Open Access Article
Open Access ArticleBeef peptides mitigate skeletal muscle atrophy in C2C12 myotubes through protein degradation, protein synthesis, and the oxidative stress pathway†
Hyeonjin
Hur‡
 a,
Hye-Jin
Kim‡
a,
Hye-Jin
Kim‡
 a,
Dongheon
Lee
a and
Cheorun
Jo
a,
Dongheon
Lee
a and
Cheorun
Jo
 *abc
*abc
aDepartment of Agricultural Biotechnology, Center for Food and Bioconvergence, and Research Institute of Agriculture and Life Science, Seoul National University, Seoul 08826, Republic of Korea. E-mail: cheorun@snu.ac.kr; Tel: +82-2-880-4820
bInstitute of Green Bio Science and Technology, Seoul National University, Pyeongchang 25354, Republic of Korea
cDepartment of Animal Product Technology, Faculty of Animal Husbandry, Universitas Padjadjaran, West Java 45363, Indonesia
First published on 25th March 2024
Abstract
This study aimed to investigate the potential of beef peptides (BPs) in mitigating muscle atrophy induced by dexamethasone (DEX) with underlying three mechanisms in vitro (protein degradation, protein synthesis, and the oxidative stress pathway). Finally, the anti-atrophic effect of BPs was enhanced through purification and isolation. BPs were generated using beef loin hydrolyzed with alcalase/ProteAX/trypsin, each at a concentration of 0.67%, followed by ultrafiltration through a 3 kDa cut-off. BPs (10–100 μg mL−1) dose-dependently counteracted the DEX-induced reductions in myotube diameters, differentiation, fusion, and maturation indices (p < 0.05). Additionally, BPs significantly reduced FoxO1 protein dephosphorylation, thereby suppressing muscle-specific E3 ubiquitin ligases such as muscle RING-finger containing protein-1 and muscle atrophy F-box protein in C2C12 myotubes at concentrations exceeding 25 μg mL−1 (p < 0.05). BPs also enhanced the phosphorylation of protein synthesis markers, including mTOR, 4E-BP1, and p70S6K1, in a dose-dependent manner (p < 0.05) and increased the mRNA expression of antioxidant enzymes. Fractionated peptides derived from BPs, through size exclusion and polarity-based fractionation, also demonstrated enhanced anti-atrophic effects compared to BPs. These peptides downregulated the mRNA expression of primary muscle atrophy markers while upregulated that of antioxidant enzymes. Specifically, peptides GAGAAGAPAGGA (MW 924.5) and AFRSSTKK (MW 826.4) were identified from fractionated peptides of BPs. These findings suggest that BPs, specifically the peptide fractions GAGAAGAPAGGA and AFRSSTKK, could be a potential strategy to mitigate glucocorticoid-induced skeletal muscle atrophy by reducing the E3 ubiquitin ligase activity.
1. Introduction
Skeletal muscles, comprising approximately 40% of total body weight, play crucial roles in facilitating movement, generating heat, protecting organs, storing glycogen, and aiding blood circulation.1 Skeletal muscle atrophy, characterized by a reduction in skeletal muscle mass, can markedly impact daily life and contribute to various metabolic problems.2 Aging is a primary driver for skeletal muscle atrophy, resulting in significant losses in muscle mass and function.3 In particular, given the anticipated rise in the population aged 60 and above, projected to increase from 11% in 2013 to 22% by 2050,4 age-related muscle atrophy (sarcopenia) is becoming a growing public health concern.Generally, muscle atrophy is also prevalent in conditions like starvation, cachexia, severe insulinopenia, and metabolic acidosis, which are often accompanied by elevated glucocorticoid (GC) levels.5 Elevated GC levels in skeletal muscle can prompt an imbalance in which protein breakdown surpasses synthesis via several pathways.5 Recent studies emphasize the pivotal role of the ubiquitin-proteasome system in protein degradation and muscle mass regulation. Muscle-specific E3 ubiquitin ligases, such as muscle RING finger 1 (MuRF1) and muscle atrophy F-box (Atrogin-1), are highly expressed during muscle atrophy and are known to induce protein degradation in skeletal muscle tissue.6 On the other hand, the Akt/mammalian target of rapamycin (mTOR) pathway plays a vital role in regulating skeletal muscle hypertrophy and mitigating muscle atrophy. Downstream effectors of mTOR, such as p70S6K and 4E-BP1, initiate translation and facilitate protein synthesis.7 Additionally, increased oxidative stress in skeletal muscle, either occurring independently or in combination with other pathways, has been shown to increase protein breakdown and contribute to muscle atrophy.8
In recent years, there has been a growing demand for natural substances and food sources that can enhance muscle development while having fewer side effects compared to pharmaceuticals. Protein hydrolysates have been recognized as potential sources of proteins and peptides with various beneficial properties.9 Over the past four decades, the health-promoting potential of beef hydrolysates has been documented10 with reports of their anti-proliferative effects on cancer cells,11 inhibitory effects on angiotensin-converting enzyme and antimicrobial activities.12,13 Beef extract has been shown to promote the proliferation of myoblasts and hypertrophy of myotubes in C2C12.14 Furthermore, beef is one of the significant contributors to branched chain amino acid (BCAA) intake. BCAAs like leucine, isoleucine, and valine have shown promise in reducing muscle atrophy by suppressing the expression of Atrogin-1 and MuRF1 mRNA,15 and in stimulating protein synthesis.16 Beef contains a high protein content of approximately 23.2%,17 with a BCAA concentration approximately 2.4 times greater than that found in soybeans.18 Despite these promising characteristics, the potential anti-muscle atrophy effects of beef or beef-derived peptides have yet to be investigated.
Therefore, the objective of this study is to investigate the potential anti-atrophic effect of beef hydrolysate and its purified peptides on DEX-induced skeletal muscle atrophy. This study seeks to understand the underlying mechanisms, particularly their influence on protein degradation, protein synthesis, and oxidative stress. Moreover, it involves the isolation and purification of specific peptide fractions with anti-atrophic potential, further elucidating their prospects as therapeutic interventions.
