Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein combined with certain dietary fibers increases butyrate production in gut microbiota fermentation†
Rachel
Jackson‡
,
Tianming
Yao‡
*,
Nuseybe
Bulut
,
Thaisa M.
Cantu-Jungles
 * and
Bruce R.
Hamaker
* and
Bruce R.
Hamaker
 *
*
Whistler Center for Carbohydrate Research, Department of Food Science, Purdue University, 745 Agriculture Mall Drive, West Lafayette, IN 47907, USA. E-mail: yao132@purdue.edu; tcantuju@purdue.edu; hamakerb@purdue.edu
First published on 3rd March 2024
Abstract
The modern diet delivers nearly equal amounts of carbohydrates and protein into the colon representing an important protein increase compared to past higher fiber diets. At the same time, plant-based protein foods have become increasingly popular, and these sources of protein are generally less digestible than animal protein sources. As a result, a significant amount of protein is expected to reach the colon and be available for fermentation by gut microbiota. While studies on diet–microbiota interventions have mainly focused on carbohydrate fermentation, limited attention has been given to the role of protein or protein–fiber mixtures as fermentation substrates for the colonic microbiota. In this study, we aimed to investigate: (1) how changing the ratio of protein to fiber substrates affects the types and quantities of gut microbial metabolites and bacteria; and (2) how the specific fermentation characteristics of different types of fiber might influence the utilization of protein by gut microbes to produce beneficial short chain fatty acids. Our results revealed that protein fermentation in the gut plays a crucial role in shaping the overall composition of microbiota communities and their metabolic outputs. Surprisingly, butyrate production was maintained or increased when fiber and protein were combined, and even when pure protein samples were used as substrates. These findings suggest that indigestible protein in fiber-rich substrates may promote the production of microbial butyrate perhaps including the later stages of fermentation in the large intestine.
Introduction
In a typical western diet, nearly equal amounts of proteins and dietary fibers are delivered to the colon daily,1 where they can be metabolized by the approximately 1012 bacterial cells present.2 Bacterial fermentation of carbohydrates and proteins produces metabolites, primarily as short-chain fatty acids (SCFAs), which are beneficial to gut and whole-body health. Protein fermentation also generates branched-chain fatty acids (BCFAs) and potentially toxic substances (e.g., ammonia), which do not occur from carbohydrate fermentation. SCFAs bind to gut epithelial free fatty acid receptors (FFARs) (e.g., metabolite-sensing G protein-coupled receptors GPR41, GPR43 and GPR109A)3 and some are absorbed through passive diffusion and transported throughout the body to influence the health or disease-state of multiple body systems.4–6 Butyrate, in particular, is the energy source for colonic enterocytes and is important in reducing localized inflammation. Butyrate also causes increased expression of tight junction proteins and maintenance of gut barrier integrity.7 Through FFARs, propionate and butyrate trigger the release of enteroendocrine hormones such as peptide YY (PYY) and glucagon-like peptide-1 (GLP-1), which regulate body energy balance.8 On the other hand, protein-bound uremic retention solutes (e.g., p-cresyl and indoxyl sulfate) from protein fermentation in the large intestine are associated with renal disease and chronic kidney disease.9The two essential nutrients for anaerobic biomass development are carbon (C) and nitrogen (N), and a C/N ratio of 20–30 to 1 was considered to be ideal for anaerobic fermentation.10 Most gut bacteria favor carbohydrates (dietary fibers and endogenous carbohydrates) for energy over proteins,11 whereas an appropriate nitrogen supply is also required for microbial protein and nucleic acids synthesis. Therefore, a nitrogen limiting condition might slow down the overall growth and substrate degradation of cells due to the lack of ready-to-use amino acids, whereas a low C/N ratio likely accumulates too much protein specific fermentation metabolites, including ammonia and BCFAs. In the human gut scenario, higher protein fermentation typically occurs in the distal colon where less fermentable carbohydrates are found,12 which is associated with several disease problems due to proliferated proteolytic metabolisms. Accordingly, when fermentable dietary fibers are readily available (e.g., fiber-rich environment in the proximal gut or a slow fermenting fiber in distal colon), bacterial protein fermentation and its production of potentially toxic products remain relatively low.13,14 Historically, protein was limiting in the diet and it was likely that the gut microbiota was exposed to high-fiber diets with low levels of undigested protein, thereby providing plentiful carbohydrates. However, that has changed in modern diets that typically are low in fiber and can have high protein levels. This is particularly exemplified in the popularity of high-protein diets for athletes, the prevalence of plant-based meat analogue, and high-protein low-carbohydrate diets for weight loss, that may provide more protein than fiber for fermentation in the gut.
Numerous studies have been conducted on the effect of dietary fiber and fat intake levels on gut microbiome health and disease, but comparatively less has been reported on protein intake levels. Studies have demonstrated that the inclusion of fiber in crude protein feeds can mitigate the adverse effects caused by protein fermentation on the gut environment of pigs.15,16 However, many of them lack precise information regarding the composition of the protein-to-fiber ratio, and they often fail to consider the effects of different types of fiber on modulating the gut microbiota. Increased protein reaching the colon is associated with dysbiosis and deleterious health outcomes such as increased risk for type II diabetes,17 renal disease9 and colorectal cancer.18,19 Due to its association with colorectal cancer, research has primarily focused on decreasing gut microbial protein fermentation and increasing dietary fiber fermentation, while comparatively little attention has been aimed at dietary protein–fiber interaction effects on metabolite production.13 In a notable paper, protein fermentation was reported as a main source of beneficial SCFAs in distal colon, with approximately 30% of the protein broken down was converted to SCFAs.20 Insight into how differing amounts of protein and carbohydrates affect the quantity and type of fermentation products, both beneficial and potentially toxic, could lead to improved dietary recommendations. Furthermore, the type of fermentable fiber may also influence these differences since fibers with diverse chemical structures and complexities are influential to fermentation speed and metabolites.
The goals of this study were two-fold: (1) to determine how a changing substrate ratio of protein-to-fiber affects the type and quantity of human gut microbial metabolites produced and bacteria; and (2) to understand how different fibers’ specific fermentation characteristics affect SCFAs and bacteria along with suppression of BCFAs and ammonia production.
Materials and methods
The experimental design consisted of 4 fiber types at 3 protein inclusion levels (i.e., 25%, 50% and 75% of protein content) plus controls (a 100% protein control and a 100% fiber control for each fiber type) and a no-substrate inoculum blank, all done in triplicate. The 100% protein control contained protein only substrate. Each fiber type had its own 100% fiber control containing a fiber only substrate. The blank did not contain any substrate and received only the fecal inoculum.Protein source and protein substrate preparation
An animal-based polypeptone [Gibco Polypeptone Peptone, Product Code (PC): B11910] consisting of equal parts pancreatic digest of casein (source: bovine) and peptic digest of animal tissue (sources: bovine, equine, porcine) was purchased from Fisher Scientific (Thermo Fisher Scientific, Waltham, MA). Soy protein acid hydrolysate (PC: S1674) was purchased from MilliporeSigma (MilliporeSigma, St. Louis, MO) (made from soy protein isolate containing no fiber). These products exist in polymer and oligomer forms, characterized by a high content of small peptides and hydrolyzed polypeptides as per the provided specifications. Their structures resemble the size of colonic proteinaceous nitrogen sources (including 2–6 unit resistant peptides and fewer large size proteins) that can pass the terminal ileum and reach the colon for fermentation.21 Full proteins were not added before the simulated upper gastrointestinal (GI) digestion, because digested products would be lost at the dialysis step. Polypeptone (85 g) and 15 g of soy protein hydrolysate were combined to create the protein portion of the protein–fiber substrate mixture for in vitro fermentation. The combination of animal-based polypeptone and soy protein acid hydrolysate in a 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio represents an omnivorous dietary pattern, that is typical of the western diet.22
3 ratio represents an omnivorous dietary pattern, that is typical of the western diet.22
Dietary fiber sources and dietary fiber substrate preparation
Four different fiber types—fructooligosaccharides (FOS), wheat bran and potato starch (WBPS), pectin, and an even mixture (EM) of the three—were selected to investigate their impact on protein fermentation. The selection was based on the following rationale: FOS was chosen as a reference for a soluble fiber with a simple chemical structure, commonly used as a prebiotic (i.e., to stimulate the growth and/or activity of beneficial colonic bacteria and promote health23); WBPS represented insoluble fiber, characterized by a complex fiber matrix structure and a slower fermentation rate compared to soluble fibers; soluble pectin was selected for its more complex structure compared to FOS, bridging the structural complexity gap between FOS and WBPS. The even mixture of these three fibers mimics the consumption of multiple types found in food together, mirroring the diversity in dietary fiber intake.Short-chain FOS (PC: 111001) was obtained from Ingredion (Ingredion Incorporated, Bridgewater, NJ). Apple pectin (PC: Classic AF 710, degree of esterification: 33%, galacturonic acid content: 83%) was gifted by Herbstreith & Fox KG (Neuenbürg/Württ, Germany) and wheat bran was gifted from the Mennel Milling Company (Fostoria, OH). An unmodified raw potato starch was used as a source of type 2 resistant starch (RS2) (“Premium Quality Unmodified Potato Starch;” Bob's Red Mill Natural Foods, Inc., Milwaukee, Oregon).
