, leading to their widespread application in the textile, fishing gear, and automotive industry. Polycaprolactam (PA6) and poly(hexamethylene adipamide) (PA6.6) are the most industrially relevant PA and are obtained by ring-opening polymerization of ε-caprolactam, that is the lactam of 6-aminohexanoate (Ahx), and polycondensation of adipic acid and 1,6-hexamethylenediamine, respectively. Despite an increasing production, PA recycling rates are below 5% contributing to the global plastics crisis as the majority of post-consumer PA is landfilled.1,2 The mostly linear cradle-to-grave lifespan of PA can be explained by the limitations of traditional recycling strategies. Mechanical recycling, performed by melting and re-extrusion, typically requires highly pure feedstocks while yielding reduced-quality products.3 Consequently, the materials undergo downcycling that ultimately leads to their disposal. Chemical recycling, on the contrary, depolymerizes the material into its monomers and oligomers, which can then be purified and re-polymerized thereby maintaining the material's properties. However, chemical depolymerization, such as for PA6, typically requires high amounts of energy as well as expensive and often non-recyclable catalysts while being sensitive towards feedstock contaminations.4 Contrary to this, enzymatic depolymerization displays a sustainable solution for the breakdown of plastics as enzymes operate at moderate conditions compared to chemical processes, catalyze selective reactions, and avoid toxic chemicals.5 For poly(ethylene terephthalate) (PET), enzymatic recycling is advancing towards commercial implementation due to the design of tailored PETase variants.6–8 Moreover, techno-economic analyses predict that enzymatic recycling could become cost-competitive with virgin PET production if key cost drivers can be reduced.9,10 Recent approaches also focused on combining chemical hydrolysis with biological catalysis, which allows metabolic funneling of mixed hydrolysates and subsequent upcycling to value-added products using engineered microorganisms. Such hybrid strategies were successfully demonstrated for PET11 and PA (de Witt et al., manuscript in preparation).Although natural PAs are ubiquitous in nature, such as in proteins or silk, enzymatic depolymerization of synthetic PAs is rare. In fact, only a small group of 6-aminohexanoate-oligomer hydrolases, designated as nylonases, is reported to hydrolyze the amide bonds of synthetic PA6 oligomers. The Ahx-cyclic-dimer hydrolase (NylA – EC 3.5.2.12) specifically converts the cyclic Ahx-dimer (Ahx2) into linear Ahx2.12 NylB (EC 3.5.1.46) acts as Ahx-oligomer exohydrolase and degrades linear Ahx-oligomers by an exo-type mechanism resulting in the sequential release of Ahx.13 The third nylonase, NylC (EC 3.5.1.117), is an Ahx-oligomer endohydrolase and degrades both cyclic and linear Ahx-oligomers with a degree of oligomerization greater than three.14 All three nylonases (NylAp2, NylBp2, and NylCp2) were discovered on plasmid pOAD2 in Paenarthrobacter ureafaciens that was isolated from wastewater of a PA-manufacturing plant.15,16 Interestingly, activity of nylonases is limited towards synthetic Ahx-oligomers while no activity was reported to a variety of natural amides.13,17
NylC is the most promising nylonase for PA depolymerization due to its endo-cleavage mechanism and specificity for larger Ahx-oligomers. Initially, NylC is expressed as inactive precursor protein. After post-translational auto-cleavage between N266 and T267, which results in the formation of an α-(27.4 kDa) and β-subunit (9.4 kDa), structural re-assembly generates an active NylC enzyme.18 Based on this characteristic, NylC is classified as a member of the N-terminal nucleophile- (N-tn) hydrolase superfamily. Besides the D308-D306-T267 catalytic triad, NylC also possesses Y146 and K189 as additional catalytic or substrate-binding residues that are required for substrate hydrolysis.19 Although NylCp2 is able to degrade linear Ahx-oligomers (n > 2), the wild type enzyme shows negligible activity towards PA6.19 Enzyme engineering of NylCp2 resulted in a quadruple mutant that showed increased thermostability and exhibited catalytic activity towards powdered and thin-layered PA6, PA6.6, and PA6.6-co-6.4.20,21 Recently, the activity of this mutant was further increased by directed evolution.22 Two highly similar homologs of NylCp2 were identified in Agromyces sp. KY5R (NylCA) and Kocuria sp. (NylCK) that share 98.6 and 95.8% sequence identity towards NylCp2.23 Until today, only a limited number of nylonases including the three highly similar NylCs were characterized in literature.24 This limited enzymological diversity severely hinders the potential of enzymatic PA depolymerization, especially with regard to machine learning-aided engineering.25
In this study, we aimed to increase the diversity of characterized NylCs with activities towards PA-related substrates including the emerging group of poly(ester amides) (PEA). The latter group gains increasing interest in the polymer industry as it combines the mechanical properties of PA with the biodegradability of polyester.26 Although PEA market volume is still relatively small, emerging polymers should be developed with their end-of-life options in mind, especially for complex polymers that contain multiple monomers. To identify novel NylC candidates, library screenings and in silico analyses were performed. NylC candidates were expressed in Bacillus sp. and subjected to activity assays using Ahx-oligomers and polymers. This study greatly increases the enzymological diversity of NylCs, thereby facilitating future enzyme engineering efforts for the development of enzymatic PA recycling processes. Further, combined enzymatic degradation and architectural design of PEA provide key insights into polymer design that allow the alignment of both biodegradability and performance, which often are opposing polymer features. This study thus facilitates the development of bio-upcycling processes in an integrated manner, enabling a shift towards a more circular plastics economy.
2 Results and discussion
2.1 Screening and expression of novel NylC candidates
To increase the diversity of NylCs, public and internal databases were screened for homologs of NylCp2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 (UniProt: Q79F77). We also performed enrichment cultures of compost samples using PEA to isolate strains with putative nylonase activities. This led to the isolation of a Rhodococcus sp. and a Gordonia sp. as revealed by 16S rDNA analysis and subsequent whole-genome sequencing (Fig. S1†). Since the characterized NylCp2, NylCA, and NylCK are closely related, we screened the obtained datasets in silico for NylC candidates with different amino acid sequences towards NylCp2 to yield a diverse set of enzyme candidates. Apart from NylCp2, six NylC candidates (NylC1–6) were successfully expressed and purified, each with varying amino acid sequence identities compared to NylCp2 (Table 1 and Fig. 1). Multiple sequence alignments revealed the presence of the typical N/T autocleavage site in all NylC candidates classifying them into the N-tn hydrolase superfamily (Fig. 1). Moreover, the D308-D306-T267 catalytic triad of NylCp2 was highly conserved in NylC1–6 (Table 1 and Fig. 1). NylC1–4 also possessed the two additional catalytic residues, namely Y148/Y130/Y137/Y120 and K191/K173/K180/K163, required for PA hydrolysis as revealed for NylCp2. In NylC5 and NylC6, these additional catalytic residues were less conserved since Y146 was replaced by F130/F129 and no K could be identified corresponding to position K189 of NylCp2 by multiple sequence alignments. Using ColabFold,27 the structures of NylC1–6 were predicted and aligned to NylCp2. These structural alignments revealed that K244 of NylC6 and K189 of NylCp2 were in close proximity indicating that NylC6 potentially harbors all catalytic residues required for PA hydrolysis. Although NylC5 contained K242, structural alignments indicated an incorrect arrangement of this residue as it was opposed to K189 of NylCp2 (Fig. 2). Overall, structural alignments revealed that the catalytic triads are highly conserved in all NylC candidates compared to NylCp2 (Fig. 2, Fig. S2 and Table S2†). Moreover, NylC5 and NylC6 differ from NylC1–4, as the additional catalytic residues were less conserved, which might influence their activity towards PA.