2. Materials and methods
2.1. Preparation of beef peptides (BPs)
A female Chikso (Korean native cattle) aged 57 months with a carcass weight of 373 kg and a quality grade of 2 was used in this study. Loin (M. longissimus lumborum) was obtained from the slaughterhouse in Dodram LPC (Anseong, Republic of Korea) at 24 h postmortem and transported to the laboratory under refrigerated conditions (4 °C). An enzymatic hydrolysis process began with the homogenization of 10 g of ground loin with 40 mL of double-distilled water (DDW). The homogenate was subjected to the sequential addition of three enzymes: 0.67% alcalase (pH 8.0, 50 °C), 0.67% ProteAX (pH 7.0, 50 °C), and 0.67% trypsin (pH 8.0, 37 °C). Each enzyme was allowed to act for 2 h, resulting in a total hydrolysis process duration of 6 h. The concentrations and following processes were developed by a preliminary study.19 After enzymatic hydrolysis, the mixture was heated to 95 °C for 10 min to inactivate the enzymes. The hydrolysate was then placed into an ultrafiltration tube (Vivaspin 20, Sartorius, Goettingen, Germany) and centrifuged at 4000g for 30 min (Combi R515, Hanil Co. Ltd, Daejeon, Korea), which facilitated the collection of peptide fractions with a molecular weight under 3 kDa, referred to as “beef peptides (BPs)”. The sample was lyophilized (PVTFD-10K, Ilshin Lab Co. Ltd, Seoul, Korea) and stored in a freezer for future analyses.2.2. Amino acid composition
The amino acid composition of both loin and BPs was analyzed following a method previously reported20 with some modifications which included conditions that result in the destruction of tryptophan. Acid hydrolysis was carried out at 130 °C for 20 h using 6 N HCl. Amino acid profiles were determined with an HPLC system (Ultimate3000, Thermo-Dionex, Sunnyvale, CA, USA) equipped with a fluorescence detector (1260FLD, Agilent Technologies, Wilmington, DE, USA).2.3. Antioxidant activity assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio and incubated in the dark at 25 °C for 16 h. The resultant mixture was then diluted with ethanol until the absorbance at 734 nm reached 0.70 ± 0.02. Subsequently, 50 μL of the sample and 950 μL of the ABTS solution were mixed and reacted for 30 min at 25 °C. The absorbance at 734 nm was measured using a UV/Vis spectrophotometer (X-ma 3100, Human Co. Ltd, Seoul, Korea). The absorbance was converted to Trolox equivalent activity (μmol TE) based on the standard curve as antioxidant activity.
1 (v/v) ratio and incubated in the dark at 25 °C for 16 h. The resultant mixture was then diluted with ethanol until the absorbance at 734 nm reached 0.70 ± 0.02. Subsequently, 50 μL of the sample and 950 μL of the ABTS solution were mixed and reacted for 30 min at 25 °C. The absorbance at 734 nm was measured using a UV/Vis spectrophotometer (X-ma 3100, Human Co. Ltd, Seoul, Korea). The absorbance was converted to Trolox equivalent activity (μmol TE) based on the standard curve as antioxidant activity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v) ratio. Subsequently, 2.7 mL of the FRAP solution, 270 μL of DDW, and 90 μL of the sample were mixed and incubated in the dark at 37 °C for 30 min. The absorbance at 593 nm was measured using a UV/Vis spectrophotometer (Human Co. Ltd). The absorbance was converted to Trolox equivalent activity (μmol TE) based on the standard curve as antioxidant activity.
1 (v/v/v) ratio. Subsequently, 2.7 mL of the FRAP solution, 270 μL of DDW, and 90 μL of the sample were mixed and incubated in the dark at 37 °C for 30 min. The absorbance at 593 nm was measured using a UV/Vis spectrophotometer (Human Co. Ltd). The absorbance was converted to Trolox equivalent activity (μmol TE) based on the standard curve as antioxidant activity.
2.4. Cell culture and muscle atrophy induction
The C2C12 mouse cell line (CRL-1772) was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in a 37 °C, 5% CO2 incubator using a growth medium composed of Dulbecco's Modified Eagle's Medium (DMEM; Welgene, Gyeongsan, Korea) supplemented with 10% Fetal Bovine Serum (FBS; Gibco, NY, USA) and 1% Antibiotic-Antimycotic (AA; Gibco, Gaithersburg, MD, USA). When the cell culture reached 100% confluency, the culture medium was replaced with a differentiation medium consisting of 2% Horse Serum (HS; Biowest, Nuaillé, France) and DMEM with 1% AA. The differentiation medium was refreshed every two days. After six days of differentiation, once myotubes were fully formed,14 the differentiation medium was replaced with serum-free media of DMEM containing 1% AA for further experiment. The cells used for the experiment were within passage numbers 4 to 6.Muscle atrophy was induced using a synthetic glucocorticoid, dexamethasone (DEX; Sigma-Aldrich, Inc., St Louis, MO, USA), in accordance with the previous cell viability results by using DEX in this study and previous research.23 The C2C12 myotubes were treated with different concentrations of BPs (0, 10, 25, 50, 100 μg mL−1) along with DEX for 24 h. The non-treated sample (control) consists of differentiated myotubes that were not subjected to any treatment with DEX or BPs. Morphological changes associated with muscle atrophy were observed using a Nikon microscope (Nikon, Tokyo, Japan).
2.5. Cell viability
The effect of BPs on the cell viability of C2C12 myotubes was assessed using the water-soluble tetrazolium salt-8 (WST-8) assay kits (Biomax, Seoul, Korea). The C2C12 cells were seeded at a density of 5 × 104 cells per well in a 48-well plate and cultured in a growth medium at 37 °C with 5% CO2. Upon reaching confluency, the culture medium was replaced with a differentiation medium, and the cells were further cultured for six days. Subsequently, BPs (0, 10, 50, 100, 250, 500 μg mL−1), DEX (0–100 μM), and a mixture of DEX (100 μM) and BPs were added to the wells. The control group remained non-treated. After 24 h incubation with each treatment, 50 μL of WST-8 reagent was added to each well. Following further 2 h incubation, cell viability was determined by measuring the absorbance at 450 nm using a spectrophotometer. The percentage of cell viability was calculated by comparing the absorbance of the treated cells to the control.2.6. Immunofluorescence analysis and myotube diameter
Immunofluorescence was performed according to a previously described method.24 C2C12 myotubes were fixed with 4% paraformaldehyde in Dulbecco's Phosphate-Buffered Saline (DPBS; Welgene, Gyeongsan, Korea) for 30 min at 4 °C. After fixation, the cells were washed twice with DPBS and permeabilized using 0.2% Triton X-100 (Sigma-Aldrich) for 15 min. To minimize non-specific binding, the cells were initially incubated with 10% goat serum (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h. The cells were then treated with a primary antibody against myosin heavy chain (MAB 4470, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; R&D Systems, Minneapolis, MN, USA) and incubated overnight at 4 °C. After washing to remove the unbound primary antibody, the myotubes were treated with a secondary antibody (A-11001, diluted 1
200; R&D Systems, Minneapolis, MN, USA) and incubated overnight at 4 °C. After washing to remove the unbound primary antibody, the myotubes were treated with a secondary antibody (A-11001, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500; Invitrogen, Waltham, MA, USA) and incubated overnight at 4 °C. Nuclei were stained using Hoechst 33342 (diluted 1
500; Invitrogen, Waltham, MA, USA) and incubated overnight at 4 °C. Nuclei were stained using Hoechst 33342 (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Molecular Probes, Eugene, OR, USA) for 10 min at 4 °C. The cells were then washed, and the medium was replaced with DPBS. Images of the stained C2C12 myotubes were captured using an inverted fluorescence microscope equipped with a digital camera (Nikon, Japan) at ×200 magnification.
