Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vitro screening of the impact of dietary prebiotic components, probiotic strains, and their symbiotic combinations on colonic microbiota in children with cystic fibrosis†
Jazmín
Viteri-Echeverría
 a,
Ana
Andrés
a,
Joaquim
Calvo-Lerma
a,
Ana
Andrés
a,
Joaquim
Calvo-Lerma
 b,
Ana
Heredia
a,
Jorge
García-Hernández
c and
Andrea
Asensio-Grau
b,
Ana
Heredia
a,
Jorge
García-Hernández
c and
Andrea
Asensio-Grau
 *a
*a
aUniversity Institute of Food Engineering (FoodUPV), Polytechnic University of Valencia, Camino de Vera s/n, 46022, València, Spain. E-mail: jazvi@alumni.upv.es; aandres@tal.upv.es; anhegu@tal.upv.es
bResearch Group in Innovative Technologies for Sustainable Food (ALISOST). University of Valencia, Avda. Vicent Andrés Estellés s/n, Burjassot, 46100, València, Spain. E-mail: joaquim.calvo@uv.es
cAdvanced Food Microbiology Centre (CAMA), Polytechnic University of Valencia, Camino de Vera s/n, 46022 València, Spain. E-mail: jorgarhe@btc.upv.es
First published on 28th May 2024
Abstract
Children with Cystic Fibrosis (CF) are more likely to have intestinal dysbiosis due to recurrent antibiotic therapy and the conventional hypercaloric diet administered to them. This study aimed at evaluating the effect of isolated prebiotic components and probiotic strains, and their combinations as potential synbiotics, on the intestinal microbiota of CF children. A static in vitro colonic fermentation model was used by colonizing vials with faecal inoculum, a culture medium, and the substrates to be tested. Post treatment, aliquots were taken to determine ammonium, lactate, and short-chain fatty acids production and to profile the microbiota composition by 16s rRNA sequencing. At genus level, Escherichia-Shigella decreased (15.8%) with the treatment pectin + L. rhamnosus, followed by the beta-glucan + L. salivarius (15.5%). Inversely, the most increase in Bacteroides (44%) was obtained by the treatment with Pectin + L. reuteri. Lactate and acetic acid production was significantly increased with prebiotics and their combinations with L. rhamnosus and L. salivarius. In conclusion, the use of beta-glucan and pectin in combination with probiotic strains from the Lactobacillaceae family suggest potential to modulate dysbiosis and metabolic activity on CF colonic microbiota, encouraging further studies in animal studies or clinical settings to confirm the findings in vivo.
Introduction
In patients with cystic fibrosis (CF) intestinal dysbiosis is thought to be the consequence of a series of factors, including the abnormal function of the transmembrane conductance regulator protein (CFTR), its mechanisms, and different acquired factors such as repeated exposure to antibiotic therapy and traditional hypercaloric diet.1 Consequently, the imbalance in the gut microbiota is characterized by increased pathogenic bacteria and reduced microbial diversity, significantly compromising patients’ health conditions.2 Most of the studies on patients with CF have focused on the lung microbiota, so there is scarce scientific evidence about the intestinal microbiota and therapeutic solutions to reverse dysbiosis.3In this context, there is a need to search for nutritional strategies that help modulating the intestinal microbiota and contribute to improve the prognosis of the disease. A study in children with CF showed that energy requirements are met through diets rich in saturated and trans fats, and poor in fibre, which contribute to gastrointestinal inflammation and microbial dysbiosis, suggesting that the impact of diets with foods rich in fibre, whole grains and resistant starch should be explored, because of the prebiotic potential.4 It is well known that both prebiotic and probiotic compounds can contribute to improve gut microbiota and the production of immunomodulatory metabolites in different situations.5 Prebiotics can be found naturally present in foods, and they are generally non-digestible compounds that serve as a substrate for host microorganisms and confer health benefits.6,7 Non-starch polysaccharides are considered as potential prebiotics. These molecules are carbohydrate fractions excluding starch, mono and disaccharides, differing in composition and structure from amylase and amylopectin. As they cannot be hydrolysed in the upper gastrointestinal tract, these compounds reach the colon where they can be eventually fermented by the gut microbiota.8 In addition, resistant starch also escapes gastrointestinal digestion and can be selectively used as substrate for gut microbiota. Different types of resistant starch have been defined,9 including retrograded starch, which is formed after a cooling period by gelatinised starch.10 Up to now, only one in vivo study on the use of prebiotics in adults with CF is available.11
On the other hand, probiotics are live microorganisms that when supplied in adequate amounts, induce beneficial effects on the host's health.12 One of the few in vivo studies in CF that evaluated the impact of probiotic supplementation on modifying colonic microbiota as a main study outcome evidenced that Lactobacillus reuteri could be effective in reducing some pathogenic bacteria in the gut.13 However, other studies on probiotic supplementation suggest contradictory results,14 due to different methodological limitations and those inherent to the multifactorial nature of CF disease.15 Likewise, a meta-analysis evaluated the efficacy and safety of probiotics in CF and showed that more research is needed to determine their clinical implications16
Therefore, an in vitro approach to evaluate the potential of different probiotic strains, prebiotic compounds, and synbiotic preparations, in correcting dysbiosis could generate a background of certainty to support the selection of the best “-biotics” supplementation strategy. Static in vitro colonic fermentation models allow for studying multiple samples and “-biotic” substrates simultaneously and evaluating the different effects on intestinal microbiota and metabolic activity.17 In this way, these models can be considered as a screening tool prior the study of long-term supplementation in dynamic in vitro models or in clinical settings.
In fact, in vitro colonic fermentation models to assess the effect of potential prebiotic compounds and probiotic strains on modulating gut microbiota in CF have been already applied. These previous studies in the field focused on Lactobacillaceae strains and beta-glucan, respectively, both suggesting promising results in improving gut microbiota in the context of CF.18,19 However, no information is available on the potential of combining a probiotic strain with a prebiotic compound.
Therefore, this study aimed at evaluating the effect of isolated prebiotic components and probiotic strains, and their combinations as potential synbiotics, on the intestinal microbiota of children with CF, using a static in vitro simulation model of the colonic fermentation.