Wheat bran was first defatted by hexane treatment (bran![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexane 1
hexane 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, w/v) for 60 min, followed with filtration and air-drying. The defatted wheat bran was sieved to a size range of 300–500 μm (Portable Sieve Shaker Model RX-24, W.S. Tyler Combustion Engineering Inc., Mentor, OH). Equal weights of defatted wheat bran and raw potato starch were used to make the WBPS samples which were then subjected to in vitro upper GI digestion (detailed below). The residual starch content in the wheat bran post upper GI digestion is known to be less than 2%,24 thereby considered negligible in comparison to the added potato starch.
7, w/v) for 60 min, followed with filtration and air-drying. The defatted wheat bran was sieved to a size range of 300–500 μm (Portable Sieve Shaker Model RX-24, W.S. Tyler Combustion Engineering Inc., Mentor, OH). Equal weights of defatted wheat bran and raw potato starch were used to make the WBPS samples which were then subjected to in vitro upper GI digestion (detailed below). The residual starch content in the wheat bran post upper GI digestion is known to be less than 2%,24 thereby considered negligible in comparison to the added potato starch.
The EM substrate was prepared by combining pectin, wheat bran, and potato starch and underwent in vitro upper GI digestion (described below), and then FOS was added. The final composition of the EM (by weight) for the in vitro fermentation was 1/3 FOS, 1/3 pectin, 1/6 wheat bran, and 1/6 potato starch.
Simulated upper GI digestion for substrates
The in vitro upper GI digestion procedure25,26 simulates the passage and digestion of starch and proteins through the upper GI tract (from stomach through small intestine) of humans. All fiber substrates underwent in vitro upper GI digestion, except FOS, which is known to be indigestible in the upper GI tract of humans. Fiber substrates (pectin, WBPS, and the EM before FOS addition) were subjected to the in vitro upper GI digestion procedure26 with slight modifications to prevent cooking/gelatinization of the raw potato starch. Briefly, fiber substrates (10 g) were suspended in 120 ml water and treated with 500 mg pepsin (P-7000, MilliporeSigma, St. Louis, MO) at pH 2.5 (30 min, 37 °C, 700 rpm stirring). Subsequently, 3125 mg pancreatin (P-7545, MilliporeSigma, St. Louis, MO) and 1 ml amyloglucosidase (E-AMGDF, Megazyme International, Wicklow, Ireland) were added at pH 6.9 (6 h, 37 °C, 700 rpm stirring). Following digestion, dialysis (6–8 kDa cutoff, #132675, Repligen Corporation, Waltham, MA) was performed for 36 h with 3 water changes to remove cleaved sugars and amino acids. The samples were freeze-dried and stored at 4 °C for further use.In vitro fecal fermentation
Fresh feces were collected from two donors with normal BMI (1 female and 1 male) who were consuming their routine omnivorous diets and had not taken antibiotics in the past 6-month period. Samples were promptly kept on ice in tightly sealed plastic tubes, immediately transferred into the anaerobic chamber, pooled, and used within 3 h of collection. Fecal samples were collected following the protocol #1510016635 approved by the Institutional Review Board at Purdue University.In vitro fermentation was carried out as previously described27 in an anaerobic chamber. Carbonate–phosphate buffer (pH 8.3)28 was prepared and sterilized by autoclaving. The buffer was then placed into an anaerobic chamber overnight and used within 24 h. The buffer contained nitrogen source of urea (6.6 mM) and ammonium (13.7 μM).
Substrates were weighed into sterile Balch tubes with 50 ± 0.3 mg weights (except the blank containing 0 mg substrate). Samples were prepared in triplicate for each time point (24 and 48 h for controls and protein–fiber mixtures, 0, 24, and 48 h for the blanks) with protein and dietary fiber amounts as shown in Table 1, and then placed into the anaerobic chamber overnight. The naming convention of substrates was based on the fiber type and percent fiber of the total substrate.
| Prepared protein substrate (mg) | Prepared dietary fiber substrate (mg) | Protein inclusion (of total substrate) |
|---|---|---|
| 0.00 | 0.00 | 0% |
| 50.00 | 0.00 | 100% |
| 37.50 | 12.50 | 75% |
| 25.00 | 25.00 | 50% |
| 12.50 | 37.50 | 25% |
| 0.00 | 50.00 | 0% |
The following day, 4 mL of carbonate–phosphate buffer was added to each tube. Fecal inoculum from two donors were evenly mixed by weight and homogenized with anaerobic carbonate–phosphate buffer [feces![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer 1
buffer 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (w/v)] followed by filtration through 4 layers of cheesecloth. Each Balch tube containing 4 mL buffer was then inoculated with 1 mL fecal slurry (total 1
3 (w/v)] followed by filtration through 4 layers of cheesecloth. Each Balch tube containing 4 mL buffer was then inoculated with 1 mL fecal slurry (total 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution of fecal input), closed with a sterilized butyl rubber stopper (Chemglass Life Sciences), sealed with an aluminum seal (Chemglass Life Sciences), and placed in a 37 °C shaking incubator.
20 dilution of fecal input), closed with a sterilized butyl rubber stopper (Chemglass Life Sciences), sealed with an aluminum seal (Chemglass Life Sciences), and placed in a 37 °C shaking incubator.
At each sampling point, tubes were removed from the incubator, and sample supernatants were taken for later quantification of metabolites (SCFAs, BCFAs, and ammonia). The seals and stoppers were then removed. Upon removal of the stopper, two separate 1 mL aliquots were immediately collected from each tube, one for SCFAs and BCFAs analysis, and another one for ammonia analysis, and then stored at −80 °C until further analysis. The remaining fermentation sample was measured for pH.
Determination of SCFAs and BCFAs using GC
SCFAs analysis was carried out as previously described29 with slight modifications. Briefly, 4-methylvaleric acid was used as the internal standard and mixed with culture supernatants to a final concentration of 10 mM. External standards of SCFAs and BCFAs were used for calibration curves. Prepared samples (4 μL) were injected into a gas chromatograph equipped with a fused silica capillary column (Nukon™, Supelco no.: 40369-03A, Bellefonte, PA) and a flame ionization detector (GC-FID 7890A, Agilent Technologies, Inc., Santa Clara, CA) with the following conditions: injector temperature at 230 °C; initial oven temperature at 100 °C with a ramp of 8 °C min−1 to 200 °C and a hold for 3 minutes at final temperature. Helium was used as a carrier gas at 0.75 mL min−1.BCFA production was also calculated on a substrate protein basis, considering that BCFAs are exclusively formed from protein fermentation, with the equation:
Determination of post fermenting ammonia
Ammonia was measured using the Megazyme Rapid Ammonia Assay Kit (PC: K-AMIAR; Megazyme International, Wicklow, Ireland) according to the manufacturer's instruction. Culture supernatants (100 μL) were removed to new tubes to be deproteinized before analysis. Samples were deproteinized by adding 100 μL of 1 M perchloric acid (PC: A2296, Thermo Fisher Scientific) with mixing and centrifuged at 1500g for 10 min. The resulting supernatant (100 μL) was neutralized with 50 μL 1 M potassium hydroxide. The ammonia assay was performed by first pipetting water, followed by the deproteinized sample, then the provided buffer (containing 2-oxoglutarate), and NADPH. After 2 min of mixing, absorbance at 340 nm was read (SpectraMax 190, Molecular Devices Corporation, Sunnyvale, CA). Finally, glutamate dehydrogenase (GIDH) was added to begin the reaction, and after 5 min of mixing, the second absorbance was recorded. A blank and a three-point calibration curve were performed concurrently. Ammonia (mg mL−1) was calculated using the calibration curve and reported on a millimolar basis.Microbiota analysis
Genomic DNA was extracted from 1 mL post-fermentation cultures using a FastDNA SPIN® kit for feces (#116570200, MP Biomedical, Santa Ana, CA, USA). Cell pellets were resuspended in lysis buffer and transferred into lysing matrix E tubes. The rest of the DNA extraction procedure was according to the manufacturer's instructions. The quality and concentration of extracted DNA was examined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technology, Thermo Fisher Scientific). High purity community DNA was then amplified targeting the V4 variable region of 16S rRNA gene with primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) and barcoded with Illumina TruSeq DNA library. Sequencing was performed using the Illumina MiSeq platform (2 × 150 bp) at the DNA Services Facility at the University of Illinois, Chicago.Raw sequencing files were processed via mothur v.1.48.0 following the online MiSeq SOP (https://www.mothur.org/wiki/MiSeq_SOP) and with modifications described previously.24 In brief, the pair-end reads were first merged into contigs and screened out bad contigs for those with errors in primer region, ambiguous bases, homopolymer larger than 8 bases and lengths longer than 291 bp (total length of the amplicon with primer regions). Filtered reads were then aligned to the SILVA bacteria reference database (v.132).30 Unique reads were merged from duplicated aligned reads and pre-clustered to the differences of 99.5% identity. Chimera reads were dropped using UCHIME algorithm in mothur. Taxonomy of reads were classified using the Ribosomal Database Project (RDP) classifier (version 18)31 with species epithets. The classification had a cut off bootstrap value of 95. Sequencies classified into bacteria and archaea domains were collected to a final OTU table and used for downstream analysis.
Alpha diversity (observed OTUs), Shannon evenness and Shannon index were executed using the “summary.single(![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) )” command in mothur. Beta diversity analyses were performed to investigate differences among sample groups using the Bray–Curtis distance matrix and visualized (PCoA plot) in R v.4.2.0 (R Foundation for Statistical Computing, Vienna, Austria) with the vegan package. Redundant analysis (RDA) and Spearman's correlation were computed by R v.4.2.0 with “rda(
)” command in mothur. Beta diversity analyses were performed to investigate differences among sample groups using the Bray–Curtis distance matrix and visualized (PCoA plot) in R v.4.2.0 (R Foundation for Statistical Computing, Vienna, Austria) with the vegan package. Redundant analysis (RDA) and Spearman's correlation were computed by R v.4.2.0 with “rda(![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) )” command and “corr.test(
)” command and “corr.test(![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) )” command.
)” command.
Statistical analysis
All statistical analyses were performed by two-way ANOVA using JMP® (version 13.2 SAS Institute Inc., Cary, NC, 1989–2019). When significant, Tukey's honestly significant difference (HSD) post hoc test was conducted to differentiate group means. All tests were conducted at the α = 0.05 level. Permutational multivariate ANOVA (PERMANOVA) test was conducted for beta-diversity statistics using R 4.2.2. Redundancy Analysis (RDA) was used to assess the variation in microbial composition and metabolites relative to treatments. To ensure the appropriateness of RDA for our dataset, we conducted detrended correspondence analysis (DCA), a preliminary step to estimate the suitability of ordination methods. The results from DCA, after Hellinger transformation, indicated that the maximum gradient length of the axes was within the preferred range for RDA (less than 3.0). Blanks containing inoculum, but no substrate, were not included in statistical analysis unless otherwise noted.Results
Fermentation metabolite profiles of fiber/protein treatments
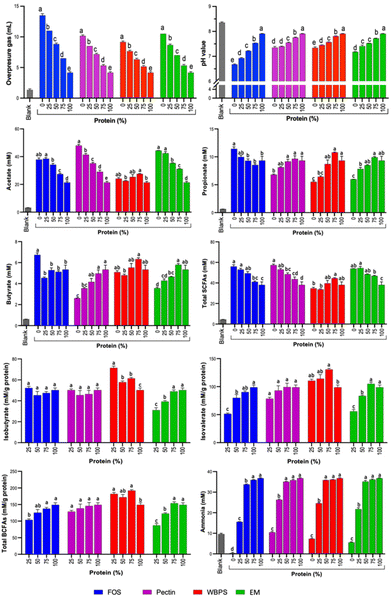
Based on our prior experience, the microbial composition and function of in vitro batch fermentation typically stabilizes within 12 to 24 h,32 depending on the fermentability of the substrates. In this study, pH variations between the 24 and 48 h fermentation demonstrated no significant differences for all substrates (Fig. 1 and ESI Fig. 1†), suggesting that the 24 h data effectively represents microbial responses reflective of substrate-specific effects. Consequently, the primary emphasis in the main results and discussions focus on the 24 h fermentation findings.Metabolites of saccharolytic and proteolytic fermentation at 24 and 48 h of in vitro fecal fermentation (SCFAs – acetate, propionate and butyrate; BCFAs – isobutyrate and isovalerate; ammonia) are shown in Fig. 1 and ESI Fig. 1.† An expected trend of a reduction in total SCFAs and an increase in BCFAs was observed for increasing proportion of protein to fiber, though this was fiber dependent as total SCFAs for WBPS did not decrease over the 24 h period. Unexpectedly, propionate and butyrate levels generally increased with protein addition, reflecting the ability of certain bacteria to ferment protein to propionate or butyrate. The response to this addition varied based on the type of fiber treatment, with FOS decreasing and the other fiber types leading to an increase in these levels. This suggests that the type of fiber can set the conditions for such a fermentation path.
After 24 h fermentation, acetate was the highest produced metabolite but ranging significantly from 21.4–52.4 mM after in vitro fecal fermentations. FOS, pectin and EM fermentations with either no protein addition or 25% protein addition led to the highest acetate levels which decreased proportionally (downward trend) with increased protein proportions up to 100% (Fig. 1). For WBPS, which presented the lowest acetate production across fiber types, no clear trend was observed at 24 h, while at 48 h WBPS with no added protein led to higher acetate production than 50–100% protein (ESI Fig. 1†). Differences in acetate production at 24 and 48 h for WBPS is likely a result of the previously reported slower fermentation profile of wheat bran and potato starch that compose WBPS.33 Moreover, at 48 hours shifts in acetate production with increased protein proportions were sharper for FOS, pectin and EM than those observed for WBPS (ESI Fig. 1†), indicating that protein addition impacts acetate production differently depending on the fiber type fermented.
In an opposite trend, propionate and butyrate production increased with increased protein content in most fiber substrates, except for FOS. 75% and 100% protein produced the highest propionate amounts for pectin, WBPS and EM fibers at both 24 and 48 h (Fig. 1 and ESI Fig. 1†). An even stronger effect was found with increased protein content and butyrate production for pectin and EM at 24 and 48 h. Contrarily, FOS and WBPS with no protein addition led to highest butyrate levels (Fig. 1 and ESI Fig. 1†). Though different in structure, both FOS and wheat bran have been shown to be butyrogenic, with the latter associated with the matrix structure of the bran fibers.34Wheat bran may also contain residual starch, which is butyrogenic, though, using the same upper GI digestion procedure in another study, WB was less than 2%. Notably, there were comparable levels of butyrate production across all protein–fiber mixtures using FOS and WBPS, which were indistinguishable from the 100% protein control (Fig. 1 and ESI Fig. 1†).
Even though butyrate and propionate production were generally better favored with higher protein content in some fiber types, lower protein content was associated with higher total SCFAs production for all fiber types, mirroring the observation for the major SCFAs produced, acetate (Fig. 1 and ESI Fig. 1†). For all SCFAs, a significant interaction effect of fiber type, fiber inclusion level, and fermentation time on cumulative response was found (ESI Table 1†), indicating that responses were specific to the fiber type tested and its level of inclusion in the protein–fiber mixtures.