14 (UniProt: Q79F77). We also performed enrichment cultures of compost samples using PEA to isolate strains with putative nylonase activities. This led to the isolation of a Rhodococcus sp. and a Gordonia sp. as revealed by 16S rDNA analysis and subsequent whole-genome sequencing (Fig. S1†). Since the characterized NylCp2, NylCA, and NylCK are closely related, we screened the obtained datasets in silico for NylC candidates with different amino acid sequences towards NylCp2 to yield a diverse set of enzyme candidates. Apart from NylCp2, six NylC candidates (NylC1–6) were successfully expressed and purified, each with varying amino acid sequence identities compared to NylCp2 (Table 1 and Fig. 1). Multiple sequence alignments revealed the presence of the typical N/T autocleavage site in all NylC candidates classifying them into the N-tn hydrolase superfamily (Fig. 1). Moreover, the D308-D306-T267 catalytic triad of NylCp2 was highly conserved in NylC1–6 (Table 1 and Fig. 1). NylC1–4 also possessed the two additional catalytic residues, namely Y148/Y130/Y137/Y120 and K191/K173/K180/K163, required for PA hydrolysis as revealed for NylCp2. In NylC5 and NylC6, these additional catalytic residues were less conserved since Y146 was replaced by F130/F129 and no K could be identified corresponding to position K189 of NylCp2 by multiple sequence alignments. Using ColabFold,27 the structures of NylC1–6 were predicted and aligned to NylCp2. These structural alignments revealed that K244 of NylC6 and K189 of NylCp2 were in close proximity indicating that NylC6 potentially harbors all catalytic residues required for PA hydrolysis. Although NylC5 contained K242, structural alignments indicated an incorrect arrangement of this residue as it was opposed to K189 of NylCp2 (Fig. 2). Overall, structural alignments revealed that the catalytic triads are highly conserved in all NylC candidates compared to NylCp2 (Fig. 2, Fig. S2 and Table S2†). Moreover, NylC5 and NylC6 differ from NylC1–4, as the additional catalytic residues were less conserved, which might influence their activity towards PA.
 | ||
| Fig. 1 Multiple sequence alignments and phylogenetic analysis. (A) T-Coffee alignment algorithm28 was used for the multiple sequence alignment using the EMBL-EBI tool.29 Amino acids are colored based on Clustal X 2.0 default coloring.30 Conserved amino acids are indicated with an asterisk (*). The D-D-T catalytic residues (red arrows) and substrate-binding residues (Y/F and K) (orange arrows) are highlighted, while the N/T autocleavage site is enclosed in a black box. The grey and black lines indicate the α- and β-subunits respectively. (B) Phylogenetic tree of NylCs enzyme within the currently known polyamide-active enzymes as described in Bell et al.24 Enzymes described in this study are printed in bold. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of trees in which the associated taxa clustered together are shown next to the branches. | ||
 | ||
| Fig. 2 Structural alignments of NylCp2 and NylC1,2,5 and enzyme purification. Protein structures of NylCs were predicted using ColabFold27 and were aligned to NylCp2. The holistic structural alignments for NylC1 (A), Nyl C2 (B) and NylC5 (C) are shown as well as the alignments of the catalytic residues (box). The N/T-autocleavage site is highlighted in red. (D) SDS-PAGE of purified NylC1–6. The lanes were loaded as followed: Marker (M), NylC1 (1), NylC5 (2), NylC6 (3), NylC3 (4), NylC4 (5), NylC2 (6). The two bands correspond to the α- and β-subunits. | ||
| Protein ID | Donor organism | Sequence identity (%) | Molecular weight (α + β) (kDa) | Putative catalytic triad | T m (°C) | Ref. |
|---|---|---|---|---|---|---|
| NylCp2 | Paenarthrobacter ureafaciens | 100 | 27.4 + 9.4 | D308, D306, T267 | 52 | Kinoshita et al.13 |
| NylCA | Agromyces sp. KY5R | 98.6 | 27.4 + 9.4 | D308, D306, T267 | 60 | Yasuhira et al.23 |
| NylCK | Kocuria sp. | 95.8 | 27.4 + 9.4 | D308, D306, T267 | 67 | Yasuhira et al.23 |
| NylC1 | Leucobacter chromiiresistens JG 31 | 84.2 | 28.0 + 10.4 | D310, D308, T269 | 58.4 | This study |
| NylC2 | Microbacterium oxydans | 62.5 | 26.0 + 10.2 | D292, D290, T251 | 67.4 | This study |
| NylC3 | Streptomyces sp. 63005 | 61.2 | 26.4 + 10.2 | D298, D296, T257 | 63.9 | This study |
| NylC4 | Variovorax boronicumulans | 48.4 | 25.9 + 9.3 | D292, D290, T251 | 82.9 | This study |
| NylC5 | Gordonia sp. | 31.7 | 24.3 + 12.6 | D289, D287, T248 | 78.1 | This study |
| NylC6 | Rhodococcus sp. | 30.0 | 25.1 + 11.7 | D291, D289, T250 | 74.9 | This study |
NylC1–6 were expressed with a C-terminal His6-tag allowing their purification. Although the ratio of cleaved (α- and β-subunit) to uncleaved (precursor) NylCp2 was reported to increase from approximately 10 to 100% within 48 h of incubation after purification,19 SDS-PAGE of purified NylC1–6 indicated that no significant amounts of precursor protein was present without prior incubation. Instead, the α- and β-subunits of NylC1–6 were detected indicating that their precursor autocleavage might be faster compared to NylCp2 (Fig. 1). Moreover, the interaction of α- and β-subunits of NylC1–6 were strong enough to allow purification of both subunits with a single C-terminal His6-tag, which was present in the β-subunits. Unfortunately, NylCp2 could not be expressed in Bacillus sp. Its heterologous expression seems to be limited to Escherichia coli until today.20,22 For commercial production of nylonases, however, expression in industrial workhorses such as Bacillus sp. is a prerequisite, and further optimization would be required.
Activity of NylCp2 towards polymeric PA6 was enabled by designing thermostable mutants as elevated reaction temperatures typically result in increased accessibility of amorphous polymer regions due to increased chain mobility.20,22 To investigate the thermostability and thus evaluate the potential activity towards polymeric substrates, purified NylC1–6 were subjected to nano differential scanning fluorimetry (DSF) to determine their melting temperature (Tm). Compared to non-engineered NylCp2 (Tm = 52 °C),21 NylC1–6 showed higher thermostabilities ranging from Tm = 58.4 °C (NylC1) to Tm = 82.9 °C (NylC4) (Table 1). The overall higher thermostability indicates that NylC1–6 might be active at higher temperatures, which could facilitate depolymerization above the glass transition temperature (Tg) of PA6 (Tg 54 °C). Thermostability is a key factor in enzymatic polymer degradation as extensively shown for PET.6,8 Thus, if active, NylC1–6 might be promising candidates for enzyme engineering or shuffling to outperform the current thermostability of engineered NylCp2 (Tm = 88 °C).21
2.2 Activity towards PA6 oligomers
To test whether the NylC1–6 candidates are true nylonases, their activities towards cyclic and linear oligomers of PA6, designated as Ahx-oligomers, were tested. Subsequent HPLC analyses were performed to detect degradation products. Although NylCp2 was reported to hydrolyze linear and cyclic Ahx-oligomers with a degree of oligomerization greater than three, none of the NylC1–6 candidates was active towards soluble linear Ahx oligomers (n = 2–7). Instead, activity towards the cyclic Ahx-oligomer fraction (n ≥ 2)31 was detected for NylC1, NylC2, and NylC5 by the release of linear Ahx-oligomers. As additional control reaction, the Ahx-cyclic-dimer hydrolase from P. ureafaciens (NylAp2)12 was incubated with the cyclic Ahx-oligomer fraction. As expected, this resulted in the conversion of cyclic Ahx2 into linear Ahx2. Activity of NylC1 and NylC2 on the cyclic Ahx-oligomers resulted in the release of linear Ahx4 and Ahx5, whereas NylC5 additionally released linear Ahx3 (Fig. 3A). The linear degradation products were not further hydrolyzed. In contrast to this, NylCp2 was reported to further degrade such linear Ahx-oligomers yielding linear Ahx2 as end-product.18 Interestingly, no decrease in the concentration of the soluble cyclic oligomers (n = 2–6) could be detected by HPLC, even though linear products were produced by NylC1, NylC2, and NylC5. This may be due to a limitation in the detection of larger, insoluble oligomers. According to this theory, the release of linear Ahx3–5 by the NylCs must originate from the hydrolysis of larger insoluble and thus undetectable cyclic oligomers (n > 6). Hence, NylC1 and NylC2 are active towards cyclic Ahx8–10, resulting in the initial ring opening to the corresponding linear oligomers, followed by subsequent degradation to linear Ahx4–5 by an endo-cleavage. The additional release of linear Ahx3 by NylC5, while not degrading cyclic Ahx6, indicates activity towards cyclic Ahx7 that is degraded to linear Ahx3–4 after initial hydrolysis to linear Ahx7. To confirm the activity towards exclusively insoluble Ahx-oligomers, only the soluble fraction of cyclic Ahx-oligomers (n ≤ 6) was tested as substrate for NylC1–6 (Fig. 3B) by filtration. Indeed, none of the enzymes showed activity towards the filtered cyclic Ahx-oligomers as no linear degradation products were detected, suggesting that the enzymes were exclusively active on insoluble cyclic and linear oligomers. | ||
| Fig. 3 Activity screenings of NylC1–6. Reactions were performed using 4 g L−1 cyclic Ahx-oligomers (Aco2)31 (A) and its soluble fraction that was obtained by filtration (B). Reactions were performed using 500 nM of purified enzyme in 50 mM phosphate buffer, pH 7.3 at 30 °C for 8 h. Cyclic and linear soluble oligomers were detected using HPLC. The mean values and standard deviations (SD) of three replicates are shown (n = 3). | ||
Overall, these results reveal that the activity and substrate specificity of NylC1, NylC2, and NylC5 differ from that of NylCp2 as no soluble cyclic or linear oligomers were hydrolyzed by the novel NylC enzymes. Instead, activity towards insoluble oligomers was observed, which might be accompanied by the ability to hydrolyze insoluble linear chains of PA. The inactivity of some NylC candidates highlights the diversity of the N-tn hydrolase superfamily, which complicates the identification of new NylCs based on in silico analyses only. The different substrate specificities of the identified NylCs compared to NylCp2 and its close homologs indicate a much greater diversity of nylonases in nature than originally anticipated.
The activity of NylC1, NylC2, and NylC5 towards insoluble Ahx-oligomers suggested that they might also be capable of hydrolyzing PA6 of higher molar masses. To test this, each NylC was incubated with powdered PA6 at temperatures between 30 and 70 °C in increments of 10 °C. However, none of the enzymes was active towards PA6 under any of the conditions tested as no soluble oligomers were detected in the supernatants. It has to be noted that the enzymatic activity towards polymeric substrates greatly depends on several factors such as crystallinity and dispersion of the substrate. Previous studies did detect NylC activity on polymeric PA, but they also demonstrated that the activities of NylC are remarkably higher when thin-layered substrate films were used. These films were approximately 1000-fold thinner than the diameter of powdered PA, resulting in a highly increased reaction rate.20,24 Moreover Nagai et al.20 discovered that even after enzymatic depolymerization of PA6, oligomers were still bound to the polymer chains through hydrogen bonding. The combination of such factors might have contributed to the apparent inactivity of NylC1–6 towards powdered PA6 in this study. Enzyme engineering might overcome these barriers as performed for NylCp2, whose non-engineered version also showed no activity towards PA6.20
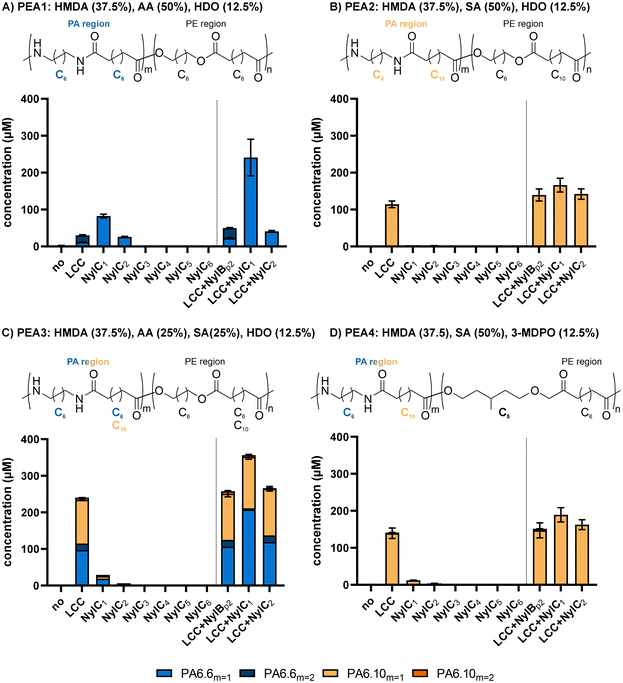
2.3 Synthesis and enzymatic degradation of poly(ester amides)
To test the activity of NylC1–6 towards other polymeric substrates, we performed activity assays using PEA. In contrast to well-established PA, PEA are an emerging group of polymers and combine the thermal and mechanical properties of PA with the biodegradability of polyesters.26,32 To investigate if NylC1–6 are able to depolymerize PEAs, we tested their activities towards powdered PA6.6-, PA6.10-, or PA6.6-co-6.10-based PEAs (PEA1–4) that contained aliphatic or branched diols to yield the desired PEAs (Fig. 5). Both the average molecular weight (Mw) and number average molar mass (Mn) differed among the synthesized statistical PEA ranging from 1846–7371 Da (Mn) and 3587–13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814 Da (Mw), respectively (Table 2). While PEA1 displayed the highest Mn and Mw values among the PEA samples (Mn = 7371 Da and Mw = 13
814 Da (Mw), respectively (Table 2). While PEA1 displayed the highest Mn and Mw values among the PEA samples (Mn = 7371 Da and Mw = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814 Da), these values still fell significantly below those of PA6 (Mn = 12
814 Da), these values still fell significantly below those of PA6 (Mn = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 Da and Mw = 67
220 Da and Mw = 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860 Da).
860 Da).
| Polymer | M n (Da) | M w (Da) | PDI | T m1 (°C) | T m2 (°C) | T rc (°C) | T c (°C) |
|---|---|---|---|---|---|---|---|
| PA 6 | 12220 ± 855 | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860 ± 4750 860 ± 4750 |
5.55 | 213 ± 2 | 220 ± 2 | 198 ± 2 | 164 ± 2 |
| PEA1 | 7371 ± 516 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814 ± 936 814 ± 936 |
1.87 | 236 ± 3 | — | 208 ± 3 | 200 ± 3 |
| PEA2 | 2099 ± 147 | 4174 ± 272 | 1.99 | 173 ± 2 | 210 ± 3 | 188 ± 3 | 179 ± 1 |
| PEA3 | 2139 ± 159 | 4880 ± 341 | 2.28 | 182 ± 2 | — | 160 ± 2 | 136 ± 2 |
| PEA4 | 1846 ± 129 | 3587 ± 231 | 1.94 | 173 ± 2 | 210 ± 3 | 189 ± 2 | 181 ± 3 |
The differential scanning calorimetry (DSC) thermograms, showing the heating and cooling curves, revealed crucial thermal properties of PEA1–4 and PA6 (Fig. 4). For PA6, two significantly distinct melting peaks were observed (Tm1 = 213 °C and Tm2 = 220 °C). In contrast, PEA2 and PEA4 showed two similar melting peaks, as both feature the first peak at a temperature of ∼173 °C and a second melting peak at 210 °C, which is lower compared to PA6. This phenomenon could be attributed to the characteristics of PEA2 and PEA4 incorporating sebacic acid as a monomer, which increases the stability of secondary crystal structures within the polymer.33 Notably, the only distinction between PEA2 and PEA4 lies in the ester monomers employed, yet their melting points exhibited similarity. Consequently, it is reasonable to anticipate that the melting point of PEA may be more significantly influenced by the amide regions rather than the ester regions. The existence of two melting peaks may reflect the presence of a secondary crystal structure or the formation of crystals of different sizes,34 but further investigation is required to define the precise cause. In contrast to PEA2 and PEA4, PEA1 and PEA 3 displayed a less sharp single melting peak at temperatures of 236 °C and 182 °C, respectively. Hence, PEA1 has the highest melting point of all PEA samples. These results suggest that based on the lower melting peaks of the PEA2–4 samples in comparison to PA6, those samples might need to be processed at lower temperatures than PA6, whereas PEA1 might demand a higher processing temperature than PA6.
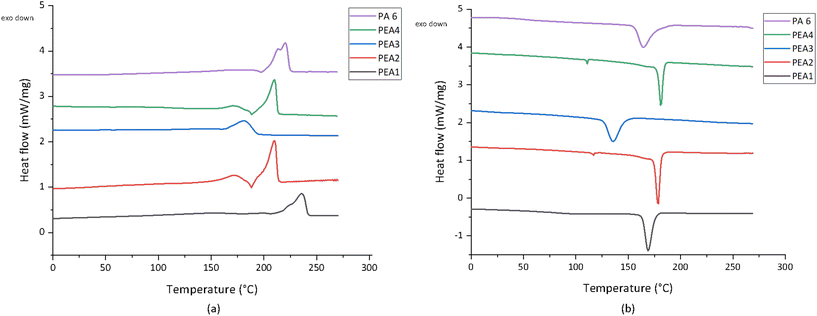 | ||
| Fig. 4 DSC thermograms of PEA1–4 and PA6 granules. The heating (a) and cooling (b) curves are shown. | ||
The DSC thermograms of both PA6 and all PEA samples revealed a recrystallization temperature (Trc) around ∼198 °C for PA6, ∼208 °C for PEA1, ∼188 °C for PEA2 and PEA4, and ∼160 °C for PEA3. During cooling, variations in crystal size or type, along with potential defects, may arise in the forming crystalline domains. Upon heating, these smaller crystalline domains (and sometimes defects in the crystal structure) can melt and recrystallize or merge into larger crystalline domains, which subsequently melt at higher temperatures. This recrystallization peak may also indicate the presence of amorphous polymer chains that gain mobility at elevated temperatures, forming a more ordered crystalline structure, as previously documented.35 In the cooling curve results, PA6 exhibited a crystallization peak remaining at approximately ∼164 °C (Fig. 4 and Table 2). Conversely, PEA1, PEA2, and PEA4 displayed peaks at higher temperatures compared to PA6, with respective values of ∼200 °C, ∼179 °C, and ∼181 °C. Only PEA3 exhibited a lower crystallization temperature of 136 °C.
No unambiguous Tg were visible in the DSC data, likely because of the limited sensitivity of the method. However, introducing more variation in the carbon-count of the monomers creates more irregularities in the crystallinity, while it also diminishes the degree of hydrogen bonding between the amide groups. Hence, the Tg of the material should become lower than in PA66 (44 °C (ref. 36)), and PA6 (54 °C (ref. 37)), for example. The relations between nylon composition, hydrogen-bonding, Tg, and Tm were neatly described previously.33
After their characterization, the activities of NylC1–6 towards PEA1–4 were investigated. NylC1 and NylC2 were active towards PEA1 and PEA3 as the release of linear PA6.6- and PA6.10-oligomers was detected (Fig. 5a and c). NylC1 showed higher activities for both substrates, as it released approximately three-fold more linear PA6.6 dimer (PA6.62) than NylC2. In contrast to this, no or only low activity was detected towards PEA2 or PEA4, respectively, which contained PA6.10 regions (Fig. 5b and d). These results indicate a preference for NylC1–2 towards amide bonds within PA6.6 regions compared to PA6.10 regions. For engineered NylCp2, a similar preference for amide bonds consisting of shorter monomers was revealed (PA6.6 and PA6.6-co-6.4) and no degradation of PA6.10 has been reported until today.20 Given the different polymer characteristics of PEA1 and PEA3, the varying biodegradability is likely related to the enzymatic specificities of NylCs rather than to the individual polymer properties of PEA1–4.
To facilitate depolymerization, the well-established leaf and branch compost cutinase (LCC, EC 3.1.1.74, EC 3.1.1.101)38 able to hydrolyze various polyester, including poly(ester-urethanes),39 was used for initial depolymerization of the PEAs. Interestingly, Bell et al.6 recently revealed minor activity of an LCC variant towards thin-layered PA6. Here, activity of LCC was expected to release amide oligomers that might be further degraded by NylC1 and NylC2. Since LCC features a high temperature optimum, initial hydrolysis of PEA1–4 was performed at 70 °C and subsequent treatment with NylC1–2 was performed at 30 °C. To track any residual activity of LCC during the subsequent incubation at 30 °C, control reactions with NylBp2 were performed. This nylonase was reported to specifically hydrolyze linear PA6-oligomers not showing any activity towards PA6.6, PA6.10, and their corresponding oligomers.13,16 Initial treatment of PEA1–4 by LCC resulted in the release of amide oligomers with primary amino groups revealing activity of LCC towards the amide regions confirming the amidase activity of LCC described by Bell et al.6 (Fig. 5). For PEA1, only containing PA6.6 regions, the release of amide oligomers by LCC was approximately 2.7-fold lower compared to NylC1 (Fig. 5a). In contrast to this, LCC treatment of PEA2–4, containing PA6.6-co-PA6.10 or PA6.10 regions, resulted in highly increased amounts of amide oligomers compared to NylC1 (Fig. 5b–d). The combination of LCC and NylC1 resulted in a synergistic effect when PEA1 and PEA3 were tested as substrates, yielding approximately 2.2- and 1.3-fold more soluble products compared to the sum of products released by individual LCC or NylC1 treatments. Hence, initial depolymerization of LCC resulted in the release of larger, undetectable PA6.6-oligomers that were subsequently cleaved by NylC1. This synergistic effect highlights the substrate specificity of NylC1 towards PA6.6-oligomers that were not a preferred substrate for LCC. In contrast to PA6-oligomers, NylC1 was active towards soluble PA6.6-oligomers, as revealed for PA6.64, which resulted in the formation of PA6.62 as the end product of hydrolysis. No synergistic effect was observed when PEA2 and PEA4 were tested confirming low activities of NylC1 towards PA6.10-oligomers. Overall, the combination of LCC and NylC1 resulted in the release of approximately 0.6, 0.5, 1,1, and 0.6% of monomer equivalents present in the tested PEA1–4 materials respectively. Although the polymer characteristics of PEA2–4 were rather similar, the biodegradability of PEA3 was remarkably higher within this group. This highlights the importance of incorporating PA6.6 regions into PEA as target hydrolysis sites for nylonases. PEA1, containing pure PA6.6-regions, showed less biodegradability compared to PEA3 (PA6.6-co-PA6.10 regions) but featured improved polymer characteristics such as increased Mn and Mw. Hence, it is crucial to design future PEA that align both counteracting parameters, as suggested with PEA1, by focusing on the incorporation of PA6.6 regions and boosting the nylonase activities. Overall, NylC1 was revealed as most active nylonase and showed activities towards not only PA6.6 oligomers but also PEA that contain PA6.6-regions (Table 3).
| Enzyme | Linear oligomers (na) | Cyclic oligomers (na) |
|---|---|---|
| a n = minimum number of monomers. Enzymes did not show activity to oligomers with less monomers. | ||
| NylC1 | PA6 (n > 6) | PA6 (n > 6) |
| PA6.6 (n ≥ 4) | ||
| NylC2 | PA6 (n > 6) | PA6 (n > 6) |
| NylC3 | No activity | No activity |
| NylC4 | No activity | No activity |
| NylC5 | PA6 (n ≥ 6) | PA6 (n ≥ 6) |
| NylC6 | No activity | No activity |
| NylA | No activity | PA6 (n = 2) |
The results from biodegradation experiments support the discussed relations of the Tg of the different materials. The higher degree of mobility of the polymer chains in material with higher monomer variety and, hence, assumed lower Tg supposedly facilitate latching of the enzymes on the polymer chains in a sterical sense, as also indicated in a recent comparison of nylon 66 and nylon 56 biodegradation.40 Notably, it is reasonable to assume that the Tg of submerged PEAs are considerably lower in the aqueous solution with water molecules interacting with the intramolecular hydrogen bond network than in the dry polymer,41,42 further facilitating enzymatic attack.
3 Conclusions and outlook
In the present study, we increased the enzymological diversity of NylCs and discovered three novel NylCs that are active towards insoluble cyclic and linear Ahx-oligomers. Overall, we greatly increased the sequence space of NylCs, facilitating future enzyme engineering to boost their activities towards PA and PEA. NylC5 has only 32% sequence identity to NylCp2, with one of the putative substrate binding residues (K242) located in an atypical position. Based on previous engineering of NylCp2,21 rational design can be performed to increase the thermostability of the novel nylonases. Moreover, the recently developed high-throughput screening system for directed evolution of nylonases can be applied to identify yet unknown mutations that increase the overall activities.22 Such mutations could for example facilitate the post-translational autocleavage, the structural re-assembly of α- and β-subunits, or the interactions between individual NylC monomers. Two of the novel NylCs were active towards polymeric PEA substrates, showing the potential to enable efficient and sustainable enzymatic recycling of PA and PEA in the future. Finally, this study paves the way for linking insights into the enzymatic biodegradability of polymers with their architectural design and highlights PEA as promising hybrid polymers that combine the contrary parameters of performance and biodegradability.4 Materials and methods
4.1 In silico tools
Public and internal databases and genome sequences of isolates were screened for putative NylC candidates by homology searches (threshold ≥30% identity with NylCp). Multiple sequence alignments were performed using the EMBL-EBI tool29 and the T-Coffee alignment algorithm28 was used chosen. Amino acids were colored based on Clustal X 2.0 default coloring.30 The phylogenetic tree construction was conducted using MEGA1143 by using the Maximum Likelihood method and JTTmatrix-based mode, with 1000 bootstraps. Protein structures were predicted using ColabFold27 and structural alignments were performed with UCSF Chimera.444.2 Enrichment cultures
Soil samples (10 g L−1) from a compost heap were used to inoculate mineral salts medium45 supplemented with 50 g L−1 of a poly(ester amide) consisting of adipic acid, 1,6-hexamethylenediamine, and 1,6-hexanediol. Shake flasks were incubated at 30 °C and 200 rpm for 3 d until an increase in turbidity was observed, after which 1% was transferred to fresh medium to enrich strains. After five re-inoculations, enriched strains were isolated on lysogeny broth (LB) agar plates. For cryogenic conservation, strains were cultivated in LB medium for 30 °C for 18 h and 500 μL of cell culture was mixed with 500 μL of 50% (v/v) glycerol. Glycerol stocks were stored at −80 °C.4.3 16S rDNA sequencing
Genomic DNA was isolated from cells obtained from LB cultures using the Monarch gDNA Purification Kit (NEB). The 16S rDNA sequence was amplified from the isolated gDNA by PCR using Q5 High-Fidelity 2× Master Mix (NEB) and the primer FD1/2 (5′-3′ AGAGTTTGATCMTGGCTCAG) and RP1/2 (5′-3′ ACGGYTACCTTGTTACGACTT).46 PCR products were purified using a Monarch PCR Cleanup Kit (NEB) and sequenced by Eurofins Genomics (Ebersberg, Germany). The obtained sequencing results were aligned to the nucleotide collection (nr/nt) of the NCBI database using BLASTn.474.4 Whole-genome sequencing
One microgram of genomic DNA was used for library preparation using the NEBNext Ultra™ II DNA Library Prep Kit for Illumina (NEB). The library was evaluated by qPCR using the KAPA library quantification kit (Peqlab, Erlangen, Germany). Afterwards, normalization for pooling was done and paired-end sequencing with a read length of 2 × 150 bases was performed on a MiSeq (Illumina). The reads of demultiplexed fastq files as the sequencing output (base calls) were trimmed and quality-filtered using the CLC Genomic Workbench software (Qiagen Aarhus A/S, Aarhus, Denmark). The filtered reads were used for de novo assembly using the CLC Genomic Workbench software.4.5 Molecular cloning
Codon-optimized genes encoding the nylonases were chromosomally integrated and expressed in Bacillus subtilis using a similar setup as described previously48 with the following modifications. A 21 bp sequence was added at the terminal gene sequences encoding a C-terminal histidine tag (His6-tag) and harboring the stop codon. Cells were made competent according to Yasbin et al.494.6 Enzyme purification
Cells of nylonase-expressing B. subtilis were cultivated in Cal18 medium (4% Yeast extract, 0.13% MgSO4·7H2O, 5% Maltodextrin, 2% Na2HPO4·12H2O, 0.67% Na2MoO4 trace metal solution and 0.01% Dowfax 63N10)48 or terrific broth (TB) medium50 for 48–72 h at 30 °C. Cell cultures were centrifuged at 7000g for 15 min and the supernatant was used for immobilized-metal affinity chromatography using His SpinTrap™ columns (Cytivia). After that, cells were desalted with HiTrap® desalting columns (Cytivia) using 50 mM HEPES buffer (pH 7.3). Purified samples were analyzed by SDS-PAGE analysis using Criterion™ Precast Gels.4.7 Nano differential scanning fluorimetry
The melting temperature (Tm) of the studied enzymes was analyzed by nano differential scanning fluorimetry (nanoDSF) using a Prometheus NT.48 (NanoTemper). For this, the enzymes were diluted to a concentration of 2 mg mL−1 in 50 mM HEPES buffer (pH 7.3). The thermal stability was tested with a heating scan range from 20 to 90 °C at a scan rate of 3 °C min−1. Data analyzes and determination of the Tm was performed using the PR ThermControl software (NanoTemper).4.8 Activity assays
All enzymatic reactions were performed at 30 °C in 50 mM phosphate buffer, pH 7.3 using 500 nM of purified enzyme for 8 h. The cyclic Ahx-oligomer fraction Aco2 (n ≥ 2)31 was used at concentrations of 4 g L−1 resulting in a turbid reaction sample due to insoluble Ahx-oligomers. For investigating the soluble cyclic Ahx-oligomers as substrate, this mixture was filtered through a 0.22 μm PES filter membrane to remove insoluble oligomers. For degradation experiments with polymeric substrates, 10 g L−1 of powdered PEA was used as substrate. Initial treatment with nylonases was performed at 30 °C, whereas LCC treatment was performed at 70 °C. Combined enzymatic degradation was initially performed at 70 °C (LCC) and subsequently at 30 °C after the addition of nylonases.4.9 High-performance liquid chromatography analysis
Samples obtained from activity assays were filtered through an AcroPrep™ 96-well filter plate (Pall Corporation, Port Washington, NY, USA) to remove any particles prior to HLPC analysis. HPLC analysis was performed using a 1260 Infinity II HPLC equipped with a fluorescence detector (FLD) and a diode array detector (DAD) (Agilent, Santa Clara, California, USA). To analyze linear PA-oligomers harboring primary amine groups, pre-column derivatization using o-phthaldialdehyde (OPA) reagent (Sigma-Aldrich, ready-to-use-mix) was performed. For separation of the derivatized molecules, the Kinetex® 2.6 μm EVO C18 100 Å column (100 × 2.1 mm) (Phenomenex, California, USA) was used. As mobile phase, 10 mM sodium-borate buffer (A) (pH 8.2) and methanol (B) was used with an initial ratio of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (A
30 (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B).51 This ratio was gradually increased to 30
B).51 This ratio was gradually increased to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 after 10 min and 0
70 after 10 min and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 after 12 min followed by 1 min of 0
100 after 12 min followed by 1 min of 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. After that, the ratio was switched to 70
100. After that, the ratio was switched to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 after 14 min and 1 min post-run was performed. The flow was adjusted to 0.4 mL min−1 at and the oven temperature was set to 40 °C. Derivatized molecules were detected using a FLD using an excitation of λ = 340 nm and an emission of λ = 450 nm. Detection of cyclic Ahx-oligomers was performed using the DAD with an absorption of λ = 210 nm (reference λ = 300 nm). Cyclic oligomers of Ahx were separated using a Zorbax Eclipse XDB-C8 column (4.6 × 150 mm) with a H2OMilliQ
30 after 14 min and 1 min post-run was performed. The flow was adjusted to 0.4 mL min−1 at and the oven temperature was set to 40 °C. Derivatized molecules were detected using a FLD using an excitation of λ = 340 nm and an emission of λ = 450 nm. Detection of cyclic Ahx-oligomers was performed using the DAD with an absorption of λ = 210 nm (reference λ = 300 nm). Cyclic oligomers of Ahx were separated using a Zorbax Eclipse XDB-C8 column (4.6 × 150 mm) with a H2OMilliQ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH ratio of 60
MeOH ratio of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 and a constant flow of 0.5 mL min−1 at 40 °C. As no analytical standards were available for cyclic Ahx-oligomers, the corresponding peak area was analyzed allowing semi-quantitative analysis.
40 and a constant flow of 0.5 mL min−1 at 40 °C. As no analytical standards were available for cyclic Ahx-oligomers, the corresponding peak area was analyzed allowing semi-quantitative analysis.
4.10 Polymer synthesis
In a typical random polycondensation procedure, the syntheses of statistical polyester-polyamide (PEA) copolymers from 1,6-hexamethylenediamine, adipic acid or sebacic acid, and 1,6-hexanediol or 3-methylpentanediol (Table 4) were conducted solvent-free and under an inert atmosphere on approximately a 0.15–0.35 kg scale in a glass polymerization reactor (500 mL size; Glasblazerij Janssen B.V.). Mechanical stirring was applied using an overhead stirrer (Heidolph, hei-TORQUE Expert) connected to a magnet stirring coupling piece with a torque restriction of 20 MPa cm−1 (Premex Glenfiz), and a 2D-round-shaped stainless steel stirring anchor. Vacuum was applied using a membrane pump (Pfeiffer MVP-15) in conjunction with a manually operated vacuum controller (Vacuubrand, Vacuu-Select). Stoichiometrically correct amounts of reagents (i.e. moldiacid = moldiamine + moldiol) were weighed into the reactor along with 0.1 wt% catalyst and 0.1 wt% antioxidant. The setup was sealed and carefully purged with five vacuum/nitrogen cycles, and subsequently set at P = 900 mbar. The reaction was initiated by heating using a silicone oil bath to reach 220 °C within 45 minutes under stirring at 50 rpm, upon which the reaction mixture turned into a viscous yellow liquid and profuse water distillation took place. Whenever the water distillation was observed to cease, all reaction conditions were gradually driven more stringent – toward 100 mbar, 240 °C, and stirring at 300 rpm – over the course of two hours. Intermediately, the reactor was filled with nitrogen gas and the flask with aqueous condensate was quickly replaced by an empty one under an outflow of nitrogen gas. The reaction was proceeded for another 3 hours at 240 °C and stirring at 300 rpm but now the vacuum was gradually decreased to 1 mbar to afford substantial buildup of polymer chains, as observed by the thickening of the reaction mixture and the gradual increase of monitored torque on the overhead stirrer. Ultimately, the vacuum was released by nitrogen filling into the reactor, and the polymeric product was cast into a silicone tray while still hot and molten.| Polymer | AA | SA | HMDA | HDO | 3-MPDO |
|---|---|---|---|---|---|
| PEA1 | 50 | 0 | 37.5 | 12.5 | 0 |
| PEA2 | 0 | 50 | 37.5 | 12.5 | 0 |
| PEA3 | 25 | 25 | 37.5 | 12.5 | 0 |
| PEA4 | 0 | 50 | 37.5 | 0 | 12.5 |
4.11 Polymer characterization
Differential scanning calorimetry was carried out using the Q2000 device (TA Instruments, Asse, Belgium). We focused on the melting temperature (Tm), the recrystallization temperature (Trc), and the crystallization temperature (Tc) within the different PEA in comparison to PA6. All samples were tested at a heating rate of 10 °C min−1, using a temperature range of 0 to 275 °C with a sample size of ∼5 mg. For each sample, we made three measurements, and the mean values are presented.The molecular weight of the samples was determined by gel permeation chromatography (GPC) using a 1260 Infinity System (Agilent Technologies, Santa Clara, CA, USA). We used hexafluor-2-isopropanol (HFIP) containing 0.19% sodium trifluoroacetate as the mobile phase, flowing at a rate of 0.33 mL min−1. GPC was used to compare the PEA samples to the polymer PA6. Solutions were prepared by dissolving 3 mg samples in HFIP for ∼3 h before passing through a 0.2 μm polytetrafluoroethylene filter and injecting them into a modified silica column filled with 7 μm particles (Polymer Standards Service, Mainz, Germany). The relative molecular weight (Mw), number average molar mass (Mn), and polydispersity index (PDI) were determined using refractive index detectors calibrated with a standard polymethyl methacrylate polymer (1.0 × 105 g mol−1). We performed GPC analysis three times with each sample and presented the mean values for comparison.
Data availability
The data supporting this article have been included as part of the ESI.† Protein sequences are available at NCBI Genbank at https://www.ncbi.nlm.nih.gov/genbank/ through the accession numbers listed in the supplement.Conflicts of interest
Kenneth Jensen works for Novonesis A/S, a major manufacturer of industrial enzymes. Christian van Slagmaat worked for B4P, a polymer synthesis company, at the time of the study. All other authors have no conflicts to declare.Acknowledgements
This project has received funding from the Bio-based Industries Joint Undertaking (JU) under the European Union's Horizon 2020 research and innovation programme under grant agreement no. 887711. The JU receives support from the European Union's Horizon 2020 research and innovation programme and the Bio-based Industries Consortium. We gratefully acknowledge Dai-ichiro Kato (Kagoshima University, Japan) for providing cyclic Ahx-oligomers as substrates.References
- Textile Exchange, Preferred Fiber and Materials Market Report, 2022 Search PubMed.
- A.-J. Minor, R. Goldhahn, L. Rihko-Struckmann and K. Sundmacher, Chem. Eng. J., 2023, 474, 145333 CrossRef CAS.
- M. Pietroluongo, E. Padovano, A. Frache and C. Badini, Sustainable Mater. Technol., 2020, 23, e00143 CrossRef CAS.
- C. Alberti, R. Figueira, M. Hofmann, S. Koschke and S. Enthaler, ChemistrySelect, 2019, 4, 12638–12642 CrossRef CAS.
- L. D. Ellis, N. A. Rorrer, K. P. Sullivan, M. Otto, J. E. McGeehan, Y. Román-Leshkov, N. Wierckx and G. T. Beckham, Nat. Catal., 2021, 4, 539–556 CrossRef CAS.
- E. L. Bell, R. Smithson, S. Kilbride, J. Foster, F. J. Hardy, S. Ramachandran, A. A. Tedstone, S. J. Haigh, A. A. Garforth, P. J. R. Day, C. Levy, M. P. Shaver and A. P. Green, Nat. Catal., 2022, 5, 673–681 CrossRef CAS.
- A. Bollinger, S. Thies, E. Knieps-Grünhagen, C. Gertzen, S. Kobus, A. Höppner, M. Ferrer, H. Gohlke, S. H. J. Smits and K.-E. Jaeger, Front. Microbiol., 2020, 11, 114 CrossRef PubMed.
- V. Tournier, C. M. Topham, A. Gilles, B. David, C. Folgoas, E. Moya-Leclair, E. Kamionka, M. L. Desrousseaux, H. Texier, S. Gavalda, M. Cot, E. Guémard, M. Dalibey, J. Nomme, G. Cioci, S. Barbe, M. Chateau, I. André, S. Duquesne and A. Marty, Nature, 2020, 580, 216–219 CrossRef CAS PubMed.
- A. Singh, N. A. Rorrer, S. R. Nicholson, E. Erickson, J. S. DesVeaux, A. F. Avelino, P. Lamers, A. Bhatt, Y. Zhang and G. Avery, Joule, 2021, 5, 2479–2503 CrossRef CAS.
- T. Uekert, J. S. DesVeaux, A. Singh, S. R. Nicholson, P. Lamers, T. Ghosh, J. E. McGeehan, A. C. Carpenter and G. T. Beckham, Green Chem., 2022, 24, 6531–6543 RSC.
- K. P. Sullivan, A. Z. Werner, K. J. Ramirez, L. D. Ellis, J. R. Bussard, B. A. Black, D. G. Brandner, F. Bratti, B. L. Buss, X. Dong, S. J. Haugen, M. A. Ingraham, M. O. Konev, W. E. Michener, J. Miscall, I. Pardo, S. P. Woodworth, A. M. Guss, Y. Román-Leshkov, S. S. Stahl and G. T. Beckham, Science, 2022, 378, 207–211 CrossRef CAS PubMed.
- S. Kinoshita, S. Kageyama, K. Iba, Y. Yamada and H. Okada, Agric. Biol. Chem., 1975, 39, 1219–1223 CAS.
- S. Kinoshita, T. Terada, T. Taniguchi, Y. Takene, S. Masuda, N. Matsunaga and H. Okada, Eur. J. Biochem., 1981, 116, 547–551 CrossRef CAS PubMed.
- S. Negoro, S. Kakudo, I. Urabe and H. Okada, J. Bacteriol., 1992, 174, 7948–7953 CrossRef CAS PubMed.
- H. Okada, S. Negoro, H. Kimura and S. Nakamura, Nature, 1983, 306, 203–206 CrossRef CAS PubMed.
- S. Negoro, Appl. Microbiol. Biotechnol., 2000, 54, 461–466 CrossRef CAS PubMed.
- S. Kinoshita, S. Negoro, M. Muramatsu, V. S. Bisaria, S. Sawada and H. Okada, Eur. J. Biochem., 1977, 80, 489–495 CrossRef CAS PubMed.
- S. Kakudo, S. Negoro, I. Urabe and H. Okada, Appl. Environ. Microbiol., 1993, 59, 3978–3980 CrossRef CAS PubMed.
- S. Negoro, N. Shibata, D. I. Kato, Y. Tanaka, K. Yasuhira, K. Nagai, S. Oshima, Y. Furuno, R. Yokoyama, K. Miyazaki, M. Takeo, K. Hengphasatporn, Y. Shigeta, Y. H. Lee and Y. Higuchi, FEBS J., 2023, 290, 3400–3421 CrossRef CAS PubMed.
- K. Nagai, K. Iida, K. Shimizu, R. Kinugasa, M. Izumi, D. Kato, M. Takeo, K. Mochiji and S. Negoro, Appl. Microbiol. Biotechnol., 2014, 98, 8751–8761 CrossRef CAS PubMed.
- S. Negoro, N. Shibata, Y. Tanaka, K. Yasuhira, H. Shibata, H. Hashimoto, Y. H. Lee, S. Oshima, R. Santa, S. Oshima, K. Mochiji, Y. Goto, T. Ikegami, K. Nagai, D. Kato, M. Takeo and Y. Higuchi, J. Biol. Chem., 2012, 287, 5079–5090 CrossRef CAS PubMed.
- H. Puetz, C. Janknecht, F. Contreras, M. Vorobii, T. Kurkina and U. Schwaneberg, ACS Sustainable Chem. Eng., 2023, 11, 15513–15522 CrossRef CAS.
- K. Yasuhira, Y. Tanaka, H. Shibata, Y. Kawashima, A. Ohara, D. Kato, M. Takeo and S. Negoro, Appl. Environ. Microbiol., 2007, 73, 7099–7102 CrossRef CAS PubMed.
- E. L. Bell, G. Rosetto, M. A. Ingraham, K. J. Ramirez, C. Lincoln, R. W. Clarke, J. E. Gado, J. L. Lilly, K. H. Kucharzyk, E. Erickson and G. T. Beckham, Nat. Commun., 2024, 15, 1217 CrossRef CAS PubMed.
- H. Lu, D. J. Diaz, N. J. Czarnecki, C. Zhu, W. Kim, R. Shroff, D. J. Acosta, B. R. Alexander, H. O. Cole, Y. Zhang, N. A. Lynd, A. D. Ellington and H. S. Alper, Nature, 2022, 604, 662–667 CrossRef CAS PubMed.
- M. Winnacker and B. Rieger, Polym. Chem., 2016, 7, 7039–7046 RSC.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, bioRxiv, 2021, DOI:10.1101/2021.08.15.456425.
- C. Notredame, D. G. Higgins and J. Heringa, J. Mol. Biol., 2000, 302, 205–217 CrossRef CAS PubMed.
- F. Madeira, M. Pearce, A. R. N. Tivey, P. Basutkar, J. Lee, O. Edbali, N. Madhusoodanan, A. Kolesnikov and R. Lopez, Nucleic Acids Res., 2022, 50, W276–W279 CrossRef CAS PubMed.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS PubMed.
- S. Negoro, D.-i. Kato, T. Ohki, K. Yasuhira, Y. Kawashima, K. Nagai, M. Takeo, N. Shibata, K. Kamiya and Y. Shigeta, in Methods in Enzymology, ed. G. Weber, U. T. Bornscheuer and R. Wei, Academic Press, 2021, vol. 648, ch. 17, pp. 357–389 Search PubMed.
- A. C. Fonseca, M. H. Gil and P. N. Simões, Prog. Polym. Sci., 2014, 39, 1291–1311 CrossRef CAS.
- F. Stempfle, P. Ortmann and S. Mecking, Chem. Rev., 2016, 116, 4597–4641 CrossRef CAS PubMed.
- Y. Furushima, M. Nakada, K. Ishikiriyama, A. Toda, R. Androsch, E. Zhuravlev and C. Schick, J. Polym. Sci., Part B: Polym. Phys., 2016, 54, 2126–2138 CrossRef CAS.
- I. Y. Phang, J. Ma, L. Shen, T. Liu and W.-D. Zhang, Polym. Int., 2006, 55, 71–79 CrossRef CAS.
- L. T. Lim, I. J. Britt and M. A. Tung, J. Appl. Polym. Sci., 1999, 71, 197–206 CrossRef CAS.
- Y. P. Khanna, W. P. Kuhn and W. J. Sichina, Macromolecules, 1995, 28, 2644–2646 CrossRef CAS.
- S. Sulaiman, S. Yamato, E. Kanaya, J.-J. Kim, Y. Koga, K. Takano and S. Kanaya, Appl. Environ. Microbiol., 2012, 78, 1556–1562 CrossRef CAS PubMed.
- J. Schmidt, R. Wei, T. Oeser, L. A. Dedavid e Silva, D. Breite, A. Schulze and W. Zimmermann, Polymers, 2017, 9, 65 CrossRef PubMed.
- K. Luo, J. Liu, K. Abbay, Y. Mei, X. Guo, Y. Song, Q. Guan and Z. You, Polymers, 2023, 15(13), 2877 CrossRef CAS PubMed.
- H. K. Reimschuessel, J. Polym. Sci., Polym. Chem. Ed., 1978, 16, 1229–1236 CrossRef CAS.
- W. G. Perkins and R. S. Porter, J. Mater. Sci., 1977, 12, 2355–2388 CrossRef CAS.
- K. Tamura, G. Stecher and S. Kumar, Mol. Biol. Evol., 2021, 38, 3022–3027 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- N. J. P. Wierckx, H. Ballerstedt, A. M. de Bont Jan and J. Wery, Appl. Environ. Microbiol., 2005, 71, 8221–8227 CrossRef CAS PubMed.
- W. G. Weisburg, S. M. Barns, D. A. Pelletier and D. J. Lane, J. Bacteriol., 1991, 173, 697–703 CrossRef CAS PubMed.
- E. W. Sayers, E. E. Bolton, J. R. Brister, K. Canese, J. Chan, D. C. Comeau, R. Connor, K. Funk, C. Kelly, S. Kim, T. Madej, A. Marchler-Bauer, C. Lanczycki, S. Lathrop, Z. Lu, F. Thibaud-Nissen, T. Murphy, L. Phan, Y. Skripchenko, T. Tse, J. Wang, R. Williams, B. W. Trawick, K. D. Pruitt and S. T. Sherry, Nucleic Acids Res., 2022, 50, D20–D26 CrossRef CAS PubMed.
- K. Jensen, P. R. Østergaard, R. Wilting and S. F. Lassen, BMC Biochem., 2010, 11, 47 CrossRef CAS PubMed.
- R. E. Yasbin, G. A. Wilson and F. E. Young, J. Bacteriol., 1975, 121, 296–304 CrossRef CAS PubMed.
- Cold Spring Harbor Laboratories, Cold Spring Harb. Protoc., 2015 DOI:10.1101/pdb.rec085894.
- R. Zuchowski, S. Schito, F. Neuheuser, P. Menke, D. Berger, N. Hollmann, S. Gujar, L. Sundermeyer, C. Mack, A. Wirtz, O. H. Weiergräber, T. Polen, M. Bott, S. Noack and M. Baumgart, Microb. Cell Fact., 2023, 22, 71 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc01662a |
| This journal is © The Royal Society of Chemistry 2024 |