Materials and methods
Subjects
The faecal samples to obtain the faecal inoculum for the colonic fermentation model (n = 4) were obtained from paediatric patients with CF recruited in the Valencian Community. The study protocol was approved by the Ethics Committe of Universitat Politècnica de València (P03_25-07-2022). The inclusion criteria were: age between 2 and 16 years old, a diagnosis of CF confirmed by a positive sweat test (>60 mEq L−1) and/or by the presence of two CF-causing mutations in the CFTR gene, and a confirmed diagnosis of exocrine pancreatic insufficiency (faecal elastase <200 μg g−1 in stool). The exclusion criteria were the presence of acute infections, acute abdominal pain, treatment with a CFTR gene modulating therapy, the absence of antibiotic treatment in the last 2 months and/or the supplementation with prebiotics or probiotics. All the subjects and their parents/legal guardians signed the informed consent.Selection of compounds with prebiotic potential and probiotic strains
Dietary fiber types with prebiotic potential were selected following the classification of Fu et al. (2022),20 including two types of non-starch polysaccharides: β-glucan (Bgl) from Neogen® (Michigan, USA) and apple pectin (Pc) from Sigma-Aldrich® (Missouri, USA); and two types of resistant starch: native potato starch granules (St) from Sigma-Aldrich® (Missouri, USA) and gelatinized and retrograde starch (rSt) modified by physical processing according to the protocol of Zhou et al. (2019).21 The selection was made under the criterium of assessing pure compounds (analytical standards) rather than commercially available supplements, as these are formulated with excipients that could interfere with the results.On the other hand, three probiotic strains from the Lactobacillaceae family were selected, based on previous proved beneficial effects in children with CF:13,18Lacticaseibacillus rhamnosus GG (ATCC 53103TM) (L. rha), Limosilactobacillus reuteri (DSM17938) (L. reu) and Lactobacillus salivarius (CECT 4063) (L. sal). L. rha and L. reu were isolated from probiotic commercial supplements containing only one microorganism strain, Kaleidon Hydro (Menarini®) and Casenbiotic (BioGaia®) respectively. L. sal was obtained from the Spanish Collection of Type Cultures (CECT). Then, the strains were grown in MRS liquid medium until obtaining a minimum concentration of 108 CFU mL−1.
Materials
Phosphate buffer (0.1M) was obtained from EMD Millipore (Massachusetts, USA). Peptone, sodium chloride, magnesium sulphate, calcium chloride hexahydrate, Tween 80, resazurin salt solution 0.25% (w/v) and bile salts were supplied from Sigma-Aldrich® (Missouri, USA.). Potassium dihydrogen phosphate was purchased from Scharlau® (Barcelona, Spain). Sodium hydrogen carbonate was obtained from Chem-Lab® (Zedelgem, Belgium). Yeast extract was obtained from Condalab® (Madrid, Spain). Hemin, vitamin K1 and cysteine were purchased from Sigma-Aldrich® (Missouri, USA).Static in vitro colonic fermentation
The experiment was conducted using the static in vitro colonic fermentation model for foods proposed by Pérez-Burillo et al. (2021)22 with some modifications to adapt the simulation to the fermentation of pre and probiotic compounds.23,24 In short, fermentation vials were prepared with the faecal inoculum, a culture medium, and the study substrate (prebiotic, probiotic or synbiotic).The faecal inoculum was prepared from the stool samples of 4 children with CF. All of them had pancreatic insufficiency and their ages were between 6 and 11 years old. None of them had taken antibiotics or supplements in two months before the study, and had not started CFTR modulator therapy. The day of the experiment, the four faecal samples were collected fresh (1–2 hours from deposition) from the house of the donors in the interior of sterile pots with anaerobiosis bags. The samples were transported to the laboratory in refrigeration and immediately processed.
The culture medium contained peptone, sodium chloride, magnesium sulphate, calcium chloride hexahydrate, Tween 80, resazurin salt solution, bile salts, potassium dihydrogen phosphate, sodium hydrogen carbonate, yeast extract were mixed, the mixture was autoclaved and then hemin, vitamin K1 and cysteine were added. Of note, the bile salts concentration was modified by reducing the final concentration to 0.05 g L−1 to better approach to the altered concentration found in children with CF.25,26 The pH of the medium was adjusted to 6.5 according to the average physiological value reported in vivo in CF,27 previously measured before sterilization in the autoclave. The samples were first pooled (1 g each) and mixed with phosphate buffer 0.1 M (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 w/v), and the blend was introduced in a stomacher for 2 minutes and the supernatant was collected to inoculate the fermentation vials.
10 w/v), and the blend was introduced in a stomacher for 2 minutes and the supernatant was collected to inoculate the fermentation vials.
To perform the colonic fermentation of dietary prebiotic components, the concentration of the study substrates was based on a previous study:28 24 mg of each compound were weighed in sterile vials, to which 5.4 mL of culture medium and 0.6 mL of faecal inoculum were added. For probiotics, 1 mL of each strain (108 CFU mL−1), 4.4 mL of culture medium and 0.6 mL of faecal inoculum were mixed. Regarding the synbiotic combinations, 12 different were prepared (3 probiotics × 4 prebiotics), for which 24 mg of the prebiotic was mixed with 1 mL of resuspended probiotic, 4.4 mL of culture medium and 0.6 mL of faecal inoculum. Additionally, a control vial was prepared (basal microbiota), including 5.4 mL of culture medium and 0.6 mL of faecal inoculum. Finally, oxygen was removed from the vials using a nitrogen gas flow for 30 seconds before sealing, and the vials were introduced into a hermetic chamber, where oxygen was removed with the use of anaerobiosis bags (Thermo Scientific™ Oxoid AnaeroGen). In total, 20 different conditions were tested in triplicate (60 assays were performed). All the samples were incubated in anaerobiosis for 20 h at 37 °C in agitation (20 rpm). After completion of colonic fermentation, different aliquots were taken for subsequent analytical determinations.