Regarding total BCFAs production, production levels increased in samples with higher protein content as expected, and the highest BCFAs were observed for 100% protein treatments (7.4 and 8.1 mM, for 24 and 48 h fermentation, respectively, ESI Fig. 1†). To better understand if the presence of fiber substrates could inhibit BCFAs formation from proteolytic fermentation, amount (mM) of total BCFAs per gram of protein was also evaluated (Fig. 1). FOS and EM were the only fiber sources to significantly inhibit BCFAs production during fermentation (Fig. 1). In the first 24 hours of fermentation, FOS (with 25% protein) and EM (with 25% and 50% protein) had significantly less total BCFAs (mM per g protein basis) than the 100% protein control (Fig. 1). FOS is a fast-fermenting fiber that was likely consumed completely within the first 8 h of fermentation. Yet, FOS had a continued suppression effect of BCFAs (mM per g protein basis) throughout the 48-hour fermentation period, likely due to significantly lowered pH compared to other protein–fiber mixture samples (ESI Fig. 1†). Interestingly, BCFAs (mM per g protein basis) at 48 h were significantly lower than at 24 h for FOS (with 25% protein), a result that did not occur for other protein–fiber mixture samples (Fig. 1). The most likely explanation is a microbial utilization of BCFAs for growth in the second 24 h period. Oddly, 24 h WBPS (with 25% protein) fermentation had a significantly higher total BCFAs concentration (mM per g protein basis) than the 100% protein control (Fig. 1), indicating an increased rate of protein metabolism at the early half of fermentation since the 48 h values were similar to that of the 100% protein control (Fig. 1). Effect of fiber suppression on isovalerate and isobutyrate was similar to that of total BCFAs. Suppression of isovalerate production was observed with higher concentrations of FOS and EM for both 24 and 48 h, and with pectin at 48 h (Fig. 1 and ESI Fig. 1†). For suppression of isobutyrate, FOS was effective at 48 h and the EM at 24 h (Fig. 1 and ESI Fig. 1†). For all BCFAs, a significant interaction effect of fiber inclusion level on cumulative response was found (ESI Table 1†).
Ammonia results are shown in Fig. 1. Even though ammonia is primarily a product of protein fermentation, there was considerable variability in amount among the pure fibers, with FOS being the fiber that led to the lowest ammonia levels at 24 and 48 h across all samples, and was much lower than the blank level. Drops in pH from FOS fermentation likely inhibited proteolysis of proteins found in the fecal matter of the blank causing reduced ammonia production observed, as has been previously reported.14 Moreover, with the growth of saccharolytic organisms, ammonia can be further recycled as simple nitrogen input reducing its levels compared to blank samples. Notably, FOS was the only fiber that significantly reduced ammonia concentration at the 50% protein level at 24 h, though the decrease was relatively small compared to the steep decline for all protein–fiber mixture samples at 25% protein inclusion. All fibers led to a decline in absolute ammonia production at a low protein inclusion level (25%) (Fig. 1). The reduction of ammonia concentration compared to the blank sample advances that pure fiber substrates, regardless of type and fermenting rate, reduced undesired protein metabolism of fecal bacteria. Such effect can be attributed to (1) pH reduction during saccharolytic fermentation since pH is critical to protease activity, and (2) ammonia can be further recycled as a nitrogen source for bacterial growth. However, when examining the normalized ammonia production per gram of protein substrate (ESI Fig. 1E†), a nearly opposite trend was observed for samples, except FOS. With higher protein content in the substrates, there was a reduction in protein's contribution to ammonia production, potentially attributed to differences in microbial fermenting biomass. Higher fiber content might support more fermenting bacterial cells, consequently resulting in accumulated metabolism for ammonia production. However, FOS exhibited a distinctive behavior – even at a low protein inclusion level of 25%, it demonstrated suppressed normalized ammonia production. Overall, low fiber inclusion in the fermentation substrate did not greatly reduce absolute ammonia response until fiber comprised over half the substrate. Thus, it is likely that the reduction of protein and not the presence of fiber had the biggest influence on reducing ammonia levels, since significant decreases in ammonia concentration generally required protein to be reduced by 75%.
Shift of microbiota composition – protein content versus fiber type
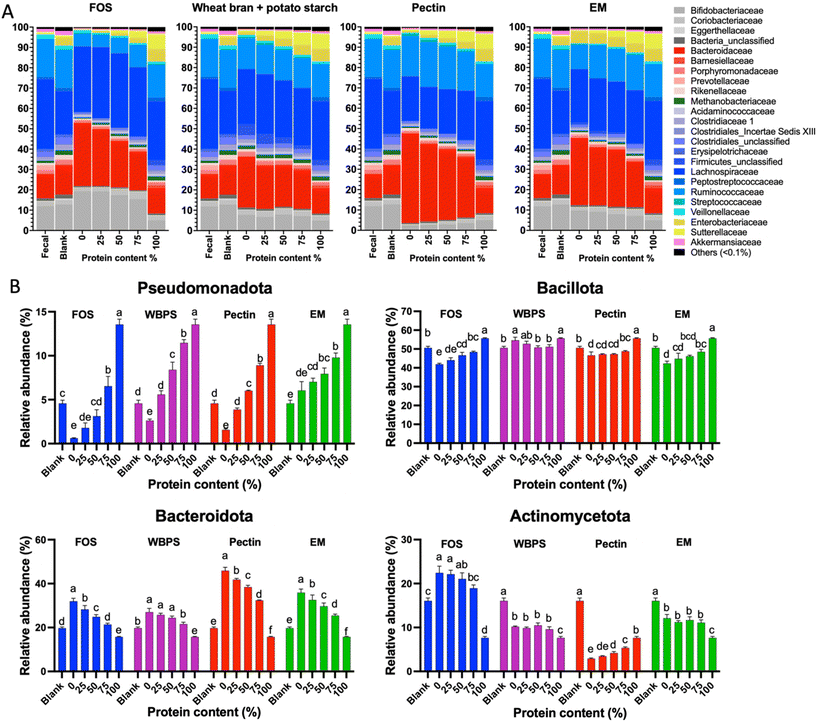
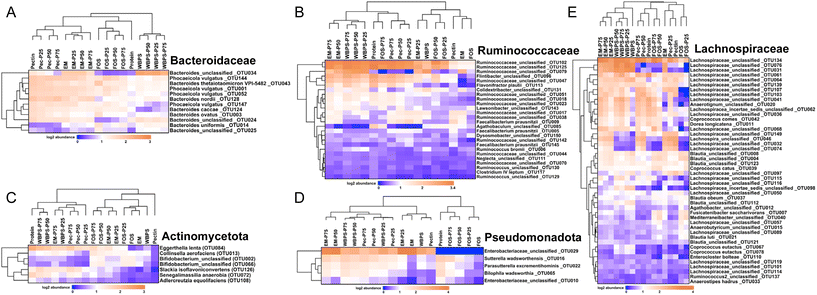
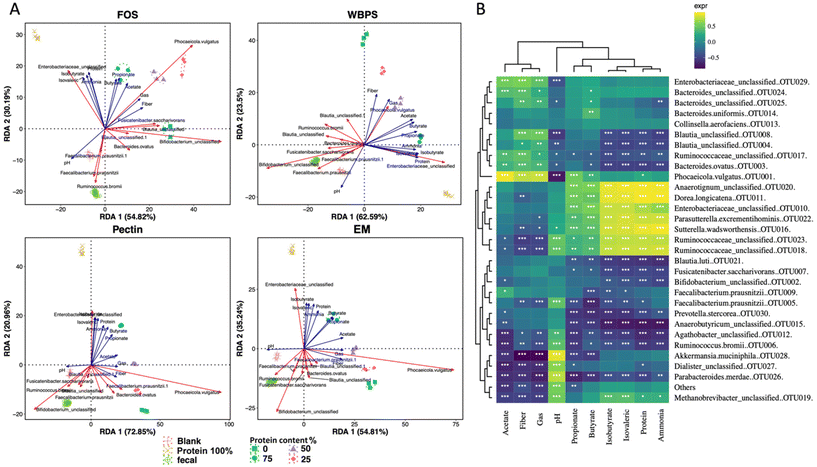
To determine the influence of protein content on fecal fermentation over diverse fiber substrates, the microbial community compositions after 24 h fermentation were measured. Family level bar plots are displayed in Fig. 2 and OTU level heatmaps are shown in Fig. 3. Pseudomonadota, Bacillota, Bacteroidota, and Actinomycetota were prevalent in the fermenting community, accounting for more than 95% of total read counts, and substantial alterations in these key phyla were observed. Fig. 3B shows a significant increasing linear trend of relative abundance of phylum Pseudomonadota, which contains some pro-inflammatory species, along with the proliferation of protein levels regardless of fiber type, demonstrating that these microbes were active responders to high protein content in fecal fermentation. Two unclassified Enterobacteriaceae spp. (OTU10 and 29) were presented in this phylum, although they behaved divergently to different fermentation conditions (Fig. 3D). OTU29 demonstrated a fiber substrate-governed shift, with significant high abundance in the EM group followed by WBPS, pectin, and no count on FOS, whereas OTU10 demonstrated a protein-governed shift, showing that sensitivity to protein or fiber substrates was down to the OTU level and differed between species.A similar trend, albeit on a smaller scale, was reported for species in the phylum Bacillota, with increased relative abundance when high protein levels were provided, except for WBPS samples (Fig. 2). Family Ruminococcaceae and Lachnospiraceae are two major groups in Bacillota and they showed different responses. Ruminococcaceae spp. displayed a robust reaction to protein level fluctuations with a distinct cluster separation (i.e., low protein samples grouped on the right clade and high protein samples on the left), which was contributed by several unclassified Ruminococcaceae OTUs (OTU102, OTU125, OTU47, OTU51, OTU18, OTU23) and organisms in Flintibacter (OTU96) and Flavonifra (OTU113) (Fig. 3B). Unlike Ruminococcaceae, there was no apparent trend with increased protein content variations in Lachnospiraceae spp., although changes in both fiber and protein impacts were seen (Fig. 3E). All WBPS samples were visibly clustered together, which was contributed by unclassified Lachnospiraceae OTUs (OTU97, OTU98, OTU76, OTU31), Blautia spp. (OTU4, OTU8, OTU123, OTU112) and Coprococcus eutactus (OTU67, OTU78), suggesting that these saccharolytic organisms preferred WBPS fiber regardless of protein levels. The increased relative abundance of these OTUs explain the high Bacillota in WBPS fermentation (Fig. 2B). Although 18 of the top 30 OTUs in the Lachnospiraceae family are likely fiber consumers (i.e., increased abundance in zero or low protein samples, such as Blautia spp.), there were also a large member size (12 OTUs) that reacted to high protein levels.