1000; Molecular Probes, Eugene, OR, USA) for 10 min at 4 °C. The cells were then washed, and the medium was replaced with DPBS. Images of the stained C2C12 myotubes were captured using an inverted fluorescence microscope equipped with a digital camera (Nikon, Japan) at ×200 magnification.
For morphological evaluation of myotubes, the average diameters of a total of 150 myotubes from each treatment group were measured across five different random fields. The measurements were performed using the NIH ImageJ software (Bethesda, MD, USA). Differentiation index: it was calculated as the ratio of the number of nuclei within myosin heavy chain-stained cells to the total number of cells with Hoechst-stained nuclei.25 Fusion index: it was determined by taking the ratio of the number of nuclei within myosin heavy chain-stained myotubes having two or more nuclei to the total number of nuclei.26 Maturation index: it was defined by the number of myotubes having five or more nuclei.26
2.7. mRNA expression analysis by reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using the EcoPURE total RNA isolation kit (E2075; EcoTech Biotechnology, Hanam, Korea) in accordance with the manufacturer's protocol. The purity of the extracted RNA was determined by measuring the absorbance ratio at 260 nm and 280 nm. An equal quantity of high-quality total RNA was subjected to cDNA synthesis using the High-Capacity RNA-to-cDNA™ Kit from Applied Biosystems (Framingham, MA, USA). For RT-qPCR, the SYBR™ Green PCR Master Mix (Applied Biosystems) was used. A 10 μL reaction volume was prepared, comprising PCR buffer, primers, and the appropriate amount of synthesized cDNA. Gene amplification was carried out using the ABI 7300 Real-Time PCR System (Applied Biosystems). The cycling conditions were set at 95 °C for 15 s (denaturation), 58.3–61.3 °C for 15 s (annealing), and 72 °C for 15 s (extension) across 40 cycles. The relative mRNA expression levels were quantified and normalized to β-actin. The detailed primer sequences utilized for amplification are listed in Table S1.†2.8. Western blot analysis
The total protein of C2C12 myotubes was lysed in RIPA buffer supplemented with a protease inhibitor cocktail (ATTO, Tokyo, Japan) for immunoblotting. Protein concentrations in the lysates were determined using a bicinchoninic acid protein assay kit (Gendepot, Barker, TX, USA), with bovine serum albumin (BSA; Sigma-Aldrich) serving as the standard, following the manufacturer's protocol. Fifteen micrograms of protein per lane were separated on a 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis at 100 V for 60–90 min and then transferred to a polyvinylidene difluoride membrane.The transferred membrane was blocked with 5% BSA in 1× Tris-buffered saline containing 1% Tween-20 (Sigma-Aldrich) for 2 h at room temperature. After blocking, the membrane was probed with primary antibodies (details provided in Table S2†) overnight at 4 °C. Following extensive washing, the membrane was then incubated with horseradish peroxidase-conjugated secondary antibodies, diluted in 5% BSA for 1–2 h at room temperature.
Antibody-bound proteins were detected using the Clarity™ Western ECL Substrate (Bio-Rad), and the resulting bands were quantified using ImageLab™ software (Bio-Rad Hercules). The relative protein abundance was assessed by normalizing the intensity of each band to that of the β-actin band.
2.9. Separation and purification of BPs
2.10. Amino acid sequencing
The amino acid sequence of the selected RP-HPLC fraction was analyzed using a mass spectrometer (Triple TOF®5600+ system, AB SCIEX, Framingham, MA, USA) connected to an HPLC (Ultimate 3000, Thermo Dinex) system, following a modified method from a previous study.19 Separation was performed on a column C8 (Zorbax 300SB-C8 RP, Agilent Technologies) using mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile), maintaining a flow rate of 0.3 mL min−1. Specific details of the linear gradient utilized are presented in Table S3.† The autosampler temperature was kept at 4 °C, and an injection volume of 5 μL was used for each sample. Mass spectra were obtained under positive electrospray ionization conditions with an ion spray voltage set at 5500 V. The source temperature was fixed at 500 °C, while the curtain gas, ion source gas 1, and ion source gas 2 pressures were set at 50, 50, and 25 psi, respectively. The MS full scan mode was employed for the acquisition of mass spectra over an m/z range of 200–2500. Data collection was executed using Analyst TF 1.7 software, with subsequent data analysis conducted using ProteinPilot 4.5 software.2.11. Statistical analysis
The experimental results, excluding the amino acid composition of Table 1, were repeated three times individually and analyzed using the general linear model procedure in the SAS program (ver. 9.4, SAS Institute Inc., Cary, NC, USA) to perform analysis of variance. To assess the significance of differences among mean values, Tukey's multiple range test was performed with a confidence level of p < 0.05. The results were presented as mean value ± SD.| Amino acids | Content (mg g−1 dry matter) | Percentage (% of total) | ||
|---|---|---|---|---|
| BL | BPs | BL | BPs | |
BCAA is the sum of isoleucine, leucine, and valine. EAA is the sum of histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, and valine. a,b![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Different letters indicate significant differences (p < 0.05). Data are expressed as the mean ± SD of two independent trials. Different letters indicate significant differences (p < 0.05). Data are expressed as the mean ± SD of two independent trials. |
||||
| Glx | 149.6a ± 3.82 | 114.8b ± 0.92 | 19.1a ± 0.00 | 14.9b ± 0.00 |
| Ser | 32.9b ± 0.64 | 36.1a ± 0.21 | 4.2b ± 0.00 | 4.7a ± 0.00 |
| His | 29.9b ± 0.42 | 32.0a ± 0.07 | 3.8b ± 0.00 | 4.2a ± 0.07 |
| Gly | 33.3a ± 0.28 | 29.2b ± 0.21 | 4.3a ± 0.07 | 3.8b ± 0.00 |
| Thr | 41.0b ± 0.28 | 45.1a ± 0.07 | 5.3b ± 0.07 | 5.9a ± 0.07 |
| Arg | 59.2a ± 1.06 | 54.9b ± 0.00 | 7.6a ± 0.07 | 7.2b ± 0.07 |
| Ala | 48.8b ± 1.20 | 54.1a ± 0.35 | 6.2b ± 0.00 | 7.1a ± 0.07 |
| Tau | 1.4 ± 0.00 | 1.6 ± 0.07 | 0.2 ± 0.00 | 0.2 ± 0.00 |
| Tyr | 30.3b ± 0.71 | 33.9a ± 0.42 | 3.9b ± 0.00 | 4.4a ± 0.00 |
| Val | 40.5b ± 1.41 | 48.1a ± 0.21 | 5.2b ± 0.07 | 6.3a ± 0.07 |
| Met | 19.5 ± 0.85 | 19.2 ± 3.18 | 2.5 ± 0.00 | 2.5 ± 0.42 |
| Phe | 34.6b ± 1.06 | 42.9a ± 0.00 | 4.4b ± 0.00 | 5.6a ± 0.07 |
| Ile | 40.8b ± 1.34 | 47.4a ± 0.14 | 5.2b ± 0.00 | 6.2a ± 0.07 |
| Leu | 69.0b ± 1.77 | 85.3a ± 0.28 | 8.8b ± 0.00 | 11.1a ± 0.14 |
| Lys | 58.8a ± 1.27 | 49.0b ± 1.98 | 7.5a ± 0.00 | 6.4b ± 0.21 |
| Pro | 17.1 ± 1.56 | 14.7 ± 1.34 | 2.2 ± 0.14 | 1.9 ± 0.14 |
| Total | 784.0 ± 20.44 | 769.3 ± 8.63 | 100.0 ± 0.00 | 100.0 ± 0.00 |
| BCAAs | 150.2b ± 4.53 | 180.7a ± 0.21 | 19.2b ± 0.07 | 23.5a ± 0.28 |
| EAAs | 334.0b ± 8.41 | 368.8a ± 5.09 | 42.6b ± 0.00 | 47.9a ± 0.14 |
3. Results and discussion
3.1. Antioxidant activities and the amino acid composition of BPs
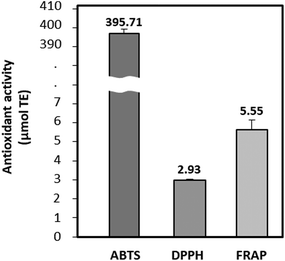
The antioxidant potential of BPs was evaluated using ABTS, DPPH radical scavenging activities, and FRAP assays. As depicted in Fig. 1, the antioxidant activities were 395.71 μmol TE for ABTS, 2.93 μmol TE for DPPH, and 5.55 μmol TE for FRAP, respectively. BPs demonstrated notably strong radical scavenging activity in the ABTS assay, outperforming substances such as egg ovalbumin, milk, and soy hydrolysate.29 This indicates BP's substantial ability to neutralize ABTS radicals by donating electrons.Table 1 reveals that both the protein in beef loin (BL) and BPs consisted of 17 amino acids, with glutamic acid and leucine being the most abundant. Notably, plant-based proteins typically exhibited a lower leucine content and digestibility than animal-derived proteins. This leads to inferior muscle anabolic effects compared to dose-matched animal-derived proteins.30 This study found that the concentrations of BCAAs and essential amino acids (EAAs) in BPs were 180.7 mg g−1 and 368.8 mg g−1, respectively. These values were notably higher than those in BL, which were 150.2 mg g−1 and 334.0 mg g−1 (p < 0.05). Upon hydrolysis of BL, the contents of valine, isoleucine, and leucine in BPs significantly increased to 48.1, 47.4, and 85.3 mg g−1, respectively, marking a 4.3% overall increase in BCAAs. This increment was similar to findings from a previous study.31 Specifically, leucine, the second most abundant amino acid in BPs, exhibited the most substantial rise of 2.3%, increasing from 8.8% to 11.1%. Leucine is known to promote protein synthesis by stimulating the phosphorylation of mTOR with S6K and 4E-BP1.32 The result highlights the potential of the augmented BCAAs content in BPs, especially leucine, to counteract muscle atrophy.
3.2. The effect of BPs on DEX-treated cell viability
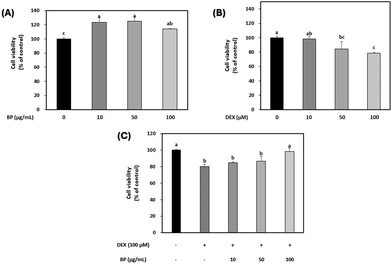
The cell viability of BPs alone (0, 10, 50, 100 μg mL−1) was evaluated initially, and it was found that BPs had no cytotoxicity on C2C12 myotubes at any concentration tested (Fig. 2A). As expected, DEX decreased the cell viability in a dose-dependent manner as shown in Fig. 2B. Based on the observed decrease in cell viability by DEX and on previous research, the concentration of DEX was set at 100 μM, at which it effectively induced protein degradation.23 Upon treatment with different concentrations of BPs (0, 10, 25, 50, 100 μg mL−1), a concentration of 100 μg mL−1 BPs effectively mitigated the reduction in cell viability induced by DEX (Fig. 2C). DEX initiates damage to DNA strands by generating reactive oxygen species, contributing to cell death and muscle wasting in skeletal muscle.33 Therefore, these results suggest that the increase in cell viability of C2C12 myotubes by BPs may have the potential to effectively mitigate DEX-induced injury to muscle myotubes. Moreover, this trend has been observed with other bioactive compounds possessing anti-atrophic properties.343.3. The effect of BPs on myotube characteristics and maturation
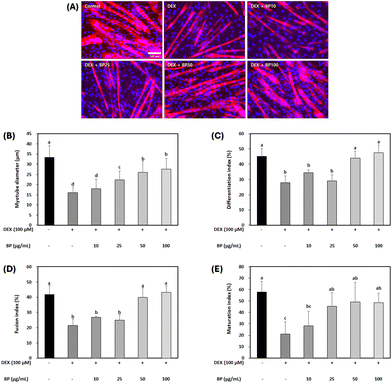
Immunocytochemistry was employed to evaluate the potential of BPs in mitigating muscle atrophy, focusing on parameters such as myotube diameter, differentiation index, fusion index, and maturation index (Fig. 3A). Under normal circumstances, myoblast differentiation into myotubes involves an increase in the number of myofibers due to cell fusion, resulting in increased myofiber diameter. However, muscle atrophy involves a decrease in both number and diameter of fibers due to muscle protein breakdown, ultimately leading to muscle mass loss.35 In this study, a marked reduction in mature myotube diameter and indices was observed in the DEX-treated C2C12 myotubes compared to the control, indicating the successful induction of myotube atrophy by 100 μM DEX (Fig. 3B–E). In contrast, BPs at concentrations above 50 μg mL−1 effectively reversed all these DEX-induced decreases in C2C12 myotubes (p < 0.05). Hence, the result suggests a protective effect of BPs on the myotube morphological parameters against muscle atrophy.During muscle atrophy induction, DEX degrades myogenin and myogenic differentiation 1 (MyoD) as transcription factors involved in muscle differentiation, leading to muscle mass loss during muscle atrophy.36 Therefore, upregulating myogenin and MyoD can help to prevent reductions in mature myotube diameter and indices in C2C12 myotubes. However, in this study, BPs did not significantly alter the mRNA expression levels of myogenin and MyoD (Fig. S1†). Similar results were observed in another study, where beef extract promoted myoblast proliferation and myotube growth in C2C12 cells but appeared to have no influence on differentiation markers like MyoD and myogenin.14 Despite its negligible effect on muscle differentiation, BP promoted muscle cell growth (Fig. 2C) and increased myotube diameter and fusion index (Fig. 3). Since DEX-induced muscle atrophy can be inhibited in several ways, BPs may have potential anti-atrophic effects through pathways other than direct modulation of muscle cell differentiation.
3.4. The effect of BPs on protein degradation in DEX-treated C2C12 myotubes
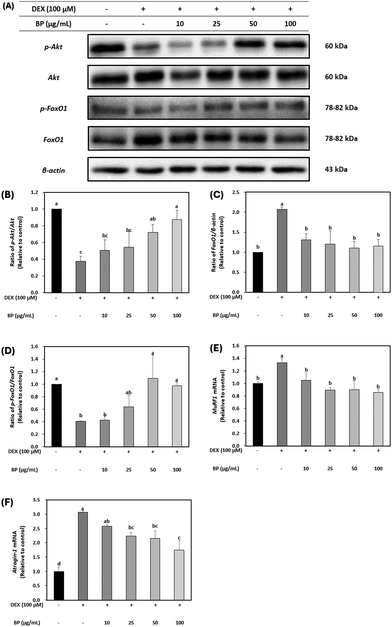
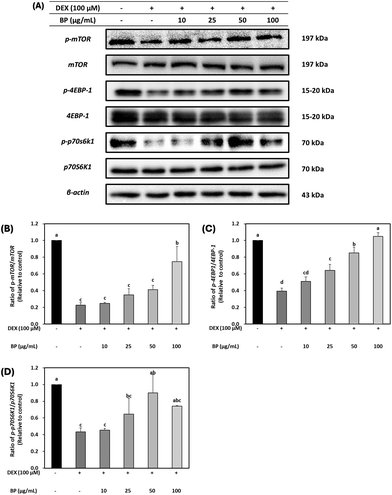
DEX is widely known for its ability to induce muscle atrophy by inhibiting the PI3K/Akt/mTOR pathway.37 Akt acts as an anabolic regulator by promoting protein translation via mTOR and its downstream effectors (4E-BP1 and S6K1). It is also involved in the suppression of protein degradation to block the upregulation of Forkhead Box O (FoxO) transcriptional factors, which have been implicated in initiating proteolysis during muscle atrophy. The dephosphorylation of FoxO family upregulates the transcriptional activation of primary muscle atrophy genes, MuRF1 and Atrogin-1.38To evaluate the effects of BPs in a DEX-induced muscle atrophy model, the protein expression of Akt was initially examined. Fig. 4A and B demonstrate that the total protein levels of Akt were not affected, but the expression of p-Akt was decreased by DEX. However, BPs led to a dose-dependent increase in p-Akt protein expression (p < 0.05), indicating the activation of Akt in myotubes. The activity of FoxO1 is influenced by its phosphorylation state, as indicated by p-FoxO1 levels.37 Protein expression of FoxO1 was significantly increased by DEX, while p-FoxO1 expression was decreased compared to the control (Fig. 4A, C, and D). However, BPs decreased the protein expression of FoxO1 and concurrently increased p-FoxO1 levels induced by DEX (p < 0.05).
Additionally, to determine the role of BPs in alleviating muscle atrophy in DEX-treated C2C12 myotubes, the mRNA expression of MuRF and Atrogin-1 was examined. It is well-established that mRNA expression patterns of MuRF1 and Atrogin-1 are heightened in muscle atrophy-inducing conditions, serving as primary muscle atrophy markers.5,6 FoxO1, acting as an upstream regulator of MuRF1 and Atrogin-1, triggers their upregulation, leading to muscle protein degradation and subsequent muscle atrophy. Fig. 4E and F illustrate that mRNA levels of MuRF1 and Atrogin-1 were significantly increased by DEX. However, BPs dose-dependently reduced the mRNA levels of these muscle atrophy markers (p < 0.05). These results suggest that BPs attenuates muscle atrophy by inhibiting protein degradation mediated by MuRF1 and Atrogin-1. This result was also correlated with enhanced maturation observed in DEX-treated myotubes in Fig. 3.
3.5. The effect of BPs on protein synthesis (hypertrophy) in DEX-treated C2C12 myotubes
The regulatory effects of BPs on protein synthesis were probed by examining mTOR, a downstream marker of Akt, in C2C12 myotubes. DEX led to a reduction in the protein expression of p-mTOR. However, BPs restored the protein expression of p-mTOR levels that was reduced by DEX, particularly at the concentration of 100 μg mL−1 BPs (Fig. 5B). Furthermore, as shown in Fig. 5C and D, BPs significantly increased the protein expression of p-4EBP1 in a dose-dependent manner, and at 50 μg mL−1, it also elevated the levels of p-p70S6K1 (Fig. 5C and D) (p < 0.05). The phosphorylation of both 4E-BP1 and p70S6K1 promotes protein synthesis by regulating protein translation initiation and phosphorylation of ribosomal protein S6.39 Therefore, these results imply that BPs not only counteract muscle atrophy by mitigating the DEX-induced reduction in protein synthesis, but may also promote protein hypertrophy. This interpretation is further substantiated by the patterns observed in the immunostaining images presented in Fig. 3A–E.3.6. The effect of BPs on antioxidant enzymes in DEX-treated C2C12 myotubes
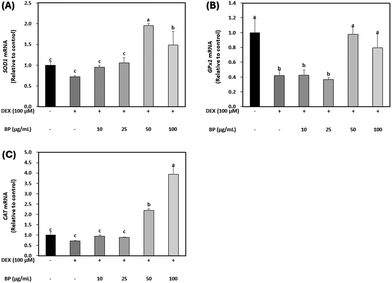
The primary antioxidant enzyme system, consisting of SOD1, GPX1, and CAT, collaboratively neutralizes ROS toxicity in oxidative stress situations induced by GC. Elevated GC levels lead to an increase in ROS production, facilitated by the dephosphorylation of FoxO, subsequently resulting in the hyperexpression of Atrogin-1, a key marker of muscle atrophy, and subsequent protein degradation.8,40 In this study, BPs significantly upregulated the mRNA expression of these antioxidant enzymes reduced by DEX, particularly at concentrations higher than 50 μg mL−1 (Fig. 6A–C). These results are supported by the strong radical scavenging activity of BPs (395.71 μmol TE; Fig. 1). Given these findings, it is suggested that BPs might function as an antioxidant, counteracting DEX-induced muscle atrophy in C2C12 myotubes.3.7. Purification of BPs and their anti-atrophic effects
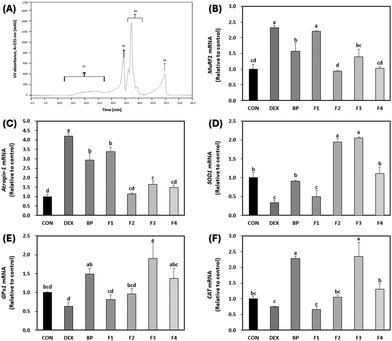
Peptide isolation and purification are crucial steps in bioanalytical studies as they enable the separation of complex mixtures into individual components, facilitating accurate characterization and examination of distinct bioactivities.41 This process significantly enhances the reliability and specificity of the results obtained, revealing the differential effects each peptide may have on a given biological process.41 With this understanding, BPs were fractionated based on peptide size to examine the potential variance in bioactivity across its constituent peptides.The 50 μg mL−1 concentration of BPs was selected for further analysis to validate the efficacy of peptides in comparison to BPs due to the significant effects observed, such as decreased protein degradation, enhanced protein synthesis, and increased activities of antioxidant enzymes. To assess the variance in bioactivity across its constituent peptides, BPs were fractionated based on peptide size using size-exclusion chromatography, resulting in four fractions labeled as F1, F2, F3, and F4, as shown in Fig. 7A. Peptide fractions F2, F3, and F4 demonstrated a greater decrease in the mRNA expression of primary muscle atrophy markers (MuRF1 and Atrogin-1) compared to BPs (p < 0.05; Fig. 7B and C). Regarding antioxidant activity, both F2 and F3 exhibited increased the mRNA expression of SOD1 (Fig. 7D). Notably, F3 showed the highest mRNA expression of GPx1 (Fig. 7E), surpassing even BPs. Additionally, both BPs and F3 demonstrated the highest mRNA expression of CAT (Fig. 7F). These results suggest that F3 may offer a strong potential for anti-atrophic effects and high antioxidant properties compared to BPs. Consequently, we decided to conduct a detailed analysis of fraction F3.