Analysis of the colonic microbiota
Sequenced read on Illumina MiSeq platform (2 × 300 bp) of FISABIO Sequencing Service were submitted to the pipeline of package dada2 (version 1.26.0)29 for R software (R version 4.3.0 (21 April 2023)) for the microbiota data processing. Only R2 reads from Illumina paired ends were truncated at 250 position and reads under 250 nucleotides were removed. Every read with maximum expected error above 2 (expected error calculated from the nominal definition of the quality score (−∑10^(−Q/10))) was also removed, the same as those which matched against the phiX genome. ASVs (Amplicon Sequence Variants) were inferred from DADA2 algorithm, and chimeras were removed with default parameters. Taxonomic assignment was performed up to genus level, based on SILVA database species train set file (version 138.1). R package phyloseq (version 1.44.0)30 was used for manipulating microbiota data. The alpha diversity (Shannon and Chao indexes) as well as beta diversity (Bray–Curtis scale) were obtained using R software.
Following the guidelines and recommendations of the manufacturer, the lactate concentration was measured using the Lactate Assay commercial enzyme kit from Sigma-Aldrich® (Missouri, USA) and the ammonia concentration was measured using the Ammonia commercial enzyme kit from R-Biopharm® (Darmstadt, Germany). Results were expressed in micromolar concentration (μM).
Statistical analyses
The statistical analysis to study the metabolic activity of the colonic microbiota was performed with the GraphPad Software® (Massachusetts, USA) version 8.4.3. This analysis included the execution of a Dunnett test for multiple comparisons (two-way ANOVA) (alpha level 0.05) using three replicates of basal microbiota samples, as well as three real replicates of each compound to assess possible significant differences. Pearson correlations were applied to assess the possible relations between metabolites and the microbial genera.Results
Response of colonic microbiota to dietary prebiotics, probiotics and synbiotics
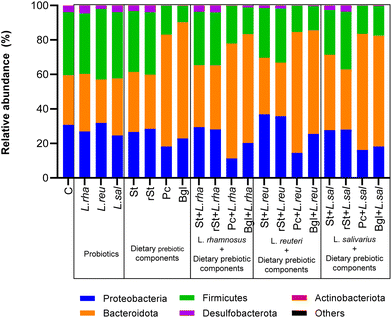
The basal microbiota of the pooled inoculum was defined by alpha diversity with Chao index of 6.8 and Shannon index of 2938. The treatments induced changes in the microbiota composition after simulated the colonic fermentation. Focusing on beta diversity (ESI Fig. 1†), the treatments that resulted in a most different diversity compared to the control, as expressed by Bray-Courtis scale, were the symbiotic combinations of pectin and beta glucan with all the prebiotics. In addition, the changes in microbiota composition were more in detail assessed by comparing the microbial abundances at different taxonomic levels.At phylum level, the basal microbiota was mainly characterised by the presence of Firmicutes (36.6% of relative abundance), followed by Proteobacteria (30.75%), Bacteroidota (28.79%), Desulfobacterota (3.67%) and Actinobacteriota (0.12%). The impact of static colonic fermentation of samples with probiotic strains, dietary prebiotic components, and their combinations on the composition of microbiota, are presented in Fig. 1. Statistically significant differences at phylum level were found between the basal microbiota versus the treatments (Table 1). The relative abundance of Firmicutes and Proteobacteria in the basal microbiota was significantly different from the most treatments to different extents and exceptions. Beta-glucan showed the highest reduction of Firmicutes (−27%), and the most reduction in Proteobacteria (−19%) was achieved by the treatment with Pectin + L. rhamnosus. Inversely, Bacteroidota was increased with pectin and beta-glucan and the synbiotics. Pectin + L. reuteri represented the highest increased of Bacteroidota (+41%).
| Phylum | Treatment | Relative abundance (%) | P value | Adjusted P value |
|---|---|---|---|---|
| Firmicutes | Basal microbiota | 36.60 | ||
| Pectin | 16.77 | <0.0001 | <0.0001 | |
| Beta-glucan | 9.44 | <0.0001 | <0.0001 | |
| Starch with L. rhamnosus | 30.91 | 0.0024 | 0.0315 | |
| Resistant starch with L. rhamnosus | 30.65 | 0.0016 | 0.0214 | |
| Pectin with L. rhamnosus | 21.88 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 15.51 | <0.0001 | <0.0001 | |
| Starch with L. reuteri | 28.68 | <0.0001 | 0.0007 | |
| Pectin with L. reuteri | 15.25 | <0.0001 | <0.0001 | |
| Beta-glucan with L. reuteri | 13.90 | <0.0001 | <0.0001 | |
| Starch with L. salivarius | 25.89 | <0.0001 | <0.0001 | |
| Pectin with L. salivarius | 16.39 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 17.17 | <0.0001 | <0.0001 | |
| Proteobacteria | Basal microbiota | 30.75 | ||
| L. salivarius | 24.60 | 0.0011 | 0.0157 | |
| Pectin | 18.23 | <0.0001 | <0.0001 | |
| Beta-glucan | 22.89 | <0.0001 | 0.0008 | |
| Pectin with L. rhamnosus | 11.33 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 20.31 | <0.0001 | <0.0001 | |
| Starch with L. reuteri | 36.87 | 0.0012 | 0.0164 | |
| Pectin with L. reuteri | 14.56 | <0.0001 | <0.0001 | |
| Pectin with L. salivarius | 16.18 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 18.18 | <0.0001 | <0.0001 | |
| Bacteroidota | Basal microbiota | 28.79 | ||
| Pectin | 64.88 | <0.0001 | <0.0001 | |
| Beta-glucan | 67.37 | <0.0001 | <0.0001 | |
| Starch with L. rhamnosus | 35.97 | 0.0002 | 0.0028 | |
| Resistant starch with L. rhamnosus | 37.18 | <0.0001 | 0.0003 | |
| Pectin with L. rhamnosus | 66.62 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 63.09 | <0.0001 | <0.0001 | |
| Pectin with L. reuteri | 70.09 | <0.0001 | <0.0001 | |
| Beta-glucan with L. reuteri | 60.07 | <0.0001 | <0.0001 | |
| Starch with L. salivarius | 43.77 | <0.0001 | <0.0001 | |
| Resistant starch with L. salivarius | 34.98 | 0.0011 | 0.0147 | |
| Pectin with L. salivarius | 67.34 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 64.25 | <0.0001 | <0.0001 |
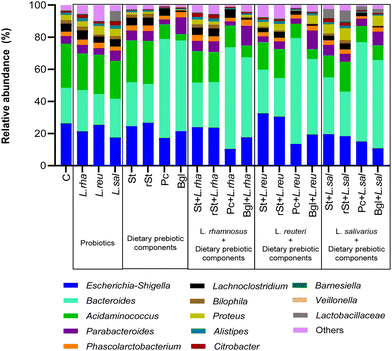
Going into the genus level, Acidaminococcus represented the highest relative abundance of the basal microbiota (27.46%), followed by Escherichia-Shigella (26.29%), Bacteroides (22.21%), Proteus (3.25%), Bilophila (2.23%), and Alistipes (1.47%) (Fig. 2). Some treatments were able to modify the proportion of the different genera of the microbiota.
Statistically significant differences at genus level between the basal microbiota versus dietary prebiotic components, probiotic strains and their combinations were found (Table 2). Some treatments changed the Acidaminococcus and Escherichia-Shigella ratio, beta-glucan alone being the one that reduced the most the relative abundance of Acidaminococcus by 23%. In the case of Escherichia-Shigella, pectin + L. rhamnosus was able to impart 15.8% decrease, followed by the beta-glucan + L. salivarius treatment (−15.5%). Inversely, the highest increase in Bacteroides (+44%) was obtained with the treatment with Pectin + L. reuteri.
| Genus | Treatment | Relative abundance (%) | P value | Adjusted P value |
|---|---|---|---|---|
| Acidaminococcus | Basal microbiota | 27.46 | ||
| Pectin | 9.13 | <0.0001 | <0.0001 | |
| Beta-glucan | 4.03 | <0.0001 | <0.0001 | |
| Starch with L. rhamnosus | 19.61 | <0.0001 | <0.0001 | |
| Resistant starch with L. rhamnosus | 18.76 | <0.0001 | <0.0001 | |
| Pectin with L. rhamnosus | 13.56 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 7.31 | <0.0001 | <0.0001 | |
| Starch with L. reuteri | 17.05 | <0.0001 | <0.0001 | |
| Resistant starch with L. reuteri | 18.09 | <0.0001 | <0.0001 | |
| Pectin with L. reuteri | 8.84 | <0.0001 | <0.0001 | |
| Beta-glucan with L. reuteri | 6.09 | <0.0001 | <0.0001 | |
| Starch with L. salivarius | 13.65 | <0.0001 | <0.0001 | |
| Resistant starch with L. salivarius | 18.62 | <0.0001 | <0.0001 | |
| Pectin with L. salivarius | 10.06 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 9.21 | <0.0001 | <0.0001 | |
| Escherichia-Shigella | Basal microbiota | 26.29 | ||
| L. rhamnosus | 21.35 | 0.0024 | 0.0319 | |
| L. salivarius | 17.50 | <0.0001 | <0.0001 | |
| Pectin | 17.32 | <0.0001 | <0.0001 | |
| Pectin with L. rhamnosus | 10.49 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 17.74 | <0.0001 | <0.0001 | |
| Starch with L. reuteri | 32.59 | 0.0001 | 0.0023 | |
| Pectin with L. reuteri | 13.47 | <0.0001 | <0.0001 | |
| Beta-glucan with L. reuteri | 19.38 | <0.0001 | 0.0006 | |
| Starch with L. salivarius | 19.51 | <0.0001 | 0.0008 | |
| Resistant starch with L. salivarius | 18.40 | <0.0001 | <0.0001 | |
| Pectin with L. salivarius | 15.05 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 10.75 | <0.0001 | <0.0001 | |
| Bacteroides | Basal microbiota | 22.21 | ||
| Starch | 27.36 | 0.0012 | 0.0169 | |
| Pectin | 61.61 | <0.0001 | <0.0001 | |
| Beta-glucan | 56.37 | <0.0001 | <0.0001 | |
| Starch with L. rhamnosus | 27.81 | 0.0006 | 0.0094 | |
| Resistant starch with L. rhamnosus | 28.34 | 0.0002 | 0.0032 | |
| Pectin with L. rhamnosus | 63.22 | <0.0001 | <0.0001 | |
| Beta-glucan with L. rhamnosus | 49.76 | <0.0001 | <0.0001 | |
| Starch with L. reuteri | 27.20 | 0.0022 | 0.0290 | |
| Pectin with L. reuteri | 65.98 | <0.0001 | <0.0001 | |
| Beta-glucan with L. reuteri | 47.04 | <0.0006 | <0.0001 | |
| Starch with L. salivarius | 35.44 | <0.0001 | <0.0001 | |
| Resistant starch with L. salivarius | 27.69 | <0.0001 | 0.0119 | |
| Pectin with L. salivarius | 61.86 | <0.0001 | <0.0001 | |
| Beta-glucan with L. salivarius | 55.01 | <0.0001 | <0.0001 |
Metabolite production during static colonic fermentation of dietary prebiotic components, probiotic strains, and their combination
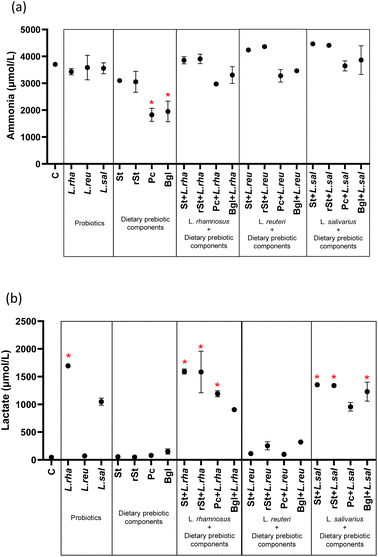
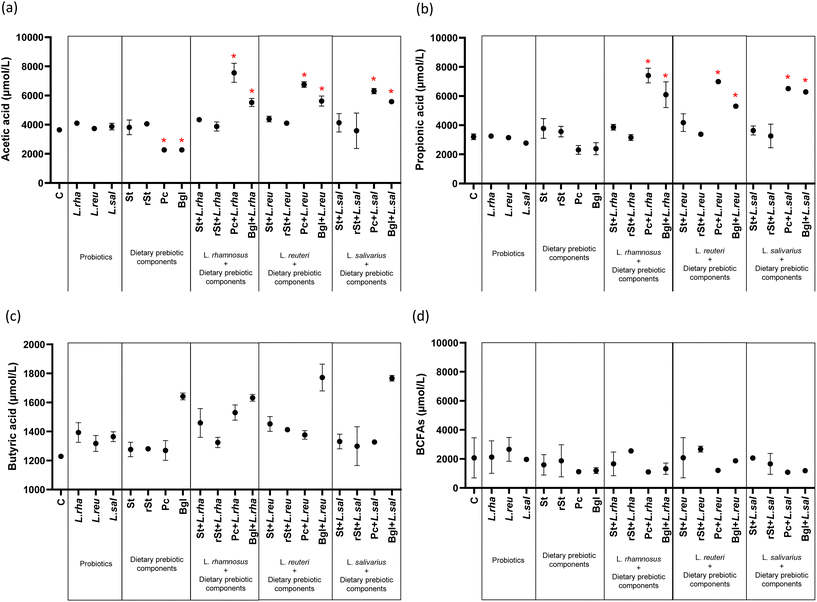
The dietary prebiotic components, probiotic strains, and their combinations influenced ammonia and lactate production (Fig. 3) (ESI Table 1†). Ammonia concentration showed a significant decrease with two dietary prebiotic components: pectin 1826.38 μmol L−1 (244.05) (adjusted p < 0.0001) and beta-glucan 1951.02 μmol L−1 (386.42) (adjusted p < 0.0001) compared to basal microbiota 3703.10 μmol L−1 (84.74) (Fig. 3a). Similarly, lactate production was significantly increased with L. rhamnosus alone and with their combinations with prebiotics. The same increased was observed with treatments combining L. salivarius with prebiotics as referred to the basal microbiota. No statistically significant differences were found with the treatments with L. reuteri (Fig. 3b).Probiotics alone, and prebiotics alone did not significantly alter the concentrations of propionic acid, butyric acid and BCFAs, with only a reduction in acetic acid obtained with pectin and beta-glucans alone (Fig. 4). Comparably, no changes occurred with the combined treatments of starch and resistant starch with probiotics, but the combinations with pectin and beta-glucan led to significant increases of acetic acid and propionic acid (Fig. 4a and b). No statistically significant differences in butyric acid and BCFAs concentrations were found with respect to the basal microbiota (Fig. 4c and d).
Correlations between genera and metabolites
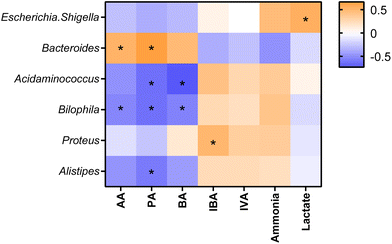
Statistically significant correlations were found between some bacterial genera and metabolite production (Fig. 5). Escherichia-Shigella was found in a positive correlation with lactate. Bacteroides showed positive correlations with AA and PA. Acidaminococcus was negatively associated with PA and BA. Bilophila showed negative associations with AA, PA, and BA, while Proteus was positively associated with IBA and, Alistipes had a negative correlation with PA.Discussion
This study evaluated the effect of 3 probiotic strains, 4 dietary prebiotic components, and their 12 combinations in terms of changes in the faecal microbiota of children with CF after colonic fermentation. The results showed that the combination of beta-glucan and pectin with any of the three probiotic strains led to the most beneficial effects both on colonic microbiota populations and metabolite production, compared to the rest of the combinations and the use of the prebiotics and probiotics alone. Complementary the assessment of beta diversity supported these observations.The first result to comment is the representativity of the pooled faecal sample (basal microbiota) of the composition and diversity of that in children with CF. Both Chao and Shannon indexes were comparable to a previous study on the in vitro simulation of colonic fermentation in CF, and lower than in the microbiota of healthy controls.19
In terms of bacterial composition, the basal microbiota was found to be reduced in Bacteroidota and increased in Firmicutes compared to previous series of healthy subjects, coinciding with the literature on altered microbiota in CF.2,15,32 Our study demonstrated that pectin and beta-glucan alone and in combination with the three probiotic strains (L. rhamnosus, L. reuteri, and L. salivarius) were effective in reducing the relative abundance of Firmicutes, which was the predominant phylum in the basal microbiota, and significantly increased Bacteroidota. The increase in Bacteroidota may be relevant as the predominance of this phylum in the gut environment would prevent from the growth and permanence of the other phyla competing for the same niche.33 In addition, species within Bacteroidota possess carbohydrate-active enzymes that degrade undigested carbohydrates into SCFA, which can be used as an additional source of energy, even for the host, after absorption in the enterocytes.34 This would be especially relevant in the case of children with CF, in which part of the dietary macronutrients are not adequately digested or absorbed, implying significant loss of energy uptake.35 On the other hand, the combinations of pectin with the three probiotic strains were able to reduce to a greater extent the Proteobacteria phylum, which is associated with pathogenic bacteria that cause intestinal inflammation.36 Therefore, these treatments could be considered effective in improving dysbiosis in the microbiota of children with CF.
With respect to the other prebiotics, beta-glucan as well as pectin alone and combined with L. reuteri significantly reduced Acidaminococcus. This opportunistic pathogen is positively associated with gastrointestinal cancer genes,37 and related to increased calprotectin levels (a marker of intestinal inflammation) in patients with CF.38 Likewise, pectin together with the three probiotic strains under study and beta-glucan with L. salivarius were significantly associated with the decrease in the relative abundance of Escherichia-Shigella; this taxon is characteristic in CF and is linked to intestinal dysbiosis.39 In turn, the combinations of pectin and the three probiotic strains, induced a significant increase in Bacteroides, which is considered a positive finding, as this genus is involved in the modulation of the immune system.40
Focusing on the production of metabolites, significant changes were found in ammonia production, which is a by-product of protein fermentation, and has been associated with negative effects on the organism, such as decreased catabolism of SCFAs and inhibition of mitochondrial oxygen consumption.41,42 Concretely, it was reduced by 50.7% and 47.3% of the concentration of the basal microbiota in the presence of pectin and beta-glucan, respectively. This suggests that the two dietary prebiotics act efficiently in the reduction of ammonia, because they stimulate carbohydrate-fermenting bacteria, which increase the colon's acidity and reduce the capacity of protein-fermenting bacteria.43 The reduction of protein-fermenting bacteria would be of special interest in the gut of children with CF, as the presence of protein in the colon is supposed to be increased as a consequence of maldigestion and malabsorption of this nutrient during the small intestine stage.35 Similar results on ammonia were reported in patients with liver cirrhosis supplemented with another prebiotic, xylooligosaccharide (XOS).44 In turn, the highest lactate production, was found in the samples treated with L. rhamnosus which coincides with a previous study of our group, in which L. rhamnosus supplementation reflected the increase in lactate after 20 days of supplementation on colonic microbiota of children with CF.18 This finding is relevant since it is known that lactate is a beneficial metabolite that exerts a positive role on the body such as regulating the biological processes of intestinal function, produces an indirect inhibition of the growth of pathogenic bacteria, and the genus Lactobacillus is related to its biosynthesis to produce propionate, butyrate, or acetate45,46
In the case of the production of acetic acid and propionic acid, the combination of pectin with L. rhamnosus allowed for a two-fold increase of the amount of these metabolites with respect to the initial content. Besides, SCFAs and the genus Bacteroides showed a positive correlation, suggesting a symbiotic effect of pectin-L. rhamnosus: the changes induced in the microbiota, such as the increase in Bacteroides, seem to modify the metabolism of pectin, resulting in higher short-chain fatty acids production. Overall, higher levels of SCFA contribute to improve the immune system, among other beneficial health effects.47–49
The relevance of the study is that new evidence on the role of different probiotics, prebiotics and their synbiotic combinations on CF gut dysbiosis has been generated, in an emerging study field where scarce or null knowledge is available.50,51 The new findings are to be interpreted with caution as the study was carried out in an in vitro setting. Besides, we acknowledge the limitation of the colonic fermentation model, which despite being adapted to the CF intestinal conditions, might not be fully representative of a CF colon. However, the results can help guiding the focus on which pre-, pro- and synbiotics could be targeted in more complex studies in the future. This aligns with the current context in which the advance in the therapies for CF have led to higher quality of life and better disease prognosis and survival. Therefore, other challenges can be addressed, such as the assessment of dietary interventions as a strategy to improve nutritional status and gut microbiota, including the supplementation with pre-, pro- and synbiotics.
In conclusion, the use of beta-glucan and pectin in combination with probiotic strains from the Lactobacillaceae family are suggested as effective approaches to revert modulate dysbiosis and metabolic activity in colonic microbiota of children with CF, future animal studies or clinical settings are encouraged to confirm the findings in vivo.
Author contributions
Jazmín Viteri-Echeverría: Methodology, validation, formal analysis, investigation, writing – original draft. Ana Andrés: Conceptualization, resources, writing – review & editing, project administration. Joaquim Calvo-Lerma: Conceptualization, writing – original draft, writing – review & editing. Ana Heredia: Writing – review & editing. Jorge García-Hernández: Writing – review & editing. Andrea Asensio-Grau: Conceptualization, resources, formal analysis, writing – review & editing, supervision, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Universitat Politècnica de València, (First Project Grant, ref. PAID-06-22). The authors also acknowledge Universitat Politècnica de València for the pre-doctoral contract given to Jazmín Viteri-Echeverría, ref. PAID-01-22, and for the post-doctoral research fostering grant awarded to Andrea Asensio-Grau, ref. PAIDPD-22. The authors also thank the families of the children with CF for their participation in the study.References
- L. R. Caley, H. White, M. C. de Goffau, R. A. Floto, J. Parkhill, B. Marsland and D. G. Peckham, Cystic Fibrosis-Related Gut Dysbiosis: A Systematic Review, Drug Delivery Syst., 2023, 68, 1797–1814 CAS.
- A. Thavamani, I. Salem, T. J. Sferra and S. Sankararaman, Impact of Altered Gut Microbiota and Its Metabolites in Cystic Fibrosis, Metabolites, 2021, 11, 1–22 CrossRef PubMed.
- Y. J. Huang and J. J. LiPuma, The Microbiome in Cystic Fibrosis, Clin. Chest Med., 2016, 37, 59–67 CrossRef CAS PubMed.
- I. McKay, J. van Dorst, T. Katz, M. Doumit, B. Prentice, L. Owens, Y. Belessis, S. Chuang, A. Jaffe, T. Thomas, M. Coffey and C. Y. Ooi, Diet and the gut-lung axis in cystic fibrosis – direct & indirect links, Gut Microbes, 2023, 15, 2156254 CrossRef PubMed.
- M. E. Sanders, D. J. Merenstein, G. Reid, G. R. Gibson and R. A. Rastall, Probiotics and prebiotics in intestinal health and disease: from biology to the clinic, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 605–616 CrossRef PubMed.
- M. K. Yadav, I. Kumari, B. Singh, K. K. Sharma and S. K. Tiwari, Probiotics, prebiotics and synbiotics: Safe options for next-generation therapeutics, Appl. Microbiol. Biotechnol., 2022, 106, 505–521 CrossRef CAS PubMed.
- G. R. Gibson, R. Hutkins, M. E. Sanders, S. L. Prescott, R. A. Reimer, S. J. Salminen, K. Scott, C. Stanton, K. S. Swanson, P. D. Cani, K. Verbeke and G. Reid, Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics, Nat. Rev. Gastroenterol. Hepatol., 2017, 14, 491–502 CrossRef PubMed.
- V. Kumar, A. K. Sinha, H. P. S. Makkar, G. de Boeck and K. Becker, Dietary Roles of Non-Starch Polysachharides in Human Nutrition: A Review, Crit. Rev. Food Sci. Nutr., 2012, 52, 899–935 CrossRef CAS PubMed.
- C. Li and Y. Hu, New definition of resistant starch types from the gut microbiota perspectives – a review, Crit. Rev. Food Sci. Nutr., 2023, 63, 6412–6422 CrossRef CAS PubMed.
- F. Gu, C. Li, B. R. Hamaker, R. G. Gilbert and X. Zhang, Fecal microbiota responses to rice RS3 are specific to amylose molecular structure, Carbohydr. Polym., 2020, 243, 116475 CrossRef CAS PubMed.
- Y. Wang, L. E. X. Leong, R. L. Keating, T. Kanno, G. C. J. Abell, F. M. Mobegi, J. M. Choo, S. L. Wesselingh, A. J. Mason, L. D. Burr and G. B. Rogers, Opportunistic bacteria confer the ability to ferment prebiotic starch in the adult cystic fibrosis gut, Gut Microbes, 2019, 10, 367–381 CrossRef CAS PubMed.
- C. F. Balthazar, J. F. Guimarães, N. M. Coutinho, T. C. Pimentel, C. S. Ranadheera, A. Santillo, M. Albenzio, A. G. Cruz and A. S. Sant'Ana, The future of functional food: Emerging technologies application on prebiotics, probiotics and postbiotics, Compr. Rev. Food Sci. Food Saf., 2022, 21, 2560–2586 CrossRef CAS PubMed.
- R. del Campo, M. Garriga, A. Pérez-Aragón, P. Guallarte, A. Lamas, L. Máiz, C. Bayón, G. Roy, R. Cantón, J. Zamora, F. Baquero and L. Suárez, Improvement of digestive health and reduction in proteobacterial populations in the gut microbiota of cystic fibrosis patients using a Lactobacillus reuteri probiotic preparation: A double blind prospective study, J. Cystic Fibrosis, 2014, 13, 716–722 CrossRef PubMed.
- S. Van Biervliet, D. Declercq and S. Somerset, Clinical effects of probiotics in cystic fibrosis patients: A systematic review, Clin. Nutr. ESPEN, 2017, 18, 37–43 CrossRef PubMed.
- J. M. van Dorst, R. Y. Tam and C. Y. Ooi, What Do We Know about the Microbiome in Cystic Fibrosis? Is There a Role for Probiotics and Prebiotics?, Nutrients, 2022, 14, 480 CrossRef CAS PubMed.
- M. J. Coffey, M. Garg, N. Homaira, A. Jaffe and C. Y. Ooi, Probiotics for people with cystic fibrosis, Cochrane Database Syst. Rev., 2020, 2020, 1 Search PubMed.
- E. Veintimilla-Gozalbo, A. Asensio-Grau, J. Calvo-Lerma, A. Heredia and A. Andrés, In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools, Appl. Sci., 2021, 11, 8135 CrossRef CAS.
- A. Asensio-Grau, J. Calvo-Lerma, M. Ferriz-Jordán, J. García-Hernández, A. Heredia and A. Andrés, Effect of Lactobacillaceae Probiotics on Colonic Microbiota and Metabolite Production in Cystic Fibrosis: A Comparative In Vitro Study, Nutrients, 2023, 15, 3846 CrossRef CAS PubMed.
- A. Asensio-Grau, A. Heredia, J. García-Hernández, R. Cabrera-Rubio, E. Masip, C. Ribes-Koninckx, M. C. Collado, A. Andrés and J. Calvo-Lerma, Effect of beta-glucan supplementation on cystic fibrosis colonic microbiota: an in vitro study, Pediatr. Res., 2023, 1–3 Search PubMed.
- J. Fu, Y. Zheng, Y. Gao and W. Xu, Dietary Fiber Intake and Gut Microbiota in Human Health, Microorganisms, 2022, 10, 2507 CrossRef CAS PubMed.
- D. Zhou, Z. Ma, J. Xu, X. Li and X. Hu, Resistant starch isolated from enzymatic, physical, and acid treated pea starch: Preparation, structural characteristics, and in vitro bile acid capacity, LWT – Food Sci. Technol., 2019, 116, 108541 CrossRef CAS.
- S. Pérez-Burillo, S. Molino, B. Navajas-Porras, Á. J. Valverde-Moya, D. Hinojosa-Nogueira, A. López-Maldonado, S. Pastoriza and J. Á. Rufián-Henares, An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality, Nat. Protoc., 2021, 16, 3186–3209 CrossRef PubMed.
- C. E. Rycroft, M. R. Jones, G. R. Gibson and R. A. Rastall, A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides, J. Appl. Microbiol., 2001, 91, 878–887 CrossRef CAS PubMed.
- A. Rubio-del-Campo, C. Alcántara, M. C. Collado, J. Rodríguez-Díaz and M. J. Yebra, Human milk and mucosa-associated disaccharides impact on cultured infant fecal microbiota, Sci. Rep., 2020, 10, 1–12 CrossRef.
- L. Humbert, D. Rainteau, N. Tuvignon, C. Wolf, P. Seksik, R. Laugier and F. Carrière, Postprandial bile acid levels in intestine and plasma reveal altered biliary circulation in chronic pancreatitis patients, J. Lipid Res., 2018, 59, 2202–2213 CrossRef CAS.
- J. T. Harries, D. P. R. Muller, J. P. K. McCollum, A. Lipson, E. Roma and A. P. Norman, Intestinal bile salts in cystic fibrosis: studies in the patient and experimental animal, Arch. Dis. Child., 1979, 54, 19–24 CrossRef CAS.
- A. Aburub, M. Fischer, M. Camilleri, J. R. Semler and H. M. Fadda, Comparison of pH and motility of the small intestine of healthy subjects and patients with symptomatic constipation using the wireless motility capsul, Int. J. Pharm., 2018, 544, 158–164 CrossRef CAS PubMed.
- S. Fehlbaum, K. Prudence, J. Kieboom, M. Heerikhuisen, T. van den Broek, F. H. J. Schuren, R. E. Steinert and D. Raederstorff, In Vitro Fermentation of Selected Prebiotics and Their Effects on the Composition and Activity of the Adult Gut Microbiota, Int. J. Mol. Sci., 2018, 10, 3097 CrossRef PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, ADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- P. J. McMurdie and S. Holmes, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8, e61217 CrossRef CAS PubMed.
- X. Zheng, Y. Qiu, W. Zhong, S. Baxter, M. Su, Q. Li, G. Xie, B. M. Ore, S. Qiao, M. D. Spencer, S. H. Zeisel, Z. Zhou, A. Zhao and W. Jia, A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids, Metabolomics, 2013, 9, 818–827 CrossRef CAS PubMed.
- D. G. Burke, F. Fouhy, M. J. Harrison, M. C. Rea, P. D. Cotter, O. O'Sullivan, C. Stanton, C. Hill, F. Shanahan, B. J. Plant and R. P. Ross, The altered gut microbiota in adults with cystic fibrosis, BMC Microbiol., 2017, 17, 1–11 CrossRef.
- K. Z. Coyte and S. Rakoff-Nahoum, Understanding Competition and Cooperation within the Mammalian Gut Microbiome, Curr. Biol., 2019, 29, R538–R544 CrossRef CAS.
- E. D. Sonnenburg, H. Zheng, P. Joglekar, S. K. Higginbottom, S. J. Firbank, D. N. Bolam and J. L. Sonnenburg, Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations, Cell, 2010, 141, 1241–1252 CrossRef CAS PubMed.
- R. Larriba, M. Roca, E. Masip, A. Cañada-Martínez, C. Ribes-Koninckx and J. Calvo-Lerma, How macronutrients and pancreatic enzyme supplements dose variability affect fat, protein and starch absorption in children with cystic fibrosis, Dig. Liver Dis., 2023, 55, 513–518 CrossRef CAS PubMed.
- M. J. Coffey, S. Nielsen, B. Wemheuer, N. O. Kaakoush, M. Garg, B. Needham, R. Pickford, A. Jaffe, T. Thomas and C. Y. Ooi, Gut Microbiota in Children With Cystic Fibrosis: A Taxonomic and Functional Dysbiosis, Sci. Rep., 2019, 9, 1–14 CrossRef.
- G. Dayama, S. Priya, D. E. Niccum, A. Khoruts and R. Blekhman, Interactions between the gut microbiome and host gene regulation in cystic fibrosis, Genome Med., 2020, 12, 1–15 CrossRef PubMed.
- R. Marsh, H. Gavillet, L. Hanson, C. Ng, M. Mitchell-Whyte, G. Major, A. R. Smyth, D. Rivett and C. van der Gast, Intestinal function and transit associate with gut microbiota dysbiosis in cystic fibrosis, J. Cystic Fibrosis, 2022, 21, 506–513 CrossRef CAS PubMed.
- A. R. Deschamp, Y. Chen, W. F. Wang, M. Rasic, J. Hatch, D. B. Sanders, S. C. Ranganathan, T. Ferkol, D. Perkins, P. Finn and S. D. Davis, The association between gut microbiome and growth in infants with cystic fibrosis, J. Cystic Fibrosis, 2023, 22, 1010–1016 CrossRef CAS PubMed.
- K. M. Antosca, D. A. Chernikova, C. E. Price, K. L. Ruoff, K. Li, M. F. Guill, N. R. Sontag, H. G. Morrison, S. Hao, M. L. Drumm, T. A. MacKenzie, D. B. Dorman, L. M. Feenan, M. A. Williams, J. Dessaint, I. H. Yuan, B. J. Aldrich, L. A. Moulton, L. Ting, A. Martinez-Del Campo, E. J. Stewart, M. R. Karagas, G. A. O’Toole and J. C. Madan, Altered stool microbiota of infants with cystic fibrosis shows a reduction in genera associated with immune programming from birth, J. Bacteriol., 2019, 201, 10 CrossRef PubMed.
- J. Zhao, X. Zhang, H. Liu, M. A. Brown and S. Qiao, Dietary Protein and Gut Microbiota Composition and Function, Curr. Protein Pept. Sci., 2018, 20, 145–154 CrossRef PubMed.
- K. Oliphant and E. Allen-Vercoe, Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health, Microbiome, 2019, 7, 1–15 CrossRef PubMed.
- J. Slavin, Fiber and Prebiotics: Mechanisms and Health Benefits, Nutrients, 2013, 5, 1417 CrossRef CAS.
- R. E. Aluko, Functional Foods and Nutraceuticals, Springer New York, New York, NY, 2012 Search PubMed.
- S. H. Duncan, P. Louis and H. J. Flint, Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product, Appl. Environ. Microbiol., 2004, 70, 5810–5817 CrossRef CAS PubMed.
- C. Huang, H. Xu, X. Zhou, M. Liu, J. Li and C. Liu, Systematic Investigations on the Metabolic and Transcriptomic Regulation of Lactate in the Human Colon Epithelial Cells, Int. J. Mol. Sci., 2022, 23, 6262 CrossRef CAS PubMed.
- D. Rios-Covian, N. Salazar, M. Gueimonde and C. G. de los Reyes-Gavilan, Shaping the metabolism of intestinal Bacteroides population through diet to improve human health, Front. Microbiol., 2017, 8, 247826 Search PubMed.
- T. J. Ashaolu, J. O. Ashaolu and S. A. O. Adeyeye, Fermentation of prebiotics by human colonic microbiota in vitro and short-chain fatty acids production: a critical review, J. Appl. Microbiol., 2021, 130, 677–687 CrossRef CAS PubMed.
- P. Markowiak-Kopeć and K. Śliżewska, The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome, Nutrients, 2020, 12, 1107 CrossRef PubMed.
- S. Esposito, I. Testa, E. Mariotti Zani, D. Cunico, L. Torelli, R. Grandinetti, V. Fainardi, G. Pisi and N. Principi, Probiotics Administration in Cystic Fibrosis: What Is the Evidence?, Nutrients, 2022, 14, 3160 CrossRef CAS PubMed.
- N. C. Williams, J. Jayaratnasingam, A. P. Prayle, S. J. Nevitt and A. R. Smyth, Prebiotics for people with cystic fibrosis, Cochrane Database Syst. Rev., 2023, 9, CD015236 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo00325j |
| This journal is © The Royal Society of Chemistry 2024 |