Bacteria of the phylum Bacteroidota, on the other hand, exhibited a strong fiber-favoring tendency (Fig. 2B). When the protein content of fermenting substrates was increased, the relative abundance of Bacteroidota spp. fell considerably. The family Bacteroidaceae dominated this phylum (>90% abundance), and they were well grouped by fiber type (Fig. 3A). For example, OTU34 and OTU24 showed a FOS-specific reduction but increased in abundance on WBPS, whereas OTU124 and OTU147 were increased by FOS, pectin and EM but not by WBPS. Such fiber specific response indicates that Bacteroidaceae spp. were more sensitive to fiber structures and less affected by protein contents. In general, the Bacteroidota phylum contains well-known saccharolytic bacteria, such as genera Bacteroides and Prevotella, which have a good ability to degrade complicated polysaccharides for energy.
In the Actinomycetota spp., the organisms’ response to protein level was trivial. The mixed fiber substrates, WBPS and EM, did not reveal significant abundance differences with protein levels ranging from 0% to 75%. In contrast, there were inconsistent but substantial changes in relative abundances for the pure fibers, FOS, and pectin. Abundance of Actinomycetota organisms in FOS samples decreased with protein content higher that 50%, but pectin samples showed an increasing abundance with high protein contents. The clustering pattern of key OTUs in Actinomycetota phylum suggests that there was a stronger influence by protein levels than fiber types (Fig. 3C). Four fiber substrates (with 0% protein) were well grouped at a right-side clade, while high protein samples were gathered at the left-side clade along with the 100% protein sample. Such clustering patterns were largely driven by Eggerthella lenta (OTU84), Slackia isoflavoniconvertens (OTU126), and Senegalimassilia anaerobia (OTU72). The genus Bifidobacterium was dominant in the phylum (>62%), and two major Bifidobacterium spp. (OTU2 and OTU66) showed reduced abundance in protein-containing samples and divergent abundance changes among fiber types, suggesting that Bifidobacteria are more fiber sensitive and less influenced by protein levels.
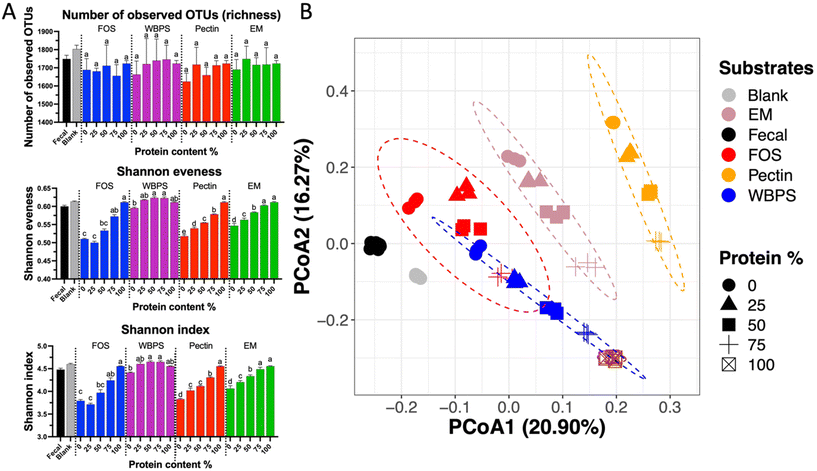
Ecological statistics indicate a significant protein influence on alpha diversity scores (Fig. 4A). Although neither protein quantity nor fiber type had a significant effect on richness, fiber fermentation decreased evenness scores relative to the blank, while adding proteins maintained a better community evenness. Reduction of evenness on high fiber substrates can be linked to the strong fiber-specific selection for organisms originally high in the fecal inoculum (e.g., Bacteroidaceae family and Bifidobacterium genus), whereas high protein content promoted initially low abundance groups, such as Enterobacteriaceae spp., that balanced relative abundance differences among taxonomic groups, leading to a higher evenness score. The beta-diversity plot displayed an obvious linear change on community structures, following a straight pattern from low protein groups (right-top) merging to high protein samples (left-bottom corner) (Fig. 4B), however such pattern was also nested within each fiber types. Comparing different fiber types, FOS and WBPS samples overlapped with each other, and they had a closer distance to the initial fecal inoculum. This proximity could potentially be attributed to the low specificity nature of both FOS and RS. As we have previously demonstrated, this trait is linked to a reduced ability to modify the community structure compared to more specific fibers to certain bacteria response.35 EM and pectin samples were well separated and far from the initial point, meaning that the different fiber types had sizable selection effect on the community structures.
Taken together, these results show that (1) protein level has substantial impact on the in vitro fecal fermentation model, which specifically influenced microbes in the phyla of Pseudomonadota and Bacillota, and (2) fiber substrates, even in low amount, promote the community selection for certain saccharolytic organisms, mainly Bacteroidaceae spp. Consequently, a suitable combination of protein and fiber content may yield a desired microbial community shift with optimized beneficial SCFAs metabolite outcomes. Beyond the fiber–protein ratio, a right fiber type also seems able to regulate microbial selection and potentially reduce hazardous proteolytic metabolites, which will be covered in the next section.
High propionate and butyrate induced by high protein-to-fiber ratios
To understand which fiber–protein combination ratio produced the best fermentation outcomes, we conducted correlation statistics including redundant analysis (RDA) and the Spearman's coefficient test (Fig. 5). The two-dimensional RDA plots on four fiber types covered good axis explanation on group variations (85.01% for FOS; 86.09% for WBPS; 93.81% for pectin and 90.05% for EM). The orientation of key metabolites showed a similar pattern across various fibers. Two branched-chain fatty acids (isobutyrate and isovalerate) and ammonia were associated with 100% protein samples, whereas beneficial SCFAs (butyrate and propionate) unexpectedly were associated with 75%, or the middle of 50–75%, protein samples, implying that a higher protein-to-fiber ratio substrate led to higher propionate and butyrate production than high fiber substrates. Acetate corresponded to 50% protein-mixed substrates, suggesting higher fiber content stimulates acetate anabolism and acid accumulation. An Enterobacteriaceae sp. (OTU10) was consistently associated with 100% protein fermentation with correlation to hazardous BCFAs and ammonia productions. Phocaeicola vulgatus (OTU1), a potential propionate producer, was highly correlated to protein substrates (50–75% protein), indicating this strain may benefit from the resource niche of mixed fiber and protein substrate. Some dominant potential butyrate producers, such as Faecalibacterium and Blautia spp., were correlated to initial inoculum and high fiber communities.Spearman correlation analysis provided statistical evidence (p < 0.05) for the possible involvement of major OTUs in metabolite production (Fig. 5B). One Enterobacteriaceae sp. (OTU29), one Ruminococcaceae (OTU17), several Bacteroidaceae OTUs (OTU24, OTU25, OTU3 and OTU1), and Blautia spp. (OTU8, OTU4) were positively related to high fiber content fermentation, which was clustered with high acetate, matching with the findings in RDA and community analyses. In contrast, different strains in Enterobacteriaceae (OTU10), Ruminococcaceae (OTU18, OTU23) along with Sutterella wadsworthensis (OTU16), Parasutterella excrementihominis (OTU22), Dorea longicatena (OTU11) and Anearotigum sp. (OTU20) were significantly correlated with high protein fermentation, which were in the same clade with ammonia production. These organisms were positively correlated with propionate and butyrate as well. The inconsistent beneficial behavior of organisms in Enterobacteriaceae and Ruminococcaceae families indicates that the protein-favoring function is species-dependent and requires thorough case-by-case examination in future protein fermentation studies.
Discussion
The purpose of this study was to elucidate how introduction of protein to dietary fiber at different substrate ratios affects in vitro gut microbial fermentation and outcome measures of metabolites (specifically as BCFAs and ammonia) and microbiota response, as well as to examine the degree to which fiber type may influence these outcomes. Three model fiber types and their mixtures were evaluated on three fiber–protein mixture levels. Fiber choices were based on differences in solubility and complexity of structure as mentioned in Materials and methods. Also, FOS and pectin have been shown to ferment rapidly and to the same extent,36 compared to wheat bran and resistant starch which ferment more slowly and to a somewhat lesser extent.24,37The outcomes of protein fermentation become more significant with higher protein to dietary fiber ratio, as observed in dietary intake in modern times.38 Moreover, with efforts being made to promote plant-based protein consumption over animal proteins, and given that the former is less digestible,39 the implications of intestinal protein fermentation are becoming even more relevant. Residual dietary proteins in the colon are digested by proteolytic bacteria and subsequently used as building blocks for microbial protein synthesis or in proteolytic fermentation for energy. Fermentation of dietary protein creates bioactive compounds of negative consequence that have been demonstrated to increase host inflammatory response, gut barrier permeability, and colitis severity in the gut. However, the outcome of protein fermentation in the gut is determined by both luminal ecology and the availability of various types of microbial nutrients. A lower amount of dietary fiber may result in higher proteolytic fermentation due to the restricted carbon supply for saccharolytic fermentation. Hence, our original thinking was aimed at incorporating dietary fibers to suppress and/or counterbalance the potentially negative outcomes of protein fermentation. We expected that protein fermentation would be reduced when fermentable dietary fiber was present, and the outcome would be dependent on the type and amount of fiber in the combination.
The addition of protein at different levels of fiber revealed several unexpected outcomes and suggests that the presence of protein as a substrate has a significant effect on gut microbial community structures. Changes in microbiota structure responded roughly linearly to increases in protein concentration in substrate mixes, with phyla Pseudomonadota and Bacillota increasing and phyla Bacteroidota and Actinomycetota decreasing (except for pectin samples). Such microbial community modulation is consistent with several observations in literature. Similarly, Holmes et al. (2017) found that species in phylum Bacteroidota were adversely affected by dietary protein intake in a mice model,40 whereas most organisms in phylum Bacillota responded positively to diets of complex carbohydrates. Bacteria in phylum Pseudomonadota are considered as good protein fermenters (such as Escherichia coli). In this phylum, Ben-Harb et al. (2019) found only eight of twenty examined E. coli isolates were able to grow on a specific pea protein,41 suggesting that responding strains could also be protein source dependent. Our fecal fermentation experiment provides evidence for a strong effect between protein levels in fermenting substrates and phylum-level gut community shifts.
Further, our results show that fermentation metabolites of protein–fiber mixtures are not simply explained by the suppression of protein fermenting organisms by carbohydrate inclusion. The choice of fiber type also largely influences fermentation outcomes. In protein–fiber substrate mixtures, total BCFAs (mM per g protein basis), which were used as a measure of protein fermentation, apparently increased proportionally with protein levels across all fiber types (Fig. 1). Indeed, only two of the tested dietary fibers (FOS and EM) inhibited protein fermentation, and the magnitude of the suppression effect was relatively small, requiring dietary fiber to make up at least half of the protein–fiber mixture to have a significant impact. The inhibition of protein fermentation was largely associated with the reduction of OTU10 – an Enterobacteriaceae species that was strongly suppressed only under the FOS and EM samples (Fig. 3D), while other high protein promoted organisms (e.g., OTU20, OTU11, OTU22, etc.) did not follow this pattern. The relative abundance of OTU10 is much higher in 25% and 50% of WBPS and pectin samples and was strongly correlated with higher BCFAs, suggesting that overall proteolytic fermentation can be influenced by a single species. Clearly, fiber types can inhibit the main protein fermenting organism in different ways. The biggest impediment to microbial protein fermentation was likely pH that is controlled by the speed of fermentation and metabolite production. This is seen in the comparison of protein-FOS samples to those of protein-WBPS samples. When FOS was included, pH and total BCFAs dropped significantly, whereas they remained relatively high in WBPS samples. In general, these findings suggest that (1) FOS, as a rapid fermenting fiber, has the strongest effect against proteolytic organisms with low BCFAs even at a low mixture ratio, (2) the EM fiber, despite containing both FOS and WBPS, retained a strong proteolytic suppression effect that was likely contributed by the FOS component, and (3) the pH drop during the rapid fiber fermentation of FOS was likely to be the main reason driving the fermentation preference towards saccharolytic rather than proteolytic fermentation, since typical bacterial proteolytic reactions occurs at the neutral pH.
The most intriguing finding in this study was that proteolytic fermentation can be butyrogenic and propiogenic, which is an overlooked outcome in gut bacterial fermentation research. The protein control sample, on its own, produced a large amount of propionate and butyrate, equal to or greater than the 50–100% WBPS and pectin samples, but not for FOS (Fig. 1). Notably, Wang et al.13 showed addition of protein to a mixture of FOS and inulin increased acetate (omnivores and vegetarians) and propionate (omnivores only), but not butyrate. Our study supports previous findings in which indigestible proteins increased butyrate in vitro,42 and high-protein diets in rats and pigs increased cecal butyrate concentrations.43,44 In contrast, several human studies have found high protein consumption to decrease fecal butyrate concentrations.45–47 The in vivo luminal SCFAs level is susceptible to the dynamics between microbial production and host absorption, making it difficult to determine how much protein fermentation in situ may account for the beneficial propionate and butyrate generation. A review of microbial physiological metabolic pathways provides insight to this question. In terms of microbial butyrate synthesis, in addition to the commonly known acetyl-CoA route that begins with saccharolytic fermentation, there are alternative catabolic pathways to produce butyrate using amino acids (i.e., arginine, glutamic acid and lysine) from protein hydrolysates (ESI Fig. 2A†).48 The glutamate and lysine pathways yield a significant amount of ammonia as a by-product in the process and such butyrogenesis has been found in some potential pathogenic gut bacteria, such as Fusobacterium. Other common pathogenic genera, such as Enterococcus, Streptococcus, and Escherichia, have not been found to produce butyrate. Similarly, microbial propionate biosynthesis can be achieved through amino acid catabolism, but with different input amino acids (i.e., threonine, methionine, isoleucine and valine) (ESI Fig. 2B†).49 Propiogenesis from amino acids, however, results in lower cell energy and cell growth, therefore this mechanism is likely activated only when environmental carbon sources are restricted, such as condition of growing on a slow-fermenting fiber, and in the later stage of fermentation in the distal colon. Our SCFAs results indicate that such amino acid-based biosynthetic pathways may be involved in the higher protein (50–100%) treatments, which were responsible for higher propionate and butyrate productions, except for the FOS samples. FOS, as a fast-fermenting fiber, may block such proteolytic-based pathways, resulting in SCFAs production patterns that are mainly driven by carbohydrate fermentation. Further in vivo study of optimized dietary fiber–protein ratios and fiber types for butyrate and propionate production may be warranted, going along with the idea that currently common high plant protein-based diets have more dietary protein entering the large intestine as a fermentation substrate to drive this response.
Additionally, increasing butyrate and other SCFAs levels in the distal colon is a target in the prevention or treatment of colon inflammation-related disorders that occurred more often at the colorectal site (e.g., ulcerative colitis).50 Our data suggests potential use of slow-fermenting fiber and low digestibility protein mixtures to promote butyrate-producing bacteria in the distal colon, despite a concern of concurrent ammonia production from protein fermentation.51 Ammonia is known to decrease butyrate uptake by colonocytes, decrease colonocyte oxidation of butyrate, and have the overall effect of disrupting epithelial cell integrity and barrier function.48,52–54 Our data shows significant ammonia inhibition by applying all tested fibers at a low protein–fiber ratio (25%) but not at higher protein inclusions, except for FOS which reduced ammonia at 50% ratio. Modulation of ammonia production was shown to be dependent on both fiber types and protein content, affecting the fermenting pH and redox balances that regulates its production.
As a proof-of-concept study, several limitations merit consideration for future investigations before applying this strategy as a possible diet regulation treatment. Single batch in vitro fermentation systems do not account for dynamic microbial ecologies that exist within the gastrointestinal tract that might be impacted differently by fast versus slow fermenting fibers. Alternatively, a multi-chamber, continuous fermenting or in vivo system could be used. Further, to draw valid conclusions about how fiber–protein mixtures impact colon health, in vivo animal model studies would be required to replicate the complexities of the human gut environment. Additionally, our use of commercial hydrolyzed casein, animal proteins, and soy protein to represent the resistant proteins might not fully mirror the actual residual proteins and peptides reaching the human colon. This is relevant since diverse protease families in the digestive tract have specific cleavage sites for protein substrates, potentially resulting in varied distributions of digested peptides. Understanding the digestion patterns and the amount of these peptides, as well as undigested proteins, reaching the colon would require a future investigation of protein digestion in the upper GI tract. Also, we chose a two fecal donor pooled design for our in vitro fermentation study, while a three-donor pooled sample or multiple individual donors could also be warranted.
Conclusions
To sum up, our study showed protein fermentation in the gut is an important factor in overall shifts of microbiota communities (at the phylum level) and resulted in unexpected outcomes of high propionate and butyrate production of fiber–protein mixtures and pure protein samples. The inclusion of protein in fiber substrates is potentially an approach to achieve increased microbial butyrate production, perhaps in the distal large intestine with slowly fermenting fibers. In addition, both fiber type and protein inclusion levels are important to regulate production of BCFAs, ammonia, and SCFAs, and the suppression of potential hazardous organisms (e.g., OTU10) in the community.Whereas deleterious effects of protein fermentation have been suggested,51,55 there is a lack of understanding regarding potential beneficial outcomes of colonic protein metabolism, and conceivably different outcomes could be obtained depending on protein source45 and combined dietary fiber types. Further studies on protein–fiber fermentation outcomes could include: (1) optimal fiber characteristics (e.g., fermentation rate, structural complexity) to reduce key species responsible for BCFAs and ammonia production, and (2) the impact of different dietary proteins (e.g., those with abundance of specific amino acids involved in butyrate and propionate pathways) on sole proteolytic or combined saccharolytic–proteolytic fermentation.
Data availability
The 16S amplicon sequences of fermented fecal microbiota have been deposited to the National Center for Biotechnology Information Sequence Read Archive (NCBI; SRA), with BioProject PRJNA985058 and BioSamples under accession numbers from SAMN35786910 to SAMN35786984.Conflicts of interest
The authors report there are no competing interests to declare.Acknowledgements
This research was supported by Whistler Center for Carbohydrate Research.References
- A. Chacko and J. H. Cummings, Gut, 1988, 29, 809–815 CrossRef CAS PubMed.
- R. Sender, S. Fuchs and R. Milo, PLoS Biol., 2016, 14, e1002533 CrossRef PubMed.
- S. Sivaprakasam, P. D. Prasad and N. Singh, Pharmacol. Ther., 2016, 164, 144–151 CrossRef CAS PubMed.
- N. I. McNeil, J. H. Cummings and W. P. James, Gut, 1978, 19, 819–822 CrossRef CAS PubMed.
- A. Tripathi, J. Debelius, D. A. Brenner, M. Karin, R. Loomba, B. Schnabl and R. Knight, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 397–411 CrossRef CAS PubMed.
- J. F. Cryan, K. J. O'Riordan, C. S. M. Cowan, K. V. Sandhu, T. F. S. Bastiaanssen, M. Boehme, M. G. Codagnone, S. Cussotto, C. Fulling, A. V. Golubeva, K. E. Guzzetta, M. Jaggar, C. M. Long-Smith, J. M. Lyte, J. A. Martin, A. Molinero-Perez, G. Moloney, E. Morelli, E. Morillas, R. O'Connor, J. S. Cruz-Pereira, V. L. Peterson, K. Rea, N. L. Ritz, E. Sherwin, S. Spichak, E. M. Teichman, M. van de Wouw, A. P. Ventura-Silva, S. E. Wallace-Fitzsimons, N. Hyland, G. Clarke and T. G. Dinan, Physiol. Rev., 2019, 99, 1877–2013 CrossRef CAS PubMed.
- D. Pérez-Reytor, C. Puebla, E. Karahanian and K. García, Front. Physiol., 2021, 12, 650313 CrossRef PubMed.
- C. B. Christiansen, M. B. N. Gabe, B. Svendsen, L. O. Dragsted, M. M. Rosenkilde and J. J. Holst, Am. J. Physiol.: Gastrointest. Liver Physiol., 2018, 315, G53–G65 CrossRef CAS PubMed.
- B. K. I. Meijers and P. Evenepoel, Nephrol., Dial., Transplant., 2011, 26, 759–761 CrossRef CAS PubMed.
- L. Lin, F. Xu, X. Ge and Y. Li, in Advances in Bioenergy, ed. Y. Li and X. Ge, Elsevier, 2019, vol. 4, pp. 121–181 Search PubMed.
- G. T. Macfarlane, G. R. Gibson and J. H. Cummings, J. Appl. Bacteriol., 1992, 72, 57–64 CrossRef CAS PubMed.
- W. Tottey, D. Feria-Gervasio, N. Gaci, B. Laillet, E. Pujos, J.-F. Martin, J.-L. Sebedio, B. Sion, J.-F. Jarrige, M. Alric and J.-F. Brugère, J. Neurogastroenterol. Motil., 2017, 23, 124–134 CrossRef PubMed.
- X. Wang, G. R. Gibson, A. Costabile, M. Sailer, S. Theis and R. A. Rastall, Appl. Environ. Microbiol., 2019, 85, e02749-18 CrossRef PubMed.
- E. A. Smith and G. T. Macfarlane, FEMS Microbiol. Ecol., 1998, 25, 355–368 CrossRef CAS.
- R. Jha, J. Bindelle, A. Van Kessel and P. Leterme, Anim. Feed Sci. Technol., 2011, 165, 191–200 CrossRef CAS.
- R. Jha and J. F. Berrocoso, Anim. Feed Sci. Technol., 2016, 212, 18–26 CrossRef CAS.
- I. Sluijs, J. W. J. Beulens, D. L. van der A, A. M. W. Spijkerman, D. E. Grobbee and Y. T. van der Schouw, Diabetes Care, 2010, 33, 43–48 CrossRef CAS PubMed.
- B. Armstrong and R. Doll, Int. J. Cancer, 1975, 15, 617–631 CrossRef CAS PubMed.
- W. Scheppach, S. Bingham, M. C. Boutron-Ruault, M. Gerhardsson de Verdier, V. Moreno, F. M. Nagengast, R. Reifen, E. Riboli, H. K. Seitz, J. Wahrendorf and H. Kasper, Eur. J. Cancer Prev., 1999, 8, 57–62 CrossRef CAS PubMed.
- G. T. Macfarlane, G. R. Gibson, E. Beatty and J. H. Cummings, FEMS Microbiol. Ecol., 1992, 10, 81–88 CrossRef.
- Y. C. Chung, Y. S. Kim, A. Shadchehr, A. Garrido, I. L. Macgregor and M. H. Sleisenger, Gastroenterology, 1979, 76, 1415–1421 CrossRef CAS.
- V. J. Clemente-Suárez, A. I. Beltrán-Velasco, L. Redondo-Flórez, A. Martín-Rodríguez and J. F. Tornero-Aguilera, Nutrients, 2023, 15, 2749 CrossRef PubMed.
- G. R. Gibson, R. Hutkins, M. E. Sanders, S. L. Prescott, R. A. Reimer, S. J. Salminen, K. Scott, C. Stanton, K. S. Swanson, P. D. Cani, K. Verbeke and G. Reid, Nat. Rev. Gastroenterol. Hepatol., 2017, 14, 491–502 CrossRef PubMed.
- Y. E. Tuncil, R. D. Thakkar, A. D. R. Marcia, B. R. Hamaker and S. R. Lindemann, Sci. Rep., 2018, 8, 16655 CrossRef PubMed.
- S. Mishra and J. A. Monro, J. Cereal Sci., 2009, 50, 61–66 CrossRef CAS.
- J. Yang, A. Keshavarzian and D. J. Rose, J. Med. Food, 2013, 16, 862–867 CrossRef CAS PubMed.
- V. Lebet, E. Arrigoni and R. Amadò, LWT – Food Sci. Technol., 1998, 31, 473–479 CrossRef CAS.
- M. Durand, C. Dumay, P. Beaumatin and M. T. Morel, Anim. Feed Sci. Technol., 1988, 21, 197–204 CrossRef.
- Y. E. Tuncil, C. H. Nakatsu, A. E. Kazem, S. Arioglu-Tuncil, B. Reuhs, E. C. Martens and B. R. Hamaker, J. Funct. Foods, 2017, 32, 347–357 CrossRef CAS.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, Nucleic Acids Res., 2013, 41, D590–D596 CrossRef CAS PubMed.
- J. R. Cole, Q. Wang, J. A. Fish, B. Chai, D. M. McGarrell, Y. Sun, C. T. Brown, A. Porras-Alfaro, C. R. Kuske and J. M. Tiedje, Nucleic Acids Res., 2014, 42, D633–D642 CrossRef CAS PubMed.
- T. M. Cantu-Jungles, T. R. Cipriani, M. Iacomini, B. R. Hamaker and L. M. C. Cordeiro, Bioact. Carbohydr. Diet. Fibre, 2017, 9, 1–6 CrossRef CAS.
- S. Karppinen, K. Liukkonen, A.-M. Aura, P. Forssell and K. Poutanen, J. Sci. Food Agric., 2000, 80, 1469–1476 CrossRef CAS.
- E. O. Ayua, A. E. Kazem and B. R. Hamaker, Bioact. Carbohydr. Diet. Fibre, 2020, 24, 100245 CrossRef CAS.
- T. M. Cantu-Jungles, N. Bulut, E. Chambry, A. Ruthes, M. Iacomini, A. Keshavarzian, T. A. Johnson and B. R. Hamaker, mBio, 2020, 12, e01028-21 CrossRef PubMed.
- T. Moro Cantu-Jungles, G. E. do Nascimento, X. Zhang, M. Iacomini, L. M. C. Cordeiro and B. R. Hamaker, Carbohydr. Polym., 2019, 206, 389–395 CrossRef CAS PubMed.
- A. Kaur, D. J. Rose, P. Rumpagaporn, J. A. Patterson and B. R. Hamaker, J. Food Sci., 2011, 76, H137–H142 CrossRef CAS PubMed.
- G. Sproesser, M. B. Ruby, N. Arbit, C. S. Akotia, M. dos Santos Alvarenga, R. Bhangaokar, I. Furumitsu, X. Hu, S. Imada, G. Kaptan, M. Kaufer-Horwitz, U. Menon, C. Fischler, P. Rozin, H. T. Schupp and B. Renner, BMC Public Health, 2019, 19, 1606 CrossRef PubMed.
- D. Chen, D. Rocha-Mendoza, S. Shan, Z. Smith, I. García-Cano, J. Prost, R. Jimenez-Flores and O. Campanella, J. Agric. Food Chem., 2022, 70, 8124–8133 CrossRef CAS PubMed.
- A. J. Holmes, Y. V. Chew, F. Colakoglu, J. B. Cliff, E. Klaassens, M. N. Read, S. M. Solon-Biet, A. C. McMahon, V. C. Cogger, K. Ruohonen, D. Raubenheimer, D. G. Le Couteur and S. J. Simpson, Cell Metab., 2017, 25, 140–151 CrossRef CAS PubMed.
- S. Ben-Harb, A. Saint-Eve, M. Panouillé, I. Souchon, P. Bonnarme, E. Dugat-Bony and F. Irlinger, Int. J. Food Microbiol., 2019, 293, 124–136 CrossRef CAS PubMed.
- C. Poelaert, X. Despret, M. Sindic, Y. Beckers, F. Francis, D. Portetelle, H. Soyeurt, A. Théwis and J. Bindelle, J. Agric. Food Chem., 2017, 65, 435–444 CrossRef CAS PubMed.
- C. L. Adam, S. W. Gratz, D. I. Peinado, L. M. Thomson, K. E. Garden, P. A. Williams, A. J. Richardson and A. W. Ross, PLoS One, 2016, 11, e0155871 CrossRef PubMed.
- Y. Peng, K. Yu, C. Mu, S. Hang, L. Che and W. Zhu, Appl. Microbiol. Biotechnol., 2017, 101, 5415–5426 CrossRef CAS PubMed.
- M. Beaumont, K. J. Portune, N. Steuer, A. Lan, V. Cerrudo, M. Audebert, F. Dumont, G. Mancano, N. Khodorova, M. Andriamihaja, G. Airinei, D. Tomé, R. Benamouzig, A.-M. Davila, S. P. Claus, Y. Sanz and F. Blachier, Am. J. Clin. Nutr., 2017, 106, 1005–1019 CrossRef CAS PubMed.
- S. H. Duncan, A. Belenguer, G. Holtrop, A. M. Johnstone, H. J. Flint and G. E. Lobley, Appl. Environ. Microbiol., 2007, 73, 1073–1078 CrossRef CAS PubMed.
- W. R. Russell, S. W. Gratz, S. H. Duncan, G. Holtrop, J. Ince, L. Scobbie, G. Duncan, A. M. Johnstone, G. E. Lobley, R. J. Wallace, G. G. Duthie and H. J. Flint, Am. J. Clin. Nutr., 2011, 93, 1062–1072 CrossRef CAS PubMed.
- S. Anand, H. Kaur and S. S. Mande, Front. Microbiol., 2016, 7, 1945 Search PubMed.
- R. A. Gonzalez-Garcia, T. McCubbin, L. Navone, C. Stowers, L. K. Nielsen and E. Marcellin, Fermentation, 2017, 3, 21 CrossRef.
- H. E. Rasmussen and B. R. Hamaker, Gastroenterol. Clin., 2017, 46, 783–795 CrossRef PubMed.
- N. E. Diether and B. P. Willing, Microorganisms, 2019, 7, 19 CrossRef CAS PubMed.
- F. Blachier, F. Mariotti, J. F. Huneau and D. Tomé, Amino Acids, 2007, 33, 547–562 CrossRef CAS PubMed.
- B. Darcy-Vrillon, C. Cherbuy, M.-T. Morel, M. Durand and P.-H. Duée, Mol. Cell. Biochem., 1996, 156, 145–151 CrossRef CAS PubMed.
- C. Villodre Tudela, C. Boudry, F. Stumpff, J. R. Aschenbach, W. Vahjen, J. Zentek and R. Pieper, Br. J. Nutr., 2015, 113, 610–617 CrossRef CAS PubMed.
- R. Hughes, E. A. Magee and S. Bingham, Curr. Issues Intest. Microbiol., 2000, 1, 51–58 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo04187e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |