Open Access Article
Open Access ArticleSupplementing infant milk formula with a multi-strain synbiotic and osteopontin enhances colonic microbial colonization and modifies jejunal gene expression in lactating piglets†
Laia
Ferreres-Serafini
a,
Susana Mª
Martín-Orúe
 *a,
Meritxell
Sadurní
a,
Jesús
Jiménez
b,
José Antonio
Moreno-Muñoz
b and
Lorena
Castillejos
a
*a,
Meritxell
Sadurní
a,
Jesús
Jiménez
b,
José Antonio
Moreno-Muñoz
b and
Lorena
Castillejos
a
aAnimal Nutrition and Welfare Service, Department of Animal and Food Science, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain. E-mail: gd.animal.aliments@uab.cat
bLaboratorios Ordesa S.L., Parc Científic de Barcelona, C/Baldiri Reixac 15-21, 08028 Barcelona, Spain. E-mail: josea.moreno@ordesa.es; susana.martin@uab.cat; Tel: +34 935811504
First published on 22nd May 2024
Abstract
A total of ninety-six weaned piglets were assigned to four dietary treatments in a 2 × 2 design. The treatments included: a standard milk formula (CTR); CTR + probiotics (6.4 × 108 cfu L−1Bifidobacterium longum subsp. infantis CECT 7210 and 1.1 × 108 cfu L−1Lactobacillus rhamnosus NH001) + prebiotics (galacto-oligosaccharides 4.36 g L−1 and human-milk-oligosaccharide 0.54 g L−1) (SYN); CTR + osteopontin (0.43 g L−1) (OPN); and CTR + SYN + OPN (CON). Daily records including feed intake, body weight, and clinical signs, were maintained throughout the 15-day trial. At the end of the study samples from blood, digestive content, and gut tissues were collected to determine serum TNF-α, intestinal fermentative activity (SCFA and ammonia), colonic microbiota (16S rRNA Illumina-MiSeq), histomorphology, and jejunal gene expression (Open-Array). No statistical differences were found in weight gain; however, the animals supplemented with osteopontin exhibited higher feed intake. In terms of clinical signs, synbiotic supplementation led to a shorter duration of diarrhoea episodes. Regarding gut health, the sequenced faecal microbiota revealed better control of potentially dysbiotic bacteria with the CON diet at day 15. In the colon compartment, a significant increase in SCFA concentration, a decrease in ammonia concentration, and a significant decrease in intraepithelial lymphocyte counts were particularly observed in CON animals. The supplemented diets were also associated with modified jejunal gene expression. The synbiotic combination was characterized by the upregulation of genes related to intestinal maturation (ALPI, SI) and nutrient transport (SLC13A1, SLC15A1, SLC5A1, SLC7A8), and the downregulation of genes related to the response to pathogens (GBP1, IDO, TLR4) or the inflammatory response (IDO, IL-1β, TGF-β1). Osteopontin promoted the upregulation of a digestive function gene (GCG). Correlational analysis between the microbiota population and various intestinal environmental factors (SCFA concentration, histology, and gene expression) proposes mechanisms of communication between the gut microbiota and the host. In summary, these results suggest an improvement in the colonic colonization process and a better modulation of the immune response when milk formula is supplemented with the tested synbiotic combined with osteopontin, benefiting from a synergistic effect.
1. Introduction
The optimal nutrition for newborns is provided by human breast milk, which offers a well-balanced combination of essential nutrients, including proteins, lipids, carbohydrates, minerals, and vitamins. Additionally, breast milk contains trace elements and bioactive components crucial for meeting infants’ nutritional requirements and ensuring proper growth and development.1,2 Among its various health benefits, breast milk plays a pivotal role in shaping the gut microbiota of newborns. The development of intestinal microbiota is a complex process that commences around birth and persists for the first 2–3 years of life. This process can be influenced by several early-life factors, such as gestational age, delivery mode, maternal weight, stress, and notably, diet, which significantly affects the relative proportion of bacteria.3–5 Due to its profound impact, breast milk is recognized as the “gold standard” for infant nutrition, recommended by the World Health Organization (WHO) as the exclusive source of nutrition for the first 6 months of life.2,6 However, some infants are unable to receive human breast milk and, milk formula becomes the alternative. Unfortunately, most conventional cow's milk-based formulas lack bioactive components crucial for promoting proper gut bacterial colonization.5 Studies have shown that exclusively breast-fed infants generally exhibit a healthier microbiome compared to exclusively formula-fed infants.7,8 Nevertheless, recent advancements in milk formulas aim to simulate the functionality of breast milk by incorporating various components, including probiotics, prebiotics and different bioactive components, to enhance early microbiota establishment and maturation in newborns.Probiotics, defined as live microorganisms that confer health benefits when consumed in adequate amounts,9 are now commonly added to infant formula. In this study, we selected two probiotic strains, Bifidobacterium longum subsp. infantis CECT 7210 and Lactobacillus rhamnosus HN001, both present in infant gut population and recognized for their beneficial role in infant health.5,10 However, the lack of long-term effects when supplemented in infant formula suggests that their inclusion as a synbiotic combination (probiotic + prebiotic) could be more effective.11 Therefore, in this trial, these probiotics were combined with two prebiotics. Prebiotics are defined as substrates selectively utilized by host microorganisms that confer health benefits.12 In this work, we tested two prebiotics naturally present in human breast milk: galacto-oligosaccharides (GOS) and a human milk oligosaccharide (HMOs). Galacto-oligosaccharides (GOS) are non-digestible short-chain oligosaccharide readily fermented by lactobacilli and bifidobacteria.13 They are well-documented to improve intestinal barrier, reduce pathogenic bacteria colonization, significantly increase short-chain fatty acid (SCFA) concentration, and modulate gene expression.14,15 Human milk oligosaccharides (HMOs) represent the third-largest solid component in human milk (following lactose and lipids), yet they are present in bovine milk at 20 times lower concentrations.16,17 Human milk contains over 200 different HMO structures, with 2′-Fucosyllactose (2′FL), 3-Fucosyllactose (3FL), 3′-sialyllactose (3′SL) and 6′-sialyllactose (6′SL), lacto-N-neotetraose (LNnT), being the most important ones.18 These human-specific milk oligosaccharides are intricate, non-digestible carbohydrates that reach the infant colon and have been linked to the growth enhancement of Bacteroides, Lactobacillus and Bifidobacterium.17,19 They play a role in modulating the immune system by inhibiting pro-inflammatory responses, altering intestinal gene expression,20 and directly influencing intestinal epithelial cells.21 According to the definition of synbiotic as “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host”,12 we could anticipate that the combination of Bifidobacterium longum subsp. infantis CECT 7210 and Lactobacillus rhamnosus HN001 with GOS and HMOs might exhibit synbiotic properties when added to the milk formula.
Osteopontin is an acidic phosphorylated glycoprotein expressed in various cell types with multiple functionalities. It is involved in processes such as activation and regulation of the immune system, biomineralization, tissue-transformative processes and interactions with bacteria, among other.22 Recently, osteopontin have been reported to promote intestinal proliferation and maturation, brain myelination, neurodevelopment and immune development.23 Human milk has the highest concentration of this bioactive protein, being 10 times higher compared to bovine milk.24 Osteopontin is believed to initiate and regulate developmental, immunological and physiological processes in infants, potentially playing a significant role in early-age development.
Despite extensive literature documenting the key roles of these breast milk components in infants, further research is needed to better understand their mode of action and their effects when combined in infant formulas. We hypothesize that the combination of these components could promote a synergic effect, resulting in earlier maturation of intestinal microbiota and improvement in intestinal development and functionality. Therefore, the aim of this study is to evaluate the impact of enhancing an infant milk formula by adding the described multi-strain synbiotic and/or osteopontin on intestinal health, using a lactating pig model.
2. Material and methods
The experiment was carried out at the Experimental Unit of Universitat Autònoma de Barcelona (UAB) and received prior approval (permit no. CEAAH 4928) from the Animal and Human Experimental Ethics Committee (DMAH 10947). The treatment, management, housing and slaughtering conditions adhered to European Union Guidelines (Directive 2010/63/EU, European Commission, 2010). All efforts were made to minimize animal suffering.2.1 Animals, housing and experimental design
A total of 96 male piglets (Large White × Landrace) × Pietrain, aged 5 days (±0.14), from a high-sanitary-status commercial farm were included in this study. The animals were transported from a commercial farm to UAB's experimental unit.The study was organized into four experimental periods of 15 days each, to ensure dedicated care of the animals during the initial days. In each period, 24 new piglets were introduced to UAB's facilities. Animals were chosen from 8 litters, selecting males of intermediate weight. Each period, animals were allocated to two rooms, each with four pens (3 animals per pen). The rooms were equipped with automatic heaters and forced ventilation. Each pen had individual heating lights, a dish with a tank for liquid dispensing (Minitainer, Rotecna, Spain), water nipples, and a plastic toy for environmental enrichment. The experiment was conducted during the autumn season (Sept-Nov).
Upon arrival, animals were distributed among treatments preserving littermates. The trial followed a 2 × 2 factorial design, encompassing four dietary groups (with/without synbiotic × with/without osteopontin): control formula (CTR), CTR supplemented with probiotics and prebiotics (SYN), CTR supplemented with osteopontin (OPN), and CTR supplemented with the synbiotic and osteopontin (SYN + OPN = CON). Each experimental group had eight replicates across the four experimental periods, with the pen considered as the experimental unit.
2.2 Probiotic strains, prebiotics, bioactive peptide and diets
The tested probiotics were Bifidobacterium longum subsp. infantis CECT 7210, supplied by Laboratorios Ordesa S.L., and Lactobacillus rhamnosus HN001 from Danisco USA Inc. Both strains were supplied in lyophilized form, with concentrations of 5 × 1010 and 3 × 1010 colony forming units (CFU) per gram of product, respectively, in a maltodextrin base. Administered in the milk formula, the final dosage was 6.36 × 108 and 1.09 × 108 CFU g−1, respectively. Before the trial, probiotic viability was confirmed through plate counting immediately after suspension in the milk formula and at various storage times on the farm, indicating good stability for both strains up to 24 h.The experimental prebiotics consisted of a mix of galacto-oligosaccharides (GOS) and human milk oligosacharide (HMO), specifically 2′-Fucosyllactose (2′FL). GOS, in a syrup form, was thoroughly mixed with the milk formula to ensure a homogenous blend with no visual lumps. The final GOS concentration was 4.36 g L−1, simulating the present amount in human milk (5–10 g L−1),25 as indicated in previous studies.14,26 HMO, in powder form, was added for a final dosage of 0.436 g L−1. The tested bioactive peptide, osteopontin, provided in powder form, was added for a final dosage of 0.436 g L−1.
The milk formula given to piglets was tailored to their nutritional requirements and manufactured ad hoc by Cargill for this study. The formula excluded acidifiers, any other probiotic strains, or derivatives different from those tested, and it reduced immunoglobulin levels. The formula's main ingredients included powdered lactose, whey powder, soybeans’ protein concentrate, porcine plasma meal, wheat hydrolysed gluten, sucrose, wheat, monopotassium phosphate, calcium carbonate, fructo-oligosaccharides and magnesium oxide. The chemical composition details are provided in Table 1. Milk was made available continuously, and fresh milk was prepared three times per day (at 7 am, 1 pm and 7 pm). Following milk reconstitution, all additives were incorporated and manually mixed until achieving complete homogeneity. Prior to introducing the probiotic strains, careful verification ensured that the milk temperature remained below 37 °C.
| Analytical constituents (%) | |
|---|---|
Nutritional additives per kg: 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 IU vitamin A; 5100 IU vitamin D3; 150 mg vitamin E; 87.0 mg Fe, 130 mg Cu, 50.0 mg Zn, 100 mg Mn, 2.0 mg I, 0.25 mg Se. 000 IU vitamin A; 5100 IU vitamin D3; 150 mg vitamin E; 87.0 mg Fe, 130 mg Cu, 50.0 mg Zn, 100 mg Mn, 2.0 mg I, 0.25 mg Se. |
|
| Crude protein | 20.9 |
| Crudes oils and fats | 10.5 |
| Crude fibre | 0.10 |
| Ash | 9.50 |
| Calcium (Ca) | 0.57 |
| Phosphorus (P) | 0.79 |
| Sodium (Na) | 0.78 |
| Lysine | 1.75 |
| Methionine | 0.50 |
2.3 Experimental procedure
Each experimental period lasted 15 days, during which animals were introduced to the experimental diets upon arrival. Daily observations were conducted, and any clinical signs such as apathy, diarrhoea, dehydration, or anorexia were recorded. No antibiotic treatment was administered.Animal performance was monitored, with individual life weight and feed intake recorded daily. Weekly calculations were made for average daily gain (ADG) and average daily feed intake (ADFI). Faecal scores were assessed per pen using the following scale: 1 = solid and dry; 2 = well formed; 3 = very soft or viscous liquid; 4 = watery greyish; 5 = watery yellowish.
For microbiota sequencing, faecal samples were collected on days 3, 9 and 15 through spontaneous defecation or digital stimulation, encompassing all animals (n = 96).
At the end of the trial, two piglets per pen, chosen for higher weight or healthier condition, were euthanized. Euthanasia and subsequent sampling occurred in the morning (between 9:00 and 13:00 h). Remaining animals were euthanized at the end of each experimental period. Deep sedation was induced by intramuscular injection of 20 mg kg−1 ketamine (Ketamidor; Wels, Austria) and 2 mg kg−1 of xylazine (Xilagesic; Les Franqueses del Vallès, Spain). Blood collection tubes without anticoagulant (Aquisel; Madrid, Spain) and with anticoagulant (Aquisel; Madrid, Spain) were used to collect 10 ml of blood from the cranial cava vein of each animal. After blood sampling, animals were euthanized with an intravenous injection of 200 mg kg−1 sodium pentobarbital (Euthasol; Le Vet B.V., Oudewater, The Netherlands). Afterwards, animals were bled, the abdomen was immediately opened, and the gastrointestinal tract was excised and transferred to a tray.
Content from the ileum and proximal colon was collected and homogenized, and their pH measured (Crison 52–32 electrode, Net Interlab; Barcelona, Spain). For ammonia determination, ileal and colonic digesta aliquots were store at −20 °C in 0.2 N H2SO4 as a preservative solution (3 ml of digesta + 3 ml of 0.2 N H2SO4). Another subsample was kept at −20 °C for SCFA and lactic acid determination.
Jejunal tissue samples were collected for gene expression study. Approximately 0.5 cm2 tissue from the jejunum was preserved in RNAlater®, initially in the fridge for 24 hours and subsequently at −20 °C.
Additionally, jejunal sections of 2–3 cm length were cut, opened longitudinally, washed thoroughly with sterile PBS, and fixed by immersion in a 4% formaldehyde solution (Panreac; Castellar del Vallès, Spain) for histomorphology.
2.4 Analytical procedures
Ammonia concentrations in ileal and colonic content were determined using a gas-sensitive electrode (Hatch Co.; CO, USA) paired with a digital voltmeter (Crison GLP 22, Crison Instruments, S.A.; Barcelona, Spain). For the determination process, preserved samples were homogenized and centrifuged at 1500× for 10 min. Subsequently, samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 in distilled water based on ammonia concentration. The ammonia released after the addition of 10 M NaOH was measured in the supernatant as a change in voltage (mV) using a digital voltmeter (Crison GLP 22, Crison Instruments, S.A.).
4 in distilled water based on ammonia concentration. The ammonia released after the addition of 10 M NaOH was measured in the supernatant as a change in voltage (mV) using a digital voltmeter (Crison GLP 22, Crison Instruments, S.A.).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) crypt depth (VH
crypt depth (VH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD), intraepithelial lymphocytes (IEL), goblet cells (GC) and mitosis (M). Between 7 and 10 intact villi were measured per animal.
CD), intraepithelial lymphocytes (IEL), goblet cells (GC) and mitosis (M). Between 7 and 10 intact villi were measured per animal.
A total of 56 genes involved in multiple physiological functions related to intestinal health were analysed and categorized into groups: (1) genes responsible for maintaining the intestinal barrier function, including OCLN, ZO1, CLDN1, CLDN4, CLDN15, MUC2, MUC13, and TFF3; (2) genes crucial for immune responses, including pattern recognition receptors, cytokines, chemokines, and stress proteins, including TLR2, TLR4, IL1B, IL6, IL8, IL10, IL17A, IL22, IFNG, TNF, TGFB1, CCL20, CXCL2, IFNGR1, HSPB1, HSPA4, REG3G, PPARGC1A, FAXDC2, and GBP1; (3) genes encoding enzymes and hormones involved in the digestive process, such as GPX2, SOD2, ALPI, SI, DAO1, HNMT, APN, IDO1, GCG, CCK, IGF1R, and PYY; (4) genes participating in nutrient transport, including SLC5A1, SLC16A1, SLC7A8, SLC15A1, SLC13A1, SLC11A2, MT1A, SLC30A1, and SLC39A4; and (5) genes associated with stress responses, specifically CRHR1, NR3C1, and HSD11B1. Detailed information regarding genes is provided in ESI (Table S1†).
For this method, DNA was extracted with the QIAamp Fast DNA Stool Mini Kit (Qiagen; West Sussex, United Kingdom), following manufacturer's protocol. Subsequently, microbiome analysis was performed by sequencing 16S (Illumina MiSeq). For this procedure, a total of 50 ng of DNA was amplified following the 16S Metagenomic Sequencing Library Illumina 15044223 B protocol (Illumina). In the first amplification step, primers were designed containing: (1) a universal linker sequence allowing amplicons for incorporation indexes and sequencing primers by Nextera XT Index kit (ILLUMINA); and (2) 16S rRNA gene universal primers.31 In the second and last assay amplification indexes were included. 16S based libraries were quantified by fluorimetry using Quant–iT™ PicoGreen™ dsDNA Assay Kit (Thermofisher). Libraries were pooled prior to sequencing on the NovaSeq platform (Illumina), 300 cycles paired reads configuration. The size and quantity of the pool were assessed on the Bioanalyzer 2100 (Agilent) and with the Library Quantification Kit for Illumina (Kapa Biosciences), respectively. PhiX Control library (v3) (Illumina) was combined with the amplicon library (expected at 20%). Image analysis, base calling and data quality assessment were performed on the NovaSeq instrument (NovaSeq Control Software (NVCS v1.7)). Raw sequences, forward (R1) and reverse (R2), were imported into QIIME2 plattform.32 Cutadapt v3.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 plugin was used to filter specific V3–V4 16S region adapters. R1 and R2 reads were processed using ‘denoise-paired’ command of DADA2 plugin.34 Low-quality reads were filtered by the function ‘filterAndTrim’, and were truncated where they started to lose quality (240 for R1, 200 for R2). Error models were generated using ‘learnErrors’ function, and DADA2 algorithm was applied using ‘dada’ function. ASVs (‘Amplicon Sequence Variants’) generated by R1 and R2 reads were merged using ‘mergepairs’ function. Quimera sequences were removed ‘removeChimeraDenovo’ function. Taxonomy of resulting ASVs was annotated using blastn v2.2.29+35 against 16S specific database from the NCBI (v. August 2021). Assigned taxonomies with an identity percentage lower than 97% were reassigned using NBAYES algorithm36 against SILVA v.138 16S database.
33 plugin was used to filter specific V3–V4 16S region adapters. R1 and R2 reads were processed using ‘denoise-paired’ command of DADA2 plugin.34 Low-quality reads were filtered by the function ‘filterAndTrim’, and were truncated where they started to lose quality (240 for R1, 200 for R2). Error models were generated using ‘learnErrors’ function, and DADA2 algorithm was applied using ‘dada’ function. ASVs (‘Amplicon Sequence Variants’) generated by R1 and R2 reads were merged using ‘mergepairs’ function. Quimera sequences were removed ‘removeChimeraDenovo’ function. Taxonomy of resulting ASVs was annotated using blastn v2.2.29+35 against 16S specific database from the NCBI (v. August 2021). Assigned taxonomies with an identity percentage lower than 97% were reassigned using NBAYES algorithm36 against SILVA v.138 16S database.
2.5 Statistical analysis
The effects of experimental treatments on various parameters (excluding microbiology) were analysed using the free R statistical analysis software version x64 4.0.3 (R Development Core Team; Franklin Lakes, NJ, USA) with the stats package.37 An ANOVA was conducted (lm function for a two-way ANOVA) to determine the main effects of synbiotic addition, osteopontin addition, or any possible interaction. The following generalized linear model was employed:| Yijkl = m + Synbiotici + Oseopontinj + (Synbiotic × Osteopontin)ij + Periodk + eijkl |
For the analysis of the evolution of ADFI and faecal scores along time, a mixed-effects model (lme function) was used following:
| Yijk = m + Dieti + Timej + (Diet × Time)ij + eijk |
When the effects of additives were established (p < 0.05), the least squares means were separated using the probability function of differences adjusted by Tukey–Kramer.
The analysis of the percentage of casualties was conducted through a frequency analysis using a Fisher test (fisher.test function) with the same statistical package.
The pen was considered the experimental unit for all parameters, and results are expressed as means with their standard errors unless otherwise stated. The α level used to determine statistical significance was p = 0.05, with statistical trend levels between p = 0.05 – p = 0.10 considered.
In the gene expression study, the Open Array data were prepared using ThermoFisher Cloud 1.0 software (Applied Biosystems). The 2 − ΔΔCt method for relative quantification was applied, using the geometric mean of 4 reference genes (ACTB, B2M, GAPDH and TBP) and a reference sample with a representative profile from the CTR group as a calibrator. The maximum allowed cycle relative threshold was set to 26, amplification score <1.240, quantification cycle confidence >0.80, and maximum allowed SD between duplicates was set to <0.38. Three samples showing inadequate amplification were removed. All data underwent a logarithmic transformation to approximate the Gaussian distribution. Statistical analyses were performed using free R statistical analysis software version x64 4.0.3 (R Development Core Team; Franklin Lakes, NJ, USA). A two-way ANOVA was conducted following the generalized linear model mentioned before, without considering period effect. Subsequent pairwise post hoc comparisons of treatments were executed using Tukey's honest significant difference test.38 Statistical differences for treatments were considered when p-values were under 0.05 for the ANOVA and Tukey's analysis.
A principal component analysis (PCA) was performed using samples as cases and gene log10-expressions as variables, following the method described by González-Solé et al. (2020).30
Microbiota sequencing data were treated as follows. The relative taxonomic abundances of the samples were displayed with collapsed histograms plotted by ‘ggplot2’ library in R (v.4.0). Data were normalized using the rarefaction technique from Phyloseq R package39 to perform alpha diversity analysis. Shannon, Simpson and Richness indexes were calculated using ‘vegan’ R package.40 Boxplots were performed by ‘ggplot2’ library in R (v.4.0), and Wilcox test was used to find significant differences in alpha diversity between groups.
To illustrate taxonomic dissimilarities based on ASVs, Principal Coordinate Analysis (PCoA) across the samples was carried out using the Bray–Curtis distance matrix. The effects of the factors on taxonomy were evaluated with a permutational multivariate analysis of variance (PERMANOVA) using a Bray–Curtis dissimilarity matrix that was previously calculated considering the relative abundances of functional categories in all samples. The Bray–Curtis dissimilarity matrix and PERMANOVA analysis were performed using ‘vegan’ R package. PCoA plot was constructed using ‘ggplot2’. DESeq2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41 was used for biomarkers identification between the different diets on each of the days. A taxon was considered differentially abundant if the corrected p-value > 0.1 and if it was present on at least 50% of the samples of one of the compared groups. Heatmaps were constructed using ‘ComplexHeatmap’ R package (v.2.10.0). MaAslin2
41 was used for biomarkers identification between the different diets on each of the days. A taxon was considered differentially abundant if the corrected p-value > 0.1 and if it was present on at least 50% of the samples of one of the compared groups. Heatmaps were constructed using ‘ComplexHeatmap’ R package (v.2.10.0). MaAslin2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 was used to study correlations between microbial abundances and organic compounds, gene expression and histology parameters in day 15 samples. A linear model test was performed for each variable, with the variable as fixed effect. The microbial taxa counts were normalized using DESeq normalization method, and the normalized counts were log-transformed. Only taxa present in more than 10% of the samples were considered.
42 was used to study correlations between microbial abundances and organic compounds, gene expression and histology parameters in day 15 samples. A linear model test was performed for each variable, with the variable as fixed effect. The microbial taxa counts were normalized using DESeq normalization method, and the normalized counts were log-transformed. Only taxa present in more than 10% of the samples were considered.
3. Results
In general terms, the trial proceeded as anticipated. Upon arrival, all animals exhibited good health. However, considering the challenges of rearing early-age piglets without the sows, the mortality rate at the end of the study reached 15.6%, with 5, 1, 3, and 6 casualties for the CTR, SYN, OPN, and CON treatments, respectively. The causes for mortality included: 2 humanitarian euthanasians due to acute meningitis (post-mortem diagnosis), from CON treatment; 1 humanitarian euthanasia due to exudative epidermitis (post-mortem diagnosis), from CON treatment; 10 casualties due to acute diarrhoea and dehydration (6 humanitarian euthanasians, 4 sudden deaths), 3 from CTR, 1 from SYN, 3 from OPN and 3 from CON; and 2 humanitarian euthanasia undiagnosed, from CTR treatment. No significant differences in mortality could be attributed to the experimental treatments (Fisher-test p-value > 0.18).In total, 8 piglets needed subcutaneous hydration: 3 for CTR (2 unsuccessful); 2 for SYN (1 unsuccessful); 2 for OPN (unsuccessful); 1 for CON (unsuccessful). No significant differences could be attributed to the treatments (Fisher-test p-value = 1).
3.1 Performance parameters
Results for live weight (LW), average daily feed (milk) intake (ADFI) and average daily gain (ADG) are presented in Table 2. Regarding the live weight of the animals, there were no differences due to treatments, but there were differences related to the experimental period at day 0 (1940, 2414, 2666, 2563 g for period 1–4 respectively, p = 0.005). This was related to the age at which animals arrived at the farm. In the first and third periods, animals arrived at 4 days of life, whereas in the second and fourth periods, they arrived at 5 days of life.| Main effects | Osteopontin − | Osteopontin + | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Synbiotic − | Synbiotic + | Synbiotic − | Synbiotic + | p-Value | |||||
| Diets | CTR | SYN | OPN | CON | RSE | Synbiotic | Osteopontin | Interaction | Period |
| Osteopontin +/− : presence/absence of osteopontin ingredient in the diet; synbiotic +/− : presence/absence of synbiotic ingredients in the diet. n = 8. *Indicates p < 0.05 and statistical difference present. RSE, residual standard error. | |||||||||
| LW (g) | |||||||||
| D0 | 2376 | 2436 | 2390 | 2383 | 367.5 | 0.839 | 0.885 | 0.798 | 0.006 |
| D7 | 3358 | 3524 | 3603 | 3425 | 325.5 | 0.692 | 0.389 | 0.313 | 0.193 |
| D15 | 4628 | 4756 | 5028 | 4794 | 536.8 | 0.225 | 0.120 | 0.954 | 0.549 |
| ADFI (ml) | |||||||||
| D0-d7 | 1075 | 1030 | 1122 | 1080 | 108.2 | 0.172 | 0.161 | 0.670 | 0.004 |
| D8-D15 | 1504 | 1397 | 1788 | 1626 | 189.5 | 0.057 | 0.002 | 0.746 | 0.604 |
| Total | 1290 | 1214 | 1450 | 1351 | 133.0 | 0.061 | 0.005 | 0.962 | 0.369 |
| ADG (g) | |||||||||
| D0-D7 | 140.4 | 155.5 | 173.3 | 148.9 | 46.49 | 0.691 | 0.389 | 0.313 | 0.193 |
| D8-D15 | 181.4 | 175.9 | 203.6 | 195.6 | 89.83 | 0.883 | 0.549 | 0.906 | 0.194 |
| Total | 160.9 | 165.7 | 188.4 | 172.2 | 38.35 | 0.680 | 0.228 | 0.452 | 0.545 |
Regarding milk intake, the animals adapted well to artificial lactation, and all animals learned how to consume milk from the plates after the first 24 hours. Milk intakes could be considered within normal limits, showing a linear increase over time (Fig. 1). Milk intakes during the first week exhibited variations related to the period, but not with the treatments or the interaction. In the second week, a higher milk intake was observed in animals supplemented with osteopontin as these animals consistently exhibited a higher milk consumption on days 12, 13, and 14 (Fig. 1).
The ADG of the animals also fell within the normal range, although with a high variability between litters (pens). No differences were observed in ADG in relation to the additives or the period.
Ultimately, no significant differences in faecal consistency were noted among treatments over time (mean values of 2.9, 2.6, 2.6, 2.7 for CTR, SYN, OPN, CON respectively, p = 0.739). However, upon examining the percentage of days when animals (pens) exhibited diarrhoea (defined as a score ≥4), a notable effect was observed with the synbiotic supplementation. Animals consuming the synbiotic additives experienced a lower percentage of days presenting diarrhoea (Table 3).
| Main effects | Osteopontin − | Osteopontin + | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Synbiotic − | Synbiotic + | Synbiotic − | Synbiotic + | p-Value | |||||
| Diets | CTR | SYN | OPN | CON | RSE | Synbiotic | Osteopontin | Interaction | Period |
| Osteopontin +/−: presence/absence of osteopontin ingredient in the diet; synbiotic +/−: presence/absence of synbiotic ingredients in the diet. n = 8. *Indicates p < 0.05 and statistical difference present. RSE, residual standard error. | |||||||||
| % of days presenting diarrhoea | 39.98 | 25.53 | 46.63 | 26.65 | 18.25 | 0.039 | 0.612 | 0.716 | 0.062 |
3.2 Intestinal fermentation
Concerning the fermentation profile in the ileum and colon, 9 piglets exhibited a substantially altered profile (outliers). Analyzing the history of these animals in the sampling book revealed that they had very little digesta content along gastrointestinal tract (almost empty) and/or presented yellowish ileal content (clear signs of diarrhoea). Therefore, these animals were excluded from the NH3 and SCFA data subset. These exclusions were as follows: 3 from the CTR group (periods 1 and 2), 2 from SYN (period 3), 2 from OPN (periods 1 and 3) and 2 from CON (period 1). No animals were excluded from the period 4.The results of fermentation products in ileal and colonic digesta after excluding these individuals are presented in Table 4. Concerning ammonia levels, there was a significant interaction for ileal concentration exhibiting lower values only when all additives were combined in the CON treatment. Significant reductions were observed in colonic ammonia concentration attributed to the synbiotic and the osteopontin supplementation that showed an additive effect in the CON treatment (no significant interaction).
| Main effects | Osteopontin − | Osteopontin + | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Synbiotic − | Synbiotic + | Synbiotic − | Synbiotic + | p-Value | |||||
| Diets | CTR | SYN | OPN | CON | RSE | Synbiotic | Osteopontin | Interaction | Period |
| SCFA, short-chain fatty acids; BCFA, branched-chain fatty acids. Osteopontin +/−: presence/absence of osteopontin ingredient in the diet; synbiotic +/−: presence/absence of synbiotic ingredients in the diet. n = 8. *Indicates p < 0.05 and statistical difference present. x,y Indicate a statistical trend between means. RSE, residual standard error.a It is not possible to provide the statistics since in many animals the values were not detectable. Number of detectable animals were 6, 5, 3 and 5 for CTR, SYN, OPN and CON respectively. | |||||||||
| Ileum digesta (mM kg −1 FM) | |||||||||
| Total SCFA | 4.02 | 4.57 | 4.66 | 5.16 | 2.485 | 0.746 | 0.340 | 0.802 | 0.039 |
| Acetate | 3.91 | 4.49 | 4.60 | 5.08 | 2.413 | 0.725 | 0.308 | 0.766 | 0.037 |
| Lactate | 20.6 | 22.3 | 19.4 | 20.2 | 7.449 | 0.148 | 0.954 | 0.495 | 0.498 |
| mmol NH3/kg MF | 2.99xy | 3.64x | 3.53xy | 2.69y | 0.91 | 0.676 | 0.453 | 0.011 | <0.001 |
| Colonic digesta (mM kg −1 FM) | |||||||||
| Total SCFA | 69.0 | 83.9 | 89.3 | 105.4 | 31.04 | 0.053 | 0.026 | 0.834 | 0.006 |
| Acetate | 44.5 | 54.4 | 58.0 | 66.5 | 20.19 | 0.067 | 0.032 | 0.958 | 0.003 |
| Propionate | 14.8 | 19.6 | 18.6 | 22.3 | 6.00 | 0.005 | 0.190 | 0.548 | 0.558 |
| Butirate | 6.90 | 8.87 | 8.54 | 12.17 | 4.53 | 0.019 | 0.059 | 0.511 | 0.056 |
| Valerate | 1.50 | 2.40 | 2.00 | 2.54 | 1.00 | 0.040 | 0.110 | 0.903 | 0.214 |
| Total BCFA | 1.50 | 1.86 | 2.11 | 1.88 | 0.92 | 0.701 | 0.292 | 0.271 | 0.041 |
| Lactatea | 13.02 | 3.70 | 10.93 | 6.04 | 21.33 | — | — | — | — |
| Molar ratio of SCFA (%) | |||||||||
| Acetate | 63.71 | 68.03 | 66.58 | 63.13 | 8.50 | 0.211 | 0.685 | 0.035 | 0.022 |
| Propionate | 21.56 | 20.35 | 19.68 | 21.83 | 5.51 | 0.153 | 0.277 | 0.715 | 0.369 |
| Butirate | 10.76 | 10.27 | 9.29 | 10.92 | 2.56 | 0.350 | 0.245 | 0.406 | 0.007 |
| Valerate | 1.88 | 2.57 | 2.26 | 2.36 | 1.00 | 0.768 | 0.329 | 0.339 | 0.346 |
| BCFA | 2.24 | 2.05 | 2.19 | 1.76 | 1.00 | 0.285 | 0.490 | 0.574 | 0.081 |
| mmol NH3/kg MF | 274.8 | 78.3 | 147.2 | 37.25 | 161.9 | 0.004 | 0.033 | 0.119 | 0.003 |
Lactic fermentation predominated in the ileum, with no significant differences noted due to treatments, neither in SCFA. In the colon, however, significant changes were evident concerning diets. The addition of the synbiotic trended to increase total SCFA and acetate and significantly increased propionate, butirate and valerate. Osteopontin supplementation significantly increased total SCFA and acetate concentrations, showing a trend for butyrate. No significant interactions were found showing additive effects for SCFA, acetate and butyrate.
Regarding molar ratio, a significant interaction was found for acetate that only was increased when both additives were added separately.
Lactic acid in the colon was only detected in some animals, with 63.5% below the minimum level of detection (1.69 mM kg−1).
3.3 Intestinal histomorphology and serum TNF-α
Histomorphometry analysis of the jejunum, ileum and colon included parameters such as villus height (VH), crypt depth (CD), ratio between villus height![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) crypt depth (VH
crypt depth (VH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD), intraepithelial lymphocytes (IEL) (only in the villus), goblet cells (GC) (only in the villus) and mitosis (only in the crypt) (Table 5).
CD), intraepithelial lymphocytes (IEL) (only in the villus), goblet cells (GC) (only in the villus) and mitosis (only in the crypt) (Table 5).
| Main effects | Osteopontin − | Osteopontin + | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Synbiotic − | Synbiotic + | Synbiotic − | Synbiotic + | p-Value | |||||
| Diets | CTR | SYN | OPN | CON | RSE | Synbiotic | Osteopontin | Interaction | Period |
Measured parameters: villous height (VH); crypt depth (CD); ratio villous height![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) crypt depth (VH crypt depth (VH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD); intraepeithelial lymphocytes (IEL); goblet cells (GC); mitosis (M). Osteopontin +/−: presence/absence of osteopontin ingredient in the diet; synbiotic +/−: presence/absence of synbiotic ingredients in the diet. n = 8. *Indicates p < 0.05 and statistical difference present. RSE, residual standard error. CD); intraepeithelial lymphocytes (IEL); goblet cells (GC); mitosis (M). Osteopontin +/−: presence/absence of osteopontin ingredient in the diet; synbiotic +/−: presence/absence of synbiotic ingredients in the diet. n = 8. *Indicates p < 0.05 and statistical difference present. RSE, residual standard error. |
|||||||||
| Jejunum | |||||||||
| VH (μm) | 236 | 319 | 319 | 291 | 67.59 | 0.344 | 0.344 | 0.098 | 0.134 |
| CD (μm) | 181 | 177 | 185 | 172 | 20.98 | 0.254 | 0.791 | 0.458 | 0.111 |
VH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD CD |
1.33 | 2.00 | 1.80 | 1.78 | 0.503 | 0.364 | 0.118 | 0.419 | 0.228 |
| IEL (cell no) | 8.53 | 14.1 | 12.9 | 11.0 | 5.046 | 0.294 | 0.655 | 0.070 | 0.339 |
| GC (cell no) | 2.20 | 3.72 | 2.95 | 4.34 | 1.784 | 0.452 | 0.920 | 0.337 | 0.010 |
| M (cell no) | 1.75 | 1.44 | 1.42 | 1.33 | 0.701 | 0.338 | 0.360 | 0.946 | 0.604 |
| Colon | |||||||||
| CD (μm) | 278 | 270 | 269 | 284 | 35.06 | 0.916 | 0.736 | 0.497 | 0.602 |
| IEL (cell no) | 2.72 | 1.98 | 1.49 | 1.32 | 0.866 | 0.263 | 0.028 | 0.688 | 0.114 |
| GC (cell no) | 26.4 | 24.3 | 24.8 | 28.3 | 5.336 | 0.697 | 0.560 | 0.168 | 0.760 |
| M (cell no) | 0.257 | 0.157 | 0.219 | 0.085 | 0.123 | 0.043 | 0.326 | 0.794 | 0.702 |
In the jejunum sections, a period effect was observed (data not shown). The number of GC showed abnormally high values with the CON treatment in the third period but not in the other periods.
In the ileum, no significant differences were observed (data not shown).
In the colon, the addition of osteopontin significantly affected the number of IEL with lower numbers. An effect of the synbiotic addition was observed regarding mitotic cells with lower mitotic cell counts.
No statistically significant effects were detected in the serum levels of TNF-α (data not shown).
3.4 Gene expression
The genes exhibiting a significant treatment effect are listed in Table 6, while comprehensive results for the entire gene set can be found in ESI (Table S1†).| Main effects | Osteopontin − | Osteopontin + | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Synbiotic − | Synbiotic + | Synbiotic − | Synbiotic + | p-Value | |||||
| Diets | CTR | SYN | OPN | CON | RSE | Synbiotic | Osteopontin | Interaction | Function |
| Osteopontin +/−: presence/absence of osteopontin ingredient in the diet; synbiotic +/−: presence/absence of synbiotic ingredients in the diet. n = 8. Gene abbreviations detailed in ESI (Table S2†). EH – enzyme/hormone; IR – immune response; BF – barrier function; NT – nutrient transport. * Indicates p < 0.05 and statistical difference present. a,b Indicate statistically significant differences between means. x,y Indicate a statistical trend between means. RSE, residual standard error. | |||||||||
| Genes | |||||||||
| ALPI | 1.140b | 2.204a | 1.859a | 2.091a | 1.588 | 0.001 | 0.084 | 0.027 | EH |
| CCK | 2.07 | 2.252 | 1.823 | 2.655 | 1.581 | 0.025 | 0.897 | 0.257 | EH |
| CXCL2 | 1.286 | 0.543 | 0.586 | 0.829 | 2.194 | 0.066 | 0.307 | 0.075 | IR |
| GBP1 | 1.415 | 0.781 | 0.9 | 0.9 | 2.289 | 0.045 | 0.347 | 0.217 | IR |
| GCG | 0.924 | 0.891 | 1.13 | 1.129 | 1.468 | 0.883 | 0.040 | 0.900 | EH |
| GPX2 | 1.826 | 1.433 | 1.868 | 1.433 | 2.094 | 0.091 | 0.846 | 0.957 | EH |
| IDO1 | 1.364a | 0.284b | 0.458ab | 0.466ab | 5.198 | 0.024 | 0.372 | 0.035 | EH |
| IFNG | 3.15 | 1.002 | 1.151 | 1.618 | 4.136 | 0.082 | 0.902 | 0.145 | IR |
| IL1beta | 1.276a | 0.406b | 0.393b | 0.560b | 2.389 | 0.036 | 0.053 | 0.027 | IR |
| IL6 | 2.899 | 1.103 | 1.379 | 1.356 | 2.304 | 0.050 | 0.356 | 0.103 | IR |
| MUC13 | 0.815 | 1.116 | 0.988 | 0.871 | 1.506 | 0.492 | 0.605 | 0.015 | BF |
| MUC2 | 1.299 | 1.008 | 1.094 | 1.291 | 1.385 | 0.447 | 0.877 | 0.040 | BF |
| OCLN | 1.202 | 1.571 | 1.383 | 1.441 | 1.302 | 0.041 | 0.815 | 0.212 | BF |
| SI | 2.286b | 4.729a | 2.771ab | 3.569ab | 1.884 | 0.004 | 0.840 | 0.026 | EH |
| SLC11A2 | 1.503 | 1.307 | 1.409 | 1.285 | 1.281 | 0.096 | 0.702 | 0.874 | NT |
| SLC13A1 | 0.666y | 1.086x | 0.934xy | 1.113x | 2.255 | 0.026 | 0.072 | 0.073 | NT |
| SLC15A1 | 1.632 | 2.440 | 1.893 | 2.171 | 1.490 | 0.010 | 0.731 | 0.166 | NT |
| SLC39A4 | 1.24 | 1.753 | 1.5 | 1.529 | 1.448 | 0.146 | 0.566 | 0.072 | NT |
| SLC5A1 | 1.694 | 2.791 | 1.844 | 2.349 | 1.668 | 0.005 | 0.909 | 0.215 | NT |
| SLC7A8 | 1.077 | 2.604 | 2.375 | 3.175 | 3.187 | 0.043 | 0.296 | 0.082 | NT |
| TGFbeta1 | 1.550a | 0.904b | 0.916b | 1.105ab | 1.583 | 0.108 | 0.163 | 0.010 | IR |
| TLR2 | 0.817 | 0.431 | 0.635 | 0.651 | 2.178 | 0.073 | 0.911 | 0.171 | IR |
| TLR4 | 2.542a | 1.297b | 1.577ab | 1.620ab | 1.755 | 0.068 | 0.465 | 0.017 | IR |
Within the category of genes associated with enzyme/hormone functions, a significant interaction was observed between additives on intestinal alkaline phosphatase (ALPI) and sucrase-isomaltase (SI) genes, with higher expression in animals only when the synbiotic was supplemented alone (SYN treatment). Significant difference associated to the synbiotic supplementation was noted in the cholecystokinin gene (CCK) gene, with higher expression observed. Among the group of digestive enzymes, an interaction effect was found for indoleamine 2,3-dioxygenase (IDO1) gene, resulting in similar lower expression levels for all three supplemented diets compared to the CTR diet. Regarding proglucagon (GCG), an effect was observed with osteopontin supplementation, resulting in higher expression. In the immune response related genes, results indicated a significant interaction for interleukin 1 beta (IL-1β) with all three experimental treatments showing similar lower values compared to the CTR group. Transforming growth beta factor 1 (TGF-β1) expression also exhibited an interaction effect with reduced levels only when the additives were supplemented separately (SYN or OPN diets). The toll-like receptor 4 (TLR4) gene also showed a significant interaction exhibiting lower expression in the SYN treatment compared to the CTR treatment, with intermediate values observed in the OPN and CON treatments. Lastly, the guanylate binding protein 1 (GBP1) presented lower expression when the synbiotic was supplemented. Regarding the expression of nutrient transport function genes, (SLC13A1, SLC15A1, SLC5A1 and SLC7A8), the synbiotic supplementation promoted a higher expression. In terms of barrier function genes, mucin 13 (MUC13) and mucin 2 (MUC2) displayed an interaction effect. Specifically, when the synbiotic was supplemented alone (SYN diet) resulted in higher values for MUC13, while lower values for MUC2. Ocludin (OCLN) was also more expressed with the synbiotic supplementation.
A principal component analysis (PCA) was conducted to assess the degree of correlation among gene expression values across samples belonging to the four different diets, as illustrated in Fig. 2. The sample identifier numbers are visually represented in the individual factor map (a), with colours corresponding to the assigned diets for each sample. In the variables factor map (b), the diets that exhibit correlations across the gene expression are depicted.
The findings reveal a discernible correlation in the patterns of gene expression within this tissue. In figure (a), samples are visually grouped by treatments in 2D space. Figure (b) clearly shows how the experimental diets with additives differ from the CTR in relation to gene expression.
3.5 Analysis of the microbiota by sequencing the 16S rRNA gene
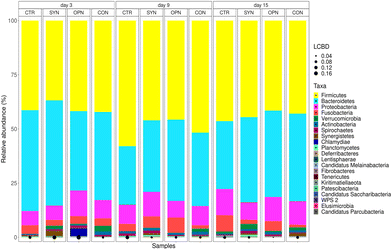
To analyse the impact of experimental diets on the microbiota of piglets, faecal samples were collected on three different days: 3, 9 and 15. The rarefaction curves for all samples reached the plateau phase, indicating comprehensive identification of bacterial species. The predominant phyla in microbial communities were Firmicutes (46.8 ± 16.65%), Bacteroidetes (36.5 ± 17.86%), and Proteobacteria (9.2 ± 10.76%) (Fig. 3). A total of 69 families were detected, with the most abundant being Bacteroidaceae (6.8 ± 6.28%), Lactobacillaceae (6.2 ± 9.03%), and Oscillospiraceae (4.4 ± 3.67%). A total of 148 genera were identified, with the 30 most abundant ones contributing to 67.6 ± 7.89% of the total abundance. Notably, Escherichia (6.2 ± 4.25%), Bacteroides (5.8 ± 3.37%), and Phocaeicola (5.8 ± 4.54%) exhibited the highest relative frequencies, although with a considerable variability among samples.Regarding alpha diversity of microbiota species, it remained stable over time, and there were no significant effects of the treatment on Richness, Simpson, or Shannon indexes. However, at the beta diversity level, the PERMANOVA test revealed statistical differences related to Treatment (R2 = 0.023 and p < 0.001), although most of the variability was explained by Animal and Day (40.7% and 4.5%, respectively).
A treatment effect was observed in the bacterial populations among the different diets. Significant differences at different taxonomic level are presented in the ESI (Fig. S1–S3†). In general, synbiotic supplementation exhibited a more pronounced effect with longer administration of the additive (day 15) across various populations. These changes were evident at different taxonomic levels, including phylum, family, and genus. For instance, the phylum Synergistetes, family Synergistaceae, and genus Cloacibacillus showed increased abundance due to synbiotic supplementation on day 15. Conversely, the family Sutterellaceae and Helicobacteraceae, along with their genera Sutterella and Helicobacter, respectively, were reduced by synbiotic supplementation on day 15. On the contrary, osteopontin supplementation resulted in a more immediate effect on day 3, which diminished as the study progressed. At the family level, there was an increase in Lactobacillaceae on day 3, which disappeared by day 15. At the genus level, on day 3, there was an increase in Oribacterium, Mucispirillum, Prevotellaceae, and Ligilactobacillus due to osteopontin supplementation. However, by day 15, the genera Mediterranea and Christensenella exhibited growth promotion, while populations of Veillonella and Faecalicatena were reduced.
Fig. 4 illustrates the most notable changes induced by the dietary interventions on specific potentially beneficial or dysbiotic genera according to the literature. As mentioned earlier, these changes gradually consolidated as the study progressed. After 15 days of supplementation, a trend was observed wherein most of the beneficial genera increased, while a notable decrease was registered in most of the potentially dysbiosis genera. However, evolution along the experimental period was not the same for all bacterial genera. For beneficial genera, supplemented diets led to a temporary decrease on day 9, which reversed by day 15, as evidenced by Enterococcus and Limosillactobacillus. However, for dysbiotic bacteria the changes had the same trend at day 9 and 15 but consolidating at day 15. Concerning potentially dysbiotic genera at day 15, the combination of synbiotic and osteopontin demonstrated superior control over this group. A significant reduction in Helicobacter, Tyzzerella, and Sutterella may be attributed to the synbiotic component, whereas osteopontin appears to impact Veillonella. The CON diet benefits from both effects, owing to an additive synergy. However, some outcomes are not solely attributable to the additive effect but suggest an unexpected interaction. For instance, a notable decrease in Campylobacter was observed exclusively with the CON diet.
4. Discussion
The benefits of additives supplemented in infant formulas, as used in this trial, are well-documented in the literature. Nevertheless, our aim was to elucidate whether the combination of the synbiotic with osteopontin would result in improvements through a synergistic approach.While there is inconsistent information about the beneficial effects of combining additives of different nature, notable achievements have been documented with the incorporation of probiotics (specifically Lactobacillus and Bifidobacterium) in conjunction with prebiotics (such as bovine milk oligosaccharides and GOS) into infant milk formulas. These combinations have successfully created an intestinal environment in infants that closely resembles that of breastfed infants, as reported by Meli et al., 2014.43 Regarding osteopontin supplementation in infant milk formulas, limited research has been conducted, especially in combination with a synbiotic. Nevertheless, important bioactive roles have been attributed to osteopontin44 and when supplemented to infant formula, it has promoted shifts in jejunal gene expression24 and functional systemic benefits45 approximating a breast-fed condition. Our hypothesis, therefore, is that each additive would endorse benefits to neonates, and the combination of all would be translated into a better response.
In this trial, suckling piglets were employed as an animal model for newborn infants. Attempting to replicate the characteristics of a neonate in an animal model has proven to be intricate. However, the suckling pig has been accepted as a suitable model for human nutritional interventions,46–48 presenting a very similar digestive system anatomy and morphology compared to human infants,49 along with similarities in digestive function and gastrointestinal fermentation profiles.48 Nevertheless, it is important to consider the distinct predominant bacterial groups that these two species host during the early stages of development. In human infants, the intestinal gut is primarily colonized by bifidobacterial, whereas in newborn piglets, the dominant microbial community comprises lactobacilli.49 Maternal passive immunity transfer also differs across species. In the case of humans, maternal antibodies are transferred to the fetus through the placenta during gestation, while in piglets, there is no passive immunity transfer. Consequently, the intake of colostrum becomes crucial for them.50 Despite these unavoidable differences, suckling piglets have already been successfully used as model for humans in nutritional research.48
Supplemented formula in this study proved to be safe. Regarding performance, animals supplemented with osteopontin registered statistical higher feed intake during the second week and overall. This effect, however, was more markedly seen in the first experimental period when animals arrived at an earlier age and lighter weights, suggesting that these additives may have a greater impact in more challenging situations. The weights at the end of the study were numerically higher in all treated groups, particularly in the animals receiving osteopontin (p = 0.120), although this difference did not reach statistical significance. The absence of statistical differences in performance could also be attributed to the limitations of the experimental design, which was primarily intended to elucidate possible mechanisms of action rather than assess the impact of diets on performance. Alternative designs, with increased numbers of replicates and extended administration periods, may have resulted in more pronounced differences in growth. Nevertheless, while animal growth remains a reliable indicator of health, other analysed parameters can also offer valuable information in this regard. For instance, the presence or absence of diarrhoea can also be considered a useful indicator. In this case, the addition of the synbiotic demonstrated a significant impact, reducing its prevalence.
The presence of beneficial microbiota in the hindgut is generally associated with higher concentrations of SCFA.5,14,21 In our study, the addition of both synbiotic and osteopontin had an effect increasing colonic SCFA concentration. Notably, the highest values were achieved when the additives were combined in the CON diet due to an additive effect. This result could indicate a greater development of colonic microbiota in these animals, promoted by a synergic effect of these additives. In connection with this finding, higher SCFA denotes a healthier gut environment by enhancing intestinal barrier and promoting an anti-inflammatory condition in the area.5,51,52 In accordance with that, colonic IEL counts were found significantly lower when all the additives were combined in CON group. Differences in gene expression related to immune response were seen as well, although they were detected in a different intestinal compartment (jejunum). The expression of the pro-inflammatory cytokine IL-1β was found statistically lower in all three supplemented diets compared to CTR. Similar results were seen in other studies where additives of the same nature helped preserve a non-inflammatory tone of the intestinal mucosa.14,15,53–56 The expression of TGFβ1, IL-1β, GBP1 and TLR4, two inflammation-modulating cytokines57,58 and two pathogen recognition receptors,59,60 respectively, was also lower in supplemented diets. In all the cases, a downregulation suggests fewer presence of pathogens and a closer approximation to a homeostatic environment.61,62
Continuing with gene expression in the small intestine, the findings suggest that the additives, particularly the synbiotic, influence the expression of various genes. Genes associated with intestinal maturation and protection against pathogens (ALPI and SI)63 increased with the synbiotic supplementation alone. The synbiotic supplementation also led to increased expression in nutrient transport genes (SCL13A1, SLC15A1, SLC5A1 and SLC7A8). Both results highlight the beneficial effect of the synbiotic combination. In terms of intestinal barrier function, existing literature suggests that both GOS and osteopontin promote higher expression in this gene group.14,15,53 In our study, synbiotic supplementation led to increased expression of OCLN.14,15,54 However, contrary to what the literature indicates,54 the addition of osteopontin did not result in increased expression of OCLN, ZO1, or mucin mRNA. Mucin 2 (MUC2) and mucin 13 (MUC13) showed an upregulation when the synbiotic was supplemented alone (SYN diet). Regarding digestive enzymes and hormones genes, such as proglucagon (GCG), osteopontin exhibited a significant upregulatory effect. This gene encodes for various peptides involved in digestive processes, with the most notable ones being glucagon and glucagon-like peptide-1,64,65 which play crucial roles in regulating individual intake and satiety. Interestingly, osteopontin supplementation affects milk consumption in animals, suggesting a potential connection with the upregulation of genes related to digestive function.
This study probably represents one of the first to provide detailed information on the early gut microbial colonization in artificially reared suckling piglets. In general, the three dominant phyla present in faecal samples, regardless of the animals’ age, were Firmicutes, Bacteroides and Proteobacteria, consistent with findings from other studies on weaning piglets.66–70 However, the taxonomic pattern observed in this study during early ages (day 3) was more akin to the pattern reported in previous studies for days 14 and 21 of age.66 Fusobacterium phyla is typically identified as the third dominant phylum in preweaning piglets, and its abundance tends to decrease around 2–3 weeks of life. In our trial, Fusobacterium, was consistently detected in low abundances from the beginning (day 3), and the three dominant phyla remained relatively stable throughout all the sampling times. This lack of variation could be attributed to the consistent composition of the artificial formula during the trial, unlike sow milk, which undergoes slight changes in composition as lactation progresses. Furthermore, it is important to note the compositional differences between sow's milk and the milk formula used in this trial, particularly regarding its wheat content, providing a certain amount of starch. Major changes in gut microbiota are typically observed when dry feed is introduced to piglets, characterized by complex carbohydrates from dietary fibre and starch.67,71 A greater diversity of microbiota species is associated with a more mature gut ecosystem and a healthier environment, contributing to resilience and stability during periods of stress.72,73 However, in our trial, no significant differences in alpha diversity were observed over time (day 7 vs. day 20 of age) in any of the treatment groups. These findings contrast with those reported by Saladrigas-García et al. (2022),66 which showed an increase in biodiversity with age. It is possible that the lack of data in the first week of life makes it difficult to discern differences. Added to the previous comment, a milk formula that does not modulate over time may not induce the same changes in gut microbiota. Regarding beta diversity, the PERMANOVA analysis revealed significant differences due to Treatment, Day, Animal, and Period, with Animal being the predominant source of variation. This aligns with previous studies,66,74 which have noted a high degree of individuality among piglets at 1–2 weeks of age. It is suggested that the gut community in young piglets is highly dynamic and tends to reach a state of greater stability around the age of 4 weeks. Regarding the influence of milk formula supplementation on microbiota composition, significant changes in family and genera abundances induced by the synbiotic were observed by day 15. In contrast, changes induced by osteopontin were predominantly noticed by day 3, diminishing later in some cases. This observation logically aligns with the distinct functions of the additives. The synbiotic, being a biotic compound, aims to gradually alter the microbiota, requiring time for noticeable effects. Conversely, osteopontin, being a peptide with multifaceted functions, appears capable of effecting more rapid changes. The most noteworthy impact induced by the additives was the improved control of potentially dysbiotic genera at day 15, predominantly observed in the CON diet group. This enhanced control could be attributed to an additive effect of the synbiotic and osteopontin. Among these potentially dysbiotic genera, it is worth noting the significant reduction observed in Campylobacter genera with the CON diet. Most Campylobacter species cannot utilise glucose and therefore rely on free amino and keto acids from the intestinal digesta for growth, making them considered proteolytic bacteria.75 The reduced levels of colonic ammonia observed in the supplemented formulae, particularly with CON, support a potential shift toward a less proteolytic and more beneficial microbiota due to supplementation. The higher colonic SCFA concentrations found in CON animals reinforces this hypothesis.
Other specific microbial groups were also significantly altered by the experimental treatments; however, drawing conclusions regarding their potential use as diet health-promoting biomarkers is challenging. Identifying which microbiota would best enhance host health remains a complex area of research. Although some progress has been made in recent years, there is still much ground to cover. We now have a better understanding of the gut colonization process in early-life piglets, which involves shifts from initial populations of lactic acid bacteria, such as genera like Ligilactobacillus or Limosilactobacillus, to other predominant populations as the animals grow.67 In this regards osteopontin supplementation was associated with increases of Ligilactobacillus at day 3. However, despite these insights, the variable environmental conditions influenced by individual hosts and their diets, along with the intricate interactions among microorganisms within the gut ecosystem, complicate the identification of specific microbial biomarkers. This makes it a challenging and likely oversimplified task.
To better understand these complex interactions and how gut microbiota impacts intestinal piglet health, we performed correlation analysis between data from SCFA concentration, histomorphology, and gene expression with sequenced microbiota at day 15. The most relevant correlations (r > 0.5; adjusted p < 0.05) are shown in Fig. 5–7. Some significant correlations between SCFA bacteria producers and SCFA concentration were detected (Fig. 5). For instance, Collinsella aerofaciens, an acetate producer,76 was positively correlated with acetate concentration; Butyricicoccus pullicaecorum, a butyrate producer,77 positively correlated with butyrate; Megasphaera elsdenii, a valerate producer,78 was positively correlated with valerate; and Ligilactobacillus salivarius, a lactate producer,79 positively correlated with lactate.
 | ||
| Fig. 5 Heatmap of correlations between colonic SCFA concentration and ASVs detected on faecal samples from day 15. *Indicates p < 0.05 and statistical difference present. | ||
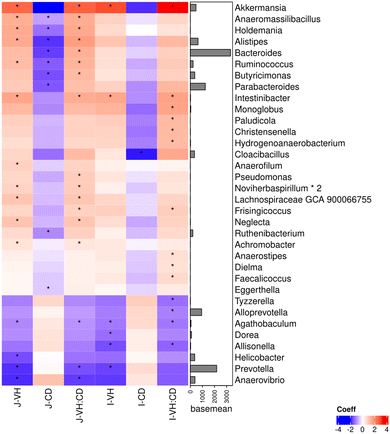
In examining the correlation with histomorphology (Fig. 6), it is noteworthy to consider the following genera due to their notable presence in the samples. The genus Prevotella, known for harboring species described as butyrate producers that tend to increase with the introduction of a solid cereal-based diet,80 showed a negative correlation with villus height in the jejunum and ileum. Bacteroides, a milk oligosaccharides fermenter genus,81 exhibited a negative correlation with crypt depths in jejunum. Lastly, Phocaeicola was negatively correlated with intraepithelial lymphocytes in the jejunum. These findings could elucidate how bacteria interact with the host, influencing variable absorption surface, villus regenerating area, or immune modulation. Although not detected in large proportion, the recently reviewed Akkermansia muciniphila, a promising probiotic well-known to improve the host metabolism function and immune responses,82 was positively correlated with jejunal and ileal villus height. In our trial, this genus was also associated with increases in CCK (digestive hormone involve in satiety)83 and PPARGC1α (related to obesity and oxidative stress)84 gene expression. These findings suggest a potential mode of action for this beneficial species. A higher presence of Akkermansia could translate into a greater surface area for intestinal absorption and enhanced communication with satiety control mechanisms. It is important to note that these data originate from distinct intestinal compartments. While the microbiota came from feces, gene expression was obtained from jejunum tissue, and histology from the jejunum, ileum and colon. The potential influence that microbiota may exert on intestinal histology and gene expression is unlikely to be only a direct local effect. Instead, it could result from the complex interaction or cross-talk of the microbiota with the host, where, for example, the metabolites produced could instigate alterations in other regions of the intestine.
Lastly, when gut microbiota was correlated with gene expression (Fig. 7), positive correlations were found between microbiota families known as disbyotic (Suturella, Tyzzerella)85 with genes well-known to be linked with a pro-inflammation condition (IDO1, TLR4, TGFβ). These same microbial groups were also found to be significantly reduced by the synbiotic supplementation, suggesting that these reductions could be associated with the lower incidence of diarrhoea registered. Similarly, microbiota families known to be beneficial (Christensenella)86 and that were increased with all the supplemented formulas, were negatively correlated with pro-inflammatory genes (IL6, TLR4, TGFβ). This discovery proposes a way of exerting effects, where the microbiota directly or indirectly seems to modulate different gene expression, developing an inflammatory habitat or not. Complementary information regarding correlation analysis can be found in ESI (Fig. S4–S9†).
5. Conclusions
In conclusion, the findings suggest that supplementing milk formulas with synbiotic and/or osteopontin during the initial days of life may impact the establishment of intestinal microbiota, digestive maturation, and neonate immune response. Synbiotic additives encourage the growth of beneficial microbiota genera and better control of dysbiotic genera in the lower digestive tract after 15 days of administration. Additionally, these additives led to reduced episodes of diarrhoea in these animals. Furthermore, they significantly influenced jejunal gene expression. Osteopontin supplementation resulted in increased milk formula consumption, alongside some promotion of beneficial microbiota genera and a reduction in dysbiotic genera in the lower digestive tract. However, more substantial results were observed with the combination of the synbiotic and osteopontin. Significant increases in SCFA concentration, reduced ammonia levels, and modulation of jejunal gene expression suggest an intriguing synergic effect of the tested products, potentially aiding in controlling the overgrowth of opportunistic pathogen and enhancing the organism's response to challenging situations. Furthermore, the correlations between gut microbiota and various intestinal parameters (SCFA concentration, histology, and gene expression) shed light on the intricate cross-talk between the microbiota and the host. For example, beneficial microbiota genera exhibited negative correlations with pro-inflammatory gene expressions, while dysbiotic genera showed contrary correlations. These findings imply a mechanism of interaction where the microbiota appears to regulate various parameters, including immune response gene expressions, either directly or indirectly.Author contributions
Conceptualization, S. M. M.-O., J. J., J. A. M.-M. and L. C.; methodology, S. M. M. and L. C.; software, L. F.-S.; validation, S. M. M.-O. and L. C.; formal analysis, L. F.-S., M. S.; investigation, L. F.-S., M. S., S. M. M.-O. and L. C.; resources, L. F.-S., S. M. M.-O., and L. C.; data curation L. F.-S., S. M. M.-O. and L. C.; writing—original draft preparation, L. F.-S., S. M. M.-O. and L. C.; writing—review and editing, L. F.-S., S. M. M.-O., J. A. M.-M. and L. C.; visualization, L. F.-S.; supervision, S. M. M.-O. and L. C.; project administration, S. M. M.-O. and L. C.; funding acquisition, J. J. and J. A. M.-M. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
J. J. and J. A. M.-M. are employees of Laboratorios Ordesa S. L. company, which provided financial support for this research study, and participated in the study design definition, writing of the manuscript, and contributing to the decision to publish the results.Acknowledgements
This research was supported by Laboratorios Ordesa S. L., through the TOLERA PROJECT (IDI-20170870) funded by the Center for Technological and Industrial Development (CDTI). L. F. S. received an FI grant from the Generalitat de Catalunya (FI SDUR 2020 Grant Number 2020 FISDU 00023).References
- A. Nomayo, et al., Infant formula with cow's milk fat and prebiotics affects intestinal flora, but not the incidence of infections during infancy in a double-blind randomized controlled trial, Mol. Cell. Pediatr., 2020, 7, 1–12 Search PubMed.
- O. Perdijk, et al., Sialyllactose and galactooligosaccharides promote epithelial barrier functioning and distinctly modulate microbiota composition and short chain fatty acid production in vitro, Front. Immunol., 2019, 10, 94 Search PubMed.
- K. Kongnum, S. Taweerodjanakarn and T. Hongpattarakere, Impacts of Prebiotic-Supplemented Diets and Breastmilk on Population and Diversity of Lactobacilli Established in Thai Healthy Infants, Curr. Microbiol., 2020, 77, 1191–1202 Search PubMed.
- S. Verkhnyatskaya, M. Ferrari, P. De Vos and M. T. C. Walvoort, Shaping the infant microbiome with non-digestible carbohydrates, Front. Microbiol., 2019, 10, 343 Search PubMed.
- M. Miqdady, J. A. Mistarihi, A. Azaz and D. Rawat, Prebiotics in the Infant Microbiome: The Past, Present, and Future, Pediatr. Gastroenterol. Hepatol. Nutr., 2020, 23, 1–14 Search PubMed.
- N. Li, W. T. Ma, M. Pang, Q. L. Fan and J. L. Hua, The commensal microbiota and viral infection: A comprehensive review, Front. Immunol., 2019, 10, 1551 Search PubMed.
- J. C. C. Davis, et al., Growth and Morbidity of Gambian Infants are Influenced by Maternal Milk Oligosaccharides and Infant Gut Microbiota, Sci. Rep., 2017, 7, 1–16 Search PubMed.
- P. D. Houghteling and W. A. Walker, Why is initial bacterial colonization of the intestine important to the infant's and child's health?, J. Pediatr. Gastroenterol. Nutr., 2015, 60, 294 Search PubMed.
- C. Hill, et al., The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic, Nat. Rev. Gastroenterol. Hepatol., 2014, 11, 506–514 Search PubMed.
- W. Turpin, C. Humblot, M. Thomas and J. P. Guyot, Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms, Int. J. Food Microbiol., 2010, 143, 87–102 Search PubMed.
- M. Bazanella, et al., Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome, Am. J. Clin. Nutr., 2017, 106, 1274–1286 Search PubMed.
- K. S. Swanson, et al., The International Scientific Association for Probiotics and Prebiotics, ISAPP) consensus statement on the definition and scope of synbiotics, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 687–701 Search PubMed.
- G. Wang, et al., Galactooligosaccharides as a protective agent for intestinal barrier and its regulatory functions for intestinal microbiota, Food Res. Int., 2022, 155, 111003 Search PubMed.
- S. Tian, J. Wang, H. Yu, J. Wang and W. Zhu, Changes in Ileal Microbial Composition and Microbial Metabolism by an Early-Life Galacto-Oligosaccharides Intervention in a Neonatal Porcine Model, Nutrients, 2019, 11, 1753 Search PubMed.
- J. Weng, S. Tian, H. Yu, J. Wang and W. Zhu, Response of Colonic Mucosa-Associated Microbiota Composition, Mucosal Immune Homeostasis, and Barrier Function to Early Life Galactooligosaccharides Intervention in Suckling Piglets, J. Agric. Food Chem., 2019, 67, 578–588 Search PubMed.
- Y. Vandenplas, et al., Human Milk Oligosaccharides: 2′-Fucosyllactose (2′-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula, Nutrients, 2018, 10(9), 1161 Search PubMed.
- P. Thomson, D. A. Medina and D. Garrido, Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization, Food Microbiol., 2018, 75, 37–46 Search PubMed.
- M. Zuurveld, et al., Specific Human Milk Oligosaccharides Differentially Promote Th1 and Regulatory Responses in a CpG-Activated Epithelial/Immune Cell Coculture, Biomolecules, 2023, 13(2), 263 Search PubMed.
- A. Marcobal, et al., Consumption of human milk oligosaccharides by gut-related microbes, J. Agric. Food Chem., 2010, 58, 5334–5340 Search PubMed.
- L. E. Carr, et al., Role of Human Milk Bioactives on Infants’ Gut and Immune Health, Front. Immunol., 2021, 12, 604080 Search PubMed.
- S. Zhang, et al., Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota, Microb. Cell Fact., 2021, 20(1), 108 Search PubMed.
- E. S. Sørensen and B. Christensen, Milk Osteopontin and Human Health, Nutrients, 2023, 15(11), 2423 Search PubMed.
- R. Jiang and B. Lönnerdal, Effects of Milk Osteopontin on Intestine, Neurodevelopment, and Immunity, Nestle Nutr. Inst. Workshop Ser., 2020, 94, 152–157 Search PubMed.
- S. M. Donovan, et al., Bovine Osteopontin Modifies the Intestinal Transcriptome of Formula-Fed Infant Rhesus Monkeys to Be More Similar to Those That Were Breastfed, J. Nutr., 2014, 144, 1910–1919 Search PubMed.
- C. Kunz, et al., Influence of Gestational Age, Secretor, and Lewis Blood Group Status on the Oligosaccharide Content of Human Milk, J. Pediatr. Gastroenterol. Nutr., 2017, 64, 789–798 Search PubMed.
- A. Alizadeh, et al., The piglet as a model for studying dietary components in infant diets: effects of galacto-oligosaccharides on intestinal functions, Br. J. Nutr., 2016, 115, 605–618 Search PubMed.
- A. J. Richardson, A. G. Calder, C. S. Stewart and A. Smith, Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography, Lett. Appl. Microbiol., 1989, 9, 5–8 Search PubMed.
- M. T. Jensen, R. P. Cox and B. B. Jensen, Microbial production of skatole in the hind gut of pigs given different diets and its relation to skatole deposition in backfat, Anim. Sci., 1995, 61, 293–304 Search PubMed.
- M. Nofrarías, et al., Effects of spray-dried porcine plasma and plant extracts on intestinal morphology and on leukocyte cell subsets of weaned pigs, J. Anim. Sci., 2006, 84, 2735–2742 Search PubMed.
- F. González-Solé, et al., Porcine Digestible Peptides (PDP) in Weanling Diets Regulates the Expression of Genes Involved in Gut Barrier Function, Immune Response and Nutrient Transport in Nursery Pigs, Animals, 2020, 10, 2368 Search PubMed.
- A. Klindworth, et al., Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 2013, 41(1), e1 Search PubMed.
- E. Bolyen, et al., Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37, 852–857 Search PubMed.
- M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet J., 2011, 17, 10–12 Search PubMed.
- B. J. Callahan, et al., DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 Search PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, Basic local alignment search tool, J. Mol. Biol., 1990, 215, 403–410 Search PubMed.
- N. A. Bokulich, et al., Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin, Microbiome, 2018, 6, 1–17 Search PubMed.
- R. Team, R: A language and environment for statistical computing, MSOR connections, 2014 Search PubMed.
- J. W. Tukey, Comparing Individual Means in the Analysis of Variance, Biometrics, 1949, 5, 99 Search PubMed.
- S. Weiss, et al., Normalization and microbial differential abundance strategies depend upon data characteristics, Microbiome, 2017, 5, 1–18 Search PubMed.
- J. Oksanen and G. L. Simpson, The vegan Package Ecological legacies in the Mesoamerican forest, 2009 Search PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 1–21 Search PubMed.
- H. Mallick, et al., Multivariable association discovery in population-scale meta-omics studies, PLoS Comput. Biol., 2021, 17, e1009442 Search PubMed.
- F. Meli, et al., Growth and safety evaluation of infant formulae containing oligosaccharides derived from bovine milk: A randomized, double-blind, noninferiority trial, BMC Pediatr., 2014, 14, 1–11 Search PubMed.
- B. Lönnerdal, Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas, Am. J. Clin. Nutr., 2014, 99(3), 712S–717S Search PubMed.
- R. Jiang and B. Lönnerdal, Osteopontin in human milk and infant formula affects infant plasma osteopontin concentrations, Pediatr. Res., 2019, 85, 502–505 Search PubMed.
- J. C. Litten-Brown, A. M. Corson and L. Clarke, Porcine models for the metabolic syndrome, digestive and bone disorders: A general overview, Animal, 2010, 4, 899–920 Search PubMed.
- A. Darragh and P. J. Moughan, The three-week-old piglet as a model animal for studying protein digestion in human infants, J. Pediatr. Gastroenterol. Nutr., 1995, 21, 387–393 Search PubMed.
- E. Roura, et al., Critical review evaluating the pig as a model for human nutritional physiology, Nutr. Res. Rev., 2016, 29, 60–90 Search PubMed.
- P. J. Moughan, M. J. Birtles, P. D. Cranwell, W. C. Smith and M. Pedraza, The Piglet as a Model Animal for Studying Aspects of Digestion and Absorption in Milk-Fed Human Infants, World Rev. Nutr. Diet., 1992, 67, 40–113 Search PubMed.
- K. Goncharova, et al., Importance of neonatal immunoglobulin transfer for hippocampal development and behaviour in the newborn pig, PLoS One, 2017, 12(16), e0180002 Search PubMed.
- K. C. Goehring, et al., Similar to Those Who Are Breastfed, Infants Fed a Formula Containing 2′-Fucosyllactose Have Lower Inflammatory Cytokines in a Randomized Controlled Trial, J. Nutr., 2016, 146, 2559–2566 Search PubMed.
- L. Peng, Z. R. Li, R. S. Green, I. R. Holzman and J. Lin, Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers, J. Nutr., 2009, 139, 1619–1625 Search PubMed.
- S. Tian, J. Wang, H. Yu, J. Wang and W. Zhu, Effects of galacto-oligosaccharides on growth and gut function of newborn suckling piglets, J. Anim. Sci. Biotechnol., 2018, 9, 1–11 Search PubMed.
- S. H. Woo, S. H. Lee, J. W. Park, D. M. Go and D. Y. Kim, Osteopontin Protects Colonic Mucosa from Dextran Sodium Sulfate-Induced Acute Colitis in Mice by Regulating Junctional Distribution of Occludin, Dig. Dis Sci., 2019, 64, 421–431 Search PubMed.
- S. Ashkar, et al., Eta-1 (osteopontin): An early component of type-1 (cell-mediated) immunity, Science, 2000, 287, 860–864 Search PubMed.
- D. Meng, et al., Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine, Pediatr. Res., 2020, 88, 209–217 Search PubMed.
- A. Vander Ark, J. Cao and X. Li, TGF-β receptors: In and beyond TGF-β signaling, Cell Signalling, 2018, 52, 112–120 Search PubMed.
- R. Bent, L. Moll, S. Grabbe and M. Bros, Interleukin-1 Beta—A Friend or Foe in Malignancies?, Int. J. Mol. Sci., 2018, 19, 2155 Search PubMed.
- S. Ghosh, et al., Elevated muscle TLR4 expression and metabolic endotoxemia in human aging, J. Gerontol., Ser. A, 2015, 70, 232–246 Search PubMed.
- C. E. Johns and L. Galam, Guanylate Binding Protein 1 (GBP1): A Key Protein in Inflammatory Pyroptosis, Cell Biochem. Biophys., 2022, 80, 295–299 Search PubMed.
- J. B. Ewaschuk, G. K. Murdoch, I. R. Johnson, K. L. Madsen and C. J. Field, Glutamine supplementation improves intestinal barrier function in a weaned piglet model of Escherichia coli infection, Br. J. Nutr., 2011, 106, 870–877 Search PubMed.
- R. Badia, R. Lizardo, P. Martinez, I. Badiola and J. Brufau, The influence of dietary locust bean gum and live yeast on some digestive immunological parameters of piglets experimentally challenged with Escherichia coli, J. Anim. Sci., 2012, 90(Suppl 4), 260–262 Search PubMed.
- K. Geddes and D. J. Philpott, A New Role for Intestinal Alkaline Phosphatase in Gut Barrier Maintenance, Gastroenterology, 2008, 135, 8–12 Search PubMed.
- D. A. Sandoval and D. A. D'Alessio, Physiology of proglucagon peptides: role of glucagon and GLP-1 in health and disease, Physiol. Rev., 2015, 95, 513–548 Search PubMed.
- D. J. Drucker, Mechanisms of Action and Therapeutic Application of Glucagon-like Peptide-1, Cell Metab., 2018, 27, 740–756 Search PubMed.
- M. Saladrigas-García, et al., An insight into the commercial piglet's microbial gut colonization: from birth towards weaning, Anim. Microbiome, 2022, 4, 1–23 Search PubMed.
- M. Saladrigas-García, et al., Understanding host-microbiota interactions in the commercial piglet around weaning, Sci. Rep., 2021, 11, 1–18 Search PubMed.
- D. B. Holman, B. W. Brunelle, J. Trachsel and H. K. Allen, Meta-analysis To Define a Core Microbiota in the Swine Gut, mSystems, 2017, 2(3) DOI:10.1128/msystems.00004-17.
- L. Chen, et al., The maturing development of gut microbiota in commercial piglets during the weaning transition, Front. Microbiol., 2017, 8, 281038 Search PubMed.
- E. A. B. Pajarillo, J. P. Chae, M. P. Balolong, H. B. Kim and D. K. Kang, Assessment of fecal bacterial diversity among healthy piglets during the weaning transition, J. Gen. Appl. Microbiol., 2014, 60, 140–146 Search PubMed.
- Y. Li, et al., Weaning Stress Perturbs Gut Microbiome and Its Metabolic Profile in Piglets, Sci. Rep., 2018, 8, 1–12 Search PubMed.
- A. Konopka, What is microbial community ecology?, ISME J., 2009, 3, 1223–1230 Search PubMed.
- S. Naeem, Z. Kawabata and M. Loreau, Transcending boundaries in biodiversity research, Trends Ecol. Evol., 1998, 13, 134–135 Search PubMed.
- C. L. Thompson, B. Wang and A. J. Holmes, The immediate environment during postnatal development has long-term impact on gut community structure in pigs, ISME J., 2008, 2, 739–748 Search PubMed.
- B. Linz, I. Sharafutdinov, N. Tegtmeyer and S. Backert, Evolution and Role of Proteases in Campylobacter jejuni Lifestyle and Pathogenesis, Biomolecules, 2023, 13, 323 Search PubMed.
- H. J. Flint, S. H. Duncan, K. P. Scott and P. Louis, Links between diet, gut microbiota composition and gut metabolism, Proc. Nutr. Soc., 2015, 74(1), 13–22 Search PubMed.
- V. Singh, et al., Butyrate producers, “The Sentinel of Gut”: Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics, Front. Microbiol., 2022, 13, 1103836 Search PubMed.
- S. Yoshikawa, et al., Valerate production by Megasphaera elsdenii isolated from pig feces, J. Biosci. Bioeng., 2018, 125, 519–524 Search PubMed.
- M. Guerrero Sanchez, S. Passot, S. Campoy, M. Olivares and F. Fonseca, Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges, Appl. Microbiol. Biotechnol., 2021, 106, 57–80 Search PubMed.
- S. Amat, H. Lantz, P. M. Munyaka and B. P. Willing, Prevotella in Pigs: The Positive and Negative Associations with Production and Health, Microorganisms, 2020, 8, 1584 Search PubMed.
- S. A. Frese, K. Parker, C. C. Calvert and D. A. Mills, Diet shapes the gut microbiome of pigs during nursing and weaning, Microbiome, 2015, 3, 1–10 Search PubMed.
- R. M. R. A. Effendi, et al. Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-Related Diseases, Microorganisms, 2022, 10(12), 2382 Search PubMed.
- J. F. Rehfeld, Cholecystokinin and the hormone concept, Endocr. Connect., 2021, 10, R139–R150 Search PubMed.
- M. Kobayashi, Y. Deguchi, Y. Nozaki and Y. Higami, Contribution of PGC-1α to Obesity- and Caloric Restriction-Related Physiological Changes in White Adipose Tissue, Int. J. Mol. Sci., 2021, 22(11), 6025 Search PubMed.
- R. G. Horne, et al., High Fat-High Fructose Diet-Induced Changes in the Gut Microbiota Associated with Dyslipidemia in Syrian Hamsters, Nutrients, 2020, 12, 1–20 Search PubMed.
- W. Mazier, et al., A New Strain of Christensenella minuta as a Potential Biotherapy for Obesity and Associated Metabolic Diseases, Cells, 2021, 10(4), 823 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo00489b |
| This journal is © The Royal Society of Chemistry 2024 |