3.8. Purification of fractions derived from BPs and their anti-atrophic effects
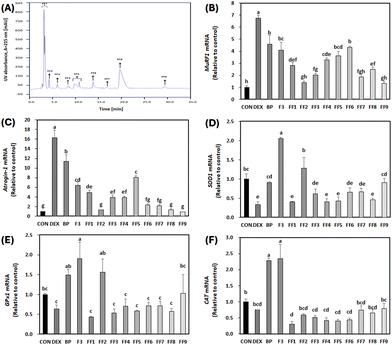
Further investigation involved polarity-based purification of F3 using RP-HPLC, resulting in its separation into nine fractions, labeled as FF1–FF9 (Fig. 8A). Remarkably, FF1–FF3 and FF7–FF9 exhibited decreased levels of primary muscle atrophy markers when compared to both BPs and F3 as depicted in Fig. 8B and C (p < 0.05). Among all the fractionated peptides, FF2 and FF9 showed the most diminished expression levels of the primary muscle atrophy markers (Fig. 8B and C). Furthermore, in terms of antioxidant properties, FF2 had the highest mRNA levels for SOD1 and GPx1 compared to other fractions (FF1, FF3–FF9) (p < 0.05) (Fig. 8D and E). There was no noticeable difference in CAT levels among the fractions relative to DEX (Fig. 8F). Therefore, given its significant anti-atrophy and antioxidant properties, the peptide sequence of FF2 was subjected to further examination.3.9. Amino acid sequencing of peptides with high anti-atrophic potential isolated from BPs
Through deconvolution of multicharged ions within the full scan mass spectra, ranging from 200 to 2500, the molecular masses of the identified peptides were determined (Fig. 9). Finally, LC-MS/MS identified two peptides in FF2, with the sequences being peptide 1: AFRSSTKK (MW 924.5; Fig. 9A) and peptide 2: GAGAAGAPAGGA (MW 826.4; Fig. 9B). The bioactivity and functionality of peptides depend on their amino acid composition, structure, polarity, and length.42 Although the literature on anti-atrophic peptides is limited, peptides reported to have an anti-atrophic effect consisted of fewer than 15 amino acids.43 Similarly, peptides 1 and 2 isolated in this study also have 8 and 12 amino acids, respectively. Such short peptides have shown potential in enhancing the proliferation or differentiation of myoblasts or myotubes in vitro.43 The ‘AF’ sequence at the N-terminal of peptide 1, due to their hydrophobic properties by the presence of alanine and phenylalanine, could indicate its potential for effective membrane interactions. Moreover, a significant portion (75%) of peptide 1 presents hydrophilic tendencies, which might enhance its solubility in aqueous media and promote interactions with polar cellular membrane components. These amphipathic characteristics exhibited by peptide 1 may imply its potential as cell-penetrating peptide (CPP) capabilities. The concept of primary amphipathic CPPs refers to chimeric peptides that possess a hydrophobic domain for attaching to cell membranes, along with a nuclear localization signal.44 Moreover, the presence of arginine (R) and two lysine (K) residues could potentially enable interactions with negatively charged cellular membranes, imparting a cationic characteristic to peptide 1. Therefore, the unique combination of hydrophobic and hydrophilic properties in peptide 1 might highlight its potential for effective membrane interactions and solubility in cell systems.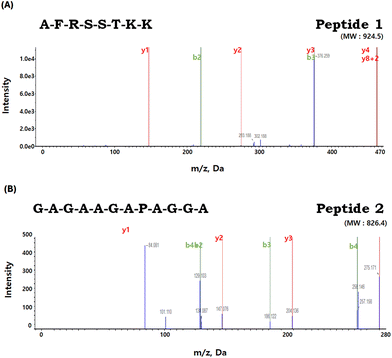 | ||
| Fig. 9 Identification of the molecular weight and amino acid sequence of the purified peptides from FF2. (A) Petide 1: AFRSSTKK (MW 924.5) and (B) Peptide 2: GAGAAGAPAGGA (MW 826.4). | ||
In contrast, peptide 2 features a balanced mix of hydrophilic and hydrophobic residues, with glycine being notably prevalent. The glycine is frequently found in previously reported anti-atrophic peptides.45–48 Studies have also demonstrated the effectiveness of glycine in preventing muscle atrophy. Caldow et al.49 further supported this by suggesting that a higher concentration of glycine might serve as a beneficial factor in preventing muscle degradation, especially in conditions of limited nutrient intake. Based on the available findings, it appears that peptide 2, particularly those characterized by a high glycine content, holds the potential to serve as a promising candidate for therapeutic interventions targeting muscle atrophy.
4. Conclusions
BPs derived from BL effectively counteracted the DEX-induced decrease in cell viability, myotube diameter, and muscle maturation in a dose-dependent manner. These beneficial effects are likely attributed to the inhibition of proteolysis, promotion of protein synthesis, and reduction in oxidative stress induced by DEX in C2C12 myotubes. Furthermore, the fractions containing the peptides ‘GAGAAGAPAGGA’ and ‘AFRSSTKK’ isolated from BPs demonstrated significant anti-atrophic and antioxidant properties compared to the parent material. Therefore, both BPs and its purified peptides exhibit promising potential as natural dietary therapeutic agents for addressing muscle atrophy.Author contributions
Hyeonjin Hur and Hye-Jin Kim: conceptualization, data curation, formal analysis, investigation, methodology, software, supervision, validation, visualization, writing – original draft, writing – review and editing. Dongheon Lee: formal analysis, investigation, methodology, writing – original draft. Cheorun Jo: funding acquisition, project administration, resources, supervision, validation, writing – original draft, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development” (Project No. PJ016201) of the Rural Development Administration, Korea.References
- J. Lopes, D. M. Russell, J. Whitwell and K. N. Jeejeebhoy, Skeletal muscle function in malnutrition, Am. J. Clin. Nutr., 1982, 36, 602–610 CrossRef CAS PubMed.
- C. English, H. McLennan, K. Thoirs, A. Coates and J. Bernhardt, Loss of skeletal muscle mass after stroke: a systematic review, Int. J. Stroke, 2010, 5, 395–402 CrossRef PubMed.
- N. Fuggle, S. Shaw, E. Dennison and C. Cooper, Sarcopenia, Best Pract. Res., Clin. Rheumatol., 2017, 31, 218–242 CrossRef PubMed.
- C. B. Newgard and N. E. Sharpless, Coming of age: molecular drivers of aging and therapeutic opportunities, J. Clin. Invest., 2013, 123, 946–950 CrossRef CAS PubMed.
- O. Schakman, S. Kalista, C. Barbé, A. Loumaye and J. P. Thissen, Glucocorticoid-induced skeletal muscle atrophy, Int. J. Biochem. Cell Biol., 2013, 45, 2163–2172 CrossRef CAS PubMed.
- S. C. Bodine and L. M. Baehr, Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1, Am. J. Physiol. Endocrinol. Metab., 2014, 307, E469–E484 CrossRef CAS PubMed.
- S. C. Bodine, T. N. Stitt, M. Gonzalez, W. O. Kline, G. L. Stover, R. Bauerlein, E. Zlotchenko, A. Scrimgeour, J. C. Lawrence, D. J. Glass and G. D. Yancopoulos, Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo, Nat. Cell Biol., 2001, 3, 1014–1019 CrossRef CAS PubMed.
- A. Bonetto, F. Penna, M. Muscaritoli, V. G. Minero, F. R. Fanelli, F. M. Baccino and P. Costelli, Are antioxidants useful for treating skeletal muscle atrophy?, Free Radicals Biol. Med., 2009, 47, 906–916 CrossRef CAS PubMed.
- R. L. Thomson and J. D. Buckley, Protein hydrolysates and tissue repair, Nutr. Res. Rev., 2011, 24, 191–197 CrossRef CAS PubMed.
- A. M. Ahhmed and M. Muguruma, A review of meat protein hydrolysates and hypertension, Meat Sci., 2010, 86, 110–118 CrossRef CAS PubMed.
- J. Lee, J. R. Park, H. Lee, S. Jang, S. M. Ryu, H. Kim, D. Kim, A. Jang and S. R. Yang, L-carnosine induces apoptosis/cell cycle arrest via suppression of NF-κB/STAT1 pathway in HCT116 colorectal cancer cells, In Vitro Cell. Dev. Biol.: Anim., 2018, 54, 505–512 CrossRef CAS PubMed.
- M. A. Maky and T. Zendo, Generation and characterization of novel bioactive peptides from fish and beef hydrolysates, Appl. Sci., 2021, 11, 10452 CrossRef CAS.
- L. Mora, M. Reig and F. Toldrá, Bioactive peptides generated from meat industry by-products, Food Res. Int., 2014, 65, 344–334 CrossRef CAS.
- S. Sawano, K. Baba, Y. Sonoda, J. I. Wakamatsu, S. Tomonaga, M. Furuse, Y. Sata, R. Tatsumi, Y. Ikeuchi and W. Mizunoya, Beef extract supplementation promotes myoblast proliferation and myotube growth in C2C12 cells, Eur. J. Nutr., 2020, 59, 3735–3743 CrossRef CAS PubMed.
- E. H. Herningtyas, Y. Okimura, A. E. Handayaningsih, D. Yamamoto, T. Maki, K. Iida, Y. Takahashi, H. Kaji and K. Chihara, Branched-chain amino acids and arginine suppress MaFbx/atrogin-1 mRNA expression via mTOR pathway in C2C12 cell line, Biochim. Biophys. Acta, Gen. Subj., 2008, 1780, 1115–1120 CrossRef CAS PubMed.
- R. R. Wolfe, Branched-chain amino acids and muscle protein synthesis in humans: myth or reality?, J. Int. Soc. Sports Nutr., 2017, 14, 30 CrossRef PubMed.
- P. Williams, Nutritional composition of red meat, Nutr. Diet., 2007, 64, S113–S119 Search PubMed.
- R. Wikandari, D. R. Tanugraha, A. J. Yastanto, R. Gmoser and J. A. Teixeira, Development of meat substitutes from filamentous fungi cultivated on residual water of Tempeh factories, Molecules, 2023, 28, 997 CrossRef CAS PubMed.
- H. J. Kim, K. W. Kim, J. W. Lee, S. H. Lee, S. S. Lee and A. Jang, Low-molecular-weight hydrolysate from black goat extract had antioxidative and anti-inflammatory effects in macrophage cells via inhibition of mapks and nf-κb pathways, J. Food Biochem., 2024, 2024, 7155015 CrossRef.
- B. A. Bidlingmeyer, S. A. Cohen and T. L. Tarvin, Rapid analysis of amino acids using pre-column derivatization, J. Chromatogr. B: Biomed. Sci. Appl., 1984, 336, 93–104 CrossRef CAS PubMed.
- J. Choe, B. Park, H. J. Lee and C. Jo, Potential antioxidant and angiotensin I-converting enzyme inhibitory activity in crust of dry-aged beef, Sci. Rep., 2020, 10, 7883 CrossRef CAS PubMed.
- H. J. Kim, H. J. Kim, K. W. Kim, J. Lee, S. H. Lee, S. S. Lee, B. H. Choi, D. J. Shin, K. H. Jeon, J. Y. Choi and A. Jang, Effect of feeding alfalfa and concentrate on meat quality and bioactive compounds in Korean native black goat loin during storage at 4 °C, Food Sci. Anim. Resour., 2022, 42, 517–535 CrossRef PubMed.
- M. K. Lee, Y. M. Kim, I. H. Kim, Y. H. Choi and T. J. Nam, Pyropia yezoensis peptide PYP1-5 protects against dexamethasone-induced muscle atrophy through the downregulation of atrogin1/MAFbx and MuRF1 in mouse C2C12 myotubes, Mol. Med. Rep., 2017, 15, 3507–3514 CrossRef CAS PubMed.
- M. Ryu, M. Kim, H. Y. Jung, C. H. Kim and C. Jo, Effect of p38 inhibitor on the proliferation of chicken muscle stem cells and differentiation into muscle and fat, Anim. Biosci., 2023, 36, 295–306 CrossRef CAS PubMed.
- X. Li, J. Baker, T. Cracknell, A. R. Haynes and G. Blanco, IGFN1_v1 is required for myoblast fusion and differentiation, PLoS One, 2017, 12, e0180217 CrossRef PubMed.
- C. Wu, C. S. Chin, Q. Huang, H. Y. Chan, X. Yu, V. A. Roy and W. J. Li, Rapid nanomolding of nanotopography on flexible substrates to control muscle cell growth with enhanced maturation, Microsyst. Nanoeng., 2021, 7, 89 CrossRef CAS PubMed.
- J. Yang, J. Huang, X. Dong, Y. Zhang, X. Zhou, M. Huang and G. Zhou, Purification and identification of antioxidant peptides from duck plasma proteins, Food Chem., 2020, 319, 126534 CrossRef CAS PubMed.
- T. L. Pownall, C. C. Udenigwe and R. E. Aluko, Amino acid composition and antioxidant properties of pea seed (Pisum sativum L.) enzymatic protein hydrolysate fractions, J. Agric. Food Chem., 2010, 58, 4712–4718 CrossRef CAS PubMed.
- L. M. E. Erik Gustavo Tovar-Perez, E. M. Yahia, B. V. C. Gonzalez-Cordova, A. H. Mendoza and E. M. González, Antioxidant capacity of egg, milk and soy protein hydrolysates and biopeptides produced by Bromelia pinguin and Bromelia karatas-derived proteases, Emirates J. Food Agric., 2018, 30, 122–130 CrossRef.
- P. T. Morgan and L. Breen, The role of protein hydrolysates for exercise-induced skeletal muscle recovery and adaptation: a current perspective, Nutr. Metab., 2021, 18, 44 CrossRef CAS PubMed.
- V. Klompong, S. Benjakul, M. Yachai, W. Visessanguan, F. Shahidi and K. D. Hayes, Amino acid composition and antioxidative peptides from protein hydrolysates of yellow stripe trevally (Selaroides leptolepis), J. Food Sci., 2009, 74, C126–C133 CAS.
- H. L. Eley, S. T. Russell and M. J. Tisdale, Effect of branched-chain amino acids on muscle atrophy in cancer cachexia, Biochem. J., 2007, 407, 113–120 CrossRef CAS PubMed.
- C. Chen, J. S. Yang, C. C. Lu, Y. J. Chiu, H. C. Chen, M. I. Chung, Y. T. Wu and F. A. Chen, Effect of quercetin on dexamethasone-induced C2C12 skeletal muscle cell injury, Molecules, 2020, 25, 3267 CrossRef CAS PubMed.
- R. Jiang, M. Wang, L. Shi, J. Zhou, R. Ma, K. Feng, X. Chen, X. Xu, X. Li, T. Li and L. Sun, Panax ginseng total protein facilitates recovery from dexamethasone-induced muscle atrophy through the activation of glucose consumption in C2C12 myotubes, BioMed Res. Int., 2019, 2019, 3719643 Search PubMed.
- S. Bhatnagar, A. Mittal, S. K. Gupta and A. Kumar, TWEAK causes myotube atrophy through coordinated activation of ubiquitin–proteasome system, autophagy, and caspases, J. Cell. Physiol., 2012, 227, 1042–1010 CrossRef CAS PubMed.
- Y. H. Son, E. J. Jang, Y. W. Kim and J.-H. Lee, Updates on global epidemiology, risk and prognostic factors of gastric cancer, Biomed. Pharmacother., 2017, 95, 1486–1149 CrossRef CAS PubMed.
- M. Sandri, C. Sandri, A. Gilbert, C. Skurk, E. Calabria, A. Picard, K. Walsh, S. Schiaffino, S. H. Lecker and A. L. Goldberg, Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy, Cell, 2004, 117, 399–412 CrossRef CAS PubMed.
- T. L. Schmitt, M. E. Martignoni, J. Bachmann, K. Fechtner, H. Friess, R. Kinscherf and W. Hildebrandt, Activity of the Akt-dependent anabolic and catabolic pathways in muscle and liver samples in cancer-related cachexia, J. Mol. Med., 2007, 85, 647–654 CrossRef CAS PubMed.
- J. G. Clohessy, M. Reschke and P. P. Pandolfi, Found in translation of mTOR signaling, Cell Res., 2012, 22, 1315–1318 CrossRef CAS PubMed.
- M. Mrowicka, J. Mrowicki, M. Mik, R. Wojtczak, L. Dziki, A. Dziki and I. Majsterek, Association between SOD1, CAT, GSHPX1 polymorphisms and the risk of inflammatory bowel disease in the Polish population, Oncotarget, 2017, 8, 109332–109339 CrossRef PubMed.
- R. J. Fitzgerald and B. A. Murray, Bioactive peptides and lactic fermentations, Int. J. Dairy Technol., 2006, 59, 118–125 CrossRef CAS.
- M. Chalamaiah, W. Yu and J. Wu, Immunomodulatory and anticancer protein hydrolysates (peptides) from food proteins: A review, Food Chem., 2018, 245, 205–222 CrossRef CAS PubMed.
- K. Yamamoto, S. Ohsumi, T. Nagashima, H. Akiyama, H. Honda and K. Shimizu, Screening of anti–atrophic peptides by using photo–cleavable peptide array and 96-well scale contractile human skeletal muscle atrophy models, Biotechnol. Bioeng., 2022, 119, 2196–2205 CrossRef CAS PubMed.
- G. Guidotti, L. Brambilla and D. Rossi, Cell-penetrating peptides: from basic research to clinics, Trends Pharmacol. Sci., 2017, 38, 406–424 CrossRef CAS PubMed.
- K. Muthuramalingam, S. Y. Kim, Y. Kim, H. S. Kim, Y. J. Jeon and M. Cho, Bigbelly seahorse (Hippocampus abdominalis)-derived peptides enhance skeletal muscle differentiation and endurance performance via activated P38MAPK/AKT signalling pathway: An in vitro and in vivo analysis, J. Funct. Foods, 2019, 52, 147–155 CrossRef CAS.
- G. Bruno, F. Cencetti, C. Bernacchioni, C. Donati, K. V. Blankenbach and D. Thomas, et al., Bradykinin mediates myogenic differentiation in murine myoblasts through the involvement of SK1/Spns2/S1P2 axis, Cell Signalling, 2018, 45, 110–121 CrossRef CAS PubMed.
- C. Vinel, L. Lukjanenko, A. Batut, S. Deleruyelle, J. P. Pradere, S. Le Gonidec, A. Dortignac, N. Geoffre, O. Pereira, S. Karaz, U. Lee, M. Camus, K. Chaoui, E. Mouisel, A. Bigot, V. Mouly, M. Vigneau, A. F. Pagano, A. Chopard, F. Pillard, S. Guyonnet, M. Cesari, O. Burlet-Schiltz, M. Pahor, J. N. Feige, B. Vellas, P. Valet and C. Dray, The exerkine apelin reverses age-associated sarcopenia, Nat. Med., 2018, 24, 1360–1371 CrossRef CAS PubMed.
- K. A. Mirza, S. M. Wyke and M. J. Tisdale, Attenuation of muscle atrophy by an N-terminal peptide of the receptor for proteolysis-inducing factor (PIF), Br. J. Cancer, 2011, 105, 83–88 CrossRef CAS PubMed.
- M. K. Caldow, D. J. Ham, J. Trieu, J. D. Chung, G. S. Lynch and R. Koopman, Glycine protects muscle cells from wasting in vitro via mTORC1 signaling, Front. Nutr., 2019, 6, 172 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo03911k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |